Abstract
Cancer metastasis, the spread of cancer cells to distant organs, is responsible for 90% of cancer-related deaths. Cancer cells need to enter and exit circulation in order to form metastases, and the vasculature and endothelial cells are key regulators of this process. While vascularized 3D in vitro systems have been developed, few have been used to study cancer, and many lack key features of vessels that are necessary to study metastasis. This review focuses on current methods of vascularizing biomaterials for the study of cancer, and three main factors that regulate intravasation and extravasation: endothelial cell heterogeneity, hemodynamics, and the extracellular matrix of the perivascular niche.
Keywords: 3D printing, angiogenesis, endothelial cells, shear stress
1. Introduction
The National Cancer Institute estimates that over 600 000 people in the United States died due to cancer.[1] The vast majority of these cancer-related deaths are a result of metastasis.[2,3] In order to metastasize, cancer cells must undergo several key steps following initial growth and malignant transformation of the cells.[4,5] Cancer cells secrete vascular endothelial growth factor (VEGF) and other proangiogenic factors[6] to recruit a capillary network to tumors. Cancer cells then become invasive, a process hypothesized by some to be related to an epithelial to mesenchymal transition, to escape the primary tumor and move into the lymphatics and vasculature and spread throughout the body via convection.[7,8]
Apart from releasing their own VEGF, cancer cells can recruit healthy cells to aid trafficking to the vasculature. Epithelial cells are one of many different cell types within the carcinoma microenvironment. In addition to epithelial cancer cells, stromal cells and a plethora of immune cells reside in or adjacent to tumors. Cancer cells can secrete chemokines to recruit many of these, including regulatory T cells, tumor-associated macrophages, dendritic cells, and neutrophils.[9,10] These immune cells are known to promote tumor progression and intravasation.[9-12]
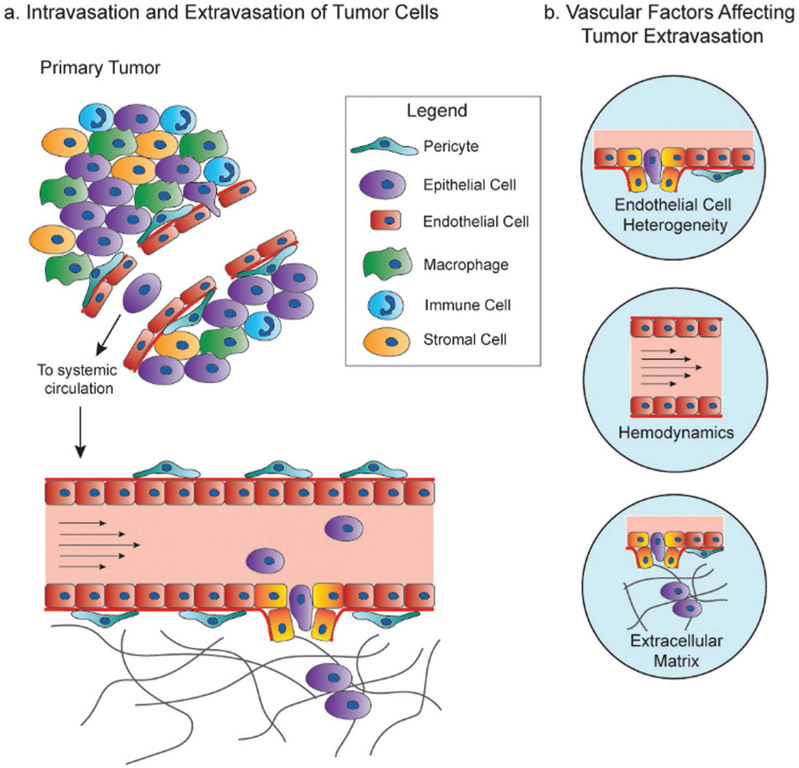
After cancer cells intravasate, they are exposed to a multitude of stressors, including loss of cell–cell and cell–matrix adhesion, shear stresses, and attacks by the immune system. Nevertheless, some cancer cells manage to survive this tortuous trip and eventually exit the vasculature to a secondary site, a process termed extravasation.[13,14] As the vasculature is involved in several key steps of metastasis, it will be the focus of this review (Figure 1).
Figure 1.
Vascular factors affecting cancer metastasis. a) Cancer cells intravasate into blood vessels near or in the tumor to enter the systemic circulation, where they then extravasate into secondary metastatic sites. b) The important factors of the vasculature and endothelium focused on in this review that affect intravasation, circulation, and extravasation are EC heterogeneity, hemodynamics, and the extracellular matrix.
Cancer cells have been shown to extravasate through the endothelial cell (EC) barrier in two ways: 1) transmigration through the EC barrier and 2) pocketing of cancer cells by the endothelium.[14-19] The transmigration of cancer cells is often compared to that of leukocytes. In fact, cancer cells exhibit some similarities to the rolling and adhesion to ECs by leukocytes.[15] However, unlike leukocytes, cancer cells have been shown to permanently disrupt the tight junction barrier of ECs during extravasation[15-17] (Figure 1a). Then, cancer cells form protrusions of their cell membrane, called invadopodia,[18] to exit the vessel. Alternatively, ECs can protrude into the lumen of the vessel to surround a circulating cancer cell, a process termed pocketing, thus trapping cells and facilitating adhesion to the vessel wall and extravasation.[14,19] This pocketing of circulating tumor cells and subsequent metastasis is hypothesized to occur at sites with specific flow conditions, discussed in detail later in this review. Furthermore, tumor cells can induce genetic changes in ECs, helping promote micrometastatic outgrowth.[20-22]
Cancer cells form metastases at specific, nonrandom sites, due to many factors, including but not limited to the stiffness and biochemical composition of the extracellular matrix (ECM) at these organs.[2,23] For example, breast cancer most commonly metastasizes to the brain, bone, lung, and liver; whereas prostate cancer predominantly metastasizes to the bone.[24,25] Paget first proposed the “seed and soil” hypothesis in 1889–the idea that the seed (cancer cell) needs a favorable soil (microenvironment) to metastasize.[26,27] In fact, specific genes have been identified that correlate to this organ-specific metastasis, or organotropism.[28] In addition to genetic factors and differences in the ECM, some literature suggests that ECs have different characteristics depending on the tissue in which they reside,[29-32] which may play a role in organotropism. It is therefore important to have model systems in place that include vasculature and potentially tissue specific ECs to study organotropism.
Although intravital imaging can capture extravasation of circulating tumor cells (CTCs), it is invasive, expensive, cumbersome, and has limited accessibility to certain anatomic sites.[33] Also, live animal imaging provides little opportunity to modulate local microenvironment to identify rate limiting factors involved in CTC extravasation. Furthermore, extravasation is a relatively rare event, making it very difficult to observe with an in vivo imaging approach. In vitro, a major gap in extravasation studies is the development of a vessel tissue model that can recapitulate physiologically relevant blood flow, while simultaneously retaining high analytical capability and experimental tunability. While there are multiple methods described in the literature to create vascularized hydrogels,[34-37] few used to study cancer attempt to recapitulate the heterogeneity of ECs, variable flow, and the makeup of the ECM of metastatic tissue sites. In this review, current and emerging models to study cancer metastasis will be discussed, as well as the importance of including attributes such as flow, EC and vascular heterogeneity, and microenvironment (Figure 1b).
2. Methods to Vascularize Biomaterials
Currently, there is no perfect model that recapitulates every aspect of human cancer biology. There have been many methods developed to study cancer, both in vivo and in vitro, and several used to specifically study cancer metastasis and extravasation. Some of these methods have been reviewed elsewhere.[38,39] Briefly, in vivo methods include both mouse and zebrafish models. In mouse models, typically metastasis is modeled by performing tail vein injections of cancer cells, and then studying metastatic sites. Unfortunately, it is not possible to monitor extravasation in real time with this method. It must be observed following necropsy and histologic examination. Zebrafish models are also becoming increasingly popular, as they are optically clear and have defined vasculature. However, in both of these methods the integrin receptors and ligands in mice and zebrafish often differ that of humans. Therefore, results gained from studies done on mouse and zebrafish models may not be completely applicable to human cancer.
In vitro, 2D plastic culture plates have historically been used for cell culture, but this involves culturing cells on a stiff, flat surface, that does not recapitulate in vivo physiology. Transwell assays bridge the gap between 2D and 3D models and use a monolayer of endothelial cells cultured on a porous membrane, and cancer cells introduced above the monolayer “extravasate” to the chemotactic medium below. A hydrogel can be incorporated on top of the membrane to better mimic the migration of cells through a 3D structure.[40] However, this method does not typically incorporate flow. Furthermore, the phenotype of endothelial cells grown on 2D surfaces differs from endothelial cells grown on 3D matrices, potentially skewing the experimental results.[41]
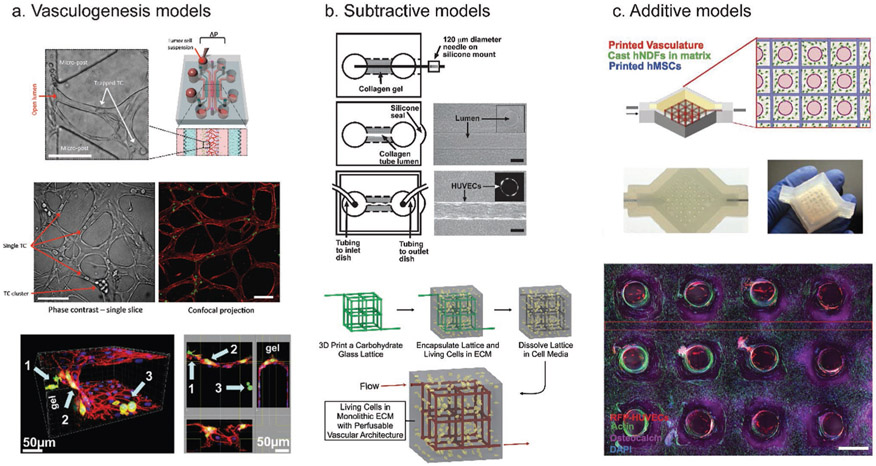
Due to these limitations, vascularized 3D biomaterials have been developed as an alternative to simplified 2D assays and difficult in vivo studies. Several methods for vascularizing biomaterials have been developed. They can be categorized broadly into three categories: 1) Vasculogenesis, where ECs are seeded and form vascularized networks within a biomaterial, 2) the subtractive method, where a hollow tube or network of tubes is created by polymerizing a material around a solid object, removing the object, and then perfusing the resulting channel with ECs, and 3) the additive method, which includes 3D bioprinting, where a vessel or network of vessels is built from the “ground-up” (Figure 2). While only the vasculogenesis method has been applied to cancer studies, each method has its advantages and disadvantages.
Figure 2.
Common methods to vascularize biomaterials. a) Vasculogenesis models induce sprouting of ECs into a biomaterial. A schematic of a commonly used vasculogenesis model is displayed in the top right. This model allows visualization of trapped tumor cells (top left and middle, with arrows indicating trapped tumor cells or clusters of tumor cells), as well as extravasated tumor cells (bottom image arrows). b) Subtractive models create an empty space in a material that can be lined with ECs. In the top image, a collagen gel was polymerized over a needle, which was then removed, and the remaining empty space was perfused with ECs to create a single channel. In the bottom image, a carbohydrate lattice was 3D printed and then encapsulated in ECM mimic. The lattice can then be dissolved, with the resulting empty space perfused and lined with ECs. c) Additive models (3D bioprinting) directly deposit the biomaterial of choice as well as multiple cell types to build a model tissue from the ground up. Here, a bioprinter was used to print ECs, fibroblasts, and stem cells to create a thick vascularized tissue. (a) Bottom: reproduced with permission.[43] Copyright 2013, PLoS One. Top and middle: reproduced with permission.[85] Copyright 2017, Springer Nature. (b) Top: reproduced with permission.[36] Copyright 2006, Elsevier. Bottom: reproduced with permission.[35] Copyright 2012, Springer Nature. (c) Reproduced with permission.[54] Copyright 2016, National Academy of Sciences.
2.1. Vasculogenesis Models
Vasculogenesis models use microfluidics and incorporate hydrogels that can be seeded with various types of media and cell cocultures. One popular method uses a microfluidic device. This device is made from a silicon wafer, which can be used as a mold to cast PDMS which serves as the device housing. In the center of this housing is a space that can be filled with a collagen gel. This is surrounded by several ports where media and cells can be introduced.[42] ECs are induced to sprout and form capillary networks across the gel. These devices have been used to successfully study cancer cell extravasation (Figure 2a).[43] Using this model, it was determined that the permeability of the EC monolayer, as measured by fluorescently labeled dextran diffusion, increased fourfold after introducing tumor cells into the microfluidic device.
In another microfluidic device, a vascular network was established using a fibrinogen-based hydrogel with ECs and fibroblasts.[44] In this system, vessels ranged from 15–50 μm in diameter and the flow velocity from 0–4000 μm s−1. This method has also successfully been used with ECs derived from human induced pluripotent stem cells.[45] Vascularized micro-tumors have also been developed using this model. Various tumor cell types are introduced in the vascularized hydrogel with stromal cells to establish tumors containing microvessels.[46] However, this model has not been used to directly study metastasis.
The major advantage of vasculogenesis models is that they result in small diameter vascular networks that more closely resemble capillaries both in size and through branching. By imaging during development, vascular branching and sprouting can be quantified. However, introducing flow into these models is often not simple, because syringe pumps cannot be attached to inlet vessels as they can in other models created with 3D printing or subtraction. Flow can be applied, often pressure-driven through gravity wells (Figure 2a). Despite this limitation, the success of these models in studying cancer extravasation is promising for future research.
2.2. Subtractive Models
Several methods have been employed to create a channel in a subtractive fashion. Chrobak et al. polymerized collagen around a needle with an initial diameter of 120 μm.[36] After polymerization of the collagen hydrogel and removal of the needle, a negative space was created, which was perfused with ECs (≈107 cells mL−1) to create a cell-seeded vessel. Final diameters after maturation ranged from 75–150 μm, with shear stresses of ≈10 dyne cm−2 (Figure 2b, top). This method was used to study the effect of cyclic AMP (cAMP) concentrations on the permeability of these vessels.[47] Lower concentrations (3 × 10−6 m) did not affect vessel permeability, but at higher concentrations (80 and 400 × 10−6 m), vessels exhibited decreased permeability and less leakage. Other work has demonstrated a similar effect with cAMP stabilizing endothelial tight junctions.[48] Conversely, VEGF appears to have the opposite effect, where VEGF increases vascular permeability, resulting in leakier vessels.[22,49] Altering cAMP and VEGF concentration in a vascularized material could thus be used to simulate different aspects of cancer biology, by simulating the leaky vessels in the primary tumor with less cAMP and more VEGF, or alternatively creating a less permeable endothelium with high cAMP to simulate metastatic sites.
Another subtractive method involves creating a tube out of a dissolvable material rather than removing a needle. Miller et al. developed a technique which used a 3D printed carbohydrate lattice structure, embedded in a fibrin hydrogel that contained fibroblasts.[35] The carbohydrate lattice was easily dissolved because it was water-soluble whilst the fibrin was not, leaving behind a complex, perfusable network that could then be lined with ECs. This network resulted in channels with diameters ranging from 150–800 μm that could be perfused with a shear stress of 1 dyne cm−2 (Figure 2b, bottom).
Subtractive methods of vascularization are attractive due to their relative simplicity and ease of tuning the exact size of vessel desired. In general, the sizes of the vessels created range from ≈75–800 μm in diameter.[36,50,51] Furthermore, flow can easily be introduced and controlled if using a single, simple vessel structure, with shear stress comparable to physiological stresses (1–10 dyne cm−2). The downside to using subtractive methods is that they typically require large amounts of ECs (on the order of 107 cells mL−1) to obtain a confluent vascular layer. In addition, it is difficult to make a subtractive model with a capillary-sized vessel, due to the physical limitations of the materials used to create the vessel. Despite this, these models can be applied to study the role of the endothelium in cancer metastasis. Because of the simplicity in design, it may be easier to track individual cancer cells extravasating in a single vessel rather than in multiple branching vessels present in vasculogenesis models.
2.3. Additive Models
Due to advancements in 3D printing technology and development of less cost-prohibitive printers, 3D bioprinting has gained traction as a method to create vascularized tissues. Using 3D bioprinting to vascularize a biomaterial can potentially combine the advantages of vasculogenesis methods and subtractive methods. Networks can be made with varying levels of complexity, and prints are reproducible and scalable.[52] Via bioprinting, larger diameter blood vessels can be directly printed (with sizes 150 μm and up), and smaller microvessels can be created by printing with bioinks that contain pro-angiogenic factors.[53] Furthermore, printing allows the ability to print an inlet and outlet vessel which can easily be hooked up to a syringe pump for controllable flow, similar to subtractive models.
3D printed vascularized biomaterials are typically thicker than those created with either vasculogenesis or subtractive methods. For instance, Kolesky et al. created 3D bioprinted tissue over 1 cm in thickness.[54] This tissue incorporated multiple cell types as well as ECM and vasculature (Figure 2c). Skylar-Scott et al. created organ-specific vascularized constructs by first creating organoids from induced pluripotent stem cells.[55] These organoids were embedded in an ECM solution made of collagen I and Matrigel. Finally, sacrificial ink made of gelatin was printed into the solution and was then evacuated and perfused with endothelial cells. Vasculature created using this method had diameters of 400–1000 μm. Others have used 3D bioprinting to print tumor cells and ECs without accompanying ECM, with a result that is similar to an actual tumor with cell deposited ECM and organization of vasculature.[56] Here, the bioink used included an alginate-based hydrogel that could be tuned to provide a desired stiffness during cell seeding but could then be removed to leave a structure containing only cells. In this way, the authors created tumor subtypes with defined architecture. While this method could be useful for studying cancer, the cost of the bioprinter used may be prohibitive to some researchers. However, prices of commercial printers are decreasing, and there are cost-effective solutions that can be utilized. Hinton et al. modified a $400 plastic 3D printer to inject hydrogel precursor solutions into a support bath, thus creating a bioprinter.[57] Characteristics of the methods discussed are summarized in Table 1.
Table 1.
Summary of the vessel characteristics of the literature discussed in Section 2.
| Method of vascularizationa) | Vessel diameter | Flow rate | Velocity | Shear (Stress or Rate) | Number of cells seeded initially | Time to create stable vasculature |
|---|---|---|---|---|---|---|
| Vasculogenesis | ||||||
| Chan et al.[37] | ≈15 μm | NS | NS | NS | 2 × 106 cells mL−1 | 6–8 days |
| Moya et al.[44] | 15–50 μm | NS | 0–4000 μm s−1 | 0–1000 1/s | 7.5 × 106 cells mL−1 (2:1 fibroblasts:ECs) | 14 days |
| Subtractive | ||||||
| Chrobak et al.[36] | 75–150 μm | ≈3 μL min−1 | NS | 10 dyne cm−2 | ≈107 cells mL−1 | ≈3 days |
| Miller et al.[35] | 150–800 μm | 10 μL s−1 | 5 mm s−1 | 1 dyne cm−2 | 35 × 106 cells mL−1 | 1 day |
| Additive | ||||||
| Kolesky et al.[54] | NS | 13–27 μL min−1 | NS | NS | 1 × 107 cells mL−1 | ≈1 day |
| Skylar-Scott et al.[55] | 400–1000 μm | 40–500 μL min−1b) | NS | 0.8 dyne cm−2 (at 500 μL min−1) | 1 × 107 cells mL−1 | ≈1 day |
| Langer et al.[56] | NS | Static | Static | Static | 1.5–2 × 108 cells mL−1 | NS |
NS = Not specified in text
Constructs with ECs were perfused at 40 μL min−1.
3. Endothelial Cell Heterogeneity
ECs make up the inner lining of all blood and lymphatic vessels and play an important role in tumor angiogenesis and metastasis.[30,31] ECs can differ phenotypically based on organ location, potentially impacting organotropism of cancer, and their gene expression can be altered by flow or by the presence of tumor cells. Furthermore, the behavior of EC lines, including sprouting, branching, and permeability, can vary based on the tissue origin of the cell line. When engineering a vascularized biomaterial, it is important to keep these differences in min d and to choose the type of cell that is relevant to the disease process being studied.
3.1. Development of the Endothelium
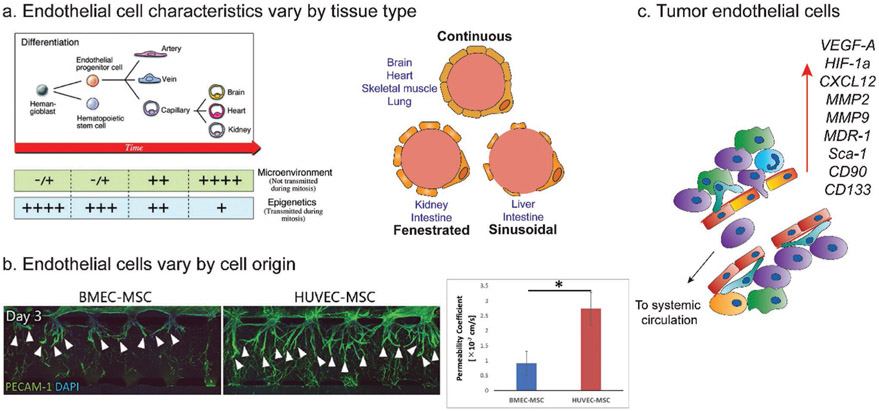
Hemangioblasts differentiate into endothelial progenitor cells (EPCs) and then into the ECs lining arteries, veins, and capillaries (Figure 3a, left). While arteries and veins contain a continuous layer of ECs bound by tight junctions, the endothelium in capillaries can vary based on the tissue type and may be continuous, fenestrated, or discontinuous/sinusoidal (Figure 3a, right).
Figure 3.
Endothelial cell heterogeneity. a) Development of end-organ EC from EPCs (left) is a multistep process affected by epigenetic factors and microenvironmental factors. Ultimately, the resulting capillary endothelium varies by tissue type. The endothelium can be continuous, fenestrated, or sinusoidal (right). b) The behavior of BMECs and HUVECs vary by source. Sprouting (left) and permeability (right) are less prominent in BMECs versus HUVECs. c) Tumor ECs associated with highly metastatic tumors have enrichment in genes corresponding to invasion, angiogenesis, drug resistance, and stemness. (a) Left: reproduced with permission.[30] Copyright 2007, Lippincott Williams & Wilkins. Right: reproduced with permission.[67] Copyright 2019, Lippincott. (b) Adapted with permission.[29] Copyright 2019, Elsevier.
Continuous and nonfenestrated endothelium is found in the capillaries of the brain, heart, lung, and skin.[30,31] Fenestrated endothelium is a subtype of continuous endothelium and is found in locations that require filtration or transport of small solutes, such as the kidney, intestine, and endocrine glands (Figure 3a, right). Discontinuous, or sinusoidal, endothelium is found in the liver, spleen, and bone marrow and contains much larger openings within individual cells allowing for the transport of large proteins. As the structure of the endothelium can vary by location, it is important to consider sourcing ECs from a particular tissue type or differentiating ECs into the desired lineage for the in vitro tissue model being studied.
3.2. Tissue Specificity of Endothelial Cells
The characteristics of ECs differ based on their origin and location in the vasculature.[30,58] For example, EC thickness can vary from 0.1 μm in capillaries to 1 μm in the aorta, and overall size and shape can vary by tissue type as well. While most ECs are flat in shape, ECs in venules are cuboidal.[30,59] In the aorta, ECs are long and narrow, but ECs in the pulmonary artery are shorter and broader.[60] EC morphology, especially in the aorta, is dictated by the blood flow environment where regions of undisturbed flow promote elongation of ECs, while regions of disturbed flow cause the more classic cobblestone shape.[61]
In addition to structural differences, the tissue origin of an EC line impacts its behavior. Using a vasculogenesis model in a microfluidic device, Uwamori et al. compared the characteristics of human brain microvascular endothelial cells (BMECs) to human umbilical vein endothelial cells (HUVECs).[29] Compared to BMECs, HUVECs developed microvasculature with greater sprout numbers, length, and branching (Figure 3b, left). Vessels derived from HUVECs also had greater permeability (Figure 3b, right), and decreased expression of tight junction proteins zonula occludens protein 1 (ZO-1) and occludin. While these differences may be due to tissue source, it is also possible they are a result of comparing a less mature (HUVEC) to a more mature (BMEC) EC. Nevertheless, it is important to keep cell source in mind when designing vascularized biomaterials.
Due to differences in EC behavior based on origin, some researchers have begun to use EPCs in their hydrogel systems in order to differentiate ECs into a desired lineage. Peters et al. seeded EPCs and HUVECs in a poly(ethylene) glycol (PEG) hydrogel system[62] and found that after 2 weeks, vascular networks originating from EPCs had longer total tubule length and branch points compared to HUVEC networks. Furthermore, tissue-specific ECs have been shown to rapidly de-differentiate and lose expression of many of the genes that define their tissue specificity, so increasingly researchers are investigating how EPCs can be differentiated into tissue specific ECs, as this process is not well understood.[63,64] This approach may be useful for creating tissue mimics for cancer metastatic sites.
3.3. Tumor Endothelial Cells
Tumor blood vessels differ from normal blood vessels in several ways. Normal blood vessels have an organized hierarchy with defined flow patterns and progress from artery to arteriole to capillary to venule to vein. In contrast, tumor blood vessels lack this defined structural hierarchy. Furthermore, compared to normal vessels, tumor blood vessels are more dilated and tortuous, with uneven and chaotic flow patterns.[20] On a cellular level, the ECs lining tumor blood vessels have larger intercellular junctions and are more fenestrated, often growing on top of one another with projections into the lumen of the blood vessel.
In addition to vessel phenotypic differences, tumor endothelial cells (TECs) differ from normal ECs at the genetic level.[21,22,65] TECs have higher proliferative rates and proangiogenic properties, and different gene expression patterns when compared to normal ECs (Figure 3c). In TECs associated with highly metastatic tumors, expression increases were observed in genes related to cell invasion (MMP2, MMP9), angiogenesis (VEGF-A, HIF-1α, CXCL12), drug resistance (MDR-1), and stemness (Sca-1, CD90, CD133). When choosing an EC type to use in a vascularized model of cancer, it would make sense to start with a less mature EC (such as EPCs) and differentiate them into a desired lineage. Unfortunately, the mechanisms of differentiation of EPCs are currently not well understood.[66,67]
4. Hemodynamic Factors
The vast majority of in vitro studies, even those incorporating ECs, are performed in static conditions on 2D substrates. 3D vascularized biomaterials can incorporate flow via several different mechanisms. In fact, including flow is necessary to build better models of cancer metastasis, as varying hemodynamics can affect the gene expression of ECs, as well as the transport and extravasation potential of cancer cells.
4.1. Role of Shear Stress on the Endothelium
Under normal conditions, human veins have average shear stress values of 5 dyne cm−2, whereas arteries have average shear stress values approaching 15 dyne cm−2. Shear stress increases with higher heart rates and can reach 30 dyne cm−2 at a heart rate of 140 beats per minute.[68] In microvessels such as capillaries, the average wall shear stress has been suggested to approximate 4 dyne cm−2.[69,70] However, the multiphase nature of blood dominates capillary blood flow and deviates from single-phase approximations.
Shear stress alters gene expression profiles in vascular ECs.[70-72] In human aortic ECs, 24 h of shear stress at 12 dyne cm−2 increased levels of Tie2 and Flk-1, which are receptors associated with EC survival and angiogenesis, as well as increased MMP1.[71] In EPCs, shear stress stimulated proliferation and increased expressions of EC-specific markers KDR, FLT-1, and VE-cadherin at early time points. Shear stress also accelerated tube formation in EPCs.[73] However, EPCs do exhibit some differences in flow sensitivity when compared to more mature ECs. Under lower shear stresses (0.1–2.5 dyne cm−2), EPCs changed morphology and aligned to the direction of flow. However, HUVECs and bovine aortic ECs do not align to flow until higher shear stresses of 8–10 dyne cm−2.[73] These differences between more mature HUVECs versus EPCs are important to keep in mind when developing in vitro models that more accurately reflect in vivo physiology.
Finally, although not a focus of this paper, lymphatic and interstitial flow has been shown to impact cell behavior. In the low-flow lymphatic system, reversing shear stress induces gene expression for further lymphatic vessel maturation and development.[74] In the brain, interstitial fluid flow increased invasion of glioma cells.[75]
4.2. Hemodynamics and Circulating Tumor Cells
Hemodynamics plays a role in cancer cell extravasation and should be considered when designing tumor models. While in the circulation, tumor cells are in a highly dynamic environment, which can differ vastly from the environment of the primary tumor. While traveling via convection through the vasculature, CTCs are subject to a variety of insults including attack by immune cells, collisions with blood cells, and shear forces, yet some CTCs manage to survive this tortuous journey and extravasate.[14] It is suggested that CTCs arrest and extravasate at sites of optimal flow, and a high shear stress of 60 dyne cm−2 has been shown to cause necrosis of CTCs.[14,76] However, the precise role of hemodynamics in extravasation of CTCs has not been widely studied. Zebrafish embryo models have been used due to the ability to easily label and image their endothelium. To study hemodynamics in vitro, it will be important to create vasculature that is long-lasting, easy to image, with flow that is controllable.
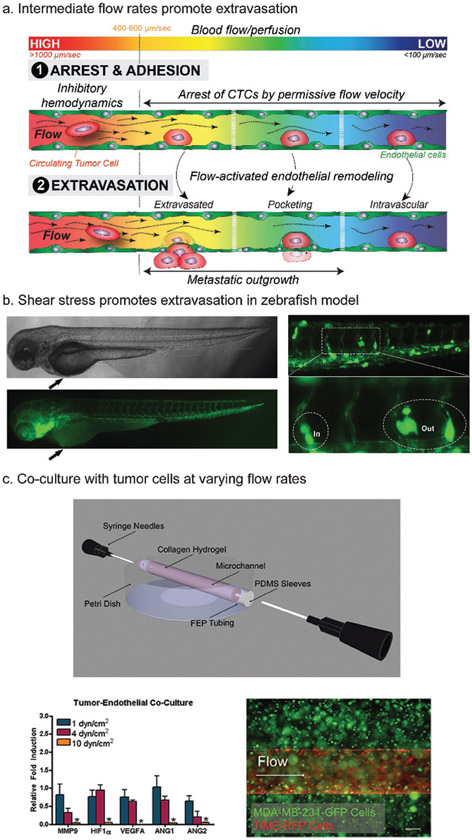
Follain et al. utilized zebrafish and mouse in vivo models and human in vitro models to study the impact of flow rates on arrest and extravasation of CTCs.[14] In the zebrafish model, while intermediate and low flow rates (below blood velocities of 600 μm s−1) allowed for arrest of CTCs and adhesion to the endothelium, extravasation occurred more often at intermediate flow rates of 400 μm s−1 (Figure 4a). They found that the majority of CTCs extravasated by inducing EC remodeling around the tumor cells, rather than via transmigration through the vessel wall. This “pocketing” of CTCs by ECs occurred more rapidly at intermediate flow rates compared to low flow rates. In an in vitro microfluidic model, ECs demonstrated protrusions under flow rates of 400 μm s−1, with or without the presence of CTCs, and these protrusions were absent under no flow, suggesting that these intermediate flow profiles may promote the adhesion of CTCs to the endothelium. The protrusions seen in the in vitro model may be precursors to the pocketing observed in the in vivo model.
Figure 4.
Hemodynamic effects on circulating tumor cells. a) Diagram of work by Follain et al., describing the pocketing of CTCs by endothelial cells that occurs at intermediate flow rates. While arrest and adhesion of CTCs to the endothelium occurs at low and intermediate flow rates, pocketing was only seen at intermediate flow rates. b) Zebrafish can be used to directly observe extravasation of CTCs. c) A subtractive method was used to create a vascularized collagen hydrogel (top) which can be perfused. This model was used with an MDA-MB-231 breast cancer and EC coculture (bottom right). Authors observed decreased gene expression of angiogenesis-related factors at high shear stress (bottom left). (a) Reproduced with permission.[14] Copyright 2018, Cell Press. (b) Reproduced with permission.[77] Copyright 2017, Elsevier. (c) Adapted with permission.[69] Copyright 2014, Taylor & Francis.
Using zebrafish, Ma et al. injected breast cancer cells into the vasculature and directly observed extravasation[77] (Figure 4b). The authors also performed transwell in vitro experiments, where various human metastatic breast cancer cell lines were exposed to shear stress. These cells exhibited higher migration capacity after fluid flow exposure, as measured by number of cells that migrated through the membrane of the transwell chamber. At a shear stress of 15 dyne cm−2, migration increased by approximately threefold.
An in vitro collagen system was used by Buchanan et al. to quantify how barrier function and gene expression of ECs varied as a function of flow rate. They used a subtractive method to create a channel in a collagen hydrogel with a 22G needle (Figure 4c, top).[69] Low (1 dyne cm−2), normal (4 dyne cm−2), and high (10 dyne cm−2) shear stresses were introduced via a syringe pump to study the impact of varying shear stresses on the endothelium. In their coculture model, low and normal shear stresses increased the gene expression of angiogenic factors by the breast cancer cell line, and increased endothelial permeability compared to high shear stress (Figure 4c, bottom).
Hemodynamic forces play an important role in the behavior of ECs and the extravasation potential of CTCs. While there have been several methods to study this, it is difficult to compare results as some studies report shear stress values and others report flow rates. Furthermore, these methods mainly use steady flow, whereas the body primarily experiences varying degrees of unsteady flow. As more researchers introduce flow into vascularized networks, it will be important that studies clearly report the hemodynamics of their systems.
5. The ECM and the Perivascular Niche
In the perivascular niche, disseminated cancer cells can lie dormant until a biochemical signal spurs their growth and the development of a symptomatic metastasis. This process has not been well studied in 3D vascularized models of cancer. In addition, the biochemical and biophysical signals from ECM can both impact angiogenesis, which has implications for how vessels may control disseminated CTCs at distant sites.
5.1. Biochemical Signals in the ECM that Promote Metastasis
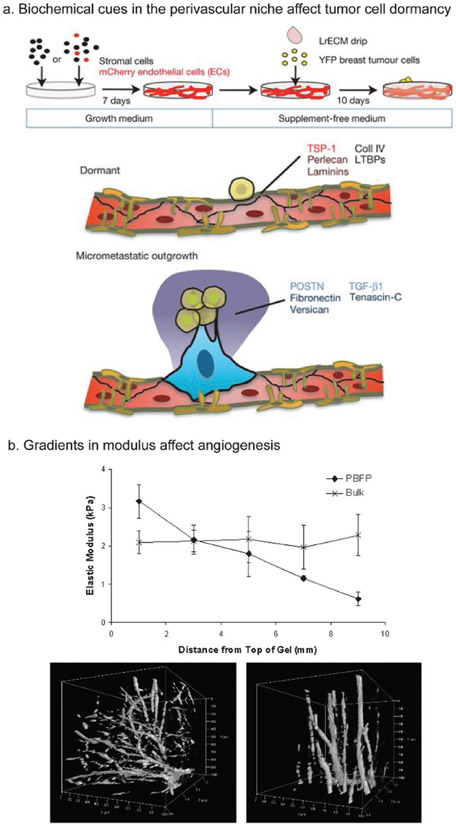
Dormant, disseminated cancer cells often lie adjacent to the endothelial layer in the perivascular niche.[78] Ghajar et al. demonstrated that biochemical cues such as thrombospondin-1, perlecan, and certain laminins promote cancer cell dormancy in the perivascular niche, while periostin, fibronectin, transforming growth factor beta 1 (TGF-β1), and others promoted outgrowth of the dormant cancer cells to form micrometastases (Figure 5a). Here, ECs were cultured with stromal cells and induced to form a microvascular niche on 2D plastic in a 96-well plate. Following this, a suspension of breast cancer cells was introduced into these same wells. Finally, a laminin-rich ECM mimic was introduced in the well to provide a 3D environment. A limitation of this method is the inability to easily control flow. Although it was not possible with Matrigel as used by Ghajar et al., one could vary the ECM makeup with a synthetic hydrogel.
Figure 5.
Biochemical and mechanical effects on vasculature. a) Interactions with the perivascular niche ECM can promote cancer cell exit from dormancy. An engineered model was used with a laminin rich ECM drip used to culture breast cancer cells on top of formed microvasculature. Biochemical cues were identified that promoted cancer cell dormancy (TSP-1, perlecan, laminins, etc.) or micrometastatic outgrowth (periostin, fibronectin, tenascin C, etc.). b) Introducing gradients in modulus can create organized vasculogenesis, which may be important when engineering 3D vascularized models. PEGDA hydrogels that had either a uniform bulk modulus or a gradient in modulus were seeded with ECs. Vascular sprouting appeared more disorganized in the uniform modulus gel (bottom left) but was more organized in the gradient gel (bottom right). (a) Adapted with permission.[78] Copyright 2013, Nature Publishing Group. (b) Adapted with permission.[81] Copyright 2013, PLoS One.
The ECM has been shown to impact cancer metastasis, particularly via integrin binding.[28,79,80] Integrins are transmembrane proteins that bind to the ECM and to the cytoskeleton and are important for mechanical and biochemical signaling. As such, elucidating the role of integrins is important in understanding the mechanisms of cancer metastasis. In a 3D vasculogenesis-based microfluidic model, tumor cells extravasated by first protruding invadopodia and associated with subendothelial basement membrane via β1 integrins.[28] Following this, invadopodia engaged with the ECM via α3β1 and α6β1 integrins. Both of these integrins bind specifically to laminins, with α3β1 specific for laminin-5 and laminin-10/11, and α6β1 specific for laminin-10/11, laminin-5, laminin-1, and laminin-2/4.[79] Altering integrin binding sites in biomaterials can also impact angiogenesis and vascular patterning. Li et al. demonstrated that by tuning integrin activation in hydrogels, vascular patterning and permeability can be altered.[80] Here, a hyaluronic acid-based hydrogel was tuned to be specific to α3/α5β1 integrin or to αvβ3 integrin. In both in vitro and in vivo experiments, vasculature in αvβ3-specific hydrogels was more disorganized, more tortuous, and had greater permeability compared to α3/α5β1-specific hydrogels. Integrin binding sites are important for tumor cell behavior and angiogenesis, and thus are an important factor to incorporate into vascularized hydrogel designs.
5.2. Altering Vasculature through ECM Modulus
Alterations in modulus have been shown to impact the sprouting of blood vessels. Turturro et al. used a PEG diacrylate (PEGDA) hydrogel with a gradient of elastic moduli ranging from 3.17 to 0.62 kPa compared to a control (no gradient) hydrogel with an elastic modulus of 2 kPa.[81] When seeded with ECs, vascular sprouts formed in an organized fashion in the gradient gels but were more disorganized in the hydrogel with a constant modulus (Figure 5b). Furthermore, changing modulus can promote formation of vasculature that is closer to in vivo tumor vasculature. Bordeleau et al. created collagen hydrogels with varying equilibrium compressive moduli from 0.18 to 1.4 kPa and polyacrylamide (PA) gels ranging from 0.2 to 10 kPa to study vessel permeability, EC migration, and EC gene expression.[82] Increased moduli resulted in more angiogenic sprouting and vasculature with increased vessel permeability and disrupted architectures. Taken together, these studies illustrate the importance of modulus on formation of vasculature. Researchers wanting to create a more tumor-like vascular phenotype should ensure their material has a modulus similar to the target tumor ECM.
Reid et al. cultured ECs on soft (0.4 kPa) or hard (22 kPa) PA gels in order to simulate normal tissue modulus or tumor modulus, respectively.[83] They found that the 22 kPa substrate resulted in upregulation of CCN1. CCN1 is upstream of N-Cadherin, which plays an important role in transendothelial migration of cancer cells.[84] Furthermore, in a mouse model, knocking out CCN1 in ECs inhibited the binding of melanoma cells to the endothelium and reduced transendothelial migration compared to control mice.[83] This literature suggests that it is important to include the relevant modulus in a 3D vascularized biomaterial system to study cancer.
6. Conclusion
Various methods have been used to create vascularized in vitro tissue models, although many of these have not yet been used to study cancer metastasis. Among the three models discussed, only the vasculogenesis model has been applied to study cancer cell extravasation. As this model involves sprouting vasculature across a growth factor gradient, the resulting vasculature is most similar to native capillaries. Since cancer metastasis occurs at the capillary level, this model is more suited to studying extravasation compared to other models. However, it can take longer to form vasculature using this method, and it is more difficult to precisely control flow across the resulting network. Subtractive methods are attractive due to their overall simplicity and ease of controlling flow. However, the size of the vasculature is limited to the size of the object used to create the channel, usually 75 μm or larger. As native capillaries are approximately 10 μm in diameter, this size limitation is a significant drawback of subtractive methods. Finally, additive methods, including bioprinting, have recently gained popularity. This method is also limited by size, as the vessel is only as small as the resolution of the printer being used. However, this method does allow for the ability to easily print potentially more complex, multicellular structures, with the ability to control ECM makeup.
No matter which method is used, researchers need to take many factors into consideration when creating these models to more adequately recapitulate in vivo tumor physiology. Specifically, the properties of and surrounding the endothelium need to be taken into account, including EC type, shear stresses, appropriate ECM proteins, and relevant moduli. EC behavior, including sprouting and permeability, can vary based on the tissue origin of the cell. Furthermore, EC behavior can be modified by shear stress and the presence of tumor cells. Through better design of vascularized biomaterials and incorporation of the properties discussed here, researchers will be able to create more accurate models of cancer, thereby better elucidating the mechanisms of metastasis.
Acknowledgements
This work was supported by a National Science Foundation CAREER (DMR1454806, 1842308) and an NIH grant (R21CA223783) to SRP, and an NSF CAREER grant (CMMI1842308) to J.M.J.
Biography

Katharine Bittner is a Ph.D. candidate of molecular and cellular biology at the University of Massachusetts, Amherst. She received her B.S.E. in bioengineering from the University of Pennsylvania and her M.D. from Robert Wood Johnson Medical School. Her research interests include applying engineering solutions to solve problems in human health, specifically in oncologic and pediatric populations. Outside of the laboratory, her interests include her automobile and her fluffy feline. Her Ph.D. work is focused on creating vasculature in 3D biomaterials to model and study cancer metastasis.

Juan M. Jiménez received a B.S. degree in mechanical engineering from the Michigan State University and an M.S. and Ph.D. in mechanical/aerospace engineering from Princeton University. His Ph.D. work is focused on turbulent wake flow measurements. As a postdoctoral fellow at the University of Pennsylvania’s Institute for Medicine and Engineering, he studied the effects of fluid flow on endothelial mechanobiology with relation to implantable biomedical devices like stents. His laboratory focuses on the effects of fluid flow in development and disease in the cardiovascular, lymphatic and skeletal systems.

Shelly Peyton is an associate professor and Graduate Program Director of Chemical Engineering at the University of Massachusetts, Amherst. She received her B.S. in chemical engineering from Northwestern University and her Ph.D. in chemical engineering from the University of California, Irvine. She was then a postdoctoral fellow in the Biological Engineering Department at MIT before starting her academic appointment at the University of Massachusetts in 2011. Shelly leads an interdisciplinary group of engineers and molecular cell biologists seeking to create and apply novel biomaterials platforms toward new solutions to grand challenges in human health.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Katharine R. Bittner, Molecular and Cellular Biology Graduate Program, University of Massachusetts, Amherst, MA 01003, USA
Juan M. Jiménez, Molecular and Cellular Biology Graduate Program, University of Massachusetts, Amherst, MA 01003, USA; Department of Mechanical and Industrial Engineering, University of Massachusetts, Amherst, MA 01003, USA
Shelly R. Peyton, Molecular and Cellular Biology Graduate Program, University of Massachusetts, Amherst, MA 01003, USA; Department of Chemical Engineering, University of Massachusetts, Amherst, MA 01003, USA
References
- [1].“Cancer Statistics – National Cancer Institute,”https://www.cancer.gov/about-cancer/understanding/statistics (accessed: October 2019).
- [2].Barney LE, Jansen LE, Polio SR, Galarza S, Lynch ME, Peyton SR, Curr. Opin. Chem. Eng 2016, 11, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Eng LG, Dawood S, Sopik V, Haaland B, Tan PS, Bhoo-Pathy N, Warner E, Iqbal J, Narod SA, Dent R, Breast Cancer Res. Treat 2016, 160, 145. [DOI] [PubMed] [Google Scholar]
- [4].Fidler IJ, Semin. Cancer Biol 2002, 12, 89. [DOI] [PubMed] [Google Scholar]
- [5].Bielenberg DR, Zetter BR, Cancer J. 2015, 21, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Folkman J, Nat. Med 1995, 1, 27. [DOI] [PubMed] [Google Scholar]
- [7].Hay ED, Cells Tissues Organs 1995, 154, 8. [DOI] [PubMed] [Google Scholar]
- [8].Lamouille S, Xu J, Derynck R, Nat. Rev. Mol. Cell Biol 2014, 15, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McAllister SS, Weinberg RA, Nat. Cell Biol 2014, 16, 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu Y, Cao X, J. Molecular Med 2016, 94, 509. [DOI] [PubMed] [Google Scholar]
- [11].Gajewski TF, Schreiber H, Fu Y-X, Nat. Immunol 2013, 14, 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kitamura T, Qian B-Z, Pollard JW, Nat. Rev. Immunol 2015, 15, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shenoy AK, Lu J, Cancer Lett. 2016, 380, 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Follain G, Osmani N, Azevedo AS, Allio G, Mercier L, Karreman MA, Solecki G, Garcia Leòn MJ, Lefebvre O, Fekonja N, Hille C, Chabannes V, Dollé G, Metivet T, Hovsepian F, Prudhomme C, Pichot A, Paul N, Carapito R, Bahram S, Ruthensteiner B, Kemmling A, Siemonsen S, Schneider T, Fiehler J, Glatzel M, Winkler F, Schwab Y, Pantel K, Harlepp S, Goetz JG, Dev. Cell 2018, 45, 33. [DOI] [PubMed] [Google Scholar]
- [15].Strell C, Entschladen F, Cell Commun. Signaling 2008, 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Heyder C, Gloria-Maercker E, Entschladen F, Hatzmann W, Niggemann B, Zänker K, Dittmar T, J. Cancer Res. Clin. Oncol 2002, 128, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aghajanian A, Wittchen ES, Allingham MJ, Garrett TA, Burridge K, J. Thromb. Haemostasis 2008, 6, 1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Leong HS, Robertson AE, Stoletov K, Leith SJ, Chin CA, Chien AE, Hague MN, Ablack A, Carmine-Simmen K, McPherson VA, Postenka CO, Turley EA, Courtneidge SA, Chambers AF, Lewis JD, Cell Rep. 2014, 8, 1558. [DOI] [PubMed] [Google Scholar]
- [19].Wirtz D, Konstantopoulos K, Searson PC, Nat. Rev. Cancer 2011, 11, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Aird WC, Cell Tissue Res. 2009, 335, 271. [DOI] [PubMed] [Google Scholar]
- [21].Maishi N, Hida K, Cancer Sci. 2017, 108, 1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hida K, Maishi N, Annan D, Hida Y, Hida K, Maishi N, Annan DA, Hida Y, Int. J. Mol. Sci 2018, 19, 1272. [Google Scholar]
- [23].Barney LE, Dandley EC, Jansen LE, Reich NG, Mercurio AM, Peyton SR, Integr. Biol 2015, 7, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lu X, Kang Y, J. Mammary Gland Biol. Neoplasia 2007, 12, 153. [DOI] [PubMed] [Google Scholar]
- [25].Arnold RS, Fedewa SA, Goodman M, Osunkoya AO, Kissick HT, Morrissey C, True LD, Petros JA, Bone 2015, 78, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Paget S, Cancer Metastasis Rev. 1989, 8, 98. [PubMed] [Google Scholar]
- [27].Ribatti D, Mangialardi G, Vacca A, Clin. Exp. Med 2006, 6, 145. [DOI] [PubMed] [Google Scholar]
- [28].Chen W, Hoffmann AD, Liu H, Liu X, npj Precis. Oncol 2018, 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Uwamori H, Ono Y, Yamashita T, Arai K, Sudo R, Microvasc. Res 2019, 122, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aird WC, Circ. Res 2007, 100, 158. [DOI] [PubMed] [Google Scholar]
- [31].Aird WC, Circ. Res 2007, 100, 174. [DOI] [PubMed] [Google Scholar]
- [32].Aird WC, Cold Spring Harbor Perspect. Med 2012, 2, a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dunn KW, Day RN, Methods 2017, 128, 1. [DOI] [PubMed] [Google Scholar]
- [34].Hasan A, Paul A, Vrana NE, Zhao X, Memic A, Hwang Y-S, Dokmeci MR, Khademhosseini A, Biomaterials 2014, 35, 7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS, Nat. Mater 2012, 11, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chrobak KM, Potter DR, Tien J, Microvasc. Res 2006, 71, 185. [DOI] [PubMed] [Google Scholar]
- [37].Chan JM, Zervantonakis IK, Rimchala T, Polacheck WJ, Whisler J, Kamm RD, PLoS One 2012, 7, e50582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bersini S, Jeon JS, Moretti M, Kamm RD, Drug Discovery Today 2014, 19, 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Reymond N, Borda B, Ridley AJ, Nat. Rev. Cancer 2013, 13, 858. [DOI] [PubMed] [Google Scholar]
- [40].Zamora PO, Danielson KG, Hosick HL, Cancer Res. 1980, 40, 4631. [PubMed] [Google Scholar]
- [41].Sacharidou A, Koh W, Stratman AN, Mayo AM, Fisher KE, Davis GE, Blood 2010, 115, 5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vickerman V, Blundo J, Chung S, Kamm R, Lab Chip 2008, 8, 1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jeon JS, Zervantonakis IK, Chung S, Kamm RD, Charest JL, PLoS One 2013, 8, e56910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Moya ML, Hsu YH, Lee AP, Christopher CWH, George SC, Tissue Eng., Part C 2013, 19, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Natividad-Diaz SL, Browne S, Jha AK, Ma Z, Hossainy S, Kurokawa YK, George SC, Healy KE, Biomaterials 2019, 194, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sobrino A, Phan DTT, Datta R, Wang X, Hachey SJ, Romero-López M, Gratton E, Lee AP, George SC, Hughes CCW, Sci. Rep 2016, 6, 31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Price GM, Chrobak KM, Tien J, Microvasc. Res 2008, 76, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Beese M, Wyss K, Haubitz M, Kirsch T, BMC Cell Biol. 2010, 11, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Traore MA, George SC, Tissue Eng., Part B 2017, 23, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bogorad MI, DeStefano J, Wong AD, Searson PC, Microcirculation 2017, 24, e12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wong AD, Searson PC, Cancer Res. 2014, 74, 4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Richards D, Jia J, Yost M, Markwald R, Mei Y, Ann. Biomed. Eng 2017, 45, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Meng F, Meyer CM, Joung D, Vallera DA, McAlpine MC, Panoskaltsis-Mortari A, Adv. Mater 2019, 31, 1806899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, Proc. Natl. Acad. Sci. USA 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Skylar-Scott MA, Uzel SGM, Nam LL, Ahrens JH, Truby RL, Damaraju S, Lewis JA, Sci. Adv 2019, 5, eaaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Langer EM, Allen-Petersen BL, King SM, Kendsersky ND, Turnidge MA, Kuziel GM, Riggers R, Samatham R, Amery TS, Jacques SL, Sheppard BC, Korkola JE, Muschler JL, Thibault G, Chang YH, Gray JW, Presnell SC, Nguyen DG, Sears RC, Cell Rep. 2019, 26, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, Ramadan MH, Hudson AR, Feinberg AW, Sci. Adv 2015, 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Florey, BMJ 1966, 2, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Girard JP, Springer TA, Immunol. Today 1995, 16, 449. [DOI] [PubMed] [Google Scholar]
- [60].Kibria G, Heath D, Smith P, Biggar R, Thorax 1980, 35, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Davies PF, Physiol. Rev 1995, 75, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Peters EB, Christoforou N, Leong KW, Truskey GA, West JL, Cell Mol. Bioeng 2016, 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lacorre D-A, Baekkevold ES, Garrido I, Brandtzaeg P, Haraldsen G, Amalric F, Girard J-P, Blood 2004, 103, 4164. [DOI] [PubMed] [Google Scholar]
- [64].Wilson HK, Canfield SG, Shusta EV, Palecek SP, Stem Cells 2014, 32, 3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ohga N, Ishikawa S, Maishi N, Akiyama K, Hida Y, Kawamoto T, Sadamoto Y, Osawa T, Yamamoto K, Kondoh M, Ohmura H, Shinohara N, Nonomura K, Shindoh M, Hida K, Am. J. Pathol 2012, 180, 1294. [DOI] [PubMed] [Google Scholar]
- [66].Peters EB, Tissue Eng., Part B 2018, 24, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Williams IM, Wu JC, Arterioscler., Thromb., Vasc. Biol 2019, 39, 1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chin LK, Yu JQ, Fu Y, Yu T, Liu AQ, Luo KQ, Lab Chip 2011, 11, 1856. [DOI] [PubMed] [Google Scholar]
- [69].Buchanan CF, Verbridge SS, Vlachos PP, Rylander MN, Cell Adhes. Migr 2014, 8, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Topper JN, Gimbrone MA Jr, Mol. Med. Today 1999, 5, 40. [DOI] [PubMed] [Google Scholar]
- [71].Chen BPC, Li YS, Zhao Y, den Chen K, Li S, Lao J, Yuan S, Shyy JYJ, Chien S, Physiol. Genomics 2002, 7, 55. [DOI] [PubMed] [Google Scholar]
- [72].Ando J, Yamamoto K, Circ. J 2009, 73, 1983. [DOI] [PubMed] [Google Scholar]
- [73].Yamamoto K, Takahashi T, Asahara T, Ohura N, Sokabe T, Kamiya A, Ando J, Yamamoto K, J. Appl. Physiol 2003, 95, 2081. [DOI] [PubMed] [Google Scholar]
- [74].Sweet DT, Jiménez JM, Chang J, Hess PR, Mericko-Ishizuka P, Fu J, Xia L, Davies PF, Kahn ML, J. Clin. Invest 2015, 125, 2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Munson JM, Bellamkonda RV, Swartz MA, Cancer Res. 2013, 73, 1536. [DOI] [PubMed] [Google Scholar]
- [76].Regmi S, Fu A, Luo KQ, Sci. Rep 2017, 7, 39975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ma S, Fu A, Chiew GGY, Luo KQ, Cancer Lett. 2017, 388, 239. [DOI] [PubMed] [Google Scholar]
- [78].Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DYR, Chen EI, Lyden D, Bissell MJ, Nat. Cell Biol 2013, 15, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Nishiuchi R, Murayama O, Fujiwara H, Gu J, Kawakami T, Aimoto S, Wada Y, Sekiguchi K, J. Biochem 2003, 134, 497. [DOI] [PubMed] [Google Scholar]
- [80].Li S, Nih LR, Bachman H, Fei P, Li Y, Nam E, Dimatteo R, Carmichael ST, Barker TH, Segura T, Nat. Mater 2017, 16, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Turturro MV, Christenson MC, Larson JC, Young DA, Brey EM, Papavasiliou G, PLoS One 2013, 8, e58897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bordeleau F, Mason BN, Lollis EM, Mazzola M, Zanotelli MR, Somasegar S, Califano JP, Montague C, LaValley DJ, Huynh J, Mencia-Trinchant N, Negrón Abril YL, Hassane DC, Bonassar LJ, Butcher JT, Weiss RS, Reinhart-King CA, Proc. Natl. Acad. Sci. USA 2017, 114, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Reid SE, Kay EJ, Neilson LJ, Henze A-T, Serneels J, McGhee EJ, Dhayade S, Nixon C, Mackey JB, Santi A, Swaminathan K, Athineos D, Papalazarou V, Patella F, Román-Fernández Á, ElMaghloob Y, Hernandez-Fernaud JR, Adams RH, Ismail S, Bryant DM, Salmeron-Sanchez M, Machesky LM, Carlin LM, Blyth K, Mazzone M, Zanivan S, EMBO J. 2017, 36, 2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Qi J, Chen N, Wang J, Siu C-H, Mol. Biol. Cell 2005, 16, 4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chen MB, Whisler JA, Fröse J, Yu C, Shin Y, Kamm RD, Nat. Protoc 2017, 12, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]