Abstract
Background
Machine learning (ML) is a subdomain of artificial intelligence that enables computers to abstract patterns from data without explicit programming. A myriad of impactful ML applications already exists in orthopaedics ranging from predicting infections after surgery to diagnostic imaging. However, no systematic reviews that we know of have compared, in particular, the performance of ML models with that of clinicians in musculoskeletal imaging to provide an up-to-date summary regarding the extent of applying ML to imaging diagnoses. By doing so, this review delves into where current ML developments stand in aiding orthopaedists in assessing musculoskeletal images.
Questions/purposes
This systematic review aimed (1) to compare performance of ML models versus clinicians in detecting, differentiating, or classifying orthopaedic abnormalities on imaging by (A) accuracy, sensitivity, and specificity, (B) input features (for example, plain radiographs, MRI scans, ultrasound), (C) clinician specialties, and (2) to compare the performance of clinician-aided versus unaided ML models.
Methods
A systematic review was performed in PubMed, Embase, and the Cochrane Library for studies published up to October 1, 2019, using synonyms for machine learning and all potential orthopaedic specialties. We included all studies that compared ML models head-to-head against clinicians in the binary detection of abnormalities in musculoskeletal images. After screening 6531 studies, we ultimately included 12 studies. We conducted quality assessment using the Methodological Index for Non-randomized Studies (MINORS) checklist. All 12 studies were of comparable quality, and they all clearly included six of the eight critical appraisal items (study aim, input feature, ground truth, ML versus human comparison, performance metric, and ML model description). This justified summarizing the findings in a quantitative form by calculating the median absolute improvement of the ML models compared with clinicians for the following metrics of performance: accuracy, sensitivity, and specificity.
Results
ML models provided, in aggregate, only very slight improvements in diagnostic accuracy and sensitivity compared with clinicians working alone and were on par in specificity (3% (interquartile range [IQR] -2.0% to 7.5%), 0.06% (IQR -0.03 to 0.14), and 0.00 (IQR -0.048 to 0.048), respectively). Inputs used by the ML models were plain radiographs (n = 8), MRI scans (n = 3), and ultrasound examinations (n = 1). Overall, ML models outperformed clinicians more when interpreting plain radiographs than when interpreting MRIs (17 of 34 and 3 of 16 performance comparisons, respectively). Orthopaedists and radiologists performed similarly to ML models, while ML models mostly outperformed other clinicians (outperformance in 7 of 19, 7 of 23, and 6 of 10 performance comparisons, respectively). Two studies evaluated the performance of clinicians aided and unaided by ML models; both demonstrated considerable improvements in ML-aided clinician performance by reporting a 47% decrease of misinterpretation rate (95% confidence interval [CI] 37 to 54; p < 0.001) and a mean increase in specificity of 0.048 (95% CI 0.029 to 0.068; p < 0.001) in detecting abnormalities on musculoskeletal images.
Conclusions
At present, ML models have comparable performance to clinicians in assessing musculoskeletal images. ML models may enhance the performance of clinicians as a technical supplement rather than as a replacement for clinical intelligence. Future ML-related studies should emphasize how ML models can complement clinicians, instead of determining the overall superiority of one versus the other. This can be accomplished by improving transparent reporting, diminishing bias, determining the feasibility of implantation in the clinical setting, and appropriately tempering conclusions.
Level of Evidence
Level III, diagnostic study.
Introduction
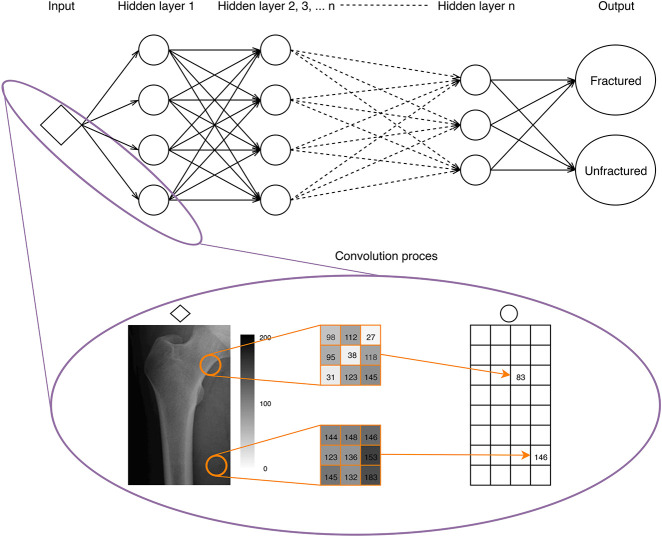
Artificial intelligence is the capability of computers to display intelligent behavior, as opposed to humans, who demonstrate natural intelligence [15, 20]. Machine learning (ML) is a subdomain of artificial intelligence that enables computers to abstract patterns from data without explicit programming [40, 42]. Machine learning applications are rapidly entering clinical practice in a variety of domains ranging from diagnostic to prognostic purposes [7, 12, 45]. The two most common types of ML used in medicine are supervised and unsupervised ML [11, 36]. Supervised learning requires both input variables and labeled outcomes. In this form of ML, the algorithms learn to map the relationships between the input variables and outcomes [2, 11]. Examples include processing the input of plain radiographs to detect the presence or absence of a fracture, often performed by convolutional neural networks (Fig. 1). Unsupervised learning, unlike supervised learning, only requires input variables [11]. The algorithm seeks to find unknown patterns in the dataset to structure the data, without reference to a known outcome.
Fig. 1.
This figure shows a basic explanation of the most frequently used supervised learning algorithm—convolutional neural networks—for diagnosing orthopaedic conditions with imaging. A convolutional neural network transforms the input (for example, a plain radiograph of the femur) into one or more classification outputs (fracture or unfractured). The expanded box is a snapshot of the convolutional process, in which the input radiograph is processed into a matrix of pixel values. After applying different filters developed in the training process, a single value is created in the output matrix (bottom right). This process is repeated in multiple hidden layers with different filters convolving across output matrices throughout hidden layers. Based on the connections and weights in the last hidden layer, the algorithm classifies the femur into fractured or not.
Several ML models and applications already exist in orthopaedics [5, 18, 50, 52, 58, 21–26, 28, 37]. Despite the number of available studies, few systematic reviews or meta-analyses have examined the quality, limitations, and potential of ML models versus clinicians. Our group conducted a similar study in a wide range of neurosurgical applications which suggested that ML outperformed humans using multiple input features including radiographic and clinical parameters [48]. However, this review lacked scrutiny of the differences in input features and subspecialties as well as an in-depth discussion of the potential of ML models in musculoskeletal imaging. The potential benefit of the implementation of ML models to assess radiographs in orthopaedics is especially worthwhile, as misinterpretation is the primary reason for malpractice claims and may lead to grave clinical consequences such as malunion or joint collapse [3]. Furthermore, the systematic neurosurgical review performed in 2016 does not reflect the current ML environment since novel techniques, new forms of knowledge, and additional explanatory methods are being developed exponentially rather than linearly. Recent nonorthopaedic high-profile studies published since 2017 such as Esteva et al. [12], Ting et al. [53], Lundberg et al. [35], Tomašev et al. [54], Liang et al. [31], Lee et al. [30], Hollon et al. [19], and Milea et al. [38], have transformed our understanding of the potential for ML to aid or replace clinicians. These studies have compared the algorithms to clinical experts and shown that these algorithms are able to diagnose or predict better than experts in a fraction of the time. Updated studies in this growing field of ML applications in medicine will help us understand if ML changes our expectations for the role of clinicians in the future. To our knowledge, no systematic reviews have compared the performance of the currently available ML models to the performance of clinicians in musculoskeletal imaging.
In this systematic review, we therefore aimed: (1) to compare performance of ML models versus clinicians on detecting, differentiating, or classifying orthopaedic abnormalities on imaging by (A) accuracy, sensitivity, and specificity, (B) input features (for example, plain radiographs, MRI scans, ultrasound), (C) clinician specialties, and (2) to compare the performance of clinicians aided versus unaided by ML models.
Materials and Methods
Systematic Study Search
We performed a systematic search in PubMed, Embase, and the Cochrane Library for studies published up to October, 2019. The search syntax was built with the guidance of a professional medical librarian using synonyms for “machine learning” and all potential orthopaedic specialties (see Appendix 1; Supplemental Digital Content 1, http://links.lww.com/CORR/A384). Two reviewers (OQG, MERB) independently screened all titles and abstracts for eligible articles based on predefined criteria (detailed below). Full-text articles were evaluated, and the references of the identified studies were examined for potentially relevant articles that were not identified by the initial search. Disagreements were solved by a discussion in which two other authors (PTO, JHS) were involved to assess article inclusion, quality assessment, and data extraction, until there was a consensus. We adhered to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines for this review [41].
Eligibility Criteria
We included articles if they compared ML models head-to-head with clinicians in applications relevant to the orthopaedic patient population. We defined the orthopaedic patient population as patients with disorders of the bones, joints, ligaments, tendons, and muscles. All application domains such as diagnosis, prognosis, treatment, and outcome were included. In ML, the “ground truth” refers to the reference standard on which the model is trained and tested. This ground truth varied by article depending on its specific domain, including surgical or histologic confirmation in a radiologic classification task or the consensus of a panel of experts. We excluded studies that did not compare ML models and human performance, nonorthopaedic specialty studies, non-English-language studies, studies with no full text available, and nonrelevant article types, such as case reports, animal studies, and letters to the editor.
Assessment of Methodological Quality
Two reviewers (OQG, MERB) independently appraised the quality of the included studies using predefined extraction sheets, based on the Methodological Index for Non-randomized Studies (MINORS) criteria [49]. We modified the seven-item MINORS checklist to make it applicable to our systematic review by including disclosure, study aim, input feature, ground truth, comparison between ML model and clinician, dataset distribution, performance metric, and description of the ML model. These eight items were scored on a two-point scale: 0 (not reported or unclear) or 1 (reported and adequate).
After screening 6531 titles and abstracts, we assessed 40 full-text studies for eligibility, and ultimately 14 studies were included for critical appraisal (Fig. 2). The study aim, inclusion and exclusion criteria for the input features, ML model used, and the human comparison group were clearly explained in all studies. The distribution of the dataset was clearly described in 11 studies; in the remainder, the dataset distribution was unclear or a test set was not used [4, 6, 17]. Disclosure was reported in 12 studies; thus, for two studies, conflicts of interest could not be evaluated [8, 44]. The ground truth was not clearly described and clear performance metrics were missing in two studies [17, 44]. This deviated considerably from existing reporting standards as it introduced bias by inadequate ground truth labeling and not providing transparent head-to-head comparison [10]. Thus, we excluded these two studies from this review. In total, 12 studies were included for quantitative synthesis (see Appendix 2; Supplemental Digital Content 2, http://links.lww.com/CORR/A385) and assessed using the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) guidelines for completeness of reporting of the ML model (see Appendix 3; Supplemental Digital Content 3, http://links.lww.com/CORR/A386). The TRIPOD guideline, which is a checklist of 22 items introduced in 2015, should be followed when reporting algorithm results [10]. This guideline is deemed essential for transparent reporting of study outcomes and guide developers of algorithms towards a more uniform reporting of their algorithm’s performance.
Fig. 2.
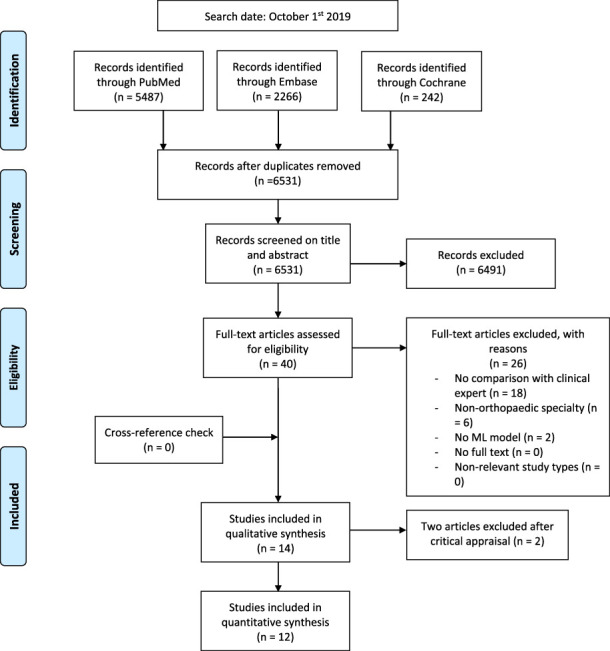
This Preferred Reporting Items for Systematic Reviews and Meta-analyses 2009 flow diagram shows how studies were systematically identified, screened, and included. After screening 6531 studies, 14 studies were critically appraised and ultimately 12 studies were included for quantitative synthesis.
Data Extraction
The data we obtained from each study were year of publication, output classes, performance measures, p value of the difference in performance, input features, outcome measures, performance of ML, performance of the clinician, ML model, level of education of the human performer and (sub)specialization of the clinician, ground truth, size of the dataset, size of training set, validation method or size of the validation set, and size of the test set. For studies comparing multiple outcome measures between artificial and natural intelligence or comparing different groups of clinicians with ML models, we extracted each separate comparison.
Study Characteristics
The median size of the training set was 1702 datapoints (interquartile range [IQR] 337 to 16,075), that of the validation set was 334 datapoints (IQR 134 to 37,481), and that of the test set was 334 datapoints (IQR 155 to 2410). Five studies used cross-validation only instead of a separate validation set [6, 9, 29, 34, 59]. Two studies did not use a test set [6, 29]. All studies used a binary assessment. No studies provided additional information (for example, physical examination findings) to either ML models or clinicians. No studies were designed as a prospective, randomized, controlled trial. None of the studies adhered to all TRIPOD checklist items.
Output classes for the 12 studies comparing ML models and humans were binary detection of fractures or other radiologic abnormalities (n = 11) [1, 4, 59, 6, 8, 14, 29, 33, 34, 43, 55] or both detection and classification of the diagnosis (n = 1) [9]. Input features used by the ML models were plain radiographs (n = 8) [1, 8, 9, 14, 33, 43, 55, 59], MRI (n = 3) [4, 29, 34], and ultrasound examinations (n = 1) [6]. A p value was provided for 91% (52 of 57) of the outcome measures. Outcome measures accompanied by a p value were used to assess the performance of ML models and clinicians, sensitivity and specificity (both 33% [17 of 52]), accuracy (31% [16 of 52]), and area under the receiver operating characteristic curve [AUC] (3.8% [2 of 52]). All ML models were supervised learning algorithms with the following two subtypes: convolutional neural networks (n = 11) [1, 4, 59, 8, 9, 14, 29, 33, 34, 43, 55] and random forest ML (n = 1) [6]. All studies used publicly available pretrained models or data augmentation methods during training. Ground truth differed by study and was established by expert agreement with the aid of a more advanced radiographic modality (n = 3) [8, 9, 14], expert agreement without the aid of a more advanced radiographic modality (n = 6) [4, 33, 34, 43, 55, 59], surgical or histologic confirmation (n = 2) [1, 29], and clinical diagnosis (n = 1) [6].
The studies were also analyzed by the type of input feature used and by the specialty of the clinician expert.
Input features could be divided into two main categories: plain radiographs and MRIs. The interpretation of plain radiographs by ML models was compared with that of clinicians in eight studies: detection of osseous abnormalities (n = 8) [1, 8, 9, 14, 33, 43, 55, 59] and fracture classification (n = 1) [9]. Detection of osseous abnormalities was the focus of seven studies, namely distal radius fractures [14, 33], femoral neck fractures [1], intertrochanteric hip fractures [55], hip osteoarthritis [59], femoral head osteonecrosis [8], or any fracture in the hand, wrist, or ankle [43]. The detection and classification of proximal humerus fractures were investigated by one study [9]. MRI interpretation by ML models was compared with that of clinicians in three studies. The first study evaluated the detection of general abnormalities in the knee, ACL tears, and meniscal tears [4]; the second study focused on the ability to differentiate between tuberculous and pyogenic spondylitis [29]; and the third study evaluated the detection of cartilage lesions of the knee [34]. Ultrasound examination as an input feature was used in one study to distinguish between lateral epicondylosis and asymptomatic elbows [6].
Assessing physicians were divided into three groups by their comparison with ML models: radiologists (6 of 12 studies) [4, 6, 8, 14, 29, 34], orthopaedic surgeons (4 of 12) [9, 14, 43, 55], and all others (5 of 12), including physiotherapists [6], general physicians [9, 59], emergency medicine clinicians (consisting of physicians assistants and medical doctors) [33], and undergraduate students with different levels of education [1].
Statistical Analysis
Given the heterogeneity of the orthopaedic applications, no quantitative meta-analysis was performed. Because all 12 studies were of comparable quality and they all clearly included six of the eight critical appraisal items (study aim, input feature, ground truth, ML versus human comparison, performance metric, and ML model description), a quantitative summarization was provided by calculating the median absolute improvement. The median absolute improvement was determined by calculating the differences in performance metrics between the ML model and clinician for the most commonly used statistical measures of performance: accuracy, sensitivity, specificity, and AUC. The absolute median represents an overview of performance where positive and negative values correspond with superior performance of the ML model and clinician, respectively. No significance of any sort can be attributed to this summary metric. Accuracy refers to the proportion of total correct predictions among the total number of predictions, sensitivity refers to the proportion of true positive cases among the total number of positive cases, and specificity refers to the proportion of true negative cases among the total number of negative cases. AUC refers to the ability of the algorithm to discriminate between two classes ranging from 0 to 1.
Superior or inferior performance of the ML model versus that of clinicians was defined as a significant better or worse performance, respectively, according to the statistical tests used in the studies (p < 0.05). Equal performance was defined as a nonsignificant performance difference (p > 0.05). The sizes of the training, validation, and test sets are reported as percentages of the total dataset. We used Microsoft Excel Version 19.11 (Microsoft Inc, Redmond, WA, USA) and Stata® 14.0 (StataCorp LP, College Station, TX, USA) for the statistical analyses, and Mendeley Desktop Version 1.19.4 (Mendeley Ltd, London, UK) as reference management software.
Results
Accuracy, Sensitivity, and Specificity
ML models slightly outperformed clinicians working alone in detecting, differentiating, or classifying orthopaedic abnormalities on musculoskeletal imaging in diagnostic accuracy and sensitivity, and were on par in specificity. The median absolute improvement values were 3% (range -12% to 19%; IQR -2.0% to 7.5%) [1, 4, 9, 14, 29, 34, 43, 55, 59] for accuracy, 0.06 (range -0.15 to 0.41; IQR -0.03 to 0.14) for sensitivity, and 0.00 (range -0.15 to 0.13; IQR -0.048 to 0.048) [4, 6, 8, 9, 14, 29, 33, 34, 55, 59] for specificity. The wide ranges and IQRs in all three performance measures narrow toward zero, which indicates that there was no strong difference between the performance of ML models and clinicians. The median absolute improvement in the AUC was not calculated because only four comparisons were provided [6, 8, 29, 55]. The ML models performed better than clinicians in 38% of all performance measures (AUC, accuracy, sensitivity, and specificity; 20 of 52) and worse than clinicians in 3.8% (2 of 52); no difference was found in 58% (30 of 52) (Table 1).
Table 1.
Performance of ML models and clinical experts
| Authora | Output | Input features | Outcome measures | ML models vs clinicians (95% CI) | p value | ML models vs clinicians | Total dataset | Training sizeb | Validation sizeb/method | Testing sizeb | Ground truthc |
| Adams et al. [1] | Detection of femoral neck fracture | Radiography | Accuracy | 91% (86 to 95) vs 91% | 1 | CNN vs BSc students | 800 | 64% | 16% | 20% | Surgically confirmed |
| Bien et al. 1d [4] | Detection of general abnormality | MRI |
|
|
|
CNN vs Radiologists | 1370 | 91% | 9% | NA | Consensus of three radiologists |
| Bien et al 2d [4] | Detection of ACL tear | MRI |
|
|
|
CNN vs seven radiologists | 1370 | 91% | 9% | NA | Consensus of three radiologists |
| Bien et al. 3d [4] | Detection of meniscal tears | MRI |
|
|
|
CNN vs seven radiologists | 1370 | 91% | 0% | NA | Consensus of three radiologists |
| Bureau et al. [6] | Differentiation of lateral epicondylosis and asymptomatic elbows | Ultrasound |
|
|
|
RF vs one MSK radiologist and one physiatrist | 54 | 100% | LOOCV | NA | Clinical diagnosis |
| Chee et al. [8] | Detection of femoral head osteonecrosis | Radiography |
|
|
|
CNN vs two radiologists | 1892 | 71% | 8% | 21% | Consensus of two radiologists and MRI |
| Chung et al. 1 [9] | Detection of proximal humerus fracture | Radiography |
|
|
|
|
1891 | 90% | 10-FCV | 10% | Consensus of two orthopaedists, one radiologist; CT for failed consensus |
| Chung et al. 2 [9] | Detection of proximal humerus fracture | Radiography |
|
|
|
CNN vs 11 general orthopaedists | 1891 | 90% | 10-FCV | 10% | Consensus of two orthopaedists, one radiologist; CT for failed consensus |
| Chung et al. 3 [9] | Detection of proximal humerus fracture | Radiography |
|
|
|
CNN vs 19 orthopaedists specialized in shoulder | 1891 | 90% | 10-FCV | 10% | Consensus of two orthopaedists, one radiologist; CT for failed consensus |
| Chung et al. 4 [9] | Classifying normal, fracture of greater tuberosity, surgical neck, three-part, or four-part | Radiography |
|
|
|
CNN vs 28 general physicians | 1891 | 90% | 10-FCV | 10% | Consensus of two orthopaedists, one radiologist; CT for failed consensus |
| Chung et al. 5 [9] | Classifying normal, fracture of the greater tuberosity, surgical neck, three-part, or four-part | Radiography |
|
|
|
CNN vs 11 general orthopaedists | 1891 | 90% | 10-FCV | 10% | Consensus of two orthopaedists, one radiologist; CT for failed consensus |
| Chung et al. 6 [9] | Classifying normal, fracture of the greater tuberosity, surgical neck, three-part, or four-part | Radiography |
|
|
|
CNN vs 19 orthopaedists specialized in shoulder | 1891 | 90% | 10-FCV | 10% | Consensus of two orthopaedists, one radiologist; CT for failed consensus |
| Gan et al. 1 [14] | Detection of distal radius fracture | Radiography |
|
|
|
CNN vs three orthopaedists | 2340 | 87% | 13% | 13% | Consensus of three orthopaedists and CT |
| Gan et al. 2 [14] | Detection of distal radius fracture | Radiography |
|
|
|
CNN vs three radiologists | 2340 | 87% | 13% | 13% | Consensus of three orthopaedists and CT |
| Kim et al. [29] | Differentiate tuberculous and pyogenic spondylitis | MRI |
|
|
|
CNN vs three MSK radiologists | 161 | 100% | 4-FCV | NA | Bacteriologic and/or histologic confirmation |
| Lindsey et al.d [33] | Detection of wrist fracture | Radiography |
|
|
|
CNN vs 39 ED clinicians (15 PAs; 24 MDs) | 135,845 | 80% | 17% | 3% | Subspecialized orthopaedist |
| Liu et al. [34] | Detection of cartilage lesions within the knee | MRI |
|
|
|
CNN versus radiology residents (2), MSK fellows (2), MSK (1) | 17,395 | 92% | 5-FCV | 8% | MSK radiologist |
| Olczak et al. [43] | Detection of fracture: hand, wrist, ankle | Radiography | Accuracy | 83% (79 to 87) vs 82% (78 to 86) | NA | CNN vs two senior orthopaedic surgeons | 256,458 | 70% | 20% | 10% | Radiology report and three orthopaedists |
| Urakawa et al. [55] | Detection of intertrochanteric hip fracture | Radiography |
|
|
|
CNN vs five orthopaedists | 3346 | 80% | 10% | 10% | Orthopaedist |
| Xue et al. [59] | Detection of hip osteoarthritis | Radiography |
|
|
|
CNN vs three physicians | 420 | 80% | 5-FCV | 20% | Consensus of two chief physicians |
Bold values indicate that the difference between the performance machine learning models and clinicians was statistically significant (p < 0.05). aSeparate comparison were extracted for Bien et al., Chung et al., and Gan et al., for comparing multiple outcome measures between machine learning models and clinicians or comparing different groups of clinicians with machine learning models. bPercentage of the total amount of the dataset. cThe definition of ground truth (reference standard for machine learning models) varied between each study. dThis study also used the measured performance of clinicians aided and unaided by machine learning models.
ML = machine learning; CNN = convolutional neural network; BSc = Bachelor of Science; NA = not available; RF = random forest; MSK = musculoskeletal; LOOCV = leave-one out cross validation; FCV = fold cross-validation; ED = emergency department; PA = physician assistant; MD = medical doctors.
Results Stratified by Input Features
ML models outperformed clinicians more frequently when interpreting plain radiographs than when interpreting MRIs. Interpretation of plain radiographs by ML models was better than that by clinicians in 17 of 34 of all performance measures (accuracy, sensitivity, and specificity) and worse in zero of 34; no difference was found in 17 of 34. On plain radiographs, ML models performed better than clinicians in terms of detecting osseous abnormalities or classifying fractures in 13 of 28 all performance measures and 4 of 9, respectively; worse in 0 of 28 and 0 of 9, respectively; and no difference was found in 15 of 28 and 5 of 9, respectively. ML models were able to interpret MRIs better than clinicians in 3 of 16 of all performance measures and worse in 2 of 16; no difference was found in 11 of 16. Only one study evaluated ultrasound interpretations [6], and it showed no difference between ML models and clinicians in distinguishing between lateral epicondylosis and asymptomatic elbows.
Results Stratified by Clinician Expert Specialty
ML models performed similarly to radiologists and orthopaedists but better than all other clinicians. ML models performed better than clinicians in two specialist groups, orthopaedics and radiology, in 7 of 19 and 7 of 23 of all performance measures, respectively, and worse in 0 of 19 and 2 of 23, respectively; no difference was found in 12 of 19 and 14 of 23, respectively. ML models performed better than all other clinicians (physiotherapists, general physicians, emergency medicine clinicians, and undergraduate students) in 6 of 10 outcome measures and worse in 0 of 10; no difference was found in 4 of 10.
Results of Studies of ML Aiding Clinicians
Two studies evaluated the performance of clinicians aided and unaided by ML models; both demonstrated that clinicians aided by ML models outperformed clinicians unaided by ML. Lindsey et al. [33] showed that clinicians aided by ML models had improved performance in detecting wrist fractures compared with their non-aided performance. On average, clinicians had a relative proportional reduction of misinterpretation when aided by ML models of 47% (95% confidence interval [CI] 37 to 54; p < 0.001), compared with their non-aided performance. Bien et al. [4] evaluated the ML-aided and ML-unaided performance of clinicians in detecting general abnormalities and specific diagnoses on MRIs of the knee and found a mean increase in specificity of 0.048 for the aided detection of ACL tears (95% CI 0.029 to 0.068; p < 0.001).
Discussion
The availability of ML applications in the orthopaedic arena is increasing rapidly, but few studies have compared the performance of these models against their human counterparts. In 2017, we compared ML models and clinicians in the neurosurgical field and found that ML generally outperformed clinicians. However, that study was performed using not only imaging but also clinical input features in a wide variety of different ML models and was performed more than three years ago. Many advancements and novel techniques have transformed our understanding of the potential for ML since that time. Frequent determination of the advancements of ML in medicine and its performance compared with clinicians is important in this rapidly growing field. In fact, none of the included studies in this review had been published before our 2017 neurosurgical review. We found that ML models again outperformed clinicians more than clinicians outperformed ML models, but in aggregate these improvements were small. Also, clinicians aided by ML models performed better and faster compared to their non-aided performance. ML models demonstrate great potential to improve the assessment of musculoskeletal imaging. However, significant hurdles–such as the lack of transparent reporting, inaccurate ground truth labeling, and transportability issues to the clinical nonresearch setting–must be overcome before clinicians can embrace ML models in daily practice.
Limitations
This review has several limitations. First, summarizing the results with medians does not provide adequate weight to each study based on quality and size. The size of studies ranged from 54 to 256,458 datapoints and no correction could be made for this imbalance. Two studies did not use a proper holdout test set [4, 29], which could overestimate model performance as the data was used for both training and testing. Three studies assessed multiple outcome measures [4, 9, 14], resulting in an overrepresentation of these performance measures. Ideally, randomized controlled trials ensure fair comparison between ML models and clinicians, but to date, only two of these randomized trials exist [32, 57]. However, to justify the data pooling, all included studies were of comparable high quality—maximum score on six of the eight critical appraisal items—and randomized clinical trials in ML models are not (yet) widely accepted. Second, our group conducted a similar review in 2017 in the field of neurosurgery [48]; there were no overlapping studies between both reviews. Third, ground-truth establishment differed throughout the studies, ranging from surgical or histologic confirmation to expert consensus. Some models could therefore have been trained on datasets containing human errors, leading to an overestimation of the clinician’s performance. For example, an incorrectly labeled ground truth can lead to incorrect training of the algorithm, thereby falsely decreasing the algorithm’s performance. If the clinician also does not assume that a fracture is present, his or her performance will falsely increase. In this review, all studies used relatively accurate ground truth labels such as data labeled by experts or histopathological confirmation compared with more error-prone radiology reports that may have been dictated by inexperienced junior residents. Therefore, the underestimation of the performance metrics of the ML models are of limited proportion. Fourth, positive publication bias may have occurred because studies that reported the favorability of ML models may have been published more frequently. Additionally, all reviewed studies included comparisons in imaging, specifically in settings where ML models currently show the most promising results in multiple disciplines and are expected to outperform clinicians [7, 48]. The superiority of ML models might therefore be overestimated and only applicable to imaging tasks, especially because it constitutes only one of the clinician’s many specific tasks. It is reasonable to expect many trials in the near future to provide a more accurate comparison between ML models and clinicians as algorithm validation, implementation, and overall acceptance is increasing in clinical care. Fifth, the performance of the ML models could have been overestimated in studies that did not use a proper independent test set. Further, studies differed in the amount of analyses and outcome measures, which could have caused overrepresentation of some studies. No uniform comparison could have been made to prevent this overrepresentation because there was heterogeneous reporting of outcome measures. Furthermore, a p value was not provided for four of 57 outcome measures. All four showed that the ML models had superior performance, and in these cases, the strength of the ML models might have been underestimated [6, 8, 33]. Sixth, the AUC was provided in only four studies with two p values, making a comparison unwarranted. However, binary predictions were made in all studies, making this limitation less problematic. Seventh, because all studies used a binary assessment, the clinician had to choose between the occurrence or nonoccurrence of an event. This meant that there was no consideration of the clinicians’ doubt—which is often the case in clinical practice—this might have underestimated the clinician’s performance. The implementation of ordinal (such as, occurrence, doubt, or nonoccurrence) or continuous (percentage of confidence of the occurrence) could mimic a more realistic environment in future comparative studies. Eighth, none of the studies adhered to the TRIPOD guideline, in particular the subitems of model specification and development. Following this statement is important to promote uniformity in presenting and developing ML models, thereby allowing future studies to be compared [10]. Lastly, no study included speed as a performance measure. In simple and repetitive tasks, the computer is increasingly expected to outperform humans on this measure.
Accuracy, Sensitivity, and Specificity
Machine learning models provided, in aggregate, only very slight improvements in diagnostic accuracy, sensitivity, and specificity compared with clinicians working alone. In the similar study by Senders et al. [48], we found an overall stronger performance of ML models compared with clinicians in neurosurgery. This might be explained by the fact that none of the included ML models in the current study used clinical input features such as age or vital parameters. The relationship between clinical parameters and outcomes such as postoperative survival is considerably more intricate, and especially in prognostication ML models, may outperform clinicians. Several nonradiology orthopaedic ML models exist but none have been compared with humans to date [7, 13, 27, 46]. In our earlier neurosurgery study, 10 of 23 studies compared ML models using clinical features as input with clinicians in predicting outcomes. All 10 demonstrated overall better performance of ML models compared with clinicians. Future studies should investigate the potential benefit of ML models using nonradiology input features to predict outcomes such as presurgical planning or survival in orthopaedic patients to determine the added value of these kind of algorithms.
Input features
Machine learning models were primarily used to interpret radiologic data with the use of neural networks. Overall, ML models outperformed clinicians more when interpreting plain radiographs than when interpreting MRIs. Studies that investigated interpretation of plain radiographs looked at single radiographs showing osseous structures, while a series of MR images were converted to a two-dimensional image showing various structures. Additionally, the availability of training data for ML models that interpret plain radiographs is much higher than for ML models that interpret MRIs. This is reflected in the size of the datasets; plain radiographs had a larger median dataset than MRIs: 2116 (IQR 1073-24,754) datapoints and 1370 (IQR 161-17,395) datapoints, respectively. As a recent study demonstrated, an increase in the size of training dataset to around 5000 images corresponded with increased performance, after which no benefit of additional training data was noticed [56]. Diversity in the predicted outcomes also influences on ML models’ performance. In Chung et al. [9], distinctive fracture lines in the greater tuberosity with low variability made detection easier compared with fractures in the more complex anatomical surgical neck site. The same applies for detecting an osseous abnormality versus soft tissue abnormality; in general osseous abnormalities are more evident on imaging resulting in a better ML models’ performance. Detection of “simple” osseous abnormalities on relatively uncomplicated plain radiographs might thus yield a higher difference in performance than complex MR images.
Clinician Specialty
Radiologists and orthopaedists generally performed similarly to ML models, while ML models mostly outperformed other nonexpert clinicians. This suggests that ML models can improve health care by assisting in well-defined tasks for non-musculoskeletal specialists or trainees and can aid clinicians in more austere or remote settings. Our neurosurgical review included studies that compared ML models and clinicians subdivided by specialty, but no separate analyses were provided to make a comparison [48].
Combining Clinicians and ML
Considerable improvements were demonstrated in the diagnostic accuracy of specialists aided by ML models. In orthopaedics, the potential benefit of lower misinterpretation rates of radiographs is especially worthwhile. In addition to potential liability issues [3], misdiagnosed radiographs may have severe clinical consequences such as joint collapse and posttraumatic osteoarthritis. Also, assessing abnormalities of the musculoskeletal system on imaging comprises a significant amount of time during daily orthopaedic practice. Clinicians must view an increasing amount of imaging studies and complexity compared with 10 to 20 years ago, making it both time consuming and more prone to error [39]. Multiple studies suggest that time devoted to imaging interpretation decreases when aided by ML models compared with non-aided time [16, 19, 33]. This emphasizes that these ML models could improve the safety and effectiveness of patient care while working in conjunction with human counterparts.
Future Prospects and Conclusions
We found that ML models have comparable performance to clinicians in assessing musculoskeletal images. ML models may enhance the performance of clinicians as a technical supplement rather than as a replacement for clinical or natural intelligence. On the other hand, there are circumstances in which ML models perform tasks that lie beyond the capacity of clinicians, such as accurately predicting complications and survival in patients with cancer [5, 21, 23, 27, 51]. Additionally, the advantages of using computers in helping make clinical decisions–such as uninterruptedly working at a high speed without fatigue–hold great potential to improve healthcare. Future studies should emphasize how ML models can complement clinicians, instead of analyzing the potential superiority of one versus the other. Substantial challenges exist before ML can be used regularly in daily practice. The sterile research environments in which algorithms are developed do not reflect the conditions observed in clinical practice. Also, ML models often reveal connections between disease characteristics and clinical outcomes in ways humans cannot understand [47]. This results in a lack of explanation or rationale for the crucial decisions ML models make, which is currently known as the “black box problem.” Clinicians could be guided toward incorrect decisions if the algorithm is not well understood. The heat map proposed by Lindsey et al. [33], could provide a solution to this issue. This heat map is overlaid on the radiograph and highlights the model’s calculated probability of a fracture—from yellow when the models is more confident to blue when less confidence—without making the binary decision of the bone being fractured or not. The optimal synergy between man and machine can be achieved by improving transparent reporting, diminishing bias, determining feasibility of application in the clinical setting, and appropriately considering conclusions. In the future, orthopaedics will likely embrace machine learning as a technical supplement rather than as a replacement for clinicians, creating a desirable synergy between “machine and man” rather than “machine versus man.”
Acknowledgments
We thank D. Hayden PhD, from Harvard Catalyst Biostatistics Consultation for his help in the design and statistics of this study.
Footnotes
Each author certifies that neither he or she, nor any member of his or her immediate family, has funding or commercial associations (consultancies, stock ownership, equity interest, patent/licensing arrangements, etc.) that might pose a conflict of interest in connection with the submitted article.
All ICMJE Conflict of Interest Forms for authors and Clinical Orthopaedics and Related Research® editors and board members are on file with the publication and can be viewed on request.
Each author certifies that his or her institution waived approval for the reporting of this investigation and that all investigations were conducted in conformity with ethical principles of research.
This work was performed at Massachusetts General Hospital - Harvard Medical School, Boston, MA, USA.
The first two authors contributed equally to this manuscript.
References
- 1.Adams M, Chen W, Holcdorf D, McCusker MW, Howe PD, Gaillard F. Computer vs human: Deep learning versus perceptual training for the detection of neck of femur fractures. J Med Imaging Radiat Oncol. 2019;63:27–32. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss L, Jones LD. The role of artificial intelligence and machine learning in predicting orthopaedic outcomes. Bone Joint J. 2019;101:1476–1478. [DOI] [PubMed] [Google Scholar]
- 3.Berlin L. Defending the “missed” radiographic diagnosis. AJR Am J Roentgenol. 2001;176:317–322. [DOI] [PubMed] [Google Scholar]
- 4.Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, Bereket M, Patel BN, Yeom KW, Shpanskaya K, Halabi S, Zucker E, Fanton G, Amanatullah DF, Beaulieu CF, Riley GM, Stewart RJ, Blankenberg FG, Larson DB, Jones RH, Langlotz CP, Ng AY, Lungren MP. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet Saria S, ed. PLOS Med. 2018;15:e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bongers MER, Thio QCBS, Karhade A V., Stor ML, Raskin KA, Lozano Calderon SA, DeLaney TF, Ferrone ML, Schwab JH. Does the SORG Algorithm Predict 5-year Survival in Patients with Chondrosarcoma? An External Validation. Clin Orthop Relat Res. 2019;477:2296–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bureau NJ, Destrempes F, Acid S, Lungu E, Moser T, Michaud J, Cloutier G. Diagnostic Accuracy of Echo Envelope Statistical Modeling Compared to B-Mode and Power Doppler Ultrasound Imaging in Patients With Clinically Diagnosed Lateral Epicondylosis of the Elbow. J Ultrasound Med. 2019;38:2631–2641. [DOI] [PubMed] [Google Scholar]
- 7.Cabitza F, Locoro A, Banfi G. Machine Learning in Orthopedics: A Literature Review. Front Bioeng Biotechnol. 2018;6:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee CG, Kim Y, Kang Y, Lee KJ, Chae H-D, Cho J, Nam C-M, Choi D, Lee E, Lee JW, Hong SH, Ahn JM, Kang HS. Performance of a Deep Learning Algorithm in Detecting Osteonecrosis of the Femoral Head on Digital Radiography: A Comparison With Assessments by Radiologists. AJR Am J Roentgenol. 2019:1–8. [DOI] [PubMed] [Google Scholar]
- 9.Chung SW, Han SS, Lee JW, Oh K-S, Kim NR, Yoon JP, Kim JY, Moon SH, Kwon J, Lee H-J, Noh Y-M, Kim Y. Automated detection and classification of the proximal humerus fracture by using deep learning algorithm. Acta Orthop. 2018;89:468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med. 2015;13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriel RA, Sharma BS, Doan CN, Jiang X, Schmidt UH, Vaida F. A Predictive Model for Determining Patients Not Requiring Prolonged Hospital Length of Stay After Elective Primary Total Hip Arthroplasty. Anesth Analg. 2019;129:43–50. [DOI] [PubMed] [Google Scholar]
- 14.Gan K, Xu D, Lin Y, Shen Y, Zhang T, Hu K, Zhou K, Bi M, Pan L, Wu W, Liu Y. Artificial intelligence detection of distal radius fractures: a comparison between the convolutional neural network and professional assessments. Acta Orthop. 2019;90:394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghahramani Z. Probabilistic machine learning and artificial intelligence. Nature. 2015;521:452–459. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert FJ, Astley SM, Gillan MGC, Agbaje OF, Wallis MG, James J, Boggis CRM, Duffy SW. Single Reading with Computer-Aided Detection for Screening Mammography. N Engl J Med. 2008;359:1675–1684. [DOI] [PubMed] [Google Scholar]
- 17.Gioftsos G, Grieve DW. The use of artificial neural networks to identify patients with chronic low-back pain conditions from patterns of sit-to-stand manoeuvres. Clin Biomech (Bristol, Avon). 1996;11:275–280. [DOI] [PubMed] [Google Scholar]
- 18.Hendrickx LAM, Sobol GL, Langerhuizen DWG, Bulstra AEJ, Hreha J, Sprague S, Sirkin MS, Ring D, Kerkhoffs GMMJ, Jaarsma RL, Doornberg JN, Machine Learning Consortium. A Machine Learning Algorithm to Predict the Probability of (Occult) Posterior Malleolar Fractures Associated With Tibial Shaft Fractures to Guide “Malleolus First” Fixation. J Orthop Trauma. 2020;34:131–138. [DOI] [PubMed] [Google Scholar]
- 19.Hollon TC, Pandian B, Adapa AR, Urias E, Save A V., Khalsa SSS, Eichberg DG, D’Amico RS, Farooq ZU, Lewis S, Petridis PD, Marie T, Shah AH, Garton HJL, Maher CO, Heth JA, McKean EL, Sullivan SE, Hervey-Jumper SL, Patil PG, Thompson BG, Sagher O, McKhann GM, Komotar RJ, Ivan ME, Snuderl M, Otten ML, Johnson TD, Sisti MB, Bruce JN, Muraszko KM, Trautman J, Freudiger CW, Canoll P, Lee H, Camelo-Piragua S, Orringer DA. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020;26:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan MI, Mitchell TM. Machine learning: Trends, perspectives, and prospects. Science. 2015;349:255–260. [DOI] [PubMed] [Google Scholar]
- 21.Karhade A V, Ahmed AK, Pennington Z, Chara A, Schilling A, Thio QCBS, Ogink PT, Sciubba DM, Schwab JH. External validation of the SORG 90-day and 1-year machine learning algorithms for survival in spinal metastatic disease. Spine J. 2020;20:14–21. [DOI] [PubMed] [Google Scholar]
- 22.Karhade A V, Bongers MER, Groot OQ, Kazarian ER, Cha TD, Fogel HA, Hershman SH, Tobert DG, Schoenfeld AJ, Bono CM, Kang JD, Harris MB, Schwab JH. Natural language processing for automated detection of incidental durotomy. Spine J. 2020;20:695–700. [DOI] [PubMed] [Google Scholar]
- 23.Karhade A V, Ogink PT, Thio QCBS, Cha TD, Gormley WB, Hershman SH, Smith TR, Mao J, Schoenfeld AJ, Bono CM, Schwab JH. Development of machine learning algorithms for prediction of prolonged opioid prescription after surgery for lumbar disc herniation. Spine J. 2019;19:1764–1771. [DOI] [PubMed] [Google Scholar]
- 24.Karhade A V, Schwab JH, Bedair HS. Development of Machine Learning Algorithms for Prediction of Sustained Postoperative Opioid Prescriptions After Total Hip Arthroplasty. J Arthroplasty. 2019;34:2272-2277.e1. [DOI] [PubMed] [Google Scholar]
- 25.Karhade A V, Thio Q, Ogink P, Kim J, Lozano-Calderon S, Raskin K, Schwab JH. Development of Machine Learning Algorithms for Prediction of 5-Year Spinal Chordoma Survival. World Neurosurg. 2018;119:e842–e847. [DOI] [PubMed] [Google Scholar]
- 26.Karhade A V, Thio QCBS, Kuverji M, Ogink PT, Ferrone ML, Schwab JH. Prognostic value of serum alkaline phosphatase in spinal metastatic disease. Br J Cancer. 2019;120:640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karhade A V, Thio QCBS, Ogink PT, Bono CM, Ferrone ML, Oh KS, Saylor PJ, Schoenfeld AJ, Shin JH, Harris MB, Schwab JH. Predicting 90-Day and 1-Year Mortality in Spinal Metastatic Disease: Development and Internal Validation. Neurosurgery. 2019;85:E671–E681. [DOI] [PubMed] [Google Scholar]
- 28.Karhade A V, Thio QCBS, Ogink PT, Shah AA, Bono CM, Oh KS, Saylor PJ, Schoenfeld AJ, Shin JH, Harris MB, Schwab JH. Development of Machine Learning Algorithms for Prediction of 30-Day Mortality After Surgery for Spinal Metastasis. Neurosurgery. 2019;85:E83–E91. [DOI] [PubMed] [Google Scholar]
- 29.Kim K, Kim S, Lee YH, Lee SH, Lee HS, Kim S. Performance of the deep convolutional neural network based magnetic resonance image scoring algorithm for differentiating between tuberculous and pyogenic spondylitis. Sci Rep. 2018;8:13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H, Yune S, Mansouri M, Kim M, Tajmir SH, Guerrier CE, Ebert SA, Pomerantz SR, Romero JM, Kamalian S, Gonzalez RG, Lev MH, Do S. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng. 2019;3:173–182. [DOI] [PubMed] [Google Scholar]
- 31.Liang H, Tsui BY, Ni H, Valentim CCS, Baxter SL, Liu G, Cai W, Kermany DS, Sun X, Chen J, He L, Zhu J, Tian P, Shao H, Zheng L, Hou R, Hewett S, Li G, Liang P, Zang X, Zhang Z, Pan L, Cai H, Ling R, Li S, Cui Y, Tang S, Ye H, Huang X, He W, Liang W, Zhang Q, Jiang J, Yu W, Gao J, Ou W, Deng Y, Hou Q, Wang B, Yao C, Liang Y, Zhang S, Duan Y, Zhang R, Gibson S, Zhang CL, Li O, Zhang ED, Karin G, Nguyen N, Wu X, Wen C, Xu J, Xu W, Wang B, Wang W, Li J, Pizzato B, Bao C, Xiang D, He W, He S, Zhou Y, Haw W, Goldbaum M, Tremoulet A, Hsu C-N, Carter H, Zhu L, Zhang K, Xia H. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat Med. 2019;25:433–438. [DOI] [PubMed] [Google Scholar]
- 32.Lin H, Li R, Liu Z, Chen J, Yang Y, Chen H, Lin Z, Lai W, Long E, Wu X, Lin D, Zhu Y, Chen C, Wu D, Yu T, Cao Q, Li X, Li J, Li W, Wang J, Yang M, Hu H, Zhang L, Yu Y, Chen X, Hu J, Zhu K, Jiang S, Huang Y, Tan G, Huang J, Lin X, Zhang X, Luo L, Liu Y, Liu X, Cheng B, Zheng D, Wu M, Chen W, Liu Y. Diagnostic Efficacy and Therapeutic Decision-making Capacity of an Artificial Intelligence Platform for Childhood Cataracts in Eye Clinics: A Multicentre Randomized Controlled Trial. EClinicalMedicine. 2019;9:52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindsey R, Daluiski A, Chopra S, Lachapelle A, Mozer M, Sicular S, Hanel D, Gardner M, Gupta A, Hotchkiss R, Potter H. Deep neural network improves fracture detection by clinicians. Proc Natl Acad Sci U S A. 2018;115:11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F, Zhou Z, Samsonov A, Blankenbaker D, Larison W, Kanarek A, Lian K, Kambhampati S, Kijowski R. Deep Learning Approach for Evaluating Knee MR Images: Achieving High Diagnostic Performance for Cartilage Lesion Detection. Radiology. 2018;289:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundberg SM, Nair B, Vavilala MS, Horibe M, Eisses MJ, Adams T, Liston DE, Low DK-W, Newman S-F, Kim J, Lee S-I. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018;2:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahadevan S. Average Reward Reinforcement Learning: Foundations, Algorithms, and Empirical Results. Mach Learn. 1996;22:159–195. [Google Scholar]
- 37.Merrill RK, Ferrandino RM, Hoffman R, Shaffer GW, Ndu A. Machine Learning Accurately Predicts Short-Term Outcomes Following Open Reduction and Internal Fixation of Ankle Fractures. J Foot Ankle Surg. 2019;58:410–416. [DOI] [PubMed] [Google Scholar]
- 38.Milea D, Najjar RP, Zhubo J, Ting D, Vasseneix C, Xu X, Aghsaei Fard M, Fonseca P, Vanikieti K, Lagrèze WA, La Morgia C, Cheung CY, Hamann S, Chiquet C, Sanda N, Yang H, Mejico LJ, Rougier M-B, Kho R, Thi Ha Chau T, Singhal S, Gohier P, Clermont-Vignal C, Cheng C-Y, Jonas JB, Yu-Wai-Man P, Fraser CL, Chen JJ, Ambika S, Miller NR, Liu Y, Newman NJ, Wong TY, Biousse V. Artificial Intelligence to Detect Papilledema from Ocular Fundus Photographs. N Engl J Med. 2020;382:1687–1695. [DOI] [PubMed] [Google Scholar]
- 39.Mirvis SE. Increasing workloads in radiology: Does it matter? Appl Radiol. 2013;42:6. [Google Scholar]
- 40.Mitchell TM. Machine Learning. Vol. 1 New York: McGraw-HillScience; 1997. [Google Scholar]
- 41.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obermeyer Z, Emanuel EJ. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. N Engl J Med. 2016;375:1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olczak J, Fahlberg N, Maki A, Razavian AS, Jilert A, Stark A, Sköldenberg O, Gordon M. Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop. 2017;88:581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piraino DW, Amartur SC, Richmond BJ, Schils JP, Thome JM, Belhobek GH, Schlucter MD. Application of an artificial neural network in radiographic diagnosis. J Digit Imaging. 1991;4:226–232. [DOI] [PubMed] [Google Scholar]
- 45.Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med. 2019;380:1347–1358. [DOI] [PubMed] [Google Scholar]
- 46.Ramkumar PN, Navarro SM, Haeberle HS, Karnuta JM, Mont MA, Iannotti JP, Patterson BM, Krebs VE. Development and Validation of a Machine Learning Algorithm After Primary Total Hip Arthroplasty: Applications to Length of Stay and Payment Models. J Arthroplasty. 2019;34:632–637. [DOI] [PubMed] [Google Scholar]
- 47.Reardon S. Rise of Robot Radiologists. Nature. 2019;576:S54–S58. [DOI] [PubMed] [Google Scholar]
- 48.Senders JT, Arnaout O, Karhade A V., Dasenbrock HH, Gormley WB, Broekman ML, Smith TR. Natural and Artificial Intelligence in Neurosurgery: A Systematic Review. Neurosurgery. 2018;83:181–192. [DOI] [PubMed] [Google Scholar]
- 49.Slim K, Nini E, Forestier D, Kwiatkowski F, Panis Y, Chipponi J. Methodological index for non-randomized studies (minors): development and validation of a new instrument. ANZ J Surg. 2003;73:712–716. [DOI] [PubMed] [Google Scholar]
- 50.Thio QCBS, Karhade A V, BJJ Bindels, Ogink PT, Bramer JAM, Ferrone ML, Calderón SL, Raskin KA, Schwab JH. Development and Internal Validation of Machine Learning Algorithms for Preoperative Survival Prediction of Extremity Metastatic Disease. Clin Orthop Relat Res. 2020;478:322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thio QCBS, Karhade A V, Ogink PT, Raskin KA, De Amorim Bernstein K, Lozano Calderon SA, Schwab JH. Can Machine-learning Techniques Be Used for 5-year Survival Prediction of Patients With Chondrosarcoma? Clin Orthop Relat Res. 2018;476:2040–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thirukumaran CP, Zaman A, Rubery PT, Calabria C, Li Y, Ricciardi BF, Bakhsh WR, Kautz H. Natural Language Processing for the Identification of Surgical Site Infections in Orthopaedics. J Bone Joint Surg Am. 2019;101:2167–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ting DSW, Cheung CY-L, Lim G, Tan GSW, Quang ND, Gan A, Hamzah H, Garcia-Franco R, San Yeo IY, Lee SY, Wong EYM, Sabanayagam C, Baskaran M, Ibrahim F, Tan NC, Finkelstein EA, Lamoureux EL, Wong IY, Bressler NM, Sivaprasad S, Varma R, Jonas JB, He MG, Cheng C-Y, Cheung GCM, Aung T, Hsu W, Lee ML, Wong TY. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA. 2017;318:2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomašev N, Glorot X, Rae JW, Zielinski M, Askham H, Saraiva A, Mottram A, Meyer C, Ravuri S, Protsyuk I, Connell A, Hughes CO, Karthikesalingam A, Cornebise J, Montgomery H, Rees G, Laing C, Baker CR, Peterson K, Reeves R, Hassabis D, King D, Suleyman M, Back T, Nielson C, Ledsam JR, Mohamed S. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572:116–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urakawa T, Tanaka Y, Goto S, Matsuzawa H, Watanabe K, Endo N. Detecting intertrochanteric hip fractures with orthopedist-level accuracy using a deep convolutional neural network. Skeletal Radiol. 2019;48:239–244. [DOI] [PubMed] [Google Scholar]
- 56.Varma M, Lu M, Gardner R, Dunnmon J, Khandwala N, Rajpurkar P, Long J, Beaulieu C, Shpanskaya K, Fei-Fei L, Lungren MP, Patel BN. Automated abnormality detection in lower extremity radiographs using deep learning. Nat Mach Intell. 2019;1:578–583. [Google Scholar]
- 57.Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyles CC, Tibbo ME, Fu S, Wang Y, Sohn S, Kremers WK, Berry DJ, Lewallen DG, Maradit-Kremers H. Use of Natural Language Processing Algorithms to Identify Common Data Elements in Operative Notes for Total Hip Arthroplasty. J Bone Joint Surg Am. 2019;101:1931–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue Y, Zhang R, Deng Y, Chen K, Jiang T. A preliminary examination of the diagnostic value of deep learning in hip osteoarthritis. PLoS One. 2017;12:e0178992. [DOI] [PMC free article] [PubMed] [Google Scholar]