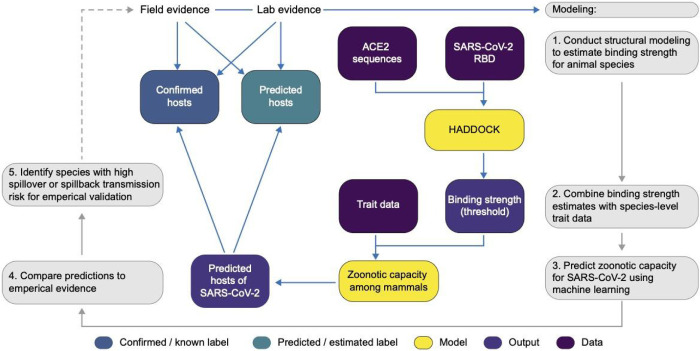
Figure 2.
A flowchart showing the progression of our workflow combining evidence from limited lab and field studies with additional data types to predict zoonotic capacity across mammals through multi-scale statistical modeling (gray boxes, steps 1–5). For all vertebrates with published ACE2 sequences, we modelled the interface of species’ ACE2 bound to the viral receptor binding domain using HADDOCK. We then combined the HADDOCK scores, which approximate binding strength, with species’ trait data and trained machine learning models for both mammals and vertebrates (yellow boxes). Mammal species predicted to have high zoonotic capacity were then compared to results of in vivo experiments and in silico studies that applied various computational approaches. Based on predictions from our model, we identified a subset of species with particularly high risk of spillback and secondary spillover potential to prioritize additional lab validation and field surveillance (dashed line).