ABSTRACT
Gut microbiome composition depends heavily upon diet and has strong ties to human health. Dietary carbohydrates shape the gut microbiome by providing a potent nutrient source for particular microbes. This review explores how dietary carbohydrates in general, including individual monosaccharides and complex polysaccharides, influence the gut microbiome with subsequent effects on host health and disease. In particular, the effects of sialic acids, a prominent and influential class of monosaccharides, are discussed. Complex plant carbohydrates, such as dietary fiber, generally promote microbial production of compounds beneficial to the host while preventing degradation of host carbohydrates from colonic mucus. In contrast, simple and easily digestible sugars such as glucose are often associated with adverse effects on health and the microbiome. The monosaccharide class of sialic acids exerts a powerful but nuanced effect on gut microbiota. Sialic acid consumption (in monosaccharide form, or as part of human milk oligosaccharides or certain animal-based foods) drives the growth of organisms with sialic acid metabolism capabilities. Minor chemical modifications of Neu5Ac, the most common form of sialic acid, can alter these effects. All aspects of carbohydrate composition are therefore relevant to consider when designing dietary therapeutic strategies to alter the gut microbiome.
KEYWORDS: Carbohydrates, glycans, gut microbiome, diet, fiber, sialic acids, neu5gc, mucin-linked O-glycans, human milk oligosaccharides
Introduction
The human gut microbiome is defined as the sum of genomic DNA of all microbes inhabiting this environment. With up to a hundred times more bacterial genes than human genes in the human body, including an especially high number of microbes in the gut, the microbial communities in our body are crucial to human life and play a key role in human development and homeostasis.1 As in every natural ecosystem, bacteria in the human gut influence the surrounding environment of their host. The human gut microbiota is involved in many essential host functions, such as the processing of nutrients to bioactive molecules like neurotransmitters, vitamins, and fatty acids and protection from pathogens.2 One of the most well-known examples of this is the breakdown of non-digestible carbohydrates found in plants. As humans do not have the metabolic capability to degrade these complex glycans in the gastrointestinal tract, they reach the colon to be fermented by gut bacteria and lead to the production of short-chain fatty acids (SCFAs), which participate in the acidification of the digestive tract.3 Through these and other similar processes, the human gut microbiota has a major impact on the host’s physiology in health and disease.
In addition to a greater number of genes and metabolic capabilities than the human genome, the composition of the gut microbiome is also highly malleable.1 Diet surpasses the role of host genetics in shaping the gut microbiome through modification of the nutritional environment of the bacteria populating the gut.4–8 Given the influence of the gut microbiome in human health, the ability to alter this microbiome through dietary changes indicates that promoting a healthy microbiome has great potential to improve human well-being and disease prevention and control. Glycans (i.e. carbohydrates) are of major importance in determining the gut microbiome composition.9 Glycans come in many forms, from long polysaccharide chains that humans are unable to digest (e.g. cellulose, pectins, resistant starch), to oligosaccharide chains attached to proteins and lipids, to individual mono – and disaccharides, such as glucose, lactose, or sialic acids.9 In this review, we detail how dietary glycans can shape the structure and function of the human gut microbiota and the impact this has on human diseases. We start with an overview of the broad impacts of carbohydrates on gut microbiota composition and metabolic activity. We then focus on the role of sialic acids, a specific monosaccharide class, in shaping the gut microbiome. Sialic acids are a prominent component of the mammalian glycosylation system, and their interactions with the human immune system make their impact on the gut microbiome of particular interest. This review of sialic acids, the gut microbiome, and impacts on health will summarize recent research and suggest directions for future studies.
Broad impact of carbohydrates on gut microbiome structure and function
Human studies have repeatedly demonstrated that dietary changes modify the relative abundance of major gut bacterial groups in a rapid and reversible manner.10,11 For example, low-carbohydrate, weight-loss, and animal-based diets reduce the proportion of the butyrate-producing phyla Firmicutes and Actinobacteria,11–13 while high animal product consumption increases the proportion of Bacteroidetes and specific Proteobacteria like Bilophila wadsworthia in the human gut.11 Lifestyle urbanization and Westernization are key factors influencing dietary behavior, with subsequent impacts on the gut microbiome and potential harmful effects on human health.14 A rural diet, typically rich in host-indigestible carbohydrates like fiber, is associated with a higher abundance of Prevotella and Xylanibacter spp., while an urbanized diet, which generally contains more saturated fat and protein from animals, is associated with an increase of Bacteroides spp. and a decrease of overall microbiome gene diversity.15–17 Interestingly, those Bacteroides-dominated, less diverse gut communities are associated with a higher incidence of obesity and metabolic syndrome.18 The loss of diversity and shift from a Prevotella – to Bacteroides-dominated microbiome has been observed in non-Western immigrants as early as 9 months after moving to the USA.19 These data demonstrate the plasticity of the human gut microbiota in response to dietary carbohydrate changes and the potential impact of these changes on human health.
Many studies examine the impact of plant carbohydrates in particular on the gut microbiome. Diets with high resistant starch intake have been associated with increased relative abundance of Firmicutes and particularly Ruminococcaceae family members, while resistant potato starch specifically has been associated with increased Bifidobacterium genera and wheat bran has been associated with increased Lachnospiraceae family members.10,12,20 A recent study also demonstrated rapid modifications of the gut microbiota in mice fed raw versus cooked plant products, due to the improvement of starch digestibility and degradation of plant-derived compounds during the cooking process. Similar observations have been made in the human population, showing that everyday nutritional habits can influence the gut microbiota.8
Within plant carbohydrates, dietary fiber is one of the most heavily studied groups. Dietary fiber is generally defined as edible carbohydrate polymers, mostly from plants and edible fungi, that are not digestible by human enzymes.21 Examples include inulin, dextrin, pectin, cellulose, resistant starch, arabinoxylans, and chitin (Figure 1A).24 Dietary fiber exists in soluble and insoluble forms, although some polymers can be soluble or insoluble depending on conditions like cooking or food processing.21 Although human metabolism cannot digest fiber, the gut microbiome often contains many enzymes capable of degrading these polymers and utilizing the sugars released for nutrition or other metabolic processes.
Figure 1.
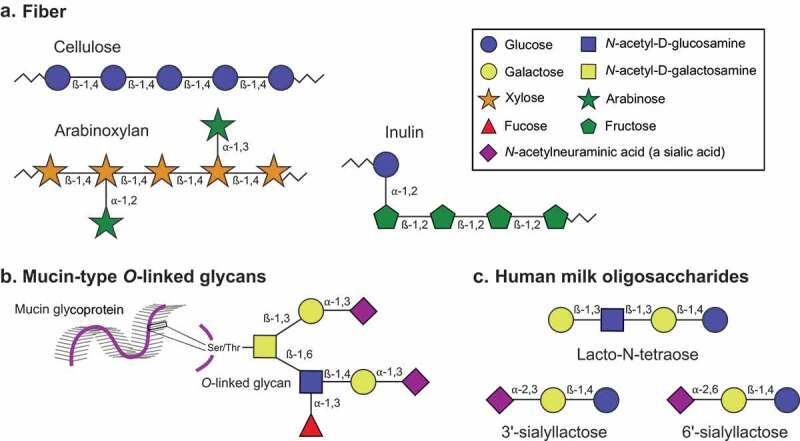
Structural composition of the main poly – and oligosaccharides discussed in this review. Monosaccharide symbols are represented as in the Symbol Nomenclature for Glycans (assignments for this figure provided in the box).22,23 The numbers between monosaccharides represent the glycosidic linkage. A) Fiber is a general classification encompassing many types of dietary polysaccharides. Examples of the polysaccharides cellulose, arabinoxylan, and inulin are provided here. B) Mucin-type O-linked glycans are host glycans linked to serine or threonine (Ser/Thr) residues on mucin proteins. Like most mammal-derived glycans, they are often tipped with sialic acids such as N-acetylneuraminic acid. An example structure is shown; many other monosaccharide and linkage compositions are possible. C) Human milk oligosaccharides (HMOs) are short oligosaccharides found in human breast milk. HMOs are composed of a lactose base (a disaccharide of glucose and galactose) with additional monosaccharides such as N-acetyl-D-glucosamine, the sialic acid N-acetylneuraminic acid, or fucose attached. Three example structures are shown
Fiber passes mostly undigested through the small intestine and is fermented in the colon by gut bacteria, leading to the production of SCFAs (Figure 2).25 The SCFAs acetate, propionate, and butyrate are the main metabolites produced during microbial fiber fermentation, and they have multiple beneficial effects on the host.14 Once produced in the colon, SCFAs are rapidly absorbed by host epithelial cells, where the great majority are directly used as an energy source. SCFAs that are not metabolized by the gut epithelium (estimated as <10% of total SCFAs produced)26,27 are then transported through the portal circulation to the liver, where they can be incorporated by hepatocytes and used as energy substrates or for the synthesis of glucose, cholesterol, and fatty acids.28
Figure 2.
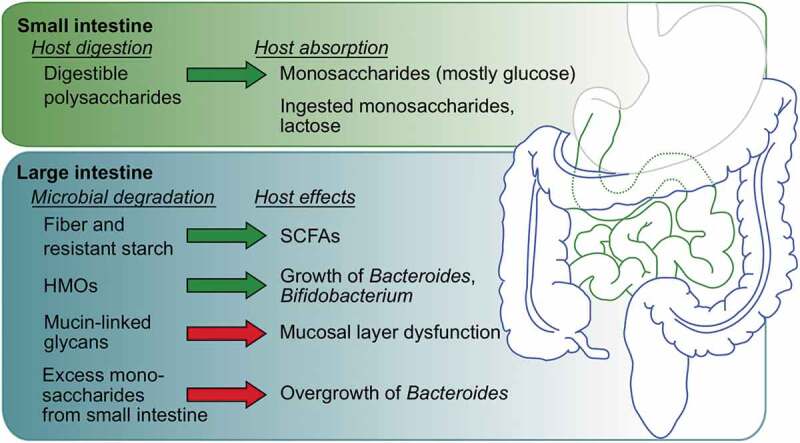
Broad overview of carbohydrate digestion and host effects discussed in this review; other effects are also possible for each glycan. Generally beneficial and detrimental effects are represented by green and red arrows, respectively. Most host digestion and nutrient absorption occurs in the small intestine. Microbes conduct most of the nutrient degradation that occurs in the large intestine, with varying effects on host health and function
A small fraction of the initial SCFAs will reach the main blood circulation and have systemic effects, particularly on the immune system.28 Notably, SCFAs downregulate the production of pro-inflammatory cytokines by colonic macrophages29 and promote the differentiation of naive CD4 + T cells into immunosuppressive regulatory T cells (Treg),30,31 by binding to G-protein coupled receptors32 or by inhibiting histone deacetylases.29,33,34 SCFAs derived by the gut microbiota from dietary fiber thus participate in the homeostasis of the immune response, with a demonstrated protective effect against inflammatory diseases, such as multiple sclerosis (MS),33 inflammatory bowel disease (IBD),31 and allergic asthma,35 as well as other pathologies, such as infection36,37 and carcinogenesis.16 Colorectal cancer (CRC) is also known to be linked with a gut microbiota dysbiosis characterized by decreased microbial diversity38 and an under-representation of SCFA-producing bacteria.39 A high-fiber dietary intake is associated with a lower risk of CRC,40 while patients with CRC precursor lesions tend to have lower fiber dietary intake than controls.41
However, not all studies have shown universal benefits from fiber intake. In a recent study, Singh et al. supplemented the diet of toll-like receptor 5 (TLR5)-deficient mice with fermentable fibers for 6 months, with the goal of demonstrating the beneficial effect of such a diet on metabolic syndrome. While the authors observed some of the expected effects (reduction of adiposity, amelioration of glycemic control), they also observed that purified fiber supplementation induced icteric hepatocellular carcinoma in 40% of the TLR5-deficient mice.42 These studies indicate that dietary supplementation with such purified compounds may have a negative effect on some individuals, and that large-scale enrichment of processed food with purified prebiotic fiber should be taken with great caution.43 For a more detailed discussion of the gut microbiota and specific health effects of dietary fiber, we refer the reader to ref. 42.44
Mono – and disaccharide dietary sugars can affect gut microbiome composition, with potential effects on human health. Fructose and glucose have been demonstrated to specifically inhibit gut colonization by Bacteroides thetaiotaomicron, a mammal gut symbiont associated with lean and healthy individuals, by silencing the Roc (regulator of colonization) protein, which promotes competitive colonization in gnotobiotic mice.45 High fructose intake has also been associated with development of nonalcoholic fatty liver disease (NAFLD) in humans46 and mouse models.47,48 The gut microbiome in general plays a causal role in NAFLD development in mouse models,49 and several studies have established correlations between NAFLD and altered abundance of taxa, such as Bifidobacterium,50 Lactobacillus,50,51 Bacteroides, and Ruminococcus.52 Supplementation of Lactobacillus rhamnosus in the gut microbiome of mice fed a high-fructose diet to induce NAFLD resulted in decreased liver inflammation and NAFLD disease development.51 This finding highlights the potential regulatory effects of dietary sugars in the small intestine on gut colonization by beneficial microbes. Later in this review, we will discuss in detail recent research on the effects of sialic acids, a biologically important class of monosaccharides, on the gut microbiome and host health.
Interplay of dietary fiber and host mucins in the gut microbiome
Although dietary glycans make up the majority of nutrients the gut microbiota consumes, restriction of carbohydrates like fiber from the diet can push microbes to consume glycans produced by the host instead.53 The colon contains a mucus gel layer composed of two parts: a loose luminal outer layer and a dense mucosal inner layer.54 The mucosal layer is composed mainly of host mucin proteins with regions of extensive O-glycosylation (forming up to 80% of the total mucin mass) (Figure 1B).55 Although microbes do not penetrate the dense inner layer in healthy subjects,37,56 microbial degradation of the outer layer is thought to be a normal part of mucin turnover and regeneration.57 For a review of how gut microbiota interact with and degrade the colonic mucosal layer, we direct the reader to ref. 56.58 Here we focus on how diet can alter the careful balance between gut microbiota and the host mucosal layer.
The section above discussed how the presence of complex polysaccharides, such as fiber, in the diet strongly affects the gut microbiome composition. Many studies have shown that fiber ingestion increases abundance of colonic bacteria capable of fermenting fiber to SCFAs,7,10,13,20 with increased diversity of plant carbohydrates believed to support greater community diversity.59 Conversely, several studies have shown that a lack of dietary fiber can push bacterial metabolism away from fiber degradation to mucin degradation. Some organisms (e.g. Bacteroides thetaiotaomicron) degrade both fiber and mucins and shift their metabolism to mucin degradation when dietary complex polysaccharides are scarce.53,60 Other organisms (e.g. Akkermansia muciniphila) are able to degrade mucins but not fiber and experience expansion of their populations upon complex polysaccharide scarcity.37,61,62
Excessive mucin degradation is associated with increased intestinal inflammation63,64 and increased penetration of bacteria into the dense mucosal mucus layer.65 In gnotobiotic mice mono-colonized with B. thetaiotaomicron, a diet lacking complex polysaccharides (including fiber) resulted in a thinner colonic mucus layer, an increased proximity of colonic microbes to the gut epithelium, and increased expression of the inflammatory marker REG3β.60 Similarly, it has been shown that dietary fiber deprivation increased the abundance of mucus-degrading bacteria like A. muciniphila and Bacteroides caccae in mice, subsequently leading to an alteration of the intestinal barrier and higher susceptibility to mucosal pathogens.37,60 Demonstrating the specific and essential role of the gut microbiome in mucus changes, antibiotic-treated mice fed a low-fiber Western diet but transplanted weekly with gut microbiota from mice fed a high-fiber chow diet had significantly lower mucus penetrability and higher mucus growth than mice transplanted with gut microbiota from Western diet-fed mice.66 These studies indicate a lack of dietary fiber leads to changes in the gut microbiome that promote dysfunction and increased microbial penetrability of the inner colonic mucus layer.
On the other hand, a recent study suggests potentially beneficial roles of microbial mucus metabolism in ulcerative colitis (UC). Certain organisms are capable of producing the SCFA n-butyrate from mucin degradation,67 and n-butyrate as well as mixed SCFAs have been shown to reduce colon inflammation in UC.68,69 Yamada et al.67 found decreased mucinase activity and decreased levels of n-butyrate in the stool of UC patients, but a significantly higher O-glycan-to-mucin protein ratio. Hypothesizing a deficiency in mucin O-glycan utilization by gut microbiota, the authors assessed the impact of feeding mice a mucin-enriched diet. After 3 weeks, they observed an increased α-diversity; increased relative abundance of Akkermansia, Allobaculum, and Bacteroidales S24-7; increased cecal SCFAs; and increased colonic Treg and IgA+ plasma cells.67 In the setting of UC, mucin degradation may therefore be an important physiologic process to promote.
Impact of the monosaccharide sialic acid on gut microbiome structure and function
Thus far, we have discussed the impact of broad dietary glycan classes on the gut microbiome and host health, including how lack of fiber promotes microbial degradation of host mucus glycans. Next, we focus on the impact of dietary sialic acids, a unique and essential class of monosaccharides, on the gut microbiome and human health. Sialic acids are essential to many physiological processes, play a large role in shaping both the infant and adult microbiome, and allow exploration of how minor chemical modifications in sugar structure can shape the microbiome. Although many authors have reviewed sialic acids in the past, to our knowledge a comprehensive review focusing specifically on dietary sialic acids and the gut microbiome has not been published. In the literature, “sialic acids” is often used to refer to both the group and its most common member, N-acetylneuraminic acid. In this review, we will refer to N-acetylneuraminic acid by its abbreviation Neu5Ac and reserve the term sialic acids for the group as a whole.
ialic acids are acidic 9-carbon monosaccharides, derivatives of neuraminic acid, and ubiquitous in all vertebrate glycosylation systems. Sialic acids often serve as the terminal sugars in N-linked and O-linked vertebrate glycans that decorate cell-surface proteins and lipids, and as such they are often some of the first monosaccharides encountered in cell–cell interactions.70,71 They play essential roles in immune system signaling, cell adhesion, membrane transport, and many other processes.71–73 The most abundant mammalian sialic acids are Neu5Ac and its close chemical cousin N-glycolylneuraminic (Neu5Gc) acid (Figure 3A).71 Humans cannot produce Neu5Gc due to loss of the CMP-N-acetylneuraminic acid hydroxylase (CMAH) enzyme,83–85 and as such Neu5Gc is perceived as a foreign antigenic sugar by the human immune system.86,87 Sialic acids are present in our diet in N – and O-linked glycans from animal-derived proteins, and Neu5Gc can be incorporated into human glycoconjugates following ingestion of certain animal-derived foods rich in Neu5Gc, chiefly red meat.88,89 Neu5Ac and Neu5Gc have drastically different effects on human health, with Neu5Ac a natural and beneficial component of human glycans and Neu5Gc an antigenic and pro-inflammatory component.90–94
Figure 3.
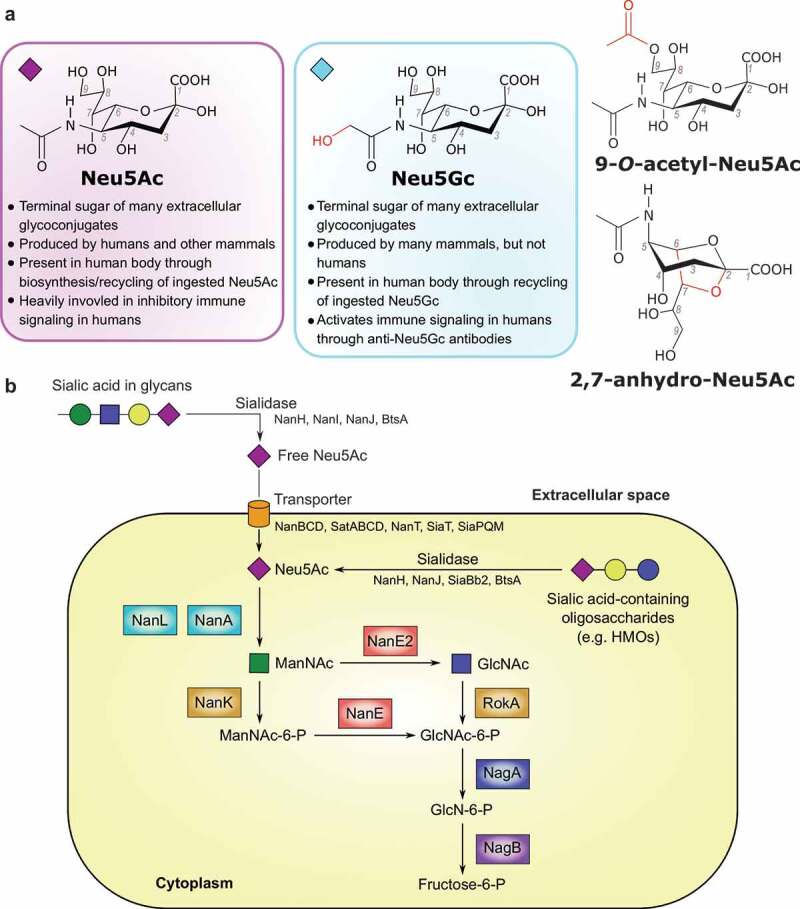
A) Structures of the sialic acids discussed in this review. Changes from the Neu5Ac structure are shown in red. Differences between Neu5Ac and Neu5Gc with regards to human physiology are listed below those structures. Diamonds by Neu5Ac and Neu5Gc structures depict the Symbol Nomenclature for Glycans symbol for each. B) Schematic of general Neu5Ac metabolism in bacterial cells. Steps with multiple characterized enzymes (sialidase and transporter) have protein names listed below the general enzyme name. Steps with one well-characterized enzyme have the enzyme name next to the arrow. The exception is the conversion of Neu5Ac to ManNAc, which has 2 well-characterized enzymes. Enzymes are color-coded based on function. References for enzyme functions: NanAEE2KL, RokA, NagAB;.74 NanHIJ;75 NanBCD;76 NanT;77 SiaBb2;78 BtsA;79 SatABCD;80 SiaT;81 SiaPQM.82
Sialic acid metabolism by gut bacteria
Human-associated bacteria, including gut microbiota, use sialic acids primarily as either a nutrient source or as a signaling molecule to interact with their host.95 For example, given the role of Neu5Ac on host cells in inhibiting autoimmune signaling through Siglec proteins,96 some pathogens evade the immune system by prominently displaying Neu5Ac on their cell surfaces.97,98 For an extensive review of sialic acids catabolism by human pathogens all over the body, we refer to ref. 87.98 Bacteria can synthesize sialic acids de novo or scavenge from the surrounding environment.72,95 Complete metabolism of sialic acids requires a sialidase to release the monosaccharide from the glycan, a transporter protein to transport the monosaccharide inside the cell, and a suite of intracellular enzymes to convert sialic acids into a sugar fed into different metabolic pathways (Figure 3B).72 Many common gut microbes contain genes for part of or for this entire pathway, affecting their role in the gut microbial community, and through that the community’s potential effects on human health.
The first full Neu5Ac metabolism pathway was described in Escherichia coli in 199999 and the ability of E. coli to metabolize Neu5Ac has since been shown to be important for gut colonization in mice.100 The Nan gene cluster in E. coli encodes the sialic acids uptake transporter NanT and three catabolic enzymes (NanA lyase, NanK kinase, and NanE epimerase) that catalyze the conversion of Neu5Ac to pyruvate and N-acetylglucosamine-6-phosphate. This is further metabolized through the N-acetylglucosamine (GlcNAc) catabolic pathway (Figure 3b). Neu5Gc is transported and catabolized by E. coli using the same NanT transporter and NanA lyase but producing glycolate instead of pyruvate.101 Similar Neu5Ac catabolic gene clusters with variations in the identity of sialic acid transporter were identified in 46 out of 1,902 bacterial genomes examined in a 2009 study.102 However, 91% of these 46 organisms were able to colonize humans, indicating the ability to metabolize sialic acids is particularly valuable for bacteria in human-associated niches. Nine of these organisms were gut commensals (Anaerotruncus colihominis, Dorea formicigenerans, D. longicatena, Faecalibacterium prausnitzii, Fusobacterium nucleatum, Ruminococcus gnavus, Lactobacillus sakei, L. plantarum, and L. salivarius), while several others were known gut pathogens (E. coli, Shigella (species unspecified), Salmonella enterica, Yersinia enterocolitica, Vibrio vulnificus, and V. cholerae).102 A similar analysis in 2015, of 4,497 genomes in NCBI at the time, found that 5.9% of species contained genes for the full pathway of Neu5Ac metabolism; again, these organisms primarily colonize humans or animals.98 An alternative Neu5Ac utilization pathway was identified in the gut commensal Bacteroides fragilis (Figure 3b) and involves a putative sialic acid transporter from the MFS superfamily (NanT), a non-orthologous Neu5Ac lyase (NanL), and two novel catabolic enzymes, epimerase NanE3 and kinase RokA.74 The nanLE2T gene cluster from B. fragilis was further identified in many colonic bacteria from the Bacteroidetes phylum, including B. vulgatus and Parabacteroides distasonis, but not in B. thetaiotaomicron, which encodes a sialidase but lacks the nanLE2T genes to fully metabolize sialic acids.103 Other microbes like Clostridioides difficile or E. coli lack a sialidase but encode a complete pathway to metabolize sialic acids.104
We analyzed the distribution of sialic acids utilization pathway and sialidase genes across a reference set of 2,662 genomes representing ~700 species and ~200 genera of bacteria from the human gut.105 For genomic identification of genes encoding sialidases, Neu5Ac transporters, and catabolic enzymes (Figure 3b), we used a subsystems-based approach implemented in the SEED platform.106 Each reference genome was assigned binary phenotypes reflecting the presence/absence of: (i) a complete Neu5Ac utilization pathway; and (ii) sialidase enzyme(s) (Figure 4). Approximately 1,040 strains were predicted as Neu5Ac-utilizing strains, representing ~80 bacterial genera. Among these, a sialidase was identified in 40% of the strains, including prominent colonic bacteria from the Akkermansia, Bacteroides, Bifidobacterium, Clostridium, Flavonifractor, Parabacteroides, and Prevotella genera. Another subgroup of strains that lack a sialidase but are capable of sialic acid utilization includes both human gut symbionts such as Anaerococcus, Blautia, Escherichia, Eubacterium, Faecalibacterium, Fusobacterium, and also a number opportunistic pathogens including Clostridioides, Staphylococcus, and Streptococcus spp. Finally, ~100 strains from 27 microbial genera possess a sialidase but apparently lack the sialic acid utilization capability. These include 18 Bacteroides strains (e.g. B. faecis, B. intestinalis, B. thetaiotaomicron), 6 Porphyromonas strains, and 6 Coprobacillus strains. The high prevalence of many of these strains in the gut microbiome suggests even strains that solely release Neu5Ac from underlying glycans contribute to the overall sialic acid degradation capability of gut communities.
Figure 4.
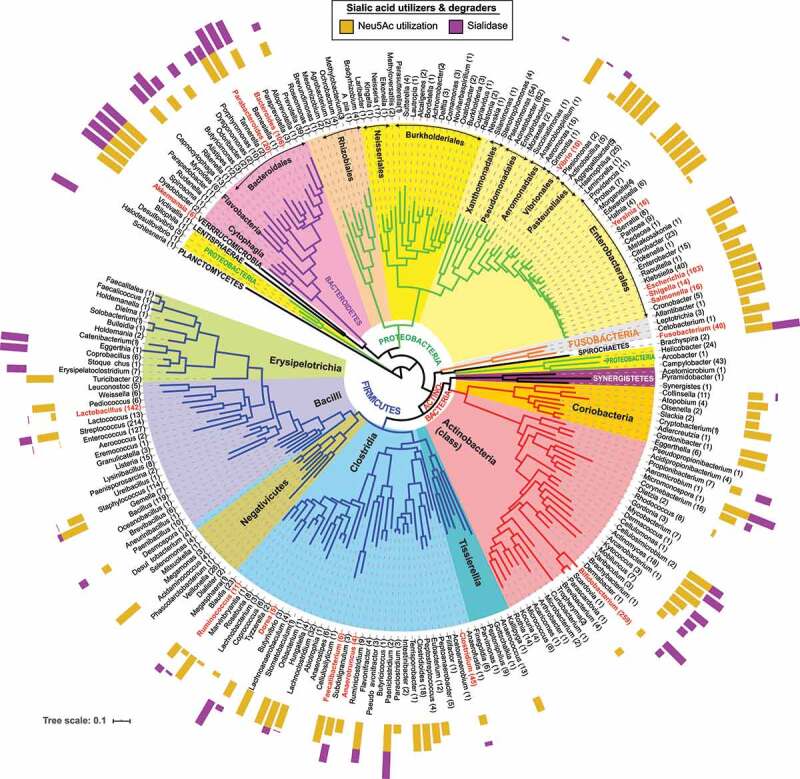
Genomic distribution of Neu5Ac utilizers and degraders in human gut microbiome strains. The phylogenetic tree was constructed using concatenated sequences of universal ribosomal proteins from approximately 2,600 human gut microbial genomes from the PATRIC genomic database107 by RAxML version 8,108 then shrunk to genus representatives and visualized via iTOL.109 Numbers next to genera in the outermost level of the tree indicate the number of analyzed genomes per genus. Bars adjacent to each genus indicate the proportion of genomes that contain genes for: a Neu5Ac transporter and full Neu5Ac catabolism pathway (gold bars); and one or more Neu5Ac sialidase genes (purple bars). Genera mentioned in this review are shown in red text
These mixed catabolic capabilities fit with studies showing ingested complex polysaccharides can be digested and metabolized by different gut organisms, in a syntrophic or synergistic interaction network.110 In support of this, recent studies of Salmonella enterica and C. difficile showed that these organisms expand following antibiotic treatment through scavenging of sialic acids liberated from ingested food by other gut microbes such as B. thetaiotaomicron.111 Colonization with B. thetaiotaomicron lacking a sialidase inhibited C. difficile expansion in the mouse gut, while feeding with exogenous Neu5Ac reversed these effects.111 Similarly, Huang et al.112 showed that increased sialidase activity from B. vulgatus drives E. coli expansion in a mouse model of colitis. Hence, sialic acids released in the gut by one organism can be scavenged and metabolized by other organisms lacking a sialidase, causing effects that ripple through the metabolic network.
Although most research has been done on Neu5Ac, microbes can also act on modifications of Neu5Ac or on other sialic acids (Figure 3a). Neu5Ac modified with an O-acetyl group is generally resistant to release by sialidases. However, recent studies of B. fragilis show the O-acetylesterase EstA removes 9-O-acetyl esterifications, allowing sialidases to release these modified Neu5Ac molecules and promote in vitro growth of E. coli.113 Although not confirmed in vivo yet, this could provide another example of bacterial interactions to share metabolic capabilities. Previous studies of the commensal anaerobe Ruminococcus gnavus showed it cannot grow on unmodified Neu5Ac alone and instead uses an intramolecular trans-sialidase to release 2,7-anhydro-Neu5Ac from α2-3-linked sialic acids.114 2,7-anhydro-Neu5Ac is then selectively transported across the Ruminococcus cell membrane and converted back to Neu5Ac for further metabolism.115 This strategy, which prevents other organisms from utilizing the uncommon 2,7-anhydro-Neu5Ac, seems designed to conserve resources for R. gnavus as opposed to the cross-talk seen in other sialic acid processing pathways. While the major part of sialidase research focuses on Neu5Ac, some recent studies have examined the activity of gut microbe sialidases on Neu5Gc. Zaramela et al.116 reported the discovery of Neu5Gc-preferential sialidases from the gut microbiome of the Hadza hunter-gatherer group,4 with four out of the five selected Bacteroides sialidases displaying preferential release of Neu5Gc over Neu5Ac in at least one of the tested conditions. Further exploration of metabolism of these and other sialic acid modifications will undoubtedly reveal more novel microbial strategies to harvest sialic acids.
Dietary sialic acids, gut microbiome composition, and human health
Sialylated HMOs and the infant microbiome
The infant gut microbiome is thought to start developing in utero through fetal ingestion of amniotic fluid.117 Peri – and post-natally, the microbiota composition is heavily influenced by mode of fetal delivery (vaginal versus Cesarean section) and infant food source (breast milk versus formula).117,118 Human milk oligosaccharides (HMOs) represent a potent source of sialic acids (and other monosaccharides) that is unique to the infant diet (Figure 1c). HMOs are a group of over 200 oligosaccharide structures present in human breast milk, making up the third most abundant component of milk at 5–15 g/L (following lactose at 70 g/L and lipids at 40 g/L).119 The composition and overall amount of HMOs in breast milk varies by woman and by time since delivery.120 The majority of HMOs are not absorbed by the infant in the small intestine for nutrition, but instead persist into the colon where they have a significant impact on infant health (Figure 2).121 For example, HMOs have been shown to directly inhibit infant gut colonization by pathogens like enterotoxic E. coli, V. cholerae toxin, Campylobacter jejuni, rotaviruses, and noroviruses.122–124
HMOs in general, and sialylated HMOs (HMOs containing sialic acid) in particular, also promote the growth of particular beneficial microorganisms in the infant gut. Of the taxa studied from the infant gut microbiome, only the Bifidobacterium and Bacteroides genera have been shown to metabolize a broad range of HMOs.125,126 The gut microbiome of breast-fed infants is typically dominated by Bifidobacterium, representing up to 70% of gut microbiota in breast-fed infants compared to 31% in formula-fed infants.127 A study of individual gut microbes in isolation showed that the sialylated HMOs 3ʹ-sialyllactose (3’SL) and 6ʹ-sialyllactose (6’SL) specifically promoted the growth of seven Bifidobacterium longum strains, as well as B. vulgatus and B. thetaiotaomicron.126 6’SL but not 3’SL promoted growth of Lactobacillus delbrueckii, although L. rhamnosus did not show appreciable growth on HMOs (Yu 2013).126 In particular, B. longum subsp. infantis is capable of fully metabolizing all HMOs studied to date and of growing on Neu5Ac alone in vitro.128 The B. longum subsp. infantis genome contains a 43-kb gene cluster (HMO1) with 16 glycoside hydrolases and many oligosaccharide transport proteins, as well as two sialidases, nanH1 in the HMO1 gene cluster and nanH2.118,128,129 Intriguingly, B. longum subsp. infantis appears to transfer oligosaccharides into its cytoplasm and digests HMOs to monosaccharides within the cell;118,130,131 by contrast other microorganisms (e.g. Bacteroides and Bifidobacterium bifidum) are thought to break down HMOs to di-/monosaccharides extracellularly and transport these components into the cytoplasm.118,132
Other B. longum strains contain genes for specific portions of the sialic acids catabolism pathway (Table 1). B. longum subsp. bifido can release monosaccharides, including Neu5Ac, from HMOs but is unable to catabolize Neu5Ac, fucose, or N-acetylglucosamine.129 In contrast, B. longum subsp. breve can ferment these monosaccharides but may or may not be able to release them from HMOs, in a strain-dependent manner.133,134 Bacteroides species also have variable sialic acids metabolic capabilities (Table 1). Similar to Bifidobacterium, B. fragilis can cleave and fully metabolize Neu5Ac from HMOs, while B. thetaiotaomicron can cleave but not metabolize Neu5Ac.103 Bacteroides and most Bifidobacterium species metabolize HMOs through the same enzymatic pathways as host mucin glycan degradation.103 However, despite its facility at HMO digestion, B. longum subsp. infantis does not appear to digest host mucins.131 These results indicate dietary Neu5Ac in HMOs is heavily involved in shaping the infant gut microbiome by promoting colonization of Bifidobacterium and Bacteroides species, potentially laying the foundation of a life-long synergy between host and gut microbes.
Table 1.
Summary of the ability of bacteria in the infant gut microbiome to release and metabolize the sialic acid Neu5Ac from HMOs
| Genus | Species | Neu5Ac release | Neu5Ac metabolism |
|---|---|---|---|
| Bifidobacterium | longum subsp. infantis | + | + |
| longum subsp. bifido | + | - | |
| longum subsp. breve | -/+ | + | |
| Bacteroides | fragilis | + | + |
| thetaiotaomicron | + | - |
As in sialic acids metabolism, studies of sialic acids and the infant microbiome focus primarily on Neu5Ac. Research on the effect of Neu5Gc on the infant microbiome is virtually nonexistent. Studies in the past have not identified Neu5Gc in human breast milk, although it is readily present in bovine milk.127,135,136 However, a recent study of human milk composition discovered that breast milk from all 16 mothers tested (split between women who consumed cow’s milk and dairy-free almond beverages) contained HMOs with Neu5Gc, indicating that diet-derived monosaccharides can be incorporated into breast milk HMOs.137 The presence of Neu5Gc in breast milk further adds another possible mechanism for the development of anti-Neu5Gc antibodies, which appears in infants within the first 6 months of life.138 Anti-Neu5Gc antibodies drive a process of chronic low-level inflammation called xenosialitis, which has been shown in animal models to contribute to inflammatory pathologies, such as liver cancer,93 atherosclerosis,94 and other autoimmune diseases.96 Other possible mechanisms for anti-Neu5Gc antibody development include the presence of Neu5Gc in commercial baby foods and exposure to Neu5Gc on the surface of bacteria like non-typeable Haemophilus influenzae.90,138
solating the impact of ingested HMOs containing Neu5Ac on the infant gut microbiome is relatively simple, arguably simpler than in adults given the stereotyped diets of infants. However, tying these changes to infant health outcomes is much more difficult. A study in 2016 provides one of the most comprehensive experimental investigations of this question. Researchers inoculated germ-free mice with a defined microbial community of 25 strains isolated from the gut microbiota of a growth-stunted Malawian infant.139 Mice were then fed a typical Malawian diet with or without purified sialylated bovine milk oligosaccharides. Mice receiving the oligosaccharides treatment showed significantly increased weight gain, lean mass, and long bone growth, compared to the control group (caloric intake was equivalent between the groups). These effects were not seen in germ-free mice treated with oligosaccharides, indicating the microbiome plays a critical role in the health benefits observed. Similar results were seen in gnotobiotic piglets.139 Intriguingly, despite the gut microbiome-dependent nature of the effects, the composition of the gut community was not significantly different between oligosaccharides and control groups after treatment. However, significant transcriptional changes were observed in B. fragilis and E. coli, including upregulation of genes in the polysaccharide utilization locus of B. fragilis. The researchers also noted that the two B. longum subsp. infantis strains included in the community failed to colonize in the gut community in both the treatment and control groups, although strains of B. longum subsp. breve, B. bifido, and B. catenulatum did colonize.139 This is surprising given the ubiquity of B. longum subsp. infantis in the gut microbiota of human infants and its superior abilities to digest and metabolize HMOs. However, recent research indicates the ability of bacterial strains to successfully colonize the infant gut is affected by many different factors.140 Follow-up studies on the mechanism of increased long bone growth with sialylated oligosaccharide treatment indicated the effect came from decreased osteoclast generation and activity, in a microbiota-dependent manner.141 Much work remains to be done to investigate the connections between the gut microbiome and infant health.
Dietary sialic acids and adult health
In contrast to studies of infants and dietary sialic acids, where studies focus on microbiome composition but often do not address direct health impacts, studies of adults and dietary sialic acids focus mainly on health impacts and rarely assess microbiome composition. The ubiquity of sialic acids in mammalian glycoconjugates gives them a role in many physiological and pathological processes, from brain development to immune regulation, infections, heart disease, and diabetes.71 Many of these pathological processes have been associated with hypo-sialylation, or low Neu5Ac levels, of relevant molecules. Several studies have therefore looked at the effect of exogenous Neu5Ac-feeding on disease development and progression. Neu5Ac-feeding in apoE−/ – mice (a model of atherosclerosis through knockout of ApoE, a protein heavily involved in lipid circulation and metabolism)142 reduced atherosclerosis plaque area, as well as lipid liver deposition, triglyceride and cholesterol levels, and expression of inflammatory cytokines and intracellular adhesion factors in aorta endothelial cells and liver cells.92 In a different study, oral supplementation of the Neu5Ac precursor N-acetyl-D-mannosamine in mice on a high-fat diet (to study type II diabetes) resulted in a restoration of IgG sialylation and preserved insulin sensitivity.143 The mechanism of action in these studies is unknown and changes in the microbiome were not investigated in either case. However, given the established connections between the gut microbiome and atherosclerosis and diabetes144–146 and the impact sialic acids can have on the microbiome, an investigation of gut microbiome composition in response to Neu5Ac in these disease models would be intriguing.
The impact of dietary sialic acids on the adult gut microbiome is often difficult to tease apart, given the varied diets of adults. In 2017, researchers analyzed the gut microbiota of the Hadza people, a community living an ancestral hunter-gatherer lifestyle in Tanzania, where diet composition is determined by seasonal food availability.4 A longitudinal analysis revealed important modifications of the microbiome over the course of a year, following shifts between dry and wet seasons that corresponded to periods of meat – and plant-based diets, respectively. Metagenomic sequencing revealed both an increased diversity and increased number (as reads per million) of carbohydrate-active enzymes (including sialidases) in dry season samples, when the Hadza diet is dominated by meat, a food rich in sialic acids.4 A different study, focusing specifically on sialidases, re-analyzed the Hadza data and found specific enrichment of an organism encoding a sialidase to release Neu5Gc from glycans in the dry season samples.116 Since Neu5Gc is not made by humans, but is specifically enriched in red meat, this finding indicates that a Neu5Gc-metabolizing microbe becomes more abundant in the Hadza gut microbiota when levels of Neu5Gc increase in the diet.
The ability of non-human mammals to produce Neu5Gc, through the functional CMAH enzyme that humans lack, has led to a great difficulty in studying the effects of anti-Neu5Gc inflammation in animal models. However, researchers have been able to work around this through the generation of Cmah−/ – animals that, like humans, produce only Neu5Ac.147,148 The presence of Neu5Gc in human glycoconjugates has been implicated in numerous disease processes, such as liver cancer and atherosclerosis.93,94 Neu5Gc-feeding in mouse models deficient in Neu5Gc exacerbates these diseases. Of particular interest, Cmah−/ – knockout in a background knockout of the low-density lipoprotein receptor (Ldlr−/-) reproduces the human-specific Neu5Gc deficiency in a classic atherosclerosis model.94 Neu5Gc-feeding in this Cmah−/ – Ldlr−/ – mouse model demonstrated significantly more atherosclerosis plaque size and necrotic core volume, compared to control groups.94 Feeding of Neu5Gc in a Cmah−/ – mouse model (without the Ldlr−/ – deletion) showed distinct changes in the gut microbiome, with Bacteroides, Barnesiella, Clostridium, Parabacteroides, Roseburia, and Turicibacter significantly enriched compared to feeding with Neu5Ac.116 Examining the effects of Neu5Gc-feeding on the microbiome of the Cmah−/ – Ldlr−/ – model and potential relationships between these changes and atherosclerosis could further our current understanding of the role the gut microbiome plays in cardiovascular disease.
Conclusion
The impact of carbohydrates on the gut microbiome is nuanced, with differences seen from alterations in large carbohydrate classes, individual monosaccharides, and even modifications of individual monosaccharides. Many studies have examined the effects of broad glycan classes, such as fiber, in animals, and humans. Many other studies have looked at the ability of bacteria common in the gut microbiome to metabolize individual monosaccharides or glycans, either in vitro or in vivo. Microbiome composition shifts rapidly and reproducibly with dietary changes, emphasizing the potential therapeutic benefits of diet modifications. However, as a genomic analysis of organisms from the Human Gut Microbiome project shows (Figure 4), we have barely scratched the surface in our studies of individual microbes that can metabolize monosaccharides like sialic acids. Studies are also lacking on the effect of dietary sialic acids on the adult gut microbiome. Given the prominent microbiome and health effects seen in infants with sialylated HMOs, we expect that dietary sialic acids could drive similarly important microbiome modifications in adults. These modifications could be a missing link to explain the changes in disease phenotypes observed with dietary sialic acids in animal models. The impact of individual dietary glycans on the gut microbiome is therefore an essential field of research as we continue to explore the relationship between the gut microbiome and human disease.
Acknowledgments
This material is based upon work supported by the NIH/NIAID Atopic Dermatitis Research Network under grant number U19AI117673, the U.S. Army Research Office under grant number W911NF1810158, the National Science Foundation under grant number CBET-1804187, the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research under Awards DE-SC0019388 and DE-SC0021234. J.C. received support from the NIH under grant 5T32GM127235-02. D.R.’s research was supported by NIH (RO1 DK030292-35).
Funding Statement
This work was supported by the National Institute of Allergy and Infectious Diseases [U19AI117673]; National Institutes of Health [5T32GM127235-02]; National Institutes of Health [R01 DK030292-35]; National Science Foundation [CBET-1804187]; U.S. Department of Energy, Office of Science [DE-SC0019388]; U.S. Deparment of Energy, Office of Science [DE-SC0021234]; US Army Research Office [W911NF1810158].
Disclosure statement
K.Z. is an inventor on a patent application related to the use of sialidases.
References
- 1.Knight R, Callewaert C, Marotz C, Hyde ER, Debelius JW, McDonald D, Sogin ML. The microbiome and human biology. Annu Rev Genomics Hum Genet. 2017;18(1):65–18. doi: 10.1146/annurev-genom-083115-022438 [DOI] [PubMed] [Google Scholar]
- 2.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336(6086):1325–1329. doi: 10.1126/science.1222195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macfarlane GT, Macfarlane S.. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012;95(1):50–60. doi: 10.5740/jaoacint.SGE_Macfarlane. [DOI] [PubMed] [Google Scholar]
- 4.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357(6353):802–805. doi: 10.1126/science.aan4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 6.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- 7.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. PNAS. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmody RN, Bisanz JE, Bowen BP, Maurice CF, Lyalina S, Louie KB, Treen D, Chadaideh KS, Maini Rekdal V, Bess EN, et al. Cooking shapes the structure and function of the gut microbiome. Nat. Microbiol. 2019;4(12):2052–2063. doi: 10.1038/s41564-019-0569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2014;10(5):323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. The ISME Journal. 2011;5(2):220–230. doi: 10.1038/ismej.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GE, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. The ISME Journal. 2014;8(11):2218–2230. doi: 10.1038/ismej.2014.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007;73(4):1073–1078. doi: 10.1128/AEM.02340-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolodziejczyk AA, Zheng D, Elinav E. Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019;17(12):742–753. doi: 10.1038/s41579-019-0256-8. [DOI] [PubMed] [Google Scholar]
- 15. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–109. doi: 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 17.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 19.Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, Lucas SK, Beura LK, Thompson EA, Till LM, et al. US immigration westernizes the human gut microbiome. Cell. 2018;175(4):962–972.e10. doi: 10.1016/j.cell.2018.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10(1):1–13. doi: 10.1128/mBio.02566-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eswaran S, Muir J, Chey WD. Fiber and functional gastrointestinal disorders. Am J Gastroenterol. 2013;108(5):718–727. doi: 10.1038/ajg.2013.63. [DOI] [PubMed] [Google Scholar]
- 22.Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, et al. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015;25(12):1323–1324. doi: 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lütteke T, O’Boyle N, Packer NH, Stanley P, Toukach P, et al. Updates to the symbol nomenclature for glycans guidelines. Glycobiology. 2019;29(9):620–624. doi: 10.1093/glycob/cwz045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Den Besten G, Van Eunen K, Groen AK, Venema K, Reijngoud D-J., Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy CC, Kien CL, Bouthillier L, Levy E. Short-chain fatty acids: ready for prime time? Nutr. Clin. Pract. 2006;21(4):351–366. doi: 10.1177/0115426506021004351. [DOI] [PubMed] [Google Scholar]
- 26.Cummings JH, Pomare EW, Branch WJ, Naylor CPE, MacFarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloemen JG, Venema K, van de Poll MC, Olde Damink SW, Buurman WA, Dejong CH. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin. Nutr. 2009;28(6):657–661. doi: 10.1016/j.clnu.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 28.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat Rev Gastroenterol Hepatol. 2019;16:461–478. [DOI] [PubMed] [Google Scholar]
- 29.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. U. S. A. 2014;111(6):2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Di Y, Schilter HC, Rolph MS, MacKay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostatsis. Science. 2013;341(6145):569–574. doi: 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sina C, Gavrilova O, Förster M, Till A, Derer S, Hildebrand F, Raabe B, Chalaris A, Scheller J, Rehmann A, et al. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J. Immunol. 2009;183(11):7514–7522. doi: 10.4049/jimmunol.0900063 [DOI] [PubMed] [Google Scholar]
- 33.Haase S, Haghikia A, Wilck N, Müller DN, Linker RA. Impacts of microbiome metabolites on immune regulation and autoimmunity. Immunology. 2018;154(2):230–238. doi: 10.1111/imm.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, Offermanns S, Ganapathy V. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J. Biol. Chem. 2010;285(36):27601–27608. doi: 10.1074/jbc.M110.102947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20(2):159–166. doi: 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- 36.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469(7331):543–549. doi: 10.1038/nature09646 [DOI] [PubMed] [Google Scholar]
- 37.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167(5):1339–1353.e21. doi: 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4(1):69. doi: 10.1186/s40168-016-0218-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6(1):70. doi: 10.1186/s40168-018-0451-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aune D, Keum N, Giovannucci E, Fadnes LT, Boffetta P, Greenwood DC, Tonstad S, Vatten LJ, Riboli E, Norat T. Whole grain consumption and risk of cardiovascular disease, cancer, and all cause and cause-specific mortality: systematic review and dose-response meta-analysis of prospective studies. BMJ. 2016;353:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen HM, Yu YN, Wang JL, Lin YW, Kong X, Yang CQ, Yang L, Liu ZJ, Yuan YZ, Liu F, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 2013;97(5):1044–1052. doi: 10.3945/ajcn.112.046607 [DOI] [PubMed] [Google Scholar]
- 42.Singh V, Yeoh BS, Chassaing B, Xiao X, Saha P, Aguilera Olvera R, Lapek JD, Zhang L, Wang WB, Hao S, et al. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell. 2018;175(3):679–694.e22. doi: 10.1016/j.cell.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golonka RM, Yeoh BS, Vijay-Kumar M. Dietary additives and supplements revisited: the fewer, the safer for gut and liver health. Curr Pharmacol Reports. 2019;5(4):303–316. doi: 10.1007/s40495-019-00187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makki K, Deehan EC, Walter J, The BF. Impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23(6):705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Townsend GE, Han W, Schwalm ND, Raghavan V, Barry NA, Goodman AL, Groisman EA. Dietary sugar silences a colonization factor in a mammalian gut symbiont. Proc. Natl. Acad. Sci. U. S. A. 2019;116(1):233–238. doi: 10.1073/pnas.1813780115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdelmalek MF, Suzuki A, Guy C, Unalp-Arida A, Colvin R, Johnson RJ, Diehl AM. Increased fructose consumption is associated with fibrosis severity in patients with NAFLD. Hepatology. 2010;51(6):1961–1971. doi: 10.1002/hep.23535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alwahsh SM, Xu M, Seyhan HA, Ahmad S, Mihm S, Ramadori G, Schultze FC. Diet high in fructose leads to an overexpression of lipocalin-2 in rat fatty liver. World J Gastroenterol. 2014;20(7):1807–1821. doi: 10.3748/wjg.v20.i7.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellmann C, Priebs J, Landmann M, Degen C, Engstler AJ, Jin CJ, Gärttner S, Spruss A, Huber O, Bergheim I. Diets rich in fructose, fat or fructose and fat alter intestinal barrier function and lead to the development of nonalcoholic fatty liver disease over time. J. Nutr. Biochem. 2015;26(11):1183–1192. doi: 10.1016/j.jnutbio.2015.05.011 [DOI] [PubMed] [Google Scholar]
- 49.Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, Martin P, Philippe C, Walker F, Bado A, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62(12):1787–1794. doi: 10.1136/gutjnl-2012-303816 [DOI] [PubMed] [Google Scholar]
- 50.Jegatheesan P, Beutheu S, Ventura G, Sarfati G, Nubret E, Kapel N, Waligora-Dupriet AJ, Bergheim I, Cynober L, De-Bandt JP. Effect of specific amino acids on hepatic lipid metabolism in fructose-induced non-alcoholic fatty liver disease. Clin. Nutr. 2016;35(1):175–182. doi: 10.1016/j.clnu.2015.01.021 [DOI] [PubMed] [Google Scholar]
- 51.Ritze Y, Bárdos G, Claus A, Ehrmann V, Bergheim I, Schwiertz A, Bischoff SC. Lactobacillus rhamnosus GG Protects against non-alcoholic fatty liver disease in mice. PLoS One. 2014;9(1):1–9. doi: 10.1371/journal.pone.0080169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, Guy CD, Seed PC, Rawls JF, David LA, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63(3):764–775. doi: 10.1002/hep.28356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051 [DOI] [PubMed] [Google Scholar]
- 54.Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol - Gastrointest Liver Physiol. 2001;280(5):922–929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 55.Baeckstrom D, Hansson GC, Nilsson O, Johansson C, Gendler SJ, Lindholm L. Purification and characterization of a membrane-bound and a secreted mucin-type glycoprotein carrying the carcinoma-associated sialyl-Lea epitope on distinct core proteins. J. Biol. Chem. 1991;266(32):21537–21547 [PubMed] [Google Scholar]
- 56.Johansson MEV, Philipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U. S. A. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Norin KE, Gustafsson BE, Lindblad BS, Midtvedt T. The establishment of some microflora associated biochemical characteristics in feces from children during the first years of life. Acta Pædiatrica. 1985;74(2):207–212. doi: 10.1111/j.1651-2227.1985.tb10951.x. [DOI] [PubMed] [Google Scholar]
- 58.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, et al. American Gut<: An Open Platform for Citizen Science. mSystems. 2018;3(3):1–28. doi: 10.1128/mSystems [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18(4):478–488. doi: 10.1016/j.chom.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Earley H, Lennon G, Balfe A, Coffey JC, Winter DC, O'Connell PR. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-51878-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010;105(11):2420–2428. doi: 10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- 63.Fu J, Wei B, Wen T, Johansson MEV, Liu X, Bradford E, Thomsson KA, Megee S, Mansour L, Tong M, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J. Clin. Invest. 2011;121(4):1657–1666. doi: 10.1172/JCI45538.is [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holmn Larsson JM, Karlsson H, Graberg Crespo J, Johansson ME.Eklund L, Sjovall H, Hansson GC. Altered O-glycsoylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17(11):2299–2307. doi: 10.1002/ibd.21625 [DOI] [PubMed] [Google Scholar]
- 65. Johansson MEV, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63(2):281–291. doi: 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, Backhed F. Bifidobacteria or fiber protect against diet-induced microbiota-mediated colonic mucus deterioration hhs public access the defects can be prevented by application of a probiotic bifidobacteria or the prebiotic fiber inulin. Cell Host Microbe. 2018;23(1):27–40. doi: 10.1016/j.chom.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada T, Hino S, Iijima H, Genda T, Aoki R, Nagata R, Han KH, Hirota M, Kinashi Y, Oguchi H, et al. Mucin O-glycans facilitate symbiosynthesis to maintain gut immune homeostasis. EBioMedicine. 2019;48:513–525. doi: 10.1016/j.ebiom.2019.09.008 [DOI] [PMC free article] [PubMed]
- 68.Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med. 1989;320(1):23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- 69.Scheppach W, Sommer H, Kirchner T, Paganelli G-M, Bartram P, Christl S, Richter F, Dusel G, Kasper H. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103(1):51–56. doi: 10.1016/0016-5085(92)91094-K [DOI] [PubMed] [Google Scholar]
- 70.Angata T, Varki A. Chemical diversity in the sialic acids and related α-keto acids: an evolutionary perspective. Chem. Rev. 2002;102(2):439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 71.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14(8):351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 2004;68(1):132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwarzkopf M, Knoebloch K, Rohde E, Hinderlich S, Wiechens N, Lucka L, Horak I, Reutter W, Horstkorte R. Sialylation is essential for early development in mice. Proc Natl Acad Sci U S A. 2002;99(8):5267–5270. doi: 10.1073/pnas.072066199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brigham C, Caughlan R, Gallegos R, Dallas MB, Godoy VG, Malamy MH. Sialic acid (n-acetyl neuraminic acid) utilization by bacteroides fragilis requires a novel n-acetyl mannosamine epimerase. J Bacteriol. 2009;191(11):3629–3638. doi: 10.1128/JB.00811-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Newstead SL, Potter JA, Wilson JC, Xu G, Chien CH, Watts AG, Withers SG, Taylor GL. The structure of clostridium perfringens NanI sialidase and its catalytic intermediates. J Biol Chem. 2008;283(14):9080–9088. doi: 10.1074/jbc.M710247200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Egan M, Motherway MO, Ventura M, van Sinderen D. Metabolism of sialic acid by bifidobacterium breve UCC2003. Appl Env Microbiol. 2014;80(14):4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nilsson I, Prathapam R, Grove K, Lapointe G, Six DA. The sialic acid transporter NanT is necessary and sufficient for uptake of 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) and its azido analog in Escherichia coli. Mol Microbiol. 2018;110(2):204–218. doi: 10.1111/mmi.14098. [DOI] [PubMed] [Google Scholar]
- 78.Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. An exo-alpha-Sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 2011;21(4):437–447. doi: 10.1093/glycob/cwq175 [DOI] [PubMed] [Google Scholar]
- 79. Park KH, Kim MG, Ahn HJ, Lee DH, Kim JH, Kim YW, Woo EJ. Structural and biochemical characterization of the broad substrate specificity of Bacteroides thetaiotaomicron commensal sialidase. Biochim Biophys Acta - Proteins Proteomics. 2013;1834(8):1510–1519. doi: 10.1016/j.bbapap.2013.04.028 [DOI] [PubMed] [Google Scholar]
- 80.Post DMB, Mungur R, Gibson BW, Munson RS. Identification of a novel sialic acid transporter in Haemophilus ducreyi. Infect Immun. 2005;73(10):6727–6735. doi: 10.1128/IAI.73.10.6727-6735.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Severi E, Hosie AHF, Hawkhead JA, Thomas GH. Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiol Lett. 2010;304(1):47–54. doi: 10.1111/j.1574-6968.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- 82.Mulligan C, Leech AP, Kelly DJ, Thomas GH. The membrane proteins siaq and siam form an essential stoichiometric complex in the sialic acid tripartite atp-independent periplasmic (trap) transporter siapqm (vc1777–1779) from vibrio cholerae. J Biol Chem. 2012;287(5):3598–3608. doi: 10.1074/jbc.M111.281030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Schoop HJ, Schauer R, Faillard H. Zur biosynthese der n-glykolyl-neuraminsäure. die oxydative entstehung von n-glykolyl-neuraminsäure aus n-acetyl-neuraminsäure. Hoppe Seylers Z Physiol Chem. 1969;350(1):155–162. doi: 10.1515/bchm2.1969.350.1.155. [DOI] [PubMed] [Google Scholar]
- 84. Schauer R. Biosynthese der N-Glykoloylneuraminsäure durch eine von Ascorbinsäure bzw. NADPH abhängige N-Acetyl-hydroxylierende „N-Acetylneuraminat: O2-Oxidoreduktase“ in Homogenaten der Unterkieferspeicheldrüse vom Schwein. Hoppe Seylers Z Physiol Chem. 1970;351(2):783–791. doi: 10.1515/bchm2.1970.351.2.783. [DOI] [PubMed] [Google Scholar]
- 85. Hayakawa T, Satta Y, Gagneux P, Varki A, Takahata N. Alu-mediated inactivation of the human CMP-N-acetylneuraminic acid hydroxylase gene. Proc. Natl. Acad. Sci. U. S. A. 2001;98(20):11399–11404. doi: 10.1073/pnas.191268198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Padler-Karavani V, Yu H, Cao H, Chokhawala H, Karp F, Varki N, Chen X, Varki A. Diversity in specificity, abundance, and composition of anti-Neu5Gc antibodies in normal humans: Potential implications for disease. Glycobiology. 2008;18(10):818–830. doi: 10.1093/glycob/cwn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Paul A, Padler-Karavani V. Evolution of sialic acids: implications in xenotransplant biology. Xenotransplantation. 2018;25(6):e12424. doi: 10.1111/xen.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Banda K, Gregg CJ, Chow R, Varki NM, Varki A. Metabolism of vertebrate amino sugars with n-glycolyl groups. J Biol Chem. 2012;287(34):28852–28864. doi: 10.1074/jbc.M112.364182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bergfeld AK, Pearce OM, Diaz SL, Pham T, Varki A. Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups. J Biol Chem. 2012;287(34):28865–28881. doi: 10.1074/jbc.M112.363549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dhar C, Sasmal A, From VA. Serum sickness’ to ‘xenosialitis’: past, present, and future significance of the non-human sialic acid neu5Gc. Front Immunol. 2019;10:807. doi: 10.3389/fimmu.2019.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang B, Brand-Miller J. The role and potential of sialic acid in human nutrition. Eur J Clin Nutr. 2003;57(11):1351–1369. doi: 10.1038/sj.ejcn.1601704. [DOI] [PubMed] [Google Scholar]
- 92. Guo S, Tian H, Dong R, Yang N, Zhang Y, Yao S, Li Y, Zhou Y, Si Y, Qin S. Exogenous supplement of N-acetylneuraminic acid ameliorates atherosclerosis in apolipoprotein E-deficient mice. Atherosclerosis. 2016;251:183–191. doi: 10.1016/j.atherosclerosis.2016.05.032 [DOI] [PubMed] [Google Scholar]
- 93. Samraj AN, Pearce OMT, Laubli H, Crittenden AN, Bergfeld AK, Banda K, Gregg CJ, Bingman AE, Secrest P, Diaz SL, et al. A red meat-derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci U S A. 2015;112(2):542–547. doi: 10.1073/pnas.1417508112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kawanishi K, Dhar C, Do R, Varki N, Gordts PLSM, Varki A. Human species-specific loss of CMP-N-acetylneuraminic acid hydroxylase enhances atherosclerosis via intrinsic and extrinsic mechanisms. Proc Natl Acad Sci U S A. 2019;116(32):16036–16045. doi: 10.1073/pnas.1902902116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153(9):2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 96.Mahajan VS, Pillai S. Sialic acids and autoimmune disease. Immunol Rev. 2016;169(1):145–161. doi: 10.1111/imr.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14(10):653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haines-Menges BL, Whitaker WB, Lubin JB, Boyd EF. Host Sialic Acids: A delicacy for the pathogen with discerning taste. Microbiol Spectr. 2015;3(4):1–17. doi: 10.1128/microbiolspec.MBP-0005-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Plumbridge J, Vimr E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by escherichia coli. J Bacteriol. 1999;181(1):47–54. doi: 10.1128/JB.181.1.47-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chang D-E, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101(19):7427–7437. doi: 10.1073/pnas.0307888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hopkins AP, Hawkhead JA, Thomas GH. Transport and catabolism of the sialic acids N-glycolylneuraminic acid and 3-keto-3-deoxy-d-glycero-d-galactonononic acid by Escherichia coli K-12. FEMS Microbiol. Lett. 2013;347(1):14–22. doi: 10.1111/1574-6968.12213. [DOI] [PubMed] [Google Scholar]
- 102.Almagro-Moreno S, Boyd EF. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 2009;9(1):118] has been updated. OK?</chg>. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Marcobal AM, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10(5):507–514. doi: 10.1016/j.chom.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38: 779–786 [DOI] [PubMed] [Google Scholar]
- 105.Rodionov DA, Arzamasov AA, Khoroshkin MS, Iablokov SN, Leyn SA, Peterson SN, Novichkov PS, Osterman AL. Micronutrient requirements and sharing capabilities of the human gut microbiome. Front Microbiol. 2019;10:1–22. doi: 10.3389/fmicb.2019.01316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Research. 2014;42(D1):206–214. doi: 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed]
- 107. Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, et al. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48(D1):D606–D612. doi: 10.1093/nar/gkz943 [DOI] [PMC free article] [PubMed]
- 108.Stamatakis A. Using rAxML to infer phylogenies. Curr Protoc Bioinforma. 2015;51:6.14.1–6.14.14. [DOI] [PubMed] [Google Scholar]
- 109.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(44):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24(1):40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502(7469):96–99. doi: 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang Y-L, Chassard C, Hausmann M, von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat Commun. 2015;6(1):8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robinson LS, Lewis WG, Lewis AL. The sialate O-acetylesterase EstA from gut Bacteroidetes species enables sialidase-mediated cross-species foraging of 9-O-acetylated sialoglycans. J Biol Chem. 2017;292(28):11861–11872. doi: 10.1074/jbc.M116.769232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Owen CD, Tailford LE, Monaco S, Suligoj T, Vaux L, Lallement R, Khedri Z, Yu H, Lecointe K, Walshaw J, et al. Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus. Nat Commun. 2017;8(1):2196. doi: 10.1038/s41467-017-02109-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Bell A, Brunt J, Crost E, Vaux L, Nepravishta R, Owen CD, Latousakis D, Xiao A, Li W, Chen X, et al. Elucidation of a unique sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut. Nat Microbiol. 2019;4(12):2393–2404. doi: 10.1038/s41564-019-0590-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zaramela LS, Martino C, Alisson-silva F, Rees SD, Diaz SL, Chuzel L, Ganatra MB, Taron CH, Secrest P, Zuniga C, et al. Gut bacteria responding to dietary changes encode sialidases the exhibit preference for red meat-associated carbohydrates. Nat. Microbiol. 2019;4(12):2082–2089. doi: 10.1038/s41564-019-0564-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res. 2015;77(1–2):229–235. doi: 10.1038/pr.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108(Supplement_1):4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R, et al. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 2006;54(20):7471–7480. doi: 10.1021/jf0615810 [DOI] [PubMed] [Google Scholar]
- 121.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 2000;71(6):1589–1596. doi: 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- 122.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25(1):37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 123.Martín-Sosa S, Martín M-J, Hueso P. The sialylated fraction of milk oligosaccharides is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. J Nutr. 2002;132(10):3067–3072. doi: 10.1093/jn/131.10.3067. [DOI] [PubMed] [Google Scholar]
- 124.Idota T, Kawakami H, Murakami Y, Sugawara M. Inhibition of cholera toxin by human milk fractions and sialyllactose. Biosci Biotechnol Biochem. 1995;59(3):417–419. doi: 10.1271/bbb.59.417. [DOI] [PubMed] [Google Scholar]
- 125.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58(9):5334–5340. doi: 10.1021/jf9044205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yu ZT, Chen C, Newburg DS. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology. 2013;23(11):1281–1292. doi: 10.1093/glycob/cwt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lis-Kuberka J, Orczyk-Pawiłowicz M. Sialylated oligosaccharides and glycoconjugates of human milk. The impact on infant and newborn protection, development and well-being. Nutrients. 2019;11(2):306. doi: 10.3390/nu11020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105(48):18964–18969. doi: 10.1073/pnas.0809584105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, Bruce German J, Chen X, Lebrilla CB, Mills DA. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem. 2011;286(14):11909–11918. doi: 10.1074/jbc.M110.193359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. Oligosaccharide binding proteins from bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One. 2011;6(3):e17315. doi: 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim JH, An HJ, Garrido D, German JB, Lebrilla CB, Mills DA. Proteomic analysis of bifidobacterium longum subsp. infantis reveals the metabolic insight on consumption of prebiotics and host glycans. PLoS One. 2013;8:e57535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect. 2012;18:12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schwab C, Gänzle M. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiol Lett. 2011;315(2):141–148. doi: 10.1111/j.1574-6968.2010.02185.x. [DOI] [PubMed] [Google Scholar]
- 134.Ruiz-Moyano S, Totten SM, Garrido DA, Smilowitz JT, Bruce German J, Lebrilla CB, Mills DA. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of bifidobacterium breve. Appl Environ Microbiol. 2013;79(19):6040–6049. doi: 10.1128/AEM.01843-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang B, Brand-Miller J, McVeagh P, Petocz P. Concentration and distribution of sialic acid in human milk and infant formulas. Am J Clin Nutr. 2001;74(4):510–515. doi: 10.1093/ajcn/74.4.510. [DOI] [PubMed] [Google Scholar]
- 136.Nwosu CC, Aldredge DL, Lee H, Lerno LA, Zivkovic AM, German JB, Lebrilla CB. Comparison of the human and bovine milk n-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res. 2012;11(5):2912–2924. doi: 10.1021/pr300008u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Quin C, Vicaretti SD, Mohtarudin NA, Garner AM, Vollman DM, Gibson DL, Zandberg WF, Hart GW. Influence of sulfonated and diet-derived human milk oligosaccharides on the infant microbiome and immune markers. J. Biol. Chem. 2020;295(12):4035–4048. doi: 10.1074/jbc.RA119.011351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Taylor RE, Gregg CJ, Padler-Karavani V, Ghaderi D, Yu H, Huang S, Sorensen RU, Chen X, Inostroza J, Nizet V, et al. Novel mechanism for the generation of human xeno-autoantibodies against the nonhuman sialic acid N-glycolylneuraminic acid. J Exp Med. 2010;207(8):1637–1646. doi: 10.1084/jem.20100575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Charbonneau MR, O’Donnell D, Blanton L V., Totten SM, Davis JCC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. 2016;164(5):859–871. doi: 10.1016/j.cell.2016.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Feng L, Raman AS, Hibberd MC, Cheng J, Griffin NW, Peng Y, Leyn SA, Rodionov DA, Osterman AL, Gordon JI. Identifying determinants of bacterial fitness in a model of human gut microbial succession. Proc. Natl. Acad. Sci. U. S. A. 2020;117(5):2622–2633. doi: 10.1073/pnas.1918951117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cowardin CA, Ahern PP, Kung VL, Hibberd MC, Cheng J, Guruge JL, Sundaresan V, Head RD, Barile D, Mills DA, et al. Mechanisms by which sialylated milk oligosaccharides impact bone biology in a gnotobiotic mouse model of infant undernutrition. Proc. Natl. Acad. Sci. U. S. A. 2019;116(24):11988–11996. doi: 10.1073/pnas.1821770116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Getz GS, Reardon CA. Animal models of atherosclerosis. Aterioscler Thromb Vasc Biol. 2012;32(5):1104–1115. doi: 10.1161/ATVBAHA.111.237693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tanigaki K, Sacharidou A, Peng J, Chambliss KL, Yuhanna IS, Ghosh D, Ahmed M, Szalai A, Vongatanasin W, Mattrey RF, et al. Hyposialylated IgG activates endothelial IgG receptor FcyRIIB to promote obesity-induced insulin resistance. J Clin Invest. 2018;128(1):309–322. doi: 10.1172/JCI89333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jonsson AL, Bäckhed F. Role of gut microbiota in atherosclerosis. Nat Rev Cardiol. 2017;14(2):79–87. doi: 10.1038/nrcardio.2016.183. [DOI] [PubMed] [Google Scholar]
- 145.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8(1):1–11. doi: 10.1038/s41467-017-00900-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, Shulzhenko N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. doi: 10.1016/j.ebiom.2019.11.051 [DOI] [PMC free article] [PubMed]
- 147.Hedlund M, Tangvoranuntakul P, Takematsu H, Long JM, Housley GD, Kozutsumi Y, Suzuki A, Wynshaw-Boris A, Ryan AF, Gallo RL, et al. N-glycolylneuraminic acid deficiency in mice: implications for human biology and evolution. Mol Cell Biol. 2007;27(12):4340–4346. doi: 10.1128/MCB.00379-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Perota A, Galli C. N-glycolylneuraminic acid (Neu5Gc) null large animals by targeting the CMP-Neu5Gc hydroxylase (CMAH). Front Immunol. 2019:10. doi: 10.3389/fimmu.2019.02396 [DOI] [PMC free article] [PubMed] [Google Scholar]


