The endonuclease III-like protein 1, encoded by NTHL1, is a bifunctional glycosylase involved in base-excision repair (BER) that recognizes and removes oxidized pyrimidines.1 Similar to biallelic loss-of-function (LoF) variants in MUTYH,2 biallelic LoF variants in NTHL1 predispose to colorectal polyps and colorectal cancer (CRC).3 Recently, a multitumor phenotype was observed in individuals diagnosed with NTHL1 deficiency.4 Carriers of monoallelic pathogenic variants in MUTYH have an increased, albeit small, risk of CRC.5 Thus far, it is unknown if monoallelic NTHL1 LoF variants also increase the risk of polyposis and/or CRC. This information is especially important for carriers of the most common LoF variant in NTHL1 (p.(Gln90*); NM_002528.5), which is heterozygous in approximately 0.28% of the general population.6 Identification of monoallelic NTHL1 LoF variants currently presents a clinical conundrum regarding how best to counsel carriers with respect to their cancer risk because of the lack of published evidence. Here, we show that monoallelic LoF variants in NTHL1 are not enriched in individuals with polyposis and/or CRC compared to the general population. Furthermore, 13 colorectal tumors from NTHL1 LoF carriers did not show a somatic second hit, and we did not find evidence of a main contribution of mutational signature SBS30, the signature associated with NTHL1 deficiency, suggesting that monoallelic loss of NTHL1 does not substantially contribute to colorectal tumor development.
Methods
A total of 5,942 individuals with unexplained polyposis, familial CRC, or sporadic CRC at young age or suspected of having Lynch syndrome with CRC or multiple adenomas were included in this study and defined as case patients (individual studies and their ascertainment are described in Supplementary Methods and Supplementary Table 1). Three independent data sets were used as controls, including (1) the non-Finnish European subpopulation of the genome aggregation database (gnomAD: n = 64,328),6 (2) a Dutch cohort of individuals without a suspicion of hereditary cancer who underwent whole-exome sequencing (WES) (Dutch WES; n = 2,329),7 and (3) a population-based and cancer-unaffected cohort from the Colon Cancer Family Registry Cohort (CCFRC; n = 1,207) (Supplementary Methods and Supplementary Table 1).
Pathogenic NTHL1 LoF variants were identified in case patients by sequencing the exonic regions of NTHL1 (n = 3,439) or by genotyping of 2 LoF variants in NTHL1 (c.268C>T, p.(Gln90*); n = 2503 and c.806G>A, p.(Trp269*); n = 261) (Supplementary Table 1). For control individuals, all pathogenic LoF variants were retrieved from gnomAD and the Dutch WES-cohort,6,7 and for the CCFRC control individuals, the exonic regions of NTHL1 were sequenced (Supplementary Table 1). Odds ratios between case patients and control groups were calculated and a Fisher exact test was performed to assess the significance of difference in carrier rates. Cosegregation analysis was performed by using Sanger sequencing. Two adenomas and 11 primary CRCs from NTHL1 LoF variant carriers were subjected to WES, and subsequently, mutational signature analysis was performed (Supplementary Methods and Supplementary Table 2). For signature analysis comparison, we included 3 CRCs from individuals with a biallelic NTHL1 LoF variant.
Results
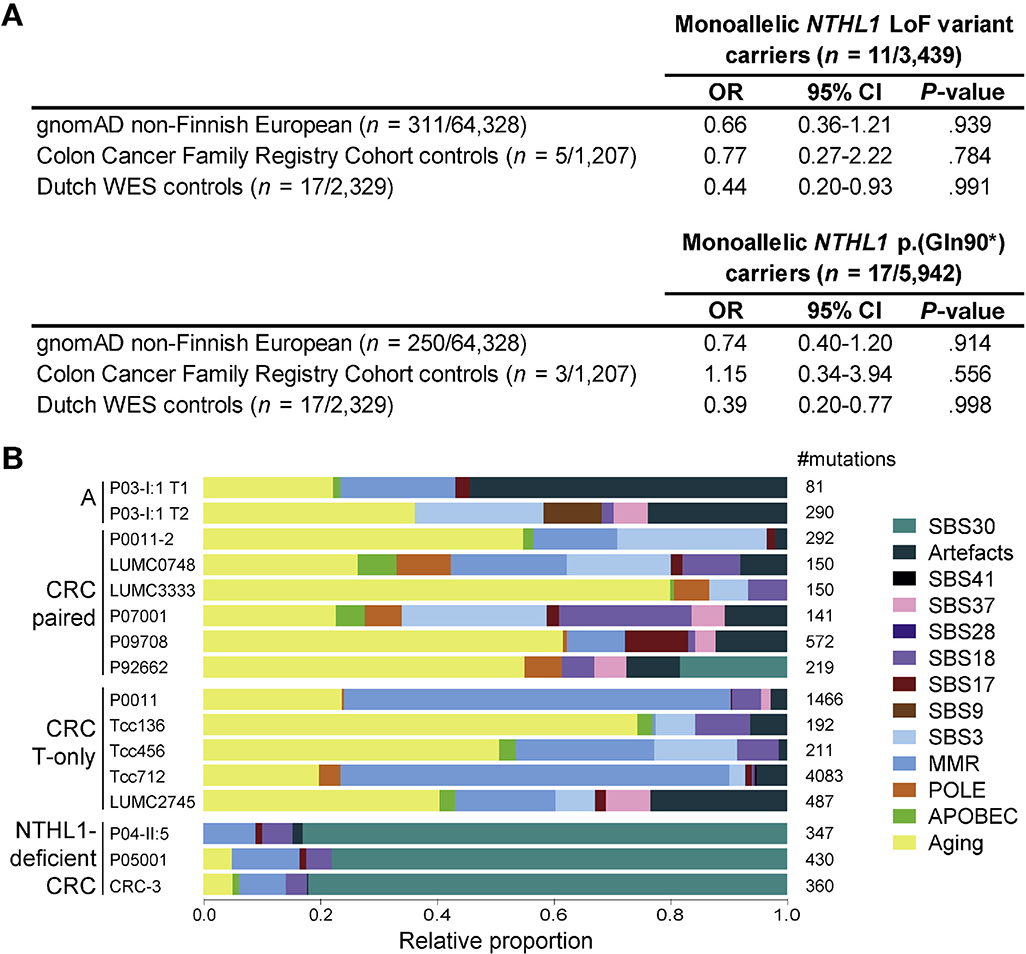
Monoallelic NTHL1 LoF variants were identified in 11 of 3,439 case patients (0.32%) and in 5 of 1,207 (0.41%) of CCFRC control individuals, indicating no significant difference (P = .784) (Figure 1A, Supplementary Table 1). Genotyping of the NTHL1 p.(Gln90*) variant in another 2,503 case patients identified 7 additional carriers (0.28%). The overall frequency of NTHL1 p.(Gln90*) in case patients was not different from the frequency in the gnomAD (17/5,942 vs 250/64,328; P = .914), CCFRC (17/5,942 vs 3/1,207; P = .556) or Dutch WES control individuals (17/5,942; vs 17/2,329; P = .998) (Figure 1A and Supplementary Table 1).
Figure 1.
Enrichment and mutational signature analysis of NTHL1 LoF variants in individuals with polyposis and/or CRC (case patients). (A) Frequencies of germline monoallelic NTHL1 LoF variants and monoallelic NTHL1 p.(Gln90*) variants in individuals with polyposis and/or CRC (case patients) compared with control populations. (B) Mutational signature analysis of tumors from carriers with a monoallelic NTHL1 LoF variant. Mutational signatures with shared etiologies were grouped for display purposes, which are the signatures associated with aging (SBS1, SBS5, and SBS40), DNA mismatch repair deficiency (SBS6, SBS15, SBS20, SBS21, SBS26, and SBS44), Polymerase Epsilon (POLE) exonuclease domain deficiency (SBS10a and SBS10b), Apolipoprotein B mRNA editing enzyme (APOBEC) activity (SBS2 and SBS13), and artifact signatures (SBS45, SBS51, SBS52, SBS54, and SBS58). Data availability: paired: tumor and normal or tumor data were available; T-only: only data from 1 tumor tissue were available. A, adenomatous polyp; CI, confidence interval; OR, odds ratio.
Via cosegregation analysis, we identified 3 additional NTHL1 p.(Gln90*) carriers. The phenotype of all carriers identified in this study is described in Supplementary Table 2. Thirteen colorectal tumors from NTHL1 LoF carriers underwent WES (details in Supplementary Table 2). The NTHL1 wild-type allele was unaffected by somatic mutations or loss of heterozygosity in all tumors tested. In contrast to NTHL1-deficient tumors, in none of the tumors of the carriers was mutational signature SBS30 the main signature, because it was only present in 1 tumor, where it had a minor contribution (Figure 1B and Supplementary Table 2).4 These observations indicate that biallelic inactivation of NTHL1 through a somatic second hit was not evident and that monoallelic inactivation of NTHL1 was insufficient to result in the accumulation of somatic mutations that are characteristic of an NTHL1-deficiency phenotype.
Discussion
In this study, the largest investigating monoallelic LoF variants in NTHL1 to date to our knowledge, we observed no evidence of an association between carriers and the risk of polyposis and/or CRC. In our case patients, the prevalence of pathogenic NTHL1 LoF variant alleles is comparable to that of the general population. However, we cannot rule out that a small risk for CRC, similar to what is observed for MUTYH carriers, still exists.
Colorectal tumors from monoallelic NTHL1 LoF variant carriers did not show evidence of a somatic second hit in NTHL1 nor of defective base-excision repair, which is typically associated with biallelic NTHL1 inactivation. Only 1 tumor showed a minor SBS30 contribution to the mutation profile, but this contribution was far less significant compared to NTHL1-deficient CRC and is likely the result of multiple testing correction. Our data suggest that inactivation of the NTHL1 wild-type allele is a rare event in colorectal tumors, which is in agreement with the observation that loss of heterozygosity of chromosome arm 16p is not frequently observed in CRC.8 We were unable to discriminate between individuals with polyposis or CRC due to the historical nature of the case collections. Therefore, differences in the frequencies of monoallelic NTHL1 LoF variants between control individuals and these 2 phenotypes were not made separately. However, because we identified NTHL1 LoF variants in individuals with polyposis or CRC, we do not consider a major difference between these 2 phenotypes. Because NTHL1 deficiency may also predispose to extracolonic tumors, the risk for these tumor types in monoallelic NTHL1 carriers still needs further assessment.
In conclusion, the evidence to date does not support an increased risk of polyposis and/or CRC for carriers of monoallelic NTHL1 LoF variants, and consequently, no additional surveillance is currently warranted beyond population screening for CRC, unless family history characteristics point to a reason for colonoscopy.
Supplementary Material
Acknowledgments
The authors thank all study participants, the CCFRC and staff, and the Dutch Parelsnoer Institute Biobank Hereditary Colorectal Cancer for their contributions to this project. Furthermore, we would like to thank Robbert Weren, Eveline Kamping, M. Elisa Vink-Börger, Riki Willems, Christian Gillissen, Peggy Manders, Dina Ruano, Ruud van der Breggen, Marina Ventayol, Sanne ten Broeke, Allyson Templeton, Maggie Angelakos, members of the Colorectal Oncogenomics Group, Sharelle Joseland, Susan Preston, Julia Como, Thomas Green, Magda Kloc, and Chris Cotsopoulos for their contributions to this project. The author(s) would further like to acknowledge networking support by the Cooperation in Science and Technology Action CA17118, supported by the European Cooperation in Science and Technology.
NTHL1 study group: Arnoud Boot, Marija Staninova Stojovska, Khalid Mahmood, Mark Clendenning, Noel de Miranda, Dagmara Dymerska, Demi van Egmond, Steven Gallinger, Peter Georgeson, Nicoline Hoogerbrugge, John L. Hopper, Erik A.M. Jansen, Mark A. Jenkins, Jihoon E. Joo, Roland P. Kuiper, Marjolijn J.L. Ligtenberg, Jan Lubinski, Finlay A. Macrae, Hans Morreau, Polly Newcomb, Maartje Nielsen, Claire Palles, Daniel J. Park, Bernard J. Pope, Christophe Rosty, Clara Ruiz Ponte, Hans K. Schackert, Rolf H. Sijmons, Ian P. Tomlinson, Carli M. J. Tops, Lilian Vreede, Romy Walker, Aung K. Win, Colon Cancer Family Registry Cohort Investigators, Aleksandar J. Dimovski, and Ingrid M. Winship.
Funding
This study was funded by research grants from the Dutch Cancer Society (KUN2015-7740), the Dutch Digestive Foundation (MLDS FP13-13 to Tom van Wezel), Instituto de Salud Carlos III and European Regional Development Fund (ERDF) (PI14/00230 to Clara Ruiz Ponte) and by grant UM1 CA167551 from the National Cancer Institute and through cooperative agreements with the following Colon Cancer Family Registry Cohort (CCFRC) sites: Australasian Colorectal Cancer Family Registry (U01 CA074778 and U01/U24 CA097735), Ontario Familial Colorectal Cancer Registry (U01/U24 CA074783), and Seattle Colorectal Cancer Family Registry (U01/U24 CA074794). Daniel B. Buchanan is a University of Melbourne Research at the Melbourne Accelerator Program (R@MAP), principal research fellow, and National Health and Medical Research Council (NHMRC) R.D. Wright Career Development Fellow. Abiram Ragunathan is a Melbourne Genomics Health Alliance Fellow.
Abbreviations used in this paper:
- CCFRC
Colon Cancer Family Registry Cohort
- CRC
colorectal cancer
- LoF
loss of function
- WES
whole-exome sequencing
Footnotes
Conflicts of interest
The authors disclose no conflicts.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Clinical Gastroenterology and Hepatology at www.cghjournal.org, and at https://doi.org/10.1053/j.gastro.2020.08.042.
References
- 1.Krokan HE, Bjørås M. Cold Spring Harb Perspect Biol 2013;5:a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Tassan N, et al. Nat Genet 2002;30:227–232. [DOI] [PubMed] [Google Scholar]
- 3.Weren RD, et al. Nat Genet 2015;47:668–671. [DOI] [PubMed] [Google Scholar]
- 4.Grolleman JE, de Voer RM, Elsayed FA, et al. Cancer Cell 2019;35:256–266. [DOI] [PubMed] [Google Scholar]
- 5.Win AK, et al. Int J Cancer 2011;129:2256–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karczewski KJ, et al. Nature 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Voer RM, Hahn MM, Mensenkamp AR, et al. Sci Rep 2015;5:14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerami E, et al. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.