Abstract
The present study was conducted to characterize microRNA-200c (miR-200c) and its regulators in adipogenic differentiation, obesity, and periodontitis in obese subjects (PiOSs), and to determine the therapeutic efficacy of plasmid DNA encoding miR-200c as a treatment for PiOSs. We report that highly expressed miR-200c in gingival tissues was downregulated in diet-induced obese (DIO) mice and during adipogenic differentiation of human bone marrow mesenchymal stromal cells (hBMSCs). Local injection of Porphyromonas gingivalis lipopolysaccharide (Pg-LPS) in the maxilla interdental gingiva of DIO mice reduced miR-200c in gingival and adipose tissues and induced periodontal inflammation associated with systemic elevation of interleukin-6 (IL-6) and impaired glucose tolerance. The inhibitory functions of Pg-LPS and IL-6 on miR-200c and their effectiveness on Zeb1 were confirmed in vitro. Injection of naked plasmid DNA encoding miR-200c into the gingiva effectively rescued miR-200c downregulation, prevented periodontal and systemic inflammation, and alleviated the impaired glucose metabolism in obese mice with LPS-induced periodontitis. Increased circulating exosomal miR-200c and its function on suppressing proinflammatory cytokines and adipogenesis explained the mechanism(s) of gingival application of miR-200c in attenuating systemic inflammation in PiOSs. These results demonstrated that miR-200c reduced by Pg-LPS and IL-6 in periodontitis and obesity might lead to the pathogenesis of PiOSs, and upregulation of miR-200c in the gingiva presents a therapeutic approach for PiOSs.
Keywords: periodontitis, obesity, microRNA-200c, gene therapy, mouse, IL-6, Pg-LPS, Zeb1, Leptin, glucose intolerance
Graphical Abstract
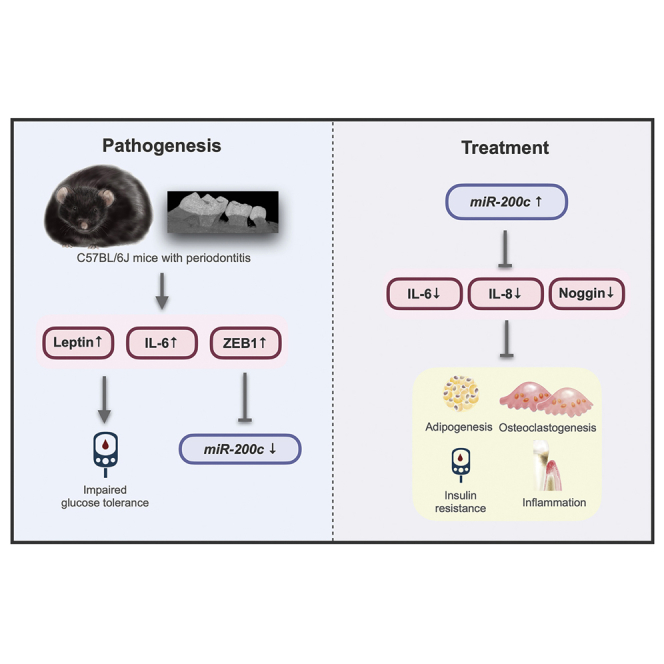
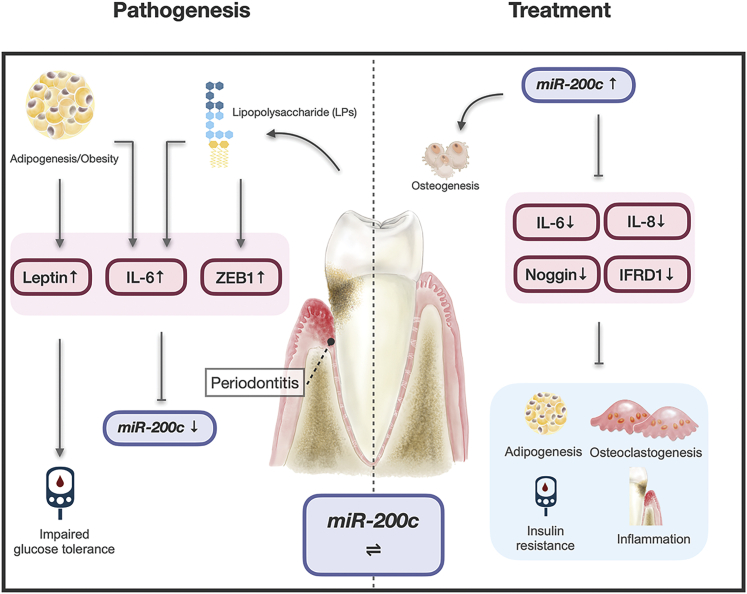
Periodontitis in obese mice downregulates miR-200c, increases systemic inflammation, and impairs glucose tolerance. miR-200c overexpression in the gingiva attenuates the periodontitis and the systemic inflammation and glucose intolerance. The interactions of miR-200c with adipogenesis, inflammation, and osteoclastogenesis indicate its roles in the pathogenesis and treatment of periodontitis in obese subjects.
Introduction
Periodontitis, a set of inflammatory diseases affecting the tissues surrounding the teeth, is linked to or considered a high-risk factor for rheumatoid arthritis, cognitive impairment, cardiovascular diseases, cancer, obesity, and diabetes.1, 2, 3, 4, 5, 6 The sustained chronic inflammatory state of obesity strongly intersects with periodontitis in the context of both pathogenesis and prognosis.7, 8, 9, 10 Adults with obesity nearly double the prevalence rate of periodontitis compared to non-obese subjects, and periodontitis in obese subjects (PiOSs) results in more severe alveolar bone (AB) loss.8,11 Obesity has been demonstrated to negatively impact periodontitis in various pathogenic aspects. Specifically, obese individuals have multiple upregulated proinflammatory molecules and processes implicated in periodontitis, including cytokines, chemokines, and T cell function.12,13 The proinflammatory environment and altered periodontal microbial composition in predisposed obese individuals increases gingival inflammatory responses and periodontal tissue destruction. Increased risk of insulin resistance in obesity also significantly promotes advanced glycation end products at the gingiva, which contributes to more significant periodontal bone loss.14 Obesity directly diminishes the effectiveness of periodontal therapy and results in inferior outcomes of non-surgical treatment,15,16 whereas dietary weight loss intervention effectively promotes a reduction in systemic inflammation and improves the outcome of periodontal treatment. Conversely, periodontitis also contributes to elevated systemic inflammatory responses in obesity. Intensive periodontal therapy has been demonstrated to effectively reduce c-reactive protein levels, interleukin-6 (IL-6), and low-density lipoprotein cholesterol in obese patients.17 Although the imbalance and dysregulation of proinflammatory cytokines, transcription factors, and bone metabolism mediators govern the pathogenic progression of the periodontitis, the underlying molecular mechanisms of a higher prevalence of periodontitis and periodontal tissue destruction in PiOSs still remain unknown. Importantly, surgical treatment is currently required for moderate to advanced periodontitis; unfortunately, the success rate is merely 50%.18 Therefore, an understanding of the pathogenic mechanism(s) underlying complications between obesity and periodontitis and the development of an efficient and safe therapy against advanced periodontitis in PiOSs are demanded.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs that promote the degradation and/or repress the translation of mRNA through sequence-specific interactions with specific mRNA targets. miRNAs have been demonstrated to actively participate in the progression and management of the inflammatory response, including those involved in the onset and development of obesity and periodontitis. Specifically, miRNAs are significantly differentially expressed between a healthy state and periodontitis.19,20 miRNAs actively regulate adipogenesis and play essential roles in obesity and obesity-associated metabolic diseases.21 A recent study discovered that miRNAs were differentially expressed in individuals with periodontitis alone, obesity alone, and in PiOSs when compared to healthy controls.22 Also, miRNAs have emerged as critical transcriptional regulators that target inflammation-related mediators, including tumor necrosis factor alpha (TNF-α), IL-1β, IL-6, and IL-8.22, 23, 24, 25 Thus, manipulating specific miRNAs that participate in the molecular pathogenesis of PiOSs may potentially be developed as a novel and efficient therapeutic tool for PiOSs.
miR-200c is a member of the miR-200 family that plays an essential role in tumor suppression by inhibiting the epithelial-mesenchymal transition (EMT). miR-200c has strong suppressive effects on cell transformation, cancer cell proliferation, migration, invasion, tumor growth, and metastasis.26, 27, 28 While the influence of obesity and PiOSs on miR-200c expression in the human gingiva remains unknown, miR-200c is significantly reduced in the gingival tissues of periodontitis patients.29 Porphyromonas gingivalis lipopolysaccharide (Pg-LPS) was reported to suppress miR-200c in human primary macrophages.30 Pg also downregulated miR-200c by upregulating Zeb1 in gingival epithelial cells.31 Additionally, miR-200c was downregulated in the circulation of obese patients with insulin resistance.32 miR-200c was reported to have a strong association with whole-body insulin sensitivity but was inversely associated with insulin resistance and basal glucose. In animal studies, Chartoumpekis et al.33 reported that diet-induced obesity (DIO) in mice significantly downregulated miR-200c in adipose tissue. Additionally, miR-200c has demonstrated strong anti-inflammatory capabilities. Specifically, miR-200c attenuated LPS-induced early pulmonary fibrosis and inhibited IL-33 in bronchial asthma.34,35 miR-200c modulated cancer inflammation by reducing nuclear factor κB (NF-κB) activation through Toll-like receptor 4 (TLR-4) and myeloid differentiation primary response 88 (MyD88)-dependent pathways.36 miR-200c reduced IL-8 expression by targeting the inhibitor of NF-κB kinase subunit beta (IKBKB) in the NF-κB signal pathway in breast cancer.37 Our previous studies have demonstrated that miR-200c directly targets 3′ UTRs of IL-6, IL-8, interferon-related developmental regulator 1 (Ifrd1), and chemokine (C-C motif) ligand 5 (CCL-5), and downregulates these proinflammatory and osteoclastogenic mediators in human periodontal ligament, gingival fibroblasts, and the periodontium of periodontitis rats.38 We have also demonstrated the capabilities of miR-200c on upregulating Wnt activity and inhibiting noggin (a novel inducer in adipogenesis) for osteogenic differentiation and bone regeneration.39,40
In the present study, we characterized miR-200c and its regulators in adipogenic differentiation, obesity, and PiOSs using human cells and mouse models and attempted to explore miR-200c as a therapeutic tool for PiOSs. We revealed the downregulation of miR-200c in DIO mice and adipogenic differentiated human bone marrow mesenchymal stromal cells (hBMSCs) in vitro. Injection of Pg-LPS into the gingiva of DIO mice effectively mimicked the clinical characteristics of PiOSs, including periodontal inflammation associated with systemic elevation of IL-6 and impaired glucose tolerance. Local injection of naked plasmid DNA encoding miR-200c in gingival tissues effectively rescued miR-200c expression, protected against periodontal and systemic inflammation, and alleviated the impaired glucose metabolism in PiOS mice. Injection of miR-200c effectively increased circulating exosomal miR-200c, probably explaining the mechanism(s) of gingival application of miR-200c in attenuating systemic and white adipose tissue (WAT) inflammation in PiOSs. The monoclonal antibody (mAb) of IL-6 can counteract the function of Pg-LPS on miR-200c and Zeb1. These data strongly indicate that the reduction of miR-200c induced by Pg-LPS and IL-6 in PiOSs might lead to an imbalance of proinflammatory cytokines and exacerbate inflammatory responses, and upregulating miR-200c expression in gingiva would serve as a potential therapeutic approach for PiOSs.
Results
miR-200c is highly expressed in oral gingiva
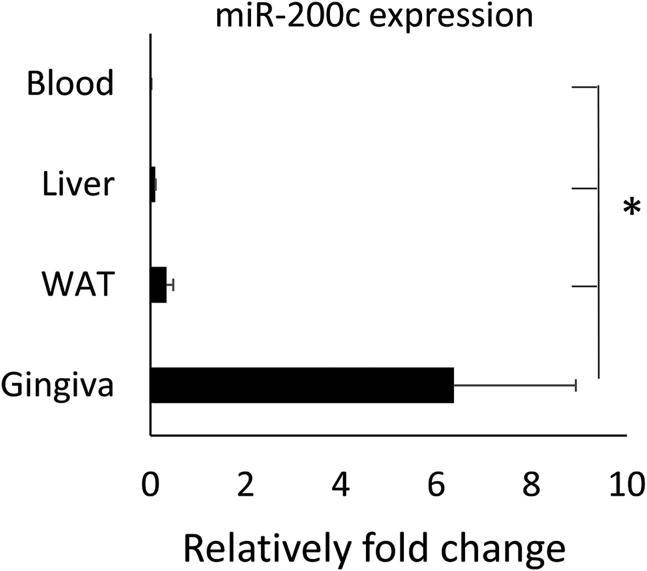
Figure 1 summarizes the tissue-specific distribution of miR-200c in mice. We observed that miR-200c was significantly higher expressed in oral gingival tissues than in the liver, WAT, and blood serum. The expression of miR-200c in WAT was higher than in the liver and blood serum. No difference between blood and liver was observed.
Figure 1.
Relative fold changes of miR-200c expression in different tissues and organs of 22-week-old C57BL/6J mice
∗p < 0.05, n = 3.
Obesity downregulated miR-200c and increased proinflammatory cytokines and insulin resistance in mice
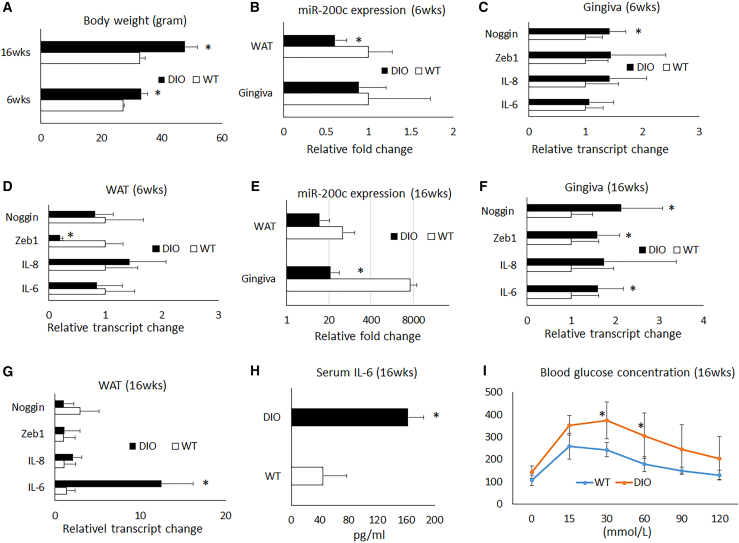
For mice, after 6 and 16 weeks, a high-fat diet (HFD) resulted in a significant increase in body weight over a regular diet (RD; Figure 2A). After 6 weeks on an HFD, a significant reduction of miR-200c was observed in WAT, while no statistically significant downregulation of miR-200c was observed in gingival tissue due to variability. Additionally, 6 weeks of an HFD did not statistically increase IL-6 and IL-8, while noggin was upregulated significantly in gingival tissues. Interestingly, Zeb1 was downregulated in WAT. However, after 16 weeks on an HFD, we found that miR-200c was significantly downregulated in the gingiva of DIO mice (Figure 2E), and transcripts of IL-6 increased in both gingiva and WAT (Figures 2F and 2G), while IL-8 was hardly changed. Both noggin and Zeb1 were significantly increased in gingival tissues. Additionally, after 16 weeks on an HFD, the protein level of IL-6 measured by ELISA and glucose concentration by glucose tolerance test (GTT) analysis in blood serum was significantly increased (Figures 2H and 2I).
Figure 2.
Characteristics of DIO mice fed with an HFD after 6 and 16 weeks
(A) Body weights of mice fed with an HFD for 6 and 16 weeks. (B) Relative fold changes of miR-200c in gingiva and WAT in mice fed with an HFD for 6 weeks. (C and D) Relative transcript level changes of noggin, Zeb1, IL-6, and IL-8 in gingival tissues and WAT in mice fed with an HFD for 6 weeks. (E) Relative fold changes of miR-200c in gingiva and WAT in mice fed with an HFD for 16 weeks. (F and G) Relative transcript change of noggin, Zeb1, IL-6, and IL-8 in gingival tissues and WAT in mice fed with an HFD for 16 weeks. (H) Concentrations of IL-6 in blood serum in mice fed with an HFD for 16 weeks. (I) Blood glucose concentrations of a GTT test in mice fed with an HFD for 16 weeks. ∗p < 0.05 versus wild-type (WT), n = 3–9.
miR-200c participates in adipogenic differentiation of hBMSCs
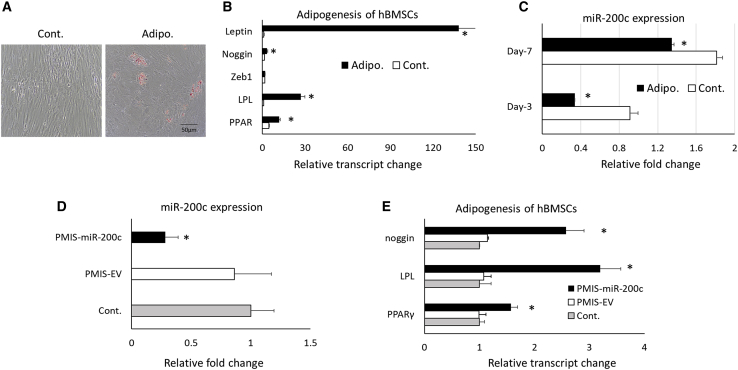
Figure 3 summarizes the mutual influence between miR-200c and adipogenic differentiation in hBMSCs. After differentiation, lipids accumulated in adipogenic differentiated hBMSCs under Oil Red O staining (Figure 3A). The transcripts of adipogenic markers, including peroxisome proliferator-activated receptor γ (PPAR-γ) and lipoprotein lipase (LPL) in the hBMSCs, were significantly increased for cells cultured in the differentiation medium than those cultured in the control medium (Figures 3B and 3C). The transcripts of noggin and leptin increased in adipogenic differentiated cells. Interestingly, the adipogenic differentiation significantly downregulated miR-200c in hBMSCs. In addition, after a plasmid-based miRNA inhibitor system (PMIS) effectively downregulated endogenous miR-200c expression, the adipogenic markers, including PPAR-γ, LPL, and noggin, were significantly increased in hBMSCs (Figures 3D and 3E).
Figure 3.
Characteristics of adipogenesis of hBMSCs on miR-200c expression
(A) Microphotographs of hBMSCs with Oil Red O staining 3 weeks after adipogenic differentiation and control. (B) Relative transcript changes of noggin, Zeb1, LPL, PPAR-γ, and leptin of hBMSCs 1 week after adipogenic differentiation compared to controls. (C) Relative fold change of miR-200c hBMSCs 3 and 7 days after differentiation and controls. (D) Normalized fold changes of miR-200c expression in hBMSCs 3 days after treatment with PMIS-miR-200c. (E) Relative transcript changes of PPAR-γ, LPL, and noggin in adipogenic differentiated hBMSCs pretreated with PMIS-miR-200c. ∗p < 0.05, performed in triplicate.
LPS-induced periodontal inflammation in DIO mice mimicked the pathophysiological variabilities of PiOSs
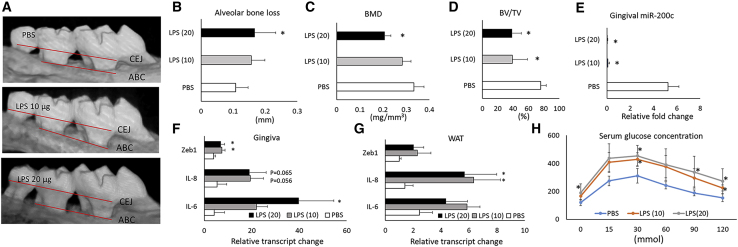
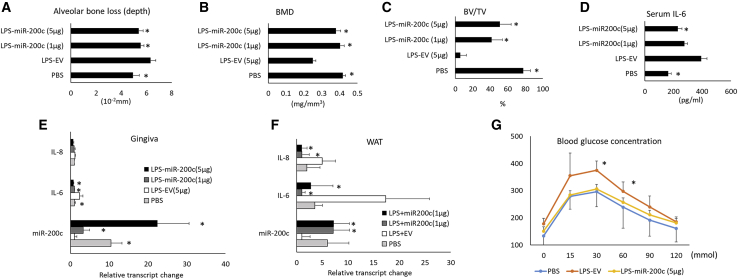
Figure 4 summarizes pathophysiological variabilities after injection of Pg-LPS into the gingival sulcus between maxillary M1/M2 of obese mice after 16 weeks of being fed an HFD. After 2 weeks, the injection of Pg-LPS induced apparent AB resorption compared to PBS as shown in micro-computed tomography (μCT) images (Figure 4A). Quantitatively, the distances between the Cementoenamel Junction (CEJ) to the AB crest (ABC) measured by μCT were significantly increased in mice with Pg-LPS injection over controls (Figure 4B). Pg-LPS injection also significantly reduced the parameter of AB microarchitecture in the maxilla, including the bone volume/tissue volume (BV/TV) and bone mineral density (BMD; Figures 4C and 4D). Pg-LPS also downregulated miR-200c in the gingiva (Figure 4E). Injection of Pg-LPS significantly increased transcripts of IL-6 and Zeb1 in gingiva and IL-8 in WAT (Figures 4F and 4G). Although both IL-8 in gingiva and IL-6 and Zeb1 in WAT were increased, no statistical difference was observed due to limited sample size. In addition, DIO mice injected with LPS have significantly increased glucose intolerance and insulin resistance than do control mice receiving PBS injection as evidenced by an increased glucose concentration through GTT analysis (Figure 4H).
Figure 4.
Pathophysiological characteristics of Pg-LPS injection into the gingival sulcus between maxillary M1/M2 of obese mice after 16 weeks of being fed an HFD
(A) μCT scan of maxillary bones on the palatal side of obese mice 2 weeks after receiving Pg-LPS at different concentrations. (B–D) Quantitative measurement of AB loss, BMD, and BV/TV at maxillary M1/M2 of mice. (E) Relative fold change of miR-200c in gingival tissues of mice with different treatments. (F and G) Relative transcript changes of IL-6, IL-8, and Zeb1 in gingiva and WAT of mice after different treatments. (H) Serum glucose concentration measured by a GTT test in mice with different treatment. ∗p < 0.05 versus PBS injection; n = 3–6.
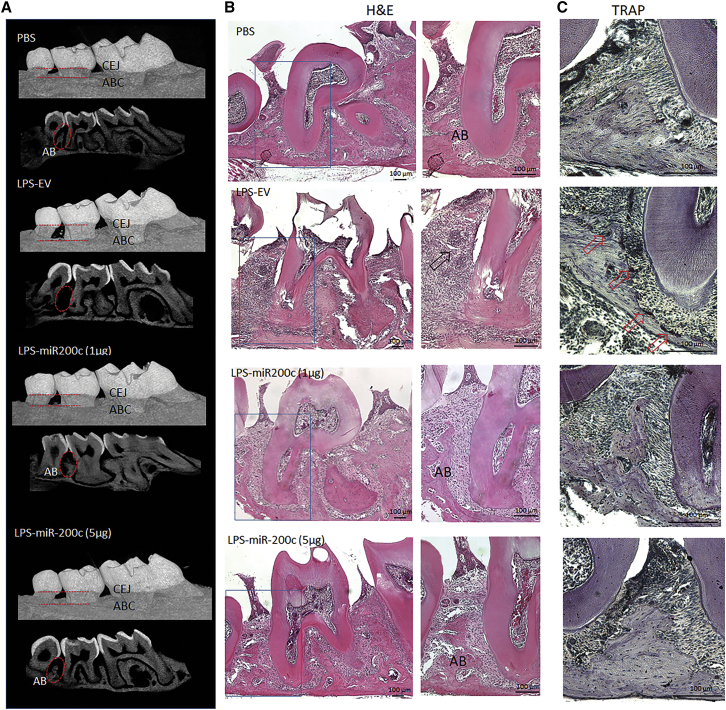
Local injection of miR-200c at gingival tissues attenuated local and systemic inflammation in a mouse model of PiOSs
We injected naked plasmid DNA encoding miR-200c into gingival tissues of obese mice with periodontitis to test the protective function of miR-200c. Pg-LPS at 20 μg was injected into the interdental region between M2/M3 of DIO mice to create a mouse model of PiOSs. PBS was injected as a control. Plasmid DNA encoding miR-200c at 1 and 5 μg was applied to treat periodontitis, and plasmid encoding empty vector (EV) at 5 μg/μL was used as a treatment control. Mice were euthanized after 2 and 4 weeks, and periodontal and systemic inflammation and glucose metabolism were analyzed. Compared to the injection with PBS, in μCT images, injection of Pg-LPS co-treated with EV increased the distances between the CEJ to the ABC and induced AB resorption between M2 and M3 (Figure 5A). The histological examination after hematoylin and eosin (H&E) and tartrate-resistant acid phosphatase (TRAP) stains confirmed that the majority of AB between M2/M3 was resorbed after injection of Pg-LPS. Inflammatory granulation tissue and activated osteoclasts were observed in mice injected with Pg-LPS co-treated with EV (Figures 5B and 5C). However, in PiOS mice treated with miR-200c, limited bone resorption and inflammation induced by Pg-LPS were observed, and ABC was comparable to mice injected with PBS (Figures 5A–5C). Quantitatively, whereas Pg-LPS injection treated with EV resulted in significantly increased distance between the CEJ to the ABC, the treatment of miR-200c effectively restored the bone loss (Figure 6A). miR-200c also significantly restored BMD and BV/TV in the AB of maxilla after challenge with Pg-LPS (Figures 6B and 6C). Compared to the EV treatment, miR-200c injection also effectively rescued miR-200c downregulated by Pg-LPS after 2 weeks (Figure 6E). Treatment of miR-200c also significantly suppressed the IL-6 transcript in the gingiva (Figure 6E). Notably, the injection of miR-200c at 5 μg significantly reduced the protein level of IL-6 in blood serum (Figure 6D) after 4 weeks. The local application of plasmid DNA encoding miR-200c also significantly increased the transcripts of miR-200c and reduced transcripts of IL-6 and IL-8 in WAT after 4 weeks (Figure 6F). In the GTT analysis, miR-200c treatment in gingival tissues reduced glucose concentration in PiOSs significantly.
Figure 5.
Representative μCT scan and microphotographs of histological cross-sections of obese mice 4 weeks after receiving Pg-LPS with miR-200c treatment and controls
(A) μCT images on the palatal side and cross-section of maxillary bones. (B) H&E-stained histological cross-section of mice after different treatments. AB, alveolar bone; arrow, granulation tissues; scale bar, 100 μm. (C) TRAP-stained histological cross-section of mice after different treatments. Arrow, activated osteoclasts; scale bar, 100 μm.
Figure 6.
Quantitative measurements of the effectiveness of miR-200c on attenuating AB loss and systemic inflammation in obese mice with periodontitis
(A–C) Quantitative μCT measurement of AB loss (A), BMD (B), and BV/TV (C) at maxillary M2/M3 of obese mice with Pg-LPS injection 4 weeks after treatment with miR-200c at different concentrations and controls. (D) Serum concentration of IL-6 in mice 4 weeks after different treatments. (E and F) Relative transcript changes of miR-200c, IL-6, and IL-8 in gingiva and WAT of mice with different treatments. (G) Serum glucose concentrations from a GTT test in mice 4 weeks after receiving different treatments. ∗p < 0.05 versus LPS-EV, n = 4–6.
Exosomal miR-200c probably mediated the anti-inflammatory function in treating PiOS mice
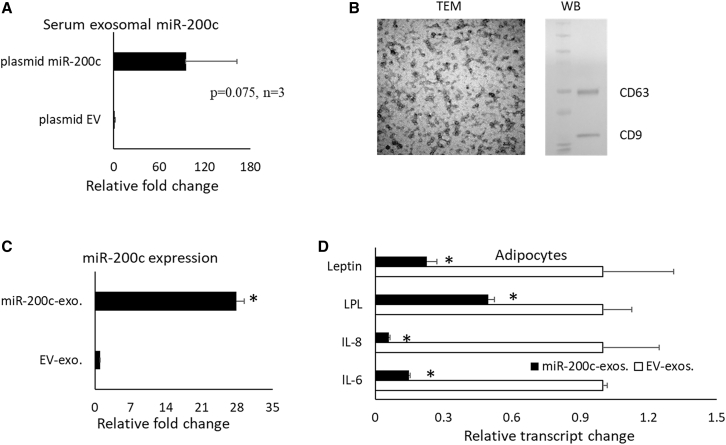
After plasmid miR-200c was injected into the gingival tissues of DIO mice, an increase of miR-200c expression in circulating exosomes was observed, compared to the injection of plasmid EV (Figure 7A, p = 0.075, n = 3; Figure 7A). In addition, we collected miR-200c-enriched exosomes from the supernatant of human embryonic palatal mesenchymal cells (HEPMs) with miR-200c overexpression. Transmission electron microscopy (TEM) and western blot were used to confirm the exosomes (Figure 7B). In order to determine the function of exosomal miR-200c on adipose tissue, human adipose derived stromal cells (hADSCs) were treated with the exosomes collected from the HEPM cells with miR-200c overexpression and the exosomes from the cells with EVs. The exosomes released from HEPM with miR-200c overexpression significantly increased miR-200c (∼27-fold; Figure 7C) and reduced the transcripts of IL-6 and IL-8 in hADSCs. Exosomal miR-200c also significantly reduced LPL (Figure 7D).
Figure 7.
Exosomal miR-200c on inflammation and adipogenesis
(A) Relative fold change of miR-200c in serum exosomes isolated from mice receiving plasmid miR-200c injection in gingival tissues. p = 0.075, n = 3. (B) TEM image of exosomes isolated from HEPM cells with an overexpression of miR-200c and western blot using anti-CD63 and CD-9. (C) Relative fold changes of miR-200c in human ADSCs after treatment with exosomes isolated from HEPM cells with miR-200c overexpression and controls. (D) Relative transcript changes of IL-6, IL-8, and LPL in human ADSCs after treatment with exosomes isolated from HEPM cells with miR-200c overexpression and controls. ∗p < 0.05, performed in triplicate.
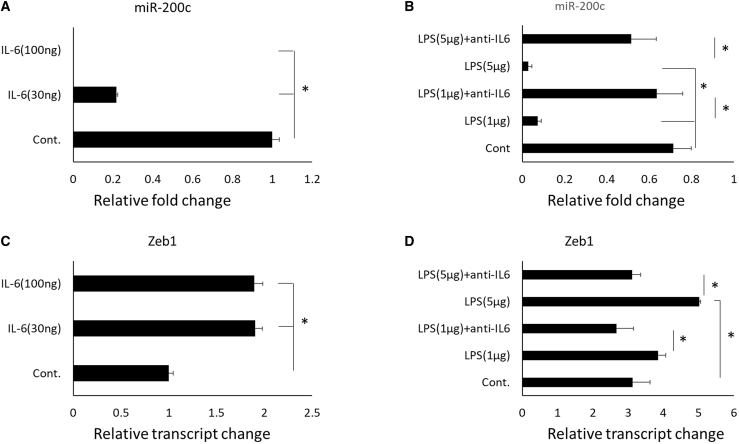
Pg-LPS and IL-6 reduced miR-200c and increased Zeb1
Figure 8 summarizes the potential functions of Pg-LPS and IL-6 on miR-200c and Zeb1. IL-6 and Pg-LPS significantly downregulated miR-200c in a dose-dependent manner in HEPM. The anti-IL-6 mAb effectively rescued the reduction induced by Pg-LPS (Figures 8A and 8B). In addition, both IL-6 and Pg-LPS upregulated Zeb1, while mAb against IL-6 effectively counteracted the function of LPS on Zeb1 (Figures 8C and 8D).
Figure 8.
Effectiveness of IL-6 and LPS on miR-200c and Zeb1
(A and B) Relative fold changes of miR-200c expression in HEPM cells treated with IL-6 and LPS at different concentrations. (C and D) Relative transcript changes of Zeb1 in HEPM cells treated with IL-6 and Pg-LPS at different concentrations. ∗p < 0.05, performed in triplicate.
Discussion
While different miRNAs and miRNA families have been reported to be predominantly expressed in certain tissues, the tissue-specific distribution of miR-200c remains unknown.41 In this study, we compared miR-200c in different tissues of mice and found a significantly higher expression of miR-200c in gingival tissues than in blood, liver, and fat tissues. This finding suggested that miR-200c may have essential roles in the physiology and pathophysiology of oral gingival tissues. Because of the extensive regulation of miR-200c in inflammation and obesity,32,33 this finding indicated that miR-200c may participate in the pathogenesis of periodontitis in obese patients.
Although many cytokines and miRs aggravate inflammation associated with periodontitis in obese subjects, the present study suggests that, for the first time, downregulation of miR-200c might play an important role in the molecular pathogenesis of PiOSs. While the variation of miR-200c in PiOSs in humans remains unknown, a significant downregulation of miR-200c was reported in gingival tissues of periodontitis patients.29 Additionally, the finding that miR-200c is downregulated in blood samples of obese patients with insulin resistance also suggests that miR-200c may be downregulated in PiOS patients.32 In this study, we have not only confirmed the previously published finding that miR-200c was downregulated in adipose tissue of obese mice,33 but also found that obesity resulting from an HFD significantly reduced miR-200c in the gingival tissues. The DIO mice experienced increased IL-6 and impaired glucose tolerance, which are consistent with pathophysiological characteristics found in obese patients.42,43 Administration of Pg-LPS in local gingival tissue not only caused AB loss and inflamed granulation tissues but also exaggerated the systemic inflammation evidenced by the upregulation of IL-6 and IL-8 and impaired glucose tolerance. These pathophysiological variations in obese mice with LPS injection were similar to variations for PiOS patients. miR-200c was significantly downregulated in gingival and adipose tissue in these mice. Thus, this strongly indicates that miR-200c most likely is downregulated in patient gingival tissues with PiOSs; however, human studies would be needed to confirm this. Additionally, because Pg-LPS represents the causal pathogen for chronic patient periodontitis, the similar pathophysiologies of PiOSs in this animal model allow us to further investigate its molecular pathogenesis and develop a treatment approach for PiOSs.
Our previous studies have reported the anti-inflammatory capabilities of miR-200c on periodontitis in vitro and in vivo. In this study, we have further confirmed that overexpression of miR-200c in gingival tissues, elicited by injecting naked plasmid DNA encoding miR-200c, could efficiently prevent periodontal inflammation and AB loss induced by LPS in obese mice. The local application of miR-200c also effectively protects the microarchitecture of AB. Moreover, we have demonstrated that the local application of miR-200c in gingival tissues could result in increased miR-200c in WAT. The latter potentially contributes to the downregulation of proinflammation cytokines, including transcripts of IL-6 and IL-8 in adipose tissues and IL-6 protein levels in blood. This indicates that local application of miR-200c in gingival tissue may also potentially improve the inflammatory status of obesity in PiOSs. In addition, we found that, for the first time, the local application of miR-200c effectively improved glucose tolerance and insulin resistance in obese mice with periodontitis. These results indicate a key potential benefit of local application of therapeutic miR-200c to alleviate both periodontitis and systemic glucose metabolism disorder associated with PiOSs. Although upregulated miR-200c found in type 2 diabetes was considered to increase apoptosis and impair pancreatic islets,44 miR-200c was reported to be downregulated in blood samples from obese patients with insulin resistance, and the level of miR-200c is highly associated with whole body insulin sensitivity but inversely associated with insulin resistance and basal glucose. Opposite miR-200c expression levels may be caused by different pathophysiological variations in diabetes and obesity. In this study, we also observed that downregulation of miR-200c is associated with impaired glucose tolerance in DIO mice. The reduction of miR-200c and impaired glucose tolerance was even worse for DIO mice with LPS-induced periodontitis, which is similar to obese patients with periodontitis. Overexpression of miR-200c by injection of plasmid DNA into gingival tissues effectively improved glucose tolerance in PiOS mice. Although the underlying mechanism(s) is not clear, modulation of inflammation induced by overexpression of miR-200c may play a major role. We speculate that attenuated systemic inflammation with reduced circulating IL-6 levels and IL-6 and IL-8 transcripts in adipose tissues may contribute to this effectiveness. IL-6 has been demonstrated to decisively induce the development of insulin resistance and pathogenesis of type 2 diabetes mellitus through the generation of inflammation by controlling differentiation, migration, proliferation, and cell apoptosis.45,46 Previous work has shown that IL-6 causes insulin resistance by impairing the phosphorylation of insulin receptor and insulin receptor substrate-1 by inducing the expression of SOCS-3, a potential inhibitor of insulin signaling.47 Thus, downregulated IL-6 in circulation and remote adipose tissue induced by overexpression of miR-200c in gingival tissues may play important roles in improving insulin resistance in PiOS mice.
In addition to the inhibitory function of obesity on miR-200c, we have also revealed the potential interactive regulation of miR-200c with adipogenesis in this study. First, we observed that the adipogenic differentiation of hBMSCs significantly downregulated miR-200c, which is consistent with downregulation of miR-200c in mice fed an HFD. We also noticed that downregulation of miR-200c could effectively promote adipogenic differentiation of hBMSCs, while overexpression of miR-200c could downregulate the adiopogenesis of hADSCs. There are several mechanism(s) probably involved in the regulation of miR-200c. miR-200c has been demonstrated previously to increase osteogenic differentiation.39,48 Due to the inverse relationship between osteogenic and adipogenic programming, this may potentially explain the inhibitory role of miR-200c in adipogenesis. In addition, the reciprocal action between miR-200c and leptin, an adipokine activating proadipogenic factor,49, 50, 51 may also explain aspects of regulation. Adipogenesis induced upregulation of leptin may potentially inhibit miR-200c while overexpression of miR-200c may act negatively on adipogenesis by suppressing noggin.52,53
In this study, we observed that circulating exosomal miR-200c in serum was upregulated after local injection of miR-200c in gingival tissues. Our in vitro test also demonstrated the anti-inflammatory function of exosomal miR-200c using human adipose cells. Exosomes are known to be key mediators in cell-cell communication and facilitate the transfer of genetic and biochemical information between distant cells. Thus, we propose that circulating exosomes may transfer the genetic signals between gingival and adipose tissues and upregulation of circulating exosomal miR-200c may play a role in systemic anti-inflammation in PiOSs. However, future studies to confirm the origin of the circulating miR-200c-enriched exosomes and track their destinations in PiOSs will provide more information for determining the mechanism(s) mediating the interaction of gingival and adipose tissue in PiOSs.
Although IL-6 was reported to mediate miR-200c suppression in NF-κB signaling in a cancer study,54 we reveal for the first time that IL-6 may directly inhibit miR-200c expression. That is, the downregulation of miR-200c in both mice and patients who are obese and have periodontitis is due to upregulated IL-6 levels. This finding provides further support for the role that miR-200c downregulation may play in PiOSs. In addition, previous studies have reported that Pg-LPS downregulates miR-200c in macrophages and gingival tissues potentially via regulation of Zeb1.30,31 In this study, we not only confirmed the function of Pg-LPS on inhibiting miR-200c and increasing Zeb1 in vitro and in vivo but also observed the effectiveness of monoclonal antibodies in counteracting IL-6 in vitro. This strongly indicates that downregulation of miR-200c and upregulation of Zeb1 induced by Pg-LPS may be mediated via the upregulation of IL-6. The upregulation of Zeb1 further contributes to the downregulation of miR-200c in vitro and in vivo. However, the mechanism(s) underlying the inhibitory function of IL-6 on the biogenesis of miR-200c is not clear. In summary, the present studies have demonstrated that downregulation of miR-200c by upregulation of IL-6, Zeb1, and leptin in periodontitis and obesity may contribute to the molecular mechanism(s) of the pathogenesis of PiOSs by further exaggerating IL-6 levels and adipogenesis. Overexpression of miR-200c in gingival tissue can effectively attenuate both local and systemic inflammation and improve glucose metabolism in obese subjects by targeting IL-6, IL-8, noggin, and leptin (Figure 9). While the inhibitory function of IL-6 on miR-200c and circulating exosomal miR-200c may function in pathogenesis and treatment of PiOSs, respectively, future studies to confirm the function and clarify the underlying mechanism(s) are needed.
Figure 9.
Potential roles of miR-200c in pathogenesis and treatment of PiOSs
Materials and methods
Characterizing miR-200c and its potential regulators in DIO mice
All in vivo animal experiments were performed with approval from the Office of Animal Resources at the University of Iowa. The surgical protocols were followed in accordance with the policies and guidelines provided by the Institutional Animal Care and Use Committee. We first measured the tissue-specific distribution of miR-200c. Gingiva, liver, subcutaneous inguinal and epididymal WAT, and blood serum were collected from 22-week-old male C57BL/6J mice (Jackson), and the transcripts of miR-200c were measured using qRT-PCR. In order to determine the influence of obesity on miR-200c and its regulators, we collected the gingiva and WAT from 12- and 22-week-old male C57BL/6J mice, respectively. The mice were fed with HFD (35.5% fat, 20% protein, and 32.7% carbohydrates) for 6 and 16 weeks. After each mouse was weighed and euthanized, the transcripts of miR-200c, IL-6, IL-8, Zeb1, and noggin were quantified using qRT-PCR. The protein level of IL-6 in blood serum was measured using ELISA according to the manufacturer’s protocol (BioLegend, San Diego, CA, USA). The GTT was performed to determine the glucose metabolism influenced by obesity. Mice of the same age on a RD were used as controls.
Evaluating miR-200c and its potential regulators in a mouse model of PiOSs
Periodontitis in obese mice was induced by Pg-LPS injection in male 22-week-old C57BL/6J mice fed with an HFD for 16 weeks. A total of 1 μL Pg-LPS (Biological Laboratories, Campbell, CA, USA) at 10 or 20 μg/μL was directly injected twice a week into the interdental region between maxillary molars using a Hamilton1700 series syringe. The same volume of PBS was injected into the same sites in controls. The mice were euthanized after 2 weeks, and the GTT test was performed before euthanization. The transcripts of miR-200c, IL-6, and IL-8 in the gingiva, and WAT were measured using qRT-PCR, and the protein level of IL-6 in blood was measured using ELISA. The AB resorption induced by Pg-LPS injection was analyzed using μCT.
Exploring the therapeutic effectiveness of miR-200c on PiOSs
To investigate the preventive and therapeutic potential of miR-200c for PiOSs, we injected Pg-LPS in gingival tissues of DIO mice to create a mouse model of PiOSs and co-treated with plasmid DNA encoding miR-200c at different doses or EV as a control. Plasmid DNA encoding miR-200c and EVs were prepared using methods described previously.38,39 The mice were divided into four groups, each receiving a different treatment, including (1) PBS injection alone; (2) 20 μg of Pg-LPS with EV at 5 μg; (3) 20 μg of Pg-LPS injection with 1 μg of miR-200c; and (4) 20 μg of Pg-LPs injection with 5 μg of miR-200c. We injected 1 μL of the Pg-LPS with miR-200c or controls into the interdental region between M2/M3 twice a week. The mice were euthanized after 2 and 4 weeks, and the transcripts of miR-200c, IL-6, and IL-8 from the gingiva, WAT, and protein levels of IL-6 in blood were measured using qRT-PCR and ELISA. GTT was performed to determine the influence of miR-200c on glucose metabolism in PiOSs after 4 weeks. The preventive function of miR-200c on AB resorption was measured using μCT and histology.
Analyzing the influence of adipogenic differentiation on miR-200c in vitro
hBMSCs were purchased and cultured in complete mesenchymal stem cell growth medium (Lonza, Switzerland) at 37°C with 5% CO2. At passage 4, the cells were seeded on a 12-well plate with a density of 105 cells/per well and cultured with adipogenic differentiation medium consisting of DMEM culture medium supplemented with 175 nM dexamethasone and 50 μM indomethacin for up to 21 days. Cells treated with DMEM culture medium containing 10% FBS and 1% antibiotics served as controls. The transcripts of miR-200c and adipogenic biomarkers, including PPAR-γ and LPL, were quantified using qRT-PCR after different time points. Oil Red O staining (Science) was performed to confirm the lipid accumulation after 21 days, as described previously.55 In order to confirm the potential function of miR-200c on adipogenic differentiation, the adipogenic biomarkers in hBMSCs were measured after endogenous miR-200c was inhibited by a PMIS56 (NaturemiRI, Iowa City, IA, USA).
Collecting and analyzing exosomal miR-200c in DIO mice and HEPM cells with miR-200c overexpression
A total of 1 μL of plasmid encoding DNA miR-200c at 5 μg/μL and PBS as a control was injected into gingival tissues at the labial vestibule of 22-week-old DIO mice twice a week. After 2 weeks, whole blood from the mice was collected from the venous sinus using a capillary tube and incubated at room temperature for 20 min. The serum was separated by centrifugation at 3,000 rpm for 15 min at 4°C and immediately aliquoted. Exosomes were subsequently extracted from serum using serum total exosome isolation kit (Invitrogen). The expression of miR-200c in serum and exosomes from the circulation of mice were measured using qRT-PCR. To determine the function of exosomal miR-200c, we collected miR-200c-rich exosomes from human embryonic palatal mesenchyme (HEPM) cells with overexpression of miR-200c. The HEPM cells with overexpression of miR-200c were prepared as previously described.23 The miR-200c-enriched exosomes were collected from the supernatant of HEPM transduced with lentiviral miR-200c using exoEasy Maxi Kit (QIAGEN) according to the manufacturer’s protocol. The exosome’s characteristics, including spherical shapes with a size of 30–100 nm and CD63 and CD9, were confirmed using TEM and western blot. In order to determine the function of exosomal miR-200c, the miR-200c enriched exosomes were added to hADSCs (Lonza). ADSCs at passage 4 were seeded on a 12-well plate at 105 cells per/well and cultured in DMEM medium. A total of 20 μL exosomes at 3.7 μg/μL were added to hADSCs in an exosome-free medium for 18 h. The exosomes from the HEPM cells transduced with lentiviral EVs were used as controls. The transcripts of miR-200c and its targets, including IL-6, IL-8, and adipogenic marker in hADSCs, LPL, were quantified after 3 days using qRT-PCR.
Analyzing inhibitory roles of IL-6 on miR-200c
HEPM cells were cultured in DMEM medium and supplemented with IL-6 at 30 and 100 ng/mL and LPS at 1 μg/mL. The range of IL-6 concentrations was based on rates among patients with chronic periodontitis.57 An anti-IL-6 mAb was used to counteract the function of Pg-LPS. The transcripts of miR-200c and Zeb1 were analyzed after 24 h using qRT-PCR.
qRT-PCR
Total RNA from mouse tissue and cultured cells was collected using miReasy mini kit (QIAGEN). The concentration and purity of the total RNA were quantified using a NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Scientific). The measurement of miR-200c expression was performed using the mirScript ll reverse transcription kit and miRScript SYBR Green PCR Kit (QIAGEN) and normalized to U6 by a comparative ΔΔCt method. mRNA expression was measured by qRT-PCR using PrimeScript Reagent Kit (Takara) to carry out reverse transcription and amplified reaction by using amplification primers with SYBR Green PCR Master Mix (PE Applied Biosystems). The comparative ΔΔCt method was used to quantify the relative level of different mRNA expression. All samples were normalized to GAPDH. Table 1 lists the primers for qRT-PCR.
Table 1.
Sequence for forward and reverse primer sets used for real-time PCR
| Gene | Forward primer (5′-3′) | Reverse primer (3′-5′) |
|---|---|---|
| PPAR-γ (H) | ACCAAAGTGCAATCAAAGTGGA | ATGAGGGAGTTGGAAGGCTCT |
| Noggin (H) | CCATGCCGAGCGAGATCAAA | TCGGAAATGATGGGGTACTGG |
| C/EBP-α (H) | TATAGGCTGGGCTTCCCCTT | AGCTTTCTGGTGTGACTCGG |
| ZEB1 (H) | GATGATGAATGCGAGTCAGATGC | ACAGCAGTGTCTTGTTGTTGT |
| IL-6 (H) | CCATCTTTGGAAGGTTCAGGTTG | ACTCACCTCTTCAGAACGAATTG |
| IL-8 (H) | AACCCTCTGCACCCAGTTTTC | ACTGAGGATTGAGAGTGGAC |
| LPL (H) | TGTGGTGGACTGGCTGTCA | CTGTCCCACCAGTTTGGTGTAG |
| GAPDH (H) | TGTGGGCATCAATGGATTTGG | ACACCATGTATTCCGGGTCAAT |
| IL-6 (M) | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| IL-8 (M) | CAAGGCTGGTCCATGCTCC | TGCTATCACTTCCTTTCTGTTGC |
| Zeb1 (M) | CCATACGAATGCCCGAACT | ACAACGGCTTGCACCACA |
| Noggin (M) | GCGTCTCGTTCAGATCCTTCTC | GCCAGCACTATCTACACA |
H, human; M, mouse
GTT measurement
For the GTT test, mice were food deprived for up to 16 h and subsequently administered up to 1 g/kg of glucose by intraperitoneal injection. Blood glucose was measured at t = 0, 15, 30, 60, 90, and 120 min, from 1–3 μL of blood obtained from a superficial nick made in the tail vein using a Breeze 2 glucose meter (Bayer).
μCT analysis
μCT analysis using SkyScan 1272 was performed to evaluate the AB resorption of mice with different treatments. Scanning was performed with a rotational angle of 360° around the longitudinal axis of the second molar tooth, utilizing a spatial resolution of 17 μm at 70 kV, 142 μA, and a 0.5° rotation step with an exposure time of 500 ms. Periodontal bone heights were measured as the distances from the CEJ to the ABC in the interdental region between M2 and M3. Volumetric measurements, including BV, TV, BV/TV, and BMD, were evaluated within the region of interest (ROI). The ROI of volumetric analysis of the interdental region was set between maxillary M2 and M3. The ROIs were drawn (size 0.19 × 0.414 = Ellipse) between the interproximal area of M2-M3 without overlap with the tooth. For image reconstruction, 2D virtual sections of maxillary molar teeth were acquired in the coronal, axial, and sagittal planes by the Skyscan CT-analyzer (CTAn) software. The CTAn software was further employed for 3D analysis and quantification of the volume of interfacial gaps and voids within the ROI. The Skyscan CT-Volume software was used for 3D visualization of periodontal bone tissues.
Histological examination
Maxillary tissue blocks in each group were fixated with 4% paraformaldehyde for 24 h at 4°C and decalcified with 10% ethylenediaminetetraacetate (EDTA) for 3 weeks. Subsequently, the samples were dehydrated in a gradient of ethanol before immersion 2 times in xylene. All samples were embedded in paraffin before sectioning. Sagittal sections of 8 μm thickness were cut perpendicular to the tooth axis. Sections were stained with H&E and TRAP staining following standard protocols. Corresponding images of the H&E- and TRAP-stained tissues were taken using a microscope to examine the AB loss.
Statistical Analysis
Descriptive statistics were conducted for both in vitro and in vivo investigations. For the in vivo study, a Student’s t test was utilized to evaluate differences between DIO mice and controls at different time points. A one-way ANOVA with post hoc Tukey’s HSD test was utilized to evaluate whether there was a significant difference in each measurement among mice with different treatments. The Shapiro-Wilks’ test was also applied to verify the assumption of normality. All statistical tests completed for the in vivo quantification used a significance level of 0.05, and statistical analyses were performed using the statistical package SAS System version 9.4 (SAS Institute, Cary, NC, USA). For the in vitro study, a Student’s t test was utilized to evaluate differences between the cells with adipogenic differentiation and controls. A one-way ANOVA with post hoc Tukey’s HSD test was utilized to evaluate whether there was a significant difference in each measurement among cells with different treatments. The Shapiro-Wilks’ test was applied to verify the assumption of normality. All statistical tests for the in vitro study utilized a significance level of 0.05, and statistical analyses were performed using commercially available SPSS 25 Statistical Software (IBM).
Acknowledgments
The authors thank Dr. J. Michael Tilley and Professor Jeff Banas for proofreading and editing the manuscript. This study was supported by the National Institute of Dental and Craniofacial Research (grant nos. R21 DE024799 and R01 DE026433) of the National Institutes of Health (NIH).
Author contributions
Investigation, T.K., M.Z., Z.Z., S.E., Y.S., A.A., D.S.; writing – original draft preparation, T.K.; supervision, T.K., B.A.A., L.Y., L.H.; Q.Q.; formal analysis, F.Q.; conceptualization, B.A.A.; methodology, L.H.; writing – review & editing, L.H.; funding acquisition, L.H.; visualization, L.H.
Declaration of interests
L.H. and B.A.A. have a US patent for miR-200c-based bone regeneration. B.A.A. is the founder of the NaturemiRI Company.
References
- 1.Chapple I.L., Bouchard P., Cagetti M.G., Campus G., Carra M.C., Cocco F., Nibali L., Hujoel P., Laine M.L., Lingstrom P. Interaction of lifestyle, behaviour or systemic diseases with dental caries and periodontal diseases: consensus report of group 2 of the joint EFP/ORCA workshop on the boundaries between caries and periodontal diseases. J. Clin. Periodontol. 2017;44(Suppl 18):S39–S51. doi: 10.1111/jcpe.12685. [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki M., Kimura Y., Ogawa H., Yamaga T., Ansai T., Wada T., Sakamoto R., Ishimoto Y., Fujisawa M., Okumiya K. Periodontitis, periodontal inflammation, and mild cognitive impairment: A 5-year cohort study. J. Periodontal Res. 2019;54:233–240. doi: 10.1111/jre.12623. [DOI] [PubMed] [Google Scholar]
- 3.Akram Z., Abduljabbar T., Abu Hassan M.I., Javed F., Vohra F. Cytokine Profile in Chronic Periodontitis Patients with and without Obesity: A Systematic Review and Meta-Analysis. Dis. Markers. 2016;2016:4801418. doi: 10.1155/2016/4801418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Rawi N.H., Shahid A.M. Oxidative stress, antioxidants, and lipid profile in the serum and saliva of individuals with coronary heart disease: is there a link with periodontal health? Minerva Stomatol. 2017;66:212–225. doi: 10.23736/S0026-4970.17.04062-6. [DOI] [PubMed] [Google Scholar]
- 5.Benguigui C., Bongard V., Ruidavets J.B., Sixou M., Chamontin B., Ferrières J., Amar J. Evaluation of oral health related to body mass index. Oral Dis. 2012;18:748–755. doi: 10.1111/j.1601-0825.2012.01940.x. [DOI] [PubMed] [Google Scholar]
- 6.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pires J.R., Dos Santos I.P., de Camargo L.F., Zuza E.P., de Toledo B.E., Monteiro S.C. Framingham cardiovascular risk in patients with obesity and periodontitis. J. Indian Soc. Periodontol. 2014;18:14–18. doi: 10.4103/0972-124X.128193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Aiuto F., Sabbah W., Netuveli G., Donos N., Hingorani A.D., Deanfield J., Tsakos G. Association of the metabolic syndrome with severe periodontitis in a large U.S. population-based survey. J. Clin. Endocrinol. Metab. 2008;93:3989–3994. doi: 10.1210/jc.2007-2522. [DOI] [PubMed] [Google Scholar]
- 9.Pischon N., Heng N., Bernimoulin J.P., Kleber B.M., Willich S.N., Pischon T. Obesity, inflammation, and periodontal disease. J. Dent. Res. 2007;86:400–409. doi: 10.1177/154405910708600503. [DOI] [PubMed] [Google Scholar]
- 10.Chaffee B.W., Weston S.J. Association between chronic periodontal disease and obesity: a systematic review and meta-analysis. J. Periodontol. 2010;81:1708–1724. doi: 10.1902/jop.2010.100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathur L.K., Manohar B., Shankarapillai R., Pandya D. Obesity and periodontitis: A clinical study. J. Indian Soc. Periodontol. 2011;15:240–244. doi: 10.4103/0972-124X.85667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falagas M.E., Kompoti M. Obesity and infection. Lancet Infect. Dis. 2006;6:438–446. doi: 10.1016/S1473-3099(06)70523-0. [DOI] [PubMed] [Google Scholar]
- 13.Boesing F., Patiño J.S., da Silva V.R., Moreira E.A. The interface between obesity and periodontitis with emphasis on oxidative stress and inflammatory response. Obes. Rev. 2009;10:290–297. doi: 10.1111/j.1467-789X.2008.00555.x. [DOI] [PubMed] [Google Scholar]
- 14.Genco R.J., Grossi S.G., Ho A., Nishimura F., Murayama Y. A proposed model linking inflammation to obesity, diabetes, and periodontal infections. J. Periodontol. 2005;76(11, Suppl):2075–2084. doi: 10.1902/jop.2005.76.11-S.2075. [DOI] [PubMed] [Google Scholar]
- 15.Akram Z., Safii S.H., Vaithilingam R.D., Baharuddin N.A., Javed F., Vohra F. Efficacy of non-surgical periodontal therapy in the management of chronic periodontitis among obese and non-obese patients: a systematic review and meta-analysis. Clin. Oral Investig. 2016;20:903–914. doi: 10.1007/s00784-016-1793-4. [DOI] [PubMed] [Google Scholar]
- 16.Gerber F.A., Sahrmann P., Schmidlin O.A., Heumann C., Beer J.H., Schmidlin P.R. Influence of obesity on the outcome of non-surgical periodontal therapy - a systematic review. BMC Oral Health. 2016;16:90. doi: 10.1186/s12903-016-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava M.C., Srivastava R., Verma P.K., Gautam A. Metabolic syndrome and periodontal disease: An overview for physicians. J. Family Med. Prim. Care. 2019;8:3492–3495. doi: 10.4103/jfmpc.jfmpc_866_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa F.O., Cota L.O., Cortelli J.R., Cortelli S.C., Cyrino R.M., Lages E.J., Oliveira A.P. Surgical and Non-Surgical Procedures Associated with Recurrence of Periodontitis in Periodontal Maintenance Therapy: 5-Year Prospective Study. PLoS ONE. 2015;10:e0140847. doi: 10.1371/journal.pone.0140847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Y.F., Shu R., Jiang S.Y., Liu D.L., Zhang X.L. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int. J. Oral Sci. 2011;3:125–134. doi: 10.4248/IJOS11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito A., Horie M., Ejiri K., Aoki A., Katagiri S., Maekawa S., Suzuki S., Kong S., Yamauchi T., Yamaguchi Y. MicroRNA profiling in gingival crevicular fluid of periodontitis-a pilot study. FEBS Open Bio. 2017;7:981–994. doi: 10.1002/2211-5463.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGregor R.A., Choi M.S. microRNAs in the regulation of adipogenesis and obesity. Curr. Mol. Med. 2011;11:304–316. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nahid M.A., Rivera M., Lucas A., Chan E.K., Kesavalu L. Polymicrobial infection with periodontal pathogens specifically enhances microRNA miR-146a in ApoE-/- mice during experimental periodontal disease. Infect. Immun. 2011;79:1597–1605. doi: 10.1128/IAI.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong L., Sharp T., Khorsand B., Fischer C., Eliason S., Salem A., Akkouch A., Brogden K., Amendt B.A. MicroRNA-200c Represses IL-6, IL-8, and CCL-5 Expression and Enhances Osteogenic Differentiation. PLoS ONE. 2016;11:e0160915. doi: 10.1371/journal.pone.0160915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du A., Zhao S., Wan L., Liu T., Peng Z., Zhou Z., Liao Z., Fang H. MicroRNA expression profile of human periodontal ligament cells under the influence of Porphyromonas gingivalis LPS. J. Cell. Mol. Med. 2016;20:1329–1338. doi: 10.1111/jcmm.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue J., Wang P., Hong Q., Liao Q., Yan L., Xu W., Chen X., Zheng Q., Zhang L., Huang D. MicroRNA-335-5p Plays Dual Roles in Periapical Lesions by Complex Regulation Pathways. J. Endod. 2017;43:1323–1328. doi: 10.1016/j.joen.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Hill L., Browne G., Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int. J. Cancer. 2013;132:745–754. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 27.Humphries B., Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472–6498. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S., Nag A., Mandal C.C. A Comprehensive Review on miR-200c, A Promising Cancer Biomarker with Therapeutic Potential. Curr. Drug Targets. 2015;16:1381–1403. doi: 10.2174/1389450116666150325231419. [DOI] [PubMed] [Google Scholar]
- 29.Stoecklin-Wasmer C., Guarnieri P., Celenti R., Demmer R.T., Kebschull M., Papapanou P.N. MicroRNAs and their target genes in gingival tissues. J. Dent. Res. 2012;91:934–940. doi: 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naqvi A.R., Zhong S., Dang H., Fordham J.B., Nares S., Khan A. Expression Profiling of LPS Responsive miRNA in Primary Human Macrophages. J. Microb. Biochem. Technol. 2016;8:136–143. doi: 10.4172/1948-5948.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sztukowska M.N., Ojo A., Ahmed S., Carenbauer A.L., Wang Q., Shumway B., Jenkinson H.F., Wang H., Darling D.S., Lamont R.J. Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cell. Microbiol. 2016;18:844–858. doi: 10.1111/cmi.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masotti A., Baldassarre A., Fabrizi M., Olivero G., Loreti M.C., Giammaria P., Veronelli P., Graziani M.P., Manco M. Oral glucose tolerance test unravels circulating miRNAs associated with insulin resistance in obese preschoolers. Pediatr. Obes. 2017;12:229–238. doi: 10.1111/ijpo.12133. [DOI] [PubMed] [Google Scholar]
- 33.Chartoumpekis D.V., Zaravinos A., Ziros P.G., Iskrenova R.P., Psyrogiannis A.I., Kyriazopoulou V.E., Habeos I.G. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE. 2012;7:e34872. doi: 10.1371/journal.pone.0034872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang X., Wu F., Fan J., Jin Y., Wang J., Yang G. Posttranscriptional Regulation of Interleukin-33 Expression by MicroRNA-200 in Bronchial Asthma. Mol. Ther. 2018;26:1808–1817. doi: 10.1016/j.ymthe.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y., Liu Y., Ping F., Yi L., Zeng Z., Li Y. miR-200b/c attenuates lipopolysaccharide-induced early pulmonary fibrosis by targeting ZEB1/2 via p38 MAPK and TGF-β/smad3 signaling pathways. Lab. Invest. 2018;98:339–359. doi: 10.1038/labinvest.2017.123. [DOI] [PubMed] [Google Scholar]
- 36.Wendlandt E.B., Graff J.W., Gioannini T.L., McCaffrey A.P., Wilson M.E. The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-κB activation. Innate Immun. 2012;18:846–855. doi: 10.1177/1753425912443903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chuang T.D., Khorram O. miR-200c regulates IL8 expression by targeting IKBKB: a potential mediator of inflammation in leiomyoma pathogenesis. PLoS ONE. 2014;9:e95370. doi: 10.1371/journal.pone.0095370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akkouch A., Zhu M., Romero-Bustillos M., Eliason S., Qian F., Salem A.K., Amendt B.A., Hong L. MicroRNA-200c Attenuates Periodontitis by Modulating Proinflammatory and Osteoclastogenic Mediators. Stem Cells Dev. 2019;28:1026–1036. doi: 10.1089/scd.2019.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akkouch A., Eliason S., Sweat M.E., Romero-Bustillos M., Zhu M., Qian F., Amendt B.A., Hong L. Enhancement of MicroRNA-200c on Osteogenic Differentiation and Bone Regeneration by Targeting Sox2-Mediated Wnt Signaling and Klf4. Hum. Gene Ther. 2019;30:1405–1418. doi: 10.1089/hum.2019.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao H., Jheon A., Li X., Sun Z., Wang J., Florez S., Zhang Z., McManus M.T., Klein O.D., Amendt B.A. The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development. 2013;140:3348–3359. doi: 10.1242/dev.089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludwig N., Leidinger P., Becker K., Backes C., Fehlmann T., Pallasch C., Rheinheimer S., Meder B., Stähler C., Meese E., Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi Y.H., McKeown R.E., Mayer-Davis E.J., Liese A.D., Song K.B., Merchant A.T. Serum C-reactive protein and immunoglobulin G antibodies to periodontal pathogens may be effect modifiers of periodontitis and hyperglycemia. J. Periodontol. 2014;85:1172–1181. doi: 10.1902/jop.2014.130658. [DOI] [PubMed] [Google Scholar]
- 43.Kumar N., Bhardwaj A., Negi P.C., Jhingta P.K., Sharma D., Bhardwaj V.K. Association of chronic periodontitis with metabolic syndrome: A cross-sectional study. J. Indian Soc. Periodontol. 2016;20:324–329. doi: 10.4103/0972-124X.183096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belgardt B.F., Ahmed K., Spranger M., Latreille M., Denzler R., Kondratiuk N., von Meyenn F., Villena F.N., Herrmanns K., Bosco D. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 2015;21:619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 45.Rehman K., Akash M.S.H., Liaqat A., Kamal S., Qadir M.I., Rasul A. Role of Interleukin-6 in Development of Insulin Resistance and Type 2 Diabetes Mellitus. Crit. Rev. Eukaryot. Gene Expr. 2017;27:229–236. doi: 10.1615/CritRevEukaryotGeneExpr.2017019712. [DOI] [PubMed] [Google Scholar]
- 46.Park S.E., Park C.Y., Sweeney G. Biomarkers of insulin sensitivity and insulin resistance: Past, present and future. Crit. Rev. Clin. Lab. Sci. 2015;52:180–190. doi: 10.3109/10408363.2015.1023429. [DOI] [PubMed] [Google Scholar]
- 47.Kim J.H., Bachmann R.A., Chen J. Interleukin-6 and insulin resistance. Vitam. Horm. 2009;80:613–633. doi: 10.1016/S0083-6729(08)00621-3. [DOI] [PubMed] [Google Scholar]
- 48.Xia P., Gu R., Zhang W., Shao L., Li F., Wu C., Sun Y. MicroRNA-200c promotes osteogenic differentiation of human bone mesenchymal stem cells through activating the AKT/β-Catenin signaling pathway via downregulating Myd88. J. Cell. Physiol. 2019;234:22675–22686. doi: 10.1002/jcp.28834. [DOI] [PubMed] [Google Scholar]
- 49.Howe E.N., Cochrane D.R., Cittelly D.M., Richer J.K. miR-200c targets a NF-κB up-regulated TrkB/NTF3 autocrine signaling loop to enhance anoikis sensitivity in triple negative breast cancer. PLoS ONE. 2012;7:e49987. doi: 10.1371/journal.pone.0049987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howe E.N., Cochrane D.R., Richer J.K. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011;13:R45. doi: 10.1186/bcr2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang C.C., Wu M.J., Yang J.Y., Camarillo I.G., Chang C.J. Leptin-STAT3-G9a Signaling Promotes Obesity-Mediated Breast Cancer Progression. Cancer Res. 2015;75:2375–2386. doi: 10.1158/0008-5472.CAN-14-3076. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 52.Blázquez-Medela A.M., Jumabay M., Rajbhandari P., Sallam T., Guo Y., Yao J., Vergnes L., Reue K., Zhang L., Yao Y. Noggin depletion in adipocytes promotes obesity in mice. Mol. Metab. 2019;25:50–63. doi: 10.1016/j.molmet.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawant A., Chanda D., Isayeva T., Tsuladze G., Garvey W.T., Ponnazhagan S. Noggin is novel inducer of mesenchymal stem cell adipogenesis: implications for bone health and obesity. J. Biol. Chem. 2012;287:12241–12249. doi: 10.1074/jbc.M111.293613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rokavec M., Wu W., Luo J.L. IL6-mediated suppression of miR-200c directs constitutive activation of inflammatory signaling circuit driving transformation and tumorigenesis. Mol. Cell. 2012;45:777–789. doi: 10.1016/j.molcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong L., Peptan I.A., Colpan A., Daw J.L. Adipose tissue engineering by human adipose-derived stromal cells. Cells Tissues Organs. 2006;183:133–140. doi: 10.1159/000095987. [DOI] [PubMed] [Google Scholar]
- 56.Cao H., Yu W., Li X., Wang J., Gao S., Holton N.E., Eliason S., Sharp T., Amendt B.A. A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Ther. 2016;23:634. doi: 10.1038/gt.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Batool H., Nadeem A., Kashif M., Shahzad F., Tahir R., Afzal N. Salivary Levels of IL-6 and IL-17 Could Be an Indicator of Disease Severity in Patients with Calculus Associated Chronic Periodontitis. BioMed Res. Int. 2018;2018:8531961. doi: 10.1155/2018/8531961. [DOI] [PMC free article] [PubMed] [Google Scholar]