Abstract
Thyroid cancers (TC) have increasingly been detected following advances in diagnostic methods. Risk stratification guided by refined information becomes a crucial step toward the goal of personalized medicine. The diagnosis of TC mainly relies on imaging analysis, but visual examination may not reveal much information and not enable comprehensive analysis. Artificial intelligence (AI) is a technology used to extract and quantify key image information by simulating complex human functions. This latent, precise information contributes to stratify TC on the distinct risk and drives tailored management to transit from the surface (population-based) to a point (individual-based). In this review, we started with several challenges regarding personalized care in TC, for example, inconsistent rating ability of ultrasound physicians, uncertainty in cytopathological diagnosis, difficulty in discriminating follicular neoplasms, and inaccurate prognostication. We then analyzed and summarized the advances of AI to extract and analyze morphological, textural, and molecular features to reveal the ground truth of TC. Consequently, their combination with AI technology will make individual medical strategies possible.
Keywords: artificial intelligence, thyroid cancer, biomarker, personalized medicine, histopathology, fine-needle aspiration biopsy, ultrasound
Introduction
Thyroid cancers (TC) have emerged in popularity over the past decades, with indolent TC accounting for the majority (1–3). For advanced TC (1, 2) and aggressive papillary thyroid carcinomas (PTC) (4), the incidence and mortality rates are also steadily increasing, which makes it imperative to adopt more effective strategies for managing such changes. In the era of personalized medicine, precise and efficient risk stratification is important before, during, and after treatment, to choose and adjust its type and intensity. The foremost step is to discover key information that reveals the biological behavior of TC. There are abundant anatomical structures (texture, internal architecture, and spatial distribution) and molecular components (gene variation, protein expression, etc.) within TC. So far, TC’s diagnosis mainly relies on image analysis (e.g., ultrasound images, cell smears, and tissue sections), but information obtained only by our naked eyes hardly enables a comprehensive analysis of the tumors (5). Given patients and their disease features, primary human cell cultures both from surgical biopsies and from fine-needle aspiration (FNA) samples foster the targeted therapies (6). However, many tough challenges still hinder a clear break of personalized treatment such as inconsistent rating ability of ultrasound (US) physicians (7), uncertainty in cytopathological diagnosis (8), difficulty in discriminating follicular neoplasms (9, 10), and inaccurate prognostication.
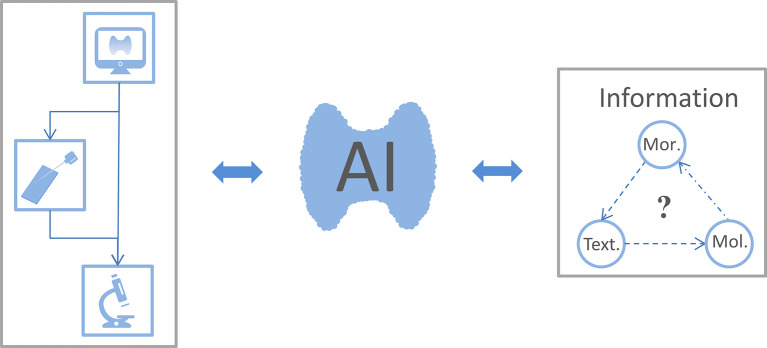
Artificial intelligence (AI) is a series of technologies combined to mimic human interaction ( Figure 1 ). In some tasks, it matches or exceeds human perception (11, 12). AI deals with various sorts of omics information in parallel, easily identifying and modeling a complicated nonlinear relationship in the image (13, 14). Several studies have demonstrated that AI classifier is comparable to radiologists while qualitatively analyzing thyroid nodules (TN) (15–18). Furthermore, AI can extract and quantify key image information, whereby image diagnosis converts from a subjective qualitative task to objective quantitative analysis. This more detailed and precise information is conducive to special risk stratification and propels tailored management to transit from the surface (population-based) to a point (individual-based).
Figure 1.
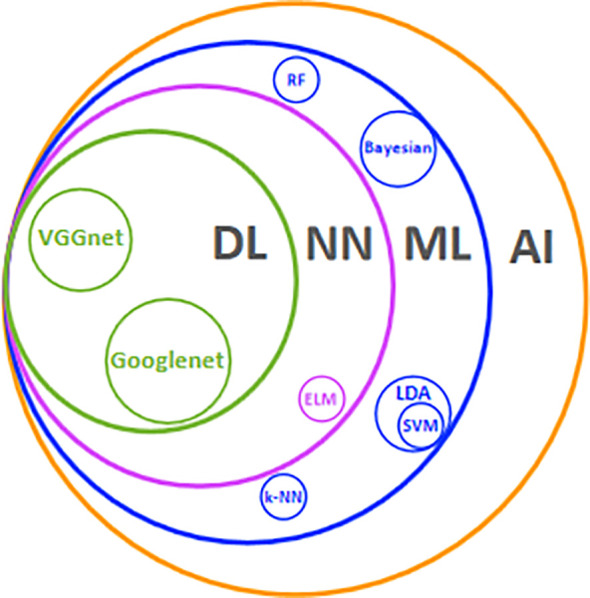
Main AI technologies and their relationships. AI. Artificial intelligence; ML, machine learning; NN, neural network; DL, deep learning; LDA, linear discriminant analysis; ELM, extreme learning machine; RF, random forest; SVM, support vector machine; k-NN, k-nearest neighbor.
In this review, we aimed to summarize the use of AI for extracting and analyzing morphological, textural, and molecular features to reveal detailed information and personalize therapies for TC patients ( Figure 2 ).
Figure 2.
The connection between the focus reviewed. Thyroid ultrasound is the preferred imaging examination for patients with thyroid nodules. When sonographers consider certain thyroid nodules as malignant, the patient could choose fine-needle aspiration biopsy or surgery for a confirmed diagnosis. Artificial intelligence (AI) uses ultrasound images, cell smears, and tissue slices to extract morphological, textural, and molecular features. This information is fed back into the AI classifier to improve its performance and thus optimize thyroid cancer diagnosis and treatment workflow. As expected, whether these morphological (Mor.), textural (Text.), and molecular (Mol.) features are related to each other warrants further study.
Applications of AI in the US Diagnosis of TN
TN with several typical ultrasound features implies an increased risk of malignancy, such as solid composition, hypoechogenicity, irregular margin, microcalcification, and taller-than-wide shape. However, these properties can neither confirm nor exclude the diagnosis of TC (19). The observer’s agreement among multiple centers is poorly satisfactory in assessing these features (7). Thyroid Imaging Reporting and Data Systems (TI-RADS) are enormously valuable to PTC as risk stratification systems, while relatively less to FTC, MTC, and other malignancies (20). Interestingly, the AI model appears to be a promising tool to facilitate a better knowledge of TN via quantitative analysis of typical US features and introduction of texture features.
Performance of Typical US Features
Wildman-Tobriner et al. (21) developed an AI TI-RADS based on the American College of Radiology (ARC) TI-RADS. This system optimized the evaluation task through reassigned values for eight ultrasound features, highlighting the status of hypoechogenicity or marked hypoechogenicity. The novel AI TI-RADS had better accuracy than ARC TI-RADS when performed by inexperienced radiologists (55% vs. 48%) and experts (65% vs. 47%). Similar to other studies, ARC TI-RADS-based classifiers had higher sensitivity and slightly lower specificity (21–24). Wu et al. (25) evaluated quantitative echoic indexes for detecting malignant TN, which showed higher accuracy than typical ultrasound hypoechogenicity (>60% vs. 54.01%). We summarized the outcomes of the ultrasound features employed by AI for classification in Table 1 and found the most widely used features were shape, margin, echogenicity, calcification, composition, and size. In other words, these discriminative features seem to be the focus for the AI model to learn (31, 37). Particularly, Choi et al. (30) demonstrated several new calcification features associated with TN malignancy, including shorter calcification distance ratio, smaller amounts of calcification, and dimmer calcification. Chen et al. (28) quantified TN malignant risk through the calcification index. These new features boosted diagnostic accuracy by combining qualitative and quantitative methods (30, 38). Current AI classifiers focus on benign and malignant TN dichotomy, and certain of them like the S-Detect series have already become commercially available (32, 34). Furthermore, they are expected to predict more tumor-biological behaviors such as lymph node metastasis (39, 40) and pathological subtypes (41).
Table 1.
Summary of key studies on the outcome of ultrasound features in artificial intelligence classifier identifying benign and malignant thyroid nodules.
| Study | Patients | Features | Classifiers | Accuracy, % | Sensitivity, % | Specificity, % | AUC |
|---|---|---|---|---|---|---|---|
| Lim et al. (26) | 96 | Size, margin, cystic change, echogenicity, and macrocalcification | ANN | 93.78 | NA | NA | 0.949 |
| Savelonas et al. (27) | 387 | Boundary features | SVM | NA | NA | NA | 0.95 |
| Chen et al. (28) | 256 | Calcification index | AmCAD-UT | NA | NA | NA | 0.746 |
| Zhu et al. (29) | 618 | Not well-circumscribed, solid, hypoechogenicity, microcalcification, taller than wide, absent peripheral halo | ANN | 83.10 | 83.80 | 81.80 | 0.828 |
| Choi et al. (30) | 85 | Quantitative calcification | NN | 82.80 | 83.00 | 82.40 | 0.83 |
| Wu et al. (25) | 333 | Quantitative echogenetic values | AmCAD-UT | 70.32 | 33.12 | 93.31 | NA |
| Xia et al. (31) | 187 | Margin, shape, composition, echogenicity, and calcification | ELM | 87.72 | 78.89 | 94.55 | 0.867 |
| Choi et al. (32) | 89 | Size, margins, shape, composition, echogenicity, orientation, and spongiform | S-Detect 1 (SVM) | 81.40 | 90.70 | 74.60 | 0.83 |
| Ouyang et al. (33) | 1036 | Size, margins, shape, composition, echogenicity, calcification, aspect ratio, capsule, hypoechoic halo, vascularity, and cervical lymph node status | RF + k-SVM | NA | NA | NA | 0.954 |
| Kim et al. (34) | 106 | Size, margins, shape, composition, echogenicity, calcification, orientation, and spongiform | S-Detect 2 (CNN) | 73.40 | 81.40 | 68.20 | NA |
| Liu et al. (35) | 4655 | Shape, context, and margin | CNN | 94.90 | 97.20 | 89.10 | NA |
| Wildman-Tobrineret et al. (21) | 1264 | ACR TIRADS | Genetic Algorithm | 65.00 | 93.30 | 64.70 | 0.93 |
| Guan et al. (36)a | 2235 | Margin size | Inception-v3 | 90.50 | 93.30 | 87.40 | 0.956 |
| Zhao et al. (22) | 822 | Size + ACR TIRADS | ML | 82.10 | 90.90 | 78.10 | 0.917 |
| Jin et al. (23) | 695 | ACR TIRADS | CNN | 80.35 | 80.64 | 80.13 | 0.87 |
| Bai et al. (24) | 13984 b | ACR TIRADS | CNN | 88.00 | 98.10 | 79.10 | NA |
ANN, artificial neural network; SVM, support vector machine; AmCAD-UT, a software integrating AI technology and clinicians’ expertise; NN, neural network; ELM, extreme learning machine; RF, random forest; k-SVM, kernel support vector machine; Inception-v3, a kind of Googlenet; CNN, convolutional neural network; ML, machine learning; NA, not available; ACR TRIADS, American College of Radiology Thyroid Imaging Reporting and Data System, the features included in this system is composition, echogenicity, shape, margin, and echogenic foci.
This study focused on the classification between papillary thyroid carcinomas and benign nodules.
Nodules, not patients.
Performance of Texture Features
A meta-analysis suggested that a taller-than-wide shape displays TN’s variation in space and orientation growth, and it is defined as the most suggestive feature for malignancy (42). Texture features refer to the characterization of spatial distribution and surface orientation with numerical features (43). Thus, texture analysis as a powerful alternative will make it possible for radiologists to comprehend the TN in depth and gain a correct diagnosis. Raghavendra et al. (44) integrated spatial and fractal texture features and screened two features with an excellent area under the curve in diagnostic practice (94.45%). Prochazka et al. (45) used AI to extract texture features from US images independent of the direction of the US probe and achieved better accuracy (94.64%). Yu et al. (46) performed a numerical transformation of two US features, unregulated shape and long/short-axis ratio into the perimeter2/area and the angle between the long axis and the horizontal axis. These new features showed excellent sensitivity and specificity (100% and 87.88%, respectively) combined with 65 texture features. Collectively, AI mode has a role in integrating typical ultrasonic and texture features, and this fusion might sharply reduce the differences in judgments among US professionals. Despite the mounting advantages of the AI model in optimizing and even creating workflows, many remarkable factors hold its ultimate practice back in the real world. The three main factors are as follows: (i) poor availability of large-high-quality datasets to guarantee great robustness (17); (ii) lack of explainability for conclusions from a black-box algorithm to solidify the trust between physicians and patients (47, 48); (iii) financial burden from specific equipment and research costs (48).
Applications of AI in Cytopathological Evaluation From FNA
FNA is a primary preoperative examination to evaluate TN. Its report system, the Bethesda System for Reporting Thyroid Cytopathology (TBSRTC), is a state-of-the-art and category-based method for clinicians’ decision-making. While TBSRTC includes six diagnostic categories on the estimated risk of malignancy (ROM) ( Table 2 ), 15%–30% of TN continues to be classified as indeterminate TN (ITN), most frequently TBSRTC categories III, IV, and V (8). Recent studies showed excellent consistency between machine learning (ML) models and cytologists in malignancy prediction (49–51), in which the ROM of TBSRTC III determined by the ML model was considerably lower than by manual classification (4.2% vs. 18.8%) (51). It’s worth noting that morphological and genetic classifications assisted by the AI model are fairly accurate at distinguishing malignancy from benign TN (52–54) ( Table 3 ).
Table 2.
The 2017 TBSRTC categories and their own risk of malignancy.
| Diagnostic category | Risk of malignancy if NIFTP≠ CA (%) |
|---|---|
|
I. nondiagnostic or unsatisfactory
Cyst fluid only Virtually acellular specimen Other (obscuring blood, clotting artifact, etc.) |
5–10 |
|
II. Benign
Consistent with a benign follicular nodule (includes adenomatoid nodule, colloid nodule, etc.) Consistent with lymphocytic (Hashimoto) thyroiditis in the proper clinical context Consistent with granulomatous (subacute) thyroiditis Other |
0–3 |
| III. atypia of undetermined significance or follicular lesion of undetermined significance | 6–18 |
|
III. follicular neoplasm or suspicious for a follicular neoplasm
Specify if Hürthle cell (oncocytic) type |
10–40 |
|
IV. suspicious for malignancy
Suspicious for papillary carcinoma Suspicious for medullary carcinoma Suspicious for metastatic carcinoma Suspicious for lymphoma Other |
45–60 |
|
V. Malignant
Papillary thyroid carcinoma Poorly differentiated carcinoma Medullary thyroid carcinoma Undifferentiated (anaplastic) carcinoma Squamous-cell carcinoma Carcinoma with mixed features (specify) Metastatic carcinoma Non-Hodgkin lymphoma Other |
94–96 |
This is an integrated table from reference (8).
Table 3.
The main performance of artificial intelligence using pathological information in different task.
| Study | Subject | Test | Feature | Task | Classifier | Accuracy,% |
|---|---|---|---|---|---|---|
| Cochand-Priollet et al. (54) | 157 | FNA | Nuclear size, shape, and texture | Classification of malignant and benign TN | FNN | 89.00 |
| Daskalakis et al. (53) | 115 | FNA | Nuclear morphology and texture | Classification of malignant and benign TN | k-NN + PNN + Bayesian | 95.70 |
| Tomei et al. (52) | 93 | FNA | mRNA expression | Classification of malignant and benign TN | BNN | 88.80 |
| Sanyal et al. (55) | 544 | FNA | Nuclear morphology and papillary structure | Classification of PTC and non-PTC | ANN | 85.06 |
| Guan et al. (56) | 279 | FNA | Nuclear contour | Classification of PTC and benign TN | VGG-16 | 97.66 |
| Savala et al. (57) | 57 | FNA | Cellular and nuclear morphology | Classification of FC and FA | ANN | 100.00 |
| Alexander et al. (58) | 249 a | FNA | RNA expression | Classification of malignant and benign ITN | SVM | 65.00 |
| Patel et al. (59) | 183 a | FNA | RNA sequencing | Classification of malignant and benign ITN | SVM | 74.00 |
| Nikiforova et al. (60) | 175 a b | FNA | Genetic alterations | Classification of malignant and benign ITN | Torrent Suite software | 90.90 |
| Lithwick-Yanai et al. (61) | 150 a b | FNA | MicroRNA expression | Classification of malignant and benign ITN | LDA + k-NN | 83.65 |
| Sun et al. (62) | 64 a | FNA | Protein | Classification of malignant and benign TN | ANN | 87.53 |
| Wang et al. (63) | 10 | Histo. | Nuclear size and chromatin concentration | Classification of FC, FA, and normal thyroid | SVM | 100.00 |
| Ozolek et al. (64) | 94 | Histo. | Nuclear morphology | Classification of five follicular lesions | LDA +k-NN | 100.00 c |
| Zhao et al. (65) | 800 | Histo. | Gene variant pathways | TC risk stratification | ANN | 77.50/86.00 d |
| Ruiz et al. (66) | 495 | Histo. | Gene signature | Prediction of lymph-node metastasis and disease-free survival | LDA | 82.63 |
FNA, fine-needle aspiration biopsy; Histo., Histopathology; TN, thyroid nodules; FNN, feedforward neural network; PNN, probabilistic neural network; BNN, Bayesian neural network; PTC, papillary thyroid carcinoma; FC, follicular carcinoma; FA, follicular adenoma; ANN, artificial neural network; SVM, support vector machine; LDA, linear discriminant analysis; k-NN, k-nearest neighbor; NA, not available.
Only the validation cohort is included, which in the study by Lithwick-Yanai et al. was specifically the set agreed upon by the three pathologists.
FNA smears, not patients.
The accuracy in the group of FA vs. FC, FA vs. NG, FC vs. NG, FA vs. FV-PTC, and FC vs. WIFC.
The accuracy of recognizing the different- risk cases was 77.50% (low-risk) and 86.00% (high-risk) respectively.
Performance of Morphological Features
PTC, the most common TC (>80%), arises from abnormal growth of thyroid epithelial cells (28, 38). In recent years, AI models with quantitative morphological features have tried to improve follicular lesions’ recognition capacity (55–57). Sanyal et al. (55) obtained the nuclear morphology and papillary structure of PTC under two magnifications (×10 and ×40). CNN model selected PTC from colloid goiter, follicular neoplasms, and lymphocytic thyroiditis by right of these features. Guan et al. (56) developed a new AI cytological classification based on nuclear size and staining information (the contours, perimeter, area, and means of pixel intensity), whose results showed high accuracy (97.66%) to differentiate PTC from benign nodules. Another research group also confirmed this performance (57). They first derived nuclear pleomorphism and area information and then reported the weight of 17 cytological and morphological features. Finally, their model successfully discriminated follicular carcinoma (FC) from follicular adenoma (FA) (57) ( Table 3 ).
The major difference between FC and FA is the occurrence of capsular or vascular invasion (67). Preoperative examinations of both US and FNA have difficulty in making a reliable diagnosis. A highly vascularized tumor protrusion on the US strongly indicates FC, which is rather rare yet (68). Seo et al. (69) took full advantage of this difference by collecting information about the tumor edge in the US images. The overall accuracy was 89.51% for distinguishing FC and FA. Yang et al. (70) segmented the whole lesions of follicular neoplasms; as a result, the classification accuracy was significantly improved to 96%. This clarified the importance of internal information and affirmed the study’s reliability by Savala et al. (57). Similarly, the diagnosis of MTC and ATC is histology dependent (71, 72), yet now no studies to our knowledge have answered the hope of AI in their ultrasound and cytopathological diagnosis.
Performance of Biomolecules
For patients with ITN, repeat FNA or lobectomy might be performed because management guidelines are more flexible (8, 73). Fortunately, molecular tests provide a noninvasive and accurate option to reduce clinical and healthy uncertainty (8, 67). Each genome contains as much information as 100,000 photographs (74). Next-generation sequencing (NGS) can perform high-speed analysis of multiple genes parallelly in a single operation, producing billions of molecular fragments (74, 75). It has always been a crucial component of big data due to its large volume of data, the astonishing velocity of the sequencing methods, and the result output’s veracity. Traditional information systems are less competent to analyze large and complex datasets (76, 77). AI as a big data algorithm can integrate multi-omic data in a different learning task, and automatically realize high-level features’ detection or classification (77). Some genetic classifiers have played their strengths in TN such as the Afirma gene expression classifier (GEC) (58), gene sequence classifier (GSC) (59), gene mutation-based classifier (ThyroSeq) (60, 78), and microRNA-based classifier (RosettaGX Reveal) (61, 79). The GEC involved 167 genes that displayed high sensitivity (92%) and positive predictive value (PPV: 93%) but limited by its relatively low specificity (52%) and negative predictive value (NPV: 47%) (58). GSC expanded the gene spectrum to 10,196 genes by RNA-enhanced NGS. Compared to GEC in the same samples, it made progress in screening for benign nodules (sensitivity: 91.1% and specificity: 68.3%) (59) ( Table 3 ). These two classifiers are the most broadly accepted methods to rule out malignant nodules. In general, ThyroSeq and RosettaGX Reveal are more like rule-in entities. Nikiforova et al. (60) achieved a robust sensitivity of 98% and a hopeful specificity of 81.8% by employing the latest version of ThyroSeq (ThyroSeq V3) to recognize a few cancers from most benign tumors. Steward et al. (78) drew similar conclusions in a prospective blinded multicenter study and reported 94% sensitivity and 82% specificity. RosettaGX Reveal showed 98% sensitivity and 78% specificity when validated in independent cases with all three pathologists’ agreement on the histopathological diagnosis (61) ( Table 3 ). However, whether the mentioned classifiers could consolidate and complement each other remains so ambiguous that we need to further investigate the precise application strategy.
The multi-gene analysis is able to enhance diagnostic performance, but it may be limited due to key genes’ deletion or their reduced expression. Of note, the number of thyroglobulins has been considered as a predictor of postoperative disease progression (67). Therefore, the key proteins might provide some added information for personalized therapy. Recent research has confirmed that proteins are more stable than RNA in clinical tissues (80). Sun et al. (62) completed a 14 protein-based ANN classifier for TN classification. This model realized the accuracy of 90.62% and 87.53% in multicenter retrospective and prospective samples respectively ( Table 3 ). Some molecular alterations such as BRAF mutations (81) are diagnostic of cancer, but most of the other alterations (82, 83) show overlap in both benign and malignant lesions. Therefore, assessing the risk of malignancy by molecular testing should depend on knowledge of the prior cytological appearance.
Applications of AI in Histopathological Analysis
Upon reliable evidence obtained by the US and FNA examination, tumor information from the resected specimens is significant for pathologists to diagnosis TC such as tumor size, pathologic types, and degree of malignancy. Molecular patterns in the tumor microenvironment like cytokines, chemokines, and adipocytokines interconnect the units of immune-inflammatory responses (e.g., macrophages, neutrophils, lymphocytes) and tumor nest (e.g., epithelial cancer cells, fibroblasts, endothelial cells) (84). The more detailed information the pathologists provide, the more precise the treatment strategies physicians take. The combination of AI, morphology, and molecular markers is expected to provide more information for TC management at a patient’s level.
Performance of Morphological Features
The morphological feature is the final station of biological behavior and genetic variation of TN. The morphological performance supported by AI might be beneficial for the accurate diagnosis of TN. Wang et al. (63) successfully classified FA, FC, and normal tissues according to nuclear size and chromatin concentration. Ozolek et al. (64) achieved nearly perfect accuracy based on nine nuclear morphological features for discriminating five thyroid follicular lesions: FA, FC, follicular variant of PTC, nodular goiter, and the widely invasive FC ( Table 3 ). However, further validation of these models is required due to tumor complicated heterogeneity, which was also turned out in a recent study for classifying TC, normal tissues, nodular goiter, and adenomas using a deep learning model (85).
Morphologically, FV-PTC is a mixed entity for typical PTC nuclear features and entirely or almost entirely follicular growth patterns. FV-PTC includes two major subtypes: encapsulated (EFV-PTC) and non-encapsulated or infiltrative variants (IFV-PTC) (86). The former generally have RAS mutations like follicular tumors, the latter often presents extrathyroidal extension (ETE), lymphatic metastasis, and BRAF mutations like classical PTC (cPTC) (87). Likewise, EFV-PTC usually appears invasive or non-invasive, and the noninvasive encapsulate tumor was redefined from carcinoma to borderline tumor, noninvasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) (86). Up to a point, the invasive EFV-PTC behaves more aggressively like FC, whereas NIFTP is with indolent clinical behaviors like FA (87). It is believed that invasive EFV-PTC might develop from NIFTP (88). Borrelli et al. (89) revealed a significant difference in miRNA expression of FA, NIFTP, and IFV-PTC. In particular, just two miRNA (miR-10a-5p and miR-320e) enable us to differentiate NIFTP from IFV-PTC. In another study by Selvaggi (90), none of the multinucleated giant cells (MGCs) were observed in 20 NIFTP cases, while the amount of MGCs varied from 1 to 4 in 88% of the FVPTC cases (both IFV-PTC and invasive EFV-PTC). When utilizing computer quantitative analysis to classify FV-PTC, Chain et al. (91) demonstrated the NIFTP nuclear area (mean, 54.8 μm2) and elongation was smaller than PTC (mean, 77.2 μm2); Hsieh et al. (92) addressed PD-L1 expression in NIFTP was lower than in invasive EFV-PTC. These quantitative morphological characteristics and definite molecular alterations contribute to FV-PTC classification.
As FV-PTC’s definition stated, the coexistence of papillary and variable follicular structures is so common in cancer nests that we hold a positive view about more transitional or intermediate categories between the cPTC and FV-PTC. Undoubtedly, the clearer the learning exemplars, the easier it is to learn for the AI model because it receives fewer error messages (13). For greater efficiency, it’s essential to accurately classify the training set and refine the output target.
Performance of Genetic Parameters
The American Thyroid Association risk stratification system and the American Joint Committee on Cancer TNM staging system are used to guide postoperative treatment and predict post-treatment outcomes, which incorporate several parameters including age, ETE, anatomic location, number, and size of metastatic lymph nodes, aggressive variants, vascular invasion, and distant metastasis. Nonetheless, these systems fail to routinely recommend a genetic determination to guide individual management (67, 93). Zhao et al. (65) selected 10 gene variant pathways that involved inflammatory and immune responses to determine the TC patients’ risk level. Based on these pathways, the patients were divided into the high-risk and low-risk groups whose survival time was significantly better than the former. Ruiz et al. (66) demonstrated a 25-gene panel related to molecular pathways, cell structure, and function was an independent prognostic factor for lymphatic metastasis and disease-free survival ( Table 3 ). Further evidence is still warranted to address the value of this genetic information to TN’s triage and biological behaviors. As AI and gene testing technology upgrade, the cooperation of traditional clinic-pathological parameters and gene molecules might yield more precise therapeutic implications.
Conclusion
The future development of personalized medicine in TC still faces several challenges like inconsistent rating ability of US physicians, uncertainty in cytopathological diagnosis, difficulty in discriminating follicular lesions, and inaccurate prognostication. AI’s application has improved the efficiency and accuracy of diagnosis and treatment in other tumors (94–96). A growing amount of medical information can be extracted and analyzed through AI technology. This review has innovatively offered ideas for the ultrasonic and pathological testing out of these dilemmas in terms of morphological, textural, and molecular features. As more key parameters are explored from the tumor and its microenvironment, the AI-aided combination of morphological and molecular features will pave the way for TC’s protocol at the individual level.
Author Contributions
L-RL: study design, literature review, article writing, and revision. BD: literature review. H-QL: literature review. CC: study design, article revision. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the grants from the Fundamental Research Funds for the Central Universities (2042019kf0229) and the Science and Technology Major Project of Hubei Province (Next-Generation AI Technologies) (2019AEA170).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Sincere thanks to the teachers, classmates, and editors who worked together on this research.
References
- 1. Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in thyroid cancer incidence and mortality in the United States, 1974-2013. JAMA (2017) 317(13):1338–48. 10.1001/jama.2017.2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jegerlehner S, Bulliard JL, Aujesky D, Rodondi N, Germann S, Konzelmann I, et al. Overdiagnosis and overtreatment of thyroid cancer: A population-based temporal trend study. PloS One (2017) 12(6):e0179387. 10.1371/journal.pone.0179387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. La Vecchia C, Malvezzi M, Bosetti C, Garavello W, Bertuccio P, Levi F, et al. Thyroid cancer mortality and incidence: a global overview. Int J Cancer (2015) 136(9):2187–95. 10.1002/ijc.29251 [DOI] [PubMed] [Google Scholar]
- 4. Ho AS, Luu M, Barrios L, Chen I, Melany M, Ali N, et al. Incidence and Mortality risk spectrum across aggressive variants of papillary thyroid Carcinoma. JAMA Oncol (2020) 6(5):706–13. 10.1001/jamaoncol.2019.6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koelzer VH, Sirinukunwattana K, Rittscher J, Mertz KD. Precision immunoprofiling by image analysis and artificial intelligence. Virchows Archiv (2019) 474(4):511–22. 10.1007/s00428-018-2485-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fallahi P, Ferrari SM, Elia G, Ragusa F, Patrizio A, Paparo SR, et al. Primary cell cultures for the personalized therapy in aggressive thyroid cancer of follicular origin. Semin Cancer Biol (2020). 10.1016/j.semcancer.2020.06.013 [DOI] [PubMed] [Google Scholar]
- 7. Persichetti A, Di Stasio E, Coccaro C, Graziano F, Bianchini A, Di Donna V, et al. Inter- and intraobserver agreement in the assessment of thyroid nodule ultrasound features and classification systems: a blinded multicenter study. Thyroid (2020) 30(2):237–42. 10.1089/thy.2019.0360 [DOI] [PubMed] [Google Scholar]
- 8. Cibas ES, Ali SZ. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid (2017) 27(11):1341–6. 10.1089/thy.2017.0500 [DOI] [PubMed] [Google Scholar]
- 9. Baloch ZW, Livolsi VA. Follicular-patterned lesions of the thyroid: the bane of the pathologist. Am J Clin Pathol (2002) 117(1):143–50. 10.1309/8vl9-ecxy-nvmx-2rqf [DOI] [PubMed] [Google Scholar]
- 10. LiVolsi VA, Baloch ZW. Follicular neoplasms of the thyroid: view, biases, and experiences. Adv Anat Pathol (2004) 11(6):279–87. 10.1097/01.pap.0000138143.34505.02 [DOI] [PubMed] [Google Scholar]
- 11. Acs B, Rantalainen M, Hartman J. Artificial intelligence as the next step towards precision pathology. J Intern Med (2020) 288(1):62–81. 10.1111/joim.13030 [DOI] [PubMed] [Google Scholar]
- 12. Cao C, Liu F, Tan H, Song D, Shu W, Li W, et al. Deep learning and its applications in biomedicine. Genomics Proteomics Bioinf (2018) 16(1):17–32. 10.1016/j.gpb.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manning T, Sleator RD, Walsh P. Biologically inspired intelligent decision making: a commentary on the use of artificial neural networks in bioinformatics. Bioengineered (2014) 5(2):80–95. 10.4161/bioe.26997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tu JV. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J Clin Epidemiol (1996) 49(11):1225–31. 10.1016/s0895-4356(96)00002-9 [DOI] [PubMed] [Google Scholar]
- 15. Sun C, Zhan Y, Chang Q, Liu T, Zhang S, Wang X, et al. Evaluation of a deep learning-based computer-aided diagnosis system for distinguishing benign from malignant thyroid nodules in ultrasound images. Med Phys (2020) 47(9):3952–60. 10.1002/mp.14301 [DOI] [PubMed] [Google Scholar]
- 16. Wang L, Yang S, Yang S, Zhao C, Tian G, Gao Y, et al. Automatic thyroid nodule recognition and diagnosis in ultrasound imaging with the YOLOv2 neural network. World J Surg Oncol (2019) 17(1):12. 10.1186/s12957-019-1558-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Zhang S, Zhang Q, Wei X, Pan Y, Zhao J, et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study. Lancet Oncol (2019) 20(2):193–201. 10.1016/s1470-2045(18)30762-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buda M, Wildman-Tobriner B, Hoang JK, Thayer D, Tessler FN, Middleton WD, et al. Management of Thyroid Nodules Seen on US Images: Deep Learning May Match Performance of Radiologists. Radiology (2019) 292(3):695–701. 10.1148/radiol.2019181343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Remonti LR, Kramer CK, Leitao CB, Pinto LC, Gross JL. Thyroid ultrasound features and risk of carcinoma: a systematic review and meta-analysis of observational studies. Thyroid (2015) 25(5):538–50. 10.1089/thy.2014.0353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trimboli P, Castellana M, Piccardo A, Romanelli F, Grani G, Giovanella L, et al. The ultrasound risk stratification systems for thyroid nodule have been evaluated against papillary carcinoma. A meta-analysis. Rev Endocr Metab Disord (2020). 10.1007/s11154-020-09592-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wildman-Tobriner B, Buda M, Hoang JK, Middleton WD, Thayer D, Short RG, et al. Using artificial intelligence to revise ACR TI-RADS risk stratification of thyroid nodules: diagnostic accuracy and utility. Radiology (2019) 292(1):112–9. 10.1148/radiol.2019182128 [DOI] [PubMed] [Google Scholar]
- 22. Zhao CK, Ren TT, Yin YF, Shi H, Wang HX, Zhou BY, et al. A comparative analysis of two machine learning-based diagnostic patterns with thyroid imaging reporting and data system for thyroid nodules: diagnostic performance and unnecessary biopsy rate. Thyroid (2020). 10.1089/thy.2020.0305 [DOI] [PubMed] [Google Scholar]
- 23. Jin Z, Zhu Y, Zhang S, Xie F, Zhang M, Zhang Y, et al. Ultrasound Computer-aided diagnosis (CAD) based on the thyroid imaging reporting and data system (TI-RADS) to distinguish benign from malignant thyroid nodules and the diagnostic performance of radiologists with different diagnostic experience. Med Sci Monit (2020) 26:e918452. 10.12659/msm.918452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bai Z, Chang L, Yu R, Li X, Wei X, Yu M, et al. Thyroid nodules risk stratification through deep learning based on ultrasound images. Med Phys (2020) 47(12):6355–65. 10.1002/mp.14543 [DOI] [PubMed] [Google Scholar]
- 25. Wu MH, Chen CN, Chen KY, Ho MC, Tai HC, Wang YH, et al. Quantitative analysis of echogenicity for patients with thyroid nodules. Sci Rep (2016) 6:35632. 10.1038/srep35632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim KJ, Choi CS, Yoon DY, Chang SK, Kim KK, Han H, et al. Computer-aided diagnosis for the differentiation of malignant from benign thyroid nodules on ultrasonography. Acad Radiol (2008) 15(7):853–8. 10.1016/j.acra.2007.12.022 [DOI] [PubMed] [Google Scholar]
- 27. Savelonas M, Maroulis D. Sangriotis M. A computer-aided system for malignancy risk assessment of nodules in thyroid US images based on boundary features. Comput Methods Prog BioMed (2009) 96(1):25–32. 10.1016/j.cmpb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 28. Chen KY, Chen CN, Wu MH, Ho MC, Tai HC, Huang WC, et al. Computerized detection and quantification of microcalcifications in thyroid nodules. Ultrasound Med Biol (2011) 37(6):870–8. 10.1016/j.ultrasmedbio.2011.03.002 [DOI] [PubMed] [Google Scholar]
- 29. Zhu LC, Ye YL, Luo WH, Su M, Wei HP, Zhang XB, et al. A model to discriminate malignant from benign thyroid nodules using artificial neural network. PloS One (2013) 8(12):e82211. 10.1371/journal.pone.0082211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi WJ, Park JS, Kim KG, Kim SY, Koo HR, Lee YJ. Computerized analysis of calcification of thyroid nodules as visualized by ultrasonography. Eur J Radiol (2015) 84(10):1949–53. 10.1016/j.ejrad.2015.06.021 [DOI] [PubMed] [Google Scholar]
- 31. Xia J, Chen H, Li Q, Zhou M, Chen L, Cai Z, et al. Ultrasound-based differentiation of malignant and benign thyroid Nodules: An extreme learning machine approach. Comput Methods Prog BioMed (2017) 147:37–49. 10.1016/j.cmpb.2017.06.005 [DOI] [PubMed] [Google Scholar]
- 32. Choi YJ, Baek JH, Park HS, Shim WH, Kim TY, Shong YK, et al. A computer-aided diagnosis system using artificial intelligence for the diagnosis and characterization of thyroid nodules on ultrasound: initial clinical assessment. Thyroid (2017) 27(4):546–52. 10.1089/thy.2016.0372 [DOI] [PubMed] [Google Scholar]
- 33. Ouyang FS, Guo BL, Ouyang LZ, Liu ZW, Lin SJ, Meng W, et al. Comparison between linear and nonlinear machine-learning algorithms for the classification of thyroid nodules. Eur J Radiol (2019) 113:251–7. 10.1016/j.ejrad.2019.02.029 [DOI] [PubMed] [Google Scholar]
- 34. Kim HL, Ha EJ, Han M. Real-world performance of computer-aided diagnosis system for thyroid nodules using ultrasonography. Ultrasound Med Biol (2019) 45(10):2672–8. 10.1016/j.ultrasmedbio.2019.05.032 [DOI] [PubMed] [Google Scholar]
- 35. Liu T, Guo Q, Lian C, Ren X, Liang S, Yu J, et al. Automated detection and classification of thyroid nodules in ultrasound images using clinical-knowledge-guided convolutional neural networks. Med Image Anal (2019) 58:101555. 10.1016/j.media.2019.101555 [DOI] [PubMed] [Google Scholar]
- 36. Guan Q, Wang Y, Du J, Qin Y, Lu H, Xiang J, et al. Deep learning based classification of ultrasound images for thyroid nodules: a large scale of pilot study. Ann Transl Med (2019) 7(7):137. 10.21037/atm.2019.04.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei X, Zhu J, Zhang H, Gao H, Yu R, Liu Z, et al. Visual interpretability in computer-assisted diagnosis of thyroid nodules using ultrasound images. Med Sci Monit (2020) 26:e927007. 10.12659/msm.927007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chambara N, Ying M. The Diagnostic Efficiency of ultrasound computer-aided diagnosis in differentiating thyroid nodules: a systematic review and narrative synthesis. Cancers (Basel) (2019) 11(11):1759. 10.3390/cancers11111759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu J, Deng Y, Liu T, Zhou J, Jia X, Xiao T, et al. Lymph node metastasis prediction of papillary thyroid carcinoma based on transfer learning radiomics. Nat Commun (2020) 11(1):4807. 10.1038/s41467-020-18497-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee JH, Baek JH, Kim JH, Shim WH, Chung SR, Choi YJ, et al. Deep Learning-Based Computer-Aided Diagnosis System for Localization and Diagnosis of Metastatic Lymph Nodes on Ultrasound: A Pilot Study. Thyroid (2018) 28(10):1332–8. 10.1089/thy.2018.0082 [DOI] [PubMed] [Google Scholar]
- 41. Wu T, Sultan LR, Tian J, Cary TW, Sehgal CM. Machine learning for diagnostic ultrasound of triple-negative breast cancer. Breast Cancer Res Treat (2019) 173(2):365–73. 10.1007/s10549-018-4984-7 [DOI] [PubMed] [Google Scholar]
- 42. Brito JP, Gionfriddo MR, Al Nofal A, Boehmer KR, Leppin AL, Reading C, et al. The accuracy of thyroid nodule ultrasound to predict thyroid cancer: systematic review and meta-analysis. J Clin Endocrinol Metab (2014) 99(4):1253–63. 10.1210/jc.2013-2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sollini M, Cozzi L, Chiti A, Kirienko M. Texture analysis and machine learning to characterize suspected thyroid nodules and differentiated thyroid cancer: Where do we stand? Eur J Radiol (2018) 99:1–8. 10.1016/j.ejrad.2017.12.004 [DOI] [PubMed] [Google Scholar]
- 44. Raghavendra U, Rajendra Acharya U, Gudigar A, Hong Tan J, Fujita H, Hagiwara Y, et al. Fusion of spatial gray level dependency and fractal texture features for the characterization of thyroid lesions. Ultrasonics (2017) 77:110–20. 10.1016/j.ultras.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 45. Prochazka A, Gulati S, Holinka S, Smutek D. Classification of thyroid nodules in ultrasound images using direction-independent features extracted by two-threshold binary decomposition. Technol Cancer Res Treat (2019) 181533033819830748. 10.1177/1533033819830748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu Q, Jiang T, Zhou A, Zhang L, Zhang C, Xu P. Computer-aided diagnosis of malignant or benign thyroid nodes based on ultrasound images. Eur Arch Otorhinolaryngol (2017) 274(7):2891–7. 10.1007/s00405-017-4562-3 [DOI] [PubMed] [Google Scholar]
- 47. Thomas J, Haertling T. AIBx, artificial intelligence model to risk stratify thyroid nodules. Thyroid (2020) 30(6):878–84. 10.1089/thy.2019.0752 [DOI] [PubMed] [Google Scholar]
- 48. Martín Noguerol T, Paulano-Godino F, Martín-Valdivia MT, Menias CO, Luna A. Strengths, Weaknesses, Opportunities, and Threats Analysis of Artificial Intelligence and Machine Learning Applications in Radiology. J Am Coll Radiol (2019) 16(9 Pt B):1239–47. 10.1016/j.jacr.2019.05.047 [DOI] [PubMed] [Google Scholar]
- 49. Zoulias EA, Asvestas PA, Matsopoulos GK, Tseleni-Balafouta S. A decision support system for assisting fine needle aspiration diagnosis of thyroid malignancy. Anal Quant Cytol Histol (2011) 33(4):215–22. [PubMed] [Google Scholar]
- 50. Margari N, Mastorakis E, Pouliakis A, Gouloumi AR, Asimis E, Konstantoudakis S, et al. Classification and regression trees for the evaluation of thyroid cytomorphological characteristics: A study based on liquid based cytology specimens from thyroid fine needle aspirations. Diagn Cytopathol (2018) 46(8):670–81. 10.1002/dc.23977 [DOI] [PubMed] [Google Scholar]
- 51. Elliott Range DD, Dov D, Kovalsky SZ, Henao R, Carin L, Cohen J. Application of a machine learning algorithm to predict malignancy in thyroid cytopathology. Cancer Cytopathol (2020) 128(4):287–95. 10.1002/cncy.22238 [DOI] [PubMed] [Google Scholar]
- 52. Tomei S, Marchetti I, Zavaglia K, Lessi F, Apollo A, Aretini P, et al. A molecular computational model improves the preoperative diagnosis of thyroid nodules. BMC Cancer (2012) 12:396. 10.1186/1471-2407-12-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Daskalakis A, Kostopoulos S, Spyridonos P, Glotsos D, Ravazoula P, Kardari M, et al. Design of a multi-classifier system for discriminating benign from malignant thyroid nodules using routinely H&E-stained cytological images. Comput Biol Med (2008) 38(2):196–203. 10.1016/j.compbiomed.2007.09.005 [DOI] [PubMed] [Google Scholar]
- 54. Cochand-Priollet B, Koutroumbas K, Megalopoulou TM, Pouliakis A, Sivolapenko G, Karakitsos P. Discriminating benign from malignant thyroid lesions using artificial intelligence and statistical selection of morphometric features. Oncol Rep (2006) 15 Spec no:1023–6. 10.3892/or.15.4.1023 [DOI] [PubMed] [Google Scholar]
- 55. Sanyal P, Mukherjee T, Barui S, Das A, Gangopadhyay P. Artificial intelligence in cytopathology: A neural network to identify papillary carcinoma on thyroid fine-needle aspiration cytology smears. J Pathol Inform (2018) 9:43. 10.4103/jpi.jpi_43_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guan Q, Wang Y, Ping B, Li D, Du J, Qin Y, et al. Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: a pilot study. J Cancer (2019) 10(20):4876–82. 10.7150/jca.28769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Savala R, Dey P, Gupta N. Artificial neural network model to distinguish follicular adenoma from follicular carcinoma on fine needle aspiration of thyroid. Diagn Cytopathol (2018) 46(3):244–9. 10.1002/dc.23880 [DOI] [PubMed] [Google Scholar]
- 58. Alexander EK, Kennedy GC, Baloch ZW, Cibas ES, Chudova D, Diggans J, et al. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. New Engl J Med (2012) 367(8):705–15. 10.1056/NEJMoa1203208 [DOI] [PubMed] [Google Scholar]
- 59. Patel KN, Angell TE, Babiarz J, Barth NM, Blevins T, Duh QY, et al. Performance of a genomic sequencing classifier for the preoperative diagnosis of cytologically indeterminate thyroid nodules. JAMA Surg (2018) 153(9):817–24. 10.1001/jamasurg.2018.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nikiforova MN, Mercurio S, Wald AI, Barbi de Moura M, Callenberg K, Santana-Santos L, et al. Analytical performance of the ThyroSeq v3 genomic classifier for cancer diagnosis in thyroid nodules. Cancer (2018) 124(8):1682–90. 10.1002/cncr.31245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lithwick-Yanai G, Dromi N, Shtabsky A, Morgenstern S, Strenov Y, Feinmesser M, et al. Multicentre validation of a microRNA-based assay for diagnosing indeterminate thyroid nodules utilising fine needle aspirate smears. J Clin Pathol (2017) 70(6):500–7. 10.1136/jclinpath-2016-204089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sun Y, Selvarajan S, Zang Z, Liu W, Zhu YJ, Zhang H, et al. Protein classifier for thyroid nodules learned from rapidly acquired proteotypes. medRxiv (2020). 10.1101/2020.04.09.20059741 [DOI] [Google Scholar]
- 63. Wang W, Ozolek JA, Rohde GK. Detection and classification of thyroid follicular lesions based on nuclear structure from histopathology images. Cytometry A (2010) 77(5):485–94. 10.1002/cyto.a.20853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ozolek JA, Tosun AB, Wang W, Chen C, Kolouri S, Basu S, et al. Accurate diagnosis of thyroid follicular lesions from nuclear morphology using supervised learning. Med Image Anal (2014) 18(5):772–80. 10.1016/j.media.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao Y, Zhao L, Mao T, Zhong L. Assessment of risk based on variant pathways and establishment of an artificial neural network model of thyroid cancer. BMC Med Genet (2019) 20(1):92. 10.1186/s12881-019-0829-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ruiz EML, Niu T, Zerfaoui M, Kunnimalaiyaan M, Friedlander PL, Abdel-Mageed AB, et al. A novel gene panel for prediction of lymph-node metastasis and recurrence in patients with thyroid cancer. Surgery (2020) 167(1):73–9. 10.1016/j.surg.2019.06.058 [DOI] [PubMed] [Google Scholar]
- 67. Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, et al. 2015 American thyroid association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The american thyroid association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid (2016) 26(1):1–133. 10.1089/thy.2015.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kobayashi K, Hirokawa M, Yabuta T, Masuoka H, Fukushima M, Kihara M, et al. Tumor protrusion with intensive blood signals on ultrasonography is a strongly suggestive finding of follicular thyroid carcinoma. Med Ultrason (2016) 18(1):25–9. 10.11152/mu.2013.2066.181.kok [DOI] [PubMed] [Google Scholar]
- 69. Seo JK, Kim YJ, Kim KG, Shin I, Shin JH, Kwak JY. Differentiation of the Follicular Neoplasm on the Gray-Scale US by Image Selection Subsampling along with the Marginal Outline Using Convolutional Neural Network. BioMed Res Int (2017) 2017:3098293. 10.1155/2017/3098293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang B, Yan M, Yan Z, Zhu C, Xu D, Dong F. Segmentation and classification of thyroid follicular neoplasm using cascaded convolutional neural network. Phys Med Biol (2020). 10.1088/1361-6560/abc6f2 [DOI] [PubMed] [Google Scholar]
- 71. Smallridge RC, Ain KB, Asa SL, Bible KC, Brierley JD, Burman KD, et al. American Thyroid Association guidelines for management of patients with anaplastic thyroid cancer. Thyroid (2012) 22(11):1104–39. 10.1089/thy.2012.0302 [DOI] [PubMed] [Google Scholar]
- 72. Wells SA, Jr., Asa SL, Dralle H, Elisei R, Evans DB, Gagel RF, et al. Revised American Thyroid Association guidelines for the management of medullary thyroid carcinoma. Thyroid (2015) 25(6):567–610. 10.1089/thy.2014.0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Misiakos EP, Margari N, Meristoudis C, Machairas N, Schizas D, Petropoulos K, et al. Cytopathologic diagnosis of fine needle aspiration biopsies of thyroid nodules. World J Clin Cases (2016) 4(2):38–48. 10.12998/wjcc.v4.i2.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Phillips KA, Trosman JR, Kelley RK, Pletcher MJ, Douglas MP, Weldon CB. Genomic sequencing: assessing the health care system, policy, and big-data implications. Health Aff (2014) 33(7):1246–53. 10.1377/hlthaff.2014.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schmidt B, Hildebrandt A. Next-generation sequencing: big data meets high performance computing. Drug Discovery Today (2017) 22(4):712–7. 10.1016/j.drudis.2017.01.014 [DOI] [PubMed] [Google Scholar]
- 76. Mayer-Schönberger V, Ingelsson E. Big Data and medicine: a big deal? J Intern Med (2018) 283(5):418–29. 10.1111/joim.12721 [DOI] [PubMed] [Google Scholar]
- 77. Hamamoto R, Komatsu M, Takasawa K, Asada K, Kaneko S. Epigenetics analysis and integrated analysis of multiomics data, including epigenetic data, using artificial intelligence in the era of precision medicine. Biomolecules (2019) 10(1):62. 10.3390/biom10010062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Steward DL, Carty SE, Sippel RS, Yang SP, Sosa JA, Sipos JA, et al. Performance of a multigene genomic classifier in thyroid nodules with indeterminate cytology: a prospective blinded multicenter study. JAMA Oncol (2019) 5(2):204–12. 10.1001/jamaoncol.2018.4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Benjamin H, Schnitzer-Perlman T, Shtabsky A, VandenBussche CJ, Ali SZ, Kolar Z, et al. Analytical validity of a microRNA-based assay for diagnosing indeterminate thyroid FNA smears from routinely prepared cytology slides. Cancer Cytopathol (2016) 124(10):711–21. 10.1002/cncy.21731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shao W, Guo T, Toussaint NC, Xue P, Wagner U, Li L, et al. Comparative analysis of mRNA and protein degradation in prostate tissues indicates high stability of proteins. Nat Commun (2019) 10(1):2524. 10.1038/s41467-019-10513-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu X, Quiros RM, Gattuso P, Ain KB, Prinz RA. High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res (2003) 63(15):4561–7. [PubMed] [Google Scholar]
- 82. Zou M, Shi Y, Farid NR, al-Sedairy ST, Paterson MC. FHIT gene abnormalities in both benign and malignant thyroid tumours. Eur J Cancer (1999) 35(3):467–72. 10.1016/s0959-8049(98)00370-0 [DOI] [PubMed] [Google Scholar]
- 83. Angell TE. RAS-positive thyroid nodules. Curr Opin Endocrinol Diabetes Obes (2017) 24(5):372–6. 10.1097/med.0000000000000354 [DOI] [PubMed] [Google Scholar]
- 84. Ferrari SM, Fallahi P, Galdiero MR, Ruffilli I, Elia G, Ragusa F, et al. Immune and inflammatory cells in thyroid cancer microenvironment. Int J Mol Sci (2019) 20(18):4413. 10.3390/ijms20184413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang Y, Guan Q, Lao I, Wang L, Wu Y, Li D, et al. Using deep convolutional neural networks for multi-classification of thyroid tumor by histopathology: a large-scale pilot study. Ann Transl Med (2019) 7(18):468. 10.21037/atm.2019.08.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kakudo K, Bychkov A, Bai Y, Li Y, Liu Z, Jung CK. The new 4th edition World Health Organization classification for thyroid tumors, Asian perspectives. Pathol Int (2018) 68(12):641–64. 10.1111/pin.12737 [DOI] [PubMed] [Google Scholar]
- 87. Daniels GH. Follicular variant of papillary thyroid carcinoma: hybrid or mixture? Thyroid (2016) 26(7):872–4. 10.1089/thy.2016.0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hodak S, Tuttle RM, Maytal G, Nikiforov YE, Randolph G. Changing the cancer diagnosis: the case of follicular variant of papillary thyroid cancer-primum non nocere and NIFTP. Thyroid (2016) 26(7):869–71. 10.1089/thy.2016.0205 [DOI] [PubMed] [Google Scholar]
- 89. Borrelli N, Denaro M, Ugolini C, Poma AM, Miccoli M, Vitti P, et al. miRNA expression profiling of ‘noninvasive follicular thyroid neoplasms with papillary-like nuclear features’ compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas. Mod Pathol (2017) 30(1):39–51. 10.1038/modpathol.2016.157 [DOI] [PubMed] [Google Scholar]
- 90. Selvaggi SM. The presence of multinucleated giant cells: Noninvasive follicular thyroid neoplasm with papillary-like nuclear features vs the follicular variant of papillary thyroid carcinoma. Diagn Cytopathol (2019) 47(10):1007–10. 10.1002/dc.24259 [DOI] [PubMed] [Google Scholar]
- 91. Chain K, Legesse T, Heath JE, Staats PN. Digital image-assisted quantitative nuclear analysis improves diagnostic accuracy of thyroid fine-needle aspiration cytology. Cancer Cytopathol (2019) 127(8):501–13. 10.1002/cncy.22120 [DOI] [PubMed] [Google Scholar]
- 92. Hsieh AM, Polyakova O, Fu G, Chazen RS, MacMillan C, Witterick IJ, et al. Programmed death-ligand 1 expression by digital image analysis advances thyroid cancer diagnosis among encapsulated follicular lesions. Oncotarget (2018) 9(28):19767–82. 10.18632/oncotarget.24833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Perrier ND, Brierley JD, Tuttle RM. Differentiated and anaplastic thyroid carcinoma: Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin (2018) 68(1):55–63. 10.3322/caac.21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gardezi SJS, Elazab A, Lei B, Wang T. Breast cancer detection and diagnosis using mammographic data: systematic review. J Med Internet Res (2019) 21(7):e14464. 10.2196/14464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lui TKL, Guo CG, Leung WK. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: a systematic review and meta-analysis. Gastrointest Endosc (2020) 92(1):11–22. 10.1016/j.gie.2020.02.033 [DOI] [PubMed] [Google Scholar]
- 96. Nagendran M, Chen Y, Lovejoy CA, Gordon AC, Komorowski M, Harvey H, et al. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. Bmj (2020) 368:m689. 10.1136/bmj.m689 [DOI] [PMC free article] [PubMed] [Google Scholar]