Abstract
The intestinal microbiota plays important roles in the maintenance of health. Strategies aiming at its modulation, such as probiotics, have received a deal of attention. Several strains have been studied in different in vitro models; however, the correlation of results obtained with the in vivo data has been limited. This questions the usefulness of such in vitro selection models, traditionally relying on over-simplified tests, not considering the influence of the accompanying microbiota or focusing on microbiota composition without considering functional traits. Here we assess the potential of six Bifidobacterium, Lactobacillus and Lacticaseibacillus strains in an in vitro model to determine their impact on the microbiota not just in terms of composition but also of functionality. Moreover, we compared the responses obtained in two different population groups: normal-weight and severely obese subjects. Fecal cultures were conducted to evaluate the impact of the strains on specific intestinal microbial groups, on the production of short-chain fatty acids, and on two functional responses: the production of gas and the interaction with human intestinal epithelial cells. The response to the different probiotics differed between both human groups. The addition of the probiotic strains did not induce major changes on the microbiota composition, with significant increases detected almost exclusively for the species added. Higher levels of gas production were observed in cultures from normal-weight subjects than in the obese population, with some strains being able to significantly reduce gas production in the latter group. Moreover, in obese subjects all the Bifidobacterium strains tested and Lacticaseibacillus rhamnosus GG were able to modify the response of the intestinal cells, restoring values similar to those obtained with the microbiotas of normal-weight subjects. Our results underline the need for the screening and selection of probiotics in a target-population specific manner by using appropriate in vitro models before enrolling in clinical intervention trials.
Keywords: in vitro model, gut microbiota, probiotics, gas production, severe obesity, Bifidobacterium, Lactobacillus, SCFA
Introduction
Recent studies have underlined the important role of the gastro-intestinal microbiota (GIM) in the maintenance of host’s health (Thursby and Juge, 2017). Alterations on this GIM, the so-called “dysbiosis,” have been identified in several diseases (Duvallet et al., 2017). To this regard, obesity is not an exception and both compositional and functional differences between normal-weight (NW) subjects and obese patients have been repeatedly reported (Gomes et al., 2018; Vallianou et al., 2019). Although studies with severely obese individuals (OB) are scarce, accumulating evidence indicates the existence of dysbiosis in this group as well (Aron-Wisnewsky et al., 2019; Cani, 2019; Nogacka et al., 2020a). As it could be expected from the GIM differences, and very likely also due to the different dietary intakes, the fecal levels of short-chain fatty acids (SCFA) in OB subjects is also modified with regard to NW individuals (Kim et al., 2019; Nogacka et al., 2020a). These SCFA are important mediators on the GIM-host interaction, playing important roles on host’s health (Ríos-Covián et al., 2016).
The GIM represents a potential target for strategies focused on health maintenance and improvement, with the use of probiotics constituting a promising approach to this end. Probiotics are “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014). Several probiotic strains are being used in different functional products and a vast array of in vitro tests have been carried out to screen different strains. Most often, the process has been based in classical tests, such as tolerance to simulated gastrointestinal transit, adhesion to intestinal epithelial cells, co-culture with pathogens or immune cells, etc. However, in these in vitro tests, frequently the effect of the accompanying GIM has not been taken into account. For assessing the impact on the microbiota, several fecal culture models and simulators of the gastrointestinal tract have been used (Williams et al., 2015; von Martels et al., 2017), with compositional changes in the GIM being the main outcome in most of these studies, without considering potential functional changes. Moreover, in most cases this in vitro screening for potential probiotic strains was not driven by the impact on a specific target population. However, several studies have reported a high inter-individual variability in the response to probiotics, pointing out to the need for a population-specific selection (van Baarlen et al., 2011; Grześkowiak et al., 2012; Arboleya et al., 2013a,b). As a result of the use of these in vitro models, the correlation between in vitro and in vivo has often been limited (Vinderola et al., 2017). This lack of correlation may be partly explained by the use of over-simplified tests that do not consider the influence of the accompanying GIM, or when considering it, focusing only on its composition regardless of functional traits. The final consequence is that in spite of the huge amount of promising in vitro studies carried out with hundreds of microbial strains, only for a very limited number of them efficacy has been finally demonstrated in human intervention trials.
Different models of GIM-host interaction, such as co-cultivation of fecal samples or isolated GIM from different population groups or added with pro/prebiotics, with epithelial and/or immune cells, have been used (Arboleya et al., 2015; Nogacka et al., 2018, 2020a; Richards et al., 2019). Interestingly, several of these studies have demonstrated differences in the response induced by microbiotas from different population groups (Nogacka et al., 2018, 2020a) and between isolated GIM or among GIM added with probiotics or prebiotics (Arboleya et al., 2015; Nogacka et al., 2020b). These suggest that the inter-population differences on the gut microbiota may partly explain the high variability observed in the response to probiotics and the low correlation between in vivo and in vitro data.
In this work we explored the impact of different probiotic strains employing previously developed in vitro models, which take into account the GIM and that allow assessing the impact of the strains not just in terms of composition but also in terms of functionality on the GIM. This was achieved by monitoring, in real-time, the production of gas in fecal cultures and the interaction with HT29 intestinal epithelial cells, in addition to the study of the microbial composition and SCFA production, in two human population groups; NW and OB individuals.
Materials and Methods
Strains and Culture Conditions
Four Bifidobacterium strains (Bifidobacterium animalis subsp lactis IF20/1 [IPLA20020], Bifidobacterium bifidum TMC3108, B. bifidum TMC3115 and Bifidobacterium longum IF14/11 [IPLA20022] as well as two lactobacilli (Lactobacillus gasseri BM7/10 [IPLA20212] and Lacticaseibacillus rhamnosus (formerly Lactobacillus rhamnosus) GG [ATCC53103]) (Zheng et al., 2020) were used in this study. Frozen stocks were reactivated weekly in MRS agar (Biokar Diagnostics, Beauvois, France) supplemented with 0.25% (w/v) L-cysteine (MRSc; Sigma Chemical Co., St. Louis, MO, United States) by 48 h incubation at 37°C in an anaerobic chamber MG500 (Don Whitley Scientific, West Yorkshire, United Kingdom) under 80% (v/v) N2, 10% (v/v) CO2, and 10% H2 atmosphere. Two overnight passages in MRSc broth before batch culture experiments were performed. The microbial suspensions for fecal cultures were obtained by inoculating (1% v/v) fresh culture medium, incubating overnight under anaerobic conditions, centrifuging and washing twice the bacterial cells with PBS and adjusting to a final concentration of 1 × 1010 CFU/mL.
Volunteers and Fecal Sample Collection
Fecal samples were obtained from nine healthy NW adults (BMI < 25 kg/m2) and six OB volunteers (BMI ≥ 40 kg/m2) recruited at the Digestive and Endocrinology and Nutrition Services, respectively, of the Asturias Central University Hospital (HUCA, Asturias, Spain). The mean age of the volunteers was 40 ± 9 and 44 ± 9 years for NW and OB subjects, respectively. All participants followed an unrestricted diet and had not taken antibiotics during the previous 6 months. The study was approved by the Bioethical Committee of CSIC and from the Regional Ethics Committee for Clinical Research of the Principality of Asturias in compliance with the Declaration of Helsinki of 1964, last revised in 2013. An informed written consent was obtained from each volunteer. Samples were collected and immediately introduced into anaerobic jars (Anaerocult A System, Merck, Darmstadt, Germany) for transportation to the laboratory within 1 h and stored at −80°C until use.
Fecal Batch Cultures
Fecal samples were thawed at 37°C under anaerobic conditions. Then the samples were diluted 1/10 (w/v) with pre-reduced PBS and homogenized in a Lab-Blender 400 stomacher (Seward Medical, London, United Kingdom) at full-speed for 5 min and used as inocula for the fecal culture experiments. Carbohydrate-free basal medium (CFBM) (Al-Tamimi et al., 2006) was prepared and reduced overnight in anaerobic chamber one day before the batch fecal experiment. Pre-reduced CFBM was inoculated (10% v/v) with the fecal homogenate described above and then distributed into 100 mL bottles of the ANKOM RF system (ANKOM Technology, United States). The fecal cultures were allowed to stabilize overnight at 37°C in anaerobic conditions.
In brief, seven independent pH-free batch fermentations were performed for each human donor. We used as a carbon source 0.3% (w/v) of the fructooligosaccharide 1-kestose (β-Food Science Co. Ltd., Japan) which in previous experiments demonstrated to be more fermentable than other fructooligosaccharides not just by bifidobacteria and other intestinal anaerobes but also by lactobacilli (Nogacka et al., 2020b). Bacterial strains were added at a final concentration of 1 × 108 CFU/mL to fecal cultures in bottles. A bottle was left without probiotic added to be used as control. The fecal cultures were then incubated under anaerobic conditions at 37°C for 24 h. Samples (1 mL) were taken in duplicate at time 0 before incubation (time 0; basal conditions) and after 24 h of incubation. These samples were centrifuged at full speed for 10 min and supernatants and pellets were stored separately at −20°C until analyses.
pH and Gas Monitoring in Fecal Cultures
The pH of the cultures was determined with a pHmeter (SensION + PH3, HACH, Barcelona, Spain) and was considered as an indicator of the progression of fermentation. The cumulative gas produced along the different fermentations was monitored in real-time by using the ANKOM RF system. This system provides the increases in pressure (psi) which can be converted to mL of gas produced using the Ideal Gas Equation:
| (1) |
where: V = gas volume at 39°C in mL, Vj = headspace of digestion jar (Glass Bottle) in mL, Ppsi = cumulative pressure recorded by Gas Monitor System software.
The data of gas production were fitted to modified-Gompertz equation, a model frequently used to fit data of bacterial, plant growth, tumor proliferation and gas production (Ware and Power, 2017), by using the formula:
| (2) |
In which variables: “A” represents the upper asymptote (mL), “μ” is the rate of gas production (mL/h) and “λ” is the time lag before exponential phase (h).
Microbiota Composition and SCFA Quantification
DNA was extracted from the bacterial pellets by using the QIAamp DNA Stool Mini kit (Qiagen GmbH, Hilden, Germany) as previously described (Nogacka et al., 2020a) and the isolated DNA was stored at −20°C until use. The absolute levels of relevant intestinal microbial groups (Bacteroides-Prevotella-Porphyromonas group, Lactobacillus-group, Akkermansia, Clostridium cluster XIVa, Bifidobacterium and Faecalibacterium genera) as well as total bacteria were determined at 0 and 24 h of fermentation by qPCR using previously described primers and conditions (Valdés et al., 2017). Variations in the levels of the species B. longum, B. bifidum, B. animalis, Bifidobacterium adolescentis and Bifidobacterium catenulatum were assessed as described elsewhere (Salazar et al., 2015; Arboleya et al., 2020). In order to investigate microbial changes as regards to the basal microbiota, the data were expressed as the log-ratio of the Fold Change before (time 0) and after 24 h incubation with different probiotic strains.
The analysis of SCFA was performed by Gas Chromatography (GC) in the fecal culture supernatants (CS) in order to determine the molar concentrations of three main compounds: acetic, propionic and butyric acids. The remaining SCFA, namely iso-butyric and iso-valeric acids were also quantified and summed up (BCFA) for further analysis. Briefly, 0.25 mL of the culture supernatants were mixed with 0.3 mL methanol, 0.05 mL of an internal standard solution (2-ethylbutyric 1.05 mg/mL), and 0.05 mL of 20% formic acid. This mixture was centrifuged and the supernatant was used for quantification of SCFA by GC as described previously (Nogacka et al., 2018). Samples were analyzed in triplicate. Increments in molar concentration of SCFA with respect to the time 0 were calculated for each fermentation batch with the different probiotic strains tested.
Monitoring the Interaction of Intestinal Microbiotas and Fecal Culture Supernatants Supplemented With the Probiotics With HT29 Cells
We also aimed at evaluating the impact of the addition of probiotic strains on the interaction between the gut microbiota and enterocytes. To this end, we purified the microbiotas of the volunteers as described by Nogacka and co-workers (2018) and added them with the probiotic strains to be tested. In order to prevent acidification, which could damage the HT29 cells, these microbial mixtures were inactivated by UV light (Nogacka et al., 2018) and the interaction with confluent HT29 cells monolayers was evaluated by using a real-time cell analyser (RTCA-DP) xCelligence apparatus (ACEA Bioscience Inc., San Diego, CA, United States). Variations in HT29 cell monolayer trans-epithelial resistance (due to changes in morphology and/or attachment of the epithelial cells) during exposure to the microbiotas and CS were assessed. The culture conditions and the maintenance of the intestinal epithelial cell line HT29 (ECACC 91072201) is detailed in a previous work where the functional model was developed (Nogacka et al., 2018). For this functional assessment, each strain was mixed with the isolated microbiota in a bacterial proportion 1:1. Then, a ratio 10:1 of the total bacteria (6.5 × 107 cells/mL) with respect to the epithelial cell was added in McCoy’s medium.
The functional assessment of the fecal CS on the behavior of HT29 cells monolayers was assessed by using filtered CS collected after 24 h of the fecal culture (pH adjusted to 7.55 ± 0.05) and diluted at 40% with McCoy’s medium. Additionally, several controls consisting on basal microbiota, and McCoy’s medium without bacteria or fecal supernatants added, were included in each experiment. Each sample was tested by duplicate using two independent E-plates. The monitoring was followed for every 10 min under standard incubation conditions. CI values recorded were normalized by the time of the sample addition and by the control sample, as previously described (Valdés et al., 2015). For statistical comparison purposes the “Area Under the Curve” (AUC), representing the CI values along 10 h of incubation for each sample, was calculated as explained in Nogacka et al. (2018).
Statistical Analyses
Unless otherwise specified, all experimental data are reported as mean ± standard deviation. Statistical analysis of results was performed using the software SPSS v.25 (SPSS Inc., Chicago, United States). Data were compared for the effect caused on the parameters analyzed by the addition of different probiotic strains in fecal cultures from each population cohort (NW and OB) at the end of fermentation (24 h). For variables with a normal distribution (Shapiro-Wilk test) and homoscedasticity (Levene test), one way ANOVA followed by post-hoc DMS comparison were conducted. In the remaining cases (variables showing non-normal distribution), a Kruskal-Wallis test followed by a post-hoc Dunn’s test of pairwise comparisons were applied when necessary. A significant p-value of 0.05 was used for the interpretation of results. For two-group comparisons between OB and NW subjects, a two-tailed Student’s t-test or Mann-Whitney’s U test was conducted for the evaluation of data by parametric or non-parametric contrast, respectively.
Results
Effect of the Probiotic Strains on pH and Gas Production in Fecal Cultures
Drops of pH observed in fecal cultures were similar between the two human populations studied, OB and NW subjects, with values ranging from 1.42 to 1.78 pH units (Table 1). However, the response to the different probiotics showed noticeable differences between both groups. Whilst in the OB group no statistically differences were observed among treatments (control or the different probiotic strains), in NW subjects the strains B. bifidum TMC3115, B. longum IF14/11 and L. gasseri BM7/10 induced significantly (p-value <0.05) higher decreases of pH than those found in the control culture. The opposite trend was observed for gas production; thus, whilst in OB subjects some strains were able to significantly (p-value <0.05) reduce gas production in comparison to the control, no significant differences among treatments were detected in fecal cultures of the NW group (Table 1). Interestingly, in all the conditions tested (control and the different probiotics) fecal cultures of NW subjects shower higher gas production ability than cultures of OB individuals, although no statistical significant differences were found between cultures of both groups. In addition, the inter-individual variability was higher in NW than in the OB group, the later subjects resulting quite homogeneous in terms of gas production in fecal cultures. The kinetic parameters analyzed confirm this observation, with lower production rates in OB than in NW subjects (Table 1).
TABLE 1.
Cumulative gas produced (mL) and decreases of pH values (Δ pH) after 24 h of incubation in fecal cultures from normal-weight (NW) and severely obesity (OB) individuals.
| Group | Probiotic | Δ pH | Cumulative gas | A | μ | R2 |
| OB | Control | −1.54 ± 0.18 | 19.47b ± 4.68 | 19.862 | 1.587 | 0.990 |
| B. animalis IF20/1 | −1.57 ± 0.24 | 12.97a ± 2.16 | 15.102 | 1.078 | 0.997 | |
| B. bifidum TMC3108 | −1.67 ± 0.11 | 14.22ab ± 0.82 | 14.004 | 1.181 | 0.996 | |
| B. bifidum TMC3115 | −1.71 ± 0.17 | 13.69a ± 2.96 | 13.555 | 1.114 | 0.997 | |
| B. longum IF14/11 | −1.78 ± 0.09 | 12.87a ± 1.62 | 12.637 | 1.127 | 0.997 | |
| L. gasseri BM7/10 | −1.74 ± 0.17 | 14.40ab ± 1.63 | 14.215 | 1.286 | 0.995 | |
| L. rhamnosus GG | −1.70 ± 0.17 | 15.54ab ± 2.80 | 15.441 | 1.335 | 0.996 | |
| NW | Control | −1.42a ± 0.18 | 24.37 ± 12.28 | 24.623 | 2.228 | 0.999 |
| B. animalis IF20/1 | −1.49ab ± 0.18 | 22.01 ± 9.84 | 21.918 | 2.192 | 0.999 | |
| B. bifidum TMC3108 | −1.63abc ± 0.07 | 20.69 ± 9.11 | 20.154 | 2.470 | 0.997 | |
| B. bifidum TMC3115 | −1.64c ± 0.07 | 19.49 ± 7.20 | 19.108 | 2.248 | 0.998 | |
| B. longum IF14/11 | −1.64bc ± 0.18 | 18.88 ± 8.46 | 18.629 | 2.121 | 0.998 | |
| L. gasseri BM7/10 | −1.63bc ± 0.13 | 16.67 ± 6.75 | 16.431 | 1.783 | 0.996 | |
| L. rhamnosus GG | −1.58abc ± 0.16 | 22.35 ± 11.07 | 22.371 | 2.168 | 0.998 |
Kinetic parameters were determined using the modified-Gompertz equation, in which “A” represents the upper asymptote (mL) and “μ” is the rate of gas production (mL/h). The values not sharing the same superscript (a, b, or c) indicate significant differences (p-value <0.05) among probiotic strains and/or control for fecal cultures from each population group (NW or OB).
Effect of the Probiotic Strains Addition to Fecal Cultures on the Intestinal Microbiota
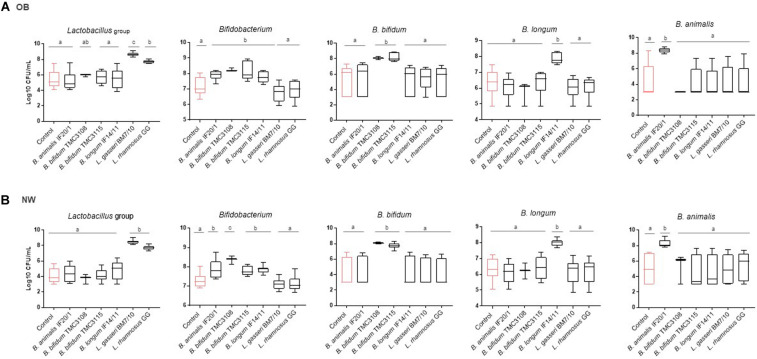
The probiotic strains tested induced changes in the microbiota of fecal cultures, mostly linked to increases in the administered bacterial group or species (Figure 1 and Supplementary Table 1). As it could be expected, the addition of Lactobacillus/Lacticaseibacillus strains increased the levels of lactobacilli in the fecal culture, whereas the administration of Bifidobacterium strains increased bifidobacteria, and specifically the species used, regardless of the human population group. Regarding the other intestinal bacterial groups assessed, no effect of the addition of the probiotics, neither in cultures of OB nor in those of NW subjects, was observed for the levels of Akkermansia, Enterobacteriaceae, Bacteroides-group, Faecalibacterium or total bacteria (Supplementary Table 1). As with regard to Clostridium cluster XIVa the levels were not affected by the probiotic strains in fecal cultures from OB subjects, but in the NW group B. bifidum TMC3115 and L. gasseri BM7/10 promoted lower levels of this bacterial group when compared to the control culture (Supplementary Table 1).
FIGURE 1.
Absolute levels (Log10 CFU/mL) of intestinal microbial groups determined by qPCR in fecal cultures of (A) OB and (B) NW subjects. For each condition, the box and whiskers plot represent median, interquartile range and minimum and maximum values obtained in each human group (NW or OB). Different letters above the boxes indicate significant differences (p-value <0.05) among probiotic strains for the microbial groups considered.
Considering the basal differences existing on the microbial composition between fecal samples of OB and NW subjects, the fold change in bacterial levels were calculated for the different probiotics in fecal cultures of both groups of individuals. Interestingly, when the response to the different probiotic strains in the cultures of the two human groups studied were compared some statistically significant differences were found (Supplementary Figure 1). B. animalis subsp lactis IF20/1 and B. bifidum TMC3115 induced significantly higher increments (p-value <0.05) in the levels of Bifidobacterium in cultures of OB than in those of NW subjects. A similar trend, although not reaching statistical significance (p-value <0.1), was observed for B. longum IF14/11. Whereas B. bifidum TMC3108 also showed a trend (p-value <0.1) toward a larger reduction on the levels of Clostridium cluster XIVa in the OB than in NW group. Regarding the Lactobacillus/Lacticaseibacillus strains tested, L. rhamnosus GG led to an increase on lactobacilli levels which resulted significantly higher (p-value <0.05) in cultures of NW than in cultures of OB, and the same trend (p-value <0.1) was also observed with L. gasseri BM7/10. These results indicate that the increases in the corresponding groups induced by the probiotic Lactobacillus/ Lacticaseibacillus and Bifidobacterium strains tested, is not limited to the increase of the administered strain. The difference in the increases induced by the same probiotic observed between OB and NW groups suggest that the probiotics may also affect the intestinal populations of lactobacilli or bifidobacteria in a way that depends on the basal microbiota, known to be different for OB and NW subjects.
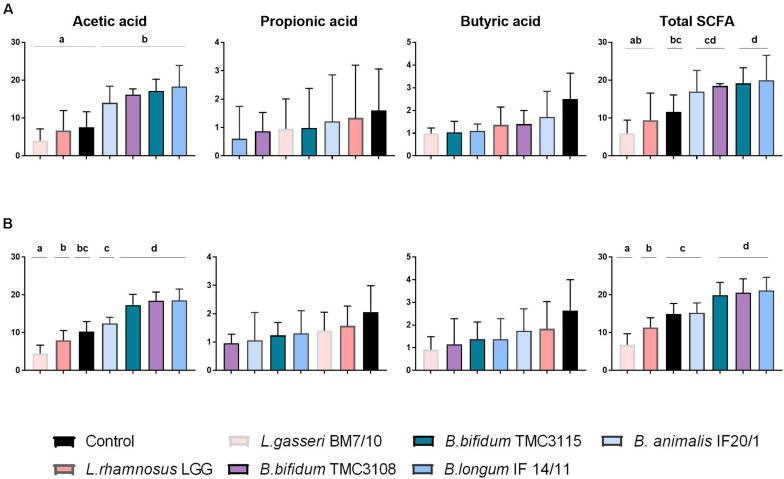
When we assessed the production of SCFA no major differences were observed in the response to the different probiotics between OB and NW fecal cultures. In cultures of both groups of individuals, acetic acid was the SCFA present at higher concentration, with propionic and butyric acids being detected at lower levels (Figure 2). No differences among probiotic strains or between these ones with respect to the control culture were observed for propionic and butyric acid. However, differences became apparent for acetic acid, with the four Bifidobacterium strains tested (B. animalis subsp lactis IF20/1, B. bifidum TMC3108, B. bifidum TMC3115 and B. longum IF14/11) inducing the production of larger amounts of acetic acid (p-value <0.05) than the lactobacilli, or the control culture, for OB individuals. Moreover, these four strains led to the production of a higher concentration of acetic acid (p-value <0.05) than the lactobacilli also in NW subjects, with the increments from B. bifidum TMC3108, B. bifidum TMC3115 and B. longum IF14/11 being significantly higher (p-value <0.05) than those obtained for all the other experimental conditions in this NW group. Given that acetic acid is clearly the predominant SCFA in the samples, these observations are mirrored as well when the total level of SCFA was considered (Figure 2).
FIGURE 2.
Increments in ascending order, with respect to time 0, in the concentration (mM) of the major short-chain fatty acids (acetic, propionic, and butyric acids) after 24 h of incubation with different probiotic strains in fecal cultures from OB (A) and NW (B) groups. Differences are shown for each SCFA, columns that do not share the same letter are significantly different (p < 0.05).
Effect of the Fecal Culture Supernatants and Microbiotas Added With Probiotic Strains on the Interaction With HT29 Cells
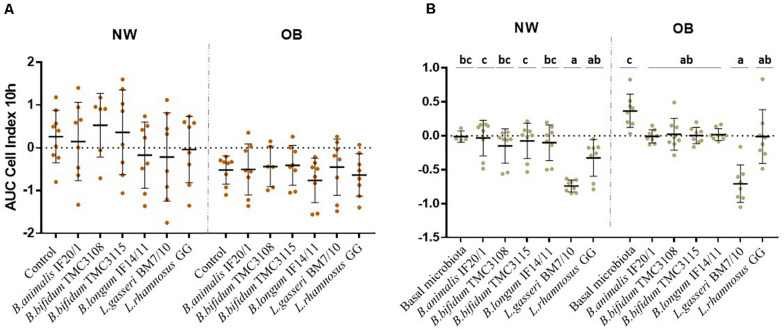
The interaction with intestinal cells monolayers was monitored in real-time using the RTCA system as a proxy for determining the impact of the probiotics on the functional response of the intestinal epithelium to the microbiota. This system allows monitoring in real time the epithelial cell monolayer structure/integrity by measuring the impedance and detecting changes in this parameter that may be due to changes in the morphology of the cells or in their attachment. We assessed first the effect of CS obtained at 24 h of incubation. The response observed with the supernatants of OB cultures displayed lower AUC values than those with cultures obtained from NW donors, which could be reflecting the significant difference found (p-value <0.05) between the control conditions from both human groups (Figure 3A). However, within cultures from each human group no statistically significant differences were observed among the different probiotics, with a similar response against all the strains tested (Figure 3A). Next, we assessed the effect of the addition of the different strains to the basal microbiota of the OB or NW individuals (Figure 3B). Again, differences (p-value <0.05) were observed between the basal microbiota of OB and NW subjects, with the microbiotas from OB subjects showing higher AUC values than that of NW individuals. When the different strains were compared, the Lactobacillus/ Lacticaseibacillus strains displayed lower AUC values than the bifidobacteria in the NW group, whereas the results reached statistical significance (p-value <0.05) only for L. gasseri BM7/10 in NW subjects. Moreover, in OB subjects all bifidobacteria and L. rhamnosus GG strains induced significantly lower (p-value <0.05) AUC values than that obtained for the basal microbiota without probiotics added (Figure 3B), reaching values similar to those obtained for the microbiota of NW subjects.
FIGURE 3.
Real-time monitoring the interaction with HT29 intestinal epithelial cells between (A) supernatants obtained after fecal culture with probiotic and (B) a mixture of a probiotic strain with the gut microbiota from NW and OB population groups. Values (media ± SD) correspond to the AUC resulting from monitoring Cell Index (CI) during 10 h. Significant differences (p-value <0.05) represent the comparison of results before and after probiotic addition in each condition.
Discussion
Restoration of the eubiotic condition of the GIM is of great interest for the prevention of different diseases and probiotics have been among the most used tools to this end (Sanders et al., 2019). However, in spite of the several beneficial effects attributed to probiotics only some of them have been substantiated by clinical evidence (Merenstein et al., 2020). Therefore, in Europe with the sole exception of the yogurt and lactose intolerance, no other probiotic-based health claim has been approved. This is in contrast to the high amount of in vitro studies claiming the probiotic potential of several strains (Vinderola et al., 2017). This lack of agreement suggests that the conventional and perhaps over-simplified in vitro models most used until now in probiotics research (adhesion to mucus or intestinal cells, tolerance to acid and bile, pathogen inhibition or immune modulation etc.) show a poor predictive value for the in vivo situation where other microorganisms are present, at very large amounts, in the ecosystem. Actually, most often the strains have been tested alone, without the accompanying microbiota which in addition may be differ among population groups. This may explain why in some cases the experiments may have failed in the identification of the most suited strains for a given target population. The use of fecal culture models, or intestinal simulators, has been also common in the search for prebiotics with microbiota modulating abilities (Roberfroid et al., 2010; Shen et al., 2011; Bajury et al., 2018; Nogacka et al., 2020b). However, such models have not been so extensively used in probiotics research. Fecal culture models, similar to those applied in this study, have been used for the selection of potentially probiotic strains for GIM modulation in newborns (Arboleya et al., 2013b) or elderly (Valdés et al., 2017), among others. However, in contrast with the current work, in most of the available studies the selection was based exclusively on the effects upon the GIM composition, without taking into consideration the functionality of the microbiota. Only a few in vitro anaerobic bacterial co-culture systems that consider host-gut microbiota interaction have been employed for that purpose (von Martels et al., 2017), moving from the simplest ones, as the Host-Microbiota Interaction (HMI) and the Human oxygen Bacteria anaerobic (HoxBan) models, to the most sophisticated ones such as the microdevices Gut-on-a-chip and the Human Microbial crosstalk model (HuMiX). In the present work, affordable and simple in vitro tests for the screening and selection of probiotics in a target-population specific manner were assessed. We monitored, in real-time, the production of gas in fecal cultures and the interaction with intestinal epithelial cells of the culture supernatants or the isolated microbiotas from NW and OB individuals supplemented with six Bifidobacterium, Lactobacillus and Lacticaseibacillus strains. Our in vitro models have taken into consideration not only the composition but also the functionality of the basal gut microbiota from NW and OB volunteers demonstrating their different response after probiotic administration.
Higher levels of gas production were generally achieved in fecal cultures from NW subjects than in the severely obese population, which is in good agreement with previous observations (Nogacka et al., 2020b). This result underlines the existence of functional differences between the GIM of NW and OB subjects and suggests a metabolically less active microbiota in OB subjects. Changes in the gas production are related with differences in the composition and metabolic activity of the basal intestinal microbiota. Additionally, prebiotics are known to affect microorganisms of the intestinal microbiota such as bifidobacteria and lactobacilli that produce acetate and lactate; these compounds could be involved in cross-feeding mechanisms with gas-producing microorganisms such as Clostridium and sulfate-reducing bacteria (Sarbini and Rastall, 2011). We screened different probiotic strains for their ability to influence in vitro the GIM composition and activity. To this end, we evaluated gas production over the fecal culture in real-time using the ANKOM RF technology. Although this method had been already applied to fecal cultures (Rotbart et al., 2018; Yao et al., 2018; Nogacka et al., 2020b), this is the first time that it is applied in the evaluation of probiotics. This real-time monitoring of gas production has made possible to discriminate between probiotics according to their different ability to modulate gas production in fecal cultures of severely OB subjects, with B. bifidum TMC3115, B. animalis IF20/1 and B. longum IF14/11 being able to reduce the production of gas as compared with the control culture with no probiotics added. The fecal cultures of the NW individuals presented greater heterogeneity than those of OB individuals, which may partly explain why we failed to obtain statistically significant differences in the NW population.
As with regard to the response of the GIM to the addition of the different strains tested in fecal cultures, in general we did not observe any major changes in the absolute levels of the microbial groups analyzed, with significant increases only detected for the species of the added probiotics. The sole exception was the microbial group Clostridium XIVa in NW individuals with the strain B. bifidum TMC3108, which promoted higher levels of this microbial group than the other strains. In contrast, B. bifidum TMC3115 and L. gasseri BM7/10 were able to reduce the levels of this microbial group when compared to the control cultures from NW subjects. Interestingly, the Lactobacillus/ Lacticaseibacillus strains tested led to higher levels of lactobacilli in cultures from NW than in those from OB subjects, whereas the contrary occurred for bifidobacteria. These differences are likely due to the distinct basal microbiota between groups. Indeed, when comparing lactobacilli and bifidobacteria levels between both groups of individuals, at time zero, we observed that the fecal cultures from OB subjects showed significantly higher levels of lactobacilli and lower of bifidobacteria (5.14 ± 1.07 and 6.44 ± 0.37 Log10 CFU/mL, respectively) than those from NW individuals (3.9 ± 0.75 and 6.91 ± 0.41 Log10 CFU/mL, respectively). Moreover, these observations suggest that, regardless of the strain, the genus to which the probiotic strain belongs may constitute a first choice for selecting the best probiotic for microbiota modulation in a certain target population, i.e., OB or NW. These different responses observed between fecal cultures from both population groups underline previous studies reporting that the composition of the basal microbiota conditions the response to the probiotic (Arboleya et al., 2013a; Maldonado-Gómez et al., 2016; Hou et al., 2020).
Regarding SCFA, we did not observe mayor differences in the response to the different probiotics between fecal cultures of both human groups. As expected in both cases, NW and OB, acetic acid was the main SCFA followed by propionic and butyric acids. All the Bifidobacterium strains tested led to greater increases in the total SCFA, and of acetic acid, than the strains of lactobacilli, with B. bifidum TMC3108, B. bifidum TMC3115 and B. longum IF14/11 being those promoting higher values.
Finally, we studied the interaction with intestinal epithelial cells of the CS or the isolated microbiotas from NW and OB individuals supplemented with probiotics. No differences among the probiotics tested was observed when the CS were assessed, whereas GIM added with the different strains showed clear differences among them. Lactobacilli showed lower AUC values than bifidobacteria. Interestingly in OB but not in NW subjects all tested strains were able to down-regulate the HT29 cells response to the basal microbiota of these subjects. This is interesting given that the response to the basal microbiota of OB individuals was higher when compared with that of NW subjects. Therefore, the addition of the probiotic strains was able to restore this elevated response observed in the OB group bringing this functional response back to the levels observed in NW subjects.
Our results from in vitro models, although performed with a low number of samples, underline the need for the study and selection of probiotics in a target-population specific manner. The effects of strains in the fecal microbiota of NW individuals may be different from those that occur in OB, as it is the case of the data reported here. This complexity is often not considered in models where the strains are studied in isolation, without taking into account the mediation of the surrounding microbiota in the final effect on the host. It is, thus, necessary to select microorganisms with large functional capacity, as a previous step to carrying out human studies that entail a high economic cost. To achieve this, affordable in vitro study models such as those used here are necessary allowing the identification of the strains of potential interest, such as B. bifidum TMC3115, for its application in a specific human population group such as severely obese subjects. Nevertheless, it has to be taken into consideration that any in vitro screening for potential probiotic strains will require of later human clinical trials with higher number of individuals to evidence efficacy.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by the Bioethical Committee of CSIC and from the Regional Ethics Committee for Clinical Research of the Principality of Asturias. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CR-G, FH, GH, AE, NS, and MG conceived and designed the study. AS and CM-F recruited the volunteers and obtained the samples. AN and PR-M conducted the research. AN, CR-G, SA, NS, and MG analyzed the data. MG wrote and prepared the original draft. AN, CR-G, and NS helped with the manuscript review. All authors have read and approved the final version of the manuscript.
Conflict of Interest
FH and GH were employed by the company Takanashi Milk Products Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as potential conflict of interest. The authors declare that this study received funding from Takanashi Milk products. The funder had the following involvement with the study: study design.
Acknowledgments
We wish to thank the volunteers participating in the study and the Scientific-Technical Service of IPLA-CSIC for carrying out the SCFA determination.
Footnotes
Funding. The present work was mainly financed by the project AGL2013-43770-R (receiving funds from the Ministry of Economy and Competitiveness—MINECO and the European Union FEDER funds), and partly by projects AGL2017-83653-R (AEI/FEDER, UE), and RTI2018-098288-B-I00 (MCIU/AEI/FEDER, UE) and by a contract with the company Takanashi Milk Products (Yokohama, Japan). AN was the recipient of a predoctoral contract granted by MINECO (reference BES-2014-068796). SA is currently financed by a postdoctoral Juan de la Cierva contract (Ministry of Science, Innovation and Universities, Spain, ref. IJCI-2017-32156) and NS is currently the recipient of a postdoctoral contract awarded by the Fundación para la Investigación y la Innovación Biosanitaria del Principado de Asturias (FINBA).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.630572/full#supplementary-material
References
- Al-Tamimi M. A. H. M., Palframan R. J., Cooper J. M., Gibson G. R., Rastall R. A. (2006). In vitro fermentation of sugar beet arabinan and arabino-oligosaccharides by the human gut microflora. J. Appl. Microbiol. 100 407–414. 10.1111/j.1365-2672.2005.02780.x [DOI] [PubMed] [Google Scholar]
- Arboleya S., Bahrami B., Macfarlane S., Gueimonde M., Macfarlane G. T., de los Reyes-Gavilán C. G. (2015). Production of immune response mediators by HT-29 intestinal cell-lines in the presence of bifidobacterium -treated infant microbiota. Benef. Microbes 6 543–552. 10.3920/BM2014.0111 [DOI] [PubMed] [Google Scholar]
- Arboleya S., Salazar N., Solís G., Fernández N., Gueimonde M., de los Reyes-Gavilán C. G. (2013a). In vitro evaluation of the impact of human background microbiota on the response to Bifidobacterium strains and fructo-oligosaccharides. Br. J. Nutr. 110 2030–2036. 10.1017/S0007114513001487 [DOI] [PubMed] [Google Scholar]
- Arboleya S., Salazar N., Solís G., Fernández N., Hernández-Barranco A. M., Cuesta I., et al. (2013b). Assessment of intestinal microbiota modulation ability of Bifidobacterium strains in in vitro fecal batch cultures from preterm neonates. Anaerobe 19 9–16. 10.1016/j.anaerobe.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Arboleya S., Saturio S., Suárez M., Fernández N., Mancabelli L., de los Reyes-Gavilán C. G., et al. (2020). Donated human milk as a determinant factor for the gut bifidobacterial ecology in premature babies. Microorganisms 8 760. 10.3390/microorganisms8050760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J., Prifti E., Belda E., Ichou F., Kayser B. D., Dao M. C., et al. (2019). Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut 68 70–82. 10.1136/gutjnl-2018-316103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajury D. M., Nashri S. M., King Jie Hung P., Sarbini S. R. (2018). Evaluation of potential prebiotics: a review. Food Rev. Int. 34 639–664. 10.1080/87559129.2017.1373287 [DOI] [Google Scholar]
- Cani P. D. (2019). Severe obesity and gut microbiota: does bariatric surgery really reset the system? Gut 68 5–6. 10.1136/gutjnl-2018-316815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C., Gibbons S. M., Gurry T., Irizarry R. A., Alm E. J. (2017). Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 8 1784. 10.1038/s41467-017-01973-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A. C., Hoffmann C., Mota J. F. (2018). The human gut microbiota: metabolism and perspective in obesity. Gut Microbes 9 308–325. 10.1080/19490976.2018.1465157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grześkowiak Ł, Grönlund M.-M., Beckmann C., Salminen S., von Berg A., Isolauri E. (2012). The impact of perinatal probiotic intervention on gut microbiota: double-blind placebo-controlled trials in Finland and Germany. Anaerobe 18 7–13. 10.1016/j.anaerobe.2011.09.006 [DOI] [PubMed] [Google Scholar]
- Hill C., Guarner F., Reid G., Gibson G. R., Merenstein D. J., Pot B., et al. (2014). The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11 506–514. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- Hou Q., Zhao F., Liu W., Lv W., Khine W. W. T., Han J., et al. (2020). Probiotic-directed modulation of gut microbiota is basal microbiome dependent. Gut Microbes 12:e1736974. 10.1080/19490976.2020.1736974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. N., Yao Y., Ju S. Y. (2019). Short chain fatty acids and fecal microbiota abundance in humans with obesity: a systematic review and meta-analysis. Nutrients 11:2512. 10.3390/nu11102512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Gómez M. X., Martínez I., Bottacini F., O’Callaghan A., Ventura M., van Sinderen D., et al. (2016). Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 20 515–526. 10.1016/j.chom.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Merenstein D. J., Sanders M. E., Tancredi D. J. (2020). Probiotics as a Tx resource in primary care. J. Fam. Prac. 69 E1–E10. [PubMed] [Google Scholar]
- Nogacka A. M., Ruas-Madiedo P., Gómez E., Solís G., Fernández N., Suárez M., et al. (2018). Real-time monitoring of HT29 epithelial cells as an in vitro model for assessing functional differences among intestinal microbiotas from different human population groups. J. Microbiol. Methods 152 210–216. 10.1016/j.mimet.2018.07.003 [DOI] [PubMed] [Google Scholar]
- Nogacka A. M., Salazar N., Arboleya S., Ruas-Madiedo P., Mancabelli L., Suarez A., et al. (2020b). In vitro evaluation of different prebiotics on the modulation of gut microbiota composition and function in morbid obese and normal-weight subjects. Int. J. Mol. Sci. 21:906. 10.3390/ijms21030906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogacka A. M. G., de los Reyes-Gavilán C., Martínez-Faedo C., Ruas-Madiedo P., Suarez A., Mancabelli L., et al. (2020a). Impact of extreme obesity and diet−induced weight loss on the fecal metabolome and gut microbiota. Mol. Nutr. Food Res. 10.1002/mnfr.202000030 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Richards A. L., Muehlbauer A. L., Alazizi A., Burns M. B., Findley A., Messina F., et al. (2019). Gut microbiota has a widespread and modifiable Effect on host gene regulation. MSystems 4 e323–e318. 10.1128/mSystems.00323-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos-Covián D., Ruas-Madiedo P., Margolles A., Gueimonde M., de los Reyes-Gavilán C. G., Salazar N. (2016). Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7:185. 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberfroid M., Gibson G. R., Hoyles L., McCartney A. L., Rastall R., Rowland I., et al. (2010). Prebiotic effects: metabolic and health benefits. Br. J. Nutr. 104 S1–S63. 10.1017/S0007114510003363 [DOI] [PubMed] [Google Scholar]
- Rotbart A., Moate P. J., Yao C. K., Ou J. Z., Kalantar-Zadeh K. (2018). A novel mathematical model for the dynamic assessment of gas composition and production in closed or vented fermentation systems. Sens. Actuators B. 254 354–362. 10.1016/j.snb.2017.07.066 [DOI] [Google Scholar]
- Salazar N., Dewulf E. M., Neyrinck A. M., Bindels L. B., Cani P. D., Mahillon J., et al. (2015). Inulin-type fructans modulate intestinal bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin. Nutr. 34 501–507. 10.1016/j.clnu.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Sanders M. E., Merenstein D. J., Reid G., Gibson G. R., Rastall R. A. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16 605–616. 10.1038/s41575-019-0173-3 [DOI] [PubMed] [Google Scholar]
- Sarbini S. R., Rastall R. A. (2011). Prebiotics: metabolism, structure, and function. Funct. Food Rev. 3 93–106. 10.2310/6180.2011.00004 [DOI] [Google Scholar]
- Shen Q., Tuohy K. M., Gibson G. R., Ward R. E. (2011). In vitro measurement of the impact of human milk oligosaccharides on the faecal microbiota of weaned formula-fed infants compared to a mixture of prebiotic fructooligosaccharides and galactooligosaccharides. Lett. Appl. Microbiol. 52 337–343. 10.1111/j.1472-765X.2011.03005.x [DOI] [PubMed] [Google Scholar]
- Thursby E., Juge N. (2017). Introduction to the human gut microbiota. Biochem. J. 474 1823–1836. 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés L., Gueimonde M., Ruas-Madiedo P. (2015). Monitoring in real time the cytotoxic effect of Clostridium difficile upon the intestinal epithelial cell line HT29. J. Microbiol. Methods 119 66–73. 10.1016/j.mimet.2015.09.022 [DOI] [PubMed] [Google Scholar]
- Valdés L., Salazar N., González S., Arboleya S., Ríos-Covián D., Genovés S., et al. (2017). Selection of potential probiotic bifidobacteria and prebiotics for elderly by using in vitro faecal batch cultures. Eur. Food Res. Technol. 243 157–165. 10.1007/s00217-016-2732-y [DOI] [Google Scholar]
- Vallianou N., Stratigou T., Christodoulatos G. S., Dalamaga M. (2019). Understanding the role of the gut microbiome and microbial metabolites in obesity and obesity-associated metabolic disorders: current evidence and perspectives. Curr. Obes. Rep. 8 317–332. 10.1007/s13679-019-00352-2 [DOI] [PubMed] [Google Scholar]
- van Baarlen P., Troost F., van der Meer C., Hooiveld G., Boekschoten M., Brummer R. J. M., et al. (2011). Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. USA. 108 4562–4569. 10.1073/pnas.1000079107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinderola G., Gueimonde M., Gomez-Gallego C., Delfederico L., Salminen S. (2017). Correlation between in vitro and in vivo assays in selection of probiotics from traditional species of bacteria. Trends Food Sci. Technol. 68 83–90. 10.1016/j.tifs.2017.08.005 [DOI] [Google Scholar]
- von Martels J. Z. H., Sadaghian Sadabad M., Bourgonje A. R., Blokzijl T., Dijkstra G., Faber K. N., et al. (2017). The role of gut microbiota in health and disease: in vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 44 3–12. 10.1016/j.anaerobe.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Ware A., Power N. (2017). Modelling methane production kinetics of complex poultry slaughterhouse wastes using sigmoidal growth functions. Renew. Energy 104 50–59. 10.1016/j.renene.2016.11.045 [DOI] [Google Scholar]
- Williams C. F., Walton G. E., Jiang L., Plummer S., Garaiova I., Gibson G. R. (2015). Comparative analysis of intestinal tract models. Ann. Rev. Food Sci. Technol. 6 329–350. 10.1146/annurev-food-022814-015429 [DOI] [PubMed] [Google Scholar]
- Yao C. K., Rotbart A., Ou J. Z., Kalantar-Zadeh K., Muir J. G., Gibson P. R. (2018). Modulation of colonic hydrogen sulfide production by diet and mesalazine utilizing a novel gas-profiling technology. Gut Microbes 9 510–522. 10.1080/19490976.2018.1451280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wittouck S., Salvetti E., Franz C. M. A. P., Harris H. M. B., Mattarelli P., et al. (2020). Taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactibacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70 2782–2858. 10.1099/ijsem.0.004107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.