Abstract
Activated osteoclasts release large amounts of small extracellular vesicles (sEVs) during bone remodeling. However, little is known about whether osteoclast-derived sEVs affect surrounding cells. In this study, osteoclasts were generated by stimulating bone marrow macrophages (BMMs) with macrophage colony stimulating factor (M-CSF) and receptor activator of nuclear actor κB ligand (RANKL). We performed microarray analysis of sEV-microRNAs (miRNAs)s secreted from osteoclast at different stages and identified four miRNAs that were highly expressed in mature osteoclast-derived sEVs. One of these miRNAs, miR-324, significantly induced osteogenic differentiation and mineralization of primary mesenchymal stem cells (MSCs) in vitro by targeting ARHGAP1, a negative regulator of osteogenic differentiation. We next fabricated an sEV-modified scaffold by coating decalcified bone matrix (DBM) with osteoclast-derived sEVs, and the pro-osteogenic regeneration activities of the sEV-modified scaffold were validated in a mouse calvarial defect model. Notably, miR-324-enriched sEV-modified scaffold showed the highest capacity on bone regeneration, whereas inhibition of miR-324 in sEVs abrogated these effects. Taken together, our findings suggest that miR-324-contained sEVs released from mature osteoclast play an essential role in the regulation of osteogenic differentiation and potentially bridge the coupling between osteoclasts and MSCs.
Keywords: extracellular vesicles, osteoclast, osteogenic differentiation
Graphical Abstract
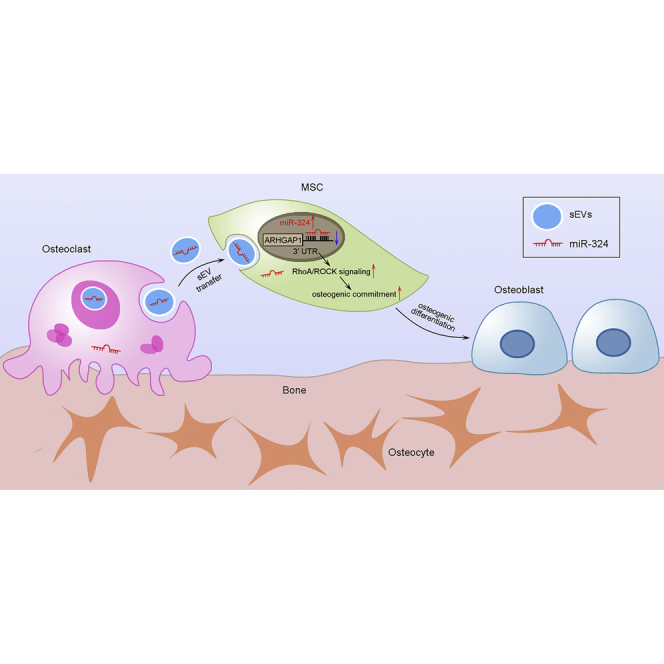
Liang et al. describe a novel mode of communication between osteoclasts and mesenchymal stem cells during bone remodeling. Specifically, miR-324-enriched sEVs derived from osteoclasts promote osteogenic differentiation via ARHGAP1/RhoA/ROCK signaling.
Introduction
Bone is a dynamic organ that undergoes constant turnover throughout the human lifespan. Bone remodeling involves the removal of old bone tissue by osteoclast bone resorption, followed by the formation of bone matrix via osteogenic differentiation that subsequently becomes mineralized.1 Bone remodeling is accomplished by basic multicellular units (BMUs), the discrete temporary anatomic structures assembled by osteoblasts and osteoclasts.2 The bone-resorbing osteoclasts play essential roles in physiological bone remodeling. Abnormal osteoclast functions lead to pathological bone disorders such as rheumatoid arthritis, postmenopausal osteoporosis, and cancer-induced osteolysis.3, 4, 5 Two essential factors, RANKL (receptor activator of nuclear actor κB ligand) and M-CSF (macrophage colony stimulating factor), are responsible for osteoclast differentiation.6, 7, 8 The intercellular communication between osteoclasts and osteoblasts is termed “coupling.”9 The coupling signals transmitted from osteoclasts to osteoblasts promote the transition from bone resorption to bone formation.10,11 Earlier studies reported that osteoblasts can regulate osteoclast differentiation through tight interplay between RANKL and osteoprotegerin, which are secreted from osteoblasts.12 Conversely, osteoclast-derived RANK also regulates osteogenesis and couples bone resorption and bone formation.13,14
MicroRNAs (miRNAs) are small non-coding RNAs that can suppress transcription activity of target mRNAs through binding to their 3′ UTR.15 In bone remodeling, miRNAs regulate the differentiation of osteoblasts and osteoclasts, as well as the orchestration of bone homeostasis.16 Recent studies demonstrated that miRNAs exist in the extracellular space, which are protected from RNase degradation mainly due to their encapsulation in extracellular vesicles (EVs).17,18 EVs are divided into small EVs (sEVs, <200 nm) and large EVs (lEVs), with specific lipid bilayer membrane structure, and they contain proteins, lipids, and a variety of RNAs such as messenger RNA (mRNAs) and miRNAs.19 Cargos of sEVs can be transferred from parental cells to recipient cells by direct fusion with the cell membrane or endocytosis to serve as essential mediators of intercellular communication.20 sEV-mediated transfer of miRNAs is widely considered to be involved in a variety of physiological and pathological conditions such as tumor development, immune response, and angiogenesis.21,22 In bone, sEVs are also considered to be essential instruments for signal transduction between bone cells. Osteoclast-derived sEVs induce the differentiation of pre-osteoblasts via RANKL reverse signaling, whereas sEVs from osteoblasts are also shown to play regulatory roles in osteogenesis.23,24
In this study, we reveal an interaction pattern between osteoclasts and mesenchymal stem cells (MSCs). Osteoclast-derived miR-324 can be transferred to MSCs and thereby promotes osteogenic differentiation through targeting ARHGAP1, a negative regulator of osteogenic differentiation and mineralization. This interplay between osteoclasts and MSCs was further confirmed in vivo.
Results
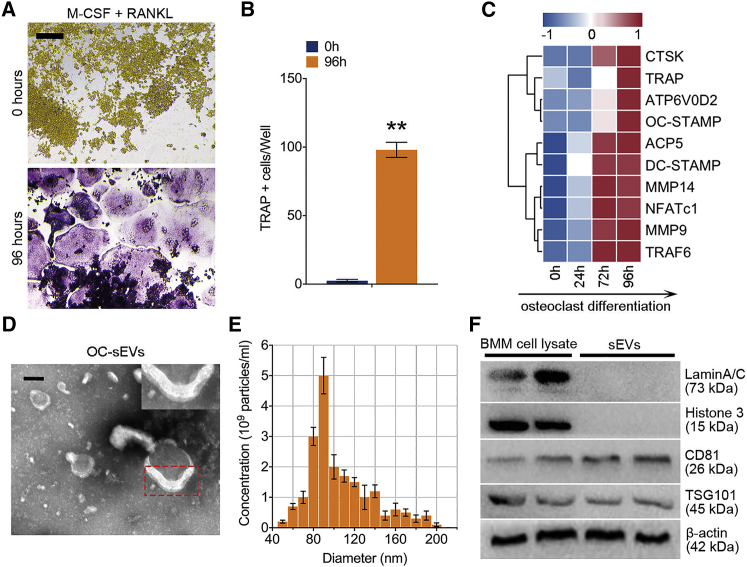
Characterization of osteoclast-derived sEVs
To investigate the sEVs derived from mature osteoclasts, bone marrow macrophages (BMMs) were isolated from 11-week-old male C57BL/6 mouse hindlimbs. BMMs were stimulated with M-CSF and RANKL for 96 h to generate mature osteoclasts. Tartrate-resistant acid phosphatase (TRAP) stain was performed, and the number of TRAP+ multinucleated cells was quantified to validate the maturation of osteoclasts (Figures 1A and 1B). The gene array analysis further revealed that the expression of osteoclast-specific genes was significantly upregulated with the increasing induction time (Figure 1C). Osteoclast-derived sEVs (OC-sEVs) were isolated through a series of microfiltration and ultracentrifugation steps. Transmission electron microscopy (TEM) revealed the presence of round-shaped vesicles surrounded by lipid bilayer membranes, the typical characteristic of sEVs (Figure 1D). Nanoparticle tracking analysis showed that most vesicles ranged from 70 to 140 nm in diameter with a peak at 90 nm (Figure 1E). sEVs were enriched with the pan-EV markers such as CD81 and TSG101 and the absence of nuclear proteins such as Lamin A/C and histone 3 (Figure 1F).
Figure 1.
Characterization of osteoclast-derived sEVs
(A) Representative images of TRAP staining of BMMs stimulated with RANKL (100 ng/mL) and M-CSF (50 ng/mL) for 96 h. Scale bar represents 200 μm. (B) Quantification analysis of the proportion of TRAP-positive cells in each well (96-well plate); n = 5. (C) A clustering heatmap shows the expression profile of RANKL-dependent specific genes of osteoclastogenesis from 0 to 96 h. (D) Transmission electron microscopy of osteoclast-derived sEVs (OC-sEVs). Red dotted box shows the lipid bilayer membrane structure of sEVs. Scale bar represents 50 nm. (E) Nanoparticle tracking analysis showed that most OC-sEVs ranged from 70 to 140 nm in diameter with a peak at 90 nm. (F) western blot analysis showed the protein levels of Lamin A/C, histone 3, CD81, TSG101, and β-actin in BMM cell lysate and extracted OC-sEVs. Data in the figures represent the averages ± SD. ∗∗p < 0.01, for differences between the treatment and control groups.
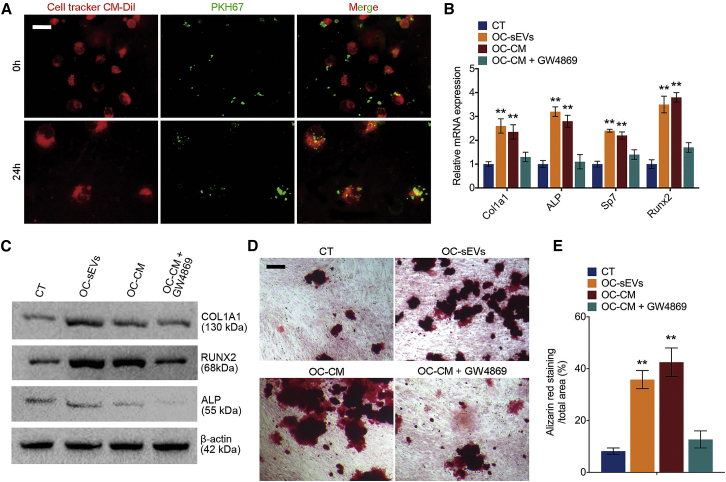
Osteoclast-derived sEVs promote osteogenic differentiation and mineralization
To explore the effect of OC-sEVs on osteogenic differentiation, we first investigated the engulfment of OC-sEVs by recipient cells. Primary MSCs labeled with CellTracker CM-Dil dye were cultured with PKH67-labeled OC-sEVs. Confocal microscopy was used to analyze the co-incubation of sEVs and recipient MSCs, and the results showed that the sEVs can be engulfed by the recipient MSCs over time (Figure 2A). After MSCs were cultured with sEVs for 7 days, qPCR was performed to detect the expression of osteogenic markers. The results revealed a significant upregulation of osteogenic regulators Runt-related transcription factor 2 (Runx2) and Osterix (OSX, also known as Sp7), as well as osteogenic markers type I collagen (Col1a1) and Alpl in OC-sEV-cultured MSCs (Figure 2B). On the protein level, western blot analysis also showed that OC-sEV culturing significantly upregulated the expression of ALP, COL1A1, and RUNX2 in MSCs (Figure 2C). GW4869 is a potent neutral sphingomyelinases inhibitor, which prevents the formation of intraluminal vesicles to further block sEV production and release in numerous cell types.25 Osteoclastic conditioned medium (OC-CM) with or without GW4869 pretreatment of cells was used to culture MSCs, subsequently followed by detection of their osteogenic potency. An ELISA assay was performed to detect the interleukin (IL)-6, transforming growth factor (TGF)-β1, and BMP2 concentrations of supernatants from osteoclasts with or without GW4869 pretreatment. We discovered that the concentration of IL-6, TGF-β1, and BMP2 did not significantly change after GW4869 treatment (Figure S1). Intriguingly, OC-CM significantly induced osteogenic differentiation characterized by upregulation of osteogenic markers. However, pretreatment with GW4869 abolished the facilitating effect of OC-CM on osteogenic differentiation, suggesting that OC-sEVs in OC-CM play a dominant role in this process (Figures 2B and 2C). To examine the effect of OC-sEVs on osteogenic mineralization, alizarin red staining was performed after culturing MSCs for 21 days with OC-sEVs (Figure 2D). The results showed that both OC-sEVs and OC-CM treatment significantly promoted the mineralization of MSCs, while GW4869 pretreatment abolished the effect of OC-CM on inducing mineralization (Figures 2D and 2E). Taken together, these results suggested that OC-sEVs promote the expression of osteogenic markers and facilitate mineralization.
Figure 2.
Osteoclast-derived sEVs promote osteogenic differentiation and mineralization
(A) Representative confocal microscopy images show MSCs labeled with CellTracker CM-Dil dye that were cultured with PKH67-labeled sEVs for 24 h. Scale bar represents 10 μm. (B) Relative mRNA expression levels of Col1a1, Alpl, Sp7, and Runx2 in MSCs cultured with OC-sEVs or osteoclast conditioned medium (OC-CM), with or without GW4869 pretreatment; n = 3. (C) Western blot analysis of COL1A1, RUNX2, ALP, and β-actin in indicated groups. (D) Representative images of alizarin red staining of MSCs cultured with OC-sEVs or OC-CM. Scale bar represents 100 μm. (E) Quantification analysis of calcium deposit of MSCs in indicated groups; n = 5. The data in the figures represent the averages ± SD. ∗∗p < 0.01, for differences between the treatment and control groups.
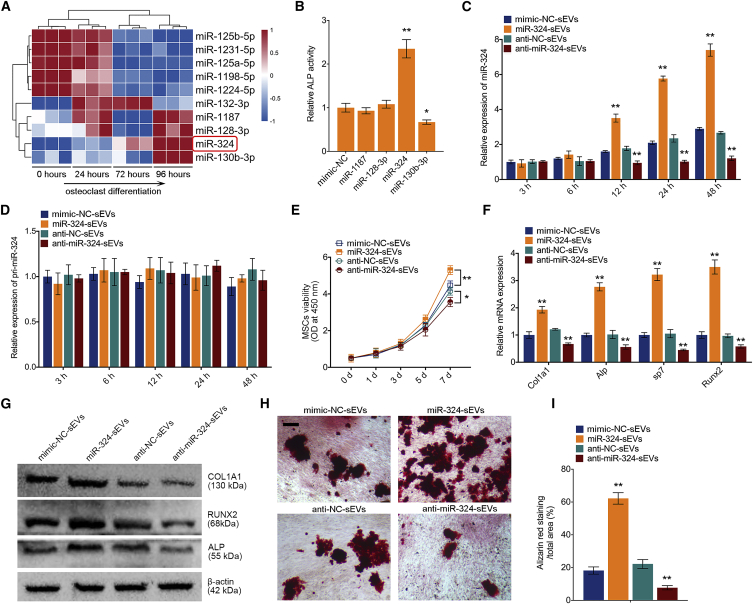
sEV-miR-324 derived from osteoclasts can be delivered into MSCs and induce osteogenic differentiation
Since miRNAs released from sEVs are demonstrated to play an important role in intercellular communications, we speculated that miRNAs contained in OC-sEVs may play a role in affecting osteogenic differentiation. To explore this idea further, we first examined the expression profile of miRNAs in sEVs secreted during osteoclastogenesis. Total RNAs, including miRNAs, were extracted from isolated sEVs and subjected to miRNA microarray analysis. The comprehensive microarray analysis revealed that miR-1187, miR-128-3p, miR-324, and miR-130b-3p were highly expressed in OC-sEVs (Figure 3A). To examine the effect of miRNAs on osteogenic differentiation, each miRNA mimic was subsequently transfected into MSCs, and cells were cultured using osteogenic medium. An alkaline phosphatase (ALP) assay was performed after 14-day osteogenic induction, and results revealed that overexpression of miR-324 significantly induced the osteogenic differentiation characterized by increasing ALP activity, whereas overexpression of miR-130b-3p adversely reduced ALP activity (Figure 3B). To further confirm the effect of miR-324 of sEVs on osteogenic differentiation, miR-324 was knocked down or overexpressed in BMMs and RAW264.7 cells by transfection. Transfection efficiency was confirmed by qPCR (Figure S2A). Knockdown or overexpression of miR-324 in parental cells resulted in a significant reduction or upregulation of miR-324 in respective sEVs (Figure S2B). Additionally, qPCR analysis showed that miR-324 in lipid-bilayer sEVs was protected from RNase degradation (Figure S2C). Next, sEVs from miR-324 knockdown (anti-miR-324-sEVs) or overexpression (miR-324-sEVs) osteoclasts were added into the osteogenic medium of MSCs. Notably, miR-324, but not primary miR-324 (pri-miR-324), was markedly upregulated in MSCs cultured with miR-324-sEVs (Figures 3C and 3C). The effect of miR-324 in sEVs on the recipient MSC cell viability was also detected by a Cell Counting Kit-8 (CCK-8) assay. Results showed that miR-324-sEVs significantly increased cell viability, whereas treatment with anti-miR-324-sEVs decreased cell viability (Figure 3E). For evaluation of osteogenic markers, qPCR analysis showed that miR-324-sEVs significantly upregulated the expression of osteogenic markers, while treatment with anti-miR-324-sEVs had adverse effects (Figure 3F). On the protein level, western blot analysis confirmed the upregulation of osteogenic markers by miR-324-sEV treatment in MSCs (Figure 3G). Similarly, alizarin red staining revealed that miR-324-sEVs significantly increased osteogenic mineralization compared with mimic-negative control (NC)-sEVs, whereas anti-miR-324-sEVs decreased osteogenic mineralization compared with anti-NC-sEV treatment (Figures 3H and 3I). These data suggested that osteoclast-derived sEV-miR-324 promotes osteogenic differentiation of MSCs.
Figure 3.
sEV-miR-324 derived from osteoclasts can be delivered into MSCs and induce osteogenic differentiation
(A) The expression profile of miRNAs in sEVs secreted during osteoclast differentiation. sEVs from BMMs were used as a normalization control. Red color represents higher expression, and blue color represents lower expression relative to the control. (B) ALP activity assay shows that overexpression of miR-324 promoted osteogenic differentiation; n = 3. (C and D) Relative expression levels of (C) miR-324 and (D) pri-miR-324 in MSCs cultured with sEVs from miR-324 overexpression (miR-324-sEVs) or knockdown (anti-miR-324-sEVs) osteoclasts; n = 3. (E) Cell viability evaluation of MSCs cultured with miR-324-sEVs or anti-miR-324-sEVs using a CCK-8 test at 1, 3, 5, and 7 days; n = 5. (F) Relative mRNA expression levels of Col1a1, Alpl, Sp7, and Runx2 in MSCs cultured with miR-324-sEVs or anti-miR-324-sEVs; n = 3. (G) Western blot analysis of COL1A1, RUNX2, ALP, and β-actin in MSCs cultured with miR-324-sEVs or anti-miR-324-sEVs. (H) Representative images of alizarin red staining of MSCs cultured with miR-324-sEVs or anti-miR-324-sEVs. Scale bar represents 100 μm. (I) Quantification analysis of calcium deposit of MSCs in indicated groups; n = 5. Data in the figures represent the averages ± SD. ∗p < 0.05, ∗∗p < 0.01, for differences between the treatment and control groups.
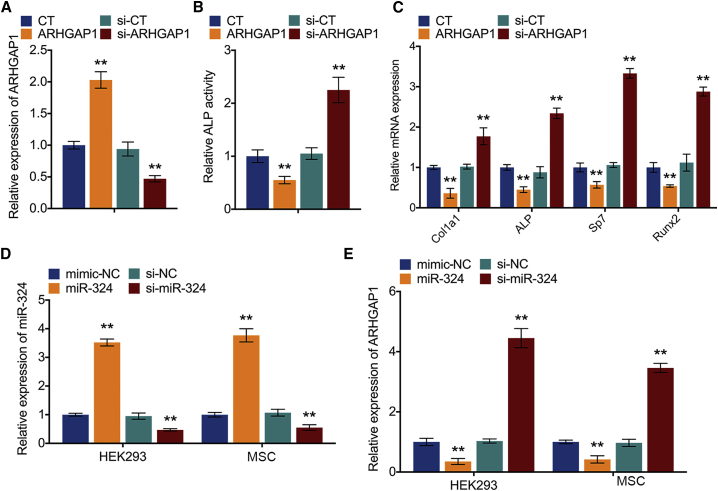
miR-324 directly binds to ARHGAP1 and induces osteogenic differentiation
To explore the target genes of miR-324 to regulate osteogenic differentiation, four mRNA target-predicting algorithms (miRanda, miRDB, miRWalk, and TargetScan) were utilized to identify the potential downstream targets of miR-324. Among the potential targets, ARHGAP1 was overlapped among all databases. It has been reported that ARHGAP1 served as a non-canonical negative regulator for osteogenic differentiation of human MSCs and was regulated by prostate cancer-derived miR-940.26 In this study, the expression of ARHGAP1 was also knocked down or overexpressed in MSCs (Figure 4A). After 14-day osteogenic induction, the knockdown of ARHGAP1 increased ALP activity of MSCs whereas overexpression of ARHGAP1 had an adverse effect (Figure 4B). Additionally, qPCR analysis showed that knockdown of ARHGAP1 increased, whereas overexpression of ARHGAP1 decreased, expression of osteogenic markers in MSCs (Figure 4C). These results suggested that ARHGAP1 is a negative regulator for osteogenic differentiation of MSCs. In addition, miR-324 was also knocked down or overexpressed in MSCs by transfection with mimics and inhibitor, respectively (Figure 4D). qPCR analysis showed that overexpression of miR-324 significantly upregulated, whereas knockdown of miR-324 (anti-miR-324) downregulated, the expression osteogenic markers (Figure S3A). To establish whether ARHGAP1 was a target of miR-324, luciferase reporter plasmid containing the wild-type 3′ UTR of ARHGAP1 was generated and cotransfected with miR-324 mimics, while Renilla luciferase plasmid was used for normalization (Figure S3B). Notably, the luciferase activities of the 3′ UTR of ARHGAP1 were suppressed by miR-324 (Figure S3C). Moreover, overexpression of miR-324 in MSCs suppressed the expression of ARHGAP1, whereas knockdown of miR-324 resulted in upregulation of ARHGAP1 (Figure 4E). These results indicate that miR-324 regulates osteogenic differentiation through silencing ARHGAP1 in MSCs.
Figure 4.
miR-324 directly binds to ARHGAP1 and induces osteogenic differentiation
(A) Relative mRNA expression levels of ARHGAP1 in MSC knockdown or overexpressed ARHGAP1; n = 3. CT represents empty control group without any treatment, and si-CT represents the random non-specific siRNA used for negative control. (B) Relative ALP activity of MSC knockdown or overexpressed ARHGAP1 after osteogenic induction for 14 days; n = 3. (C) Relative mRNA expression levels of Col1a1, Alpl, Sp7, and Runx2 in MSC knockdown or overexpressed ARHGAP1; n = 3. (D) Relative expression levels of miR-324 in MSC knockdown or overexpressed miR-324; n = 3. (E) Relative expression levels of ARHGAP1 in MSC knockdown or overexpressed miR-324; n = 3. Data in the figures represent the averages ± SD. ∗∗p < 0.01, for differences between the treatment and control groups.
Osteoclast-secreted miR-324 silences ARHGAP1 during osteogenic differentiation
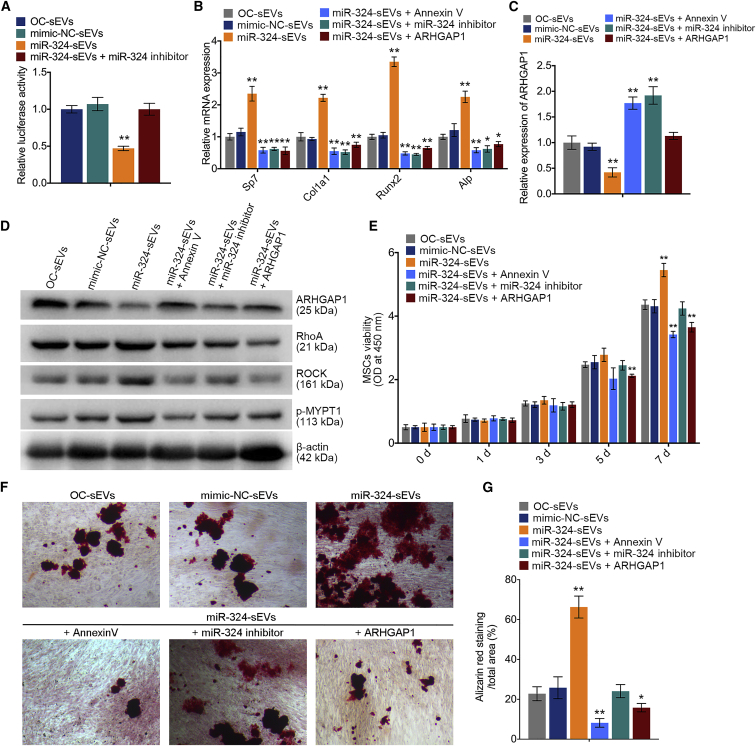
We then examined the luciferase activities of the ARHGAP1 3′ UTR in MSCs cultured with miR-324-sEVs. Notably, the luciferase activities of the ARHGAP1 3′ UTR were markedly decreased by miR-324-sEVs compared with mimic-NC-sEVs, whereas application of miR-324 inhibitor abrogated these effects (Figure 5A). These results suggested that ARHGAP1 expression in MSCs can be suppressed by miR-324 of sEVs released from osteoclasts. Additionally, miR-324-sEVs dramatically decreased ARHGAP1 expression and increased the expression levels of osteogenic markers (Figures 5B and 5C). It has been suggested that ARHGAP1 negatively regulates the downstream RhoA/ROCK signaling to inhibit cytoskeletal tension and reorganization during osteoblast differentiation, which determines the osteogenic commitment of stem cell lineage.27 Notably, western blot analysis revealed that miR-324-sEVs markedly decreased the expression of ARHGAP1, while they increased the expression levels of downstream RhoA, ROCK, and the MYPT1 phosphorylation by ROCK kinase (Figure 5D). It has been reported that annexin V is an inhibitor of EV internalization.28 Treatment with annexin V or miR-324 inhibitor in miR-324-sEVs abolished their pro-osteogenic effects and suppression of ARHGAP1 (Figures 5B and 5C). Adenovirus overexpressed ARHGAP1 was designed and used to transfect MSCs, while these ARHGAP1-overexpresssing MSCs did not show a strong mineralization after culturing with miR-324-sEVs, suggesting that the positive effect of miR-324-sEVs on osteogenesis was abrogated by the restoration of ARHGAP1 (Figures 5B and 5C). Additionally, application of annexin V, miR-324-inhibitor, or restoration of the ARHGAP1 expression suppressed the positive regulation of miR-324-sEVs in the RhoA/ROCK pathway (Figure 5D). A CCK-8 assay was performed, and results showed that either treatment with annexin V or miR-324 inhibitor, or restoration of the expression of ARHGAP1, significantly reduced the recipient MSC viability (Figure 5E). Alizarin red staining and quantification analysis showed that transfection of miR-324-sEVs with miR-324 inhibitor or pretreatment of miR-324-sEVs with annexin V alleviated their function in facilitating osteogenic mineralization. Additionally, ARHGAP1 restoration in recipient MSCs suppressed these effects (Figures 5F and 5G). Taken together, the findings demonstrate that miR-324 of sEVs released from osteoclasts is sufficient to promote osteogenic differentiation by targeting ARHGAP1.
Figure 5.
Osteoclast-secreted miR-324 silences ARHGAP1 during osteogenic differentiation
(A) Relative luciferase activity of reporter containing the 3′ UTR of ARHGAP1 in MSCs upon culturing with OC-sEVs, miR-NC-sEVs, miR-324-sEVs, and miR-324-sEVs + miR-324 inhibitor; n = 5. (B) Relative mRNA expression levels of Col1a1, Alpl, Sp7, and Runx2 in MSCs cultured with OC-sEVs, miR-NC-sEVs, miR-324-sEVs, miR-324-sEVs + annexin V, miR-324-sEVs + miR-324 inhibitor, and miR-324-sEVs + ARHGAP1; n = 3. (C) Relative expression levels of ARHGAP1 in indicated groups; n = 3. (D) Western blot analysis of ARHGAP1, RhoA, ROCK, p-MYPT1, and β-actin expression in MSCs cultured with OC-sEVs, miR-NC-sEVs, miR-324-sEVs, miR-324-sEVs + annexin V, miR-324-sEVs + miR-324 inhibitor, and miR-324-sEVs + ARHGAP1; n = 3. (E) Cell viability evaluation of MSCs in indicated groups at 0, 1, 3, 5 and 7 days; n = 3. (F) Representative images of alizarin red staining of MSCs cultured with OC-sEVs, miR-NC-sEVs, miR-324-sEVs, miR-324-sEVs + annexin V, miR-324-sEVs + miR-324 inhibitor, and miR-324-sEVs + ARHGAP1. Scale bar represents 100 μm. (G) Quantification analysis of calcium deposit of MSCs in indicated groups; n = 5. The data in the figures represent the averages ± SD. ∗p < 0.05, ∗∗p < 0.01, for differences between the treatment and control groups.
miR-324 of sEVs released from osteoclasts facilitates bone defect healing in vivo
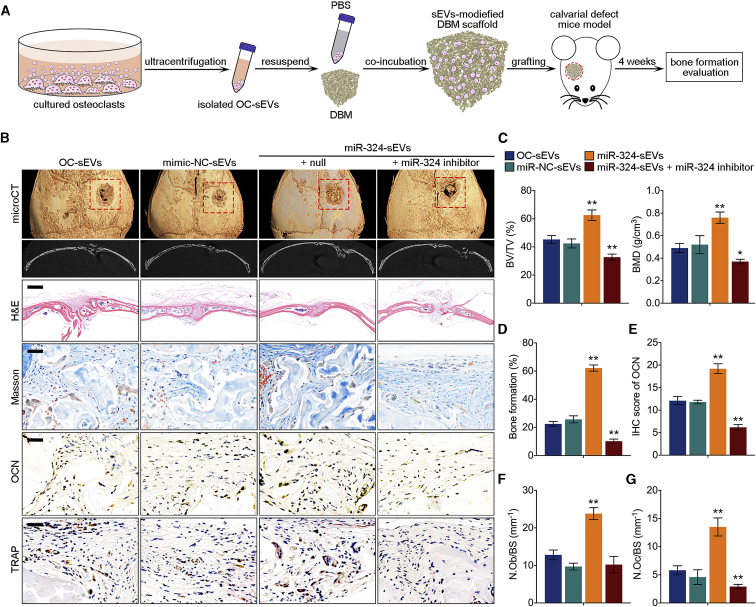
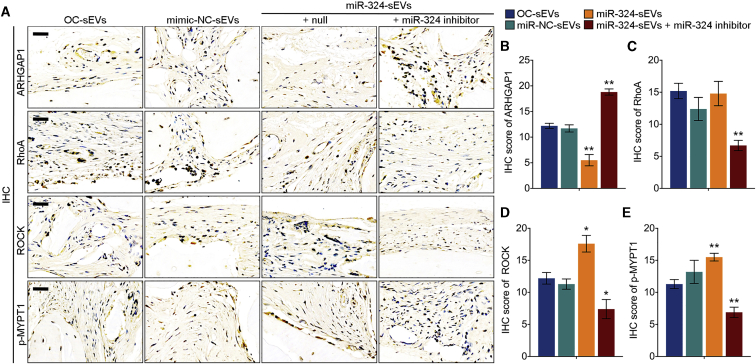
To explore the pro-osteogenic effects of miR-324 of sEVs in vivo, we fabricated a novel sEV-modified scaffold by coating decalcified bone matrix (DBM) with isolated sEVs, and the pro-osteogenic potential of sEV-modified scaffolds was evaluated using a mouse calvarial defect model generated by cranial drilling (Figure 6A). This mouse model is useful to evaluate the impacts of cell- or EV-modified scaffolds on osteogenesis.29,30 sEV-modified DBM was implanted in the defect area, followed by normal keeping for 4 weeks before euthanasia. To assess the bone regeneration after implantation of sEV-modified DBM, mouse calvarias in all groups were harvested and scanned by micro-computed tomography (micro-CT). The results revealed that DBM modified with miR-324-sEVs significantly increased bone regeneration, bone volume density (bone volume [BV]/total volume [TV]), and bone mineral density (BMD) compared with OC-sEV-modified DBM, whereas transfection of miR-324 inhibitor in miR-324-sEVs not only diminished the effect of miR-324-sEVs, but even reduced osteogenic activity of sEVs to a lower level compared with OC-sEVs (Figures 6B and 6C). qPCR analysis also revealed that compared with OC-sEVs, the miR-324 level in miR-324-sEVs was markedly reduced after transfection (Figure S4). We then performed histological analysis of the calvarial section. H&E staining displayed an overview of the defect repair area, while Masson staining revealed a significant increase of bone formation rate in mice grafted with DBM modified with miR-324-sEVs (Figures 6B and 6D). We also performed TRAP staining and immunohistochemistry (IHC) of osteoblast-specific marker osteocalcin (OCN) of the calvarial section. The results showed that mice grafted with miR-324-sEV-modified DBM showed a higher OCN-positive cell percentage, as well as the number of osteoblasts and osteoclasts, suggesting that miR-324-sEVs enhanced the bone remodeling of the cranial defect area (Figures 6B, 6F, and 6G). However, transfection of miR-324 inhibitor abrogated these effects characterized by decreased bone formation rate, IHC score of OCN, and the number of osteoblasts and osteoclasts (Figures 6B–6G). These data suggested the facilitating effects of miR-324 in sEVs on osteogenic differentiation and calvarial defect healing. To further confirm the mechanism of sEVs facilitating bone defect healing in vivo, we also detected the expression level of ARHGAP1 and the downstream RhoA/ROCK pathway in decalcified bone sections. Notably, mice grafted with miR-324-sEV-modified DBM showed a significantly lower expression of ARHGAP1 compared with OC-sEV-modified DBM, whereas transfection of miR-324 inhibitor rescued ARHGAP1 expression marked by the highest ARHGAP1 score (Figures 7A and 7B). Additionally, modification with miR-324-sEVs significantly decreased the expression levels of downstream RhoA, ROCK, and phosphorylated MYPT1 (p-MYPT1), while transfection of miR-324 inhibitor reversed these effects (Figures 7A and 7C–7E). Taken together, these data indicated that miR-324 of sEVs released from osteoclasts facilitated osteogenic differentiation in calvarial defect mice through regulating ARHGAP1/RhoA/ROCK signaling.
Figure 6.
miR-324 of sEVs released from osteoclast facilitates bone defect healing in vivo
(A) Schematic diagram shows the fabrication of sEV-modified scaffold and grafting of calvarial defect mice. (B) Representative general and coronal micro-CT images, H&E, Masson, and TRAP staining, and IHC of OCN in decalcified bone sections from mice treated with OC-sEVs, miR-NC-sEVs, miR-324-sEVs, and miR-324-sEVs + miR-324 inhibitor. Scale bars represents 2 mm in H&E staining, 50 μm in Masson, TRAP, and IHC staining. (C) Quantitative micro-CT analysis shows the bone volume density (BV/TV) and bone mineral density (BMD) of total defect repair area in indicated groups; n = 8. (D) Quantification analysis of bone formation ratio in indicated groups; n = 8. (E) Semiquantitative analysis of OCN in indicated groups; n = 8. (F and G) Quantitative analysis of the number of (F) osteoblasts (N.Ob) and (G) osteoclasts (N.Oc) on the cortical bone surface (BS) using IHC staining of OCN and TRAP staining in indicated groups. The data in the figures represent the averages ± SD. ∗p < 0.05, ∗∗p < 0.01, for differences between the treatment and control groups.
Figure 7.
miR-324 of sEVs released from osteoclast facilitates osteogenic through regulating ARHGAP1/RhoA/ROCK signaling
(A) Representative IHC staining images of ARHGAP1, RhoA, ROCK, and p-MYPT1 of mice in indicated groups. (B–E) Semiquantitative analysis of (B) ARHGAP1, (B) RhoA, (D) ROCK, and (E) p-MYPT1 in indicated groups; n = 8. The data in the figures represent the averages ± SD. ∗p < 0.05, ∗∗p < 0.01, for differences between the treatment and control groups.
Discussion
Bone remodeling is a programmed process that is precisely regulated by physiological coupling of osteoclasts and osteoblasts, so as to regenerate new bone to replace old bone at the relevant area of bone injury.31 Physiological bone turnover generally consists of three stages: (1) an osteoclast-mediated bone resorption phase for approximately 3 weeks; (2) an osteoblast-mediated bone formation phase for about 3–4 months; and (3) a 5-week reversal phase of transition from bone resorption to bone formation, which is almost unclear.32,33 The coupling between osteoclasts and osteoblasts may play a dominant role in this transition phase between bone resorption and formation.23 In addition to the in-depth studies of osteoblast-derived molecules regulating osteoclast differentiation and activity, growing numbers of reports have shown that osteoblast progenitors are also capable of sensing osteoclast-derived signals either secreted from osteoclasts or expressed on the cell membrane to promote osteogenic differentiation.34 However, it is still unclear whether and how osteoclast-derived sEVs play a role in the transition from bone resorption to bone formation in whole-bone turnover. Our study filled this gap by demonstrating the significant osteogenic ability of osteoclast-derived sEVs.
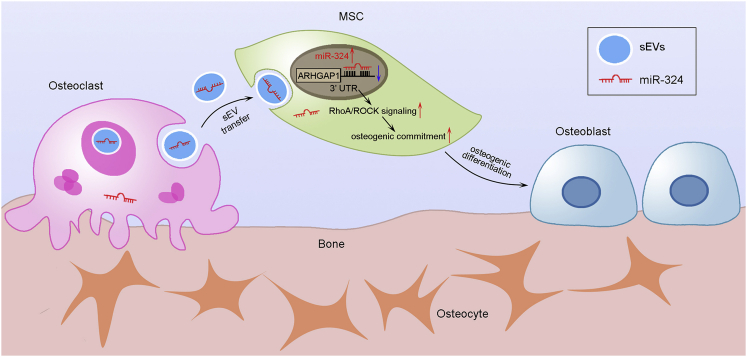
It is well known that osteocytes and osteoblasts are the predominant providers of RANKL and osteoprotegerin, and therefore are not only critical for regulation of osteoclast formation, but they simultaneously coordinate and maintain the balance between bone resorption and bone formation.12,35 However, the regulatory role of osteoclasts in these processes is still unclear. In this study, we performed a series of in vitro and in vivo studies to determine that miR-324 of sEVs released from osteoclasts could transfer to MSCs to promote osteogenic differentiation, proposing a novel mode of miRNA-mediated osteoclast-osteoblast coupling. Osteoclasts release large amounts of miR-324-enriched sEVs, and these sEVs can be internalized into MSCs to further promote their activity. Therefore, we speculate that active osteoclasts release large numbers of sEVs at the bone resorption phase, which in turn facilitate osteogenesis of MSCs to better stabilize and bridge the transition between bone resorption and formation (Figure 8).
Figure 8.
Schematic diagram shows that osteoclast-derived sEVs bridge the osteoclast-osteoblast coupling
In the bone resorption phase, active osteoclasts release large amounts of sEV-miR-324, which can be transferred into MSCs, and subsequently promote osteogenic commitment and differentiation via regulating the ARHGAP1/RhoA/ROCK axis.
sEVs regulate a series of intracellular or intercellular signal transductions in a paracrine or autocrine manner. Increasing evidence indicates that sEVs derived from bone cells contain important factors that may be involved in bone remodeling.36 For instance, osteoblast-derived sEVs contain specific osteogenic factors, including BMP1–BMP7, ALP, and non-collagenous matrix protein osteoprotegerin and OCN,37 whereas sEVs derived from osteoclasts contain osteoclast-associated proteins such as RANK and RANKL.38 Additionally, studies have also suggested that sEVs derived from bone cells have regulatory effects on bone remodeling through targeting adjacent cells. For instance, both osteoclast-derived exosomal miR-214-3p and osteocyte-derived exosomal miR-218 can regulate osteogenic differentiation.39,40 Furthermore, sEVs from MSCs have been shown to have the capacity to upregulate the expression of BMP9 and TGF-β1, both of which significantly promote osteogenic differentiation.41,42 Studies have shown that MSC-derived sEVs protect cartilage and bone from degradation in osteoarthritis.43 A recent study suggested that bone marrow-derived MSC (BMSC)-derived miR-151-5p targets adjacent BMSCs to rescue damaged osteogenic differentiation and decrease adipogenic differentiation.44 In bone remodeling, the delicate coupling of bone resorption and bone formation determines bone homeostasis. Studies have demonstrated that osteoclast bone resorption results in the release of TGF-β from bone matrix, leading to the recruitment of BMSCs for further osteogenesis.45 In our study, we demonstrated that osteoclast-derived sEVs promote osteogenesis via delivering miR-324 to recipient MSCs. Other than the well-studied osteoclast-osteoblast coupling, the involvement of osteoclast-derived sEVs further bridged the transition from bone resorption to formation in bone remodeling.
miRNAs also serve as essential intercellular communication tools, as horizontal transfer of miRNAs mediated by sEVs affects biological functions of recipient cells.46 A number of miRNAs have been identified in the regulation of osteoblast differentiation.47, 48, 49 These findings raise the possibility that osteogenic differentiation during bone remodeling can be regulated by miRNAs derived from bone cells. In our study, we identified that miR-324 was highly expressed in osteoclast-derived sEVs using miRNAs microarray analysis. Both in vitro and in vivo studies suggested that miR-324 significantly promoted osteogenic differentiation. So far, several studies have reported that miR-324 plays diverse roles in multiple pathological and physiological conditions, including tumor development, vascular remodeling, and neuropathic pain.50,51 However, the role of miR-324 in osteogenic differentiation and bone metabolism has not been reported. Our study uncovers the osteogenic potency of miR-324 of sEVs released from osteoclasts. In this study, ARHGAP1 was identified as target of miR-324 to regulate osteogenic differentiation. ARHGAP1 is a founding member of the RhoGAP family containing GTPase-activating proteins, which result in enhancement of intrinsic GTPase activity and inactivation of G protein.52 It has been suggested that ARHGAP1 regulates the epithelial-to-mesenchymal transition by inhibiting RhoA/ROCK signaling.53 RhoA/ROCK signaling is considered to be involved in the proliferation and differentiation of distinctive cell types, including osteogenic commitment and differentiation, since RhoA/ROCK pathway-mediated cytoskeletal tension and reorganization are essential for osteogenic commitment.27 A recent study has been reported that cancer-derived miR-940 induces osteoblastic destruction via targeting ARHGAP1 in human MSCs.26 In the present study, we also found that knockdown of ARHGAP1 in MSCs increased the expression of osteogenic markers and promoted osteogenic mineralization, which is consistent with previous studies. One step farther, our study suggested that this reverse effect that determined osteogenic commitment is also associated with activation of the RhoA/ROCK pathway.
In conclusion, our study suggested that miR-324 of sEVs released from osteoclasts promotes the osteogenic differentiation of MSCs in vitro and facilitates bone defect healing in vivo through ARHGAP1/RhoA/ROCK signaling. This study provides a novel mode of sEV-mediated osteoclast-MSC coupling.
Materials and methods
Reagents
Primary cultured BMMs were used to study osteoclast differentiation. Mouse bone marrow cells were isolated from 11-week-old male C57BL/6 mouse hindlimbs (femur and tibia) and incubated with M-CSF (50 ng/mL) for 96 h to obtain BMMs. The macrophage RAW264.7 cell line and human embryonic kidney 293 (HEK293) cell line were obtained from the American Type Culture Collection (Rockville, MD, USA). Cells were cultured with α-minimal essential medium (α-MEM; Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin-streptomycin (HyClone, Thermo Scientific, MA, USA) in a 37°C incubator containing 5% CO2-enriched atmosphere. Recombinant mouse RANKL and recombinant mouse M-CSF were purchased from R&D Systems (Minneapolis, MN, USA). A TRAP stain kit was obtained from Sigma-Aldrich (NY, USA). An alizarin red stain kit was obtained from Solarbio Life Sciences (Beijing, China). Antibodies against histone 3 (bs-17422R), CD81 (bs-2489R), TSG101 (bs-1365R), Lamin A/C (bs-1839R), COL1A1 (bs-10423R), RUNX2 (bs-20003R), ALP (bsm-52252R), and β-actin (bs-0061R) were all purchased from Bioss Antibodies (Beijing, China).
Osteoclast differentiation
For osteoclast differentiation, 5 × 103 BMMs were incubated in 96-well plates and cultured with osteoclast differentiation medium consisting of α-MEM supplemented with 10% FBS, 1% penicillin-streptomycin, 50 ng/mL M-CSF, and 100 ng/mL RANKL for 96 h to generate osteoclasts. For TRAP staining, cells were fixed in 4% paraformaldehyde for 20 min and then stained with TRAP staining solution (0.1 mg/mL naphthol phosphate disodium salt, 0.3 mg/mL fast red violet zinc chloride stain) according to the manufacturer’s instructions. Relative TRAP activity was analyzed by colorimetry.
Isolation of osteoclast-derived sEVs
Osteoclast-derived sEVs were collected by differential centrifugations as previously described with some modifications.54 Briefly, osteoclasts were cultured in sEV-free medium for 48 h before sEV isolation. The supernatant was collected and centrifuged at 2,000 × g for 15 min to remove dead cells and apoptotic bodies. Subsequently, the supernatant was centrifuged at 12,000 × g for 30 min to further remove microvesicles. sEVs were pelleted via ultracentrifugation at 100,000 × g for 70 min. The pellet was washed with 50 mL of PBS and ultracentrifuged again at 100,000 × g for 70 min. Finally, the sEVs were resuspended in PBS and set aside for further experiments. Ultracentrifugation operations were conducted using an Optima XE-90 (Beckman Coulter) at 4°C. The sEV concentration was estimated using an exosome counting system (Sysmex). The supernatant from 5 × 106 cells was collected and centrifuged, and the final number of sEVs separated from the supernatant was about 1011.
TEM
Purified sEVs were processed at 21°C for TEM analysis. In brief, sEVs were diluted with 1× PBS to 1:5 and then fixed in 2% paraformaldehyde. After fixation, the samples were applied to formvar copper grids and stained with uranyl acetate (Electron Microscopy Sciences) for 10 min at room temperature. Samples were naturally dried and observed in an FEI Tecnai 110-kV microscope at an accelerating voltage of 80 kV, and digital images were obtained.
miRNA microarray analysis
To perform microarray analysis, total RNA from the isolated sEVs during osteoclast differentiation was extracted using the miRNeasy mini kit (QIAGEN). sEVs from each differentiation stage have three different clones. An Agilent 2100 Bioanalyzer was used to examine the purity of the extracted RNAs. Microarray analysis was then performed using a mouse miRNA microarray kit (Agilent Technologies, v3); in this study, we used a sample input of 100 ng of each total RNA. Briefly, calf intestine phosphatase was used to dephosphorylate the samples. Then, T4 RNA ligase was used to label the 3′ end of the dephosphorylated sample with cyanine 3-cytidine biphosphate. The labeled miRNA was hybridized to microarray glass slides in a hybridization rotator with 20 rotations/min at 55°C for 20 h. After that, the glass was washed. Then, the glass was scanned with an Agilent microarray scanner (Agilent Technologies) according to the manufacturer’s instructions. Feature Extraction software v11.0.1.1 (Agilent Technologies) was used to extract the data. GeneSpring GX software (Agilent Technologies) was used for normalizing and analysis. Finally, R v3.6.0 software was used to plot heatmaps for visualizations of data.
Osteogenic differentiation
MSCs were generated from 11-week-old male C57BL/6 mice. Briefly, mice were sacrificed by cervical dislocation, and 75% alcohol was used for disinfection. Under sterilized conditions, the femora and tibia of mice were separated. Bone marrow cells were isolated by washing the marrow cavity with α-MEM until the cavity became white. Bone marrow cells were collected and screened with a 200-mesh sieve. Then, cells were centrifuged at 300 × g for 5min. The cell pellets were plated on culture dishes and MSCs were separated by plastic adherence. For osteogenic differentiation, MSCs were incubated (1 × 104 per well) in 24-well plates. Osteogenic induction was performed when cells reached 60% contact and stopped proliferating. Every 2 × 107 sEVs were added into 1 mL of osteogenic induction medium, and about 105 MSCs (per well of 24-well plates) were incubated with 0.5 mL of sEV medium containing 107 sEVs for 14 days (for ALP assay) and 21 days (for alizarin red stain). Osteogenic medium consisted of 2 mM β-glycerophosphate (Aladdin), 100 μM ascorbic acid 2-phosphate (Aladdin), and 10 nM dexamethasone (Aladdin). After 14 days for osteogenic induction, total RNA or protein was extracted from cultured cells and the expression of osteogenic markers was assessed by qPCR and western blot analysis. For detection of ALP activity, cells were fixed in 4% paraformaldehyde and then treated according to the protocol by an ALP assay kit (Beyotime Biotechnology) after 14 days of osteogenic induction. To assess the deposition of calcium, cells were fixed in 4% paraformaldehyde and then stained with 1% alizarin red-S (Solarbio Life Sciences) after 14 days of osteogenic induction.
Fabrication of sEV-modified DBM scaffolds
In our study, DBM was prepared from bovine limbs as previously reported.55 The DBM was cut into 2.5-mm cubes to fit the calvarial defect size. After being immersed in 75% ethanol for 2 h, the DBM was washed three times with PBS and then coated with 10 μg/mL fibronectin (Sigma) overnight at 37°C. As for incubation, 50 μg of isolated sEVs was resuspended in 50 μL of control medium, which was followed by the incubation of DBM with sEV-containing media for 6 h at 37°C. The DBM was then air-dried and stored at −80°C until they were used in experiments.
Mice
The bone regeneration potential of decalcified bone matrix pre-incubated with sEVs was evaluated using a critical size of cranial bone defects. The C57BL/6 mice aged 6–8 weeks were used. After mice were anesthetized, a sagittal median incision of 1.5 cm was made and a 2.5-mm-sized defect was created on the right side of the calvarial bone using a dental micro-drill. Afterward, the DBM pre-incubated with sEVs for 6 h was implanted into the bone defect area. After normal feeding of 4 weeks, mice were euthanized and the calvarial bones were removed and set aside for further detection. We maintained all animals in the Animal Facility of the Third Military Medical University. All experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the Third Military Medical University.
Histology and IHC evaluation
Calvarial bone was obtained from mice at the time of sacrifice, and bones were fixed in 10% neutral-buffered formalin, washed, and decalcified in a solution of 10% EDTA for 2 weeks, then embedded in paraffin. Histological sections (5 μm) were prepared for subsequent H&E staining. For immunohistochemical assessment, the expression of OCN and ARHGAP1 was detected according to the following procedure. Sections were first incubated for 12 h at 4°C with primary antibody for OCN (1:200) and ARHGAP1 (1:200). Then, biotinylated secondary antibody was used with the EnVision+ system horseradish peroxidase (HRP) kit (Dako, Sweden), and nuclei were counterstained with hematoxylin. Then, the IHC-stained cells were washed and observed using light microscopy. In this study, we used a German semiquantitative scoring system that considered the intensity and area of staining as follows: 0 for not staining, 1 for weak staining, 2 for moderate staining, and 3 for strong staining. Additionally, the staining percentage was given a score of 0 (<5%), 1 (5%–25%), 2 (25%–50%), 3 (51%–75%), or 4 (>75%). These two scores were multiplied as the final score.
Cell viability assay
We performed a CCK-8 assay to assess cell proliferation. Briefly, MSCs were seeded (2 × 103 per well) in 96-well plates overnight. Cells were cultured with sEVs for 7 days. According to the manufacturer’s instructions, cell viability was examined using a CCK-8 (Dojindo) at 1, 3, 5, and 7 days. Cell absorbance was measured by a 450-nm 96-hole plate reader and cell viability was evaluated.
Luciferase reporter assay
Cells were seeded in quintuplicate into 24-well plates (1 × 105 cells per well) and then cultured for 24 h. The constructed pGL3-basic luciferase reporter plasmid (1.5 μg, Promega) or the control luciferase plasmid (1.5 μg, Promega) was cotransfected into the cells with the pRL-SV40 plasmid (0.15 μg, Promega) using Lipofectamine 2000 reagent (Invitrogen). After transient transfection with constructed luciferase plasmids and miR-324 mimics for 48 h, luciferase and Renilla activities were detected with a Dual-Luciferase reporter assay system (Promega, USA) according to the manufacturer’s protocol.
Statistical analysis
All data are representative of at least three experiments of similar results performed in triplicate unless otherwise indicated. The data are presented as means ± standard deviation. Comparisons between two groups were analyzed using independent unpaired two-tailed Student’s t tests, and comparisons between more than two groups were analyzed using one-way ANOVA followed by Student-Newman-Keuls post hoc tests. The values were considered significant at p <0.05. Statistically significant differences between the treatment and control groups are indicated as ∗p < 0.05 or ∗∗p < 0.01.
Acknowledgments
This work was supported by the State Key Program of National Natural Science of China (no. 81930067).
Author contributions
Q.M. and S.D. designed the study. M.L., X.Y., and H.A. performed experiments. Q.M. and M.L. collected data and analyzed images. Q.M., M.L., and S.Z. bred and maintained the mice. Q.M., C.D., F.L., and J.X. drafted and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2021.01.031.
Contributor Information
Ce Dou, Email: lance.douce@gmail.com.
Shiwu Dong, Email: dongshiwu@tmmu.edu.cn.
Qinyu Ma, Email: mqy32473069@gmail.com.
Supplemental information
References
- 1.Hadjidakis D.J., Androulakis I.I. Bone remodeling. Ann. N Y Acad. Sci. 2006;1092:385–396. doi: 10.1196/annals.1365.035. [DOI] [PubMed] [Google Scholar]
- 2.Jilka R.L. Biology of the basic multicellular unit and the pathophysiology of osteoporosis. Med. Pediatr. Oncol. 2003;41:182–185. doi: 10.1002/mpo.10334. [DOI] [PubMed] [Google Scholar]
- 3.Steffen U., Schett G., Bozec A. How autoantibodies regulate osteoclast induced bone loss in rheumatoid arthritis. Front. Immunol. 2019;10:1483. doi: 10.3389/fimmu.2019.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang N., Agrawal A., Jørgensen N.R., Gartland A. P2X7 receptor regulates osteoclast function and bone loss in a mouse model of osteoporosis. Sci. Rep. 2018;8:3507. doi: 10.1038/s41598-018-21574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang M., Ma Q., Ding N., Luo F., Bai Y., Kang F., Gong X., Dong R., Dai J., Dai Q. IL-11 is essential in promoting osteolysis in breast cancer bone metastasis via RANKL-independent activation of osteoclastogenesis. Cell Death Dis. 2019;10:353. doi: 10.1038/s41419-019-1594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasuda H., Shima N., Nakagawa N., Yamaguchi K., Kinosaki M., Mochizuki S., Tomoyasu A., Yano K., Goto M., Murakami A. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 8.Lacey D.L., Timms E., Tan H.L., Kelley M.J., Dunstan C.R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 9.Howard G.A., Bottemiller B.L., Turner R.T., Rader J.I., Baylink D.J. Parathyroid hormone stimulates bone formation and resorption in organ culture: evidence for a coupling mechanism. Proc. Natl. Acad. Sci. USA. 1981;78:3204–3208. doi: 10.1073/pnas.78.5.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sims N.A., Martin T.J. Coupling the activities of bone formation and resorption: a multitude of signals within the basic multicellular unit. Bonekey Rep. 2014;3:481. doi: 10.1038/bonekey.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florencio-Silva R., Sasso G.R., Sasso-Cerri E., Simões M.J., Cerri P.S. Biology of bone tissue: structure, function, and factors that influence bone cells. BioMed Res. Int. 2015;2015:421746. doi: 10.1155/2015/421746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suda T., Takahashi N., Udagawa N., Jimi E., Gillespie M.T., Martin T.J. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 13.Chen X., Zhi X., Wang J., Su J. RANKL signaling in bone marrow mesenchymal stem cells negatively regulates osteoblastic bone formation. Bone Res. 2018;6:34. doi: 10.1038/s41413-018-0035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Q., Liang M., Wu Y., Ding N., Duan L., Yu T., Bai Y., Kang F., Dong S., Xu J., Dou C. Mature osteoclast-derived apoptotic bodies promote osteogenic differentiation via RANKL-mediated reverse signaling. J. Biol. Chem. 2019;294:11240–11247. doi: 10.1074/jbc.RA119.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Lian J.B., Stein G.S., van Wijnen A.J., Stein J.L., Hassan M.Q., Gaur T., Zhang Y. MicroRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrie C.H., Gal S., Dunlop H.M., Pushkaran B., Liggins A.P., Pulford K., Banham A.H., Pezzella F., Boultwood J., Wainscoat J.S. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 19.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horibe S., Tanahashi T., Kawauchi S., Murakami Y., Rikitake Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer. 2018;18:47. doi: 10.1186/s12885-017-3958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiteside T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Invest. 2016;126:1216–1223. doi: 10.1172/JCI81136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Z., Li Y., Pan Y., Lan X., Song F., Sun J., Zhou K., Liu X., Ren X., Wang F. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikebuchi Y., Aoki S., Honma M., Hayashi M., Sugamori Y., Khan M., Kariya Y., Kato G., Tabata Y., Penninger J.M. Coupling of bone resorption and formation by RANKL reverse signalling. Nature. 2018;561:195–200. doi: 10.1038/s41586-018-0482-7. [DOI] [PubMed] [Google Scholar]
- 24.Cui Y., Luan J., Li H., Zhou X., Han J. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016;590:185–192. doi: 10.1002/1873-3468.12024. [DOI] [PubMed] [Google Scholar]
- 25.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto K., Ochi H., Sunamura S., Kosaka N., Mabuchi Y., Fukuda T., Yao K., Kanda H., Ae K., Okawa A. Cancer-secreted hsa-miR-940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting ARHGAP1 and FAM134A. Proc. Natl. Acad. Sci. USA. 2018;115:2204–2209. doi: 10.1073/pnas.1717363115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Liu K., Liu Y., Xu Y., Zhang F., Yang H., Liu J., Pan T., Chen J., Wu M. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat. Immunol. 2013;14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y., Cao L., Xia L., Wu Q., Wang J., Wang X., Xu L., Zhou Y., Xu Y., Jiang X. Evaluation of osteogenesis and angiogenesis of icariin in local controlled release and systemic delivery for calvarial defect in ovariectomized rats. Sci. Rep. 2017;7:5077. doi: 10.1038/s41598-017-05392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng S.J., Xie Z.J., Wu Z.Y., Yan D.Y., Tang J.H., Shen Z.J., Li H., Bai B.L., Boodhun V., Eric Dong X.D., Yang L. Effects of combined menaquinone-4 and PTH1–34 treatment on osetogenesis and angiogenesis in calvarial defect in osteopenic rats. Endocrine. 2019;63:376–384. doi: 10.1007/s12020-018-1761-7. [DOI] [PubMed] [Google Scholar]
- 31.Boyce B.F., Li J., Xing L., Yao Z. Bone remodeling and the role of TRAF3 in osteoclastic bone resorption. Front. Immunol. 2018;9:2263. doi: 10.3389/fimmu.2018.02263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito M., Amizuka N., Nakajima T., Ozawa H. Ultrastructural and cytochemical studies on cell death of osteoclasts induced by bisphosphonate treatment. Bone. 1999;25:447–452. doi: 10.1016/s8756-3282(99)00197-0. [DOI] [PubMed] [Google Scholar]
- 33.Tran Van P., Vignery A., Baron R. An electron-microscopic study of the bone-remodeling sequence in the rat. Cell Tissue Res. 1982;225:283–292. doi: 10.1007/BF00214682. [DOI] [PubMed] [Google Scholar]
- 34.Sims N.A., Martin T.J. Coupling signals between the osteoclast and osteoblast: how are messages transmitted between these temporary visitors to the bone surface? Front. Endocrinol. (Lausanne) 2015;6:41. doi: 10.3389/fendo.2015.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakashima T., Hayashi M., Fukunaga T., Kurata K., Oh-Hora M., Feng J.Q., Bonewald L.F., Kodama T., Wutz A., Wagner E.F. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 2011;17:1231–1234. doi: 10.1038/nm.2452. [DOI] [PubMed] [Google Scholar]
- 36.Liu M., Sun Y., Zhang Q. Emerging role of extracellular vesicles in bone remodeling. J. Dent. Res. 2018;97:859–868. doi: 10.1177/0022034518764411. [DOI] [PubMed] [Google Scholar]
- 37.Xiao Z., Camalier C.E., Nagashima K., Chan K.C., Lucas D.A., de la Cruz M.J., Gignac M., Lockett S., Issaq H.J., Veenstra T.D. Analysis of the extracellular matrix vesicle proteome in mineralizing osteoblasts. J. Cell. Physiol. 2007;210:325–335. doi: 10.1002/jcp.20826. [DOI] [PubMed] [Google Scholar]
- 38.Deng L., Wang Y., Peng Y., Wu Y., Ding Y., Jiang Y., Shen Z., Fu Q. Osteoblast-derived microvesicles: a novel mechanism for communication between osteoblasts and osteoclasts. Bone. 2015;79:37–42. doi: 10.1016/j.bone.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Qin Y., Peng Y., Zhao W., Pan J., Ksiezak-Reding H., Cardozo C., Wu Y., Divieti Pajevic P., Bonewald L.F., Bauman W.A., Qin W. Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: a novel mechanism in muscle-bone communication. J. Biol. Chem. 2017;292:11021–11033. doi: 10.1074/jbc.M116.770941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li D., Liu J., Guo B., Liang C., Dang L., Lu C., He X., Cheung H.Y., Xu L., Lu C. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat. Commun. 2016;7:10872. doi: 10.1038/ncomms10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao L., Jiang S., Hantash B.M. Transforming growth factor β1 induces osteogenic differentiation of murine bone marrow stromal cells. Tissue Eng. Part A. 2010;16:725–733. doi: 10.1089/ten.TEA.2009.0495. [DOI] [PubMed] [Google Scholar]
- 42.Luther G., Wagner E.R., Zhu G., Kang Q., Luo Q., Lamplot J., Bi Y., Luo X., Luo J., Teven C. BMP-9 induced osteogenic differentiation of mesenchymal stem cells: molecular mechanism and therapeutic potential. Curr. Gene Ther. 2011;11:229–240. doi: 10.2174/156652311795684777. [DOI] [PubMed] [Google Scholar]
- 43.Cosenza S., Ruiz M., Toupet K., Jorgensen C., Noël D. Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci. Rep. 2017;7:16214. doi: 10.1038/s41598-017-15376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C., Wang D., Moshaverinia A., Liu D., Kou X., Yu W., Yang R., Sun L., Shi S. Mesenchymal stem cell transplantation in tight-skin mice identifies miR-151-5p as a therapeutic target for systemic sclerosis. Cell Res. 2017;27:559–577. doi: 10.1038/cr.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Y., Wu X., Lei W., Pang L., Wan C., Shi Z., Zhao L., Nagy T.R., Peng X., Hu J. TGF-β1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 2009;15:757–765. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kosaka N., Yoshioka Y., Fujita Y., Ochiya T. Versatile roles of extracellular vesicles in cancer. J. Clin. Invest. 2016;126:1163–1172. doi: 10.1172/JCI81130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng Y.C., Lin T., Jiang H., Zhang Z., Shu L., Yin J., Ma X., Wang C., Gao R., Zhou X.H. miR-122 exerts inhibitory effects on osteoblast proliferation/differentiation in osteoporosis by activating the PCP4-mediated JNK pathway. Mol. Ther. Nucleic Acids. 2020;20:345–358. doi: 10.1016/j.omtn.2019.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R., Zhang H., Ding W., Fan Z., Ji B., Ding C., Ji F., Tang H. miR-143 promotes angiogenesis and osteoblast differentiation by targeting HDAC7. Cell Death Dis. 2020;11:179. doi: 10.1038/s41419-020-2377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moura S.R., Bras J.P., Freitas J., Osório H., Barbosa M.A., Santos S.G., Almeida M.I. miR-99a in bone homeostasis: regulating osteogenic lineage commitment and osteoclast differentiation. Bone. 2020;134:115303. doi: 10.1016/j.bone.2020.115303. [DOI] [PubMed] [Google Scholar]
- 50.Sindi H.A., Russomanno G., Satta S., Abdul-Salam V.B., Jo K.B., Qazi-Chaudhry B., Ainscough A.J., Szulcek R., Jan Bogaard H., Morgan C.C. Therapeutic potential of KLF2-induced exosomal microRNAs in pulmonary hypertension. Nat. Commun. 2020;11:1185. doi: 10.1038/s41467-020-14966-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang S.B., Lin S.Y., Liu M., Liu C.C., Ding H.H., Sun Y., Ma C., Guo R.X., Lv Y.Y., Wu S.L. CircAnks1a in the spinal cord regulates hypersensitivity in a rodent model of neuropathic pain. Nat. Commun. 2019;10:4119. doi: 10.1038/s41467-019-12049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barfod E.T., Zheng Y., Kuang W.J., Hart M.J., Evans T., Cerione R.A., Ashkenazi A. Cloning and expression of a human CDC42 GTPase-activating protein reveals a functional SH3-binding domain. J. Biol. Chem. 1993;268:26059–26062. [PubMed] [Google Scholar]
- 53.Clay M.R., Halloran M.C. Rho activation is apically restricted by Arhgap1 in neural crest cells and drives epithelial-to-mesenchymal transition. Development. 2013;140:3198–3209. doi: 10.1242/dev.095448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshioka Y., Kosaka N., Konishi Y., Ohta H., Okamoto H., Sonoda H., Nonaka R., Yamamoto H., Ishii H., Mori M. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z.X., Chen C., Zhou Q., Wang X.S., Zhou G., Liu W., Zhang Z.Y., Cao Y., Zhang W.J. The treatment efficacy of bone tissue engineering strategy for repairing segmental bone defects under osteoporotic conditions. Tissue Eng. Part A. 2015;21:2346–2355. doi: 10.1089/ten.TEA.2015.0071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.