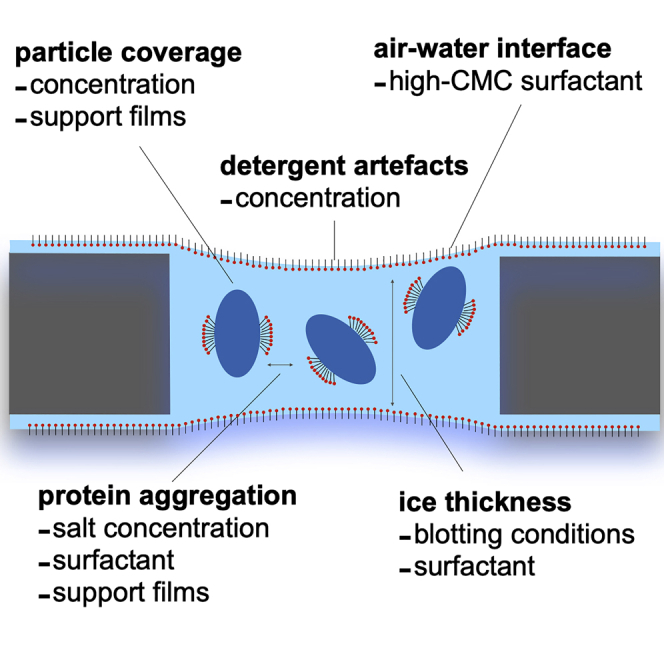
Summary
Cryo-EM grid preparation is an important bottleneck in protein structure determination, especially for membrane proteins, typically requiring screening of a large number of conditions. We systematically investigated the effects of buffer components, blotting conditions and grid types on the outcome of grid preparation of five different membrane protein samples. Aggregation was the most common type of problem which was addressed by changing detergents, salt concentration or reconstitution of proteins into nanodiscs or amphipols. We show that the optimal concentration of detergent is between 0.05 and 0.4% and that the presence of a low concentration of detergent with a high critical micellar concentration protects the proteins from denaturation at the air-water interface. Furthermore, we discuss the strategies for achieving an adequate ice thickness, particle coverage and orientation distribution on free ice and on support films. Our findings provide a clear roadmap for comprehensive screening of conditions for cryo-EM grid preparation of membrane proteins.
Subject areas: Biochemistry, Structural Biology
Graphical abstract
Highlights
-
•
Cryo-EM grid optimization is a limiting factor in protein structure determination
-
•
Membrane proteins require additional optimization of parameters such as surfactants
-
•
Aggregation and intactness of proteins depend on detergent and salt concentrations
-
•
Support films can help preserve protein intactness and improve particle coverage
Biochemistry; Structural Biology
Introduction
Cryo-EM technology has revolutionized structural biology in the past decade, an era that has been termed the resolution revolution (Kühlbrandt, 2014). Numerous technological advances have allowed the resolutions achievable with cryo-EM to rival those of X-ray crystallography and the number of cryo-EM structures deposited into the Protein DataBank has been exponentially increasing. From 439 structures available in 2012, there are now 4884 structures deposited and ever higher resolutions of ever smaller proteins are being achieved. Combined with heavy investments into the cryo-EM infrastructure, cryo-EM is now the method of choice for membrane structural biology.
The most obvious advantage of cryo-EM is the removal of the need for ordered crystals, which are very hard to obtain for many proteins, especially membrane proteins. Furthermore, proteins are imaged in vitreous ice in a state virtually identical to their native state in solution, as opposed to tightly packed crystals. However, cryo-EM grids suitable for high-resolution data collection must satisfy a number of criteria and preparing an adequate sample is still a serious bottleneck in the cryo-EM workflow. Most importantly, the protein needs to be pure, monodisperse, sufficiently concentrated, show random orientations, and not be disassembled or aggregated (Orlova and Saibil, 2011). The ice thickness needs to be low to minimize background noise, particularly important for small proteins, and to limit the distribution of protein particles along the z axis (Herzik et al., 2017).
To achieve this, the grid preparation protocol needs to be iteratively optimized and a number of different conditions screened. The first crucial component is isolating the protein of interest under the right conditions, especially with respect to the choice and concentration of detergent, buffer and protein concentration. The second crucial component is the optimization of conditions during the vitrification process itself, e.g. the grid type and blotting time (Bloch et al., 2020). Many optimized parameters will be unique for each protein examined but nevertheless, common patterns are starting to emerge.
Previous work on cryo-EM grid preparation has mainly focused on the choice of support foils and foil treatments, to minimize charging effects and movement (Vinothkumar and Henderson, 2016; Sgro and Costa, 2018). However, for membrane proteins there is another layer of complexity as detergents or other surfactants need to be added to stabilize the proteins in solution. In addition to classic detergents, membrane proteins can also be stabilized using amphipols, or in small patches of lipid bilayer such as in nanodiscs or styrene maleic acid lipid particles (SMALPs) (Bayburt and Sligar, 2010; Popot et al., 2011; Lee et al., 2016). In these preparations, membrane protein samples behave more like soluble proteins as the buffers do not contain any free detergents.
Here, we discuss the patterns that have emerged during optimizing cryo-EM grids for five different membrane proteins (complex I, F1Fo-ATP synthase and transhydrogenase from ovine mitochondria, E. coli complex I and Mrp antiporter from Anoxybacillus flavithermus), all of which are large multi-subunit complexes involved in energy metabolism. The most common problems encountered were aggregation, problems with ice thickness, disassembly of protein complexes, presence of detergent artefacts and heterogeneity within or between grids. These were tackled by a range of methods, especially changing of buffer conditions and adding support films.
Results
Reproducibility of grid preparation
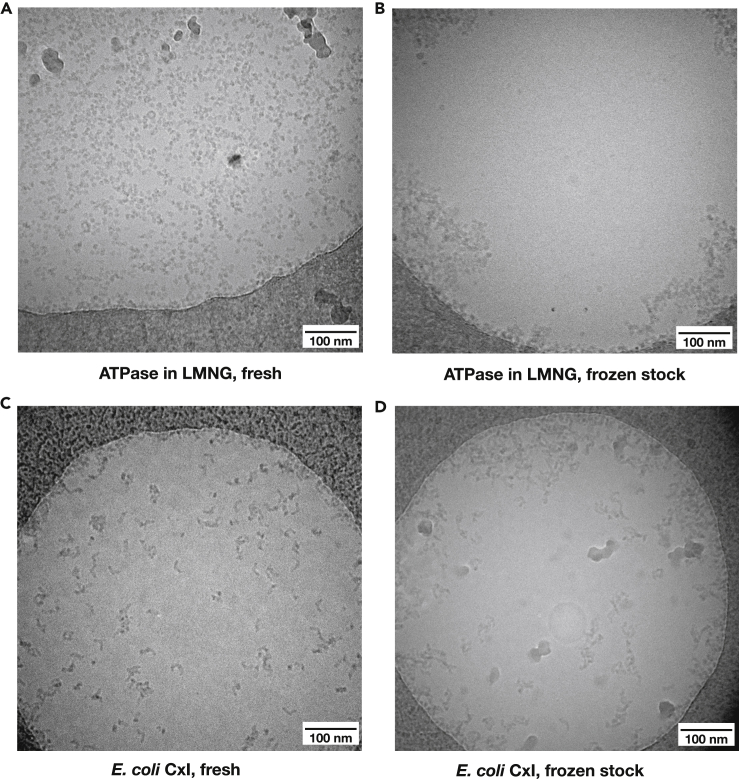
The first requirement for reproducible and consistent grids is a pure, stable, and fresh protein sample that behaves in a predictable way when concentrated and applied on a cryo-EM grid. Grid preparation was always done from freshly gel-filtrated sample (see Transparent methods). Diluting and then re-concentrating protein samples from thawed glycerol-containing stocks resulted in aggregation of protein on the cryo-EM grids (Figure 1). In general, the fewer the handling steps of protein between gel filtration and grid freezing, the better the grids.
Figure 1.
Effects of gel filtration on cryo-EM grids of ATPase (top) and E. coli complex I (bottom)
(A and B) Comparison of grids of ovine F1Fo-ATPase at 1.5 mg/mL in LMNG prepared (A) immediately after gel filtration without any further concentration and (B) prepared from a frozen stock with 30% glycerol that has been diluted and reconcentrated to remove glycerol.
(C and D) Comparison of E. coli complex I at 5 mg/mL in DDM prepared (C) freshly after gel filtration and concentrating it and (D) prepared from a frozen stock with 30% glycerol that has been diluted and reconcentrated to remove glycerol.
The other critical step for achieving consistency is blotting conditions. Care was taken to operate the Vitrobot as consistently as possible across different sessions. This included setting the temperature, blotting force and blotting time parameters, maintaining the same orientation of the tweezers and insertion of blotting paper immediately before the actual grid preparation and not during Vitrobot equilibration. This achieved a satisfactory reproducibility but a representative grid from the same batch was still always checked in-house before the other grids were used for high-resolution data collection elsewhere.
Sometimes it was observed that particle coverage varied markedly within the same grid. Ultimately, this seems to be an effect of too low protein concentration since a higher protein concentration solved the variability.
Protein aggregation
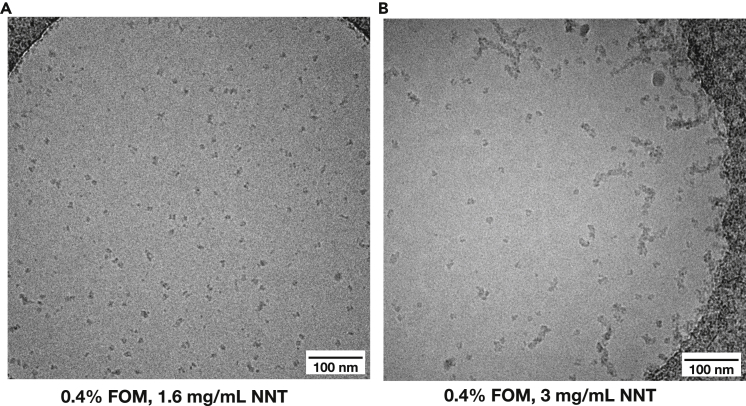
Aggregation was one of the most persistent problems encountered during cryo-EM grid preparation. Membrane proteins in particular tend to aggregate easily because of the exposed hydrophobic regions normally embedded in the membranes. The most obvious solution for aggregation is therefore a change of detergent. Typically, detergents in which proteins easily aggregate can be discarded already during the purification optimization step, by stability and activity screening. However, that is not always the case as demonstrated by NNT in fluorinated octyl-maltoside (FOM). Despite the protein having a high activity in FOM and giving a symmetric monodisperse peak on a gel filtration column, concentrating NNT in FOM above ∼2.5-3 mg/mL resulted in aggregation (Figure 2). Because the UV-Vis spectra of purified and concentrated samples also showed evidence of aggregates (A260/280 > 1) (Porterfield and Zlotnick, 2010; Fiedorczuk, 2017) we concluded that aggregation proceeds fast and happens before grid preparation. Ultimately, the aggregation was overcome by switching from FOM to LMNG, in which NNT did not aggregate even at high concentrations. It is worth pointing out that FOM is by virtue of its fluorinated alkyl chain a very mild detergent that can preserve many protein-lipid interactions and is a good detergent to screen in any membrane protein cryo-EM optimization protocol (Frotscher et al., 2015).
Figure 2.
Aggregation of NNT in FOM depends on the protein concentration
(A) At 1.6 mg/mL NNT in FOM is monodisperse.
(B) At 3 mg/mL NNT in FOM aggregates.
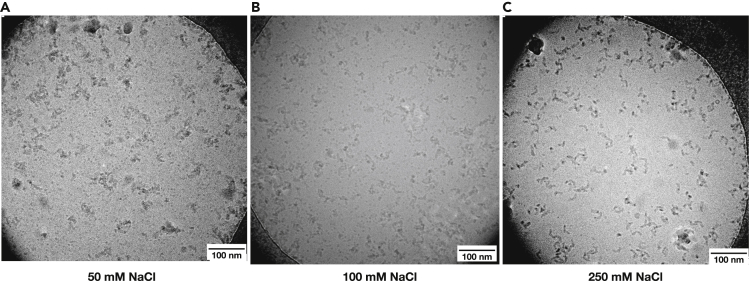
Secondly, salt concentration affects the aggregation of particles in solution because it shields charges: cations arrange themselves around negatively charged protein surfaces and anions arrange around positively charged protein surfaces. Salt can have two opposing effects on aggregation behavior. Simple colloidal particles can be induced to aggregate by increasing the salt concentration as the dissolved ions shield repelling charges on their surfaces (Dill and Bromberg, 2010). Proteins, however, have large and heterogeneous surfaces with both positively and negatively charged patches. Salt will not only shield equally charged repelling surfaces but also oppositely charged surfaces which will associate less readily at increased salt concentrations. Moderate concentrations of salt therefore increase the solubility of proteins, a phenomenon known as “salting in”. The latter effect of salt on protein aggregation was documented for E. coli complex I, which was prone to aggregate at 50 mM NaCl, but when this was increased to 250 mM NaCl, aggregation was drastically diminished (Figure 3) and an increase to 400 mM did not improve monodispersity further. Other proteins investigated were sufficiently soluble at 50 mM NaCl, demonstrating that salt concentration needs to be adjusted for each specific protein. None of the proteins investigated seemed to demonstrate “salting out” (aggregation at very high salt concentrations) in the physiological ranges of NaCl tried for grid preparation (50-250mM NaCl).
Figure 3.
Aggregation of E. coli complex I depends on salt concentration
(A–C) Increasing salt concentration from 50 mM (A) to 100 mM (B) and 250 mM (C) diminished aggregation of particles due to the “salting in” effect.
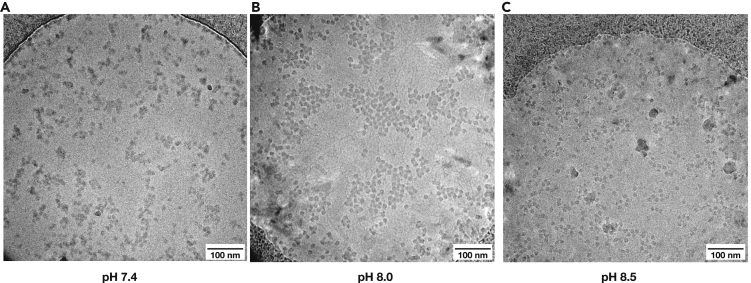
Thirdly, protein surface charges are affected by pH which affects the protonation states of amino acid residues. The effects of pH change on the aggregation of F1Fo-ATPase sample are not easy to explain, although a higher pH was beneficial in this case (Figure 4). Surface charge properties of large protein assemblies are complex and consist of regions of opposing charges which will behave in unpredictable ways at different pH values. Varying pH could also affect protein conformation in unpredictable ways and so the protein activity should be verified at each pH value.
Figure 4.
Aggregation of F1Fo-ATPase depends on pH
(A–C) Aggregation of ATPase depends on the buffer pH, which modulates surface charges on the protein.
Denaturation and disassembly of particles
Large multi-subunit membrane protein complexes can easily disassemble into smaller pieces or denature (D’Imprima et al., 2019). It has been described that this is due to the denaturing effects at the air-water interface, the area of which is massively enlarged in relation to the volume during the grid preparation and allows protein particles to interact with it (Glaeser et al., 2016). This can be avoided by using a high-CMC secondary detergent at below CMC concentrations. Such a detergent forms a layer at the air-water interface and prevents protein adsorption and denaturation there. This was observed to be particularly useful for samples without primary surfactant (nanodiscs, amphipols) or with primary surfactants with very low CMC values (LMNG). Some partitioning of secondary high-CMC detergent into primary detergent micelles also probably happens and contributes to different sample properties (Chen et al., 2019).
Previously, Fos-choline-8 and FOM have been described to improve grid preparation (Efremov et al., 2015; Johnson and Chen, 2017). In our hands, secondary detergents with high CMC values (octylglucoside and especially CHAPS) at concentrations between 0.05 and 0.2% also improved cryo-EM grids and prevented particle disintegration (Figure 5). The addition of secondary detergents also seemed to make the ice thickness less variable across the grids. On the negative side, as described below, higher concentrations of detergent also require higher concentrations of protein to achieve the same particle coverage.
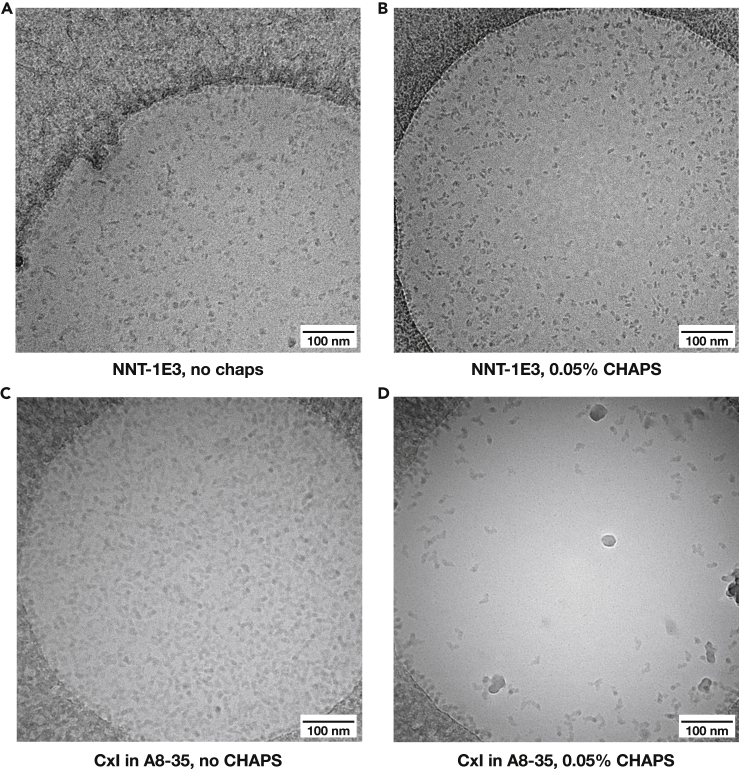
Figure 5.
High-CMC detergent additives improve particle distribution and protect them from denaturation
(A and B) Comparison of NNT in MSP1E3D1 nanodiscs without (A) and with added CHAPS (B). Particle distribution and intactness are much better when CHAPS is included.
(C and D) Comparison of complex I in A8-35 amphipols without (C) and with added CHAPS (D). Particle distribution and intactness are much better when CHAPS is included, although particle coverage is lower at the same protein concentration.
Detergent artefacts
Protein samples after gel filtration are rather diluted and need to be concentrated several-fold before they can be used for grid preparation. Transhydrogenase in particular, for which the yield was not high, had to be concentrated 20-30-fold after gel filtration to reach ∼5 mg/mL suitable for grid preparation. For samples without detergent this is not a problem (in amphipols or nanodiscs) but concentrating of samples with detergent also concentrates detergent even on 100 kDa-cutoff filters because detergents exist in the form of large molecular weight micelles in aqueous solutions.
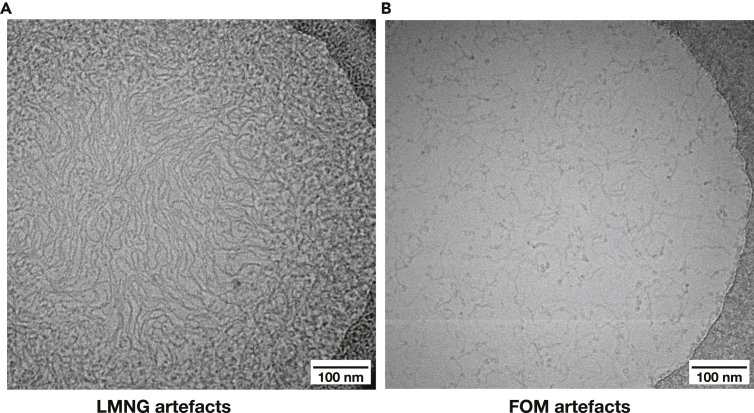
The optimal concentration of total detergent for cryo-EM grids is around 0.2–0.4%. Anything above this results in artefacts: either free micelles visible or long threads (Figure 6). For some detergents, such as LMNG, artefacts were apparent even at lower concentrations, above 0.1%. Too low concentration of free detergent (not in micelles or bound to protein), however, resulted in denaturation of protein sample at the air-water interface.
Figure 6.
Detergent artefacts observed at high detergent concentrations
(A) Sample of complex I in ∼1% LMNG.
(B) Sample of NNT in ∼1% FOM.
For samples purified in LMNG, the final concentration of detergent was controlled by gel filtration at 2xCMC = 0.002%, so even several-fold concentration of samples did not result in [LMNG] > 0.1%. For NNT purified in FOM, however, excess detergent was removed using detergent removal columns (ThermoFisher Ltd.) that bind detergents and other hydrophobic contaminants. Typically, 750 μL NNT was concentrated to 20 μL ([FOM] ∼ 2%), and after a single passage through a detergent removal column according to the manufacturer's instructions, 0.075% remaining FOM was measured (Urbani and Warne, 2005). It was important to measure the detergent concentration of each preparation prepared this way and adjust the FOM concentration back to 0.2–0.4%. Detergent removal columns did not decrease activity or cause aggregation. Detergent removal by dilution and re-concentration or by using spin desalting Zeba columns did not remove more than ∼25% of the detergent.
Ice thickness
The most important factor to tune for achieving thin ice is blotting time. This varied 5–10 s for most samples. Blotting times up to 20–30 s also worked sometimes, especially for samples with high concentrations of detergent. Blotting force did not seem to have as pronounced an effect on the ice thickness and was chosen initially such that solution was blotted uniformly and concentrically from the entire surface of the grid. “Over-blotting” leads either to no ice in holes or to no visible protein in very thin ice. Samples with little detergent were more prone to “over-blotting”, as were the samples set up on gold grids. Samples without detergent and grids covered with support generally work well with short blotting times of 2-3 s. A persistent problem of many preparations was partial over-blotting, where the centers of the holes became too thin and excluded the particles from that region. The presence of detergent leads to a “meniscus effect”, when ice in the middle of the hole can be much thinner than at the edges. This was partly avoided by increasing protein concentration and concentration of the secondary detergent. Furthermore, consistently thin ice with good particle spread was achieved much more easily using Quantifoil 0.6/1 grids, which were less prone to over-blotting and “meniscus effect” than Quantifoil 1.2/1.3 grids or other grids with larger hole size.
Ice thickness was qualitatively assessed based on the contrast between the particles and the background in the acquired images. Another semi-quantitative method for estimating ice thickness is measuring electron fluence over an empty hole and a full hole, the ratio of which is approximately related to the ice thickness according to d/λ = ln (Ii/Io), where d is the mean electron path in vitreous ice at 120kV (∼600 Å), λ is the ice thickness, Ii is the incident fluence, and Io is the fluence after the sample (Feja and Aebi, 1999). However, this method gives reasonable results only for a relatively thick ice (>50 nm). Another semi-quantitative method for measuring ice thickness is the channel-method from tomography, which involves burning a narrow channel into the vitreous ice when the grid is tilted at 45° (Feja and Aebi, 1999). When the sample is not tilted anymore, the edges of the channel reveal the thickness of the sample. The method was useful for ice thicknesses above 50 nm but became increasingly imprecise below that as the vitreous ice around the channel became less stable. Ultimately, each cryo-EM grid displayed a range of ice thicknesses at different grid squares and also within the same grid square and it was important to choose the suitable holes during the collection itself. One useful criterion is to look for grid holes where the protein “just about” makes it into the center of the hole, as compared to holes with slightly thinner ice which exclude protein in the center. In the case of grids with continuous support the measurement methods above would work only for very thick ice (due to higher background) and so are not useful. One has to rely on particles' contrast to find thin ice and to avoid freeze-dried protein areas.
Particle coverage
An ideal outcome of cryo-EM grid preparation is a single layer of tightly packed particles embedded in thin ice. The packing can be as tight as possible, to the point where proteins seemingly touch. As long as the particles are not overlapping the data set will be processable and a higher number of particles will be extracted from the same number of cryo-EM images. This required a high concentration of protein, reaching 5 mg/mL in most conditions and seemed to be dependent on the concentration of detergent present in the sample: the more detergent, the higher the protein concentration required to reach the desired coverage. The requirement for a higher protein concentration in the presence of detergents has been observed previously. Interestingly, the protein concentrations typically used for detergent-containing samples reflect the actual density of particles in the solution, whereas detergent-free samples can be used at lower protein concentrations because they adsorb preferentially to the air-water interface (Schmidt-Krey and Rubinstein, 2011).
As such a high protein concentration may not always be achievable due to the propensity for aggregation and to avoid a concomitant concentration of detergent, we investigated the use of support films. As proteins often adsorb on support films such as graphene oxide, graphene or amorphous carbon, an order or two of magnitude lower protein concentrations are required to achieve the same coverage as on grids without support films (Figure 7). This comes at the expense of a lower signal-to-noise ratio due to the support film, as it contributes to the background. This has been reported to be quite minimal for “home-made” grids covered with continuous carbon (Steiner and Sazanov, 2020) as compared to commercially produced continuous carbon covered grids and should not interfere with image analysis of at least large (>0.4 MDa) protein complexes. The presence of a continuous support also helps with initial contrast transfer function (CTF) parameter estimations and should limit beam-induced movement (Russo and Passmore, 2016).
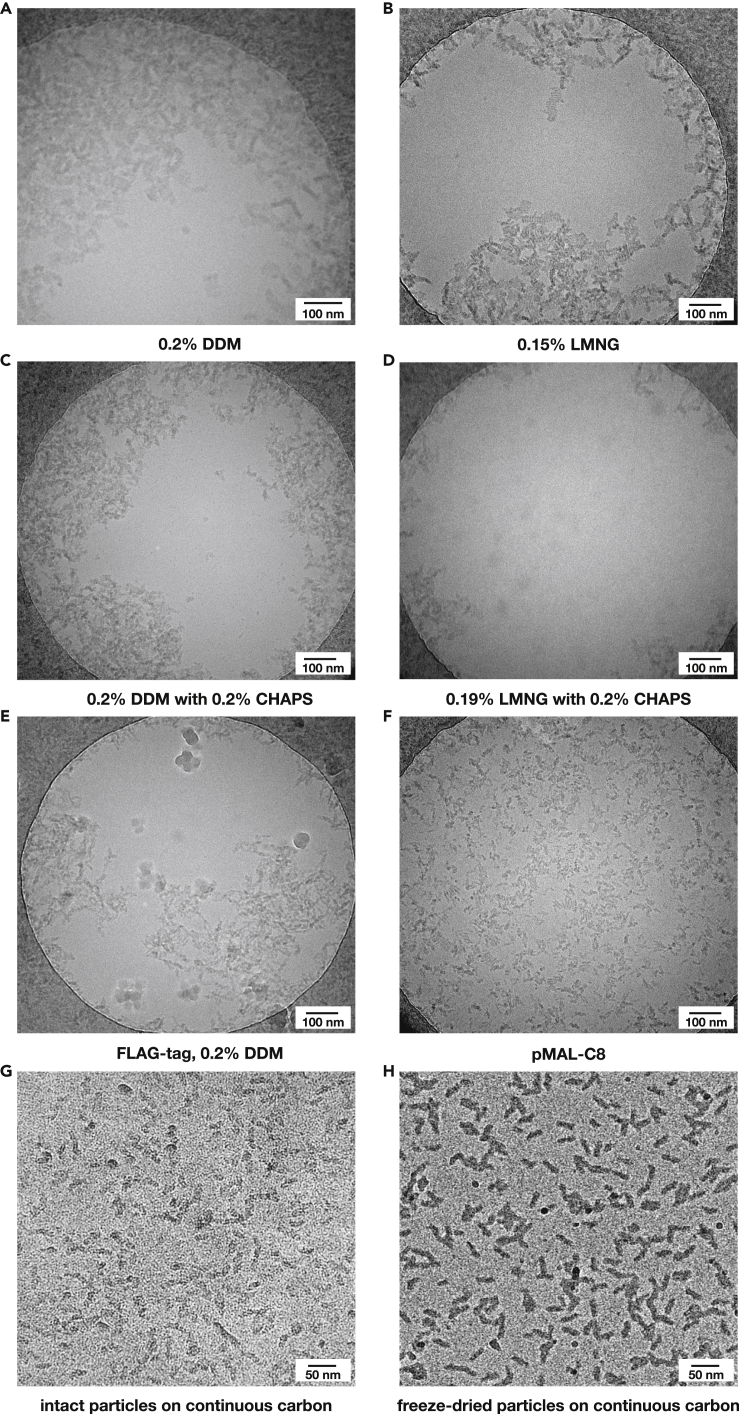
Figure 7.
Comparison of particle distribution of Mrp in free-standing ice and on continuous carbon support
Cryo-EM grids of the Mrp complex under different conditions as labeled.
(A) 0.2% DDM; 3 mg/mL Mrp.
(B) 0.15% LMNG; 2 mg/mL Mrp.
(C) 0.2% DDM +0.2% CHAPS; 1.5 mg/mL Mrp.
(D) 0.19% LMNG +0.2% CHAPS; 2 mg/mL Mrp.
(E) 0.2% DDM; 2.2 mg/mL of FLAG tag Mrp.
(F) Mrp in pMAL-C8.
(G) Intact Mrp particles in LMNG on continuous carbon support.
(H) Freeze-dried Mrp particles on carbon support.
Graphene oxide grids were however not very evenly covered and often contained overlapping layers of material over some grid squares, which may detrimentally affect the image quality. In our hands, this was the case with either the original protocol (Bokori-Brown et al., 2016) or the modified procedure using methanolic graphene oxide solution (Palovcak et al., 2018). Additionally, over-blotting of grids with continuous support is quite common and results in dehydrated and freeze-dried protein, precluding high resolution data reconstruction. Nevertheless, when freeze-dried areas were avoided during collection, complex I from a small data set on graphene-oxide-covered grids reached 4 Å. The Mrp (Multi resistance and pH) complex is a cation/H+ antiporter found in many Bacteria and Archaea. It consists of seven subunits and forms a dimer with 100 transmembrane helices in total (Steiner and Sazanov, 2020). It does not have any significant hydrophilic domains and it is perhaps this extreme hydrophobicity which made it an especially challenging target for cryo-EM grid optimization. Although the protein eluted as a monodisperse peak during size-exclusion chromatography, in all trials it was heavily aggregated in ice holes of cryo-EM grids. An extensive screening of different primary and secondary detergents, additives, buffer conditions and salt concentrations, as well as changing the affinity tag from a His-tag (which may cause aggregation) to an FLAG tag, the usage of alternatives to detergents, such as amphipols or the application of PEGylated gold grids in combination with a lower protein concentration, did not prevent aggregation (Figure 7).
However, as soon as a support film was used, the aggregation was prevented. This is not likely due to the 10 times lower protein concentration used for the preparation of continuous carbon grids, as the protein was also aggregated in a wide range of protein concentrations over ice holes. The most likely reason is that the highly hydrophobic Mrp tended to accumulate at the air-water interface, leading to protein damage and aggregation. This was avoided with a continuous support, as the protein became attached to the support film, completely avoiding absorption at the damaging air-water interface. Particles were reasonably spread on graphene and graphene oxide-covered grids; however, the problems of uneven coverage of grids by the support material, mentioned above, led to sub-optimal grid preparations. Graphene sheets can be obtained commercially and attached to the grid as a monolayer, but this renders each grid very expensive. Furthermore, it is a highly hydrophobic material and rendering it hydrophilic using published chemical modification techniques (D’Imprima et al., 2019) leaves a lot of crystalline artefacts on the surface. This made such grids unusable for data collection unless the grids were washed extensively with ethanol (Figure 8).
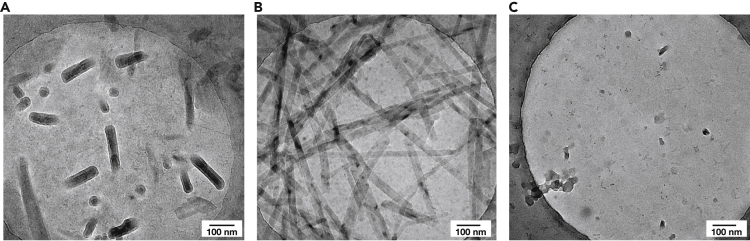
Figure 8.
Graphene grids
(A) Example of a grid covered with residual crystals.
(B) Example of a grid covered with residual long fibers.
(C) Example of a grid which had been washed with ethanol after hydrophilization. Particles have low contrast and are not suitable for collection.
The continuous carbon layer of commercially produced grids is too thick to be used for high-resolution cryo-EM data collection. However, a very thin (∼1–1.4 nm) layer of continuous carbon can be produced in-house using any coating apparatus and then deposited on standard Quantifoil grids. This procedure is very reproducible and results in an even monolayer of support film. Particles on such carbon were easily observable with a high contrast, showing that an extra background due to the layer of thin carbon is tolerable. Particles did not show any signs of aggregation and were evenly distributed, suitable for data collection (Figure 7G). One major problem when working with support films is judging the ice thickness, as the particle distribution does not inform us about that anymore. Even if there is no ice due to over-blotting, resulting freeze-dried particles will still be present. However, such areas can be distinguished by a very high protein contrast (Figure 7H), and hence can be avoided during data collection. As noted above, ice thickness measurements are of limited use for thin ice. Therefore, to collect from as thin as possible ice, which is especially important in the presence of carbon, one has to look for areas with as thin ice and light background as possible, but making sure not to stray into freeze-dried areas. Additionally, freeze-drying can be observed by the appearance of fuzzy ice rings in Fourier transformed images, and such images can be excluded during data processing.
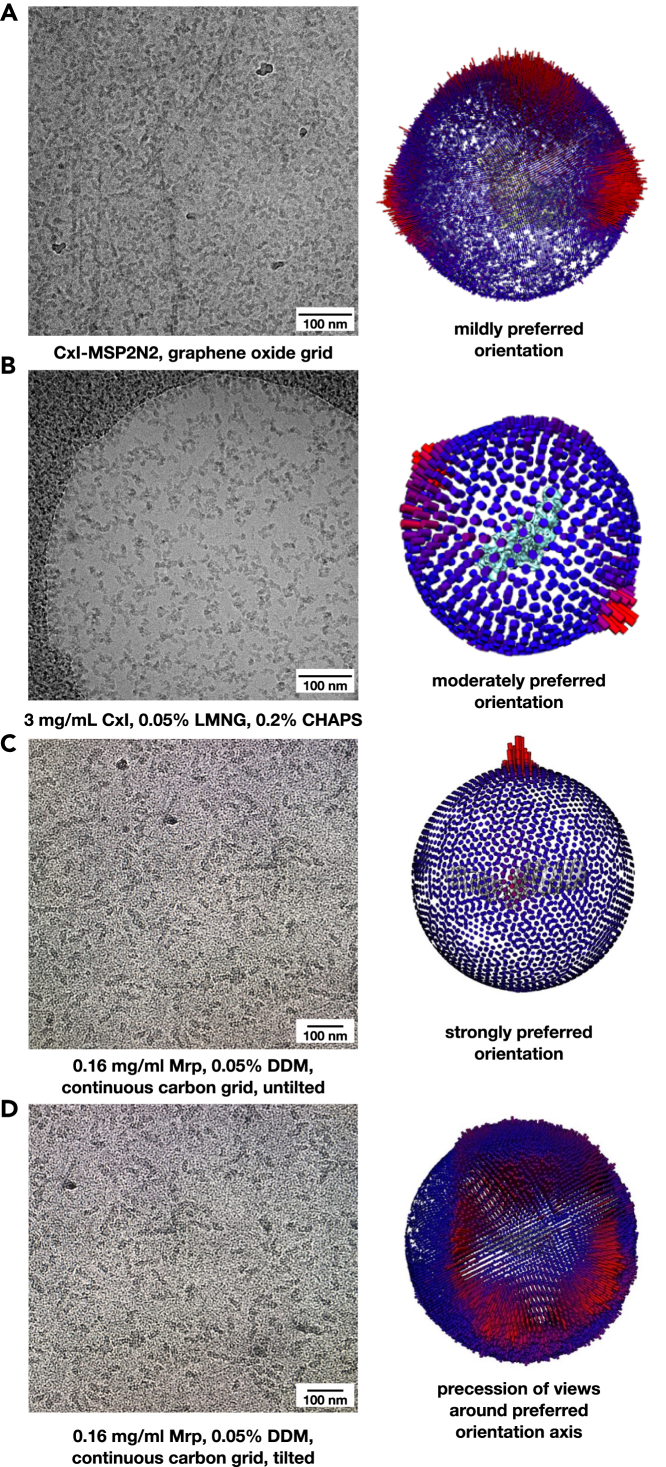
Angular distribution
Detergent-containing samples frozen in free ice often do not display preferential orientation, whereas the use of support films can sometimes lead to a preferential orientation of particles because molecules tend to adsorb to the film with the largest or the most charged surface. A preferential orientation complicates downstream processing and needs to be addressed when devising the data collection strategy. Support films, however, do not necessarily lead to a preferential orientation and can also be used to change the orientation of particles. In the case of complex I, which has a large soluble domain, graphene oxide support film actually improved the angular distribution compared to the free ice (Figure 9).
Figure 9.
Support films affect particle coverage and angle distribution
(A and B) Left: Representative micrographs of cryo-EM grids of ovine complex I in free-standing ice and on graphene oxide support film. Right: Comparison of angular distribution of ovine complex I on graphene oxide (A) and in free-standing ice (B). Angular distribution of particles is improved on graphene oxide support.
(C and D) Left: Representative micrographs of the Mrp complex on continuous carbon collected without tilting (C) and with tilting (D). Right: Comparison of angular distribution of Mrp on continuous carbon without tilt (C) and with 35° tilt (D).
In the case of the Mrp complex a severe preferential orientation was observed, with just one main view of the dimer, attached flat to the continuous carbon with the periplasmic surface. This leads to a Fourier component under-sampling within the final reconstruction and to an anisotropic resolution which becomes apparent through an elongation of the map into the direction of the missing data (“smearing” effect). A strategy to overcome this problem is to tilt the grid during data collection. Tilts result in a more uniform coverage of Fourier space and hence an improved reconstructed volume, with the problem mostly resolved at about 40° tilt (Zi Tan et al., 2017). Higher tilts often lead to unacceptable loss of high-resolution information due to increased background noise from the tilted ice/support and also lead to challenges in automatic data collection from such highly tilted grids.
We opted for a 35° tilt for the Mrp data collection. In the case of the tilted collection, it is important to estimate initial CTF parameters on a per-particle basis, as the usual per-micrograph estimates are often too far off to allow reliable 2D and 3D classification. Therefore, after an initial gross per-micrograph CTFFIND software run (Rohou and Grigorieff, 2015) and auto-picking of all particle candidates we performed a per-particle estimation with the gCTF software (Zhang, 2016). Due to a very strong signal from the supporting carbon, this estimation could be run on a smaller area around each particle (--local_radius 512) and provided very precise values, so that a CTF refinement later did not improve the results. This strategy resulted in well-resolved and nearly isotropic maps at 2.98 Å resolution, including some water molecules. Thus, preferential orientation per se does not represent a problem and the use of a thin carbon support could be a nearly universal recipe to resolve problems such as aggregation or insufficient protein concentration. As the Mrp complex is about 400 kDa in size, it remains to be established whether such a strategy is also applicable to much smaller proteins, where an increased background noise from carbon could represent a bigger problem.
Discussion
We have discussed the main issues during cryo-EM grid preparation of membrane proteins and analyzed the methods that can be used to overcome them. While every sample behaves slightly differently, we found that the optimal values for some parameters are in the similar ranges for all of them. In particular, a primary detergent concentration of around 0.2%, including high-CMC secondary detergents and varying salt concentration was used to control protein aggregation, denaturation, and ice thickness. For samples for which a high protein concentration was not possible to achieve, or those prone to aggregation, the use of support films was beneficial, even though it sometimes led to a preferential orientation. We hope that this provides a roadmap for fast and efficient screening of conditions to optimize cryo-EM grid preparation for a variety of membrane protein samples.
Limitations of the study
It is important to emphasize that the membrane protein complexes examined here all belong to a family of energy-transducing complexes and three of them are found in the inner mitochondrial membrane. Comparing the conditions examined here to the rest of the membrane structural biology field it seems that digitonin is the most widely used detergent and was used in one-third of all the structures solved in the last ten years (Choy et al., 2021). Digitonin is useful because it preserves the native lipid environment in membrane protein complexes, but also it has important drawbacks, such as high-CMC, batch-to-batch variability, and tendency to precipitate. For these reasons, digitonin has often been replaced by its synthetic analog glyco-diosgenin (GDN) or mixtures of cholesteryl hemisuccinate with DDM or LMNG (Guo et al., 2017; Garaeva et al., 2018; Velazhahan et al., 2020). Integrity of some protein complexes appears absolutely dependent on the presence of a steroid moiety in the surfactant, probably by preserving important protein-protein or protein-lipid interactions. This may depend on the presence of cholesterol in the protein's native membrane. However, fragile multi-complex assemblies, such as mitochondrial respiratory supercomplexes, also appear to be best solubilized and preserved in digitonin, as exemplified by the respirasome (Gu et al., 2016; Letts et al., 2016; Sousa et al., 2016) and ATP synthase tetramer (Gu et al., 2019; Flygaard et al., 2020; Pinke et al., 2020; Mühleip et al., 2021). In some cases digitonin or other mild detergent can be exchanged for amphipols, still preserving the supercomplex assembly. This can help with cryo-EM grid optimization and structure determination, since one does not have to deal with very large and dense digitonin micelles (Sousa et al., 2016; Letts et al., 2019).
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Leonid A. Sazanov (sazanov@ist.ac.at).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Cryo-EM density maps of Mrp, ovine F1Fo-ATPase, ovine complex I, and ovine transhydrogenase are deposited in the EMDB with the accession numbers EMD-11027, EMD-10573, EMD-4635, and EMD-11246.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank the Electron Microscopy Facilities at the Institute of Science and Technology Austria and at the Vienna Biocenter for providing access and training for the electron microscopes. This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant Agreement no. 665385.
Author contributions
D.K. prepared and screened cryo-EM grids with ovine transhydrogenase, complex I, ATP synthase, and E. coli complex I. J.S. prepared and screened grids with Mrp. D.K. wrote the initial draft of the manuscript, with input from all authors. L.A.S. supervised the project and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 19, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2021.102139.
Supplemental information
References
- Bayburt T.H., Sligar S.G. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch M., Santiveri M., Taylor N.M.I. Methods in Molecular Biology. Humana Press Inc.; 2020. Membrane protein Cryo-EM: cryo-grid optimization and data collection with protein in detergent; pp. 227–244. [DOI] [PubMed] [Google Scholar]
- Bokori-Brown M., Martin T.G., Naylor C.E., Basak A.K., Titball R.W., Savva C.G. Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein. Nat. Commun. 2016;7:1–7. doi: 10.1038/ncomms11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Noble A.J., Kang J.Y., Darst S.A. Eliminating effects of particle adsorption to the air/water interface in single-particle cryo-electron microscopy: Bacterial RNA polymerase and CHAPSO. J. Struct. Biol. X. 2019;1:100005. doi: 10.1016/j.yjsbx.2019.100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy B.C., Cater R.J., Mancia F., Pryor E.E. A 10-year meta-analysis of membrane protein structural biology: detergents, membrane mimetics, and structure determination techniques. Biochim. Biophys. Acta - Biomembranes. 2021;1863:183533. doi: 10.1016/j.bbamem.2020.183533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K., Bromberg S. Second edition. Garland Science; 2010. Molecular Driving Forces: Statistical Thermodynamics in Chemistry, Physics, Biology, and Nanoscience. [Google Scholar]
- D’Imprima E., Floris D., Joppe M., Sánchez R., Grininger M., Kühlbrandt W. Protein denaturation at the air-water interface and how to prevent it. eLife. 2019;8:e42747. doi: 10.7554/eLife.42747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov R.G., Leitner A., Aebersold R., Raunser S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature. 2015;517:39–43. doi: 10.1038/nature13916. [DOI] [PubMed] [Google Scholar]
- Feja B., Aebi U. Determination of the inelastic mean free path of electrons in vitrified ice layers for on-line thickness measurements by zero-loss imaging. J. Microsc. 1999;193:15–19. doi: 10.1046/j.1365-2818.1999.00436.x. [DOI] [PubMed] [Google Scholar]
- Fiedorczuk K. University of Cambridge; 2017. Cryo-electron Microscopy Studies on Ovine Mitochondrial Complex I.https://www.repository.cam.ac.uk/handle/1810/270318 [Google Scholar]
- Flygaard R.K., Mühleip A., Tobiasson V., Amunts A. Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization. Nat. Commun. 2020;11:5342. doi: 10.1038/s41467-020-18993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frotscher E., Danielczak B., Vargas C., Meister A., Durand G., Keller S. A fluorinated detergent for membrane-protein applications. Angew. Chem. Int. Ed. 2015;54:5069–5073. doi: 10.1002/anie.201412359. [DOI] [PubMed] [Google Scholar]
- Garaeva A.A., Oostergetel G.T., Gati C., Guskov A., Paulino C., Slotboom D.J. Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nat. Struct. Mol. Biol. 2018;25:515–521. doi: 10.1038/s41594-018-0076-y. [DOI] [PubMed] [Google Scholar]
- Glaeser R.M., Han B.G., Csencsits R., Killilea A., Pulk A., Cate J.H.D. Factors that influence the formation and stability of thin, cryo-EM specimens. Biophysical J. Biophysical Soc. 2016:749–755. doi: 10.1016/j.bpj.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Wu M., Guo R., Yan K., Lei J., Gao N., Yang M. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- Gu J., Zhang L., Zong S., Guo R., Liu T., Yi J., Wang P., Zhuo W., Yang M. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science. 2019;364:1068–1075. doi: 10.1126/science.aaw4852. [DOI] [PubMed] [Google Scholar]
- Guo H., Bueler S.A., Rubinstein J.L. Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science. 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzik M.A., Wu M., Lander G.C. Achieving better-than-3-Å resolution by single-particle cryo-EM at 200 keV. Nat. Methods. 2017;14:1075–1078. doi: 10.1038/nmeth.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Z.L., Chen J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell. 2017;168:1075–1085.e9. doi: 10.1016/j.cell.2017.01.041. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- Lee S.C., Knowles T.J., Postis V.L.G., Jamshad M., Parslow R.A., Lin Y.P., Goldman A., Sridhar P., Overduin M., Muench S.P., Dafforn T.R. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11:1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- Letts J.A., Fiedorczuk K., Degliesposti G., Skehel M., Sazanov L.A. Structures of respiratory supercomplex I+III2 reveal functional and conformational crosstalk. Mol. Cell. 2019;75:1131–1146.e6. doi: 10.1016/j.molcel.2019.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts J.A., Fiedorczuk K., Sazanov L.A. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- Mühleip A., Kock Flygaard R., Ovciarikova J., Lacombe A., Fernandes P., Sheiner L., Amunts A. ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria. Nat. Commun. 2021;12:120. doi: 10.1038/s41467-020-20381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova E.V., Saibil H.R. Structural analysis of macromolecular assemblies by electron microscopy. Chem. Rev. 2011;111:7710–7748. doi: 10.1021/cr100353t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovcak E., Wang F., Zheng S.Q., Yu Z., Li S., Betegon M., Bulkley D., Agard D.A., Cheng Y. A simple and robust procedure for preparing graphene-oxide cryo-EM grids. J. Struct. Biol. 2018;204:80–84. doi: 10.1016/j.jsb.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinke G., Zhou L., Sazanov L.A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol. 2020;27:1077–1085. doi: 10.1038/s41594-020-0503-8. [DOI] [PubMed] [Google Scholar]
- Popot J.-L., Althoff T., Bagnard D., Banères J.-L., Bazzacco P., Billon-Denis E., Catoire L.J., Champeil P., Charvolin D., Cocco M.J. Amphipols from A to Z∗. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- Porterfield J.Z., Zlotnick A. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology. 2010;407:281–288. doi: 10.1016/j.virol.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo C.J., Passmore L.A. Progress towards an optimal specimen support for electron cryomicroscopy. Curr. Opin. Struct. Biol. 2016;37:81–89. doi: 10.1016/j.sbi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Krey I., Rubinstein J.L. Electron cryomicroscopy of membrane proteins: specimen preparation for two-dimensional crystals and single particles. Micron. 2011;42:107–116. doi: 10.1016/j.micron.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgro G.G., Costa T.R.D. Cryo-EM grid preparation of membrane protein samples for single particle analysis. Front. Mol. Biosci. 2018;5:74. doi: 10.3389/fmolb.2018.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa J.S., Mills D.J., Vonck J., Kühlbrandt W. Functional asymmetry and electron flow in the bovine respirasome. eLife. 2016;5:e21290. doi: 10.7554/eLife.21290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J., Sazanov L. Structure and mechanism of the Mrp complex, an ancient cation/proton antiporter. eLife. 2020;9:e59407. doi: 10.7554/eLife.59407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbani A., Warne T. A colorimetric determination for glycosidic and bile salt-based detergents: applications in membrane protein research. Anal. Biochem. 2005;336:117–124. doi: 10.1016/j.ab.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Velazhahan V., Ma N., Pándy-Szekeres G., Kooistra A.J., Lee Y., Gloriam D.E., Vaidehi N., Tate C.G. Structure of the class D GPCR Ste2 dimer coupled to two G proteins. Nature. 2020;589:148–153. doi: 10.1038/s41586-020-2994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar K.R., Henderson R. Single particle electron cryomicroscopy: trends, issues and future perspective. Q. Rev. Biophys. 2016;49:e13. doi: 10.1017/S0033583516000068. [DOI] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi Tan Y., Baldwin P.R., Davis J.H., Williamson J.R., Potter C.S., Carragher B., Lyumkis D. Addressing preferred specimen orientation in single-particle cryo-EMthrough tilting. Nat. Methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density maps of Mrp, ovine F1Fo-ATPase, ovine complex I, and ovine transhydrogenase are deposited in the EMDB with the accession numbers EMD-11027, EMD-10573, EMD-4635, and EMD-11246.