Abstract
As interest in space exploration rises, there is a growing need to quantify the impact of microgravity on the growth, survival, and adaptation of microorganisms, including those responsible for astronaut illness. Motility is a key microbial behavior that plays important roles in nutrient assimilation, tissue localization and invasion, pathogenicity, biofilm formation, and ultimately survival. Very few studies have specifically looked at the effects of microgravity on the phenotypes of microbial motility. However, genomic and transcriptomic studies give a broad general picture of overall gene expression that can be used to predict motility phenotypes based upon selected genes, such as those responsible for flagellar synthesis and function and/or taxis. In this review, we focus on specific strains of Gram-negative bacteria that have been the most studied in this context. We begin with a discussion of Earth-based microgravity simulation systems and how they may affect the genes and phenotypes of interest. We then summarize results from both Earth- and space-based systems showing effects of microgravity on motility-related genes and phenotypes.
Subject terms: Biophysics, Microbiology, Fluid dynamics
Background
Earth-based organisms, including microorganisms, have developed under the influence of gravity. As interest in spaceflight grows, it is important to understand the effects of microgravity on single-celled organisms including Bacteria, Archaea, and eukaryotes such as yeast and microalgae. In the microgravity environment particles experience weightlessness, such as in the case of constant free-fall in orbit aboard the International Space Station (ISS). The effects of microgravity exposure on microorganisms are of intense interest for medical and bioengineering applications, and many studies have been conducted over the past few decades, of which there are several comprehensive reviews1–6. However, a systematic summary of the effects of microgravity on microbial motility has not yet been done. Microbial motility is important for normal function of the human microbiome (e.g., in the gut and oral cavity)7,8 as well as for pathogenesis of some bacteria involved in common food- and water-borne infections9–12. Alterations in motility affect the distribution of microorganisms in tissues, encourage or inhibit bacterial invasiveness, and affect biofilm formation11,13. These changes may have important implications for astronaut health, especially combined with host factors; studies suggest that astronauts aboard the ISS suffer from compromised immune systems14,15 and altered microbiota16, potentially making them susceptible to opportunistic bacterial infections.
Microorganisms rapidly adapt to their environment by altering their gene expression to increase survivability. While earlier studies of microgravity effects on microorganisms were largely phenotypic, more recently “omics” techniques have become practical: genomics, transcriptomics, and proteomics, quantifying DNA, mRNA, and proteins, respectively17–19. Genomic studies are the most prevalent and in general the easiest to perform for bacteria, since many organisms have been fully sequenced and DNA-sequencing technology is widely available and relatively inexpensive20. RNA sequencing remains costly for transcriptomic studies, although less expensive microarray technologies may also be used19. Proteomics is a rapidly emerging field, but fractionation of bacteria for proteomic studies, particularly Gram-negative strains, is challenging21,22. Beginning with microarray studies of simulated microgravity responses in 200223, gene expression in microbes has been studied in both ground-based24–26 and spaceflight studies27,28. Transcriptomic and proteomic studies have also begun to appear29,30. Development of technologies for “omics” studies in space has been progressing but is not yet routinely used31. Sorting through the plethora of information available is rapidly becoming increasingly difficult, with numerous databases and software tools devoted to the task32–35.
An important consideration is the relationship between the presence of motility genes (and their transcripts) and phenotype. The genes of the flagellar regulon are expressed as a cascade, and transcriptional activators influence expression of motility genes based upon environmental conditions36,37. This illustrates the importance of performing linked transcriptomic/proteomic and phenotypic studies when working with a test strain to study changes over short time periods (minutes to hours). Despite this complexity, the overall presence of flagellar and chemoreceptor genes has been used to predict community-wide motility behaviors38,39; combined “omics” studies (genomic/transcriptomic) can capture diel and seasonal variations in motility40.
Spaceflight analog devices used on Earth, such as those shown in Fig. 1, mimic specific hallmarks of the microgravity environment including low fluid shear, lack of sedimentation, and low turbulence41. Thus, we will refer to the culture environment in spaceflight analog devices as ‘simulated microgravity.’ The impact of simulated microgravity studies goes beyond prediction of microbial changes during spaceflight. In fact, low fluid shear conditions also exist in vivo under special circumstances, such as in the microvilli of the intestines42. In these environments, organisms colonize and infect their hosts. Therefore, simulated microgravity devices can also be an invaluable tool to understanding mechanisms of the infection process43–45.
Fig. 1. Ground-based systems for simulating microgravity.
HARV, STLV, and RWPV images from Synthecon, Houston, TX with permission from Bill Anderson. RPM image reprinted from Wuest et al.139 with no modification under the CC BY 3.0 License (https://creativecommons.org/licenses/by/3.0/).
This review begins by summarizing the technologies available for simulating microgravity, with some recent analysis of the fluid mechanics of spaceflight analog devices. The possible effects of the different types of vessels on microorganisms apart from their simulation of weightlessness are discussed, as well as other complications that may arise from attempting to compare ground-based with ISS-based microbial gene expression studies. We then detail recent studies in gene expression regarding motility and chemotaxis in selected Gram-negative bacterial strains: Escherichia coli, Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, and Vibrio fischeri.
Simulated microgravity and fluid mechanics
Due to the difficulty of conducting experiments in space, ground-based bioreactors were developed by the NASA Johnson Space Center (Houston, TX) to study simulated microgravity on Earth46. These devices are generally called rotating wall vessels (RWV), though this terminology can be confusing2,47 as it includes different configurations such as: high aspect ratio vessels (HARVs)48, slow turning lateral vessels (STLVs)46,49, and rotating wall perfused vessels (RWPVs)50. Other devices used to simulate microgravity and of slightly different design are clinostats51 and random positioning machines (RPMs)52. Though they have been used to study simulated microgravity on plants51, and human cells and tissues53–55, this review’s focus is limited to effects on microbes.
Fluid behavior under simulated microgravity is characterized by low fluid shear, often referred to as low shear modeled microgravity (LSMMG)23. Low fluid shear values associated with the simulated microgravity environment are of the order ~10–2 dyne/cm243,50. The low fluid shear condition is dependent on many factors including vessel geometry, particle size, vessel rotation speed, and fluid properties. Though all vessel types create simulated microgravity, analysis differs slightly among types. Flow characterization can be approximated with the Reynolds number (Re), which is a ratio of the inertial to viscous forces. For example, for flow in a cylindrical vessel, the Reynolds number is given by Eq. (1):
| 1 |
where ρ is the fluid density, V is the fluid velocity, D is the particle diameter, and μ is the kinematic viscosity.
Common principles and the development of RWVs have been outlined in many sources41,50,56. Ground-based systems replicate weightlessness by rotating cells such that gravity vectors are nullified, and organisms do not have the opportunity to adapt to a specific gravity orientation. In other words, gravity is not altered; rather, the summation of gravity effects cancels out41,57. When the vessel is filled completely (zero headspace), the chamber contents resemble a rigid body. Fluid within the vessel is assumed both incompressible and Newtonian with approximately uniform density and viscosity. Vessel orientation determines whether microbes are under normal gravity (vertical axis) or simulated microgravity (horizontal axis) as seen in Fig. 2. A common method of control is to inoculate two chambers and grow cells in both configurations. However, caution should be exercised when comparing differences between these two configurations. It is possible that there are fluid dynamic effects under simulated microgravity that do not appear in the normal gravity configuration58,59. Different studies have employed both analytical and numerical techniques such as computational fluid dynamics (CFD) to elucidate the fluid mechanical effects and when they must be considered. This review aims to provide a brief overview of these analyses specific to device type when possible, focusing on those devices primarily used in microbial-simulated microgravity studies.
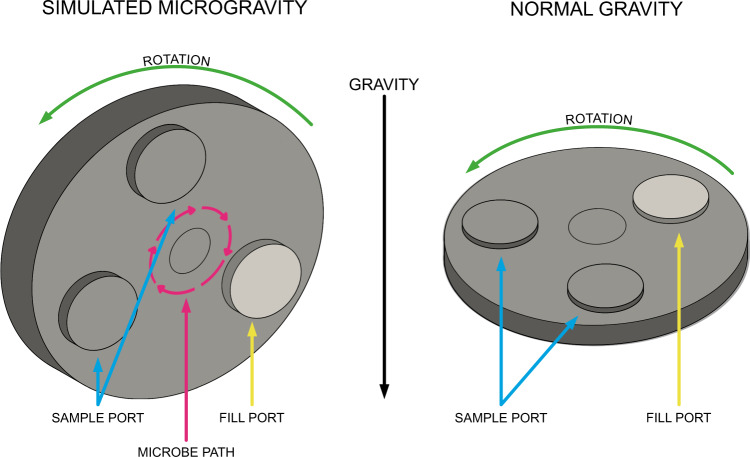
Fig. 2. 3D orientation of a rotating wall vessel (RWV) with the direction of the gravity vector included.
Left: Simulated microgravity showing a typical microbe path with gravity vectors canceling in the completely vertical orientation. Right: Normal gravity control with a horizontal orientation.
We begin with HARVs48, as these devices were used for most simulated microgravity studies presented in this review. These devices are cylindrical in shape with a membrane on the back to facilitate gas exchange60. Ayyaswamy and Mukundakrishnan59 outlined experimental conditions necessary for simulated microgravity in HARVs and STLVs including low fluid shear at the cell surface, adequate mass transport, and if microcarriers are used, such as when studying biofilm development61, to exercise careful thought about size and shape so as to minimize collisions with the vessel walls and adverse fluid effects.
The STLV49 consists of an inner and an outer cylinder, both of which may rotate along a horizontal axis, in either the same or opposite directions. Oxygenation is provided through a membrane along the inner column. The fluid behavior was initially reviewed in Hammond and Hammond41 in 2001, and further studies have been done since then. Gao et al.62 showed that particles tended to migrate radially when the particle density and fluid density differed. Liu et al.63 later replicated and expanded on this work, providing a comprehensive analysis of the forces on the particle. They also proposed rotational speeds consistent with simulated microgravity on inert particles and reaffirmed difficulties present with larger particle sizes, reporting fluid shear stress values several orders of magnitude higher than simulated microgravity.
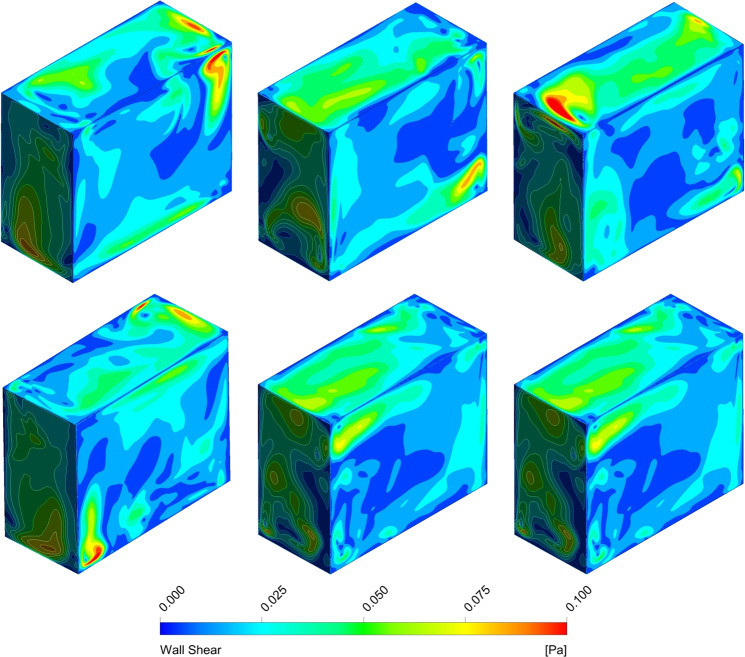
The RPM was developed from the clinostat52, and consists of two frames that can be independently driven. Therefore, an RPM can be operated in different modes64: clinostat mode (only one axis rotates), 3D clinostat mode (two axes rotated at constant speed), and 3D Random mode (two axes rotated at different speeds). Wuest et al.58 studied the fluid motion using a numerical approach, comparing the classic clinostat rotation with the 3D clinostat mode. They found that the fluid motion in the cell flask varied as a function of position and never seemed to reach a steady state, as seen in Fig. 3. Fluid shear stress values were found to be highest at the walls and could reach up to a 100 mPa. They urge that experimental design on microbes or tissues must consider fluid dynamic effects.
Fig. 3. Simulated shear stress values along the walls in an RPM.
Shear stress values vary throughout with no clear steady state. Reprinted from Wuest et al.58 with no alterations under the CC BY 4.0 License (https://creativecommons.org/licenses/by/4.0/).
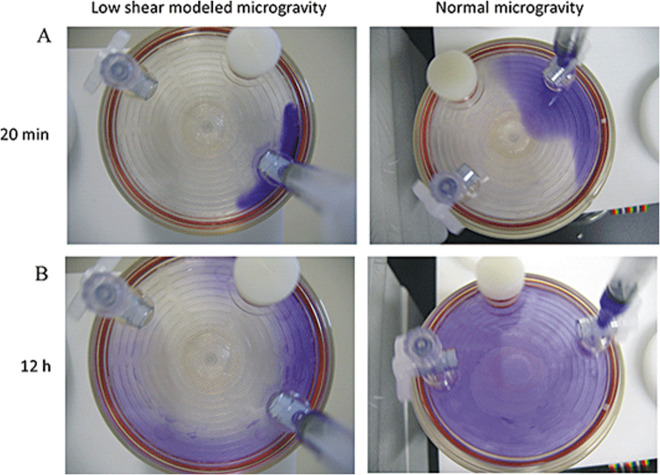
Although each device simulates microgravity, they do not all do so in the exact same manner. Consequently, organisms grown in one device may not react the same as another device47. When conducting a study of P. aeruginosa, Crabbe et al.65 found differing levels of gene expression when culturing in a HARV vs. in an RPM. They probed this difference by injecting dye into one HARV port as seen in Fig. 4. In their supplemental videos, they show how the dye spread to the center of chamber more slowly in the HARV than in the RPM indicating a subtle difference in fluid behavior. This could be a possible reason for the difference in gene expression.
Fig. 4. HARV bioreactors injected with crystal violet.
(Left) Under simulated microgravity, the dye stays along the outer wall before gradually migrating inward. (Right) The dye spread under normal gravity. Reprinted from Crabbe et al. 65 with permission from John Wiley and Sons with no alterations.
Simulated microgravity and imaging
Experimental calculation of fluid shear stress requires estimation of particle velocities41,50,66, but imaging under simulated microgravity conditions on Earth is challenging. Although any of the traditional methods described above can be used to study simulated microgravity, changes in gene expression can occur in minutes. This is particularly relevant for observing dynamic phenotypic changes in motility and chemotaxis. To address this, one approach is to attach a microscope to a simulated microgravity device, as was done with the clinostat microscope67 developed by the German Aerospace Research Establishment in 1996. The microscope is positioned to rotate horizontally and was used to study the behavior of Paramecium biaurelia in both simulated microgravity and spaceflight68. Another example of this was when Pache et al.69 attached a digital holographic microscope to an RPM, upgraded by Toy et al.70,71 to include a widefield epifluorescence microscopy module. Yew et al.72 developed a lab-on-a-chip clinorotation system as a more cost-effective alternative to the systems previously described. It uses 2D clinostat rotation and requires pauses for imaging.
Simulated microgravity, spaceflight and gene expression
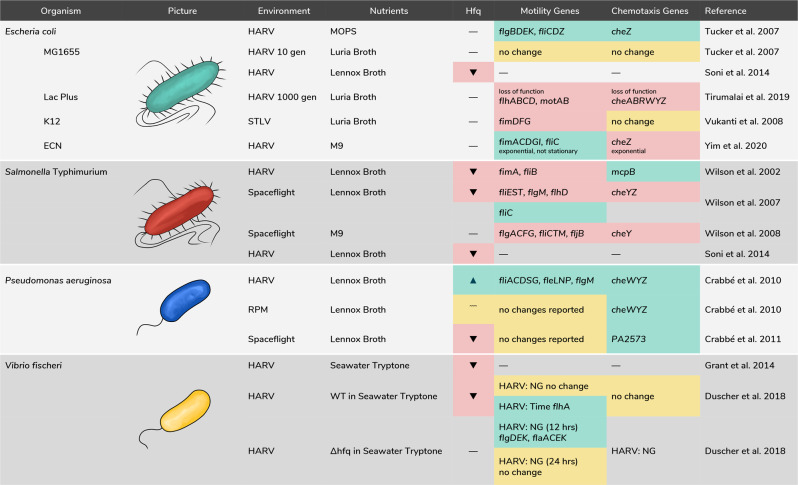
Gene regulation is a process in which gene expression is upregulated or downregulated and can be influenced by environmental factors. Here we report the influence of simulated microgravity and spaceflight on gene expression involving hfq regulation23,27,29,65,73–75, motility-related systems (flagella or fimbriae)23,26,27,29,30,65,74,76–79, and chemotaxis-related systems (chemical sensing fimbriae, pH sensing, media sensing)23,26,27,29,30,65,74,76–79. Results are illustrated in Fig. 5 with more details to follow. First, we provide a brief overview of hfq, motility and chemotaxis followed by organism-specific spaceflight and simulated microgravity studies.
Fig. 5. Motility and chemotaxis gene expression categorized by study and organism.
Color indicators: Green: upregulation, Yellow: no change reported, Red: downregulation. Dashes indicate no data available. Normal Gravity Rotation control is represented by NG.
Overview of hfq, motility and chemotaxis hfq
hfq
Hfq has emerged as an important post-transcriptional factor that facilitates the pairing of small RNAs with their target mRNAs; its role in bacteria has been recently reviewed80–82. To highlight its importance, in some organisms it can impact expression of up to 20% of all genes83. In a landmark study in 2007, Wilson et al.27 first identified Hfq as a global regulator in response to the spaceflight and simulated microgravity environment based on their global microarray and proteomic analyses. Additionally, changes in hfq gene expression can influence virulence of bacterial pathogens83 including S. Typhimurium83,84 thus making this gene a recent focus in simulated microgravity studies27,29,30,44.
Motility and chemotaxis
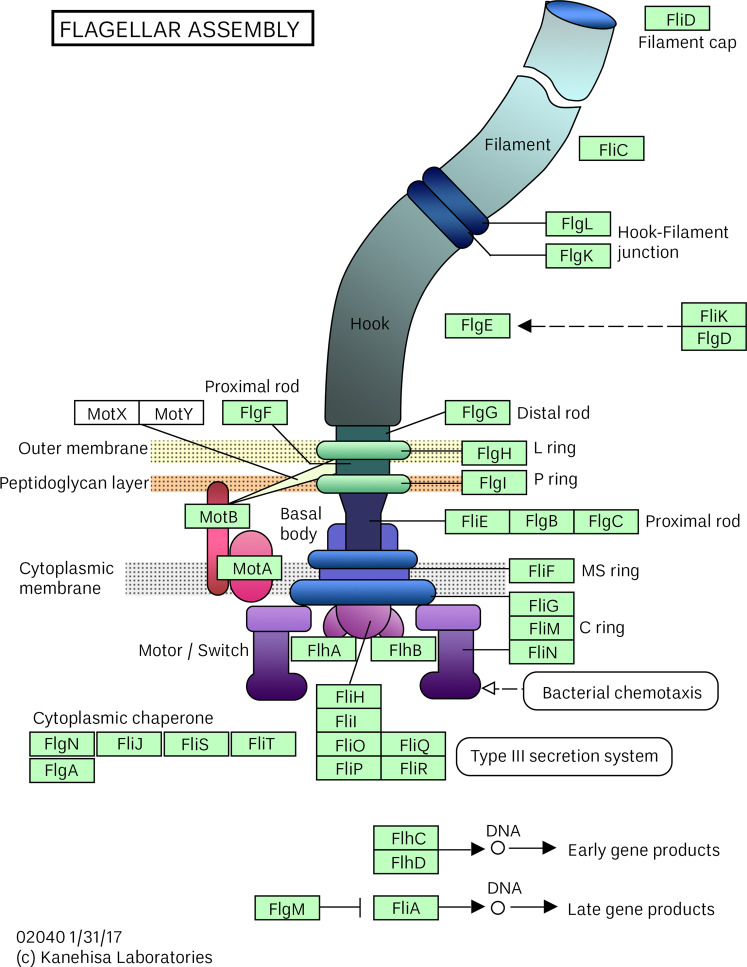
Changes in microbial motility can result from alterations in gene expression of motility machinery including flagella, fimbriae, and pili. Motor assembly and chemosensory machinery requires about 50 genes in E. coli and S. Typhimurium85. Studies have been conducted to identify genes86 and proteins87 involved in bacterial motility, with the E. coli flagellar assembly shown in Fig. 688. Flagellar genes are organized into regulatory hierarchies of classes depending on the organism, with each hierarchy affecting the next37,89,90. Flagella are not only important for motility85; they have also been shown to play a role in pathogenicity9,91–93 and biofilm formation13,94.
Fig. 6. Bacterial flagellar motor (E. coli) with associated protein components.
Eco02040 reprinted with modification under the CC BY 4.0 License (https://creativecommons.org/licenses/by/4.0/) based on KEGG, Kanehisa et al.88.
Motility and cell shape have been shown to influence cell growth95. Interestingly, transmission electron microscope (TEM) images have shown phenotypic changes in E. coli after spaceflight, with decreased size and increased presence of outer membrane vesicles as seen in Fig. 796. Benoit and Klaus3 reviewed microbial motility as it related to final cell populations, comparing motile and non-motile cultures during spaceflight, simulated microgravity and normal gravity controls. They revealed that spaceflight and simulated microgravity compared to normal gravity controls resulted in higher final cell counts in non-motile bacteria grown in liquid. Microbial motility is governed by an electromotive gradient of ions across the cell membrane, so concentration differences or nutrient depletion due to microgravity or simulated microgravity can affect cell motility85.
Fig. 7. Thin-section transmission electron microscopy images of E. coli.
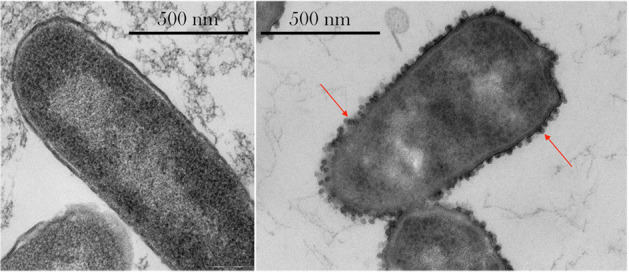
Left: Sample cultures on Earth. Right: Sample cultured in space exhibiting an irregular cell shape. Red arrows indicate extracellular vesicles. Images taken with a Phillips CM 100 TEM at an accelerating voltage of 80 kV. Figure and modified caption reprinted from Zea et al.96 under the CC BY 4.0 License (https://creativecommons.org/licenses/by/4.0/).
Chemotaxis is the response of a cell to a chemical gradient. Adler’s pioneering work97 in E. coli chemotaxis showed that movement towards attractants is independent of the mechanistic benefit of the compound itself. Although taxis components can vary between species, there are core components to all prokaryotic chemotactic systems98,99. Genomic analysis of chemotaxis shows that methyl-accepting chemotaxis proteins (MCPs), CheA and CheW are present in >95% of prokaryotic genomes that contain at least one chemotaxis gene, and CheBYR are common in ~90% of prokaryotic genomes100.
E. coli: hfq, motility, and chemotaxis
E. coli is an organism with motile and non-motile strains and is the most well studied and most thoroughly characterized organism on Earth101. E. coli K12 MG 1655 is motile with peritrichous flagella and fully sequenced102. E. coli Nissle (EcN) 1917 is another fully sequenced103, probiotic, non-pathogenic strain of E. coli lacking pathogenic adhesion factors; it carries genes that help limit the proliferation of other bacteria104. E. coli K12 MG 1655 has shown increased biofilm formation under simulated microgravity61.
E. coli and hfq
Tsui et al.105 showed that an hfq insertion mutation caused pleiotropic phenotypes, thus highlighting the importance of hfq expression in E. coli. Soni et al.73 studied the response of hfq and trp genes under simulated microgravity for various enterobacteria. Cultures of E. coli MG1655, DH5α, and AS11 were grown in Lennox broth using HARVs and compared with normal gravity rotation. They found hfq expression to be downregulated.
E. coli: motility and chemotaxis
In E. coli, motility is critical for biofilm formation94. Tucker et al.76 cultured E. coli MG1655 in both minimal MOPS medium and Luria broth, comparing cultures in HARVs vs. normal gravity rotation. In the MOPS media, flagellar genes flgBDEK and fliCDZ were upregulated, whereas no changes were observed with the Luria broth compared to normal gravity controls, respectively. Regarding chemotaxis, they found an upregulation of cheZ in the MOPS media vs. normal gravity rotation. Motility and chemotaxis are used by bacteria to identify and obtain nutrients; thus, expression levels vary with the level of nutrients in the environment. This study underlines the importance of this factor in designing studies of motility changes under simulated microgravity.
Tirumalai et al. conducted two studies77,79 analyzing gene mutation following different HARV cleaning protocols. In one study79, the HARVs were cleaned using steam sterilization; in a later study77, they used chloroamphenicol to prevent contamination. In both studies, they cultured 1000 generations of lac+ and lac− cultures of E. coli 1655, chosen because they could be visually distinguished. Following the chloramphenicol treatment, the lac+ cultures showed mutations in the predicted fimbriae-like adhesion proteins yadL. Flagellar and motility proteins also had loss of function mutations flhABCD, motAB. Since these changes were not present in the steam sterilization study, they were thought to aid in antibiotic resistance under simulated microgravity conditions. Additionally, they found loss of function mutations in the chemotaxis-related genes cheABRWYZ. Although they did not look at gene expression changes in motility specifically, Lynch et al.61 found that biofilm coverage of E. coli on microcarrier particles cultured in Luria Bertani broth in 10 mL HARVs was more pronounced than under normal gravity. Vukanti et al.26 cultured E. coli in Luria broth in an STLV and found a downregulation of fimDFG. Yim et al.106 grew EcN in M9 minimal media in 10 mL HARVs. Sample collection followed exponential and stationary growth. They found upregulation of fliC, fimACDGI after exponential growth, but changes did not persist in samples taken from stationary growth. Regarding chemotaxis, they also found a downregulation of cheZ following the same conditions.
S. Typhimurium: hfq, motility and chemotaxis
S. Typhimurium is motile with peritrichous flagella, a fully sequenced107 pathogenic organism shown to have increased virulence after simulated microgravity exposure108 and spaceflight27,74.
S. Typhimurium and hfq
In non-space-related studies, Sittka et al.109 showed that in S. Typhimurium Hfq influences 87% of genes in the flagellar system and 84% of genes in the chemotaxis system. Monteiro et al.110 showed that Hfq controls biofilm formation through regulation of CsgD in S. Typhimurium. Since changes in hfq expression have been shown to impact expression of genes related to motility109, chemotaxis109, and biofilm formation110 in S. Typhimurium in Earth-based studies, studies were conducted to ascertain whether these changes persisted under simulated microgravity and spaceflight. In a study conducted by Wilson et al. of bacterial gene expression after spaceflight27, hfq expression was down-regulated. Noting that simulated microgravity induced acid resistance in a previous study at late log phase23, they cultured both the wild type and an hfq-deficient mutant in Lennox broth in HARVs, comparing survivability. The wild type showed greater survivability in normal gravity rotation vs. simulated microgravity. However, this difference in survivability was not observed between simulated microgravity and normal gravity rotation for the hfq-deficient strain. Pacello et al.111 cultured S. Typhimurium in Luria Bertani broth in 10 mL HARVs using normal gravity as a control. They found that simulated microgravity increased acid resistance even in the absence of hfq as compared to normal gravity. Soni et al.73 cultured S. Typhimurium in Lennox broth in HARVs using normal gravity rotation as a control. They observed a downregulation of hfq.
S. Typhimurium: motility and chemotaxis
S. Typhimurium motility has been shown to increase invasiveness9, therefore making changes in motility important from a health perspective. Wilson et al.23 compared cultures of S. Typhimurium grown in Lennox broth in HARVs and normal gravity, finding 163 genes differentially expressed under simulated microgravity, including downregulation of fimA, fliB. They reported an upregulation of mcpB, a gene identified in chemotaxis in S. Typhimurium112. In a later study27, they compared spaceflight S. Typhimurium samples with controls on the ground kept under similar temperature and nutrient conditions. They showed downregulation in fliEST, flgM, flhD; however, fliC was upregulated. Regarding chemotaxis, they found a downregulation of cheYZ. In another study74, they cultured S. Typhimurium in M9 media during spaceflight and showed eight genes in flgACFG, fliCMT, fljB downregulated compared to ground-based controls. Additionally, they found a downregulation of cheY.
P. aeruginosa: hfq, motility and chemotaxis
P. aeruginosa is motile with a single polar flagellum, fully sequenced113, and occasionally a part of human flora. In spaceflight114 and simulated microgravity29, it has shown increased biofilm formation.
P. aeruginosa and hfq
Crabbe et al.65 investigated global changes in gene expression including the hfq-dependent response under simulated microgravity using both a HARV and an RPM in P. aeruginosa PAO1. Hfq was upregulated using the HARV as compared to normal gravity rotation, but not differentially expressed when comparing the RPM to normal gravity rotation. In P. aeruginosa, the loss of hfq can result in reduction in growth115. Interestingly, comparison of total bacterial counts showed no significant differences between HARV, RPM, and normal gravity. Crabbe et al.29 also investigated the hfq-dependent response after spaceflight, comparing their results with their earlier simulated microgravity study. In contrast to the HARV results, they found a downregulation of hfq after spaceflight. This finding was significant because following S. Typhimurium, it showed Hfq as a regulator acting across bacterial species.
P. aeruginosa: motility and chemotaxis
P. aeruginosa is associated with infections in immunocompromised hosts. Motility, particularly the presence of fliC, plays a key role in its pathogenesis91. Crabbe et al.65 cultured P. aeruginosa in Lennox broth in HARVS and RPMs and cataloged changes in motility, finding upregulation of motility genes fliACDGS, fleLNP, flgM but no apparent changes under the RPM. No significant changes in motility gene expression were reported after spaceflight. In contrast to what is seen in E. coli and S. Typhimurium, the chemotactic response pathway in P. aeruginosa involves more than 20 che genes and 26 MCP-like genes116. Crabbe et al.65 showed upregulation of chemotaxis genes cheWYZ in the HARV and to a lesser extent in the RPM compared to normal gravity. Spaceflight29 showed an upregulation of PA2573, an MCP homolog117 compared to ground controls. Kim et al.114,118 compared biofilm formation in P. aeruginosa PA14 during spaceflight with ground controls finding that spaceflight not only increased biofilm formation but also revealed a different biofilm architecture than normally appears on Earth that they termed “column-and-canopy”. Additionally, they investigated biofilm formation in the wild type, mutants deficient in flagella-driven motility, ΔmotABCD and type IV pili-driven motility ΔpilB. The wild type and ΔpilB made the column-and-canopy architecture, while the ΔmotABCD did not, thus underscoring the importance of motility in the formation of this architecture.
V. fischeri: hfq, motility and chemotaxis
V. fischeri is a motile marine bacterium with a single polar flagellum, completely sequenced119. It forms a symbiotic relationship with the bobtail squid Euprymna scolopes and as such is a good model for understanding how such relationships can change in simulated microgravity120.
V. fischeri and hfq
Hfq expression was also studied in V. fischeri and its resulting effect on E. scolopes. Grant et al.75 placed both symbiotic partners in HARVs and examined changes in V. fischeri’s colonization of E. scolopes at different stages of development, using both the wild type and a mutant with a non-functioning hfq gene. Under simulated microgravity, both the wild type and mutant reached higher cell counts than under normal gravity. Hfq was also downregulated under simulated microgravity. No differences in the ability of V. fischeri to colonize E. scolopes were observed between the wild type and hfq mutant, though colonization occurred more quickly under simulated microgravity with both strains. V. fischeri is also critical for morphogenesis of the light organ in E. scolopes by triggering developmental events121. The wild type, during simulated microgravity, negatively impacted these developmental events, while the hfq mutant did not. This coupled with the fact that the hfq mutant under normal gravity also negatively impacted these developmental events showed that hfq while not necessary for colonization, was still necessary for light organ morphogenesis.
V. fischeri: motility and chemotaxis
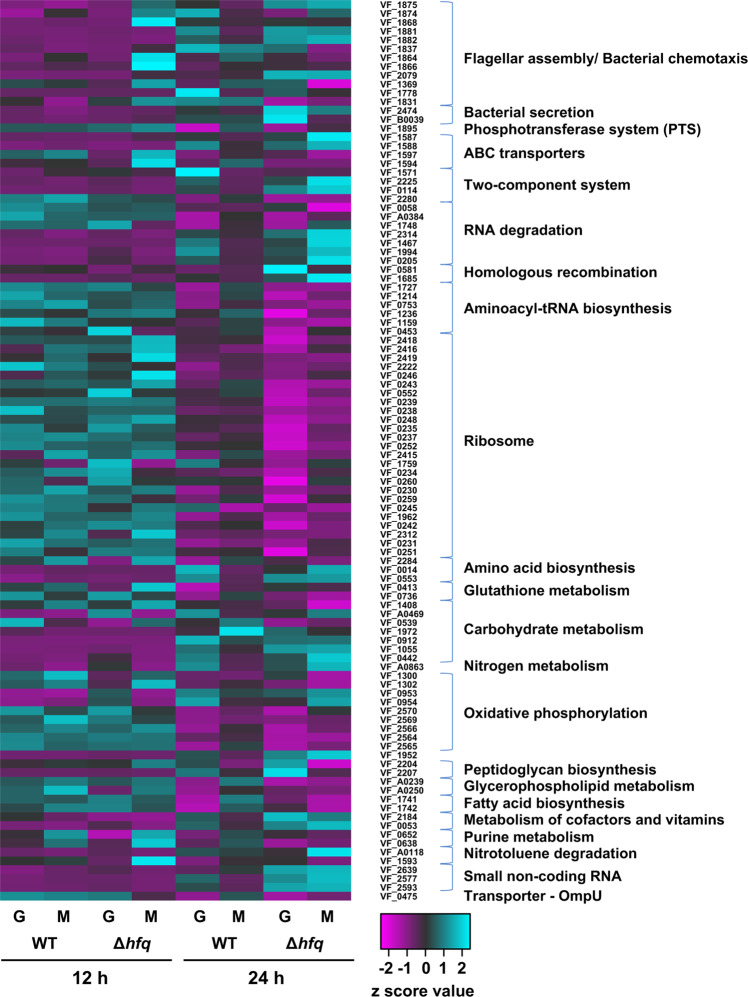
Motility is necessary for V. fischeri to successfully colonize E. scolopes122. Duscher et al.30 investigated V. fischeri exposure to simulated microgravity using HARVs. The 12- and 24-h cultures of V. fischeri (wild type) and associated global regulator hfq protein knockout strains (∆hfq) were grown in seawater tryptone. They used a heat map to qualitatively depict changes in gene expression as shown in Fig. 830. When comparing the wild type grown in simulated microgravity vs. normal gravity after 12 and 24 h, no genes showed significant differences. However, comparison of the wild type after 12- and 24-h exposure to simulated microgravity showed an upregulation in flhA similar to an upregulation of flgEM when viewing changes under normal gravity. The ∆hfq strain after 12 h under simulated microgravity showed upregulation of flgDEK, flaACEK as compared to normal gravity. After 24 h under simulated microgravity, these changes did not seem to persist. The ability to chemotax is an advantage for V. fischeri when colonizing E. scolopes, though not strictly necessary123. Duscher et al.30 did not report many changes to chemotaxis gene expression. Under simulated microgravity, only the ∆hfq showed a downregulation of V. fischeri chemotaxis genes.
Fig. 8. Heat map depicting the clustering patterns of the eight treatments by KEGG pathways associated with the proposed function of V. fischeri genes at 12 and 24 h.
Gene changes governing flagellar assembly and bacterial chemotaxis labeled at the top. Colors represent the differential abundance of individual genes listed by V. fischeri identification number (VF-ID) for both wild type (WT) and Δhfq mutants under simulated microgravity (M) and normal gravity (G) conditions. Figure and modified caption reprinted from Duscher et al.30 under the conditions of the CC BY 4.0 License (https://creativecommons.org/licenses/by/4.0/).
Conclusions
There is still much work needed to understand microbial response to microgravity. Gene expression can vary based on environmental factors including temperature, nutrients, and fluid shear stress ranges. A recent study argued that there is no identifiable common bacteria “spaceflight response,”124 although another suggested Hfq as a general spaceflight regulon27.
The two gravity-dependent processes believed to most influence bacteria are indirect, namely (a) settling (of both cells and nutrients) and (b) buoyant convection125. Both affect non-motile microorganisms more than motile cells, since flagellar motility stirs the liquid surrounding the cell as well as permitting the cell to avoid settling. Thus, increased growth seen under microgravity conditions was initially hypothesized to be restricted to non-motile cells3; however, later studies showed that this was not universally the case. Instead, phosphate and/or oxygen availability are decreased in microgravity due to lack of convection, leading to altered microbial behavior, such as in P. aeuginosa118. Reduced convection leading to substrate concentration gradients has been proposed as a general mechanism underlying all microbial alterations seen in microgravity126. These models are supported by studies using diamagnetic levitation to simulate weightlessness. Levitation prevents settling but increases convection because of the diamagnetic properties of oxygen127.
What is known about low-oxygen and low-phosphate environments can help inform microgravity studies, but careful attention to experimental and strain differences is essential. Low-nutrient environments have different effects on motility depending upon the motility features (e.g., run-and-tumble vs. forward-reverse128) and stress responses of the particular strain. A significant number of studies have been done with human pathogens, since limiting environments are frequently encountered during the infection process, for example in intestinal villi, and can trigger the transition from planktonic to biofilm in many Gram-negatives129. Reduced phosphate leads to increased virulence and swarming motility in P. aeruginosa130. Oxygen availability influences microbial pathogenicity at all stages of the infection process131; S. Typhimurium grown in low oxygen environments shows greater adhesion to and invasion of epithelial cells132. In the mutualistic V. fischeri, which transitions from motile to non-motile as it enters stationary phase, the low-nutrient microenvironments in microgravity may simulate the transition to stationary phase30.
Downregulation of hfq expression was the most consistent finding in the studies we focused on here and is a common theme in stress-response studies as well133–136. As mentioned previously, Hfq is a global transcriptional regulator that has been found in approximately half of all known bacterial genomes. It is an RNA chaperone which can serve as both a positive and a negative regulator. Hfq stabilizes small RNAs (sRNAs) and acts as a platform for sRNA–mRNA interaction; regulation by sRNAs requires Hfq. Hfq-dependent sRNAs play a key role in regulation of flagellar genes by acting on the master regulator, FlhD, as well as other factors (refer again to Fig. 6)137. Almost 90% of flagellar genes are Hfq-regulated in common Gram-negative pathogens138. The general downregulation of hfq expression seen in spaceflight and simulated microgravity studies may be related to oxygen and micronutrient availability.
Understanding changes in gene expression is an important step in understanding phenotypic changes. The results shown here suggest that swimming speeds and patterns of microbes could be altered under simulated microgravity conditions. Motility and chemotaxis have evolved to provide some microbes an evolutionary advantage. It remains to be seen whether prolonged exposure to simulated microgravity could fundamentally alter both motility gene expression and swimming phenotypes. Experiments involving imaging during and after exposure to simulated microgravity and spaceflight to quantify motility and chemotaxis behaviors remain to be performed.
Acknowledgements
The authors acknowledge the support of the National Science Foundation Grant Number 1828793. Portions of this work were supported under a contract from the Jet Propulsion Laboratory, California Institute of Technology, under a contract with the National Aeronautics and Space Administration.
Author contributions
All authors contributed towards the writing of this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rosenzweig JA, et al. Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl. Microbiol. Biotechnol. 2010;85:885–891. doi: 10.1007/s00253-009-2237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL. Microbial responses to microgravity and other low-shear environments. Microbiol. Mol. Biol. Rev. 2004;68:345–361. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit, M. R. & Klaus, D. M. Microgravity, bacteria, and the influence of motility. Adv. Space Res.39, 1225-1232 (2007).
- 4.Horneck G, Klaus DM, Mancinelli RL. Space microbiology. Microbiol. Mol. Biol. Rev. 2010;74:121–156. doi: 10.1128/MMBR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenzweig, J. A., Ahmed, S., Eunson, J. & Chopra, K.A. Low-shear force associated with modeled microgravity and spaceflight does not similarly impact the virulence of notable bacterial pathogens. Appl. Microbiol. Biotechnol.98, 8797–8807 (2014). [DOI] [PMC free article] [PubMed]
- 6.Higginson, E. E., Galen, J. E., Levine, M. M. & Tennant, S. M. Microgravity as a biological tool to examine host-pathogen interactions and to guide development of therapeutics and preventatives that target pathogenic bacteria. Pathog. Dis.74, ftw095 (2016). [DOI] [PMC free article] [PubMed]
- 7.De Angelis, M. et al. Diet influences the functions of the human intestinal microbiome. Sci. Rep.10, 4247 (2020). [DOI] [PMC free article] [PubMed]
- 8.Shrivastava A, et al. Cargo transport shapes the spatial organization of a microbial community. Proc. Natl Acad. Sci. USA. 2018;115:8633–8638. doi: 10.1073/pnas.1808966115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stecher, B. et al. Flagella and chemotaxis are required for efficient induction of Salmonella enterica Serovar Typhimurium Colitis in streptomycin-pretreated mice. Infect. Immun.72, 4138–4150 (2004). [DOI] [PMC free article] [PubMed]
- 10.Kao CY, et al. The complex interplay among bacterial motility and virulence factors in different Escherichia coli infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:2157–2162. doi: 10.1007/s10096-014-2171-2. [DOI] [PubMed] [Google Scholar]
- 11.Duan Q, Zhou M, Zhu L, Zhu G. Flagella and bacterial pathogenicity. J. Basic Microbiol. 2013;53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 12.Josenhans C, Suerbaum S. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 2002;291:605–614. doi: 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- 13.Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013;37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akiyama, T. et al. How does spaceflight affect the acquired immune system? npj Microgravity6, 14 (2020). [DOI] [PMC free article] [PubMed]
- 15.Cervantes JL, Hong BY. Dysbiosis and immune dysregulation in outer space. Int. Rev. Immunol. 2016;35:67–82. doi: 10.3109/08830185.2015.1027821. [DOI] [PubMed] [Google Scholar]
- 16.Voorhies, A. A. et al. Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Sci. Rep.9, 9911 (2019). [DOI] [PMC free article] [PubMed]
- 17.Grimm D, et al. The impact of microgravity-based proteomics research. Expert Rev. Proteom. 2014;11:465–476. doi: 10.1586/14789450.2014.926221. [DOI] [PubMed] [Google Scholar]
- 18.Milojevic, T. & Weckwerth, W. Molecular mechanisms of microbial survivability in outer space: a systems biology approach. Front. Microbiol.11, 923 (2020). [DOI] [PMC free article] [PubMed]
- 19.Manzoni C, et al. Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief Bioinforma. 2018;19:286–302. doi: 10.1093/bib/bbw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raskin DM, Seshadri R, Pukatzki SU, Mekalanos JJ. Bacterial genomics and pathogen. Evolution. 2006;124:703–714. doi: 10.1016/j.cell.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Thein M, Sauer G, Paramasivam N, Grin I, Linke D. Efficient subfractionation of Gram-negative bacteria for proteomics studies. J. Proteome Res. 2010;9:6135–6147. doi: 10.1021/pr1002438. [DOI] [PubMed] [Google Scholar]
- 22.Pérez-Llarena, F. J. & Bou, G. Proteomics as a tool for studying bacterial virulence and antimicrobial resistance. Front. Microbiol.7, 410 (2016). [DOI] [PMC free article] [PubMed]
- 23.Wilson JW, et al. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc. Natl Acad. Sci. USA. 2002;99:13807–13812. doi: 10.1073/pnas.212387899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abshire CF, et al. Exposure of Mycobacterium marinum to low-shear modeled microgravity: effect on growth, the transcriptome and survival under stress. npj Microgravity. 2016;2:16038. doi: 10.1038/npjmgrav.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosado, H. et al. Rotating wall vessel exposure alters protein secretion and global gene expression in Staphylococcus aureus. Int. J. Astrobiology11, 71–81 (2012).
- 26.Vukanti R, Mintz E, Leff L. Changes in gene expression of E. coli under conditions of modeled reduced gravity. Microgravity—Sci. Technol. 2008;20:41–57. doi: 10.1007/s12217-008-9012-9. [DOI] [Google Scholar]
- 27.Wilson JW, et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl Acad. Sci. USA. 2007;104:16299–16304. doi: 10.1073/pnas.0707155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aunins, T. R. et al. Spaceflight modifies Escherichia coli gene expression in response to antibiotic exposure and reveals role of oxidative stress response. Front. Microbiol.9, 310 (2018). [DOI] [PMC free article] [PubMed]
- 29.Crabbe A, et al. Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl. Environ. Microbiol. 2011;77:1221–1230. doi: 10.1128/AEM.01582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duscher, A. A. et al. Transcriptional profiling of the mutualistic bacterium Vibrio fischeri and an hfq mutant under modeled microgravity. npj Microgravity4, 25 (2018). [DOI] [PMC free article] [PubMed]
- 31.Karouia F, Peyvan K, Pohorille A. Toward biotechnology in space: High-throughput instruments for in situ biological research beyond Earth. Biotechnol. Adv. 2017;35:905–932. doi: 10.1016/j.biotechadv.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 32.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaughan, L. K. & Srinivasasainagendra, V. Where in the genome are we? A cautionary tale of database use in genomics research. Front. Genet.4, 38 (2013). [DOI] [PMC free article] [PubMed]
- 34.Pinu FR, et al. Systems biology and multi-omics integration: viewpoints from the metabolomics research community. Metabolites. 2019;9:76. doi: 10.3390/metabo9040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray S, et al. GeneLab: omics database for spaceflight experiments. Bioinformatics. 2019;35:1753–1759. doi: 10.1093/bioinformatics/bty884. [DOI] [PubMed] [Google Scholar]
- 36.Zhao K, Liu M, Burgess RR. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 2007;35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica Serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000;64:694–708. doi: 10.1128/MMBR.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandre G, Greer-Phillips S, Zhulin IB. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 2004;28:113–126. doi: 10.1016/j.femsre.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Delong EF. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert JA, et al. The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic’ study of seasonal and diel temporal variation. PLoS ONE. 2010;5:e15545. doi: 10.1371/journal.pone.0015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond TG, Hammond JM. Optimized suspension culture: the rotating-wall vessel. Am. J. Physiol. Ren. Physiol. 2001;281:F12–F25. doi: 10.1152/ajprenal.2001.281.1.F12. [DOI] [PubMed] [Google Scholar]
- 42.Guo P, Weinstein AM, Weinbaum S. A hydrodynamic mechanosensory hypothesis for brush border microvilli. Am. J. Physiol.-Ren. Physiol. 2000;279:F698–F712. doi: 10.1152/ajprenal.2000.279.4.F698. [DOI] [PubMed] [Google Scholar]
- 43.Nauman EA, et al. Novel quantitative biosystem for modeling physiological fluid shear stress on cells. Appl. Environ. Microbiol. 2007;73:699–705. doi: 10.1128/AEM.02428-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castro SL, Nelman-Gonzalez M, Nickerson CA, Ott CM. Induction of attachment-independent biofilm formation and repression of hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl. Environ. Microbiol. 2011;77:6368–6378. doi: 10.1128/AEM.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen CA, Niesel DW, Torres AG. The effects of low-shear stress on adherent-invasive Escherichia coli. Environ. Microbiol. 2008;10:1512–1525. doi: 10.1111/j.1462-2920.2008.01567.x. [DOI] [PubMed] [Google Scholar]
- 46.Ray P. Schwarz, D. A. W. Rotating bio-reactor cell culture apparatus. United States patent 4988623 (1991).
- 47.Herranz R, et al. Ground-based facilities for simulation of microgravity: organism-specific recommendations for their use, and recommended terminology. Astrobiology. 2013;13:1–17. doi: 10.1089/ast.2012.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prewett, T. L., Goodwin, T. J. & Spaulding, G. F. Three-dimensional modeling of T-24 human bladder carcinoma cell line: a new simulated microgravity culture vessel. J. Tiss. Cult. Meth.15, 29–36 (1993).
- 49.Schwarz RP, Goodwin TJ, Wolf DA. Cell culture for three-dimensional modeling in rotating-wall vessels: an application of simulated microgravity. J. Tissue Cult. Methods. 1992;14:51–57. doi: 10.1007/BF01404744. [DOI] [PubMed] [Google Scholar]
- 50.Begley CM, Kleis SJ. The fluid dynamic and shear environment in the NASA/JSC rotating-wall perfused-vessel bioreactor. Biotechnol. Bioeng. 2000;70:32–40. doi: 10.1002/1097-0290(20001005)70:1<32::AID-BIT5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 51.Hoson T, Kamisaka S, Masuda Y, Yamashita M. Changes in plant growth processes under microgravity conditions simulated by a three-dimensional clinostat. Bot. Mag. = Shokubutsu-gaku-zasshi. 1992;105:53–70. doi: 10.1007/BF02489403. [DOI] [Google Scholar]
- 52.van Loon JJWA. Some history and use of the random positioning machine, RPM, in gravity related research. Adv. Space Res. 2007;39:1161–1165. doi: 10.1016/j.asr.2007.02.016. [DOI] [Google Scholar]
- 53.Barrila J, et al. Organotypic 3D cell culture models: using the rotating wall vessel to study host–pathogen interactions. Nat. Rev. Microbiol. 2010;8:791–801. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- 54.Barrila, J. et al. Modeling host–pathogen interactions in the context of the microenvironment: three-dimensional cell culture comes of age. Infect. Immunity86, 00282-18 (2018). [DOI] [PMC free article] [PubMed]
- 55.Grimm D, et al. Growing tissues in real and simulated microgravity: new methods for tissue engineering. Tissue Eng. Part B. 2014;20:555–566. doi: 10.1089/ten.teb.2013.0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolf, D. A. K. S. J. Principles of Analogue and True Microgravity Bioreactors to Tissue Engineering. In C.A. Nickerson, N.R. Pellis, M.C. Ott (Eds.), Effect of Spaceflight and Spaceflight Analogue Culture on Human and Microbrial Cells, 39–60 (2016).
- 57.Klaus DM. Clinostats and bioreactors. Gravit. Space Biol. Bull. 2001;14:55–64. [PubMed] [Google Scholar]
- 58.Wuest SL, Stern P, Casartelli E, Egli M. Fluid dynamics appearing during simulated microgravity using random positioning machines. PLoS ONE. 2017;12:e0170826. doi: 10.1371/journal.pone.0170826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayyaswamy PS, Mukundakrishnan K. Optimal conditions for simulating microgravity employing NASA designed rotating wall vessels. Acta Astronaut. 2007;60:397–405. doi: 10.1016/j.actaastro.2006.09.008. [DOI] [Google Scholar]
- 60.Barnes, S. J. & Harris, L. P. Tissue Engineering: Roles, Materials, and Applications (Nova Science Publishers, 2008).
- 61.Lynch SV, Mukundakrishnan K, Benoit MR, Ayyaswamy PS, Matin A. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl. Environ. Microbiol. 2006;72:7701–7710. doi: 10.1128/AEM.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao H, Ayyaswamy P, Ducheyne P. Dynamics of a microcarrier particle in the simulated microgravity environment of a rotating-wall vessel. Microgravity Sci. Technol. 1997;10:154–165. [PubMed] [Google Scholar]
- 63.Liu T, Li X, Sun X, Ma X, Cui Z. Analysis on forces and movement of cultivated particles in a rotating wall vessel bioreactor. Biochem. Eng. J. 2004;18:97–104. doi: 10.1016/S1369-703X(03)00171-2. [DOI] [Google Scholar]
- 64.Borst AG, Van Loon JJWA. Technology and developments for the random positioning machine, RPM. Microgravity Sci. Technol. 2009;21:287–292. doi: 10.1007/s12217-008-9043-2. [DOI] [Google Scholar]
- 65.Crabbé, A. et al. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol.12, 1545–1564 (2010). [DOI] [PubMed]
- 66.Ramirez LES, Lim EA, Coimbra CFM, Kobayashi MH. On the dynamics of a spherical scaffold in rotating bioreactors. Biotechnol. Bioeng. 2003;84:382–389. doi: 10.1002/bit.10778. [DOI] [PubMed] [Google Scholar]
- 67.Friedrich ULD, Joop O, Pütz C, Willich G. The slow rotating centrifuge microscope NIZEMI—a versatile instrument for terrestrial hypergravity and space microgravity research in biology and materials science. J. Biotechnol. 1996;47:225–238. doi: 10.1016/0168-1656(96)01371-5. [DOI] [PubMed] [Google Scholar]
- 68.Hemmersbach R, Voormanns R, Häder DP. Graviresponses in Paramecium biaurelia under different accelerations: studies on the ground and in space. J. Exp. Biol. 1996;199:2199. [PubMed] [Google Scholar]
- 69.Pache C, et al. Digital holographic microscopy real-time monitoring of cytoarchitectural alterations during simulated microgravity. J. Biomed. Opt. 2010;15:026021. doi: 10.1117/1.3377960. [DOI] [PubMed] [Google Scholar]
- 70.Toy, M. F. et al. Dual-mode Digital Holographic and Fluorescence Microscopy for the Study of Morphological Changes in Cells Under Simulated Microgravity. In J.Conchello, C.J. Cogswell, T. Wilson (Eds.), Three-Dimensional and Multidimensional Microscopy: Image Acquisition and Processing XVII, 7570 (SPIE, 2010).
- 71.Toy MF, et al. Enhanced robustness digital holographic microscopy for demanding environment of space biology. Biomed. Opt. Express. 2012;3:313–326. doi: 10.1364/BOE.3.000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yew AG, Atencia J, Hsieh AH. Lab-on-chip clinorotation system for live-cell microscopy under simulated microgravity. Cell. Mol. Bioeng. 2013;7:165–170. doi: 10.1007/s12195-013-0319-2. [DOI] [Google Scholar]
- 73.Soni A, et al. Conservation of the low-shear modeled microgravity response in Enterobacteriaceae and analysis of the trp genes in this response. Open Microbiol. J. 2014;8:51–58. doi: 10.2174/1874285801408010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson, J. W. et al. Media ion composition controls regulatory and virulence response of Salmonella in spaceflight. PLoS One3, e3923 (2008). [DOI] [PMC free article] [PubMed]
- 75.Grant KC, Khodadad CLM, Foster JS. Role of Hfq in an animal–microbe symbiosis under simulated microgravity conditions. Int. J. Astrobiol. 2014;13:53–61. doi: 10.1017/S1473550413000359. [DOI] [Google Scholar]
- 76.Tucker DL, et al. Characterization of Escherichia coli MG1655 grown in a low-shear modeled microgravity environment. BMC Microbiol. 2007;7:15. doi: 10.1186/1471-2180-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tirumalai, M. R. et al. Evaluation of Acquired antibiotic resistance in Escherichia coli exposed to long-term low-shear modeled microgravity and background antibiotic exposure. mBio10, 02637-18 (2019). [DOI] [PMC free article] [PubMed]
- 78.Yim J, et al. Transcriptional profiling of the probiotic Escherichia coli Nissle 1917 strain under simulated microgravity. Int. J. Mol. Sci. 2020;21:2666. doi: 10.3390/ijms21082666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tirumalai MR, et al. The adaptation of Escherichia coli cells grown in simulated microgravity for an extended period is both phenotypic and genomic. NPJ Microgravity. 2017;3:15. doi: 10.1038/s41526-017-0020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 81.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011;3:a003798–a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 84.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Derosier DJ. The turn of the screw: the bacterial flagellar. Motor. 1998;93:17–20. doi: 10.1016/s0092-8674(00)81141-1. [DOI] [PubMed] [Google Scholar]
- 86.Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:e154. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rajagopala SV, et al. The protein network of bacterial motility. Mol. Syst. Biol. 2007;3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kanehisa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 90.Anderson JK, Smith TG, Hoover TR. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol. 2010;18:30–37. doi: 10.1016/j.tim.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feldman M, et al. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 1998;66:43–51. doi: 10.1128/IAI.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin. Cell Dev. Biol. 2015;46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 93.Ottemann KM, Miller JF. Roles for motility in bacterial–host interactions. Mol. Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 94.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 95.Portela, R., Almeida, P. L., Sobral, R. G. & Leal, C. R. Motility and cell shape roles in the rheology of growing bacteria cultures. Eur. Phys. J. E42, 26 (2019). [DOI] [PubMed]
- 96.Zea, L. et al. Phenotypic changes exhibited by E. coli cultured in space. Front. Microbiol.8, 1598 (2017). [DOI] [PMC free article] [PubMed]
- 97.Adler J. Chemoreceptors in bacteria. Science. 1969;166:1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- 98.Hazelbauer GL. Bacterial chemotaxis: the early years of molecular studies. Annu. Rev. Microbiol. 2012;66:285–303. doi: 10.1146/annurev-micro-092611-150120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 100.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in Prokaryotes. Sci. Signal. 2010;3:ra50–ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blount, Z. D. The unexhausted potential of E. coli. eLife4, e05826 (2015). [DOI] [PMC free article] [PubMed]
- 102.Hayashi, K. et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol.2, 2006.0007 (2006). [DOI] [PMC free article] [PubMed]
- 103.Grozdanov L, et al. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 2004;186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sonnenborn U, Schulze J. The non-pathogenic Escherichia coli strain Nissle 1917—features of a versatile probiotic. Microb. Ecol. Health Dis. 2009;21:122–158. [Google Scholar]
- 105.Tsui H-CT, Leung H-CE, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 106.Yim J, et al. Transcriptional profiling of the probiotic Escherichia coli Nissle 1917 strain under simulated microgravity. Int. J. Mol. Sci. 2020;21:2666. doi: 10.3390/ijms21082666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McClelland M, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 108.Nickerson CA, et al. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 2000;68:3147–3152. doi: 10.1128/IAI.68.6.3147-3152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sittka A, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Monteiro C, et al. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 2012;9:489–502. doi: 10.4161/rna.19682. [DOI] [PubMed] [Google Scholar]
- 111.Pacello F. Low-shear modeled microgravity enhances Salmonella Enterica resistance to hydrogen peroxide through a mechanism involving KatG and KatN. Open Microbiol. J. 2012;6:53–64. doi: 10.2174/1874285801206010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frye J, et al. Identification of new flagellar genes of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 114.Kim W, et al. Spaceflight promotes biofilm formation by Pseudomonas aeruginosa. PLoS ONE. 2013;8:e62437. doi: 10.1371/journal.pone.0062437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hill, I. T., Tallo, T., Dorman, M. J. & Dove, S. L. Loss of RNA chaperone Hfq unveils a toxic pathway in Pseudomonas aeruginosa. J. Bacteriol.201, e00232-19 (2019). [DOI] [PMC free article] [PubMed]
- 116.Kato J, Kim H-E, Takiguchi N, Kuroda A, Ohtake H. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J. Biosci. Bioeng. 2008;106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 117.Francis, V. I., Stevenson, E. C. & Porter, S. L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett.364, fnx104 (2017). [DOI] [PMC free article] [PubMed]
- 118.Kim W, et al. Effect of spaceflight on Pseudomonas aeruginosa final cell density is modulated by nutrient and oxygen availability. BMC Microbiol. 2013;13:241. doi: 10.1186/1471-2180-13-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ruby, E. G. et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci.102, 3004–3009 (2005). [DOI] [PMC free article] [PubMed]
- 120.Foster, J., Wheeler, R. & Pamphile, R. Host–microbe interactions in microgravity: assessment and implications. Life4, 250–266 (2014). [DOI] [PMC free article] [PubMed]
- 121.McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal–bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 122.Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 1994;176:6986–6991. doi: 10.1128/JB.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deloney-Marino CR, Visick KL. Role for cheR of Vibrio fischeri in the Vibrio–squid symbiosis. Can. J. Microbiol. 2012;58:29–38. doi: 10.1139/w11-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Morrison, M. D. & Nicholson, W. L. Meta-analysis of data from spaceflight transcriptome experiments does not support the idea of a common bacterial “spaceflight response”. Sci. Rep.8, 14403 (2018). [DOI] [PMC free article] [PubMed]
- 125.Klaus D, Simske S, Todd P, Stodieck L. Investigation of space flight effects on Escherichia coli and a proposed model of underlying physical mechanisms. Microbiology. 1997;143:449–455. doi: 10.1099/00221287-143-2-449. [DOI] [PubMed] [Google Scholar]
- 126.Zea, L. et al. A molecular genetic basis explaining altered bacterial behavior in space. PLoS One11, e0164359 (2016). [DOI] [PMC free article] [PubMed]
- 127.Dijkstra, C. E. et al. Diamagnetic levitation enhances growth of liquid bacterial cultures by increasing oxygen availability. J. R. Soc. Interface8, 334–344 (2011). [DOI] [PMC free article] [PubMed]
- 128.Mitchell JG, Kogure K. Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol. Ecol. 2006;55:3–16. doi: 10.1111/j.1574-6941.2005.00003.x. [DOI] [PubMed] [Google Scholar]
- 129.Rossi E, Paroni M, Landini P. Biofilm and motility in response to environmental and host-related signals in Gram negative opportunistic pathogens. J. Appl. Microbiol. 2018;125:1587–1602. doi: 10.1111/jam.14089. [DOI] [PubMed] [Google Scholar]
- 130.Bains, M., Fernandez, L. & Hancock, R. E. W. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol.78, 6762–6768 (2012). [DOI] [PMC free article] [PubMed]
- 131.Marteyn, B., Scorza, F. B., Sansonetti, P. J. & Tang, C. Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell. Microbiol.13, 171–176 (2011). [DOI] [PubMed]
- 132.Lee, C. A. & Falkow, S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci.87, 4304–4308 (1990). [DOI] [PMC free article] [PubMed]
- 133.Deng Y, et al. Complete genome sequence of Vibrio alginolyticus ZJ-T. Genome Announc. 2016;4:e00912. doi: 10.1128/genomeA.00912-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gangaiah, D. et al. Haemophilus ducreyi Hfq contributes to virulence gene regulation as cells enter stationary phase. Mbio5, e01081-01013-e01081 (2014). [DOI] [PMC free article] [PubMed]
- 135.Kim S, et al. hfq plays important roles in virulence and stress adaptation in Cronobacter sakazakii ATCC 29544. Infect. Immun. 2015;83:2089–2098. doi: 10.1128/IAI.03161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang, C. et al. Hfq, a RNA chaperone, contributes to virulence by regulating plant cell wall–degrading enzyme production, Type VI secretion system expression, bacterial competition, and suppressing host defense response in Pectobacterium carotovorum. Mol. Plant–Microbe Interact.31, 1166–1178 (2018). [DOI] [PubMed]
- 137.Osterman IA, Dikhtyar YY, Bogdanov AA, Dontsova OA, Sergiev PV. Regulation of flagellar gene expression in bacteria. Biochemistry. 2015;80:1447–1456. doi: 10.1134/S000629791511005X. [DOI] [PubMed] [Google Scholar]
- 138.Caldelari I, Chao Y, Romby P, Vogel J. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb. Perspect. Med. 2013;3:a010298–a010298. doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wuest SL, Richard S, Kopp S, Grimm D, Egli M. Simulated microgravity: critical review on the use of random positioning machines for mammalian cell culture. BioMed. Res. Int. 2015;2015:971474. doi: 10.1155/2015/971474. [DOI] [PMC free article] [PubMed] [Google Scholar]