Abstract
Objective
We have developed and validated a novel EEG-based signal processing approach to distinguish PD and control patients: Linear-predictive-coding EEG Algorithm for PD (LEAPD). This method efficiently encodes EEG time series into features that can detect PD in a computationally fast manner amenable to real time applications.
Methods
We included a total of 41 PD patients and 41 demographically-matched controls from New Mexico and Iowa. Data for all participants from New Mexico (27 PD patients and 27 controls) were used to evaluate in-sample LEAPD performance, with extensive cross-validation. Participants from Iowa (14 PD patients and 14 controls) were used for out-of-sample tests. Our method utilized data from six EEG leads which were as little as 2 minutes long.
Results
For the in-sample dataset, LEAPD differentiated PD patients from controls with 85.3±0.1% diagnostic accuracy, 93.3±0.5% area under the receiver operating characteristics curve (AUC), 87.9±0.9% sensitivity, and 82.7±1.1% specificity, with multiple cross-validations. After head-to-head comparison with state-of-the-art methods using our dataset, LEAPD showed a 13% increase in accuracy and a 15.5% increase in AUC. When the trained classifier was applied to a distinct out-of-sample dataset, LEAPD showed reliable performance with 85.7% diagnostic accuracy, 85.2% AUC, 85.7% sensitivity, and 85.7% specificity. No statistically significant effect of levodopa-ON and levodopa-OFF sessions were found.
Conclusion
We describe LEAPD, an efficient algorithm that is suitable for real time application and captures spectral EEG features using few parameters and reliably differentiates PD patients from demographically-matched controls.
Keywords: EEG, Parkinson’s Disease, Diagnosis, Classifier
1. INTRODUCTION
Parkinson’s disease (PD) is a progressive neurodegenerative disorder associated with the death of the dopaminergic neurons of the substantia nigra resulting in slow movement, rigidity, tremor, postural instability, and non-motor symptoms [1, 2]. Correct diagnosis of PD is crucial from both prognostic and therapeutic perspectives. For example, accurate diagnosis is critical for selecting patients for advanced therapies like adaptive deep brain brain stimulation (aDBS) [3]. However, the accuracy of the diagnosis of PD relies on clinical features and has not improved significantly in 25 years [4]. In a meta-analysis of 20 studies, the accuracy of clinical diagnosis using pathologic examination was 80.6%, and by non-specialists 73.8% [4].
Non-invasive electroencephalography (EEG) could help non-specialists diagnose PD. EEG records electric fields produced by neuronal ensembles, reflecting the functional state of cortical layers and corresponding subcortical structures. Both structures can be abnormal in PD. Our prior work has documented cortical EEG abnormalities associated with PD [5, 6].
Most EEG-based approaches to PD diagnosis use isolated spectral features (e.g. absolute and relative power, amplitude and cross-frequency coupling in different frequency bands) and nonlinear features based on higher-order spectra or differential equations in recorded EEG [7–9] or source localized EEG [10]. Although these capture many aspects of EEG, they are either inefficient in capturing PD-related characteristics requiring complex machine learning techniques to identify PD-related markers for diagnosis or require large computational power for generating them [9, 11, 12]. Hence, classification methods with these features are moderately accurate in PD detection but are both computationally expensive and highly sensitive to the medications [12]. This imposes significant challenges for real time applications. Hence, a computationally efficient PD vs. control classification method capable of PD detection regardless of the medication state and amenable to real time application is of significant interest.
We present a novel feature based on linear predictive coding (LPC) [13] that efficiently captures PD-related changes in EEG and provide an algorithm for PD detection suitable for real time application that performs reliably regardless of the medication state. Our algorithm, termed Linear-predictive-coding Electroencephalography Algorithm for PD (LEAPD), codes EEG spectrum into a few parameters known as LPC coefficients. We have empirically found that LPC coefficients for PD patients and controls lie in distinct hyperplanes. We use these hyperplanes to generate a diagnostic index and evaluate LEAPD performance. Our method works with as little as two minutes of EEG data and is amenable to fast processing making it well suited to real time applications like aDBS.
2. METHODS
2.1. Patient, participants and experimental design
We used EEG recordings of 27 PD patients and 27 controls, from a study at the University of New Mexico (UNM; Albuquerque, New Mexico), along with 14 PD patients and 14 controls from the University of Iowa (UI; Iowa City, Iowa). Additionally, we used EEG recordings from OFF medication sessions for the 27 PD patients from UNM which were recorded in the practically defined OFF levodopa period, 12 hours after the last dose of dopaminergic medication. Control participants were demographically matched for age and sex with PD patients and did not differ in any measurements of education or premorbid intelligence (supplementary materials: Table A1).
All procedures were approved by the UNM or UI Office of the Institutional Review Board. All participants provided written informed consent and were paid $20/hour in UNM and $40/hour in Iowa. The PD patients completed neuropsychological and questionnaire assessments in their ON state. A neurologist administered United Parkinson’s Disease Rating Scale (UPDRS) motor scores. Previous studies have reported data from some of these participants [5, 6].
Resting state EEG recordings of the UNM subjects were gathered under both eyes-open and eyes-closed conditions, while resting state EEG data for the 28 Iowa subjects were recorded only with eyes-open. We wished to generate EEG data from a highly generalizable and reproducible protocol not requiring any additional training or equipment. Eyes-open and eyes-closed are standard elements of clinical EEG assessment protocols [14], are collected in every major hospital, aren’t biased by any specific ability and generate a range of features for classification [7, 8, 10–12, 15, 16].
2.2. EEG recordings
EEG was recorded from sintered Ag/AgCl electrodes across 0.1 to 100 Hz with a sampling rate of 500 Hz on a 64-channel Brain Vision system, with online reference set to channel CPz (UNM) and Pz (Iowa) as baselines. Hence, CPz and Pz data were missing from UNM and Iowa datasets, respectively. We used EEG data from the 62 channels that were common to both datasets. Eye blinks were identified and removed following independent component analysis. In the UNM dataset, each subject has an EEG recording that includes one session of eyes-closed condition, followed by one of eyes-open condition. EEG recordings under these sessions were separated into two additional variants of the EEG data: eyes-open dataset and eyes-closed dataset. These two datasets and the original with both conditions (eyes open + closed) were analyzed separately to determine the best variant (eyes-open / eyes-closed / eyes open + closed) for classification.
2.3. Feature Identification
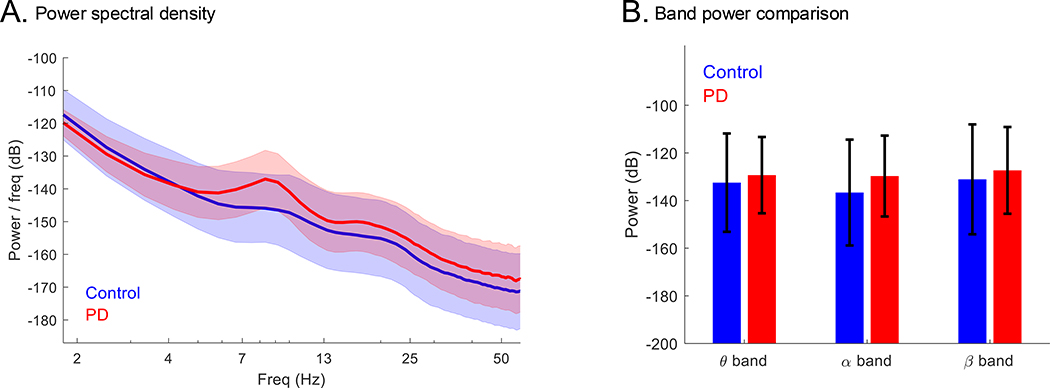
LEAPD uses the entire power spectral density (PSD) of EEG recordings, rather than powers of different frequency bands. Figure 1A illustrates a distinct difference in the shape of the mean PSD of PD and control subjects (UNM data) under eyes open + closed condition in the 2.5–14 Hz frequency range. This difference is consistent among all PD and control subjects and can also be seen in PSD plots from other studies [10, 17, 18]. Figure 1B compares the differences in power between PD and control subjects (UNM data) in 2.5–14 Hz range, showing changes in theta, alpha and beta bands with large standard deviations. These justify capturing the differences in PSD in a holistic fashion: Rather than combining features from separate frequency bands in an ad hoc fashion, LPC implicitly combines them in a principled fashion.
Figure 1.
Comparison of PSD shapes and band power between PD and control: (A) Comparison of EEG recordings (eyes open + closed) highlighting the difference in PSD shapes at 2.5–14 Hz between PD (n = 27) and control (n = 27) subjects (UNM data). (B) Comparison of theta (4–8 Hz), alpha (8–12 Hz) and beta (12–32 Hz) band power for the same data set. All plots: mean ± standard deviation.
2.4. Feature extraction using LPC
We used LPC to capture PSD differences in PD subjects. We have shown this as effective in rodent models of PD [19]. LPC predicts time series behavior [13], and has applications in speech coding and analyzing myoelectric signals. The core idea is to encode the entire EEG time series data by a vector of LPC coefficients. These coefficients represent the pertinent features of the EEG data, while preserving key aspects of the PSD.
We used Burg’s method [13] for calculating LPC coefficients. It reduces spectral loss and provides better frequency resolution [13] and has been applied to EEG for migraine, epileptic and PD [15, 16, 20]. Details are in the supplementary materials (Methodology and Figure A3).
2.5. Classification
2.5.1. Classifier design
The rationale for our approach was the observation that vectors of LPC coefficients from PD and healthy subjects fall on two distinct hyperplanes. As multi-dimensional hyperplanes cannot be visualized, the validity of our hypothesis was tested using quantitative separation of these hyperplanes. The hyperplanes can be identified by principal component analysis (PCA), where principal components represent a set of orthogonal basis functions maximizing variance in multi-dimensional data [21]. After identifying these hyperplanes during training, new subjects were classified using vector projections. We formulated the LEAPD index between 0 and 1, as a diagnostic index. An index value < 0.5 represents controls and the opposite PD. This threshold 0.5 was obtained mathematically by vector projection without further fine-tuning. Details of classifier design are in the supplementary materials (Methodology).
2.5.2. Single-channel classification
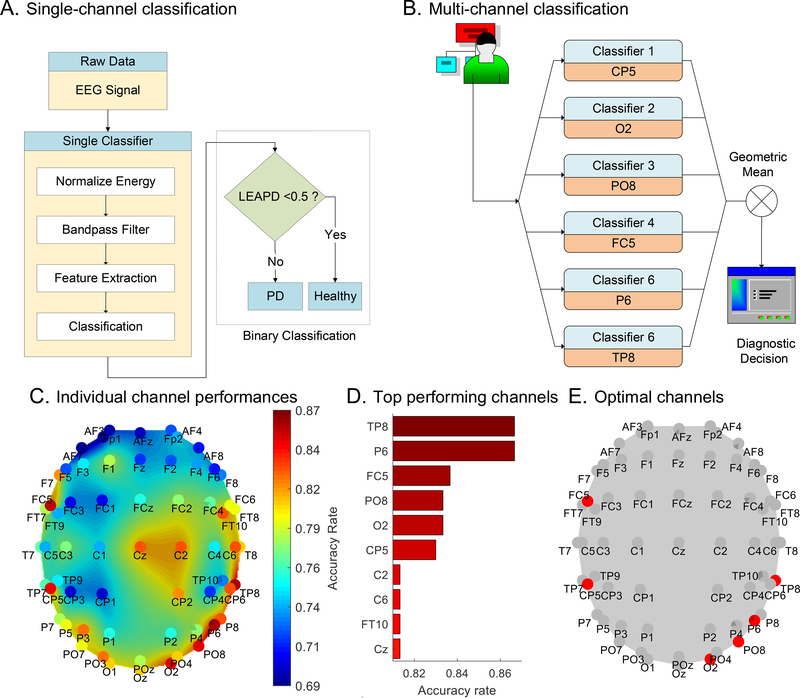
Our single-channel classification method calculates the LEAPD index utilizing single-channel EEG. It consists of four steps: normalization of the energy of EEG data, band-pass filtering, feature extraction and classification (Figure 2A). Details are in the supplementary materials (Methodology).
Figure 2.
Classification method:(A) Detailed flow chart of a single-channel PD vs. control classifier. (B) Simplified diagram for the multi-channel classification with six single-classifier collaboration. (C) Performance comparison of 62 channels (UNM data). (D) Comparison of the 10 top-performing channels. (E) Channel positions of the 6 top-performing channels; red indicates selected or multi-channel classification.
2.5.3. Multi-channel Classification
Multiple single-channel classifiers were selected to make collaborative decisions for improving performance (Figure 2B). Each classifier used EEG data from one specific channel to independently generate LEAPD indices. Their geometric mean was used as a collective index.
2.5.4. Parameter selection
Parameters were chosen in the training phase in a principled manner to maximally separate PD and control. Key parameters were: EEG data channel, bandpass filter, number of principal components forming the hyperplane, and LPC order. We performed an exhaustive search on the UNM data to choose these parameters (see supplementary material: Methodology). Figure 2C compares 62 channels with individual best performances. The central electrodes (Cz, C2 and CP2) had 80%+ accuracy. The most effective channels were TP8, P6, FC5, PO8, O2, and CP5 with 83%+ accuracy (Figure 2D). These were selected for multi-channel classification (Figure 2E). The best parameters for these channels are in supplementary materials: Table A2. Optimal parameters were obtained with EEG data from eyes open + closed condition.
2.6. Measurements for performance evaluation
PD patients were diagnosed by a movement-disorders clinician [6]. Performance was measured by receiver operating characteristics (ROC), area under ROC curve (AUC), sensitivity, specificity, accuracy rate, positive predictive value (PPV), negative predictive value (NPV), odds ratio (OR) and positive likelihood ratio (LR+) for classification indices.
2.7. Performance evaluation and validation schemes
For validation, we used 10 rounds of 5 and 10-fold cross-validations and leave-one-out cross-validations (LOOCV) on the UNM dataset. We tested the Iowa dataset as an out-of-sample set after full training with the UNM dataset with eyes open + closed. We investigated whether we can classify OFF sessions as PD after training with PD ON sessions and control. These validation schemes are discussed in the supplementary materials (Methodology and Figure A4).
Because our PCA-based classification is computationally simple, once trained, LEAPD index can be generated in real time using incoming EEG data with extremely low computational latency. We measured the real-time performance of our method by training with the whole dataset from UNM and then feeding the UNM and Iowa data into the classifier using an expanding time window. Finally, we compared our approach with three state-of-the-art methods [7, 8, 10]. Details of these methods and our real time performance measurement procedure are in the supplementary material (Methodology). Our goal was to compare our novel LPC-based feature with other feature design techniques. Therefore, these state-of-the-art methods were selected to compare the efficiency and accuracy of our classification feature with other features based on spectral power [7], source localized EEG [10] and nonlinear higher-order spectral characteristics [8] for PD detection. As we were mainly interested in a computationally efficient PD classification method, we excluded studies that either require computationally expensive machine learning techniques [11, 12] or generate a large number of features and hundreds of thousands of possible models [9].
2.8. Statistical methods
For statistical significance of the correlation between LEAPD and classification, we used a two-sided Wilcoxon rank-sum test and univariate analysis of variance (ANOVA). We calculated 95% confidence intervals for all outcomes measured. Details are in the supplementary material (Methodology).
2.9. Data availability
Individual deidentified data, including the necessary codes and detailed outcomes will be shared upon publication at: http://narayanan.lab.uiowa.edu and PRED+CT (http://predict.cs.unm.edu/).
3. RESULTS
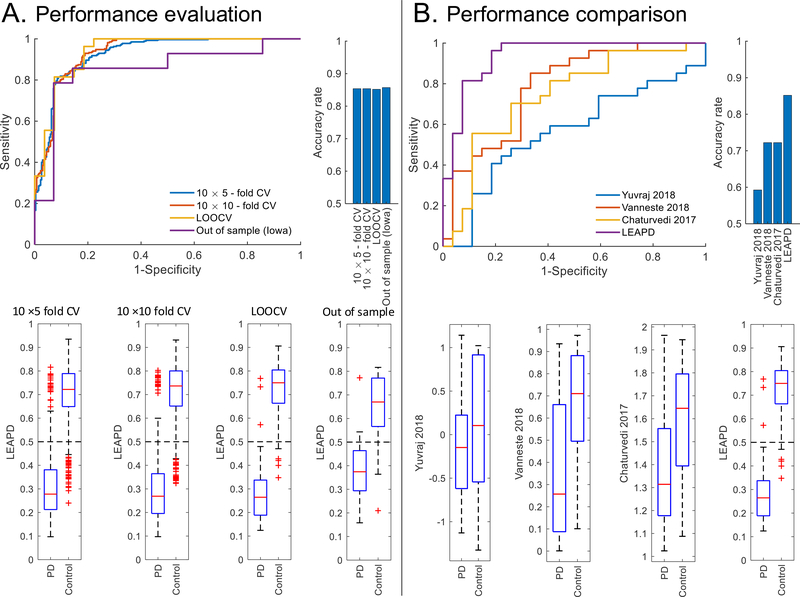
We compared the three variants EEG data (eyes-open, eyes-closed, and eyes open + closed) and found that the accuracy of eyes open + closed condition was 85.3±0.1% for in-sample data (from UNM). This is superior to the in-sample accuracy obtained from either eyes-open (78.7±0.9%) or eyes-closed condition (82.2±1%). Therefore, we performed in-sample analysis of the eyes open + closed condition. LEAPD robustly classified demographically-matched PD and control subjects in the in-sample dataset with 93.3±0.5% AUC, 85.3±0.1% accuracy, 87.9±0.9% sensitivity, 82.7±1.1% specificity and 5.1±0.2 LR+ using 5-fold, 10-fold and leave one out cross-validations (Figure 3A and Table 1).Impressively, LEAPD-based classifiers also extended to out-of-sample data, with 85.2% AUC, 85.7% accuracy, 85.7% sensitivity, and 85.7% specificity (14 PD and 14 control subjects from Iowa; Figure 3A and Table 1).
Figure 3.
Classifier performance: (A) Performance evaluation of the proposed method with receiver-operative characteristic (ROC) curve (top left), accuracy (top right), and boxplots (bottom). (B) Performance comparison of the proposed method (LEAPD) with traditional approaches (Yuvaraj 2018 [8], Vanneste 2018 [10] and Chaturvedi 2017 [7]) using ROC curve (top left), accuracy (top right), and boxplots (bottom).
Table 1.
Performance of the proposed classification method
| Proposed method | Other methods | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| New Mexico | Iowa | New Mexico (LOOCV) | |||||
|
| |||||||
| 10 × 5-fold CV | 10 × 10-fold CV | LOOCV | Out-of-sample test | Chaturvedi et al. [7] | Vanneste et al. [10] | Yuvaraj et al. [8] | |
|
| |||||||
| AUC % | 92.7 (90.5–94.9) |
93.3 (91.2–95.4) |
93.8 (87.4–100) |
85.2 (69.5–100) |
74.3 (60.8–87.9) |
78.3 (66–90.7) |
56.4 (40.6–72.1) |
| ACC% | 85.4 (82.1–88.2) |
85.4 (82.1–88.2) |
85.2 (72.9–93.4) |
85.7 (67.3–96) |
72.2 (58.4–83.5) |
72.2 (58.4–83.5) |
59.3 (45–72.4) |
| SEN% | 87 (82.4–90.8) |
87.8 (83.3–91.4) |
88.9 (70.8–97.6) |
85.7 (57.2–98.2) |
74.1 (53.7–88.9) |
70.4 (49.8–86.3) |
63 (42.4–80.6) |
| SPC% | 83.7 (78.7–87.9) |
83 (77.9–87.2) |
81.5 (61.9–93.7) |
85.7 (57.2–98.2) |
70.4 (49.8–86.3) |
74.1 (53.7–88.9) |
55.7 (35.3–74.5) |
| PPV% | 84.2 (80.2–87.5) |
83.7 (79.8–87.1) |
82.8 (68.3–91.5) |
85.7 (62–95.7) |
71.4 (57.3–82.3) |
73.1 (57.8–84.3) |
58.6 (45.9–70.3) |
| NPV% | 86.6 (82.5–89.8) |
87.2 (83.1–90.4) |
88 (71.3–95.6) |
85.7 (62–95.7) |
73.1 (57.8–84.3) |
71.4 (57.3–82.3) |
60 (45.2–73.1) |
| OR | 34.5 (21.3–55.7) |
35 (21.6–56.7) |
35.2 (7.5–164.8) |
36 (4.3–299) |
6.8 (2.1–22.4) |
6.8 (2.1–22.4) |
2.1 (0.7–6.3) |
| LR+ | 5.3 (4.1–7) |
5.1 (3.9–6.7) |
4.8 (2.1–10.7) |
6 (1.6–22.3) |
2.5 (1.3–4.7) |
2.7 (1.4–5.4) |
1.42 (0.8–2.4) |
| p-value Wilcoxon | 5.6 × 10−66 | 7 × 10−68 | 3.4 × 10−8 | 6 × 10−4 | 2.2 × 10−3 | 3.6 × 10−4 | 0.42 |
| p-value ANOVA | 1.5 × 10−101 | 1 × 10−106 | 4.1 × 10−12 | 1.6 × 10−3 | 9.6 × 10−4 | 1 × 10−4 | 0.4 |
Summary of the performance of the proposed classification method with cross-validation schemes and the out-of-sample test and the performance of the other state-of-the-art methods.
Abbreviations: SEN = Sensitivity; SPC = Specificity; AUC = Area under the receiver operating characteristics curve; ACC = Accuracy; PPV = Positive Predictive Value; NPV = Negative Predictive Value; OR = Odds Ratio; LR+ = Positive Likelihood Ratio; 95% Confidence interval included in braces; LOOCV = leave one out cross-validation; CV = Cross-validation
We investigated whether the index differences between PD and control subjects were statistically significant. Both Wilcoxon rank-sum and univariate ANOVA tests provided significant p-values (« 0.05) for LEAPD indices that were obtained in the cross-validations and the out-of-sample test (Table 1). LEAPD outperformed three prior state-of-the-art classification methods, resulting in a 13% increase in accuracy and a 15.5% increase in AUC (Figure 3B and Table 1).
A mainstay of therapy for PD is levodopa, having the potential to affect scalp EEG [18], though our previous research found these effects to be minimal [6, 22, 23]. LEAPD indices of PD patients between OFF and ON medication had statistically similar accuracy (81.5% vs. 88.9%; p-value > 0.15 for univariate ANOVA; Supplementary Figure A5), demonstrating no significant impact of levodopa on LEAPD index.
We investigated how much data was required for reliable classification in a real time scenario. There was a gradual increase in accuracy in the UNM data, reaching ~85% accuracy with just two minutes of incoming data (supplementary Figure A6: A and C). In the out-of-sample testing, over 70% accuracy was obtained with 2 minutes of data (supplementary Figure A6: A and B). This result along with the previous finding of ~85% accuracy in the out-of-sample full-length data (which includes the entire EEG data of each subject with range = 2–5.7 minutes, average = 3.09 minutes) suggest that EEG data longer than 2 minutes is better for accuracy in real time scenarios. Hence, LEAPD is useful for rapidly determining PD-related neurophysiology and amenable to real time applications [3, 24], although a more in-depth analysis is needed with longer EEG recordings to evaluate performance. Taken together, our results demonstrate that LEAPD can robustly discriminate EEG signals of PD subjects from those of demographically-matched controls in as little as 120 seconds.
4. DISCUSSION
We designed an efficient feature for PD detection and developed and assessed LEAPD, a computationally efficient novel real-time algorithm for distinguishing EEG activity in PD patients vs. demographically-matched controls. This approach rapidly codes filtered EEG data between 2.5 and 12 Hz into a few parameters to generate classification features. Our results showed that these features are very efficient for PD detection. We proposed a PCA-based classification method (LEAPD) capable of obtaining high accuracy in PD detection. The algorithm had a principled design with classification threshold being selected using vector projection formula. Parameter selection of our classifier used exhaustive search. As our LPC based features were efficient in PD detection, no further fine-tuning was necessary as tuning the classifier often reduces generality and increases complexity. Our work is novel because LEAPD is distinct from prior EEG approaches. It holistically captures EEG-derived PSDs using a few parameters and a computational speed that is amenable to real time classification. These results suggest a new EEG-based diagnostic direction for neurodegenerative diseases such as PD.
We found that LEAPD can rapidly and robustly distinguish between PD and control patients. Our work is an advance for several reasons: (1) We showed a 13% increase in accuracy compared to other methods in a head-to-head test on our dataset. While some of the studies reported moderate accuracy performance (78% by Chaturvedi et al. [7]), there are high accuracies in past works (94.34% by Vanneste et al. [10] and 99.62% by Yuvaraj et al. [8]) but these studies included longer datasets (4–5 minutes for Vanneste et al. [10] and Yuvaraj et al. [8]), unbalanced classes (31 PD subjects and 264 controls for Vanneste et. al. [10]) across a wide age range (20–75 years) without age/sex-matched controls [7, 8, 10]. (2) We extensively cross-validated our results; to our knowledge prior work only performed one round of cross-validation [7, 8, 10]. (3) We did not manually tune our classifier to optimize classification, which can result in overfitting. (4) We performed out-of-sample testing, which was not done in previous studies. (5) Our results are consistent despite medication state, unlike prior reports [12]. (6) Finally, our proposed method is amenable to real time application and achieved stable adequate accuracy with only two minutes of data even in our emulated real time scenarios, which we are not aware of in prior works. Our algorithm has the potential to lead to rigorous, reliable, and reproducible performance in future real time applications.
The frequency bands were obtained for maximal separation between PD and control subjects in all channels. The 2.5–12 Hz bound spans frequency bands like delta (0.5 Hz-4 Hz), theta (4 Hz-8 Hz), and low alpha (8 Hz-12 Hz) that are known to be abnormal in PD [18, 25, 26]. We used resting EEG data for our analysis. In future studies, procedures can be included to improve the classification performance, e.g. by adding a simple stereotyped motor task (e.g. UPDRS movements).
One limitation of our work is that the gold-standard diagnosis of neurodegenerative diseases such as PD requires post-mortem assessment. The accuracy of clinical vs. pathology-confirmed diagnosis in PD is ~90%, close to the LEAPD accuracy reported here. However, our data are not verified by pathological data, as this would take several years to acquire. Secondly, there are other movement disorders diagnoses including essential tremor, multiple-systems atrophy, or other Parkinson’s plus disorders. We have not yet compared our PD diagnosis with these related brain diseases, although this is our planned future direction. Finally, we selected patients with a limited range of cognitive function and without any brain or psychiatric disease and minimized the other variabilities between PD and control subjects by matching age, sex and education. As mentioned previously, any feature that differs between groups will be capitalized upon by classification. While we controlled for the most relevant biological and socio-demographic features between groups, only future work with a very large sample will demonstrate the true generalizability of the LEAPD index.
We developed a novel LEAPD index that rapidly and robustly separates PD patients from controls. The LEAPD approach encodes PSD with very few parameters and has the potential to generate PD diagnostics or contribute to control algorithms for real time applications. It may also be useful in refining predictions of motor progression or PD-related subtypes. It can contribute to cost-effective diagnostic tools and real time control signals for PD and other neurodegenerative diseases.
Supplementary Material
A novel machine-learning approach to diagnose Parkinson’s disease with EEG
Cross-validation and out-of-sample tests yield more than 85% accuracy
Outperformes other state-of-the-art EEG methods
Computationally effiecient and amenable to real-time implementation
Acknowledgements
This work was supported by the National Institutes of Health [Grant: NIH R01NS100849-01A1].
Footnotes
Declarations of interest:
None
A provisional patent application for LEAPD (United States Patent Application No. 62/899,915) has been filed on September 13, 2019.
Appendix A. Supplementary data
Supplementary materials for dataset, methodology and results are provided.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Fahn S, The history of dopamine and levodopa in the treatment of Parkinson’s disease, 23(S3) (2008) S497–S508. [DOI] [PubMed] [Google Scholar]
- [2].Chaudhuri KR, Schapira AHV, Non-motor symptoms of Parkinson’s disease: dopaminergic pathophysiology and treatment, The Lancet Neurology 8(5) (2009) 464–474. [DOI] [PubMed] [Google Scholar]
- [3].Little S, Pogosyan A, Neal S, Zavala B, Zrinzo L, Hariz M, Foltynie T, Limousin P, Ashkan K, FitzGerald J, Green AL, Aziz TZ, Brown P, Adaptive deep brain stimulation in advanced Parkinson disease, Ann Neurol 74(3) (2013) 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rizzo G, Copetti M, Arcuti S, Martino D, Fontana A, Logroscino G, Accuracy of clinical diagnosis of Parkinson disease, A systematic review and meta-analysis 86(6) (2016) 566–576. [DOI] [PubMed] [Google Scholar]
- [5].Singh A, Cole RC, Espinoza AI, Brown D, Cavanagh JF, Narayanan NS, Frontal theta and beta oscillations during lower-limb movement in Parkinson’s disease, Clinical Neurophysiology 131(3) (2020) 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Singh A, Richardson SP, Narayanan N, Cavanagh JF, Mid-frontal theta activity is diminished during cognitive control in Parkinson’s disease, Neuropsychologia 117 (2018) 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chaturvedi M, Hatz F, Gschwandtner U, Bogaarts JG, Meyer A, Fuhr P, Roth V, Quantitative EEG (QEEG) Measures Differentiate Parkinson’s Disease (PD) Patients from Healthy Controls (HC), Frontiers in Aging Neuroscience 9(3) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yuvaraj R, Rajendra Acharya U, Hagiwara Y, A novel Parkinson’s Disease Diagnosis Index using higher-order spectra features in EEG signals, Neural Computing and Applications 30(4) (2018) 1225–1235. [Google Scholar]
- [9].Lainscsek C, Hernandez M, Weyhenmeyer J, Sejnowski T, Poizner H, Non-Linear Dynamical Analysis of EEG Time Series Distinguishes Patients with Parkinson’s Disease from Healthy Individuals, Frontiers in Neurology 4(200) (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vanneste S, Song J-J, De Ridder D, Thalamocortical dysrhythmia detected by machine learning, Nature Communications 9(1) (2018) 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Oh SL, Hagiwara Y, Raghavendra U, Yuvaraj R, Arunkumar N, Murugappan M, Acharya UR, A deep learning approach for Parkinson’s disease diagnosis from EEG signals, Neural Computing and Applications 32(15) (2020) 10927–10933. [Google Scholar]
- [12].Lee S, Hussein R, McKeown MJ, A Deep Convolutional-Recurrent Neural Network Architecture for Parkinson’s Disease EEG Classification, 2019 IEEE Global Conference on Signal and Information Processing (GlobalSIP), 2019, pp. 1–4. [Google Scholar]
- [13].Kay SM, Marple SL, Spectrum analysis-A modern perspective, Proceedings of the IEEE 69(11) (1981) 1380–1419. [Google Scholar]
- [14].Sinha SR, Sullivan L, Sabau D, San-Juan D, Dombrowski KE, Halford JJ, Hani AJ, Drislane FW, Stecker MM, American Clinical Neurophysiology Society Guideline 1: Minimum Technical Requirements for Performing Clinical Electroencephalography, Journal of Clinical Neurophysiology 33(4) (2016) 303–307. [DOI] [PubMed] [Google Scholar]
- [15].Han C-X, Wang J, Yi G-S, Che Y-Q, Investigation of EEG abnormalities in the early stage of Parkinson’s disease, Cognitive Neurodynamics 7(4) (2013) 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yi G-S, Wang J, Deng B, Wei X-L, Complexity of resting-state EEG activity in the patients with early-stage Parkinson’s disease, Cognitive Neurodynamics 11(2) (2017) 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stoffers D, Bosboom JLW, Deijen JB, Wolters EC, Berendse HW, Stam CJ, Slowing of oscillatory brain activity is a stable characteristic of Parkinson’s disease without dementia, Brain 130(7) (2007) 1847–1860. [DOI] [PubMed] [Google Scholar]
- [18].Jackson N, Cole SR, Voytek B, Swann NC, Characteristics of Waveform Shape in Parkinson’s Disease Detected with Scalp Electroencephalography, eneuro 6(3) (2019) ENEURO.0151-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Md Fahim A, Joshua H, Stephanie AL, Soura D, Raghuraman M, Morgan KA, Nandakumar N, Linear Predictive Approaches Separate Field Potentials in Animal Model of Parkinson’s Disease, Frontiers in Aging Neuroscience (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Akben SB, Subasi A, Tuncel D, Analysis of EEG Signals Under Flash Stimulation for Migraine and Epileptic Patients, Journal of Medical Systems 35(3) (2011) 437–443. [DOI] [PubMed] [Google Scholar]
- [21].Jolliffe IT, Cadima J, Principal component analysis: a review and recent developments, Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 374(2065) (2016) 20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cavanagh JF, Kumar P, Mueller AA, Richardson SP, Mueen A, Diminished EEG habituation to novel events effectively classifies Parkinson’s patients, Clinical Neurophysiology 129(2) (2018) 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brown DR, Richardson SP, Cavanagh JF, An EEG marker of reward processing is diminished in Parkinson’s disease, Brain Research 1727 (2020) 146541. [DOI] [PubMed] [Google Scholar]
- [24].Little S, Tripoliti E, Beudel M, Pogosyan A, Cagnan H, Herz D, Bestmann S, Aziz T, Cheeran B, Zrinzo L, Hariz M, Hyam J, Limousin P, Foltynie T, Brown P, Adaptive deep brain stimulation for Parkinson’s disease demonstrates reduced speech side effects compared to conventional stimulation in the acute setting, Journal of Neurology, Neurosurgery & Psychiatry 87(12) (2016) 1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tanaka H, Koenig T, Pascual-Marqui RD, Hirata K, Kochi K, Lehmann D, Event-Related Potential and EEG Measures in Parkinson’s Disease without and with Dementia, Dementia and Geriatric Cognitive Disorders 11(1) (2000) 39–45. [DOI] [PubMed] [Google Scholar]
- [26].Caviness JN, Hentz JG, Belden CM, Shill HA, Driver-Dunckley ED, Sabbagh MN, Powell JJ, Adler CH, Longitudinal EEG changes correlate with cognitive measure deterioration in Parkinson’s disease, J Parkinsons Dis 5(1) (2015) 117–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual deidentified data, including the necessary codes and detailed outcomes will be shared upon publication at: http://narayanan.lab.uiowa.edu and PRED+CT (http://predict.cs.unm.edu/).