Abstract
Lung cancer is the most prevalent cancer globally. It is also the leading cause of cancer-related death because of the late diagnosis and the frequent resistance to therapeutics. Therefore, it is impending to identify novel biomarkers and effective therapeutic targets to improve the clinical outcomes. Identified as a new class of RNAs, circular RNAs (circRNAs) derive from pre-mRNA back splicing with considerable stability and conservation. Accumulating research reveal that circRNAs can function as microRNA (miRNA) sponges, regulators of gene transcription and alternative splicing, as well as interact with RNA-binding proteins (RBPs), or even be translated into proteins directly. Currently, a large body of circRNAs have been demonstrated differentially expressed in physiological and pathological processes including cancer. In lung cancer, circRNAs play multiple roles in carcinogenesis, development, and response to different therapies, indicating their potential as diagnostic and prognostic biomarkers as well as novel therapeutics. In this review, we summarize the multi-faceted functions of circRNAs in lung cancer and the underlying mechanisms, together with the possible future of these discoveries in clinical application.
Keywords: circular RNAs (circRNAs), microRNAs (miRNAs), lung cancer, biomarker, prognosis
Introduction
Nowadays, lung cancer is the most frequent cause of cancer-related mortality (Siegel et al., 2017). Non-small cell lung cancer (NSCLC), including lung adenocarcinoma, squamous cell lung carcinoma, and large cell lung carcinoma, constitutes about 85% of lung cancer cases. Although the novel therapeutic strategies such as targeting drugs toward the epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), and immune checkpoints programmed cell death-1 (PD-1) and/or programmed cell death-1 ligand (PD-L1) have led to an great progress of advanced NSCLC patients among the past decades, the long-term survival of lung cancer remains unfavorable because of the late diagnosis and the frequent resistance to therapeutics (Chansky et al., 2017; Kris et al., 2017). Therefore, the identification of sensitive biomarkers for early detection and prognosis estimation, as well as effective therapeutic targets is urgently needed to improve the clinical outcomes.
Circular RNAs (circRNAs) are identified as a new class of endogenous RNAs derived from back splicing. Lacking the 3′-poly(A) tails and 5′-end caps (Suzuki and Tsukahara, 2014), circRNAs have closed loop structures generated from the ligation of exons, introns, or both (Wang et al., 2019b), thus are divided into the three main subtypes as exonic circRNAs (EcircRNAs), intronic circRNAs (ciRNAs), and exonic circRNAs with introns (EIciRNAs), respectively. EcircRNAs exist in the eukaryotic cytoplasm, while ciRNAs and EIciRNAs are mainly in the nucleus. Other subtypes of circRNAs include intergenic circRNAs, tRNA intronic circRNAs (tricRNAs), antisense circRNAs, overlapping circRNAs, circRNA rRNAs (circrRNAs), and intragenic circRNAs (Liang et al., 2020). Owing to the absence of the 3′ and 5′ ends, circRNAs exhibit much more stability and conservation than the linear RNAs and are insusceptible to RNA exonuclease or RNase R-induced degradation. In general, circRNAs are expressed at lower levels than the host genes (Enuka et al., 2016). Although discovered half a century ago (Sanger et al., 1976), circRNAs are recently considered as the by-products from pre-mRNA back splicing without important biological functions. Currently, a large body of circRNAs have been demonstrated differentially expressed in physiological and pathological states including cancer due to the development of next-generation sequencing and bioinformatic technologies (Dragomir and Calin, 2018). In lung cancer, circRNAs reveal multiple roles in carcinogenesis, development, and response to therapies, implying their potential roles as not only the diagnostic and prognostic biomarkers but also novel therapeutics.
Biogenesis of circRNAs
RNA alternative splicing is a basic gene expression event in eukaryotic cells. Unlike conventional splicing of mRNA, circRNAs are mainly produced from back-splicing process by ligating a downstream 5′ site with an upstream 3′ site and forming a single-strand closed loop (Jeck et al., 2013). After that, all or part of introns will be removed by the spliceosome and the rest of sequences are to be connected, generating the corresponding subtypes of circRNAs. Exon-skipping is another mechanism for circRNA circularization. It is reported that exon-skipping promotes the shaping process of the spliced lariat containing the circularized exon (Kelly et al., 2015). Furthermore, RNA-binding proteins (RBPs) are demonstrated to be able to induce circRNAs formation. For example, splicing factor muscleblind (MBL) has binding sites on flanking introns of its pre-mRNA and is able to bring the two splicing sites close together and facilitate circularization (Ashwal-Fluss et al., 2014). Enzyme adenosine deaminase 1 (ADAR1) can inhibit circRNAs’ expression by Adenosine-to-Inosine editing to diminish RNA pairing structure of flanking introns and diminish the back-splicing efficiency (Ivanov et al., 2015). RNA-binding protein quaking (QKI) is also proved to regulate circRNAs’ biogenesis by binding to sites flanking circRNAs forming exons to induce exon circularization during epithelial–mesenchymal transition (EMT) (Conn et al., 2015).
Functions and Mechanisms of circRNAs
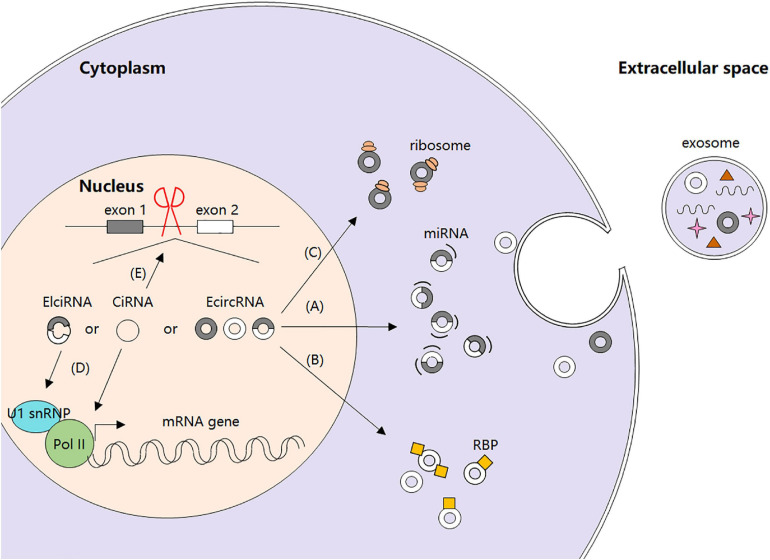
Recently, increasing studies have focused on circRNAs’ biological functions and their regulation. It is confirmed that circRNAs can function as microRNA (miRNA) sponges to stop miRNAs from regulating gene expression via a circRNA–miRNA–mRNA pathway (Hansen et al., 2013). Moreover, circRNAs can act as regulators of gene transcription and alternative splicing, as well as interact with RBPs, or even be translated into peptides or full-length proteins directly (Liu et al., 2017) (Figure 1).
FIGURE 1.
Biogenesis and regulatory functions of circRNAs. (A) circRNAs as miRNA sponges. (B) circRNA-protein interactions. (C) circRNAs and protein translation. (D) circRNAs and gene transcription. (E) circRNAs and alternative splicing.
circRNAs as miRNA Sponges
miRNAs are small non-coding RNAs that post-transcriptionally regulate gene expression by base paring with specific mRNA target sequences, thereby leading to translational inhibition or mRNA degradation (Salmanidis et al., 2014). They can be endogenously sponged by long non-coding RNAs (lncRNAs) owing to the presence of the miRNA response element (MRE) in lncRNA sequences (Cesana et al., 2011). It has been shown that some circRNAs located in cytoplasm also have complementary binding sites of miRNAs and can thus function as competing endogenous RNAs (ceRNAs) to compete with miRNAs and further regulate cellular functions (Zhong et al., 2018). For instance, EcircRNA ciRS-7 is reported to harbor more than 60 conserved miRNA seed match segments for miR-7, thereby antagonizing miR-7 biological activity and functions (Hansen et al., 2013). Subsequent studies confirm the importance of ciRS-7 as miR-7 sponges in many pathological processes including myocardial infarction, insulin secretion, and carcinogenesis (Pan et al., 2018).
circRNA–Protein Interactions
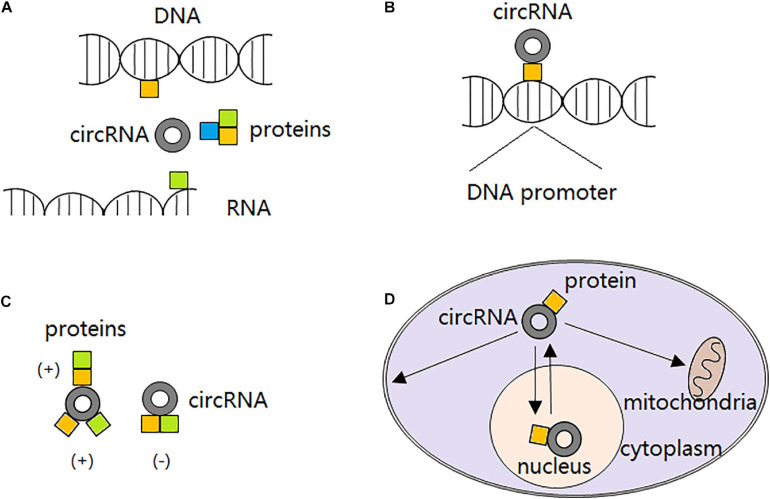
Recently, it is demonstrated that certain circRNAs can serve as protein decoys through binding to RBPs and regulate their interaction with DNAs, RNAs, and/or other proteins. For example, with several MBL binding sites, circMbl is able to sponge out the excessive MBL proteins and maintain its expression balance (Ashwal-Fluss et al., 2014). circANRIL can bind to pescadillo homolog 1 (PES1) and restrain exonuclease-mediated pre-rRNA (ribosomal RNA) processing (Holdt et al., 2016). circPABPN1 binds to HuR and prevents its binding to PABPN1 mRNA, resulting in PABPN1 translation attenuation (Abdelmohsen et al., 2017). circ-Foxo3 participates in the composition of a ternary complex by binding to cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (CDKN1), leading to impaired function of CDK2 and cell cycle arrest (Du et al., 2016). However, not all the circRNAs interacting with RBPs inhibit proteins functions. Particularly, circ-Amotl1 is reported to interact with and stabilize oncogene c-myc and upregulate c-myc targets, thereby promoting tumorigenesis (Yang Q. et al., 2017). The second action mode of circRNAs in interacting with proteins is to serve as protein recruiters. They can recruit not only transcription factors (Wang et al., 2019c), but also chromatin remodelers (Ding et al., 2019) and DNA or histone modifying enzymes (Chen N. et al., 2018) to the promoters and alter transcription either positively or negatively. Furthermore, circRNAs are able to alter interactions between proteins. In detail, circRNAs can strengthen interactions between proteins through direct binding to both of them (Huang S. et al., 2019) or just one of them (Fang et al., 2018), or dissociate interactions between proteins originally combined together by direct binding to both of them (Fang et al., 2019). Interestingly, it is reported that some circRNAs can also serve as protein transporters. circRNAs transport proteins between nucleus and cytoplasm (Yang Z. et al., 2017; Wang et al., 2019e), and to the mitochondria (Liu et al., 2020) or membrane (Du et al., 2020) (Figure 2).
FIGURE 2.
circRNA-protein interactions. (A) circRNAs block proteins from interacting with DNAs, RNAs or other proteins, thereby compromising their original functions. (B) circRNAs recruit transcription factors, chromatin remodelers, and DNA or histone modifying enzymes to the promoters, altering transcription either positively or negatively. (C) circRNAs strengthen interactions between proteins through direct binding to both of them or just one of them; or dissociate interactions between proteins originally combined together by direct binding to both of them. (D) circRNAs transport proteins between nucleus and cytoplasm, and to the mitochondria or membrane.
circRNAs and Protein Translation
Although initially considered as non-coding RNAs (Jeck et al., 2013), circRNAs are shown to be translatable in human transcriptome in vivo by ribosome footprinting and mass spectrometry analysis recently (Pamudurti et al., 2017). With the help of internal ribosome entry site (IRES) which is a non-circle structure, circRNA can be translated into either small peptides or full-length proteins. For example, circ-SHPRH is reported to encode the protein SHPRH-146aa, protect full-length SHPRH from degradation by the ubiquitin proteasome, and ultimately inhibit glioma tumorigenesis (Zhang et al., 2018a). An endogenous circFBXW7 can be encoded into a 21 kDa protein with suppressive roles in malignant phenotypes of human glioblastoma (Yang et al., 2018). Moreover, extensive N6-methyladenosine modification can drive the cap-independent translation of circRNAs together with m6A reader YTHDF3 and translation initiation factors eIF3A and eIF4G2 (Yang Y. et al., 2017).
Known circRNAs and their protein production are listed in Table 1.
TABLE 1.
circRNAs and protein translation.
| circRNA | Peptide/protein | References |
| circSHPRH | SHPRH-146aa | Zhang et al., 2018a |
| circFBXW7 | FBXW7-185aa | Yang et al., 2018 |
| HPV16 circE7 | E7 oncoprotein | Zhao et al., 2019 |
| circβ-catenin | β-catenin-370aa | Liang et al., 2019 |
| circAKT3 | AKT3-174aa | Xia et al., 2019 |
| circPPP1R12A | PPP1R12A-73aa | Zheng et al., 2019 |
| circPIN-Texon2 | PINT87aa | Zhang et al., 2018b |
| circGprc5a | circGprc5a-peptide | Gu et al., 2018 |
| circLgr4 | circLgr4-peptide | Zhi et al., 2019 |
circRNAs and Gene Transcription
It is demonstrated that certain ElciRNAs and ciRNAs may act as transcriptional regulators. For instance, ElciRNAs circEIF3J and circPAIP2 are reported to promote their parental genes promotion by a specific U1 small nuclear RNA (snRNA)-ElciRNA interaction. Mechanically, EIciRNAs bind to U1 small nuclear ribonucleoprotein (snRNP) through interaction with U1 snRNA and to form EIciRNA–U1 snRNP complexes, which will further interact with RNA polymerase II (Pol II) transcription complex of the parental gene and enhance gene transcription (Li et al., 2015). Besides, ciRNA ci-ankrd52 is able to accumulate to its transcription sites and positively regulate elongation Pol II machinery, suggesting a cis-regulatory role of ciRNAs in expression of their parental genes (Zhang et al., 2013).
circRNAs and Alternative Splicing
RNA alternative splicing is a basic gene expression event in eukaryotic cells. During this process, the backsplicing of circRNAs competes with the linear splicing of pre-mRNAs for splicing sites. For example, derived from the second exon of splicing factor MBL, circMbl itself and its flanking introns have conserved MBL binding sites, indicating that MBL may have effects on alternative splicing and modulate the balance between the backsplicing of circRNAs and the linear splicing of pre-mRNAs (Ashwal-Fluss et al., 2014). Meanwhile, circRNAs can act as “mRNA traps” during the back-splicing process by sequestering the translation start site to prevent translation of certain normal linear transcripts and reduce the expression of the very proteins. For instance, EcircRNAs generated from Formin (Fmn) and Duchenne muscular dystrophies traps (DMD) genes are able to cause inactivation of RNA transcripts with certain deletion mutations, thereby diminishing the expression levels of the corresponding functional proteins (Chao et al., 1998; Gualandi et al., 2003).
circRNAs and Lung Cancer
Emerging evidence suggests that circRNAs are abnormally expressed and playing endogenous regulatory roles in carcinogenesis and development of lung cancer, including cell viability, apoptosis, autophagy, invasion, migration, tumor microenvironment (TME) regulation (such as tumor immunosuppression, angiogenesis, hypoxia, and metabolic abnormalities), and therapeutic sensitivities. Moreover, circRNAs own relative specificity and stability compared to other non-coding RNAs, making them more attractive as novel diagnostic and prognostic biomarkers as well as promising therapeutics in lung cancer management. With the development of high-throughput sequencing technology, circRNA expressions in cell lines, tissue samples, and liquid biopsies (especially blood) from lung cancer patients have been detected (Wang et al., 2019a; Zhang et al., 2020). Functional validation assays as well as bioinformatic analysis have been performed to reveal the interaction network of circRNAs and other regulatory factors in lung cancer tumorigenesis and development.
circRNAs as Diagnostic and Prognostic Biomarkers for Lung Cancer
With advantages such as non-invasion, specificity, reproducibility, and sensitivity, circRNAs can act as biomarkers for lung cancer pathological subtyping. For example, hsa_circ_0013958 is indicated to be used as a potential biomarker for screening and early detecting lung adenocarcinoma (Zhu et al., 2017). In a recent work, circRNA expressions are profiled in both lung adenocarcinoma and squamous cell carcinoma and the result indicates that the two subtypes exhibit distinct circRNA expression signatures (Wang et al., 2019a). Similarly, circNOL10 expression varies significantly between lung adenocarcinoma and lung squamous cell carcinoma and has close relationship with the degree of differentiation (Nan et al., 2018). Another study confirms that circ-STXBP5L is selectively expressed in small cell lung cancer (SCLC) samples compared with NSCLC (Zhang et al., 2020). These results highlight the important diagnostic value of circRNAs in pathological classification of lung cancer.
Moreover, mounting studies demonstrate that circRNAs can also serve as prognostic biomarkers for lung cancer occurrence, development, tumor node metastasis (TNM) staging, pathological grade, and lymphatic metastasis. Generally, many circRNAs are upregulated in lung cancer and are considered to be “onco-circRNAs” due to their positive roles in cancer cell proliferation, invasion, and migration, as well as the negative regulation on cancer cell apoptosis. Vice versa, circRNAs that are downregulated in lung cancer with tumor-suppressive functions are considered as “tumor-suppressive circRNAs.”
Onco-circRNAs in Lung Cancer
To date, a multitude of onco-circRNAs have been identified in lung cancer and proposed as potential biomarkers for prognosis.
As is well-known that metastasis is one of the main characteristics of malignant tumors including lung cancer. It is reported that friend leukemia virus integration 1 (FLI1) exonic circRNA FECR1 can promote SCLC metastasis by increasing rho-associated coiled-coil kinase 1 (ROCK1) expressions through direct inhibition of miR584-3p (Li L. et al., 2019). It is described above that circ-STXBP5L participates in the carcinogenesis of SCLC as an onco-circRNA by sponging miR-224-3p and miR-512-3p and regulating a subset of target genes, including Akts, NFκB and Pik3ca (Zhang et al., 2020). As for NSCLC, the data are more detailed. For instance, over-expressed in EGFR-resistant H1975 cells, circRNA CCDC66 is regulated by HGF/c-Met to increase EMT process and drug resistance of lung adenocarcinoma (Joseph et al., 2018). Produced from the EML4-ALK fusion, circRNA F-circEA-2a reveals positive effect on cell invasion and migration in NSCLC, highlighting its critical role in EML4-ALK-positive NSCLC (Tan et al., 2018). circRNA_102231 promotes cellular proliferation, invasion, and migration in lung cancer. Highly expressed circRNA_102231 may serve as a biomarker for both diagnosis and prognosis for lung cancer patients (Zong et al., 2018). It is demonstrated that circHIPK3 can restore lung cancer cell survival and proliferation via sponging miR-124 and regulating expression of its potential targets such as SphK1, STAT3, and CDK4 (Yu et al., 2018). Particularly, circHIPK3 also functions as a negative autophagy regulator in lung cancer through the miR124-3p-STAT3-PRKAA pathway which is dependent on STK11 status (Chen et al., 2020). circHIPK3 regulates the EMT progress of NSCLC through miR-149-mediated Forkhead Box transcription factor FOXM1 expression regulation, closely correlated with the aggressive potential and unfavorable prognosis (Lu et al., 2020). CircPVT1 positively regulates NSCLC cell proliferation, invasion, and metastasis by sponging miR-125b and activating the corresponding E2F2 signaling pathway (Li X. et al., 2018). It can also act as a competing endogenous RNA for miR-497 and indirectly increase the expression of Bcl-2, leading to promoted NSCLC progression and predicting poor survival of the patients (Qin et al., 2019).
Hypoxia caused by the instability of the tumor-associated microvasculature is one of the key reasons for cancer progression. It is reported that circ_0000376 can promote NSCLC progression by regulating the miR-1182/NOVA2 axis and is relative to the poor overall survival of NSCLC patients. Hypoxia enhances circ_0000376 expression and promotes the glycolysis, viability, invasion and migration of NSCLC cells. Manipulated inhibition of circ_0000376 suppresses the progressive activities of hypoxia-induced NSCLC cells both in vitro and in vivo (Li C. et al., 2020).
Several circRNAs are verified to effect both cell proliferation and apoptosis in lung cancer. For instance, upregulated hsa_circ_0000064 demonstrates effect on cell proliferation, metastasis, and apoptosis by regulating target genes such as cell cycle regulators p21WAF1, cyclin D1, and CDK6, as well as apoptotic factors caspase-3, caspase-9, and bax (Luo et al., 2017). Serving as a sponge for miR-503, circ-BANP promotes proliferation, invasion, and migration while attenuates apoptosis of lung cancer cells through promotion of LARP1 expression (Han et al., 2018). circ-FOXM1 works as a ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in NSCLC. It promotes cellular proliferation, invasion, and migration, and suppresses apoptosis, thus playing oncogenic roles in progression of NSCLC. The elevation of circ-FOXM1 in NSCLC is proved to be strongly linked to advanced TNM stages, lymph node invasion, as well as dismal prognosis (Liu G. et al., 2019). Commonly upregulated in NSCLC tissues and cell lines, circ_0026134 facilitates NSCLC proliferation and metastatic properties and weakens cell apoptosis, dependent on the sponge and down-regulation of miR-1287 and miR-1256 (Chang et al., 2019). Similarly, circRNA 100146 is highly expressed and playing an oncogenic role in the progression of NSCLC. It enhances NSCLC cell proliferation and invasion and inhibits cell apoptosis through direct binding to miR-361-3p and miR-615-5p and affections on multiple downstream molecules such as NFAT5, COL1A1, TRAF3, and MEF2C (Chen L. et al., 2019).
It is shown that lung cancer progression is also promoted by increased glycolysis. For example, Enolase 1 (ENO1) is a glycolysis enzyme which performs crucial roles in glucose metabolism and contributes to progression of lung cancer. Recently, circ-ENO1 and its host gene ENO1 are reported to be upregulated in lung adenocarcinoma. Mechanistically, circ-ENO1 acts as a ceRNA of miR-22-3p and upregulates ENO1 expression, promoting glycolysis and tumor progression in lung adenocarcinoma. Silencing of circ-ENO1 inhibits glycolysis, cell proliferation, migration and EMT of lung adenocarcinoma (Zhou J. et al., 2019).
Tumor immune microenvironment is another pivotal factor for the development of lung cancer where cancer cells interact with immune cells to facilitate immune evasion. For example, NSCLC-derived intracellular and extracellular PD-L1 can not only promote cancer progression and drug resistance but also facilitate tumor immune evasion (Li Y. et al., 2019). It is recently demonstrated that circ-CPA4 is high-expressed in NSCLC and can regulate cell growth, metastasis, stemness and drug resistance as well as inactivate CD8+ T cells in tumor immune microenvironment through miRNA let-7/PD-L1 regulatory axis. Inhibition of circ-CPA4 suppresses NSCLC cell growth, mobility and EMT, while enhances cell death via downregulation of let-7/PD-L1 axis. Furthermore, circ-CPA4 can positively regulate the expression of exosomal PD-L1 which promotes NSCLC cell stemness and increases the resistance toward cisplatin (Hong et al., 2020).
Dysregulated onco-circRNAs in lung cancer, their functions, and the underlying mechanisms are listed in Table 2.
TABLE 2.
Dysregulated onco-circRNAs in lung cancer.
| circRNA | Function | Mechanism | References |
| circFECR1 | Metastasis of SCLC | miR584-3p/ROCK1 | Li L. et al., 2019 |
| circSTXBP5L | Carcinogenesis of SCLC | miR-224-3p and miR-512-3p/Akts, NFκB, and Pik3ca | Zhang et al., 2020 |
| circ_100876 | Prognosis values | ? | Yao et al., 2017 |
| circCCDC66 | EMT and drug resistance of lung adenocarcinoma | ? | Joseph et al., 2018 |
| circF-circEA-2a | Invasion and migration of NSCLC | ? | Tan et al., 2018 |
| circRNA_102231 | Proliferation, invasion, and migration of lung cancer | ? | Zong et al., 2018 |
| hsa_circRNA_103809 | Proliferation and invasion of lung cancer | miR-4302/ZNF121/MYC | Liu W. et al., 2018 |
| circHIPK3 | Cell survival and proliferation of lung cancer | miR-124/SphK1, STAT3, and CDK4 | Yu et al., 2018 |
| Autophagy of lung cancer | miR124-3p/STAT3/PRKAA | Chen et al., 2020 | |
| EMT and aggressiveness of NSCLC | miR-149/FOXM1 | Lu et al., 2020 | |
| circPVT1 | Proliferation, invasion, and metastasis of NSCLC | miR-125b/E2F2 | Li X. et al., 2018 |
| NSCLC progression | miR-497/Bcl-2 | Qin et al., 2019 | |
| circMAN2B2 | Proliferation and invasion of lung cancer | miR-1275/FOXK1 | Ma et al., 2018 |
| circ_0067934 | Tumorigenesis, EMT, and metastasis of NSCLC | ? | Wang and Li, 2018 |
| hsa_circ_0020123 | Proliferation, invasion, and migration of NSCLC | miR-144/ZEB1 and EZH2 | Qu et al., 2018 |
| Growth, invasion, migration, and apoptosis of NSCLC | miR-488e3p/ADAM9 | Wan et al., 2019 | |
| circFADS2 | Proliferation and invasion of lung cancer | miR-498/FOXO1/KLF6 | Zhao et al., 2018 |
| circ_0016760 | Progression of NSCLC | miR-1287/GAGE1 | Li Y. et al., 2018 |
| circPRKCI | Proliferation and tumorigenesis of lung adenocarcinoma | miR-545 and miR-589/E2F7 | Qiu et al., 2018 |
| circCMPK1 | Cell proliferation of NSCLC | miR-302e/cyclin D1 | Cui et al., 2020 |
| circFGFR1 | Proliferation, migration, invasion, and immune evasion abilities of NSCLC | miR-381-3p/CXCR4 | Zhang P. et al., 2019 |
| circP4HB | EMT and metastasis of NSCLC | miR-133a-5p/vimentin | Wang et al., 2019f |
| hsa_circ_000984 | Proliferation and metastasis of NSCLC | Wnt/β-catenin signaling | Li X. et al., 2019 |
| circ_0003645 | NSCLC progression | miR-1179/TMEM14A | An et al., 2019 |
| circZFR | NSCLC progression | miR-101-3p/CUL4B | Zhang H. et al., 2019 |
| hsa_circ_0023404 | Proliferation, invasion, and migration of NSCLC | miR-217/ZEB1 | Liu C. et al., 2019 |
| F-circSR1 and 2 | Migration and invasion of NSCLC | ? | Wu et al., 2019 |
| circPRMT5 | NSCLC progression | miR-377, miR-382, and miR-498/EZH2 | Wang et al., 2019h |
| circPIP5K1A | Proliferation and metastasis of NSCLC | miR-600/HIF-1α | Chi et al., 2019 |
| circATXN7 | Proliferation and invasion abilities of NSCLC | ? | Huang Q. et al., 2019 |
| circFGFR3 | Proliferation and invasion of NSCLC | miR-22-3p/Gal-1, p-AKT and p-ERK1/2 | Qiu et al., 2019 |
| circ_0043278 | Proliferation, invasion and migration of NSCLC | miR-520f/ROCK1, CDKN1B and AKT3 | Cui et al., 2019 |
| circRAD23B | Progression of NSCLC | miR-593e3p/CCND2 and miR-653e5p/TIAM1 | Han et al., 2019 |
| circNT5E | Cell growth, proliferation, and migration of NSCLC | miR-134 | Dong et al., 2020 |
| hsa_circ_0013958 | Proliferation, invasion, and apoptosis of lung adenocarcinoma | miR-134/cyclin D1 | Zhu et al., 2017 |
| circ_0000376 | Glycolysis, cell viability, invasion and migration of NSCLC | miR-1182/NOVA2 | Li Y. et al., 2020 |
| hsa_circ_0000064 | Proliferation, apoptosis, and metastasis of lung cancer | Cell cycle regulators (p21WAF1, cyclin D1, and CDK6) and apoptotic factors (caspase-3, caspase-9, and bax) | Luo et al., 2017 |
| hsa_circ_0007385 | NSCLC progression | miR-181/Bcl-2 and CDK1 | Jiang et al., 2018 |
| circBANP | Proliferation, invasion, migration, and apoptosis of lung cancer | miR-503/LARP1 | Han et al., 2018 |
| circVANGL1 | Proliferation, migration, invasion, and apoptosis of NSCLC | miR-195/Bcl-2 and Bax | Wang et al., 2019d |
| circFOXM1 | Proliferation, invasion, migration, and apoptosis of NSCLC | miR-1304-5p/PPDPF and MACC1 | Liu G. et al., 2019 |
| circ_0026134 | Proliferation, metastasis, and apoptosis of NSCLC | miR-1287/PIK3R3 and miR-1256/TCTN1 | Chang et al., 2019 |
| circRNA 100146 | Proliferation, invasion, and apoptosis of NSCLC | miR-361-3p and miR-615-5p/NFAT5, COL1A1, TRAF3, and MEF2C | Chen L. et al., 2019 |
| circCDR1 | Cell viability, migration, invasion, and apoptosis of NSCLC | miR-219a-5p/SOX5 | Li Y. et al., 2020 |
| circ-ENO1 | Glycolysis, proliferation, apoptosis, migration and EMT of lung adenocarcinoma | miR-22-3p/ENO1 | Zhou J. et al., 2019 |
| circ-CPA4 | Cell growth, mobility, stemness, drug resistance, and CD8+ T cell inactivation in the tumor immune microenvironment of NSCLC | let-7/PD-L1 | Hong et al., 2020 |
Tumor-Suppressive circRNAs in Lung Cancer
A series of circRNAs have been found downregulated in lung cancer and thus considered as tumor suppressors. For instance, downregulated in SCLC and chemo-resistant NSCLC cells, circRNA cESRP1 enhances drug sensitivity by directly binding to and repressing miR-93-5p, thereby up-regulating the expression of Smad7 and p21, forming a negative feedback loop to regulate EMT process dependent of transforming growth factor-β (TGF-β) (Huang W. et al., 2019).
In NSCLC, circRNA ITCH is reported markedly decreased in cancer tissues with negative regulation on the proliferation of cancer cells by down-regulating oncogenic miR-7 and miR-214 as well as up-regulating T-cell factor, β-catenin, c-Myc, and cyclin D1, thereby enhancing the activation of the Wnt/β-catenin signaling pathway (Wan et al., 2016). circ_0001649 is demonstrated to have a decreased expression in NSCLC tissues and cell lines with suppressive functions on cell growth and metastasis both in vitro and in vivo partially by sponging out miR-331-3p and miR-338-5p. The down-regulation of circ_0001649 is highly interrelated with advanced TNM stage, positive lymph node metastasis, and poor prognosis of NSCLC patients (Liu T. et al., 2018). circPTK2 promotes TIF1γ expression and inhibits TGF-β-induced EMT and metastasis in NSCLC dependent on miR-429/miR-200b-3p sponging (Wang et al., 2018). circRNA-FOXO3 expression is also found decreased in NSCLC and correlated with clinical outcomes. circRNA-FOXO3 inhibits NSCLC cell proliferation, invasion, and migration by acting as a ceRNA to sponge miR-155 and release FOXO3 expression (Zhang et al., 2018c).
Cell apoptosis is also regulated by certain tumor-suppressive circRNAs in lung cancer. For example, acting as an endogenous sponge for miR-1252, has_circ_0043256 can upregulate the expression of ITCH and finally inhibit Wnt/β-catenin pathway, leading to suppression of cell proliferation and enhancement of apoptosis in NSCLC (Tian et al., 2017). circNOL10 is downregulated in both lung cancer tissues and cells and conducive in differentiating lung adenocarcinoma and lung squamous cell carcinoma. Furthermore, circNOL10 inhibits lung cancer by enhancing transcriptional regulation of the HN polypeptide family, which exerts pivotal functions on biological processes such as proliferation, apoptosis, and cell cycle progression (Nan et al., 2018).
Dysregulated tumor-suppressive circRNAs in lung cancer, their functions, and the underlying mechanisms are listed in Table 3.
TABLE 3.
Dysregulated tumor-suppressive circRNAs in lung cancer.
| circRNA | Function | Mechanism | References |
| circESRP1 | EMT and drug sensitivity of SCLC | miR-93-5p/Smad7/p21(CDKN1A)/TGF-β | Huang W. et al., 2019 |
| circITCH | proliferation of NSCLC | miR-7 and miR-214/T-cell factor, β-catenin, c-Myc, and cyclin D1 | Wan et al., 2016 |
| circ_0001649 | growth and metastasis of NSCLC | miR-331-3p and miR-338-5p | Liu T. et al., 2018 |
| circPTK2 | EMT and metastasis of NSCLC | miR-429 and miR-200b-3p/TIF1γ/TGF-β | Wang et al., 2018 |
| circFOXO3 | Proliferation, invasion, and migration of NSCLC | miR-155/FOXO3 | Zhang et al., 2018c |
| hsa_circ_100395 | Proliferation, invasion, and migration of lung cancer | miR-1228/TCF21 | Chen D. et al., 2018 |
| hsa_circ_0033155 | Proliferation and migration of NSCLC | PTEN | Gu et al., 2019 |
| circSMARCA5 | Proliferation and chemo-sensitivity of NSCLC | miR-19b-3p/HOXA9 | Wang et al., 2019g; Tong, 2020 |
| hsa_circ_0007059 | Proliferation and EMT of lung cancer | miR-378/Wnt/β-catenin and ERK1/2 | Gao et al., 2019 |
| circARHGAP10 | NSCLC progression | miR-150-5p/GLUT1 | Jin et al., 2019 |
| circPTPRA | EMT and metastasis of NSCLC | miR-96-5p/RASSF8 | Wei et al., 2019 |
| has_circ_0043256 | Proliferation and apoptosis of NSCLC | miR-1252/ITCH/Wnt/β-catenin | Tian et al., 2017 |
| circNOL10 | Proliferation, apoptosis, and cell cycle progression of lung cancer | SCLM1/the HN polypeptide family | Nan et al., 2018 |
circRNAs and the Therapeutic Response of Lung Cancer
Except for chemotherapy, targeted therapies and immunotherapies have revolutionized the lung cancer management within the last decades. However, drug-resistance still develops after the treatment. Considering the multiple roles of circRNAs in lung cancer progression, it is not surprising to apply them as predictive biomarkers for the follow-up of patients. Furthermore, the detection of circRNAs in liquid biopsies has provided a more convenient method for the management of post-treatment follow-up.
Chemotherapy
With the assistance of a high-throughput circRNA microarray, a significant upregulation of 2,909 circRNAs as well as downregulation of 8,372 circRNAs are discovered in taxol-resistant A549 lung adenocarcinoma cells compared with the parental cells (Xu et al., 2018). Functional validation assays highlight the circRNA/miRNA networks in this context. The most pronouncedly enriched pathways for aberrant circRNA-related host genes include VEGFR, EGFR, integrin, and rho GTPase signaling, which are all involved in the progression of chemo-resistance.
Eukaryotic initiation factor 3 (EIF3) is one of the largest translation initiation factors. Previous studies have suggested the involvement of EIF3a in tumorigenesis and drug resistance of lung cancer. Recently, it is found that the expression of hsa_circ_0004350 and hsa_circ_0092857, both derived from EIF3a, varies prominently in cisplatin-resistant lung cancer cell line and the parental cell line. Manipulated regulation of the two circEIF3as affects cisplatin sensitivity of lung cancer cells. Further bioinformatic analysis indicates that the two circEIF3as are not only related to translational regulation, but also showing functional synergy with their parental gene EIF3a, thus serving as potential therapeutic targets during lung cancer management (Huang M.S. et al., 2019).
Targeted Therapy
EGFR-tyrosine kinase inhibitors (EGFR-TKIs) have become important constituents for NSCLC treatment these years. Despite the good initial responses, tumor progression is systematically observed due to the emergence of acquire resistance. Furthermore, detection for EGFR driver mutation is hindered by problems such as cancer heterogeneity and lack of cancer tissues. By screening circRNAs expression profile via circRNA microarray, it is found that hsa_circ_0109320 and hsa_circ_0134501 are upregulated in gefitinib-effective NSCLC patients. Moreover, hsa_circ_0109320 expression is associated with better progression-free survival (PFS), indicating its potential role as a prognostic biomarker for gefitinib-treated NSCLC patients (Liu Y. et al., 2019). Similarly, hsa_circ_0004015 is identified to be highly expressed in NSCLC cells and tissues. Patients with high expression of hsa_circ_0004015 often have a worse overall survival rate. Further study indicates that hsa_circ_0004015 promotes NSCLC progression and gefitinib resistance through sponge for miR-1183 and induction of 3-phosphoinositide-dependent protein kinase 1 (PDPK) as well as the downstream AKT pathway (Zhou Y. et al., 2019). Recently, circRNA expression profiles have been explored in Osimertinib (AZD9291)-resistant NSCLC cells and the result shows that the most modulated circRNAs are involved in regulation of cancer-related pathways including proliferation, invasion, apoptosis, and resistance to chemotherapeutic drugs as well as γ-radiation (Chen T. et al., 2019).
Another role of circRNAs during tumorigenesis is the formation of fusion circRNAs (f-circRNAs) derived from chromosomal translocations (Guarnerio et al., 2016). It is demonstrated that f-circEA-2a which derived from back splicing of the EML4-ALK fusion gene promotes cell invasion and migration but not cell proliferation in NSCLC. Interestingly, f-circEA-2a is detected in tumor tissues but not plasma of EML4-ALK-positive NSCLC patients (Tan et al., 2018).
Immunotherapy
Anti-PD-1-based immunotherapy has led to an effective response in multiple advanced cancers, lung cancer included. However, more than half of NSCLC patients lack a long-term response to this immunotherapy (Melosky et al., 2019). Emerging evidence show that dysregulated chemokine receptor expression is one of the critical intrinsic reasons for tumor-promotion and immune system evasion (Adrover et al., 2019). Recently, it is found that circFGFR1 may act as a sponge for miR-381-3p, thereby promoting NSCLC progression and resistance to anti-PD-1 therapy by up-regulating CXCR4 expression. CircFGFR1 is upregulated in NSCLC tissues with its expression closely correlated with unfavorable prognosis of NSCLC patients. Manipulated upregulation of circFGFR1 promotes the proliferation, invasion, migration, and immune evasion of NSCLC cells, while knockdown of CXCR4 resensitizes NSCLC cells to anti-PD-1 immunotherapy (Zhang P. et al., 2019). As is mentioned above, circ-CPA4 can regulate cell growth, mobility, stemness and drug resistance in NSCLC cells and inactivates CD8+ T cells in the tumor immune microenvironment through the let-7 miRNA/PD-L1 axis. On the one hand, PD-L1 selfregulates NSCLC cell malignant activities. On the other hand, secreted PD-L1 by exosomes inactivates CD8+ T cells by activating extracellular and intracellular pathways and mediates cell death to facilitate immune evasion (Hong et al., 2020).
circRNAs as Promising Therapeutics for Lung Cancer
Regarding the onco- and tumor suppressive- roles in lung cancer, circRNAs provide insight into the exploration of novel strategies in lung cancer management. Moreover, as mentioned above, circRNAs are more suitable for targeted molecular therapy because of their stable, tissue specific, and ceRNA-equivalent characteristics. Further investigation is needed to translate circRNAs into clinics and provide a foundation for developing novel potential therapeutic strategies for lung cancer and improve the prognosis of the patients.
Discussion
Despite significant advances in diagnosis and treatment, lung cancer remains the leading cause of death worldwide with its underlying mechanisms remaining largely undiscovered. A major obstacle of improving clinical outcomes is to identify sensitive biomarkers and novel therapeutics for individualized diagnosis and treatment of lung cancer. In recent years, thousands of circRNAs have been identified with the rapid development of NGS technology and bioinformatics. Although considered as splicing by-products initially, circRNAs are now becoming a hotspot in the field of cancer owing to their conservation across species, the relative high stability and abundance, and the accessibility in body fluids, especially blood. They reveal diverse regulatory functions on genes and proteins involved in cancer cell proliferation, invasion, migration, cell cycle, apoptosis, and drug sensitivity. Moreover, recent studies have shown that the role of circRNAs in cancer is mainly dependent on the circRNA–miRNA–mRNA regulatory network, indicating their potential functions in the regulation of transcriptional and post-transcriptional levels. Other mechanisms include interacting with RBPs, translating into either peptides or full-length proteins, and regulating transcription. This provides novel biomarkers for lung cancer prognosis prediction, especially considering the lack of reliable clinical biomarkers in SCLC. In addition, engineered circRNAs can be applied to effectively sequester not only RNAs (including miRNAs), but also DNAs and RBPs with specific sequences both in vitro and in vivo, providing promising molecular targets for the therapy of lung cancer (Lasda and Parker, 2014).
However, it should be noted that the study on circRNAs in lung cancer is still in the early stage. The functions and underlying mechanisms of circRNAs in the regulatory network of tumorigenesis and progression remains largely unknown and needs to be further studied in more cell lines and clinical samples. Moreover, problems such as the high cost of experiment, difficulties in detection and monitoring, and the potential side effects, are still limiting the application of circRNAs in clinic. We hope that in the future, with the rapid development of bioinformatic technology and universal studies in patient tissue samples, circRNAs could help to achieve better individualized diagnosis and treatment of lung cancer.
Author Contributions
RW conceptualized the review. BF and HZ were the major contributors in writing the manuscript. TW designed the figures. XL and YL prepared the tables. XC critically reviewed and edited the manuscript. All the authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- ADAR1
adenosine deaminase 1
- CDK2
cyclin-dependent kinase 2
- CDKN1
cyclin-dependent kinase inhibitor 1
- ceRNA
competing endogenous RNA
- circRNAs
circular RNAs
- circrRNAs
circRNA rRNAs
- ciRNAs
intronic circRNAs
- CXCR4
C-X -C motif chemokine receptor 4
- EcircRNAs
exonic circRNAs
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EIF3
eukaryotic initiation factor 3
- ElciRNAs
exonic circRNAs with introns
- EMT
epithelial-to-mesenchymal transition
- f-circRNAs
fusion circRNAs
- FLI1
friend leukemia virus integration 1
- FOXM1
Forkhead Box (Fox) transcription factor 1
- HIF
hypoxia-inducible factor
- IRES
internal ribosome entry site
- lncRNA
long non-coding RNA
- m6A
N6-methyladenosine
- MBL
splicing factor muscleblind
- miRNA
microRNA
- NSCLC
non-small cell lung cancer
- ORF
open reading frame
- PD-1
programmed cell death-1
- PD-L1
programmed cell death-1 ligand
- PDPK
phosphoinositide-dependent protein kinase
- PES1
pescadillo homolog 1
- PFS
progression-free survival
- Pol II
RNA polymerase II
- PTEN
phosphatase and tensin homolog deleted on chromosome 10
- QKI
RNA-binding protein quaking
- RBP
RNA-binding protein
- ROCK1
Rho-associated coiled-coil kinase 1
- rRNA
ribosomal RNA
- SCLC
small cell lung cancer
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein
- TGF- β
transforming growth factor- β
- TKI
tyrosine kinase inhibitor
- TME
tumor microenvironment
- TNM
tumor node metastasis
- tricRNAs
tRNA intronic circRNAs.
Footnotes
Funding. This study was supported by grants from the National Natural Science Foundation of China (Nos. 81772995 and 81472266); the Excellent Youth Foundation of Jiangsu Province, China (No. BK20140032); Jiangsu Province’s Key Provincial Talents Program (No. ZDRCA2016090); Jiangsu Provincial Medical Youth Talent, The Project of Invigorating Health Care through Science, Technology and Education (No. QNRC2016886).
References
- Abdelmohsen K., Panda A. C., Munk R., Grammatikakis I., Dudekula D. B., De S., et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14 361–369. 10.1080/15476286.2017.1279788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrover J. M., Fresno C. D., Crainiciuc G., Cuartero M. I., Casanova-Acebes M., Weiss L. A., et al. (2019). A neutrophil timer coordinates immune defense and vascular protection. Immunity 51 966–967. 10.1016/j.immuni.2019.01.002 [DOI] [PubMed] [Google Scholar]
- An J., Shi H., Zhang N., Song S. (2019). Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 511 921–925. 10.1016/j.bbrc.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 358–369. 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Qu J., Wang J., Liang X., Sun W. (2019). Circular RNA circ_0026134 regulates non-small cell lung cancer cell proliferation and invasion via sponging miR-1256 and miR-1287. Biomed. Pharmacother. 112:108743. 10.1016/j.biopha.2019.108743 [DOI] [PubMed] [Google Scholar]
- Chansky K., Detterbeck F. C., Nicholson A. G., Rusch V. W., Vallières E., Groome P., et al. (2017). The IASLC lung cancer staging project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J. Thorac. Oncol. 12 1109–1121. 10.1016/j.jtho.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Chao C., Chan D., Kuo A., Leder P. (1998). The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med. 4 614–628. [PMC free article] [PubMed] [Google Scholar]
- Chen D., Ma W., Ke Z., Xie F. (2018). CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 17 2080–2090. 10.1080/15384101.2018.1515553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhao G., Yan X., Lv Z., Yin H., Zhang S., et al. (2018). A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 19:218. 10.1186/s13059-018-1594-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Nan A., Zhang N., Jia Y., Li X., Ling Y., et al. (2019). Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol. Cancer. 18:13. 10.1186/s12943-019-0943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Luo J., Gu Y., Huang J., Luo Q., Yang Y. (2019). Comprehensive analysis of circular RNA profiling in AZD9291-resistant non-small cell lung cancer cell lines. Thorac. Cancer 10 930–941. 10.1111/1759-7714.13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Mao R., Su W., Yang X., Geng Q., Guo C., et al. (2020). Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy 16 659–671. 10.1080/15548627.2019.1634945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Luo Q., Song Y., Yang F., Wang Y., Jin M., et al. (2019). Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J. Cell Biochem. 120 19019–19030. 10.1002/jcb.29225 [DOI] [PubMed] [Google Scholar]
- Conn S. J., Pillman K. A., Toubia J., Conn V. M., Salmanidis M., Phillips C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160 1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Cui D., Qian R., Li Y. (2020). Circular RNA circ-CMPK1 contributes to cell proliferation of non-small cell lung cancer by elevating cyclin D1 via sponging miR-302e. Mol. Genet. Genomic Med. 8:e999. 10.1002/mgg3.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li W., Liu G., Chen X., Gao X., Lu H., et al. (2019). A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif. Cells Nanomed. Biotechnol. 47 810–821. 10.1080/21691401.2019.1575847 [DOI] [PubMed] [Google Scholar]
- Ding L., Zhao Y., Dang S., Wang Y., Li X., Yu X., et al. (2019). Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer. 18:45. 10.1186/s12943-019-1006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Zheng J., Gao Y., Zhou X., Song W., Huang J. (2020). The circular RNA NT5E promotes non-small cell lung cancer cell growth via sponging microRNA-134. Aging 12 3936–3949. 10.18632/aging.102861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragomir M., Calin G. A. (2018). Circular RNAs in cancer - lessons learned from microRNAs. Front. Oncol. 8:179. 10.3389/fonc.2018.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Yang W., Li X., Fang L., Wu N., Li F., et al. (2020). The circular RNA circSKA3 binds integrin β1 to induce invadopodium formation enhancing breast cancer invasion. Mol. Ther. 28 1287–1298. 10.1016/j.ymthe.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44 2846–2858. 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuka Y., Lauriola M., Feldman M. E., Sas-Chen A., Ulitsky I., Yarden Y. (2016). Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44 1370–1383. 10.1093/nar/gkv1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Du W., Awan F., Dong J., Yang B. (2019). The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett. 459 216–226. 10.1016/j.canlet.2019.05.036 [DOI] [PubMed] [Google Scholar]
- Fang L., Du W., Lyu J., Dong J., Zhang C., Yang W., et al. (2018). Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell Death Differ. 25 2195–2208. 10.1038/s41418-018-0115-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Yu Y., Liu L., Meng J., Li G. (2019). Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 233:116692. 10.1016/j.lfs.2019.116692 [DOI] [PubMed] [Google Scholar]
- Gu C., Zhou N., Wang Z., Li G., Kou Y., Yu S., et al. (2018). CircGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol. Ther. Nucleic Acids 13 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Wang G., Shen H., Fei X. (2019). Hsa_circ_0033155: a potential novel biomarker for non-small cell lung cancer. Exp. Ther. Med. 16 3220–3226. 10.3892/etm.2018.6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualandi F., Trabanelli C., Rimessi P., Calzolari E., Toffolatti L., Patarnello T., et al. (2003). Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J. Med. Genet. 40:e100. 10.1136/jmg.40.8.e100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnerio J., Bezzi M., Jeong J. C., Paffenholz S. V., Berry K., Naldini M. M., et al. (2016). Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165 289–302. 10.1016/j.cell.2016.03.020 [DOI] [PubMed] [Google Scholar]
- Han J., Zhao G., Ma X., Dong Q., Zhang H., Wang Y., et al. (2018). CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem. Biophys. Res. Commun. 503 2429–2435. 10.1016/j.bbrc.2018.06.172 [DOI] [PubMed] [Google Scholar]
- Han W., Wang L., Zhang L., Wang Y., Li Y. (2019). Circular RNA circ-RAD23B promotes cell growth and invasion by miR-593-3p/CCND2 and miR-653-5p/TIAM1 pathways in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 510 462–466. 10.1016/j.bbrc.2019.01.131 [DOI] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Holdt L. M., Stahringer A., Sassm K., Pichler G., Kulak N. A., Wilfert W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7:12429. 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W., Xue M., Jiang J., Zhang Y., Gao X. (2020). Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J. Exp. Clin. Cancer Res. 39:149. 10.1186/s13046-020-01648-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. S., Yuan F. Q., Gao Y., Liu J. Y., Chen Y. X., Wang C. J., et al. (2019). Circular RNA screening from EIF3a in lung cancer. Cancer Med. 8 4159–4168. 10.1002/cam4.2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Wang S., Li X., Yang F., Feng C., Zhong K., et al. (2019). Circular RNA ATXN7 is upregulated in non-small cell lung cancer and promotes disease progression. Oncol. Lett. 17 4803–4810. 10.3892/ol.2019.10168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Li X., Zheng H., Si X., Li B., Wei G., et al. (2019). Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation 139 2857–2876. 10.1161/CIRCULATIONAHA.118.038361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Yang Y., Wu J., Niu Y., Yao Y., Zhang J., et al. (2019). Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling. Cell Death Differ. 27 1709–1727. 10.1038/s41418-019-0455-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A., Memczak S., Wyler E., Torti F., Porath H. T., Orejuela M. R., et al. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10 170–177. 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Mai Z., Wan S., Chi Y., Zhang X., Sun B., et al. (2018). Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J. Cancer Res. Clin. Oncol. 144 667–674. 10.1007/s00432-017-2576-2 [DOI] [PubMed] [Google Scholar]
- Jin M., Shi C., Yang C., Liu J., Huang G. (2019). Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in NSCLC through regulation of the miR-150-5p/GLUT-1 Axis. Mol. Ther. Nucleic Acids 18 219–231. 10.1016/j.omtn.2019.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph N. A., Chiou S. H., Lung Z., Yang C., Lin T., Chang H., et al. (2018). The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J. Hematol. Oncol. 11:74. 10.1186/s13045-018-0557-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S., Greenman C., Cook P. R., Papantonis A. (2015). Exon skipping is correlated with exon circularization. J. Mol. Biol. 427 2414–2417. 10.1016/j.jmb.2015.02.018 [DOI] [PubMed] [Google Scholar]
- Kris M. G., Gaspar L. E., Chaft J. E., Kennedy E. B., Azzoli C. G., Ellis P. M., et al. (2017). Adjuvant systemic therapy and adjuvant radiation therapy for Stage I to IIIA completely resected non-small-cell lung cancers: american society of clinical oncology/cancer care ontario clinical practice guideline update. J. Clin. Oncol. 35 2960–2974. 10.1200/JCO.2017.72.4401 [DOI] [PubMed] [Google Scholar]
- Lasda E., Parker R. (2014). Circular RNAs: diversity of form and function. RNA 20 1829–1842. 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Liu H., Niu Q., Gao J. (2020). Circ_0000376, a Novel circRNA, promotes the progression of non-small cell lung cancer through regulating the miR-1182/NOVA2 network. Cancer Manag. Res. 12 7635–7647. 10.2147/CMAR.S258340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang J., Pan S., Zhou J., Diao X., Liu S. (2020). CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thorac. Cancer 11 537–548. 10.1111/1759-7714.13274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li W., Chen N., Zhao H., Xu G., Zhao Y., et al. (2019). FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin. Cancer Res. 25 1302–1317. 10.1158/1078-0432.CCR-18-1447 [DOI] [PubMed] [Google Scholar]
- Li X., Liu Y., Zhou J., Li W., Guo H., Ma H. (2019). Enhanced expression of circular RNA hsa_circ_000984 promotes cells proliferation and metastasis in non-small cell lung cancer by modulating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 23 3366–3374. 10.26355/eurrev_201904_17700 [DOI] [PubMed] [Google Scholar]
- Li Y., Yu J., Liu Z., Yang H., Tang J., Chen Z. (2019). Programmed death Ligand 1 indicates pre-existing adaptive immune response by tumor-infiltrating CD8+ T Cells in non-small cell lung cancer. Int. J. Mol. Sci. 20:5138. 10.3390/ijms20205138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Z., Jiang H., Li Q., Wang R., Pan H., et al. (2018). Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell Physiol. Biochem. 51 2324–2340. 10.1159/000495876 [DOI] [PubMed] [Google Scholar]
- Li Y., Hu J., Li L., Cai S., Zhang H., Zhu X., et al. (2018). Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem. Biophys. Res. Commun. 503 2089–2094. 10.1016/j.bbrc.2018.07.164 [DOI] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Liang W. C., Wong C. W., Liang P. P., Shi M., Cao Y., Rao S. T., et al. (2019). Translation of the circular RNA circ(β-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 20:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Guo C., Zou M., Meng P., Zhang T. (2020). CircRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 20:173. 10.1186/s12935-020-01245-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhang Z., Qi D. (2019). Circular RNA hsa_circ_0023404 promotes proliferation, migration and invasion in non-small cell lung cancer by regulating miR-217/ZEB1 axis. Onco. Targets Ther. 12 6181–6189. 10.2147/OTT.S201834 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu G., Shi H., Deng L., Zheng H., Kong W., Wen X., et al. (2019). Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 513 207–212. 10.1016/j.bbrc.2019.03.213 [DOI] [PubMed] [Google Scholar]
- Liu Y., Han X., Xing P., Hu X., Hao X., Wang Y., et al. (2019). Circular RNA profiling identified as a biomarker for predicting the efficacy of Gefitinib therapy for non-small cell lung cancer. J. Thorac. Dis. 11 1779–1787. 10.21037/jtd.2019.05.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang J., Khanabdali R., Kalionis B., Tai X., Xia S. (2017). Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 14 1715–1721. 10.1080/15476286.2017.1367886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Song Z., Gai Y. (2018). Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem. Biophys. Res. Commun. 503 1503–1509. 10.1016/j.bbrc.2018.07.070 [DOI] [PubMed] [Google Scholar]
- Liu W., Ma W., Yuan Y., Zhang Y., Sun S. (2018). Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem. Biophys. Res. Commun. 500 846–851. 10.1016/j.bbrc.2018.04.172 [DOI] [PubMed] [Google Scholar]
- Liu X., Wang X., Li J., Hu S., Deng Y., Yin H., et al. (2020). Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 63 1429–1449. 10.1007/s11427-020-1631-9 [DOI] [PubMed] [Google Scholar]
- Lu H., Han X., Ren J., Ren K., Li Z., Sun Z. (2020). Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol. Ther. 21 113–121. 10.1080/15384047.2019.1669995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zhu X., Huang K., Zhang Q., Fan Y., Yan P., et al. (2017). Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed. Pharmacother. 96 892–898. 10.1016/j.biopha.2017.12.015 [DOI] [PubMed] [Google Scholar]
- Ma X., Yang X., Bao W., Li S., Liang S., Sun Y., et al. (2018). Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem. Biophys. Res. Commun. 498 1009–1015. 10.1016/j.bbrc.2018.03.105 [DOI] [PubMed] [Google Scholar]
- Melosky B., Juergens R., McLeod D., Leighl N., Brade A., Card P. B., et al. (2019). Immune checkpoint-inhibitors and chemoradiation in stage III unresectable non-small cell lung cancer. Lung Cancer 134 259–267. 10.1016/j.lungcan.2019.05.027 [DOI] [PubMed] [Google Scholar]
- Nan A., Chen L., Zhang N., Jia Y., Li X., Zhou H., et al. (2018). Circular RNA circNOL10 inhibits lung cancer development by promoting SCLM1-mediated transcriptional regulation of the humanin polypeptide family. Adv. Sci. 6:1800654. 10.1002/advs.201800654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., et al. (2017). Translation of CircRNAs. Mol. Cell. 66 9.e7–21.e7. 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Li T., Jiang Y., Pan C., Ding Y., Huang Z., et al. (2018). Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J. Cell Biochem. 119 440–446. 10.1002/jcb.26201 [DOI] [PubMed] [Google Scholar]
- Qin S., Zhao Y., Lim G., Lin H., Zhang X., Zhang X. (2019). Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed. Pharmacother. 111 244–250. 10.1016/j.biopha.2018.12.007 [DOI] [PubMed] [Google Scholar]
- Qiu B., Zhang P., Xiong D., Xu J., Long X., Zhu S., et al. (2019). CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non-small cell lung cancer by regulating Galectin-1-AKT/ERK1/2 signaling. J. Cell Physiol. 234 11256–11264. 10.1002/jcp.27783 [DOI] [PubMed] [Google Scholar]
- Qiu M., Xia W., Chen R., Wang S., Xu Y., Ma Z., et al. (2018). The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 78 2839–2851. 10.1158/0008-5472.CAN-17-2808 [DOI] [PubMed] [Google Scholar]
- Qu D., Yan B., Xin R., Ma T. (2018). A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am. J. Cancer Res. 8 1387–1402. [PMC free article] [PubMed] [Google Scholar]
- Salmanidis M., Pillman K., Goodall G., Bracken C. (2014). Direct transcriptional regulation by nuclear microRNAs. Int. J. Biochem. Cell Biol. 54 304–311. 10.1016/j.biocel.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2017). Cancer statistics, 2017. CA Cancer J. Clin. 67 7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Tsukahara T. (2014). A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 15 9331–9342. 10.3390/ijms15069331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Sun D., Pu W., Gou Q., Guo C., Gong Y., et al. (2018). Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol. Cancer 17:138. 10.1186/s12943-018-0887-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Yu C., Ye W., Wang Q. (2017). Cinnamaldehyde induces cell apoptosis mediated by a novel circular RNA hsa_circ_0043256 in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 493 1260–1266. 10.1016/j.bbrc.2017.09.136 [DOI] [PubMed] [Google Scholar]
- Tong S. (2020). Circular RNA SMARCA5 may serve as a tumor suppressor in non-small cell lung cancer. J. Clin. Lab Anal. 34:e23195. 10.1002/jcla.23195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Hao L., Zheng X., Li Z. (2019). Circular RNA circ_0020123 promotes non-small cell lung cancer progression by acting as a ceRNA for miR-488-3p to regulate ADAM9 expression. Biochem. Biophys. Res. Commun. 515 303–309. 10.1016/j.bbrc.2019.05.158 [DOI] [PubMed] [Google Scholar]
- Wan L., Zhang L., Fan K., Cheng Z., Sun Q., Wang J. (2016). Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-Catenin pathway. Biomed. Res. Int. 2016:1579490. 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tong X., Zhou Z., Wang S., Lei Z., Zhang T., et al. (2018). Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol. Cancer 17:140. 10.1186/s12943-018-0889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Tan S., Liu W., Lei Q., Qiao W., Wu Y., et al. (2019a). RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol. Cancer 18:134. 10.1186/s12943-019-1061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhu M., Pan J., Chen C., Xia S., Song Y. (2019b). Circular RNAs: a rising star in respiratory diseases. Respir. Res. 20:3. 10.1186/s12931-018-0962-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Long H., Zheng Q., Bo X., Xiao X., Li B. (2019c). Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol. Cancer 18:119. 10.1186/s12943-019-1046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ma H., Kong W., Liu B., Zhang X. (2019d). Up-regulated circular RNA VANGL1 contributes to progression of non-small cell lung cancer through inhibition of miR-195 and activation of Bcl-2. Biosci. Rep. 39:BSR20182433. 10.1042/BSR20182433 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang S., Zhang Y., Cai Q., Ma M., Jin L., Weng M., et al. (2019e). Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol. Cancer 18:145. 10.1186/s12943-019-1078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wang X., Du Q., Wu N., Liu X., Chen Y., et al. (2019f). The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem. Biophys. Res. Commun. 513 904–911. 10.1016/j.bbrc.2019.04.108 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li H., Lu H., Qin Y. (2019g). Circular RNA SMARCA5 inhibits the proliferation, migration, and invasion of non-small cell lung cancer by miR-19b-3p/HOXA9 axis. Onco. Targets Ther. 12 7055–7065. 10.2147/OTT.S216320 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Y., Li Y., He H., Wang F. (2019h). Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene 720:144099. 10.1016/j.gene.2019.144099 [DOI] [PubMed] [Google Scholar]
- Wang J., Li H. (2018). CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur. Rev. Med. Pharmacol. Sci. 22 3053–3060. 10.26355/eurrev_201805_15063 [DOI] [PubMed] [Google Scholar]
- Wei S., Zheng Y., Jiang Y., Li X., Geng J., Shen Y., et al. (2019). The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 44 182–193. 10.1016/j.ebiom.2019.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Liao X., Gong Y., He J., Zhou J., Tan S., et al. (2019). Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol. Cancer 18:98. 10.1186/s12943-019-1028-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Li X., Li F., Wu X., Zhang M., Zhou H., et al. (2019). A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol. Cancer 18:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Chen S., Liu Y., Li W., Liu Z., Bian X., et al. (2018). Profiles and bioinformatics analysis of differentially expressed circrnas in taxol-resistant non-small cell lung cancer cells. Cell Physiol. Biochem. 48 2046–2060. 10.1159/000492543 [DOI] [PubMed] [Google Scholar]
- Yang Q., Du W., Wu N., Yang W., Awan F. M., Fang L., et al. (2017). A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 24 1609–1620. 10.1038/cdd.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Awan F., Du W., Zeng Y., Lyu J., Wu D., et al. (2017). The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol. Ther. 25 2062–2074. 10.1016/j.ymthe.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110 304–315. 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Zhao S., Liu Q., Lv M., Zhou D., Liao Z., et al. (2017). Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol. Res. Pract. 213 453–456. 10.1016/j.prp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Yu H., Chen Y., Jiang P. (2018). Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem. Biophys. Res. Commun. 506 455–462. 10.1016/j.bbrc.2018.10.087 [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhang B., Yuan B., Chen C., Zhou Y., Zhang Y., et al. (2020). RNA-Seq profiling of circular RNAs in human small cell lung cancer. Epigenomics 12 685–700. 10.2217/epi-2019-0382 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang X., Hu B., Zhang F., Wei H., Li L. (2019). Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif. Cells Nanomed. Biotechnol. 47 3410–3416. 10.1080/21691401.2019.1652623 [DOI] [PubMed] [Google Scholar]
- Zhang P., Pei X., Li K., Jin L., Wang F., Wu J., et al. (2019). Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol. Cancer 18:179. 10.1186/s12943-019-1111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., et al. (2018a). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37 1805–1814. 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., et al. (2018b). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9:4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhao H., Zhang L. (2018c). Identification of the tumor-suppressive function of circular RNA FOXO3 in non-small cell lung cancer through sponging miR-155. Mol. Med. Rep. 17 7692–7700. 10.3892/mmr.2018.8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Chen T., Xiang J., Yin Q., Xing Y., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zhao F., Han Y., Liu Z., Zhao Z., Li Z., Jia K. (2018). circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci. Rep. 38:BSR20180570. 10.1042/BSR20180570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Lee E. E., Kim J., Yang R., Chamseddin B., Ni C., et al. (2019). Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 10:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Chen L., Zhou Y., Wang Q., Zheng Z., Xu B., et al. (2019). A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 18:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X., Zhang J., Cheng Z., Bian L., Qin J. (2019). CircLgr4 drives colorectal tumorigenesis and invasion through Lgr4-targeting peptide. Int. J. Cancer [Epub ahead of print]. 10.1002/ijc.32549 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Du Y., Yang X., Mo Y., Fan C., Xiong F., et al. (2018). Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 17:79. 10.1186/s12943-018-0827-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhang S., Chen Z., He Z., Xu Y., Li Z. (2019). CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 10:885. 10.1038/s41419-019-2127-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zheng X., Xu B., Chen L., Wang Q., Deng H., et al. (2019). Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem. Biophys. Res. Commun. 508 527–535. 10.1016/j.bbrc.2018.11.157 [DOI] [PubMed] [Google Scholar]
- Zhu X., Wang X., Wei S., Chen Y., Chen Y., Fan X., et al. (2017). hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. Febs J. 284 2170–2182. 10.1111/febs.14132 [DOI] [PubMed] [Google Scholar]
- Zong L., Sun Q., Zhang H., Chen Z., Deng Y., Li D., et al. (2018). Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed. Pharmacother. 102 639–644. 10.1016/j.biopha.2018.03.084 [DOI] [PubMed] [Google Scholar]