Abstract
Background
Although the pathophysiology of coronavirus disease 2019 (COVID-19) is not clearly defined, among the proposed mechanisms, immune system dysfunction is more likely than others. The aim of this study was to clarify the characteristics and clinical significance of dynamic changes of lymphocyte subsets in the course of COVID-19.
Methods
In this prospective study, the levels of peripheral lymphocyte subsets including CD4+, CD8+, CD4+CD25+FOXP3+, CD38+, CD3+HLA-DR+, CD19+, CD20+, and CD16+CD56+ cells were measured by flow cytometry in 52 confirmed hospitalized patients with COVID-19 at the day of admission and after 7 days of care. Clinical response was defined as improvement in symptoms (fever, dyspnea, and cough as well as blood oxygen saturation), and patients who met these criteria after 1 week of admission were classified as early responders; others who survived and finally discharged from the hospital were classified as late responders and patients who died were categorized as nonresponders. Immunophenotyping of studied cell changes on the first day of admission and 7 days after treatment were compared. Besides, the correlation between cellular subset variation and clinical response and outcome were analyzed.
Results
Total counts of white blood cell, T cells, CD4+ T cells, CD8+ T cells, CD38+ lymphocytes, and CD3+HLA-DR+ lymphocytes were significantly increased in both early and late responders. No statistically significant difference was observed in CD4+/CD8+ ratio, B cells, FOXP3+T<sub>reg</sub> lymphocytes, and FOXP3 median fluorescence intensity among studied groups. According to the multivariate analysis, an increase in CD4+ T cells (p = 0.019), CD8+ T cells (p = 0.001), and administration of interferon (p < 0.001) were independent predictors of clinical response.
Conclusion
We found an increasing trend in total T cells, T helpers, cytotoxic T cells, activated lymphocytes, and natural killer cells among responders. This trend was not statistically significant among nonresponders. The findings of this study may enhance our knowledge about the pathogenesis of COVID-19.
Keywords: Coronavirus disease 2019, Cellular immunity, Lymphocyte, Flow cytometry
Introduction
The coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus officially designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The first cases occurred in December 2019 in China, and then the virus spread rapidly in numerous countries throughout the world [1, 2]. It has led to a public health emergency of international concern, and the World Health Organization on January 30, 2020, introduced COVID-19 as a pandemic disease [3].
The pathophysiology of COVID-19 has not yet been clearly defined [4]. Existing data from severe forms of Middle East respiratory syndrome coronavirus and SARS-CoV and recent accumulating evidence concerning SARS-CoV-2 suggest that the host immunity response is contributing to the pathogenesis of COVID-19 [2, 5, 6].
Lymphocytes in peripheral blood including T cells, B cells, and natural killer (NK) cells are involved in the humoral and cellular immunity against viral infections [7], and effective immune response against a viral infection is dependent on the activation of cytotoxic CD8+ T cells through the killing of virus-infected cells [8]. Also, regulatory T cells (Treg) lymphocytes have an important role in the prevention of excessive and harmful immune reactions to the pathogens [4]. Thus, a better understanding of the characteristics of cellular immunity in COVID-19 could provide novel insights into the pathogenesis of the disease. An increasing number of studies concerning changes of lymphocyte subsets, such as CD4+ T cells, CD8+ T cells, B cells, and NK cells in COVID-19 patients and their correlation with the severity and outcome of the disease have been performed [9, 10, 11], but our knowledge about dynamic of lymphocyte subsets during the course of the disease is limited to few studies [12, 13]. Moreover, kinetic changes in cellular response may be correlated to clinical outcome. Therefore, to obtain a clearer picture of this entity, we designed a prospective, single-center study. In this study, we aimed to clarify the characteristics and clinical significance of changes of lymphocyte subsets during the course of COVID-19 and determine if these changes are associated with the prognosis, which might help in better understanding of the pathogenesis of the disease and lead to the discovery of new therapeutic options.
Materials and Methods
The Ethics Committee of the National Research Institute of Tuberculosis and Lung Diseases approved the protocol of this study (reference number: IR.SBMU.NRITLD.REC.1399.037IR.SBMU.NRITLD.REC.1399.037). Also, informed consent was obtained from all the participants.
Patients
This prospective study was conducted at Masih Daneshvari Hospital, Tehran, Iran. In the period of study, confirmed cases of COVID-19 who were symptomatic and admitted to the infectious disease ward were recruited to the study. Patients with a history of autoimmune disease, immunocompromised state, or immune suppression therapy were excluded. The presence of SARS-CoV-2 was detected by the real-time reverse transcription PCR (RT-PCR) method using a nasopharyngeal swab, as previously described [14]. Two blood samples were obtained from all patients, one on the first day of admission, and the second after 7 days of care.
Concerning the severity of the disease, the patients were categorized into 3 groups: moderate, severe, and critical. At rest, oxygen saturation in room air at the time of sampling was used to determine the severity of the disease. Patients with abnormal chest imaging and O2 saturation >93% were categorized as cases with moderate disease. Cases with O2 saturation equal or <93% were classified as severe cases, and patients who needed critical care, noninvasive or mechanical ventilation, or in a shock state were categorized as critical cases [15].
Therapeutic Interventions
All the patients received supportive care, supplemental oxygen, and intravenous fluid. Considering the treatment protocol released by the Iranian national scientific committee for management of COVID-19, lopinavir/ritonavir 400/100 mg was prescribed for 7 days [16]. Also, 3 doses of subcutaneous interferon beta-1a (12 million units) every other day were used for some cases and intravenous dexamethasone (4 mg BID) for limited patients.
Assessment of Clinical Response
Clinical response was defined as improvement in symptoms (fever, dyspnea, and cough) as well as blood oxygen saturation. Patients who met these criteria after 1 week of admission were classified as early responders; others who survived and finally discharged from the hospital were classified as late responders and patients who died were categorized as nonresponders.
Method of Flow Cytometry
In both sampling, 2 mL of whole blood was obtained, collected in tubes containing EDTA, and tested up to 6 h after sampling. Total cell blood counts and percentage and absolutes counts (cells/μL) of CD3+, CD4+, and CD8+ T lymphocytes; CD19+ and CD20+ B lymphocytes; CD16+ CD56+ NK cells; and CD4+CD25+FOXP3+Treg were measured.
For this purpose, after centrifuging of the samples, RBCs were removed using lysis buffer. Then white blood cells (WBCs) were harvested and washed with cold PBS. Before staining, the cell-surface Fc receptors were blocked with 2.4G2 (PharMingen, San Diego, CA, USA). Phycoerythrin-conjugated anti-human CD4, CD19, and CD56 antibodies (PharMingen) were used to stain CD4+ T cells, CD19+ B cells, and CD56+ NK cells. Allophycocyanin-conjugated anti-human CD8 and CD16 antibodies were used to stain CD8+ and CD16+ cells; and fluorescein isothiocyanate-conjugated antibody for CD3+ T cells according to the manufacturer's instructions. The cells were analyzed on a FACSCaliburTM flow cytometer (Becton Dickinson, San Jose, CA, USA). Dead cells were gated out by staining with propidium iodide. FlowJo software version 8 was used for the final analysis.
Statistical Analysis
Categorical variables were described as number and percentage, and continuous variables, as median and interquartile range. Categorical data were compared using the χ2 test or Fisher's exact test. The Kolmogorov-Smirnov and Shapiro-Wilk tests were used to test the normality of data. For comparison of medians between different groups of severity, the Kruskal-Wallis test was used and differences between the 2 groups were analyzed using the Mann-Whitney test. SPSS Statistics version 21.0 software was used for statistical analyses. All reported p values are two sided and a value of <0.05 was considered statistically significant. The generalized estimating equation model was used to estimate and find how factors affect the response variable as a crude and adjusted model.
Immunophenotyping of studied cell changes on the first day of admission and 7 days after treatment were compared. Besides, the correlation between cellular subset variation and clinical response were analyzed.
Results
Demographics and Baseline Characteristics
From June 8 to June 15, 2020, 52 confirmed COVID-19 cases were recruited in this study. Infection with SARS-CoV-2 was confirmed by RT-PCR. The mean age of the cases was 51.3 ± 14.8 years, including 24 men (46.2%) and 28 women. Among them, 51.9% had minimally 1 comorbidity; diabetes mellitus (26.9%), and hypertension (19.2%) were the most common. The median time from onset of symptoms to first sampling was 10.4 ± 5.2 days. Twenty-seven (51.9%) and 8 cases (15.4%) were categorized as experiencing severe illness and a critical state on admission. Demographics, basic characteristics, treatment, and the outcome of the patients are listed in Table 1.
Table 1.
Demographic and clinical characteristics of 52 confirmed cases of COVID-19
| Parameter | N (%) | |||
|---|---|---|---|---|
| Age | ||||
| Mean ± SD, years | 51.3±14.8 | |||
| Gender | ||||
| Male | 24 (46.2) | |||
| Female | 28 (53.8) | |||
| Comorbidity | 27 (51.9) | |||
| Diabetes mellitus | 14 (26.9) | |||
| Hypertension | 10 (19.2) | |||
| Chronic heart diseases | 6 (11.5) | |||
| Chronic obstructive lung diseases | 3 (5.8) | |||
| BMI ≥30 | 4 (7.7) | |||
| Other risk factors | 3 (5.8) | |||
| From the onset of illness to 1st sample | ||||
| Mean ± SD, days | 10.4±5.2 | |||
| Severity at admission | ||||
| Moderate | 17 (32.7) | |||
| Severe | 27 (51.9) | |||
| Critical | 8 (15.4) | |||
| Treatment modality | ||||
| Lopinavir/ritonavir | 52 (100) | |||
| Interferon | 19 (36.5) | |||
| Steroid | 6 (11.5) | |||
| Clinical response and outcome | ||||
| Cure | 48 (92.3) | |||
| Early response | 30 (57.7) | |||
| Late response | 18 (34.6) | |||
| Death | 4 (7.7) | |||
| Total | 52 | |||
COVID-19, coronavirus disease 2019.
Based on the clinical response, the patients were divided into 3 groups: early responders (30 cases, 57.7%), late responders (18 cases, 34.6%), and nonresponders (4 cases, 7.7%). All 3 groups were similar concerning age, gender, duration from onset of symptom, and steroid therapy. There was a significant correlation between the severity of the disease and the clinical response (p < 0.0001). All the patients with moderate disease, as well as 13 cases with severe disease (48.1%) on admission, had a clinical response on the seventh day. Twenty-two patients had a stable condition (no improvement and no deterioration) after 1 week, among them, 18 cases finally survived and 4 died.
Dynamic of Cellular Subsets in the Course of the Disease
We compared WBC and lymphocyte subset counts at days 0 of admission and following 1 week posttreatment for all patients. Table 2 shows the distribution of WBC and lymphocyte subsets between days 0 and 7 of admission and the significance of their difference in detail. Except for Treg T cells, B cells, and FOXP3+ median fluorescent intensity (MFI), other parameters were significantly different between the 2 times. Moreover, the CD4+/CD8+ ratio did not statistically change during the study time. WBC, relative and absolute count of lymphocytes, absolute count of T cells, CD4+ T cells, CD8+ T cells, CD27+ lymphocytes, CD3+HLA DR+ lymphocytes, CD38+ lymphocytes, and NK cells were significantly increased. In addition, the relative count of polymorphonuclears after 7 days of treatment was decreased. Change in cellular subsets among patients under interferon therapy was limited to WBC, total lymphocyte, and T cells (see online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000514202).
Table 2.
Comparison of WBC and lymphocyte subsets between day 0 and 7
| Parameter* | Day 0 |
Day 7 |
p value | ||
|---|---|---|---|---|---|
| median | IQR | median | IQR | ||
| WBC | 5,880 | (4,702–8,417) | 7,130 | (5,230–9,755) | 0.001** |
| PMN, % | 63.5 | (53.0–73.3) | 56.4 | (48.3–67.5) | 0.004** |
| Lymphocyte, % | 25.1 | (17.3–31.3) | 30.0 | (22.3–37.3) | 0.019** |
| Total lymphocyte | 1,416 | (1,035–1,936) | 1,808 | (1,498–2,391) | <0.0001** |
| T cells | 863 | (551–1,237) | 1,255 | (932–1,548) | <0.0001** |
| CD4+ T cells | 598 | (347–821) | 787 | (576–920) | <0.0001** |
| CD8+ T cells | 256 | (141–357) | 394 | (248–478) | <0.0001** |
| CD4+/CD8+ (ratio) | 2.7 | (1.6–3.4) | 2.2 | (1.5–3.2) | 0.132 |
| Treg cells# | 49 | (6–108) | 55 | (0–186) | 0.286 |
| FOXP3+ MFI# | 6.5 | (5.1–14.5) | 4.2 | (3.6–14.1) | 0.859 |
| CD27+ lymphocytes | 275 | (126–507) | 344 | (202–611) | 0.003** |
| CD38+ lymphocytes | 166.5 | (112.5–225.5) | 273.4 | (196.6–370) | <0.0001** |
| CD3+HLA DR+ | 44.3 | (0–86) | 82.9 | (9.6–216) | 0.003** |
| CD19+ B cells | 146 | (87–300) | 184 | (100–304) | 0.189 |
| CD20+ B cells | 136 | (85–280) | 171 | (102–284) | 0.188 |
| NK cells | 201 | (149–302) | 265 | (183–380) | 0.014** |
IQR, interquartile range; WBC, white blood cell; PMN, polymorphonuclear; Treg, regulatory T lymphocyte (CD3+CD25+FOXP3+); MFI, median fluorescent intensity; NK, natural killer.
All parameters are counts (cells per cubic millimeter) other than specified.
Statistically significant.
FOXP3 marker was checked for 31 samples on the 1st day and 11 samples on the 7th day.
Cellular Subsets and Outcome of the Disease
Patients who finally died had a higher neutrophil percentage (p = 0.013), lower lymphocyte percentage (p = 0.032), and lower counts of total T cells, CD4+ and CD8+ T cells, and NK cells at day 0 of admission. On the seventh day of hospitalization, in addition to the above factors, lower counts of CD27+ and CD3+HLA DR+ lymphocytes were associated with death (online suppl. Tables 2, 3). After using multiple adjusted logistic regression analysis, interferon therapy was the only parameter associated with early response and none of the parameters was a predictor of mortality (online suppl. Tables 4, 5).
Dynamic Changes in Cellular Subsets concerning Clinical Response
Table 3 indicates the detailed dynamic changes of the WBC and lymphocyte subsets according to the clinical response. Total counts of WBC, T cells, CD4+ T cells, CD8+ T cells, CD3+ HLA DR+ lymphocytes, and CD38+ lymphocytes were significantly increased in both early and late responders. An increase in the counts of CD27+ lymphocytes and NK cells was correlated with early response. No statistically significant difference was observed in CD4+/CD8+ ratio, B cells, FOXP3+Treg lymphocytes, and FOXP3 MFI among studied groups.
Table 3.
Comparison of WBC and lymphocyte subsets concerning clinical response
| Early response (N = 30) |
Late response (N = 18) |
No response (N = 4) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Median | IQR | p value | median | IQR | p value | median | IQR | p value | |
| WBC | |||||||||
| Day 0* | 5,605 | (4,710–8,560) | 0.019** | 6,375 | (4,580–7,160) | 0.028** | 7,600 | (4,870–14,280) | 0.144 |
| Day 7* | 6,640 | (5,220–9,010) | 7,175 | (5,160–10,780) | 10,190 | (7,545–18,930) | |||
| PMN, % | |||||||||
| Day 0 | 60.4 | (51–66) | 0.012** | 66.5 | (59–74.2) | 0.028** | 81 | (71.9–86.7) | 0.068 |
| Day 7 | 51.7 | (46–59.5) | 60 | (56.1–65) | 87.2 | (85–90.4) | |||
| Lymphocyte, % | |||||||||
| Day 0 | 27 | (21.7–36) | 0.021** | 24 | (18–31) | 0.055 | 12.5 | (8.65–21.05) | 0.068 |
| Day 7 | 32.8 | (28–38.2) | 25.8 | (22.5–33.1) | 7.3 | (6.5–8.7) | |||
| Total lymphocyte | |||||||||
| Day 0 | 1,589.5 | (1,080–2,198) | 0.004** | 1,375.5 | (1,110–1,733) | 0.001** | 1,198 | (636–1,719.5) | 0.465 |
| Day 7 | 1,955.5 | (1,712–2,652) | 1,728 | (1,331–1,953) | 869.5 | (602–1,362.5) | |||
| T cells | |||||||||
| Day 0 | 1,020.5 | (659–1,428) | 0.003** | 856 | (355–951) | 0.001** | 547 | (396–637) | 0.465 |
| Day 7 | 1,338.5 | (1,129–1,910) | 1,224 | (799–1,370) | 295.5 | (187–617) | |||
| CD4+ T cells | |||||||||
| Day 0 | 678.5 | (383–926) | 0.006** | 598.5 | (312–680) | 0.005** | 326 | (260–381.5) | 0.715 |
| Day 7 | 803.5 | (707–1,226) | 756.5 | (527–914) | 228 | (130–416) | |||
| CD8+ T cells | |||||||||
| Day 0 | 310.5 | (179–421) | 0.004** | 197 | (145–272) | 0.005** | 119.5 | (90.5–178) | 0.273 |
| Day 7 | 438.5 | (343–585) | 322 | (221–422) | 65 | (50–176) | |||
| CD4/8 ratio | |||||||||
| Day 0 | 2.4 | (1.4–3.3) | 0.309 | 2.8 | (2.2–4.2) | 0.199 | 3.2 | (2.1–3.5) | 0.715 |
| Day 7 | 2.2 | (1.4–2.8) | 2.3 | (1.5–3.3) | 2.6 | (1.9–4.1) | |||
| Treg T cells# | |||||||||
| Day 0 | 72 | (22–108) | 0.465 | 20 | (4–144) | 0.225 | 0 | (0–39) | 0.655 |
| Day 7 | 29 | (1.5–103) | 186 | (96–194) | 2.5 | (0–5) | |||
| FOXP3+ MFI# | |||||||||
| Day 0 | 8 | (5.5–14.8) | 0.465 | 6.5 | (5.3–14.5) | 0.500 | 4.1 | (4–4.6) | 0.180 |
| Day 7 | 4.15 | (2.95–5.95) | 14.1 | (6.5–16.8) | 3.65 | (3.6–3.7) | |||
| CD27+ lymphocytes | |||||||||
| Day 0 | 259 | (126–533) | 0.006** | 282 | (127–408) | 0.058 | 247.5 | (110.5–399) | 0.465 |
| Day 7 | 437 | (239–712) | 299.5 | (200–396) | 185 | (147.5–226.5) | |||
| CD38+ lymphocytes | |||||||||
| Day 0 | 171 | (125–227) | 0.011** | 168 | (116–193) | 0.006** | 126.5 | (97–278) | 0.715 |
| Day 7 | 253.4 | (195–366) | 307.2 | (236.2–373.9) | 100.9 | (56.3–317.1) | |||
| CD3+HLA DR+ | |||||||||
| Day 0 | 30.8 | (0–84.2) | 0.011** | 50.7 | (0–100) | 0.044** | 45.3 | (18.3–133.5) | 0.285 |
| Day 7 | 76.9 | (19.3–168.1) | 189.7 | (52.5–265.8) | 0 | (0–33.7) | |||
| CD19+ B cells | |||||||||
| Day 0 | 141.5 | (87–319) | 0.111 | 152 | (105–237) | 0.906 | 229 | (56–408) | 0.715 |
| Day 7 | 197 | (150–315) | 133 | (88–205) | 231 | (95–400) | |||
| CD20+ B cells | |||||||||
| Day 0 | 129.5 | (85–312) | 0.102 | 152 | (104–237) | 0.723 | 228.5 | (49–408) | 1.000 |
| Day 7 | 188 | (149–284) | 124 | (84–205) | 223.5 | (92.5–395.5) | |||
| NK cells | |||||||||
| Day 0 | 221 | (171–338) | 0.045** | 166 | (153–279) | 0.193 | 43 | (33–110) | 0.999 |
| Day 7 | 309 | (246–411) | 218.5 | (137–298) | 79 | (49.5–100) | |||
IQR, interquartile range; WBC, white blood cell; PMN, polymorphonuclear; Treg, regulatory T lymphocyte (CD3+CD25+FOXP3+); MFI, median fluorescent intensity; NK, natural killer.
Day of hospitalization.
Statistically significant.
FOXP3 marker was checked for 31 samples on the 1st day and 11 samples on the 7th day.
Dynamic Changes in Cellular Subsets concerning the Outcome
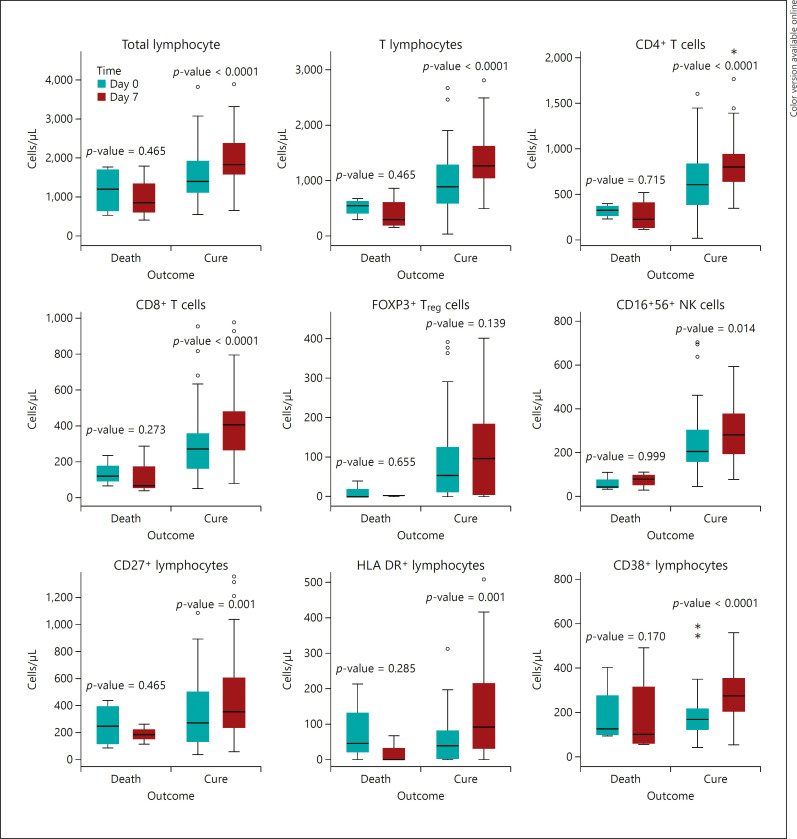
Four cases eventually died, all of them were nonresponders at the time of the second sampling. A significant increase of total lymphocyte count, T cells, CD4+ T cells, CD8+ T cells, CD27+ lymphocytes, CD3+HLA DR+ lymphocytes, and NK cells were seen among patients who survived. This trend was not statistically significant among the nonresponders (death group). There was no significant difference in the CD4+/CD8+ ratio, counts of B cells, and FOXP3+Treg cells, as well as FOXP3 MFI.
The most important findings of the correlation between cellular subsets and outcome were shown in Figure 1.
Fig. 1.
Relation between the outcome of COVID-19 and dynamic changes of cellular subsets between day 0 and 7 of admission p value <0.05 is significant. COVID-19, coronavirus disease 2019; Treg, regulatory T lymphocyte (CD3+CD25+FOXP3+); NK, natural killer.
According to the multivariate analysis, an increase in CD4+ T cells (p = 0.019) and CD8+ T cells (p = 0.001) both after 1-week posttreatment, and administration of interferon (p < 0.001) as independent predictors of clinical response, with adjusting the factors of age, sex, total lymphocyte count, and counts of NK cells was found (Table 4).
Table 4.
Multiple analysis of dynamic changes of cellular subsets between day 0 and 7 of admission and early clinical response
| Parameters | p value | Odds ratio | 95% CI |
|---|---|---|---|
| Older age 0.129 | 1.055 | 0.985–1.131 | |
| Male | 0.063 | 0.142 | 0.018–1.107 |
| Total lymphocyte increase | 0.549 | 1.000 | 0.999–1.002 |
| CD4+ T cells increase | 0.027* | 1.004 | 1.000–1.008 |
| CD8+ T cells increase | 0.004* | 0.989 | 0.981–0.996 |
| CD4/8 ratio increase | 0.074 | 0.292 | 0.076–1.127 |
| NK cell increase | 0.212 | 0.992 | 0.979–1.005 |
| Interferon therapy | <0.001* | 70.886 | 9.649–518.345 |
Statistically significant. (p value <0.05). NK, natural killer; CI, confidence interval
Discussion
Since December 2019, when COVID-19 was first reported from China, numerous studies were published to clarify the pathogenesis of the disease and various hypotheses have been proposed in this regard [17, 18, 19]. Among the proposed mechanisms underlying the worsening of the disease, the strongest is immune system dysfunction [20]. Recent findings hypothesize that virus-induced immune system dysregulation can lead to T cell exhaustion [4, 21] and cytokine storm [22, 23, 24]. Lymphocyte and the subsets play an important role in the performance of the adaptive immune system and flow cytometry is a valuable tool for monitoring the maintenance of cellular immunity [25, 26]. A growing list of studies regarding lymphocyte subset counts among patients with COVID-19 has been published, most of them focusing on the prognostic value and correlation of cellular subsets with disease severity among COVID-19 patients [10, 11, 27, 28, 29, 30]. However, a limited number of studies have concentrated on the dynamic and longitudinal changes of these parameters during the course of COVID-19 [12, 13, 31, 32]. Available data indicates that lymphopenia, particularly in CD4+ and CD8+ T cells is common during SARS-CoV-2 infection [2, 33] especially in severe or critical cases [34, 35].
In this study, we found a significant increasing trend in the counts of WBC, total lymphocytes, total T cells, CD4+ T cells, and CD8+ T cells, and the improvement of COVID-19. This correlation was observed among patients with both the early (in the first week of admission) and late (after 7 days of admission) clinical response. These results are consistent with the work of Moratto et al. [12] in Italy. They evaluated the longitudinal values of lymphocyte subsets in 26 patients with COVID-19 and observed improvement in the counts of CD3+, CD4+, and CD8+ cells among patients who eventually recovered, while in cases who progressed to a worse condition, all of these subsets remained very low. Also, Weiskopf and colleagues performed a study to evaluate the kinetics of cellular immunity response in 19 patients with COVID-19 during the acute phase of respiratory distress syndrome. They showed that SARS-CoV-2 specific T cells (both CD4+ and CD8+) appeared during 2 weeks after onset of the symptoms, and a direct negative correlation between viral loads and CD4+ T cells was found [32].
Wang et al. [13] assessed dynamic changes of CD4+ cells, CD8+ cells, B cells, and NK cells in a 1-week interval in 60 patients with COVID-19. They detected no significant changes in any lymphocyte subset in nonresponder patients. In responder group patients, total lymphocytes, CD8+ T cells, and B cells increased significantly after 7, and no statistically significant alteration was found in CD4+ T cells, CD4+/CD8+ ratio, and NK cells. Also, Huang and coworkers showed that T lymphocytes (total, CD4+, and CD8+ cells) had a downward trend until the fourteenth day and then gradually returned to the normal levels except in the unimproved patients with COVID-19. They concluded that irreversible damage of cellular immunity occurs in the early stages of COVID-19 in the unimproved group [36]. In a similar work, Deng et al. [37] measured lymphocyte subset counts weekly and found a decreasing trend during the first week, while a stable trend was seen during the second week after the onset of symptoms. After that, these markers increased gradually and rose to the normal levels in the fifth week although they were lower in patients who finally died.
Our results showed an association between the decreasing trend of neutrophil percentage and clinical response. This finding is consistent with the work of Wang et al. [38] They showed that excessive neutrophils were associated with disease severity.
In the present study, patients with good outcomes including early and late responders displayed a similar trend in the lymphocyte subset. These findings are consistent with the work published recently by Mathew and coworkers [31]. They found that some patients with COVID-19 had dynamic changes in the lymphocyte subset during the first week of admission, but others remained stable; this finding confirms the different immune responses among the patients [31]. On the other hand, hospitalized patients are in different phases in the course of the disease. Therefore, more frequent measurement of the elements (e.g., sampling every week) until normalization gives us more information.
HLA DR+ and CD38+ T cells are the markers of T lymphocyte activation. Although the total lymphocyte counts and the subsets decrease in COVID-19, deep analysis of T cells revealed a correlation between the activation of T cells and antiviral responses [31]. In our study, CD3+ HLA DR+ and CD38+ lymphocytes were significantly increased during the first week of admission in responders.
Treg cells are essential for immune homeostasis. They suppress autoimmune reactions and inhibit exaggerated inflammatory responses following viral infections [39]. The available evidence suggests that the Treg count in peripheral blood is reduced in patients with COVID-19 especially in severe cases [9, 28, 40]. The reason for this phenomenon is not clearly defined and may be one of the causes of immune system hyperactivation that lead to lung damage in the severe form of COVID-19 [41]. We did not find any significant differences concerning the percent and count of Treg T cells and FOXP3 MFI during the first week. Frequent measurements can give us more accurate information about the condition of the Treg in the course of the disease. Since the Treg-based therapies had been proposed as a potential tool for control of the severe form of COVID-19 [41, 42], more investigation about the role and kinetics of Treg is recommended.
Our data about the role of NK cells in the pathogenesis of COVID-19 are limited. In one study there were no differences in the levels of NK cells among responders and nonresponders before and following the treatment [13]. In another study, this trend was increasing among survivors and decreasing among non-survivors [37]. In our study, increasing of NK cells during the first week of admission correlated with clinical response.
Some of the treatments experimentally used in the management of COVID-19 might affect total lymphocyte counts and the subsets. For example, corticosteroids have lymphocytolytic effects and could reduce the lymphocytes, while in one study corticosteroid treatment of patients with COVID-19 increased the total number of lymphocytes significantly [13]. In our study, 19 patients received interferon beta-1a that may cause cytopenia [43]. In multivariate analysis, we showed a posttreatment increase in CD4+ and CD8+ T cell counts as predictors of good response, after adjusting the effects of other factors.
Conclusion
In the current study after a follow-up of the dynamic changes of lymphocyte subsets among COVID-19 patients, we found an increasing trend in WBC, total T cells, CD4+ T cells, CD8+ T cells, CD38+ lymphocytes, and CD3+HLA DR+ lymphocytes among responders. This trend was not statistically significant among nonresponders. The finding of this study may enhance our knowledge about the pathogenesis of COVID-19 that could help to design therapeutic options for control of severe cases.
Statement of Ethics
The research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki. The Ethics Committee of the National Research Institute of Tuberculosis and Lung Diseases approved the protocol of this study (reference number: IR.SBMU.NRITLD.REC.1399.037IR.SBMU.NRITLD.REC.1399.037). Also, informed consent was obtained from all participants.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
The authors did not receive any funding.
Author Contributions
M.R. and M.M. had the idea for and designed the study and had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. M.R. and S.M. contributed to the writing of the manuscript. All authors contributed to data acquisition, data interpretation, and reviewed and approved the final version.
References
- 1.Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395((10223)):514–23. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395((10223)):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang Z, et al. Epidemiology of COVID-19 among children in China. Pediatrics. 2020;145((6)):e20200702. doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- 4.Mahmoudi S, Rezaei M, Mansouri N, Marjani M, Mansouri D. Immunologic features in coronavirus disease 2019: functional exhaustion of T cells and cytokine storm. J Clin Immunol. 2020;40((7)):974–6. doi: 10.1007/s10875-020-00824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136((1)):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagadeesh A, Prathyusha AMVN, Sheela GM, Bramhachari PV. T cells in viral infections: the myriad flavours of antiviral immunity. In: Bramhachari PV, editor. Dynamics of immune activation in viral diseases. Singapore: Springer Singapore; 2020. pp. p.139–48. [Google Scholar]
- 8.Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181((8)):5490–500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130((5)):2620–9. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Long W, Tu M, Chen S, Huang Y, Wang S, et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J Infect. 2020;81((2)):318–56. doi: 10.1016/j.jinf.2020.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W, Berube J, McNamara M, Saksena S, Hartman M, Arshad T, et al. Lymphocyte subset counts in COVID-19 patients: a meta-analysis. Cytometry A. 2020;97((8)):772–6. doi: 10.1002/cyto.a.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moratto D, Chiarini M, Giustini V, Serana F, Magro P, Roccaro AM, et al. Flow cytometry identifies risk factors and dynamic changes in patients with COVID-19. J Clin Immunol. 2020 Oct;40((7)):970–973. doi: 10.1007/s10875-020-00806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Nie J, Wang H, Zhao Q, Xiong Y, Deng L, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221((11)):1762–9. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekbatani MS, Hassani SA, Tahernia L, Yaghmaei B, Mahmoudi S, Navaeian A, et al. Atypical and novel presentations of coronavirus disease 2019: a case series of three children. Br J Biomed Sci. 2020 Jul 15;:1–6. doi: 10.1080/09674845.2020.1785102. [DOI] [PubMed] [Google Scholar]
- 15.Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323((15)):1502–3. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marjani M, Tabarsi P, Moniri A, Hashemian MR, Nadji SA, Abtahian Z, et al. NRITLD protocol for the management of patients with COVID-19 admitted to hospitals. Tanaffos. 2020;19((2)):91–9. [PMC free article] [PubMed] [Google Scholar]
- 17.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20((6)):363–74. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leisman DE, Deutschman CS, Legrand M. Facing COVID-19 in the ICU: vascular dysfunction, thrombosis, and dysregulated inflammation. Intensive Care Med. 2020;46((6)):1105–8. doi: 10.1007/s00134-020-06059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein S, Herbert JA, McNamara PS, Hedrich CM. COVID-19: Immunology and treatment options. Clin Immunol. 2020;215:108448. doi: 10.1016/j.clim.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mamishi S, Heydari H, Aziz-Ahari A, Shokrollahi MR, Pourakbari B, Mahmoudi S, et al. Novel coronavirus disease 2019 (COVID-19) outbreak in children in Iran: atypical CT manifestations and mortality risk of severe COVID-19 infection. J Microbiol Immunol Infect. 2020 Aug 6; doi: 10.1016/j.jmii.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17((5)):533–5. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahmudpour M, Roozbeh J, Keshavarz M, Farrokhi S, Nabipour I. COVID-19 cytokine storm: the anger of inflammation. Cytokine. 2020;133((May)):155151. doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368((6490)):473–4. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 24.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39((5)):529–39. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cossarizza A, De Biasi S, Guaraldi G, Girardis M, Mussini C, Modena Covid-19 Working Group SARS-CoV-2, the virus that causes COVID-19: cytometry and the new challenge for global health. Cytometry A. 2020;97((4)):340–3. doi: 10.1002/cyto.a.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reifel KM, Swan BK, Jellison ER, Ambrozak D, Baijer J, Nguyen R, et al. Procedures for flow cytometry-based sorting of unfixed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infected cells and other infectious agents. Cytometry A. 2020;97((7)):674–80. doi: 10.1002/cyto.a.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020;58((7)):1021–8. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 28.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71((15)):762–8. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganji A, Farahani I, Khansarinejad B, Ghazavi A, Mosayebi G. Increased expression of CD8 marker on T-cells in COVID-19 patients. Blood Cells Mol Dis. 2020;83:102437. doi: 10.1016/j.bcmd.2020.102437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827–7. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369((6508)):eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5((48)):1–14. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323((11)):1061–9. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan L, Wang Q, Zhang D, Ding J, Huang Q, Tang YQ, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5((1)):33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Q, Meng M, Kumar R, Wu Y, Huang J, Deng Y, et al. Lymphopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a systemic review and meta-analysis. Int J Infect Dis. 2020;96:131–5. doi: 10.1016/j.ijid.2020.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang M, Wang Y, Ye J, Da H, Fang S, Chen L. Dynamic changes of T-lymphocyte subsets and the correlations with 89 patients with coronavirus disease 2019 (COVID-19) Ann Transl Med. 2020;8((18)):1145. doi: 10.21037/atm-20-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng Z, Zhang M, Zhu T, Zhili N, Liu Z, Xiang R, et al. Dynamic changes in peripheral blood lymphocyte subsets in adult patients with COVID-19. Int J Infect Dis. 2020;98:353–8. doi: 10.1016/j.ijid.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Li Q, Yin Y, Zhang Y, Cao Y, Lin X, et al. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front Immunol. 2020;11:2063. doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133((5)):775–87. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Hou H, Luo Y, Tang G, Wu S, Huang M, et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5((10)):e137799. doi: 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephen-Victor E, Das M, Karnam A, Pitard B, Gautier JF, Bayry J. Potential of regulatory T-cell-based therapies in the management of severe COVID-19. Eur Respir J. 2020;56((3)):3–6. doi: 10.1183/13993003.02182-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Qi G, Bellanti JA, Moser R, Ryffel B, Zheng SG. Regulatory T cells: a potential weapon to combat COVID-19? MedComm. 2020 doi: 10.1002/mco2.12. PubMed PMID: 32838397; PubMed Central PMCID: PMCPMC7436572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rieckmann P, O'Connor P, Francis GS, Wetherill G, Alteri E. Haematological effects of interferon-beta-1a (Rebif) therapy in multiple sclerosis. Drug Saf. 2004;27((10)):745–56. doi: 10.2165/00002018-200427100-00005. [DOI] [PubMed] [Google Scholar]