Abstract
Background & Aims
Acute liver failure (ALF) represents an unmet medical need in Western countries. Although the link between intestinal dysbiosis and chronic liver disease is well-established, there is little evidence for a functional role of gut-liver interaction during ALF. Here we hypothesized that intestinal dysbiosis may affect ALF.
Methods
To test this hypothesis, we assessed the association of proton pump inhibitor (PPI) or long-term antibiotics (ABx) intake, which have both been linked to intestinal dysbiosis, and occurrence of ALF in the 500,000 participants of the UK BioBank population-based cohort. For functional studies, male Nlrp6-/- mice were used as a dysbiotic mouse model and injected with a sublethal dose of acetaminophen (APAP) or lipopolysaccharide (LPS) to induce ALF.
Results
Multivariate Cox regression analyses revealed a significantly increased risk (odds ratio, 2.3–3) for developing ALF in UK BioBank participants with PPI or ABx. Similarly, dysbiotic Nlrp6-/- mice displayed exacerbated APAP- and LPS-induced liver injury, which was linked to significantly reduced gut and liver tissue microbiota diversity and correlated with increased intestinal permeability at baseline. Fecal microbiota transfer (FMT) from Nlrp6-/- mice into wild-type (WT) mice augmented liver injury on APAP treatment in recipient WT mice, resembling the inflammatory phenotype of Nlrp6-/- mice. Specifically, FMT skewed monocyte polarization in WT mice toward a Ly6Chi inflammatory phenotype, suggesting a critical function of these cells as sensors of gut-derived signals orchestrating the inflammatory response.
Conclusions
Our data show an important yet unknown function of intestinal microbiota during ALF. Intestinal dysbiosis was transferrable to healthy WT mice via FMT and aggravated liver injury. Our study highlights intestinal microbiota as a targetable risk factor for ALF.
Keywords: Acute Liver Failure, Gut-Liver-Axis, Microbiota, Dysbiosis
Abbreviations used in this paper: ABx, antibiotics; ALF, acute liver failure; ALI, acute liver injury; ALT, alanine aminotransferase; APAP, acetaminophen; APRI, aspartate aminotransferase to platelet ratio index; AST, aspartate aminotransferase; BMI, body mass index; DILI, drug-induced liver injury; FMT, fecal microbiota transfer; GLDH, glutamate dehydrogenase; GSH, glutathione; ICD-10, International Classification of Diseases; 10th Revision, IP; intraperitoneal, JNK; Jun N-terminal kinase, LEfSe; linear discriminant analysis effect size, LPS; lipopolysaccharide, MAMP; microbiota-associated molecular pattern, MoMF; monocyte-derived macrophage, NAPQI; N-acetyl-p-benzoquinone imine, NASH; nonalcoholic steatohepatitis, NLRP6; NACHT, LRR and PYD domains protein 6; NMDS, non-metric multidimensional scaling; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PPI, proton pump inhibitor; rDNA, recombinant DNA; TBST, tris-buffered saline tween; WT, wild-type
Graphical abstract
Summary.
Proton pump inhibitor or long-term antibiotics intake, which have been linked to intestinal dysbiosis, are associated with increased risk of acute liver failure in the 500,000 participants of the UK BioBank population-based cohort. In mice, APAP intoxication prompts intestinal dysbiosis, barrier impairment, and bacterial translocation. Dysbiotic microbiota of Nlrp6-/- mice induces a Ly6Chi phenotype of hepatic monocyte-derived macrophages and amplifies acute liver injury, a phenotype that is transferable to WT mice by fecal microbiota transfer.
Acute liver failure (ALF) is a syndrome with rapidly progressing hepatic dysfunction often leading to multi-organ failure and death. According to the American Association for the Study of Liver Diseases, ALF includes evidence of coagulation abnormality, hepatic encephalopathy in a patient without preexisting cirrhosis and with illness <26 weeks’ duration.1 Acetaminophen (APAP) poisoning represents the leading cause of ALF in Western countries and remains a condition with poor outcome and high mortality.2,3 At therapeutic doses, APAP is safely used as an analgesic and antipyretic drug. However, intake of an overdose can lead to ALF.4 After absorption, APAP is metabolized in hepatocytes into its toxic metabolite N-acetyl-p-benzoquinone imine (NAPQI), which induces oxidative stress and finally hepatocyte death.5 Although ALF is usually caused by consumption of an overdose of more than 150 mg/kg, interindividual susceptibility to the hepatotoxic effects of APAP varies, and severe liver failure can occur rarely when doses as low as 3–4 g/day are taken.1 This cannot be fully explained by known risk factors such as co-consumption of alcohol, herbals, other medications, age, and history of chronic liver disease.6, 7, 8, 9 Thus, the dose of APAP does not necessarily correlate with the outcome of ALF, indicating that other environmental and host-derived factors may influence susceptibility to APAP.10
In recent years, the reciprocal interaction between liver and gut, termed gut-liver axis, has emerged as a field of intense research activities. Studies revealed a close interaction between the 2 organs with regard to normal physiology and disease. Recent studies have shown an involvement of the gut-liver axis in various liver diseases ranging from nonalcoholic fatty liver disease,11 alcoholic steatohepatitis (NASH),12 to liver fibrosis13 as well as hepatocellular carcinoma.14,15 In several studies, intestinal dysbiosis and loss of intestinal barrier integrity have been linked to translocation of microbiota-associated molecular patterns (MAMPs) such as lipopolysaccharide (LPS) through the portal vein into the liver, where they trigger inflammation via pathogen recognition receptors.16,17
Factors associated with modern Western lifestyle as well as medication and other environmental factors contribute to intestinal dysbiosis, which is characterized by qualitative and quantitative changes in bacterial communities. Long-term proton pump inhibitor (PPI) or antibiotics (ABx) intake results in loss of taxonomic and functional microbiota diversity and barrier impairment.18, 19, 20, 21, 22
Beside environmental and host-derived factors, loss of the NACHT, LRR and PYD domains protein 6 (NLRP6) inflammasome sensor molecule leads to disturbed intestinal barrier function as well as changes in microbiota composition, fueling NASH progression in mice.23,24 NLRP6 is mainly expressed in intestinal epithelial cells and regulates host microbiota interaction at the gut mucosal surface. Loss of NLRP6 results in intestinal dysbiosis.25,26 Therefore, Nlrp6-/- mice are a useful tool to study how intestinal dysbiosis affects host physiology.
Recent data demonstrate that activation of the innate immune system after APAP-induced hepatocyte necrosis plays a central role in perpetuating liver injury.27,28 On experimental APAP overdose, monocyte-derived macrophages (MoMFs) are recruited in a CCR2-dependent fashion and exacerbate liver inflammation by forming dense clusters around necrotic areas.27 Ly6Chi MoMFs in particular exert proinflammatory functions because of their high expression of proinflammatory cytokines and pathogen recognition receptors. On the basis of their receptor repertoire, MoMFs are well-equipped to respond rapidly to LPS and other gut-derived MAMPs.29
Whereas intestinal dysbiosis has been identified as an important driver of the innate immune response fueling progression of chronic liver diseases, insights into an involvement of the gut-liver axis during ALF remain limited. Here we hypothesized that intestinal dysbiosis may affect ALF. Long-term PPI or ABx treatments are strong inducers of intestinal dysbiosis and barrier impairment.18, 19, 20,22 We therefore investigated the risk for developing ALF in patients with PPI or ABx intake. To test the hypothesis that gut dysbiosis increases the risk of ALF, we performed functional experiments using Nlrp6-/- mice. Altogether, our data show an important yet unknown function of the intestinal microbiota during ALF. Intestinal dysbiosis was associated with an increased risk for developing ALF. In mice dysbiotic microbiota was transferrable to healthy wild-type (WT) mice via fecal microbiota transfer (FMT) and aggravated liver injury. Our study highlights intestinal microbiota as a targetable risk factor for ALF.
Results
Medication With PPI or Long-term ABx Treatment Is Associated With an Increased Risk of Developing ALF
Long-term medication with PPI and ABx is strongly associated with intestinal dysbiosis.18, 19, 20,30 To study the relevance of PPI and ABx for developing ALF, we analyzed data from the 502,511 participants of the UK BioBank population-based study. Patients with heavy alcohol consumption as well as chronic hepatitis B and C were excluded (Figure 1A). During follow-up (mean, 9.8 years) of 39,582 patients (364,154 patient-years) with long-term PPI intake, 117 patients developed ALF (incidence, 0.03% per patient-year), whereas during follow-up of 458,143 patients (4,489,801 patient-years) without PPI, 433 patients developed ALF (incidence, 0.01% per patient-year). Overall, patients with long-term PPI intake showed a significantly higher cumulative incidence of ALF compared with those without (0.3% vs 0.1%; P < .0001). The number of participants who reported at baseline to take long-term ABx was markedly lower than for long-term PPI treatment (Figure 1A), which has a considerable impact on statistical power of the analyses. Of 594 patients with long-term ABx intake (5821 patient-years), 20 patients developed ALF (incidence, 0.03% per patient-year), whereas of 496,577 patients (4,866,455 patient-years) without long-term ABx, 530 patients developed ALF (incidence, 0.01% per patient-year; Table 1). Overall, patients with long-term ABx intake showed a significantly higher risk of ALF compared with those without (0.3% vs 0.1%; P < .006; Table 2).
Figure 1.
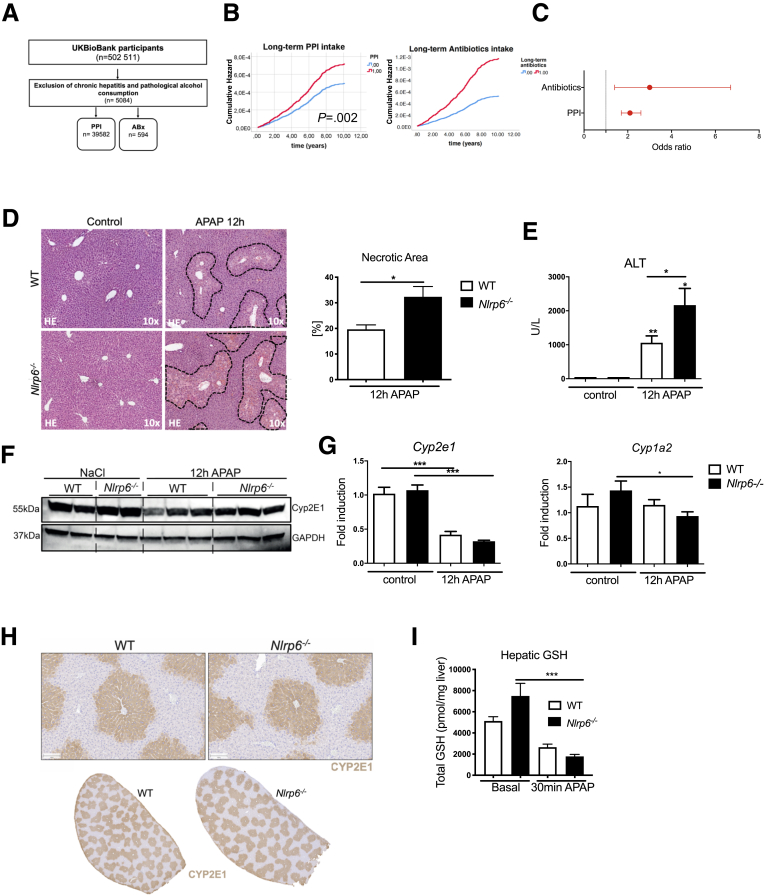
Intestinal dysbiosis is associated with increased risk and severity of ALF. (A) Overview of the analyzed population. Population-based study analyzing UK BioBank participants. (B) Multivariate Cox regression analysis showing cumulative risks for developing ALF in patients with long-term ABx or PPI medication compared with patients without medication. (C) Odds ratio for developing ALF in patients with PPI or ABx. Corrected for age, sex, APRI, Charlson comorbidity index. (D) Representative liver histopathology (hematoxylin-eosin staining) showing necrotic areas, which were quantified on high-resolution liver scans of WT and Nlrp6-/- mice 12 hours after treatment (n = 6). (E) Liver injury assessed by serum ALT levels after APAP (n = 13) or control (n = 5) treatment. (F) Cytochrome Cyp2E1 protein expression in liver samples of representative NaCl (n = 2) and APAP (n = 3) treated WT and Nlrp6-/- mice. (G) Quantitative PCR of liver samples showing mRNA levels of cytochromes Cyp1A2 and Cyp2E1 after NaCl (n = 5) or APAP treatment (n = 13). (H) Immunohistochemistry staining against CYP2E1 in livers of WT and Nlrp6-/- mice. (I) Total hepatic GSH levels (pmol/mg liver tissue) in control (n = 7) liver and 30 minutes after APAP (n = 4) administration. All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t test). ∗Significantly different from respective control group.
Table 1.
Baseline Characteristics of UKBioBank Participants With Long-term ABx Medication
| No ABx PPI (n = 496,577) | Long-term ABx (n = 594) | P value | |
|---|---|---|---|
| Characteristics | |||
| Age (y) | 56.6 ± 8.1 | 59.9 ± 8.1 | .369 |
| Women (%) | 54 | 55 | .716 |
| BMI (kg/m2) | 27.4 ± 4.8 | 28.2 ± 5.8 | .001 |
| Charlson comorbidity index (units) | 1.4 ± 1.0 | 1.7 ± 1.2 | <.0001 |
| APRI (units) | 0.28 ± 0.51 | 0.29 ± 0.22 | .473 |
NOTE. Statistical significance was determined by unpaired Student t test or χ2 test. All significant covariates were included in the multivariate model.
Table 2.
Risk Factors for ALF: Results of Multivariate Logistic Regression Analysis in the UKBioBank Cohort
| Adjusted odds ratio | 95% Confidence interval | P value | |
|---|---|---|---|
| Long-term ABx intake | 3.00 | 1.37-6.70 | .006 |
| Female sex | 2.20 | 1.98–2.44 | <.0001 |
| BMI (per 1 kg/m2) | 1.04 | 1.02–1.05 | <.0001 |
| APRI (per unit) | 1.03 | 1.02–1.04 | <.0001 |
| Charlson comorbidity index (per unit) | 1.95 | 1.86–2.03 | <.0001 |
NOTE. Boldface highlights antibiotic treatment.
It is known that patients with PPI medication or long-term ABx treatment may suffer from an increased number of comorbidities31 (Table 3). To adjust for these differences, we assessed the comorbidities by calculating the Charlson disease severity index,32 which is commonly used to adjust for the effect of comorbidities in hazard models. This index includes 14 different categories of comorbidities at baseline including different organ systems (see Materials and Methods). In the multivariate Cox regression analysis adjusted for Charlson index as well as known risk factors for ALF (univariate analysis, Tables 4 and 5), long-term PPI intake was associated with a relative risk of 1.44 (95% confidence interval, 1.15–1.80; P = .002) for developing ALF during follow-up (mean time to event, 4 years; Figure 1B, Table 5), and long-term ABx intake showed an increased hazard ratio for ALF during follow-up, but hazard ratio did not reach significance because of the low number of person-years (Table 6). However, the odds ratio for risk of ALF was still significant in a multivariable logistic regression model including the Charlson comorbidity index, age, sex, body mass index (BMI), and aspartate aminotransferase to platelet ratio index (APRI) score (Figure 1C, Table 2). Together, these data demonstrate that long-term medication with PPI and ABx is associated with an increased risk for ALF. The increase in ALF is independent of comorbidities or known risk factors for ALF.
Table 3.
Baseline Characteristics of UKBioBank Participants With Long-term PPI Medication
| No long-term PPI (n = 458,143) | Long-term PPI (n = 39,582) | P value | |
|---|---|---|---|
| Characteristics | |||
| Age (y) | 56.3 ± 8.1 | 59.9 ± 7.1 | <.0001 |
| Women (%) | 54 | 55 | .213 |
| BMI (kg/m2) | 27.3 ± 4.7 | 29.4 ± 5.2 | <.0001 |
| Charlson comorbidity index (units) | 1.4 ± 1.0 | 2.0 ± 1.1 | <.0001 |
| APRI (units) | 0.28 ± 0.51 | 0.29 ± 0.23 | <.0001 |
NOTE. Statistical significance was determined by unpaired Student t test or χ2 test. All significant covariates were included in the multivariate model.
Table 4.
Risk Factors for ALF: Results of Univariate Cox Proportional Hazard Analysis in the UKBioBank Cohort
| Unadjusted hazard ratio | 95% Confidence interval | P value | |
|---|---|---|---|
| Long-term PPI intake | 3.2 | 2.6–3.9 | <.0001 |
| Female sex | 2.2 | 1.8–2.6 | <.0001 |
| BMI (per 1 kg/m2) | 1.07 | 1.06–1.09 | <.0001 |
| APRI (per unit) | 1.03 | 1.02–1.04 | <.0001 |
| Charlson comorbidity index (per unit) | 2.8 | 2.7–2.9 | <.0001 |
| Age (per year) | 1.05 | 1.04–1.06 | <.0001 |
Table 5.
Risk Factors for ALF: Results of Multivariate Cox Proportional Hazard Analysis in the UKBioBank Cohort
| Adjusted hazard ratio | 95% Confidence interval | P value | |
|---|---|---|---|
| Long-term PPI intake | 1.44 | 1.15–1.80 | .002 |
| Female sex | 1.80 | 1.49–2.16 | <.0001 |
| BMI (per 1 kg/m2) | 1.03 | 1.02–1.05 | <.0001 |
| APRI (per unit) | 1.03 | 1.02–1.04 | <.0001 |
| Charlson comorbidity index (per unit) | 2.70 | 2.57–2.83 | <.0001 |
Table 6.
Risk Factors for ALF: Results of Multivariate Cox Proportional Hazard Analysis in the UKBioBank Cohort
| Adjusted hazard ratio | 95% Confidence interval | P value | |
|---|---|---|---|
| Long-term ABx intake | 2.31 | 0.56–9.01 | .24 |
| Female sex | 1.78 | 1.46–2.14 | <.0001 |
| BMI (per 1 kg/m2) | 1.04 | 1.02–1.05 | <.0001 |
| APRI (per unit) | 1.03 | 1.02–1.04 | <.0001 |
| Charlson comorbidity index (per unit) | 2.76 | 2.63–2.89 | <.0001 |
Lack of NLRP6 Expression Aggravates Liver Damage in Two Independent Models of Acute Liver Injury
To study the working hypothesis that intestinal dysbiosis is causally linked to ALF and to identify the responsible mechanisms, we used Nlrp6-/- mice, a well-established mouse model of intestinal dysbiosis.24 WT and Nlrp6-/- mice were treated with a high, acutely hepatotoxic dose of APAP. Nlrp6-/- mice showed massive hepatocyte death compared with WT mice 12 hours after APAP intoxication (Figure 1D). Quantification of dead cell area by image analysis revealed significantly enlarged zones of necrosis in Nlrp6-/- mice compared with WT controls (Figure 1D). This was still evident 24 hours after APAP administration (Figure 2A). In line, Nlrp6 deficient mice showed significantly higher serum levels of liver injury markers compared with WT mice, eg, alanine aminotransferase (ALT) (Figure 1E), aspartate aminotransferase (AST), and glutamate dehydrogenase (GLDH) (Figure 2B).
Figure 2.
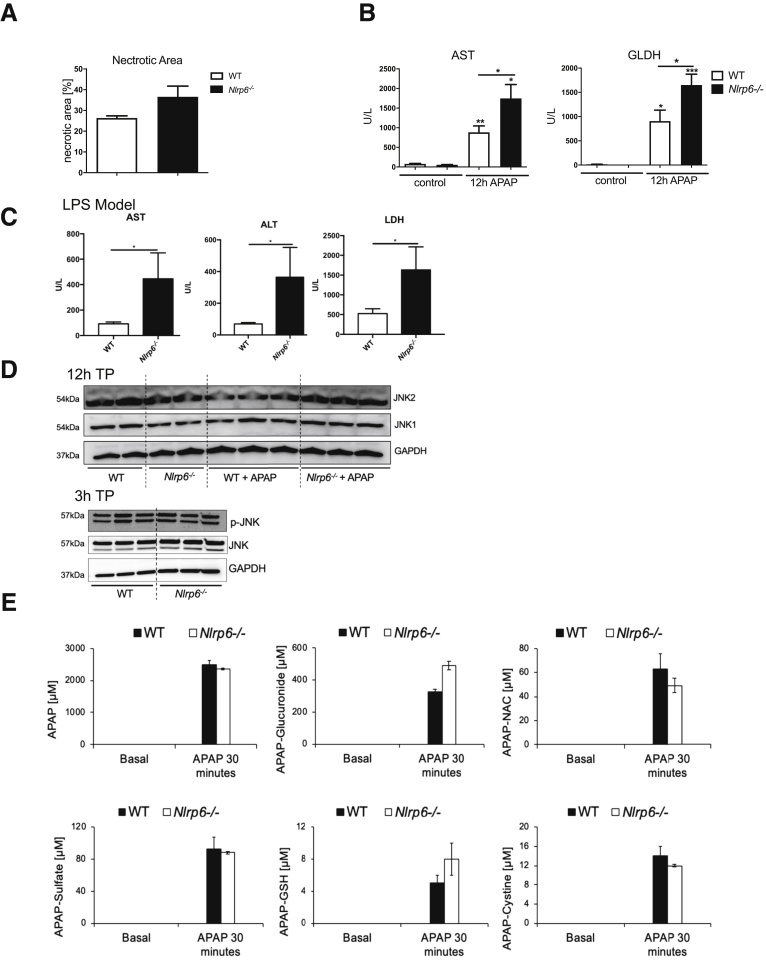
Nlrp6-/-do not show increased apoptosis, JNK activation, or altered APAP metabolism. (A) Quantitative measurement of necrotic areas on high-resolution liver scans of WT (n = 3) and Nlrp6-/- mice (n = 3) 24 hours after treatment. (B) Liver injury assessed by serum AST and GLDH levels after 12-hour APAP (n = 13) or control (n = 5) treatment. (C) Liver injury assessed by serum AST, ALT, and LDH levels 6 hours after LPS injection in WT (n = 6) or Nlrp6-/- (n = 5) mice. (D) Western blot analysis of JNK1 and JNK2 protein in WT and Nlrp6-/- mice 12 hours and 3 hours after control or APAP treatment. (E) High-performance liquid chromatography analysis of APAP serum metabolites in WT (n = 4) and Nlrp6-/- (n = 4) 30 minutes after APAP administration. All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t test). ∗Significantly different from respective control group. AILI, acetaminophen-induced liver injury; NAC, N-acteylcysteine; TP, time point.
To investigate the general implications of these findings in a second model, we treated WT and Nlrp6-/- mice with a sublethal dose of LPS. Similar to APAP, acute intoxication with LPS induced exacerbated liver injury in Nlrp6-/- compared with WT mice, as evidenced by significantly increased serum ALT, AST, and GLDH concentrations (Figure 2C).
To better understand aggravated liver damage in Nlrp6-/- mice after ALF, Jun N-terminal kinase (JNK) activation was analyzed at 3 as well as 12 hours after APAP intoxication in Nlrp6-/- and age-matched WT controls (Figure 2D). Interestingly, we did not observe differences between WT and Nlrp6-/- mice in p-JNK levels 3 hours after APAP treatment. In addition, JNK1 and JNK2 protein levels were also not increased 12 hours after APAP administration (Figure 2D). These data indicate that exacerbated liver damage in Nlrp6-/- mice was not associated with increased JNK activation.
Next we investigated the relevance of hepatic APAP metabolism for the observed phenotype. We determined RNA and protein expression levels of cytochrome p450 2E1 (CYP2E1) and 1A2 (CYP1A2). Both enzymes are involved in converting APAP into the toxic metabolite NAPQI. WT and Nlrp6-/- animals showed equal hepatic Cyp2E1 and Cyp1A2 expression levels (Figure 1G). Also immunostaining showed no differences in Cyp2E1 protein distribution between WT and Nlrp6-/- livers (Figure 1F). The differences in APAP metabolism were studied between WT and Nlrp6-/- livers; we measured APAP and its metabolites in the systemic circulation before as well as 30 minutes after APAP intoxication and saw the following constellation: (1) no statistically significant difference in basal glutathione (GSH) concentrations was found between Nlrp6-/- and WT mice, although a nonsignificant trend toward higher GSH was seen in the Nlrp6-/- mice (Figure 2E); (2) after APAP intoxication GSH concentrations were significantly lower in the Nlrp6-/- than in the WT mice, although the difference was quite small (Figure 2E); and (3) Nlrp6-/- mice showed higher APAP GSH adduct levels after APAP administration; however, this difference did not reach statistical significance (Figure 2E). Taken together, the results indicate a slightly higher capacity of the Nlrp6-/- mice to metabolically activate APAP to NAPQI; however, it is unlikely that these very small differences have a major impact on the extent of APAP-induced tissue damage.
Nlrp6-/-Mice Display an Aggravated Inflammatory Response After APAP Administration
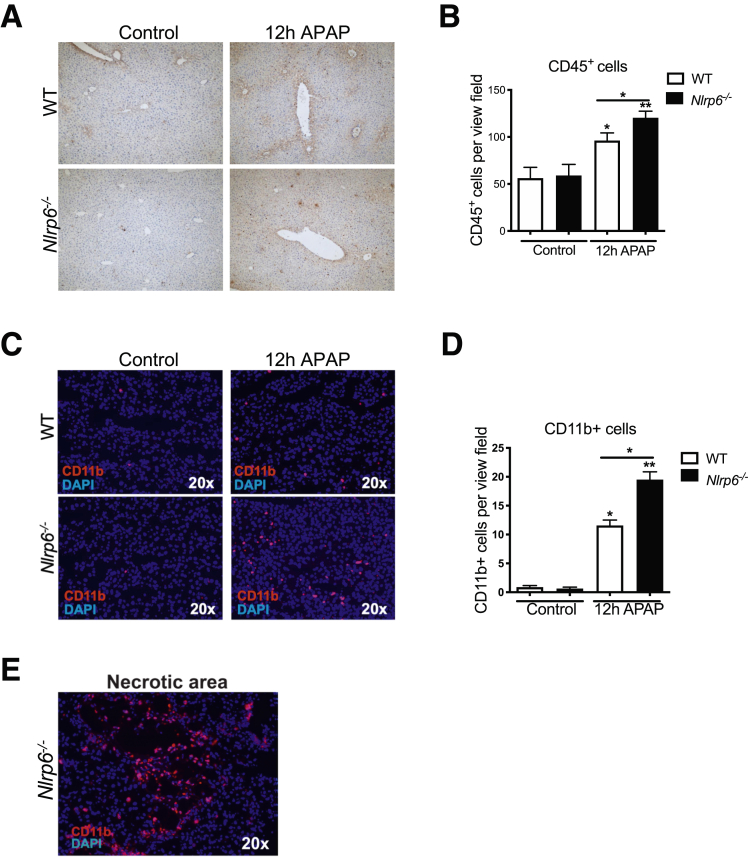
Besides APAP-induced toxic injury, recruitment of inflammatory monocytes is an important driver of liver damage.5 Previously we have demonstrated that intestinal dysbiosis affects polarization of hepatic MoMF modulating NASH progression.11 The contribution of intestinal dysbiosis to the inflammatory response after APAP intoxication is unclear. Immunohistochemistry staining against CD45 revealed increased leukocyte infiltration in livers of WT and Nlrp6-/- mice 12 hours after APAP administration (Figure 3A and B).
Figure 3.
NLRP6 modulates the inflammatory response after APAP administration. (A) Immunohistochemistry staining against CD45 and (C) immunofluorescent staining against CD11b in livers of WT and Nlrp6-/- mice on 12-hour NaCl (control) or APAP challenge. (B) Corresponding quantitative analysis of CD45 positive stained cells in livers of WT and Nlrp6-/- mice upon 12 hours of treatment (Control: n = 2, APAP: n ≥ 6). (D) Quantification of CD11b positive stained cells in livers of WT and Nlrp6-/- mice (Control: n = 3, APAP: n ≥ 10). (E) Exemplary immunofluorescent staining against CD11b on frozen liver sections depicting a necrotic area. All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t test). ∗Significantly different from respective control group.
Immunofluorescence staining of CD11b displayed a significant almost 2-fold higher infiltration of CD11b+ cells in Nlrp6-/- mice compared with WT controls after APAP intoxication (Figure 3C and D). These cells formed dense clusters in proximity to necrotic foci (Figure 3E).
APAP-Induced Shift in Microbiota Composition Is Restricted to WT Mice
After we found increased leukocyte infiltration in Nlrp6-/- mice upon ALF, we next concentrated on APAP-induced ALF to characterize the functional mechanism mediating the increased liver damage and inflammatory response in these mice. To this end, we investigated intestinal microbiota composition in WT and Nlrp6-/- mice. DNA from cecal samples was used to generate amplicons of the V4 hypervariable region of the 16S ribosomal RNA (rRNA) gene, which were subsequently sequenced using Illumina technology (San Diego, CA). Non-metric multidimensional scaling (NMDS) analysis based on UniFrac beta diversity metric displayed distinct clustering in WT versus Nlrp6-/- mice (Figure 4A). In line with previous data,28 Nlrp6-/- animals in our colony harbored a significantly less diverse microbiota indicative of intestinal dysbiosis (Figure 4C). Nlrp6-/- microbiota diversity did not change upon ALF. In contrast, APAP treatment induced a significant shift in microbiota composition in WT mice (Figure 4A).
Figure 4.
APAP-induced shift in microbiota composition is restricted to WT mice. (A) NMDS analyses based on UniFrac beta diversity metric from cecal stool samples of WT and Nlrp6-/- mice 12 hours after NaCl (n = 5) or APAP (n ≥ 6) injection. Every point represents cecal microbiota composition of 1 mouse. Similar microbiota composition is reflected in closer ordination. (B) Permutational multivariate analysis of variance (adonis) considering the factors genotype and treatment. (C) Shannon diversity of microbiota species in cecum stool of WT and Nlrp6-/- mice taken after 12 hours of NaCl (WT: n = 5, Nlrp6-/-: n = 5) or APAP (WT: n = 8, Nlrp6-/-: n = 6) treatment. (D) Computational LEfSe analysis comparing NaCl-treated with APAP-treated WT mice. (E) Relative abundance of Lachnospiraceae and Firmicutes in WT and Nlrp6-/- mice. All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t test). ∗Significantly different from respective control group. OTU, operational taxonomic unit.
To evaluate the relative contribution of APAP treatment on the observed differences between both groups, we performed permutational multivariate analysis of variance (adonis) considering the factor “treatment”. These analyses revealed that about 25% of gut microbiota variability in WT mice could be explained by treatment (Figure 4B). APAP treatment resulted in a significant decrease in the species richness in the microbiota of WT mice as evidenced by different alpha diversity metrics including Shannon index and observed species (Figure 4C). After APAP treatment microbiota diversity of WT mice was similar to Nlrp6-/- mice. Specifically, computational linear discriminant analysis effect size (LEfSe) analysis33 revealed a significant expansion of the bacterial species Akkermansia muciniphila in APAP-treated WT mice, whereas the abundance of the phylum Firmicutes decreased. This was mainly due to a loss of the family Lachnospiraceae (Figure 4D and E).
Altogether, these data indicate that healthy WT microbiota undergo a robust change upon drug-induced liver injury (DILI) whereas this response is absent in dysbiotic Nlrp6-/- animals. Whereas Nlrp6-/- microbiota already displayed characteristics of dysbiosis at baseline, WT microbiota changed into a dysbiotic state on APAP treatment.
APAP Intoxication Results in Intestinal Barrier Impairment
Recent data highlighted the importance of an intact intestinal microbiota in maintaining intestinal barrier integrity in the context of NASH and ASH,11,12 whereas its implication for ALF is unclear. APAP intoxication often leads to multi-organ failure, which dictates disease outcome. We hypothesized that ALF may negatively affect intestinal barrier integrity and promote systemic bacterial translocation.
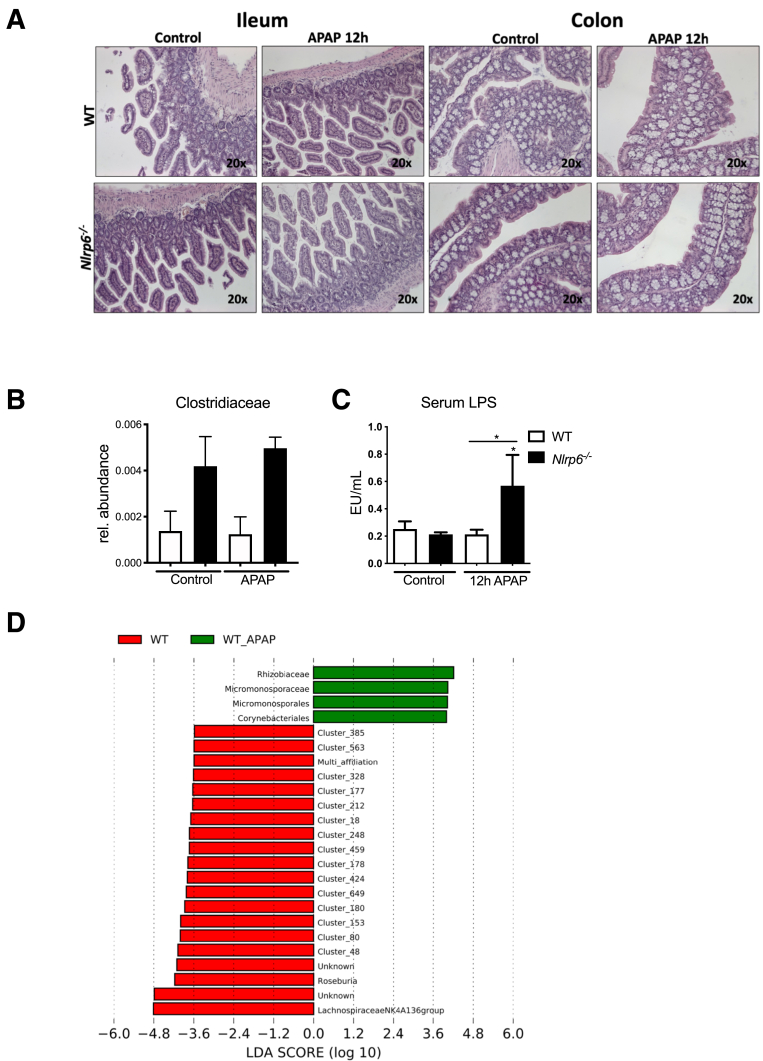
To assess the impact of APAP intoxication on the intestine, we investigated the gut epithelium. H&E-stained tissue sections did not reveal obvious differences between the different groups (Figure 5). However, specific immunofluorescence staining against the tight junction protein zonula occludens-1 revealed a marked reduction of colon zonula occludens-1 expression on APAP treatment (Figure 6A). In line, Western blot analyses of ileum tissue homogenates showed a significant suppression of the tight junction protein occludin on APAP treatment in both WT and Nlrp6-/- mice (Figure 6B and C). Because APAP treatment resulted in an expansion of the bacterial species Akkermansia muciniphila (Figure 4D), which has been linked to intestinal mucus production,34 colonic mucus layers were investigated in more detail. Interestingly, Carnoy’s-fixed colon specimen revealed a marked increase in thickness of colonic mucus layers in WT mice, whereas this response did not appear in Nlrp6-/- mice (Figure 6D and E).
Figure 5.
APAP treatment results in intestinal barrier impairment and shapes hepatic tissue microbiota. (A) Representative ileum and colon histology (hematoxylin-eosin staining) of WT and Nlrp6-/- mice 12 hours after APAP administration. (B) Hepatic abundance of Clostridiaceae DNA after 12 hours of NaCl (WT: n = 6, Nlrp6-/-: n = 6) or APAP (WT: n = 5, Nlrp6-/-: n = 5) challenge. (C) LPS concentrations in portal serum of control (WT: n = 4, Nlrp6-/-: n = 4) and APAP-treated (WT: n = 6, Nlrp6-/-: n = 6) WT and Nlrp6-/- mice. (D) Computational LEfSe analysis comparing hepatic microbiota composition of NaCl-treated with APAP-treated WT mice. All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t test). ∗Significantly different from respective control group.
Figure 6.
Intestinal barrier impairment on APAP challenge. (A) Representative immunofluorescence of colon of WT and Nlrp6-/- mice staining against zonula occludens-1. (B) Western blot analyses and (C) quantification of occludin protein expression in ileum of representative WT and Nlrp6-/- mice after 12 hours of NaCl (n = 3) or APAP (n = 5) treatment. (D) Colonic mucus layers and bacterial colonization shown by immunofluorescent staining against Muc2 and fluorescence in situ hybridization using EuBac-probe for eubacteria in colon of WT and Nlrp6-/- mice. (E) Enzyme-linked immunosorbent assay of fecal albumin concentrations in freshly collected fecal pellets from APAP-treated WT (n = 9) and Nlrp6-/- (n = 7) mice and controls (WT: n = 4, Nlrp6-/-: n = 3). (F) Quantitative analysis of thickness of colonic mucus layers in immunofluorescent stainings (NaCl: WT: n = 4, Nlrp6-/-: n = 2; APAP: WT: n = 3, Nlrp6-/-: n = 5) 24 hours after APAP administration. All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t test).
Using fluorescence in situ hybridization probes against eubacteria, we next investigated the relation of microbiota, mucus layers, and intestinal epithelium (Figure 6D). We found bacteria in the upper mucus layers of WT and Nlrp6-/- animals, with close proximity to intestinal epithelial cells in Nlrp6-/- mice because of reduced mucus thickness (Figure 6D and E). Of note, neither WT nor Nlrp6-/- mice harbored bacteria below the epithelium.
Finally, to assess whether impaired tight junction expression results in loss of barrier function, we measured fecal albumin concentrations by enzyme-linked immunosorbent assay. Here, NaCl-treated Nlrp6-/- mice already displayed clear signs of an impaired intestinal barrier as evidenced by about 3-fold increase in fecal albumin concentrations (Figure 6F). On APAP treatment WT mice displayed a significant increase in fecal albumin concentrations, whereas a further increase was not observed in Nlrp6-/- mice (Figure 6E).
Together, these data demonstrate that in WT mice APAP intoxication triggered marked changes in microbiota composition, followed by loss of intestinal barrier integrity. In contrast, intestinal dysbiosis in Nlrp6-/- mice was associated with intestinal barrier impairment at baseline, which did not worsen 12 hours after APAP intoxication.
Intestinal Barrier Impairment Prompts Bacterial Translocation and Shapes Hepatic Tissue Microbiota
To further investigate the impaired barrier function and bacterial translocation on APAP treatment, we next isolated liver bacterial DNA and measured total 16 recombinant DNA (rDNA) copies/mg liver tissue by using an optimized protocol of purification and amplification.35,36 These analyses showed a significant increase in total bacterial DNA in both APAP-treated Nlrp6-/- and WT mice. LPS levels were significantly increased in portal serum of APAP-treated Nlrp6-/- mice compared with WT mice (Figure 5C). However, we did not observe significantly higher levels of bacterial DNA in Nlrp6-/- mice compared with WT controls (Figure 7A). On the basis of these data we hypothesized that not only the amount of bacterial rDNA but rather specific bacteria and microbiota composition might augment the inflammatory response on APAP intoxication. We therefore studied the taxonomic profiles, alpha and beta diversity of the bacterial DNA by 16s targeted metagenomic sequencing in regard to the inflammatory response upon APAP treatment. At baseline, tissue microbiota composition of Nlrp6-/- mice displayed significantly reduced alpha diversity compared with WT control mice (Figure 7B). This finding was interesting because we also saw significantly reduced gut microbiota diversity in Nlrp6-/- mice compared with WT controls, where reduced diversity has been linked to intestinal dysbiosis25 (Figure 4C). APAP-induced liver injury was followed by a significant reduction of hepatic microbiota diversity in WT mice, which was again similar to gut microbiota, whereas it only slightly decreased in Nlrp6-/- mice (Figures 7B and 4C).
Figure 7.
Intestinal barrier impairment prompts bacterial translocation and shapes hepatic tissue microbiota. (A) Hepatic 16s rDNA copies/mg liver tissue of NaCl (WT: n = 6, Nlrp6-/-: n = 6) or APAP (WT: n = 5, Nlrp6-/-: n = 5) treated mice. (B) Hepatic bacterial DNA alpha diversity (Chao1 index) in control (n = 6) or APAP (n = 5) treated Nlrp6-/- and WT mice. (C) Hepatic 16s rDNA microbiota profiles of WT and Nlrp6-/- mice shown by ordination plots based on generalized UniFrac distances. Every dot represents 1 mouse. (D) Hepatic abundance of LachnospiraceaeNK4A136 and Firmicutes DNA after 12 hours of NaCl (WT: n = 6, Nlrp6-/-: n = 6) or APAP (WT: n = 5, Nlrp6-/-: n = 5) challenge. (E) Relative abundance of hepatic Clostridiaceae DNA correlates with serum GLDH levels (Spearman). All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t). ∗Significantly different from respective control group.
Next we performed beta diversity analyses based on the generalized UniFrac distances to further discriminate whether microbiota composition based on hepatic 16s rDNA profiles differs between WT and Nlrp6-/- mice. Whereas WT and Nlrp6-/- liver microbiota showed moderate shift in the NaCl-treated control mice, this difference disappeared on APAP treatment (Figure 7C). APAP intoxication resulted in a marked decrease of phylum Firmicutes, which was mainly due to a loss in the genus Lachnospiraceae NK4A136 in both WT and Nlrp6-/- mice (Figure 7D), again resembling the pattern observed in gut microbiota. In LEfSe analysis the genus Lachnospiraceae NK4A136 could be confirmed as the most differentially abundant genus between NaCl- and APAP-treated WT mice (linear discriminant analysis score –4.8; Figure 5D).
We finally asked whether the abundance of certain bacteria is linked to the severity of liver injury. Interestingly, hepatic Clostridiales (family level) proportions were low but significantly higher in APAP-treated Nlrp6-/- mice compared with WT controls and showed a strong correlation with GLDH serum levels (r = 0.82, P = .0064) (Figures 7E and 5B).
Together, these data demonstrate that Nlrp6-/- mice show baseline differences in gut microbiota configuration, which are reflected in hepatic tissue microbiota. Alterations in hepatic tissue microbiota could be linked to liver injury.
FMT From Nlrp6-/-Mice Into WT Mice Changes Their Gut Microbiota Toward a Nlrp6-/-Phenotype
We subsequently assessed the causal role of Nlrp6-/- microbiota for the outcome of ALF. To this end, repetitive FMTs of Nlrp6-/- microbiota into WT mice were performed for 2 weeks before ALF induction (Figure 8A). The effect of FMT on gut microbiota was studied by using bacterial DNA isolated from cecal stool samples. Bacterial DNA was sequenced on an Illumina platform, and structures of microbial communities were compared on their phylogenetic content by UniFrac.
Figure 8.
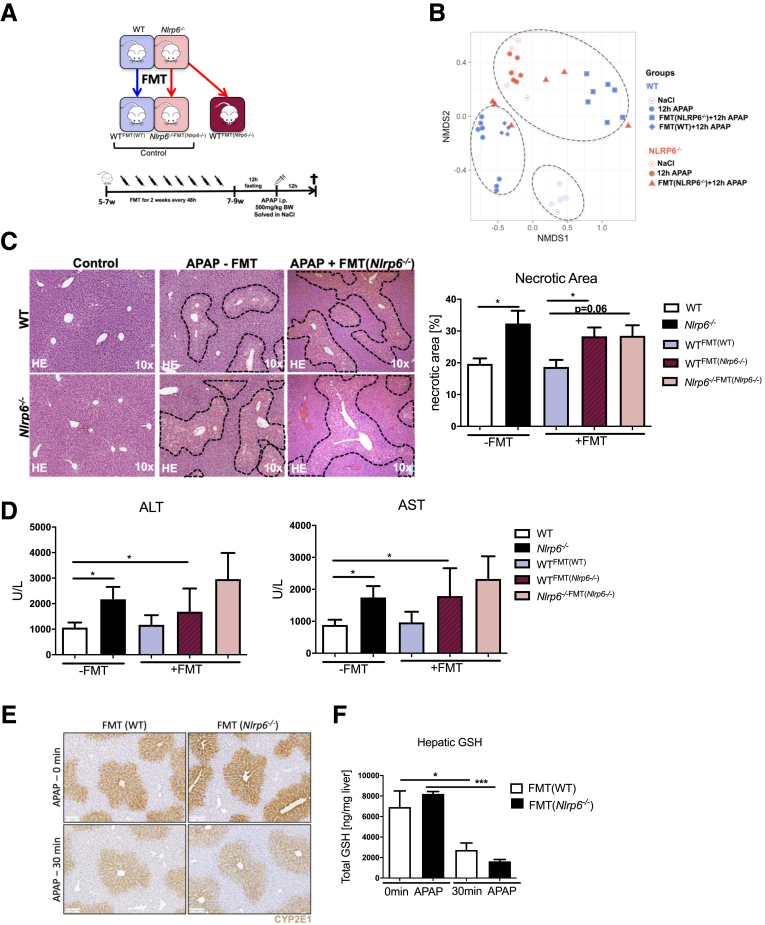
Increased severity of DILI is transmissible to wild-type animals via FMT. (A) Experimental setup: FMT was performed from WT to WT and Nlrp6-/- to WT as well as Nlrp6-/- animals. WT and Nlrp6-/- mice were treated with fresh Nlrp6-/- or WT feces every 48 hours via oral gavage for 2 weeks, fasted 12 hours before injection of APAP (500 mg/kg) or NaCl, and were analyzed 12 hours after injection. (B) NMDS analysis based on UniFrac beta diversity metric displaying microbiota composition in cecal stool samples taken after 12 hours of NaCl or APAP treatment from WT and Nlrp6-/- mice as well as FMT-treated WT and Nlrp6-/- mice. (C) Representative liver histology (hematoxylin-eosin staining) showing necrotic areas and inflammation in 12-hour NaCl (Control) challenged and either non–FMT-treated or with Nlrp6-/- feces FMT-treated APAP challenged WT and Nlrp6-/- mice. Quantitative measurement of necrotic areas on high-resolution liver scans of WT, Nlrp6-/- (n = 6) and FMT-treated WT and Nlrp6-/- mice (n ≥ 4) after 12 hours of APAP treatment. (D) Liver damage indicated by serum ALT and AST levels of 12-hour APAP challenged WT, Nlrp6-/- (n = 13), WTFMT(Nlrp6-/-) (n = 6), WTFMT(WT) (n = 4), and FMT-treated Nlrp6-/-FMT(Nlrp6-/-) mice (n = 6). (E) Immunohistochemistry staining against CYP2E1 in mouse livers before or 30 minutes after APAP injection. (F) Total GSH levels in liver tissue before or 30 minutes after APAP injection (n = 4). All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t test).
Interestingly, NMDS analyses based on UniFrac distances revealed that WT mice treated with microbiota of Nlrp6-/- mice (WTFMT(Nlrp6-/-)) formed a separate cluster close to Nlrp6-/- mice in the NMDS graph, which was significantly different from WT animals (Figure 8B). In contrast, microbiota composition of WT control mice treated with WT microbiota (WTFMT(WT)) clustered together with WT mice (Figure 8B). These data demonstrated that WT mice partially adopted the microbiota of NLRP6 deficient animals after FMT. These changes were not due to the FMT procedure itself, because the transfer of WT microbiota did not significantly change microbial community structure in WT mice.
Increased ALF Severity Is Transmissible to WT Animals via FMT
To investigate the effect of Nlrp6-/--associated microbiota on the outcome of DILI in WTFMT(Nlrp6-/-) mice, DILI was induced after FMT and followed by comprehensive analyses of the liver phenotype. Strikingly, FMT resulted in a significant increase in hepatic centrilobular necrosis in WTFMT(Nlrp6-/-) compared with WT mice after APAP treatment (Figure 8C). After FMT, WTFMT(Nlrp6-/-) animals presented similar sizes of hepatic necrosis as found in Nlrp6-/- mice (Figure 8C). Liver histology translated into transaminase levels, showing increased ALT and AST levels in WTFMT(Nlrp6-/-) compared with WTFMT(WT) animals (Figure 8D); however, because of high variance this difference did not reach statistical significance. Importantly, necrotic area size remained unchanged in WTFMT(WT) compared with WT mice (Figure 8C), demonstrating that the observed phenotype was not due to the FMT procedure itself. Together these data demonstrate that increased severity of ALF is transmissible to WT mice via FMT.
Gut microbiota may influence Cyp2E1 expression and/or hepatic GSH levels and thus APAP metabolism.37 However, also the inflammatory response after APAP administration is a key driver of subsequent liver damage.5 Both mechanisms are important for the extent of liver damage after APAP intoxication. We therefore assessed the impact of the dysbiotic Nlrp6-/- microbiota on hepatic APAP metabolism. First, we analyzed Cyp2E1 protein levels by immunostaining in WT littermate mice treated with WT or Nlrp6-/- microbiota. No differences in Cyp2E1 staining between WT and Nlrp6-/- mice before or 30 minutes after APAP treatment were observed (Figure 8E, Figure 9A). Moreover, we could not detect significant differences between hepatic GSH levels in WT mice treated with WT or Nlrp6-/- microbiota before or after APAP treatment (Figure 8F). WTFMT(Nlrp6-/-) displayed a slightly increased GSH depletion compared with WTFMTWT) mice. Together, these data demonstrate that FMT only had a very small influence on APAP metabolism.
Figure 9.

FMT does not affect hepatic Cyp2E1 expression. (A) Immunohistochemistry staining against CYP2E1 of whole liver scans of WT mice treated with WT microbiota (FMT(WT)) or Nlrp6-/- microbiota (FMT(Nlrp6-/-)) before and 30 minutes after APAP treatment.
Liver Injury Upon FMT of Nlrp6-/- Microbiota Is Orchestrated by Ly6Chi Inflammatory Monocytes
Increased liver damage in Nlrp6-/- mice was linked to a stronger hepatic inflammatory response. Thus, we investigated the effect of FMT on the innate immune response in the liver. Strikingly, FMT of Nlrp6-/- microbiota strongly influenced infiltration of CD11b+ inflammatory cells, clustering in close proximity to hepatic necrosis (Figure 10A). Quantification of immunofluorescence staining against CD11b revealed a significant increase of CD11b+ cells in WTFMT(Nlrp6-/-) compared with WTFMT(WT) mice (Figure 10B). To further characterize the inflammatory infiltrates, we performed flow cytometry analyses of all groups of mice. These analyses revealed a pronounced infiltration of MoMFs (defined as CD11bhiF4/80low) in WTFMT(Nlrp6-/-) compared with WTFMT(WT). Transfer of the dysbiotic Nlrp6-/- community promoted an inflammatory Ly6Chi polarization of freshly infiltrated monocytes (Figure 10C). Accordingly, the frequency of MoMFs with high expression of Ly6C was significantly higher in WTFMT(Nlrp6-/-) compared with WTFMT(WT) mice (Figure 10D). Although transfer of WT microbiota into WT mice (WTFMT(WT) control) did not result in increased MoMF abundance compared with untreated WT mice, Nlrp6-/-FMT(Nlrp6-/-) displayed significantly increased MoMFs compared with untreated Nlrp6-/-, indicating that in the absence of NLRP6 the FMT procedure itself may aggravate intestinal dysbiosis in these mice and fuel the inflammatory response (Figure 10B and C).
Figure 10.
Liver injury on FMT of Nlrp6-/-microbiota is orchestrated by Ly6Chiinflammatory monocytes. (A) Immunofluorescence staining against CD11b and (B) quantitative analysis of CD11b+ stained cells in liver samples of WT and Nlrp6-/- mice either with (n ≥ 3) or without (n ≥ 10) previous FMT after 12 hours of APAP treatment. (C) Quantification of MoMFs (defined as CD11bhiF4/80low) as percent of living liver leukocytes of either non–FMT-treated (–FMT, WT: n = 10, Nlrp6-/-: n = 11) or FMT(Nlrp6-/-)-treated (+FMT, WT: n = 7, Nlrp6-/-: n = 3) WT and Nlrp6-/- mice determined by fluorescence-activated cell sorter analysis. (D) Flow cytometry analysis of Ly6Chi expression of MoMFs in liver samples of WTFMT(WT) (n = 4) or WTFMT(Nlrp6-/-) (n = 7) mice after 12-hour APAP treatment. (E) NMDS analyses based on UniFrac beta diversity metric. Every point represents cecal microbiota composition of 1 mouse. Similar microbiota composition is reflected in closer ordination. (F) Microbiota alpha diversity shown by Shannon index in APAP- and NaCl-treated WT and Nlrp6-/- mice without (WT/Nlrp6-/-: n ≥ 5) and with (WT NaCl: n = 2; WT APAP: n = 4, Nlrp6-/- APAP: n = 3) ABx treatment. (G) Representative liver histology showing necrotic areas and inflammation in ABx-treated mice. (H) Quantitative measurement of necrotic areas on high-resolution liver scans of APAP-treated WT (n = 5) and Nlrp6-/- (n = 7) mice 12 hours after treatment. (I) Liver injury assessed by serum ALT and GLDH levels after APAP (WT: n = 5, Nlrp6-/-: n = 7). (J) Liver-to-bodyweight ratio of ABx-treated WT and Nlrp6-/- mice. Every dot represents 1 mouse. All data are presented as mean ± standard error of mean and considered significant at ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, respectively (unpaired Student t test). ∗Significantly different from respective control group.
Collectively, these data demonstrate that increased severity of ALF in Nlrp6-/- animals is transmissible to WT animals via FMT. Transfer of dysbiotic Nlrp6-/- microbiota skewed MoMF polarization in WT mice toward a Ly6Chi inflammatory phenotype. Hence, after FMT, WTFMT(Nlrp6-/-) mice resemble the hepatic inflammatory phenotype of Nlrp6-/- animals.
Broad-Spectrum ABx Treatment Equalizes Liver Injury in WT and Nlrp6-/-Mice
Nlrp6-/- mice demonstrated increased susceptibility toward APAP-induced liver injury. This susceptibility was transmissible to WT mice via FMT. To better dissect the impact of NLRP6 on microbiota from other potential NLRP6-mediated mechanisms, we depleted gut microbiota with broad-spectrum ABx in drinking water. WT and Nlrp6-/- mice received ABx water for 2 weeks before APAP administration. After antibiotic treatment, WT and Nlrp6-/- mice formed a common cluster in the NMDS graph based on UniFrac distances. ABx treatment explained a large fraction of about 58% of gut microbiota variability (adonis, R = 0.58, ∗∗∗) (Figure 10E). Shannon diversity was significantly reduced in WT control mice without ABx treatment compared with ABx-treated WT mice. Differences in Shannon diversity between the genotypes and treatment groups disappeared on ABx treatment (Figure 10F). Interestingly, ABx treatment equalized liver injury between WT and Nlrp6-/- mice, which was reflected in necrosis formation and liver transaminase levels (Figure 10G, H and I). Nlrp6-/- mice on ABx even showed a trend toward lower liver injury as evidenced by GLDH and a lower liver-to-bodyweight ratio (Figure 10I and J).
Together with the FMT experiments, these data demonstrate that increased susceptibility toward APAP-induced ALF of Nlrp6-/- mice compared with WT mice is mediated by the gut microbiota.
Discussion
ALF is a condition with poor outcome, rapid progression, and high mortality.6,38,39 APAP intoxication is one of the leading causes of ALF, and strong differences in susceptibility to the hepatotoxic effect of APAP have been reported.40,41 Together, these data point to host-derived and environmental factors modulating host susceptibility to APAP. Intestinal microbiota is a host-environmental interface that serves important regulatory functions in immunity and metabolism.25 Although unfavorable intestinal microbiota may drive various chronic liver diseases,11, 12, 13 data on the role of the gut-liver axis and intestinal dysbiosis for ALF remain limited.
Because ALF is a rare condition, it is challenging to study the influence of intestinal dysbiosis on ALF susceptibility. The ideal study would require data on microbiota before the onset of ALF because liver failure by itself may strongly shape microbiota composition. However, such a prospective population-based study including microbiota samples for a sufficiently large number of patients does not exist. There is strong evidence on the basis of clinical as well as preclinical data highlighting long-term medication with PPI or ABx as a strong inducer of intestinal dysbiosis and barrier impairment.18, 19, 20, 21,30 Therefore, we used PPI and ABx intake as surrogate markers of intestinal dysbiosis. We are aware that there are limitations for this approach because patients with PPI or long-term ABx may have various comorbidities affecting the risk for ALF. We therefore calculated the Charlson comorbidity index, which accounts for multiple comorbidities including all main organ systems and especially liver disease.32 Because chronic liver disease is a major risk factor for ALF, we also calculated APRI as a serum biomarker for chronic liver disease. After excluding patients with heavy alcohol consumption as well as viral hepatitis and correcting for age, gender, APRI, and Charlson comorbidity index, medication with PPI was associated with 1.4-fold increase in risk of ALF in our multivariate model. However, this population-based study is limited in that we cannot exclude that differences in the patient population that have not been fully captured in the statistical model may explain the observed association. On the basis of this analysis we aimed to test the hypothesis whether intestinal dysbiosis is causally involved in ALF and continued our analyses using mouse models of ALF.
Similar to what has been described in patients with PPI or ABx medication, NLRP6-deficient mice displayed intestinal dysbiosis reflected in reduced microbiota diversity, lack of beneficial bacteria (eg, Lachnospiraceae), as well as barrier impairment.25,42, 43, 44 Hence, these mice were used as an appropriate model to study whether intestinal dysbiosis may modulate host susceptibility to ALI.
Nlrp6-/- mice showed more severe APAP as well as LPS-induced liver injury compared with WT mice. To uncouple the role of the dysbiotic microbiota of these mice from a potential influence of the constitutive Nlrp6 knockout, we performed FMT experiments. Strikingly, on transfer of the dysbiotic community, liver injury in the recipient WT mice was similar to Nlrp6-/- mice. To further uncouple microbiota-dependent from potential microbiota-independent functions of NLRP6, we depleted the gut microbiota of WT and Nlrp6-/- mice with broad-spectrum ABx. Importantly, this treatment equalized liver injury between WT and Nlrp6-/- mice, which then showed even less liver injury compared with ABx-treated WT mice. Together, these data demonstrate that microbiota-dependent functions in Nlrp6-/- mice account for increased susceptibility toward ALI.
Next we assessed the molecular mechanisms by which gut microbiota affect ALF. We studied microbiota composition, gut epithelium, and barrier function in WT and Nlrp6-/- mice before and after APAP intoxication.
At baseline, Nlrp6-/- mice revealed pronounced reduction in microbiota alpha diversity and intestinal barrier function. Interestingly, reduced microbiota diversity in Nlrp6-/- mice was reflected in the liver microbiota signature, showing the intimate interaction between gut and liver. Dysbiotic Nlrp6-/- mice displayed a markedly reduced rDNA abundance of Lachnospiraceae NK4A136 in gut and liver, which was among the most abundant taxa in WT mice. Interestingly, recent data associated hepatic abundance of Lachnospiraceae NK4A136 DNA with protection against liver inflammation.45 Lachnopiraceae have the important ability to form secondary bile acids by 7α-dehydroxylation and thus serve immune regulatory functions via bile acid mediated signaling pathways.46,47 In WT mice, the APAP-induced loss of gut microbiota alpha diversity was paralleled by a decrease in the hepatic microbiota diversity, linked to intestinal barrier impairment and increased translocation of bacterial DNA. On the basis of our current data, it is tempting to speculate whether loss of gut-microbiota diversity in patients on APAP intoxication may have prognostic implications.
In line with another recent study,48 we observed an expansion of the bacterium Akkermansia muciniphila, which was accompanied by an increased thickness of colonic mucus layers. Protective functions for intestinal barrier integrity have been suggested in metabolic syndrome49,50 and nonalcoholic fatty liver disease.11 Future studies involving colonization experiments are needed to further define its role in ALI.
Recent data demonstrate that microbiota control hepatic metabolism via the cytochrome P450 system.51 At therapeutic doses, APAP is metabolized mainly by glucuronidation and sulfation, but overdoses are metabolically activated, mainly via Cyp2E1 enzyme, into the highly reactive intermediate NAPQI, which is normally detoxified by GSH.38 After GSH depletion, NAPQI binds to cellular and mitochondrial proteins, leading to oxidative stress, mitochondrial dysfunction, and finally cell death culminating in liver inflammation and hepatic injury.5
To assess whether APAP metabolism was different in Nlrp6-/- mice, we performed analyses of APAP as well as its metabolites in the blood. Nlrp6-/- mice displayed a trend toward higher GSH levels and increased GSH depletion compared with WT mice, which was in line with increased liver damage observed in these mice. However, differences in GSH levels of Nlrp6-/- mice were not recapitulated in WTFMT(Nlrp6-/-) mice, suggesting that changes in APAP metabolism are not essential to explain increased liver damage upon microbiota transfer.
NAPQI toxicity as well as the resulting innate immune response contribute to ALI, and therefore dissecting the impact of both mechanisms is highly relevant for understanding the pathogenesis of ALI and ultimately for defining novel molecular therapeutic targets.5,38 In our study, Nlrp6-/- associated microbiota prompted increased infiltration and a Ly6Chi phenotype of infiltrating MoMFs, which are essential drivers of DILI. Interestingly, genome-wide transcriptional profiling demonstrated that microbiota depletion by ABx treatment dampened the expression of innate immune response genes in macrophages.52 Similarly a recent study demonstrated that differences in composition and function of gut microbial communities contribute to interindividual variation in cytokine responses to microbial stimulation.53 Correspondingly, we have shown that hepatic MoMFs of mice treated with broad-spectrum ABx displayed increased expression of markers associated with restorative macrophage function.11
These data suggest that constant diffusion of microbiota-derived products into the bloodstream calibrate the activation threshold of macrophage-mediated innate immunity. Similarly, in the context ALF, LPS, and other MAMPs might be involved in the basal tuning of the immune system because mice resistant to LPS/toll-like receptor 4 signaling are partially protected against ALF.54,55 Accordingly, WT mice transplanted with dysbiotic Nlrp6-/- microbiota presented significantly increased infiltration of MoMF, which displayed proinflammatory Ly6Chi polarization. Together, these data point to the relevance of the gut-liver axis during ALI and identify MoMF as important sensors of gut derived signals.
APAP intoxication often leads to multi-organ dysfunction with systemic complications. Interestingly, patients who have undergone liver transplantation for APAP-induced liver injury have a worse prognosis compared with those with other etiologies.56 Toxic effects beyond the liver may explain this. In our study APAP intoxication resulted in loss of microbiota diversity, intestinal barrier impairment, and translocation of 16s rDNA in WT mice, which may critically contribute to the development of multi-organ dysfunction. Although our current study mainly focuses on the role of microbiota in the early phase of APAP-induced liver injury, it will be equally important to investigate whether changes in microbiota composition and intestinal barrier function may determine the short-term outcome of ALF.
In summary, we describe an important yet unknown function of the intestinal microbiota during ALF. Intestinal dysbiosis, as seen in Nlrp6-/- mice and transferrable to healthy WT controls via FMT, rendered these mice susceptible to ALF. Specifically, transfer of dysbiotic microbiota skewed monocyte polarization in WT mice toward a Ly6Chi inflammatory phenotype, suggesting a critical function of these cells as sensors of gut-derived signals orchestrating the hepatic inflammatory response.
Our current study identifies gut microbiota as a potentially modifiable risk factor of ALF. Moreover, we demonstrate that ALF itself alters gut microbiota composition, leading to gut barrier impairment and bacterial translocation, thereby providing the basis for further translational studies aiming to better define gut-liver crosstalk in patients with ALF.
Materials and Methods
In Vivo Experiments
All animal experiments were approved by the appropriate German authorities (LANUV, Recklinghausen, North Rhine-Westphalia, Germany; Az 84-02.04.2012.A260 to C.T., Az 84-02.04.2017.A327 to C.T., Az 84-02.04.2016.A279). All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86-23 revised 1985). C57BL/6J WT and Nlrp6 deficient (Nlrp6-/-) mice (C57BL/6J background) were housed in the same conditions in individually ventilated cages with free access to a standard food diet and water. Mouse lines for WT and Nlrp6-/- mice were created from an initial heterozygous Nlrp6+/- breeding pair. Homozygous offspring of this line were used to create individual WT and Nlrp6-/- breeding. These lines were held in individually ventilated cages in the same room of our animal facility. To avoid transmission of the dysbiotic Nlrp6-/- microbiota to WT mice, we did not allow any exchange of mice, cage, or nesting material between these 2 lines.
For induction ALI, 7- to 9-week-old male mice were fasted for 12 hours before intraperitoneal (IP) injection of 500 mg/kg APAP (Sigma-Aldrich, St Louis, MO) (WT: n = 16, Nlrp6-/-: n = 16) or sodium chloride (NaCl) (WT: n = 5; Nlrp6-/-: n = 5). In addition, LPS (4 mg/kg, IP) (Sigma-Aldrich) (WT: n = 6, Nlrp6-/-: n = 5) was used as a second model. Blood as well as tissue samples were collected at 12 and 24 hours after APAP injection and at 6 hours after LPS injection.
For FMT, WT (n = 12) or Nlrp6-/- (n = 6) mice were treated for 2 weeks every 48 hours via oral gavage with 200 μL fecal dilution before induction of ALI. To prepare this dilution, 20 mg freshly harvested stool from Nlrp6-/- or WT mice was solved in 200 μL phosphate-buffered saline (PBS) and centrifuged at 350g for 5 minutes before gavage of the supernatant.
Collection of tissue and blood specimens, RNA isolation, complementary DNA synthesis, quantitative polymerase chain reaction (PCR), and serum parameters (transaminases, GLDH, and alkaline phosphatase) were determined as described previously.57,58
UK BioBank Population-based Study
The UK BioBank is a population-based cohort including 502,511 European participants between ages 37 and 73 years. All volunteers, who received a postal invitation, were recruited in 22 assessment centers in England, Scotland, and Wales. The baseline assessment was performed between 2006 and 2010 and includes demographics, lifestyle, disease history, and current medication as well as a physical examination and baseline laboratory values. Only long-term medication was recorded. The volunteers gave informed consent for genotyping and data linkage to medical reports. At baseline a questionnaire recorded their responses to prevalent diseases. In addition, ongoing inpatient hospital records beginning in 1996 were used to identify diagnoses according to International Classification of Diseases, 10th Revision (ICD-10) codes. All reported ICD-10 codes were connected with the date of their first diagnosis. For follow-up, hospital inpatient data, national cancer registries, or death registration were used. End of follow-up was defined as death or end of hospital inpatient data collection in March 2018, whichever occurred first. The study has been approved by the UKB Access Committee (Project #59657).
Sample selection and outcome definition
We excluded participants with prevalent chronic hepatitis B or C (302 cases) or pathologic alcohol consumption (>60 g alcohol/d for men or >40 g alcohol/d for women, 4782 cases). PPI medication at baseline was defined as long-term pantoprazole and/or omeprazole and/or esomeprazole and/or lansoprazole intake. Long-term ABx was defined as long-term intake of tetracycline, aminoglycoside, glycopeptide, and macrolide antibiotics at baseline. Hepatic failure was defined as hospital inpatient record ICD-10 code K72 that occurred after the initial baseline assessment.
To assess the comorbidities of the UK BioBank participants we calculated the Charlson disease severity index.32 This index is commonly used to adjust for the effect of comorbidities in hazard models. The Charlson comorbidity index includes 14 different categories that we used to assess the comorbidities at baseline: age (baseline questionnaire), myocardial infarction (baseline questionnaire), chronic heart failure (baseline questionnaire), peripheral vascular disease (baseline questionnaire), transient ischemic attack (baseline questionnaire), dementia (baseline questionnaire), chronic obstructive pulmonary disease (baseline questionnaire), connective tissue disease (baseline questionnaire), peptic ulcer (baseline questionnaire), liver disease (baseline questionnaire plus ICD-10 codes to distinguish between mild and moderate disease), diabetes mellitus (baseline questionnaire), hemiplegia (ICD-10 codes, as not given in the baseline questionnaire), chronic kidney disease (baseline questionnaire), leukemia (ICD-10 codes, as not given in the baseline questionnaire), lymphoma (ICD-10 codes, as not given in the baseline questionnaire), and acquired immunodeficiency syndrome (baseline questionnaire). APRI was calculated as a noninvasive marker of fibrosis as previously described.59
Mouse Model
The aim of our study was to investigate the role of intestinal dysbiosis during APAP-induced liver injury. To this end, WT and Nlrp6-/- mice were subjected to a well-established model of IP APAP-induced liver injury. IP injection of LPS was used as a second model of ALI. Liver injury was assessed by analyzing necrotic areas, liver function tests, and inflammatory cell infiltration. For statistical power calculation, effect size estimation was based on prior experiments of our group. All key experiments were replicated at least 2 times. No animals were excluded from analyses unless stated otherwise.
Broad-Spectrum ABx Treatment
Intestinal microbiota were eradicated in mice using 4 broad-spectrum ABx (1 g/L ampicillin, 160 mg/L gentamycin, 1 g/L metronidazole, 1 g/L vancomycin). Male mice received this cocktail continuously in the drinking water for 2 weeks. ABx water was changed every second day.
Histology
Conventional H&E stainings were performed according to standard protocols,11 and necrotic areas were quantified on high-resolution liver scans by area fraction analysis (ImageJ; National Institutes of Health, Bethesda, MD).
Staining of Mucus and Gut Bacteria
Colon tissue sections containing feces were fixed using the Carnoy’s fixation method (60% absolute methanol, 30% chloroform, 10% glacial acetic acid). After paraffin embedding, mucus and gut bacteria were stained using an anti-Muc2 primary antibody (Santa Cruz Biotechnology, Dallas, TX; sc-15334) and a fluorescence in situ hybridization probe against eubacteria (16S rRNA: 5'-GCTGCCTCCCGTAGGAGT-3'). Staining was performed according to an established protocol.60
Quantitative Real-time PCR
RNA was isolated from liver tissue specimen using Trizol Reagent (Life Technologies, Carlsbad, CA), concentration was measured by using NanoDrop Lite (Thermo Scientific, Dreieich, Germany), and reverse transcription was performed using an Omniscript kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s protocol. Fast SYBR GreenER Master Mix (Thermo Fisher, Waltham, MA) was used for the assembling of real-time PCR reactions according to manufacturer’s recommendations. Following primer sequences were used for GAPDH (3'-TGT TGA AGT CAC AGG AGA CAA CCT-5', 5'- AAC CTG CCA AGT ATG ATG ACA TCA-3'), Cyp1A2 (3'- GAG GCG AAC AGG CTA CCT AC -5', 5'-CTG AGT TGT TTT GCC CGC TC -3'), Cyp2E1 (3'-GCT GTC AAG GAG GTG CTA CTG AAC-5', 5'-CTC CGC ACG TCC TTC CAT GTG G-3'), and occludin (3'-GCT GTG ATG TGT GTT GAG CT-5', 5'-GAC GGT CTA CCT GGA GGA AC-3'). For analysis QuantStudio Flex software (Thermo Fisher) was used. Expression of mRNA was calculated using the 2−ΔΔCT method, which determines the relative quantification of a target gene in comparison to the GAPDH gene.
Immunoblotting
The protein samples (1 μg/μL) were separated electrophoretically on precast 4%–12% polyacrylamide gel (Bio-Rad, Hercules, CA) in sodium dodecyl sulfate running buffer at 200 V for approximately 30 minutes. For immunologic detection, the separated proteins in the gel were transferred to a nitrocellulose membrane using Trans-Blot Turbo Transfer System (Bio-Rad). Staining with Ponceau Red was used to confirm protein transfer. Nonspecific binding sites were blocked by incubation for 1 hour in 5% nonfat dry milk diluted in tris-buffered saline tween (TBST) (0.5%). Afterwards the membrane was incubated overnight at 4°C with primary antibody diluted in 5% bovine serum albumin TBST. After 3× washing for 5 minutes with TBST, the membrane was incubated with the horseradish peroxidase–conjugated secondary antibody diluted 1:5000 in 5% nonfat dry milk diluted in TBST for 1 hour at room temperature. Next, the membrane was again washed 3× for 5 minutes in TBST and then incubated in ECL Substrate (Pierce, Waltham, MA) for 5 minutes. To detect specific signals the membrane was exposed to a LAS mini 4000 developing machine (Fuji, Tokyo, Japan). The following antibodies were used in this study: Cyp2E1 (Abcam, Cambridge, UK; ab28146), beta-actin (Sigma; A2066), occludin (Invitrogen, Carlsbad, CA; 71-1500), p-JNK/p-SAPK (Cell Signaling, Danvers, MA; #9251S), JNK/SAPK (#9252S), GAPDH (Bio-Rad; AHP1628).
Immunofluorescence Staining
Immunofluorescence staining was performed on cryopreserved 5-μm tissue sections. The sections were dried in air for 20–30 minutes, fixed with 4% PFA for 8–10 minutes at room temperature, and washed with PBS 3× for 5 minutes each. They were blocked with PBS containing 5% goat serum for 45 minutes, followed by incubation with primary antibody against CD11b (BD, Franklin Lakes, NJ; 550282) diluted 1:200 to 1:400 in blocking solution at 4°C overnight in a humidified chamber. After washing with PBS for 3 × 5 minutes, slides were incubated with a fluorescence-linked secondary antibody 1:400 in blocking buffer for 1 hour at room temperature in a humidified chamber. Slides were then again washed with PBS 3× for 5 minutes. Nuclei were counterstained with Vectashield Antifade Mounting Medium with DAPI (Vector, Burlingame, CA).
GSH Assay
Total GSH concentrations were measured in liver tissue homogenate using liquid chromatography-tandem mass spectrometry, as described in Sezgin et al.61
Analyses of APAP, APAP Metabolites, and APAP Adducts in Blood
APAP as well as its metabolites (APAP-glucuronide, APAP-sulfate) and adducts (APAP-glutathione, APAP-cysteine, APAP-N-acetylcysteine) were measured in systemic blood plasma using high-performance liquid chromatograpy-mass spectrometry, as described in detail in Sezgin et al.61
Immunohistochemistry
Immunohistochemical staining was performed on paraffin-embedded sections, as described previously.58 The sections were blocked with peroxidase-conjugated avidin-biotin method (ABC Reagent Kit; Vector). To block unspecific binding sites, slides were further incubated for 30 minutes at room temperature in 50% fetal calf serum + 50% PBS containing 1% bovine serum albumin. Slides were then incubated with primary antibody in blocking solution at optimized dilutions at 4°C overnight in a humidified chamber. Next day slides were washed twice with PBS, and the secondary antibody (1:200 diluted in Phospate-Buffered Saline 0,1% Tween 20) was applied for 1 hour at room temperature. To make the visualization of the signal, the enzyme substrate 3,3'-diaminobenzidine (DAB) Substrate Kit (Vector) was used. Counterstaining was performed with hematoxylin. Primary antibodies against CD45 (BD; 550539) and CYP2E1 (Sigma; HPA009128) were used.
Flow Cytometry Analysis of Intrahepatic Leukocytes
Same amounts of livers were digested by collagenase type IV for 2 hours (Worthington Biochemical Corporation, Lakewood, NJ), and intrahepatic immune cells were isolated by multiple differential centrifugation steps as detailed.62 Immune cell isolates were incubated with blocking buffer for 30 minutes to block the unspecific binding sites of cell surface, then divided into 2 subgroups, and stained with fluorochrome-conjugated antibodies of either a monocyte panel against Ly6G, CD11b, CD11c, F4/80, Gr1.1, and CD45 (1:200) or a lymphocyte panel against CD3, CD4, CD8, CD19, Nk1.1, and CD45 (1:200). All of the antibodies were purchased from eBioscience (Frankfurt, Germany). All samples were acquired by flow cytometry (FACS Canto II; BD Biosciences, Heidelberg, Germany) and analyzed using the Flowjo software version 9 (Ashland, OR).
DNA Isolation and 16S rRNA Amplicon Sequencing
For 16S rRNA gene sequencing, DNA was isolated from fecal samples by using an established protocol.63 Briefly, each sample (around 200 mg) was resuspended in 500 μL extraction buffer (200 mmol/L Tris, 20 mmol/L EDTA, 200 mmol/L NaCl, pH 8.0), 200 μL 20% sodium dodecyl sulfate, 500 μL phenol:chloroform:isoamyl alcohol (24:24:1), and 100 μL of zirconia/silica beads (0.1-mm diameter). Samples were homogenized twice with a bead beater (BioSpec Products, Bartlesville, OK) for 2 minutes. After precipitation of DNA, crude DNA extracts were resuspended in TE buffer with 100 μg/mL RNase I and column purified to remove PCR inhibitors.
Amplification of the V4 region (F515/R806) of the 16S rRNA gene was performed according to previously described protocols.64 Briefly, for amplicon sequencing 25 ng DNA was used per PCR reaction (30 μL). The PCR conditions consisted of initial denaturation for 30 seconds at 98°C, followed by 25 cycles (10 seconds at 98°C, 20 seconds at 55°C, and 20 seconds at 72°C). Each sample was amplified in triplicates and subsequently pooled. After normalization PCR amplicons were sequenced on an Illumina MiSeq platform (PE250).
Quantitative Insights into Microbial Ecology (QIIME) version 1.9.1 was used for data analysis.65 After de-multiplexing, an open-reference operational taxonomic unit picking protocol66 was performed by searching reads against the greengenes database. The observed operational taxonomic units (observed richness) and Shannon index (estimated richness), which are metrics of species richness and diversity within communities (alpha diversity), were computed. NMDS analysis based on UniFrac beta diversity metric was performed. The LEfSe algorithm was used to characterize statistically significant and biologically relevant differences between the different groups as previously described.33
16S rDNA Quantitation and Taxonomic Profiling in Liver Tissue
DNA from liver biopsies were extracted, and quantitative real-time PCR amplification was performed by using 16S universal primers targeting the hypervariable V3-V4 region of the bacterial 16S ribosomal gene, with a protocol carefully designed to minimize any risk of contamination between samples or from the experimenters, environment as described previously.35,36,67 The quality and quantity of extracted nucleic acids were controlled by gel electrophoresis (1% w/w agarose in TBE 0.5×) and absorbance spectroscopy using a NanoDrop 2000 UV spectrophotometer (Thermo Scientific). The quantitative PCR was performed on a ViiA 7 PCR system (Life Technologies) using Sybr Green technology. The microbial populations based on rDNA present in liver samples were determined by using next-generation high-throughput sequenced using the Illumina-MiSeq technology as described previously.35,36,67 We performed numerous controls both in vitro and in silico to ensure the absence of artefacts such as bacterial DNA contaminants from reagents or nonspecific amplification of eukaryotic DNA.35,68 Alpha and beta diversity was tested after clustering reads into operational taxonomic units, and taxonomic assignment was performed using FROGS v.1.4.069 and PhyloSeq v1.22.3 with the databank Silva 132 Parc.
Fecal Albumin Assay
To assess intestinal permeability the albumin concentration in the feces was measured by enzyme-linked immunosorbent assay (E99-134; Bethyl Laboratories, Montgomery, TX). Freshly collected feces were homogenized and diluted in dilution buffer (75 mg/mL). After centrifugation at 10,000g the supernatant was diluted 1:50, and samples were analyzed following the manufacturer’s instructions.
Endotoxin Measurement
Endotoxin concentrations in serum were measured as described previously.70 Briefly, they were determined by an endpoint enzymatic assay based on limulus amebocyte lysate for a concentration range of 0.015–1.2 EU/mL (LAL Kit; Charles River, Ecully, France).
Statistical Analyses
For analyses of UKBioBank data, all categorical variables were described as absolute (n) and relative (%) frequencies. Continuous variables were displayed as mean ± standard deviation. Contingency tables were analyzed with χ2 tests, and continuous variables were analyzed by unpaired, two-tailed t tests. The occurrence of ALF was modeled as the outcome variable in a Cox proportional hazard survival analysis that examined the risk associated with PPI while controlling for Charlson comorbidity index, APRI, BMI, and sex. Hazard ratios were presented with their corresponding 95% confidence intervals given in brackets. Models were adjusted for all tested variables with P <.25 (Tables 1 and 3). Nominal P values were given for all statistical tests. Differences were considered to be statistically significant when P < .05. The data were analyzed using SPSS Statistics version 25 (IBM, Armonk, NY). All murine data are expressed as mean ± standard error of the mean. Significant differences were calculated with GraphPad Prism (San Diego, CA) software version 8. Statistical significance was determined by one-way analysis of variance followed by Student t test. Data were considered significant between experimental groups as ∗P < .05, ∗∗P < .01, or ∗∗∗P < .001.
Acknowledgments
The authors thank Ms Sonja Strauch and Ms Bettina Jansen, University Hospital RWTH Aachen, Aachen Germany for their excellent technical assistance.
The study has been approved by the UKB Access Committee (Project #59657).
CRediT Authorship Contributions
Kai Markus Schneider, MD, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Supporting; Methodology: Lead; Project administration: Lead; Supervision: Equal; Validation: Equal; Writing – original draft: Lead; Writing – review & editing: Equal)
Carsten Elfers (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Lead; Investigation: Equal; Writing – original draft: Equal; Writing – review & editing: Equal),
Ahmed Ghallab, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting),
Carolin Victoria Schneider, MD (Formal analysis: Supporting; Writing – review & editing: Supporting)
Eric J. C. Galvez, PhD (Formal analysis: Supporting)
Antje Mohs, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Supervision: Supporting)
Wenfang Gui (Data curation: Supporting)
Lena Susanna Candels (Data curation: Supporting; Formal analysis: Supporting),
Theresa Hildegard Wirtz, MD (Writing – review & editing: Supporting)
Sebastian Zuehlke (Data curation: Supporting)
Michael Spiteller (Data curation: Supporting)
Maiju Myllys (Data curation: Supporting)
Alain Roulet, PhD (Data curation: Supporting)
Amirouche Ouzerdine, Msc (Data curation: Supporting)
Benjamin Lelouvier, PhD (Data curation: Supporting)
Konrad Kilic (Data curation: Supporting)
Lijun Liao, MD (Data curation: Supporting)
Anika Nier (Data curation: Supporting; Writing – review & editing: Supporting)
Eicke Latz, Prof Dr (Writing – review & editing: Supporting)
Ina Bergheim, Prof Dr (Writing – review & editing: Supporting)
Christoph A. Thaiss, Prof PhD (Resources: Supporting; Writing – review & editing: Supporting)
Jan G. Hengstler, Prof Dr (Conceptualization: Supporting; Formal analysis: Supporting; Project administration: Supporting; Resources: Supporting; Supervision: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Till Strowig, Prof Dr (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Christian Trautwein, Prof Dr (Conceptualization: Equal; Formal analysis: Equal; Funding acquisition: Lead; Project administration: Equal; Resources: Lead; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the German Research Foundation CRC1382 and SFB/TRR 57 to C.T. (TR285/10-2), the Federal Ministry of Education and Research (ObiHep grant #01KU1214A to C.T.), the Liver-LiSyM grant (BMBF) to C.T., AG (FKZ 031L0052), and to J.G.H. (031L0045); the HDHL-INTIMIC Di-Mi-Liv to C.T. and K.M.S., the Interdisciplinary Centre for Clinical Research (START Grant #691438) within the Faculty of Medicine at RWTH Aachen University, the Helmholtz Association (VH-NG-933 to T.S.), German Research Foundation (DFG SCHN 1626/1-1) to K.M.S., and the German National Academic Foundation (to C.E. and K.M.S.).
References
- 1.Lee W.M. AASLD position paper: update. Hepatology. 2005;41:1179–1197. doi: 10.1002/hep.20703. [DOI] [PubMed] [Google Scholar]
- 2.Sandhu N., Navarro V. Drug-induced liver injury in GI practice. Hepatol Commun. 2020 doi: 10.1002/hep4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade R.J., Aithal G.P., Björnsson E.S., Kaplowitz N., Kullak-Ublick G.A., Larrey D., Karlsen T.H. EASL clinical practice guidelines: drug-induced liver injury. J Hepatol. 2019;70:1222–1261. doi: 10.1016/j.jhep.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Davidson D.G.D., Eastham W.N. Acute liver necrosis following overdose of paracetamol. Br Med J. 1966;2:497–499. doi: 10.1136/bmj.2.5512.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krenkel O., Mossanen J.C., Tacke F. Immune mechanisms in acetaminophen-induced acute liver failure. Hepatobiliary Surg Nutr. 2014;3:331–343. doi: 10.3978/j.issn.2304-3881.2014.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larson A.M., Polson J., Fontana R.J., Davern T.J., Lalani E., Hynan L.S., Reisch J.S., Schiødt F.V., Ostapowicz G., Shakil A.O., Lee W.M. Acute Liver Failure Study Group. Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology. 2005;42:1364–1372. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt L.E., Dalhoff K., Poulsen H.E. Acute versus chronic alcohol consumption in acetaminophen-induced hepatotoxicity. Hepatology. 2002;35:876–882. doi: 10.1053/jhep.2002.32148. [DOI] [PubMed] [Google Scholar]
- 8.Whitcomb D.C., Block G.D. Acetaminophen hepatotoxicity, fasting, and ethanol. JAMA. 1995;274:302. doi: 10.1001/jama.1994.03520230055038. [DOI] [PubMed] [Google Scholar]
- 9.Abebe W. Herbal medication: potential for adverse interactions with analgesic drugs. J Clin Pharm Ther. 2002;27:391–401. doi: 10.1046/j.1365-2710.2002.00444.x. [DOI] [PubMed] [Google Scholar]
- 10.Gregory B., Larson A.M., Reisch J., Lee W.M. Acetaminophen dose does not predict outcome in acetaminophen-induced acute liver failure. J Investig Med. 2010;58:707–710. doi: 10.231/JIM.0b013e3181db8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider K.M., Bieghs V., Heymann F., Hu W., Dreymueller D., Liao L., Frissen M., Ludwig A., Gassler N., Pabst O., Latz E., Sellge G., Penders J., Tacke F., Trautwein C. CX3CR1 is a gatekeeper for intestinal barrier integrity in mice: limiting steatohepatitis by maintaining intestinal homeostasis. Hepatology. 2015;62:1405–1416. doi: 10.1002/hep.27982. [DOI] [PubMed] [Google Scholar]
- 12.Yan A.W., Fouts D.E., Brandl J., Stärkel P., Torralba M., Schott E., Tsukamoto H., Nelson K.E., Brenner D.A., Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53:96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki E., Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol. 2012;590:447–458. doi: 10.1113/jphysiol.2011.219691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dapito D.H., Mencin A., Gwak G.Y., Pradere J.P., Jang M.K., Mederacke I., Caviglia J.M., Khiabanian H., Adeyemi A., Bataller R., Lefkowitch J.H., Bower M., Friedman R., Sartor R.B., Rabadan R., Schwabe R.F. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshimoto S., Loo T.M., Atarashi K., Kanda H., Sato S., Oyadomari S., Iwakura Y., Oshima K., Morita H., Hattori M., Hattori M., Honda K., Ishikawa Y., Hara E., Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 16.Macpherson A.J., Heikenwalder M., Ganal-Vonarburg S.C. The liver at the nexus of host-microbial interactions. Cell Host and Microbe. 2016;20:561–571. doi: 10.1016/j.chom.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Liao L., Schneider K.M., Galvez E.J.C., Frissen M., Marschall H.-U., Su H., Hatting M., Wahlstrom A., Haybaeck J., Puchas P., Mohs A., Peng J., Bergheim I., Nier A., Hennings J., Reissing J., Zimmermann H.W., Longerich T., Strowig T., Liedtke C., Cubero F.J., Trautwein C. Intestinal dysbiosis augments liver disease progression via NLRP3 in a murine model of primary sclerosing cholangitis. Gut. 2019;68:1477–1492. doi: 10.1136/gutjnl-2018-316670. [DOI] [PubMed] [Google Scholar]
- 18.Horvath A., Rainer F., Bashir M., Leber B., Schmerboeck B., Klymiuk I., Groselj-Strele A., Durdevic M., Freedberg D.E., Abrams J.A., Fickert P., Stiegler P., Stadlbauer V. Biomarkers for oralization during long-term proton pump inhibitor therapy predict survival in cirrhosis. Sci Rep. 2019;9:12000. doi: 10.1038/s41598-019-48352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llorente C., Jepsen P., Inamine T., Wang L., Bluemel S., Wang H.J., Loomba R., Bajaj J.S., Schubert M.L., Sikaroodi M., Gillevet P.M., Xu J., Kisseleva T., Ho S.B., Depew J., Du X., Sørensen H.T., Vilstrup H., Nelson K.E., Brenner D.A., Fouts D.E., Schnabl B. Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal Enterococcus. Nat Commun. 2017;8:837. doi: 10.1038/s41467-017-00796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imhann F., Bonder M.J., Vila A.V., Fu J., Mujagic Z., Vork L., Tigchelaar E.F., Jankipersadsing S.A., Cenit M.C., Harmsen H.J.M., Dijkstra G., Franke L., Xavier R.J., Jonkers D., Wijmenga C., Weersma R.K., Zhernakova A. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y., Huang Y., Wang Y., Wang P., Song H., Wang F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS One. 2019 doi: 10.1371/journal.pone.0218384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holota Y., Dovbynchuk T., Kaji I., Vareniuk I., Dzyubenko N., Chervinska T., Zakordonets L., Stetska V., Ostapchenko L., Serhiychuk T., Tolstanova G. The long-term consequences of antibiotic therapy: role of colonic short-chain fatty acids (SCFA) system and intestinal barrier integrity. PLoS One. 2019;14 doi: 10.1371/journal.pone.0220642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W.Z., Strowig T., Thaiss C.A., Kau A.L., Eisenbarth S.C., Jurczak M.J., Camporez J.-P., Shulman G.I., Gordon J.I., Hoffman H.M., Flavell R.A. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy M., Shapiro H., Thaiss C.A., Elinav E. NLRP6: a multifaceted innate immune sensor. Trends in Immunology. 2017;38:248–260. doi: 10.1016/j.it.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Levy M., Kolodziejczyk A.A., Thaiss C.A., Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 26.Elinav E., Henao-Mejia J., Strowig T., Flavell R. NLRP6 and dysbiosis: avoiding the luring attraction of over-simplification. Immunity. 2018;48:603–604. doi: 10.1016/j.immuni.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Mossanen J.C., Krenkel O., Ergen C., Govaere O., Liepelt A., Puengel T., Heymann F., Kalthoff S., Lefebvre E., Eulberg D., Luedde T., Marx G., Strassburg C.P., Roskams T., Trautwein C., Tacke F. Chemokine (C-C motif) receptor 2–positive monocytes aggravate the early phase of acetaminophen-induced acute liver injury. Hepatology. 2016;64:1667–1682. doi: 10.1002/hep.28682. [DOI] [PubMed] [Google Scholar]
- 28.Huebener P., Pradere J.P., Hernandez C., Gwak G.Y., Caviglia J.M., Mu X., Loike J.D., Jenkins R.E., Antoine D.J., Schwabe R.F. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J Clin Invest. 2015;125:539–550. doi: 10.1172/JCI76887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krenkel O., Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17:306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 30.Francino M.P. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Frontiers in Microbiology. 2016 doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heidelbaugh J.J., Kim A.H., Walker P.C. Overutilization of proton-pump inhibitors: what the clinician needs to know. Therap Adv Gastroenterol. 2012;5:219–232. doi: 10.1177/1756283X12437358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlson M.E., Pompei P., Ales K.L., MacKenzie C.R. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 33.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B., Guiot Y., Derrien M., Muccioli G.G., Delzenne N.M., de Vos W.M., Cani P.D. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lluch J., Servant F., Païssé S., Valle C., Valière S., Kuchly C., Vilchez G., Donnadieu C., Courtney M., Burcelin R., Amar J., Bouchez O., Lelouvier B. The characterization of novel tissue microbiota using an optimized 16S metagenomic sequencing pipeline. PLoS One. 2015 doi: 10.1371/journal.pone.0142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lelouvier B., Servant F., Païssé S., Brunet A.C., Benyahya S., Serino M., Valle C., Ortiz M.R., Puig J., Courtney M., Federici M., Fernández-Real J.M., Burcelin R., Amar J. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: a pilot analysis. Hepatology. 2016;64:2015–2027. doi: 10.1002/hep.28829. [DOI] [PubMed] [Google Scholar]
- 37.Gong S., Lan T., Zeng L., Luo H., Yang X., Li N., Chen X., Liu Z., Li R., Win S., Liu S., Zhou H., Schnabl B., Jiang Y., Kaplowitz N., Chen P. Gut microbiota mediates diurnal variation of acetaminophen induced acute liver injury in mice. J Hepatol. 2018 doi: 10.1016/j.jhep.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon E., Babar A., Choudhary M., Kutner M., Pyrsopoulos N. Acetaminophen-induced hepatotoxicity: a comprehensive update. J Clin Transl Hepatol. 2016;4:131–142. doi: 10.14218/JCTH.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chun L.J., Tong M.J., Busuttil R.W., Hiatt J.R. Acetaminophen hepatotoxicity and acute liver failure. J Clin Gastroenterol. 2009;43:342–349. doi: 10.1097/MCG.0b013e31818a3854. [DOI] [PubMed] [Google Scholar]
- 40.Clark R., Fisher J.E., Sketris I.S., Johnston G.M. Population prevalence of high dose paracetamol in dispensed paracetamol/opioid prescription combinations: an observational study. BMC Clin Pharmacol. 2012;12:11. doi: 10.1186/1472-6904-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiødt F.V., Rochling F.A., Casey D.L., Lee W.M. Acetaminophen toxicity in an urban county hospital. N Engl J Med. 1997;337:1112–1117. doi: 10.1056/NEJM199710163371602. [DOI] [PubMed] [Google Scholar]
- 42.Antharam V.C., Li E.C., Ishmael A., Sharma A., Mai V., Rand K.H., Wang G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol. 2013;51:2884–2892. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brüssow H. Problems with the concept of gut microbiota dysbiosis. Microbial Biotechnology. 2020;13:423–434. doi: 10.1111/1751-7915.13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukui H. Role of gut dysbiosis in liver diseases: what have we learned so far? Diseases. 2019;7:58. doi: 10.3390/diseases7040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sookoian S., Salatino A., Castaño G.O., Landa M.S., Fijalkowky C., Garaycoechea M., Pirola C.J. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut. 2020;69:1483–1491. doi: 10.1136/gutjnl-2019-318811. [DOI] [PubMed] [Google Scholar]
- 46.Schneider K.M., Albers S., Trautwein C. Role of bile acids in the gut-liver axis. J Hepatol. 2018;68:1083–1085. doi: 10.1016/j.jhep.2017.11.025. [DOI] [PubMed] [Google Scholar]
- 47.Vital M., Rud T., Rath S., Pieper D.H., Schlüter D. Diversity of bacteria exhibiting bile acid-inducible 7α-dehydroxylation genes in the human gut. Comput Struct Biotechnol J. 2019;17:1016–1019. doi: 10.1016/j.csbj.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coelho I., Duarte N., Macedo M.P., Penha-Gonçalves C. Trem-2 modulation of gut microbiota is blunted during hepatotoxic injury and uncoupled from liver repair responses. bioRxiv. 2019:857078. [Google Scholar]
- 49.Plovier H., Everard A., Druart C., Depommier C., Van Hul M., Geurts L., Chilloux J., Ottman N., Duparc T., Lichtenstein L., Myridakis A., Delzenne N.M., Klievink J., Bhattacharjee A., van der Ark K.C.H., Aalvink S., Martinez L.O., Dumas M.-E., Maiter D., Loumaye A., Hermans M.P., Thissen J.-P., Belzer C., de Vos W.M., Cani P.D. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 50.Depommier C., Everard A., Druart C., Plovier H., Van Hul M., Vieira-Silva S., Falony G., Raes J., Maiter D., Delzenne N.M., de Barsy M., Loumaye A., Hermans M.P., Thissen J.P., de Vos W.M., Cani P.D. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claus S.P., Ellero S.L., Berger B., Krause L., Bruttin A., Molina J., Paris A., Want E.J., de Waziers I., Cloarec O., Richards S.E., Wang Y., Dumas M.E., Ross A., Rezzi S., Kochhar S., van Bladeren P., Lindon J.C., Holmes E., Nicholson J.K. Colonization-induced host-gut microbial metabolic interaction. mBio. 2011;2 doi: 10.1128/mBio.00271-10. e00271–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abt M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J., Wherry E.J., Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Jansen T., Jacobs L., Bonder M.J., Kurilshikov A., Fu J., Joosten L.A.B., Zhernakova A., Huttenhower C., Wijmenga C., Netea M.G., Xavier R.J. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;1674:1125–1136. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su G.L., Gong K.Q., Fan M.H., Kelley W.M., Hsieh J., Sun J.M., Hemmila M.R., Arbabi S., Remick D.G., Wang S.C. Lipopolysaccharide-binding protein modulates acetaminophen-induced liver injury in mice. Hepatology. 2005;41:187–195. doi: 10.1002/hep.20533. [DOI] [PubMed] [Google Scholar]
- 55.Possamai L.A., Mcphail M.J., Khamri W., Wu B., Concas D., Harrison M., Williams R., Cox R.D., Cox I.J., Anstee Q.M., Thursz M.R. The role of intestinal microbiota in murine models of acetaminophen-induced hepatotoxicity. Liver Int. 2015;35:764–773. doi: 10.1111/liv.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooper S.C., Aldridge R.C., Shah T., Webb K., Nightingale P., Paris S., Gunson B.K., Mutimer D.J., Neuberger J.M. Outcomes of liver transplantation for paracetamol (acetaminophen)-induced hepatic failure. Liver Transplant. 2009;15:1351–1357. doi: 10.1002/lt.21799. [DOI] [PubMed] [Google Scholar]
- 57.Kroy D.C., Schumacher F., Ramadori P., Hatting M., Bergheim I., Gassler N., Boekschoten M.V., Müller M., Streetz K.L., Trautwein C. Hepatocyte specific deletion of c-Met leads to the development of severe non-alcoholic steatohepatitis in mice. J Hepatol. 2014;61:883–890. doi: 10.1016/j.jhep.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 58.Hatting M., Zhao G., Schumacher F., Sellge G., Al Masaoudi M., Gaßler N., Boekschoten M., Müller M., Liedtke C., Cubero F.J., Trautwein C. Hepatocyte caspase-8 is an essential modulator of steatohepatitis in rodents. Hepatology. 2013;57:2189–2201. doi: 10.1002/hep.26271. [DOI] [PubMed] [Google Scholar]
- 59.Wai C.T., Greenson J.K., Fontana R.J., Kalbfleisch J.D., Marrero J.A., Conjeevaram H.S., Lok A.S.F. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003 doi: 10.1053/jhep.2003.50346. [DOI] [PubMed] [Google Scholar]
- 60.Johansson M.E.V., Hansson G.C. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol Biol. 2012;842:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- 61.Sezgin S., Hassan R., Zühlke S., Kuepfer L., Hengstler J.G., Spiteller M., Ghallab A. Spatio-temporal visualization of the distribution of acetaminophen as well as its metabolites and adducts in mouse livers by MALDI MSI. Arch Toxicol. 2018;92:2963–2977. doi: 10.1007/s00204-018-2271-3. [DOI] [PubMed] [Google Scholar]
- 62.Heymann F., Hammerich L., Storch D., Bartneck M., Huss S., Rüsseler V., Gassler N., Lira S.A., Luedde T., Trautwein C., Tacke F. Hepatic macrophage migration and differentiation critical for liver fibrosis is mediated by the chemokine receptor C-C motif chemokine receptor 8 in mice. Hepatology. 2012;55:898–909. doi: 10.1002/hep.24764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., Egholm M., Henrissat B., Heath A.C., Knight R., Gordon J.I. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pẽa A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rideout J.R., He Y., Navas-Molina J.A., Walters W.A., Ursell L.K., Gibbons S.M., Chase J., McDonald D., Gonzalez A., Robbins-Pianka A., Clemente J.C., Gilbert J.A., Huse S.M., Zhou H.-W., Knight R., Caporaso J.G. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Païssé S., Valle C., Servant F., Courtney M., Burcelin R., Amar J., Lelouvier B. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56:1138–1147. doi: 10.1111/trf.13477. [DOI] [PubMed] [Google Scholar]
- 68.Schierwagen R., Alvarez-Silva C., Servant F., Trebicka J., Lelouvier B., Arumugam M. Trust is good, control is better: technical considerations in blood microbiome analysis. Gut. 2019 doi: 10.1136/gutjnl-2019-319123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Escudié F., Auer L., Bernard M., Mariadassou M., Cauquil L., Vidal K., Maman S., Hernandez-Raquet G., Combes S., Pascal G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics. 2018;34:1287–1294. doi: 10.1093/bioinformatics/btx791. [DOI] [PubMed] [Google Scholar]
- 70.Nier A., Engstler A.J., Maier I.B., Bergheim I. Markers of intestinal permeability are already altered in early stages of non-alcoholic fatty liver disease: studies in children. PLoS One. 2017 doi: 10.1371/journal.pone.0183282. [DOI] [PMC free article] [PubMed] [Google Scholar]