Abstract
Despite showing a great promise in the field of nanomedicine, nanoparticles have gained a significant attention from regulatory agencies regarding their possible adverse health effects upon environmental exposure. Whether those nanoparticles are generated through intentional or unintentional means, the constant exposure to nanomaterials can inevitably lead to unintended consequences based on epidemiological data, yet the current understanding of nanotoxicity is insufficient relative to the rate of their emission in the environment and the lack of predictive platforms that mimic the human physiology. This calls for a development of more physiologically relevant models, which permit the comprehensive and systematic examination of toxic properties of nanoparticles. With the advancement in microfabrication techniques, scientists have shifted their focus on the development of an engineered system that acts as an intermediate between a well-plate system and animal models, known as organ-on-a-chips. The ability of organ-on-a-chip models to recapitulate in vivo like microenvironment and responses offers a new avenue for nanotoxicological research. In this review, we aim to provide overview of assessing potential risks of nanoparticle exposure using organ-on-a-chip systems and their potential to delineate biological mechanisms of epidemiological findings.
Keywords: Organ-on-a-chip, Toxicity, Nanoparticles
Graphical abstract
Highlights
-
•
The current understanding of nanotoxicity is insufficient relative to the rate of their emission in the environment and the lack of predictive platforms that mimic the human physiology.
-
•
This calls for a development of more physiologically relevant models, which permit the comprehensive and systematic examination of toxic properties of nanoparticles.
-
•
With the advancement in microfabrication techniques, scientists have shifted focused to develop an engineered system that act as an intermediate between well-plate system and animal models, known as organ-on-a-chips.
-
•
Organ-on-a-chip systems offer the potential to delineate biological mechanisms of epidemiological findings.
1. Introduction
Environmental protection agencies and regulators are facing unprecedented challenges in assessing the potential toxicity of nanoparticles. With a rapid pace of economic development in the past few decades along with increase in population, it is not a surprise that humans have generated intentional and unintentional artificial byproducts which are capable of facilitating health hazards to humans. Ultrafine particles, one of the subsets of air pollution, are the major environmental risk factors contributing to pathogenesis of pulmonary [1] and cardiovascular disease [2,3], increased hospitalization [4], and even premature death [5]. World Health Organization estimates that more than 7 million deaths across the globe have been caused by air pollution exposure, making it the fourth greatest risk of death [6]. Nanoplastics have also emerged as a new class of materials which may possess health risks to humans [7,8]. With increase in plastic production and with the lack of global plastic disposal capability, a large amount of plastics is being introduced into the natural environment, which naturally increases exposure to humans through various means. While the presence of nanoparticles is becoming common, the current understanding of nanoparticle toxicity is relatively insufficient despite increasing human exposure. As a matter of fact, approximately 95% of publications in the field of nanotechnology have been aimed at exposing advantages of nanomaterials [9], and remaining 5% of the literature addresses their toxicity. With the scarcity in nanotoxicological data along with the increase in public awareness of potential toxicity of nanomaterials, there has been a surge in the literature that uses pre-existing models to understand the effects of nanoparticles on humans at the cellular, organ, and systemic levels.
Up until now, approaches for evaluating nanoparticle toxicity have mainly involved using pre-existing in vivo animal models to understand the biodistribution of nanoparticles and their subsequent systemic responses [[10], [11], [12], [13]], complemented with utilizing 2 dimensional (2D) in vitro system to understand the effects of nanoparticles at the cellular level [14]. Researchers have identified a wide array of effects which are believed to be associated with nanoparticle exposure in different biological cell lines, predominantly through mechanisms involving oxidative stress-mediated pathways [14,15]. While these scientific data, been generated from conventional systems are relatively robust, reproducible, easy to analyze, and more suitable for high-throughput toxicity screening given the vast variations in nanoparticle size, charge, shape, and composition, it is questionable if those methods can provide insightful interpretation to regulators on the potential nano-toxic effects on human tissues and organs. Unlike our native human organs, traditional 2D cell culture methods still lack the complex three-dimensional (3D) cell-cell and cell-matrix interactions, and they are restricted to the use of immortalized human cell lines that do not exhibit phenotypes similar to the human primary cells [16,17], resulting in inaccuracy and discrepancy in the prediction of toxicity. Although animal models are useful in predicting nanoparticle toxicity at the systemic level, it is notably challenging to extract the precise molecular mechanisms due to the discrepancy in physiological responses between animal models and humans [[18], [19], [20]]. Thus, to extend the current knowledge of nano-toxic effects based on epidemiological and clinical work, it is becoming apparent that there is an urgent need to develop a complementary experimental approach that can reconstitute complex organ-level physiological functions relying on clinically relevant cell sources, in order to permit for a more accurate prediction of the complex responses in in vivo settings while simultaneously addressing shortcomings of a 2D systems.
Organ-on-a-chip systems offer an alternative approach for nanoparticle toxicity screening that can fill the gap between the conventional pre-clinical models, specifically 2D cell culture and animal models, and human population studies. The inherent advantage of using organ-on-a-chip systems is their ability to reverse engineer native microenvironment (extracellular matrix [21], geometry [22,23], mechanical stiffness [24,25], and flow [26,27]) and responses by generating high fidelity models of human tissues and organs, which can potentially provide the new and exciting opportunity to systemically dissect nanoparticle toxicity in models with human physiology using established detection assays. This complementary experimental approach represents a major advance from traditional 2D culture systems and is readily applicable to the development of models of various organs in the body including vasculature [28], lungs [29], heart [25], gastrointestinal tract [30,31], and brain [32,33]. Although advancing diverse applications, there is still a significant lag in using organ-on-a-chip systems to understand biological mechanisms attributed to the nanoparticle environmental exposure and majority of studies remain in the proof-of-principle stage. The slow process is partly attributed to the lack of standardization in nanoparticle dosage, characterization, and analysis, all of which prevent true comparisons between results from one system to the other. In this review, we aim to provide an overview of the literature focusing on the use of organ-on-a-chip systems to analyze nanoparticle induced toxicity. We will also introduce some representative organ-on-a-chip systems as well as other engineered models which have potential to be used for studies of environmental nanotoxicity in the future. Finally, we will discuss limitations of the current system and provide general guidelines for the engineering design requirements which will assist us in translating biological plausibility of epidemiological findings.
2. Sources of nanoparticles
2.1. Ultrafine and fine particulates
Accumulating evidence indicates that air pollution contributes to serious, even fatal damage to human health and development [2,[34], [35], [36], [37]]. With increase in pace of economic growth over the past decades, thousands of artificial-sourced chemicals have entered our air, reaching our bodies through inhalation. Air pollution, which is commonly derived from fuel combustion [38] and road traffic [39,40], is a mixture of gas and microscopic particulates that has been implicated in a wide range of adverse health effects (Fig. 1A). Metal particles including iron, copper, zinc, and aluminum are commonly emitted from exhaust stream of diesel engines [39], power stations [41], or inside the subway stations [42], exhibiting much higher toxicity compared to the non-metal based nanoparticles, as they cause higher cytotoxic effects through generation of reactive oxygen species and lipid peroxidation [14]. Inhalation of construction dust or coal mine dust, which is primarily composed of SiO2 nanoparticles, may also cause the development of symptoms such as pulmonary fibrosis, pneumoconiosis, and lung dysfunction [43,44], yet the mechanisms by which particulate matter instigates adverse health effects is less known due to its heterogenous combination of compounds of varying concentration, size, chemical composition, surface area, and origin.
Fig. 1.
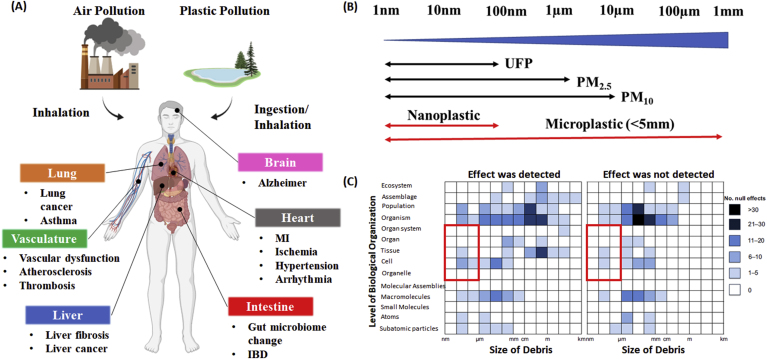
Presence of nano and micro particles in the environment and their possible effects on human health. (A) Routes of exposure, distribution, and the subsequent effects of air pollution and plastic pollution on human health outcome. (B) Size comparisons for air pollution particulate matter and plastic pollution. Microplastic can undergo continuous fragmentation to form nanoplastic. (C) The matrix representing the number of impacts of pollution debris in peer reviewed literature. Red squared region represents lack of toxicological studies in biological settings on the nano-meter scale. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Reproduced with permission [89]. Copyright 2020, John Wiley and Sons.
The release of nanoparticles into the environment is not limited to an artificial source. Many engineered materials have found a niche in the consumer market, and it is expected these novel materials will likely become a mainstream in a few years. Carbon nanotubes and graphene, for example, exhibit excellent electrical and mechanical characteristics which have potential to be used in a structural composite and sensing applications. Fullerene nanomaterials have been applied in the cosmetic products owning to their antioxidant potential. While these engineered materials are versatile and promising, as large-scale production and applications of these nanomaterial increase, the general population and manufacturing workers are likely to be exposed to the nanoparticles which may pose health concerns due to their small size. A study of the National Institute of Occupational Safety and Health on carbon nanotubes demonstrated that fine particles of respirable size ranging from 10 nm to 1000 nm could be generated from the bulk, increasing the risk of unintentional occupational or environmental exposure [45]. Based on the data from acute toxicity studies, no conclusions on carcinogenic or genotoxic effects to humans can be made. It is expected that the risk of chronic inhalation and dermal exposure to engineered nanomaterials can be expected when exposure is not controlled. Indeed, animal inhalation studies demonstrate toxic nature of carbon nanotubes and graphene, as marked by the inflammatory responses, fibrotic reactions, and formation of granulomas, which raises a possible concern about human exposure risks related to these materials [46,47].
The size of particles is the primary factor that is linked to their ability to cause health problems, with smaller particles posing the greatest risk [[48], [49], [50]]. Air pollution particulates are traditionally categorized into three different groups based on their aerodynamic diameter, namely, (i) particulates less than or equal to 10 μm in diameter, referred to as particulate matter 10 (PM10), (ii) a subset of PM10 with particulates less than or equal to 2.5 μm, referred to as PM2.5, and (iii) particulates less than 100 nm in diameter, referred to as ultrafine particles (UFP) (Fig. 1B). The World Health Organization provides air quality guideline for PM2.5 to be 10 μg/m3 for an annual average concentration and 25 μg/m3 daily mean. This is based on a large population long-term studies in which mortality rate at the annual mean concentration in the range of 11–15 μg/m3 consistently showed an adverse health impact on humans [51]. While there is a large body of scientific evidence suggesting the potential health impact of PM2.5, much of what we know about the consequence of UFP exposure still relies on the use of animal studies and World Health Organization has not released air quality guidelines regarding the concentration at which adverse health effects have been demonstrated due to a paucity of information on the long-term effects of UFP and the lack of solid experimentation framework. However, several epidemiological studies suggest UFP concentration above 10,000 particles/cm3 induces significant health impact to humans [3,52,53]. For instance, Dutch studies found that long-term exposure to UFP was associated with an increased risk of cardiovascular disease, myocardial infarction, and heart failure at the annual predicted concentration of 11,110 particles/cm3 [3]. Once inhaled, majority of PM10 tend to accumulate in the bronchi of the lungs, however, smaller particles including PM2.5 and UFP deposit at the alveoli, the region that is responsible for gas exchange between lungs and blood stream [54]. The deposited particles can then cause significant burden on the cells through mechanisms involving oxidative stress or pulmonary inflammation. For example, intratracheal instillation of 20 nm TiO2 nanoparticles in mice induces the upregulation of interleukin (IL)-1, tumor necrosis (TNF)-α, and IL-6 [55], which are cytokines that are commonly found in chronic obstructive pulmonary disease patients [56]. Another major concern regarding the UFP is its high number of particles per given volume. Given the small size, it is intuitive that the number of particles per given volume is much higher than the number of particles of micro-meter size due to its high surface to volume ratio. As a matter of fact, it is estimated that 90% of the total number of particulate matter is below 300 nm, and 70% of the particles are in the range of nano-meter size (100 nm), highlighting toxicological concerns when these particles interact with cells upon inhalation [57]. UFP-mediated pulmonary complications are further aggravated by the duration of nanoparticle residence in the lungs. As nanoparticles are sometimes left unrecognized from alveolar macrophages for more than 100 days [58], the concerns regarding the long-term cumulative pulmonary complications require more scientific investigation.
The toxic effect of UPF is not limited to the lungs. Recent controlled human study reveals that at least 0.2% of inhaled gold nanoparticles are capable of translocating from the lungs and gaining access in the circulation [59]. Crossing of air-blood barrier allows nanoparticles to distribute through the secondary organs including vasculature [59], heart [12,60], liver [10,11], and kidney [10,11]. Given the nanoparticle biodistribution data in animal studies, it is not a surprise that clinical and epidemiological evidence suggests the correlation of UFP exposure and pathogenesis of extrapulmonary organs. Both acute and chronic exposure to UFP has been implicated with exacerbation of cardiovascular diseases including ischemic heart disease [61], vascular dysfunction [62], thrombosis [63], hypertension [64], and arrhythmia [65]. Air pollution may also gain access to the gut, causing the change of the composition of the gut's resident microbial population [66,67]. Besides an increasing incidence of gut and cardiovascular dysfunction, researchers also started to raise the alarm about possible links to the brain function [68], kidneys [69,70], and liver [71], yet the biological mechanisms that account for the extrapulmonary dysfunction remain largely unclear. A summary of air pollution-based nanoparticle induced toxicity featuring both in vitro and in vivo studies is shown in Table 1.
Table 1.
Effect of nanoparticles on modulating cellular and organ functions.
| Findings | Nanoparticle | Model | Results | References |
|---|---|---|---|---|
| Nanoparticle Translocation | 20–29 nm Carbon Black | Rats Inhalation |
|
[100] |
| 3.8 nm Gold nanoparticles | Human Inhalation |
|
[59] | |
| 10–250 nm Gold nanoparticles | Rats Intravenous |
|
[60] | |
| 200 nm PS particles | Mice Inhalation |
|
[101] | |
| 100–3000 nm PS particles | Rats Gavage |
|
[102] | |
| Lung Injury | UFP | Mice Inhalation |
|
[103] |
| 20 nm Nickel Oxide | Rats Intratracheal instillation |
|
[104] | |
| 46.5 nm CuO nanoparticles | Mice Intranasal |
|
[105] | |
| ~30 nm Polyacrylate nanoparticles | Human |
|
[106] | |
| 15- and 46 nm SiO2 nanoparticles | A549 |
|
[107] | |
| 10- and 50 nm SiO2 nanoparticles | BEAS-2B HBEC3-KT |
|
[108] | |
| Findings | Nanoparticle | Model | Results | References |
| Vascular Dysfunction | 14 nm Carbon black nanoparticles | HUVEC Aorta Segments |
|
[109] |
| 100 nm TiO2 nanoparticles | Rats Inhalation |
|
[110] | |
| UFP <180 nm | Mice Inhalation |
|
[111] | |
| 22 nm Fe2O3 | HAEC U937 Cells |
|
[112] | |
| 23.5 nm TiO2 nanoparticles | HMVEC |
|
[113] | |
| Various nanoparticles | HUVEC |
|
[16] | |
| Heart Injury | 38 nm TiO2 nanoparticles | Rats Intratracheal Instillation |
|
[65] |
| 62 nm SiO2 nanoparticles | Zebrafish |
|
[114] | |
| 7- and 670 nm SiO2 nanoparticles | Isolated rat cardiomyocytes |
|
[115] | |
| 60 nm SiO2 nanoparticles | H9C2 Rat Cardiomyocytes |
|
[116] | |
| 25–35 nm TiO2 nanoparticles | Rats Intratracheal Instillation |
|
[117] | |
| TiO2 SiO2 | Guinea Pig Langendorff Heart |
|
[118] | |
| Findings | Nanoparticle | Model | Results | References |
| Gut Damage | UFP | Mice Oral Administration |
|
[67] |
| PM2.5 | Mice Gavage |
|
[119] | |
| 50 nm carboxylated PS nanoparticles | Caco2/HT29-MTX Chicken Oral Administration |
|
[86] | |
| 500 nm PS particles | Mice Oral Administration |
|
[83] | |
| 44- and 100 nm PS nanoparticles | Human gastric adenocarcinoma cells |
|
[87] | |
| Liver Injury | PM2.5 | Murine Inhalation |
|
[120] |
| PM2.5 | Mice Inhalation |
|
[121] | |
| 80 nm Cu nanoparticles | Rats Oral Administration |
|
[122] | |
| TiO2 nanoparticles | Mice Intragastric Administration |
|
[123] |
2.2. Nanoplastics
Plastic particles have emerged as a new class of material shown to facilitate human health hazards. The plastic is thought to be inert, with highly diverse and desirable properties including the anti-corrosion, chemical inertness, electrically insulating properties, as well as the low production cost [72]. Those attractive properties of plastics lead to the increase in global production, from 2 million metric tons in 1950 to 380 million tons in 2015, and it is expected to continue growing exponentially in next 30 years [73]. Despite the plastics becoming essential material in our life, it is also the source of the worst artificially made environmental disaster of our time. Up until now, it is estimated that only 9% of all produced plastics has been recycled and two-third of all plastics ever produced has been released into the environment in the form of debris, plastic particles or microfibers [73]. Because of the intrinsic long-term stability of plastic materials in parallel with the lack of plastic disposal capabilities, there has been a global accumulation of plastics in the environment, which will be naturally introduced into human body through air, dust, drinking water, and consumption of food.
The source of plastic particles is primarily derived from the pre-existing larger plastic objects, which are commonly manufactured from polystyrene (PS), polyethylene (PE), polypropylene (PP), polyvinylchloride (PVC), polyethylene terephthalate (PET), and polyurethane resins (PUR). These objects then experience degradation and fragmentation process by UV-photodegradation, oxidation, and mechanical and hydrolytic degradation. The presence of microplastics, particles less than or equal to 5 μm, has been reported all around the globe including polar regions [74,75], rain [76,77], and deep sea [78]. Studies also raise the possible concerns regarding the presence of microplastics in drinking water, as 93% of commercially available bottled water contains 10.4 particles/L on average [79]. Recentl studies confirm that the degradation process of plastics is not limited to the micro-meter level, and microplastic particles continue to degrade to form nanoplastics with size less than 100 nm (Fig. 1C). Based on the current evidence, ingestion of nanoplastic particles is likely to represent the primary port of entry due to their presence in sea food and drinking water. For example, based on nanoparticle tracking analysis, Lambert et al. reveals a commonly used PS disposable coffee cup releases 1.26 × 108 particles/ml of nano-meter size particles with the average particle diameter of 224 nm, under constant UV exposure for 56 days [80]. Ingested or inhaled nanoplastics can cross gut/lung epithelial barrier and potentially penetrate deeper into the systemic circulation. For example, Jani et al. uses PS particles size ranging from 50 nm to 3 μm to model plastics translocation through gastrointestinal track in mice, with particle size between 50 nm and 100 nm are found in the liver, blood, and bone marrow [81]. The translocation of nanoplastic particles has also been demonstrated in inhalation studies in which 200 nm fluorescent PS latex particles were observed in the kidney and liver of mice [82]. It appears that concerns regarding acute and chronic effects of nanoplastics that can lead to significant health impact on humans are not well-documented. Animal models have provided some clear illustration of the microplastic/nanoplastic induced gut toxicity, which include a reduction in mucus secretion [83], gut barrier dysfunction [84], intestinal inflammation [85], iron absorption effects [86], and gut microbiota dysbiosis [[83], [84], [85]] in a concentration range of 1.5 × 103~1.5 × 104 particles/cm3. While the concentration in those models could be too high for the prediction of human exposure, it is expected that humans could accumulate nanoparticles in the gut from different food sources, which eventually reach the threshold concentration that can instigate significant mortality and morbidity.
Several in vitro studies have also investigated the adverse effects of nanoplastics, yet there are some conflicting results. For example, Forte et al. uses gastric adenocarcinoma cells to show 44 nm PS nanoparticles strongly induce the upregulation of IL-1β, IL-6, and IL-8 compared to 100 nm PS nanoparticles, while no significant variation of the level of genes involved in proliferation is observed [87]. Yet, Cortes et al. show plastic nanoparticles cause little or no effects in term of cytotoxicity, genotoxicity, and oxidative stress despite the internalization in human colon adenocarcinoma [88]. Based on a meta-analysis published by Rochman and co-workers, authors caution the current evidence of plastic toxicity remains split between detected and non-detected results [89] (Fig. 1C). The primary concern associated with plastic toxicity is the lack of analytic methods to characterize nanoplastics. Unlike UFP in which the composition of nanoparticles can be characterized using elemental composition analysis, the conventional instruments are unable to identify overall chemical composition and specific concentration of plastics particles effectively at present, since they cannot be sieved easily and separated from other naturally occurring micro and nanoplastics [90]. In many microplastic studies, visual examination of the isolated microplastic sample is necessary to sort the plastics from the other materials, which is done by the direct examination of the samples with the aid of a microscope [[91], [92], [93]]. However, visual isolation of individual nanoplastics is virtually unfeasible due to their small nature. It is possible to classify nanoplastics based on their sizes through various techniques such as ultrafiltration [[94], [95], [96]], ultracentrifugation [97], and field flow fraction [94], however the methodologies are still lacking for the isolation of specific nanoplastics from the environmental samples [7,98,99]. Since nanoplastics are mostly carbon-based materials they require a complete identification of particle composition in terms of the polymer type and molecular structure instead of an elemental analysis to obtain complete profile of the environmental sample. This illustrates that for the analysis of sub-micron and nanoplastics we are facing a major methodology gap to develop a solid experimental framework to characterize the toxicity of nanoplastics to humans, and a new approach in the analytical methodology must be taken coupled with a reliable method to classify mixture of nanoplastics. Due to the lack of realistic concentration data available from the natural environment, the nanoplastic toxicity experiment is usually not carried out at the concentration that simulates human exposure settings. As nanoparticle toxicity varies depending on the size, surface charge and material composition, the extrapolation of findings from solely PS materials should also be made with caution, and our understanding of the health effects of nanoplastics are likely just the tip of an iceberg.
3. Organ-on-a-chip platforms for assessment of nanoparticle toxicity
3.1. Endothelium-on-a-chip
As nanoparticles gain access to the blood circulation through lung or intestinal barriers, nanoparticles are first encountered by the layer of endothelial cells. There is a plethora of evidence that supports the detrimental effects of nanoparticles on vascular function in both animals and humans, as both acute and chronic exposure of particulate air pollution is associated with the progression of vascular dysfunction [124]. The generation of reactive oxygen species (ROS) and upregulation of pro-inflammatory cytokines following air pollution exposure is implicated in reduction of vascular nitric oxide (NO) levels [125], which is a soluble gas that maintains vascular homeostasis. The decreased production of NO under pathological conditions can lead to several abnormalities in blood vessel function including activation of platelets and increased stimulation of inflammation in blood vessels, thus promoting atherosclerosis [126]. The effect of nanoparticles on endothelial cells has been extensively studied in vitro. Treatment of endothelial cells with nanoparticles, such as TiO2 and SiO2 nanoparticles, results in the change in cytoskeletal structures, leading to disruption of endothelial junction proteins and increase in vascular permeability [16,113,127]. Nanoparticle exposure also stimulates monocytic cell adhesion on human aortic endothelial cells through upregulation of intracellular cell adhesion molecule-1 (ICAM-1) and IL-8, which are considered as early signs of atherosclerosis [112].
Most recently, Ewejeie et al. developed a minimalistic model in which 2 endothelial cells are seeded on micro-patterned substrate to systematically quantify and compare the effect of 10 different nanomaterials in terms of cellular viability, cytoskeletal structural changes, internuclear separation, nuclear eccentricity, and junction protein expression using a “similarity scoring” method [128]. While 2D systems, such as this one, allow us to gain basic understanding of toxic effects of nanomaterials, it is becoming evident that static culture systems may not be suitable for evaluation of nanoparticle toxicity as they do not reflect the recognized effect of fluid flow on endothelial cells and nanoparticles. To fill this gap, microfluidic models featuring endothelial cell coated vascular lumens have been engineered as an alternative in vitro platform to capture complex progression of vascular dysfunction upon nanoparticle exposure.
The classical approach is to create a single micro-channel from biocompatible polydimethylsiloxane (PDMS) via replica molding technique. The PDMS containing a channel on the order of tens of micrometers is then bonded to a substrate such as glass, or another piece of PDMS, to form “vascular” like lumen or an engineered blood vessel by culturing endothelial cells. Variety of perfusable platforms have been created to understand the biological mechanisms of interest, including utilizing 3D printing techniques [129], 3D stamping technique [130,131], or manual extrusion methods [23]. Following the fabrication of the devices, an extracellular matrix protein (ECM) is typically coated to the inner surface of the microfluidic channel to promote the adhesion and differentiation of endothelial cells. Most commonly, naturally derived materials including collagen [132], fibronectin [22,26,133,134], gelatin [131], and laminin [28] have been extensively used to support endothelial cell attachment. Other strategies featuring chemical modification with type 1 collagen nanofibers have also been reported to improve the growth of engineered micro-vessels [21]. After 3–7 days of cellular proliferation, endothelial cells eventually form a uniform monolayer across the inner lumen of the microfluidic channel, at which point, the devices are ready for further experimentation. The proper endothelial attachment and growth on the ECM is important to understand overall mechanisms governing barrier function and molecular mechanisms regulating cellular toxicity upon nanoparticle treatment, as minute discontinuity can drastically influence subsequent functional readouts including vascular permeability and examination of adherens/tight junction integrity.
Using microfluidic systems, one of the most extensively studied topics is the role of fluid flow as it provides an important benchmark for understanding the crucial role of shear stress in regulating the uptake of nanoparticles by endothelial cells and their subsequent toxicity (Table 2). Unlike traditional drug screening methods in which chemical/drug concentration in the cell culture media is assumed to be well-dispersed, lack of flow causes nanoparticles to sediment on the cell surface, thus increasing nanoparticle uptake [135]. The nanoparticle sedimentation is resolved by the introduction of continuous perfusion, allowing nanoparticles to disperse homogenously in the cell culture media. Many studies featuring microfluidic systems have been published to investigate the effect of nanoparticle interaction, uptake and translocation under continuous perfusion, yet there are some conflicting results.
Table 2.
Organ-on-a-chip system for studies of nanoparticle induced toxicity.
| Model | Nanoparticle | Device Materials | Cell Type | Key Findings/Significance | References |
|---|---|---|---|---|---|
| Endo-thelium | 100 nm Au nanoparticles | Ibidi polymer coverslip | HUVEC |
|
[27] |
| 40–60 nm inhaled atmospheric nanoparticles (ANP) | PDMS | HUVEC/human pulmonary fibroblast |
|
[159] | |
| 6.5 nm Au nanoparticles | PDMS | HUVEC |
|
[26] | |
| Lung | 12 nm Silica nanoparticle | PDMS | Human alveolar epithelial cells/microvascular endothelial cells |
|
[29] |
| Cigarette Smoke | PDMS | Primary human small airway epithelial cells |
|
[183] | |
| Carbon nanotubes | PDMS | BEAS-2B lung epithelial cells |
|
[186] | |
| Heart | 20 nm TiO2 nanoparticles | Gelatin/PDA + PCL nanofiber | Neonatal rat ventricular myocyte |
|
[208] |
| 50 nm CuO and SiO2 nanoparticles | PDMS | HUVEC/iPSC-derived cardiomyocytes |
|
[214] | |
| Gut | 50 nm carboxylated PS nanoparticles | Silicon/plexiglass | Caco-2/HT29-MTX + HepG2/C3A |
|
[236] |
| Liver | 10 nm Fe3O4 nanoparticles | PDMS/Glass | Rat hepatocyte |
|
[235] |
For example, Fede et al. have demonstrated the addition of gold nanoparticles into the microfluidic channel coated with human umbilical vein endothelial cells (HUVEC) significantly reduced sedimentation of nanoparticles onto the cells which reduced cytotoxicity and nanoparticle accumulation in the cytoplasm compared to the level measured in a static platform. This suggests that the presence of flow plays pivotal role in translating actual toxicity in human body [26]. Yet, Park et al. argues that the continuous flow of cell culture media with constant level of nanoparticles delivers a higher dose of nanoparticles to the cells due to the continuous replenishment of nanoparticles, which in turn induces more cytotoxicity by accelerating G2 cell cycle arrest and subsequent apoptosis [136]. The discrepancy of these results may be attributed to the lack of uniformity and standardization in nanoparticle treatment methods. As majority of literature reports nanoparticle dose concentration in the unit of mass of nanoparticles over the volume of cell culture media, the number of nanoparticles that are applied to the cells significantly varies if the volume of cell culture media used in experiment is different. Therefore, one must treat and compare toxicity results from different studies with caution.
It is important to note that vascular microfluidic models should also take into account physiological stress that the endothelial cells are experiencing in vivo. Endothelial cells that line the luminal surface of the entire vascular system are constantly exposed to various level of shear stress ranging between 10 and 70 dyn/cm2 in the arteries, 5–20 dyn/cm2 in the microvasculature, and 1–6 dyn/cm2 in the veins [137]. To illustrate the role of shear stress on nanoparticle uptake, Chen et al. prepared single channel microfluidic device cultured with HUVEC at varying flow rates (1.5, 3, 5 ml/min). They found that gold nanoparticle uptake is flow rate dependent, with increased flow rates leading to a decreased nanoparticle uptake (Fig. 2A) [27]. Shear stress not only plays a critical role in governing the uptake of nanoparticles by endothelial cells and their subsequent toxicity [27,138,139], but also endothelial cells are known to perceive such hemodynamic stress as a potential mechanical signal, therefore changing cellular morphology [[140], [141], [142]], expression of pro-inflammatory genes [143], actin alignment [141,144], and the secretion of endocrine factors [142].
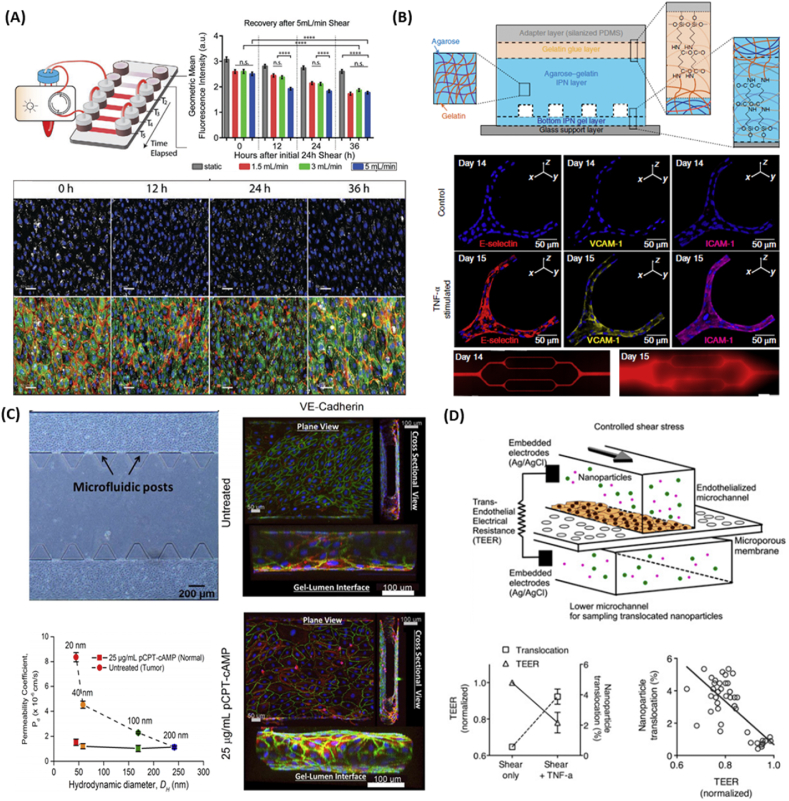
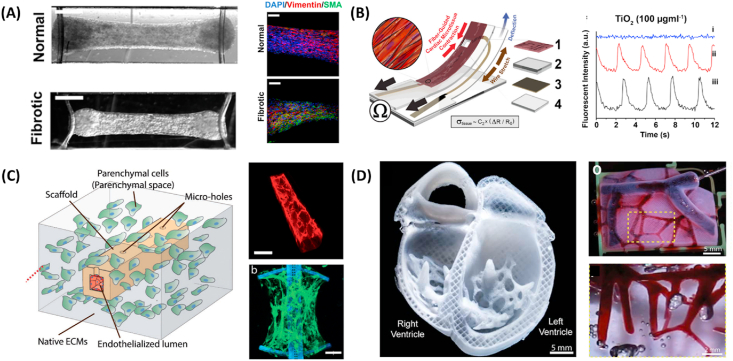
Fig. 2.
Development of microfluidic devices to understand nanoparticle-induced vascular dysfunction. (A) A single microfluidic device connected to a peristatic pump for controlled perfusion (top left). Increased shear stress reduces nanoparticle uptake (top right). Fluorescent images of nanoparticles (white) revealed that nanoparticle cellular uptake stabilized after 36 h of exposure at a flow rate of 5 ml/min (bottom). Reproduced with permission [27]. Copyright 2020, John Wiley and Sons. (B) Hydrogel based microvasculature-on-a-chip system for investigation of different modes of endothelial barrier dysfunction (top). Upon TNF-α stimulation, adhesion molecules such as E-selectin, VCAM-1, and ICAM-1 were upregulated (middle), which increased the permeability of engineered microvasculature as indicated by leakage of BSA-AF594 (bottom). Reproduced with permission [28]. Copyright 2018, Springer Nature. (C) Microfluidic device featuring microfluidic posts to permit the diffusion of nanoparticles (top left). pCRT-cAMP was added to reduce vascular paracellular permeability by promoting expression of adherens junctions (right) to model normal vasculature. Upon exposure of fluorescent PS nanoparticles, the permeability of nanoparticles across untreated leaky vasculature decreased with increased particle size, while permeability did not change for pCRT-cAMP treated vasculature [150]. Copyright 2017, Springer Nature. (D) A microfluidic device integrated TEER sensor to probe nanoparticle translocation (top). TNF-α treatment caused decrease in vascular barrier function as marked by decrease in TEER, which in turn increased nanoparticle translocation (bottom left). TEER measurement and nanoparticle translocation is inversely correlated (bottom right).
Reproduced with permission [114].
While there is a growing number of papers demonstrating the role of shear stress on endothelial cell growth and subsequent nanoparticle-induced toxicity, it is important to mention endothelial cells also respond to their biophysical environment which modulates cell-nanoparticle interactions. Many microfluidic devices in biological settings feature linear rectangular channel, yet it is undeniable that the use of non-circular channel poorly mimics the shape of the native blood vessesl. Non-circular surfaces or tortuous vessels may limit the cell growth by exposing cells to sharp corners and non-uniform shear stress, which can influence their cytoskeletal alignment, differentiation, and gene expression [145]. Using a PDMS microfluidic device featuring circular channel, Mannino et al. demonstrated the expression of VCAM-1 correlated with shear stress, dictated by the differences in vascular geometry, with higher VCAM-1 consistently upregulated in areas of lower shear stress, as demonstrated in the vascular geometry that corresponds to aneurysm [133]. As the number of nanoparticle interactions with the cells is inversely correlated with shear stress, low shear stress at the rectangular edges of the microfluidic device or disturbance of flow at the bifurcation regions may lead to the increased nanoparticle collision, which may result in higher nanoparticle accumulation and their subsequent toxicity compared to the regions with higher shear stress [146]. Since these local shear stresses play a pivotal role in governing functional phenotype and gene expression in endothelial cells, the extent to which experiments replicate geometries of the physiological settings must be considered for nanotoxicity studies.
In conjunction with shear stress, another key parameter to consider for nanotoxicity assessment in the vasculature is the type of endothelial cells as cells in different tissues vary greatly in their morphology and vascular permeability in accordance to their specific roles in the tissue [147]. For example, the vasculature in the brain forms a strong barrier to protect brain from exposure to potentially toxic substances, whereas endothelial cells in liver display specialized nanoscopic fenestrations on the cell membrane to allow rapid molecular exchange. To highlight how different endothelial cells have different sensitivity towards nanoparticle exposure, Setyawati and his co-workers applied gold nanoparticles within the range of 10 nm to human mammary endothelial cells (HMMEC), human skin microvascular endothelial cells (HMVEC), and HUVEC under static condition. They have found that both HMMEC and HMVEC were sensitive to gold nanoparticles as illustrated by the increase in permeability and disruption of VE-cadherin, while HUVEC were insensitive to those changes [148]. The use of HUVEC to recreate endothelial barrier for nanotoxicity research may cause measurement variability throughout the studies due to their unrealistically high barrier function, therefore it is important to select endothelial cells in accordance with the specific study goals.
Overall, studies on the progression of vascular dysfunction in vivo often suffer from the lack of physiological readout or rely on invasive procedures. Microfluidic platforms, on the other hand, not only allow for the fine tuning of geometry and shear stress, but they also enable the real-time monitoring of vascular function. With advancement in microfluidic techniques, modified microfluidic systems have been developed to quantitatively study the change in endothelial cell permeability by analyzing transport of fluorescently-tagged proteins. As microfluidic platforms featuring PDMS material exhibit low permeability towards fluorescently-tagged proteins, microfluidic channel platform in gels [149] or vascular lumen featuring sub-micron holes have been utilized to allow fluorescently-tagged proteins to permeate [131,150], enabling quantitative measurement of endothelial barrier function based on the protein transport. For instance, Qui et al. developed a microvasculature-on-a-chip featuring an interpenetrating-polymer-network hydrogel composed of agarose and gelatin to investigate different modes of vascular dysfunction [28]. Upon 10 μM TNF-α exposure, it not only stimulated upregulation of adhesion molecules including E-selectin, VCAM-1, and ICAM-1, but it also resulted in approximately 20-fold increase in permeability based on BSA-AF594 diffusion from vascular lumen to the hydrogel (Fig. 2B). In another study by Ho et al., HUVEC were grown in a microfluidic device to investigate extravasation rate of PS nanoparticles between healthy and tumor-like vasculature [150]. As the size of PS nanoparticles increased, the permeability coefficient decreased in tumor vasculature, while the permeability remained unchanged in healthy vasculature, demonstrating tumor selectivity for smaller nanoparticles (Fig. 2C).
Alternatively, the implementation of transepithelial/transendothelial electrical resistance (TEER) sensors into the microfluidic system also permits the quantification of barrier integrity in real time by measuring electrical resistance across a cellular monolayer. Using TEER sensor integrated microfluidic system, Kim et al. applied TNF-α in such a way to recreate a vessel that closely mimics vascular permeability observed in in vivo atherosclerotic vessels and studies the nanoparticle transport behavior across the engineered atherosclerotic vessel (Fig. 2D) [151]. Treatment with TNF-α resulted in disrupted intercellular junctions and increased endothelial permeability, permitting nanoparticles to travel across vascular barrier. These findings are consistent with in vivo rabbit atherosclerosis models, suggesting that this blood vessel on a chip could provide a useful tool to examine nanoparticle translocation form the microvessels.
The microfluidic systems have also been used to study the different aspects of blood vessel physiologies. A successful engineered blood vessel device should not only be able to capture in vivo physiological characteristics, but it should also enable the co-culture of blood cells, including leukocytes and platelets, to understand the implication of nanoparticle exposure in local action of pro-inflammatory cytokines in inducing endothelium dysfunction. For instance, Kim et al. exploited the use of PDMS-based device to explore the impact of nanoparticles on platelets and endothelial cells, in which an increased platelet adhesion to endothelial cells was observed without compromising platelet viability upon silica nanoparticle exposure [152]. Human 3D vascular network can also be created using a combination of vasculogenesis-driven methods with the aid of stroma cells in a microfluidic system [[153], [154], [155]]. Fibroblasts, in particular, are known for their potential to secrete a variety of ECM proteins (collagen, elastin, and fibronectin) and stimulating factors that are necessary in the stabilization and formation of the vascular network [156,157]. Vessel formation by co-culturing endothelial cells with fibroblasts displays excellent barrier function and vascular stability as illustrated by the confinement of microbeads [153,155] and fluorescent dye [154,158] in the lumen of the microvasculature. Utilizing a vasculogeneis-driven vascular network featuring both endothelial cells and pulmonary fibroblasts, Li et al. were able to show different mode of action of inhaled atmospheric nanoparticle exposure in endothelial dysfunction, including increased vessel permeability by disruption of cell tight junctions, increased calcium influx, particle-induced inflammation, and imbalance of vasoactive substances [159]. Given the ability to control the shear stress, spatial and temporal microenvironment, in conjunction with the built-in readouts of the endothelial functional changes, ppresent the microfluidic systems as an ideal platform to study the complex interaction of nanoparticles with endothelial cells.
3.2. Lung-on-a-chip
With rapid advancement in technology, man-made environmental toxicants, which are poorly understood and/or not yet identified, are accumulating in the air. As we are constantly breathing 11,000L of air on a daily basis and the lung is one of the main routes of entry for particulate matters into the body, the deposition of particles in the respiratory system is inevitable. Studies in both in vitro and in vivo models have shown exposure to nanoparticles not only instigates significant oxidative-stress mediated cellular dysfunction [160,161], but it also consistently induces upregulation of pro-inflammatory cytokines such as MCP-1 [162,163], IL-33 [164], IL-6 [108,165], TNF- α [165,166], and IL-1β [167] that are responsible for pathogenesis of pulmonary complications, including pulmonary fibrosis [168], chronic obstructive pulmonary disease [56,169], asthma [170,171], and pulmonary edema [172]. Nanoparticle-mediated pulmonary complications are further aggravated by the lack of clearance mechanisms by alveolar macrophages, which depends on the rate of particle deposition and the rate of macrophage clearance. Under the steady state, in which the rate of alveolar macrophage clearance is at a level above the particle deposition, the retention half-time is approximately 70 days in rats. The retention half-time is significantly increased up to 100–500 days if the deposition rate of the inhaled particles exceeds the clearance rate [173]. The cumulative burden of particles on the lungs due to pulmonary inflammation causes pulmonary wall thickening, macrophage infiltration, and cystic lesions as demonstrated by a sub-chronic 3-month inhalation exposure of mice to UFP [103].
As the effects of environmental particles on human cells and tissues are not fully recognized and understood until they cause serious, even irreversible consequences, researchers and environmental protection agencies have shown constant interest in understanding the effect of nanoparticles on the pulmonary system. For decades, traditional 2D in vitro well-plate systems and animal models have served as the gold standard for predicting nanoparticle toxicity. However, it is becoming increasingly appreciated that traditional approaches fall short in predicting the pathophysiology of disease. A major limitation of existing in vitro systems is that they are unable to recapitulate complex pulmonary microenvironment, requiring an organ-on-a-chip approach (Table 2). Investigation of adverse biological effects of nanoparticles in the lungs are usually performed under the condition in which nanoparticles are suspended in the culture media. This approach does not reflect native environment where epithelial cells in the lungs are exposed to the air-liquid interface (ALI). ALI has been proven to play a pivotal role in recreating the respiratory airway niche for the epithelial cells, thus promoting apical/basolateral polarity as well as in vivo like functionality. Epithelial cells, such as tracheal and bronchial epithelial cells, cultured at the ALI recapitulate the key hallmark of the cellular morphology observed in vivo as illustrated by the formation of pseudostratified cells with tight junctions, formation of cilia, production of mucin, and steady decline in barrier function [[174], [175], [176]]. Additionally, culturing cells at the ALI allows nanoparticle dosimetry in an experimental set-up that reflects, realistic cell-particle interactions similar to the native scenarios in which air pollution interacts with the lungs. Exposure of nanoparticles directly into the cell culture medium increases the possibly of particle agglomeration, which is directly connected to the alteration of particle toxicity. It is for this reason, that the comparison of cells cultured at the ALI shows considerable differences in terms of biological responses such as the release of higher proinflammatory markers (IL-1β, IL-6, IL-8, TNF-α and GM-CSF), cellular toxicity, and the level of oxidative stress compared to the cell growth at the submerged conditions upon nanoparticle exposure [[177], [178], [179], [180]]. In addition to the ALI the conventional 2D system lacks rhythmic cyclic strain that cells are constantly experiencing during breathing motion. The resulting mechanical stretching of the alveolar epithelium causes the alveoli and adjacent endothelium to expand, allowing for efficient oxygen and carbon dioxide diffusion. Thus, to best model pulmonary interface in vitro, it is important to capture these key micro-environmental and mechanical strains of the lungs to advance our understanding towards nanoparticle-mediated toxicity.
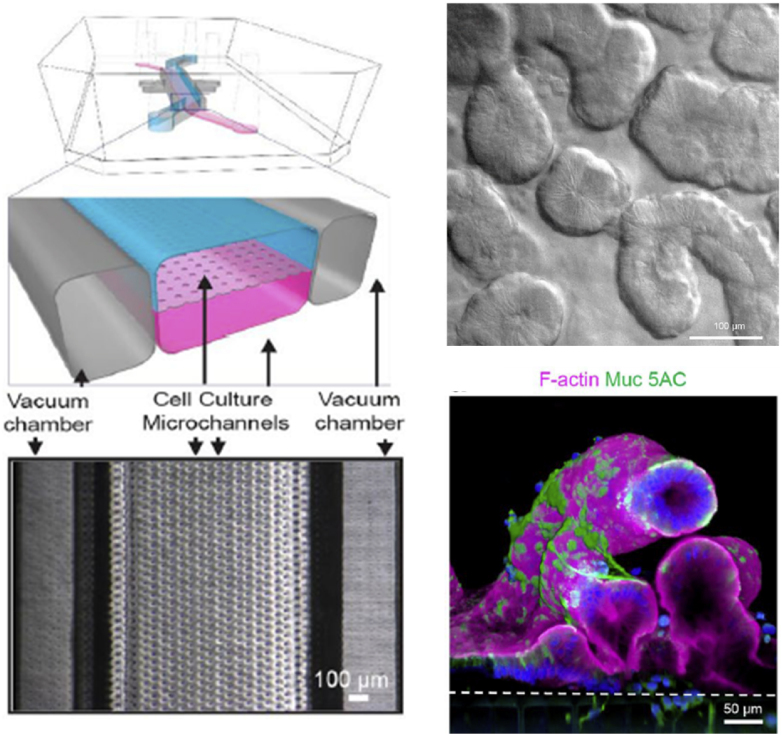
To address those shortcomings, Huh and his colleagues have defined the field of organ-on-a-chip by developing alveolus-capillary interface that experiences cyclic mechanical strain cultured at the air-liquid interface [29]. This system is achieved by fabricating two PDMS pieces containing a single microfluidic channel, which is separated by a permeable thin, porous membrane coated with the ECM. Subsequently, human epithelial cells are cultured on the apical compartment in the presence of air and microvascular endothelial cells are cultured submerged on the basal compartment to establish the ALI in the system. The mechanical strain is achieved through the integration of lateral side chambers, which can be tightly controlled by applying/releasing vacuum to mimic mechanical movement of the alveolar-capillary interface caused by the breathing motion. To mimic the delivery of airborne pollution into the lungs, 12 nm silica nanoparticles solution is perfused into the alveolar compartment followed by the removal of the solution to leave a layer of nanoparticles covered on the epithelial cell surface in the air. Authors have demonstrated that silica nanoparticle exposure induces significantly higher cellular toxicity as indicated by increased ROS production and the amount of nanoparticle translocation from the epithelium to the endothelial cell compartment compared to the cells experiencing no cyclic motion (Fig. 3A). Additionally, application of silica nanoparticles in the epithelial compartment stimulates the activation of adhesion proteins as marked by the ICAM-1 expression and neutrophil attachment in the endothelium, elucidating the complex multi-step intercellular epithelial-endothelial crosstalk. This is the first study that demonstrates physiological mechanical stress due to the breathing motion acting in concert with nanoparticles to induce higher degree of toxic effects and accelerate onset of nanoparticle toxicity in the lungs, which is not traditionally observed in static 2D culture system.
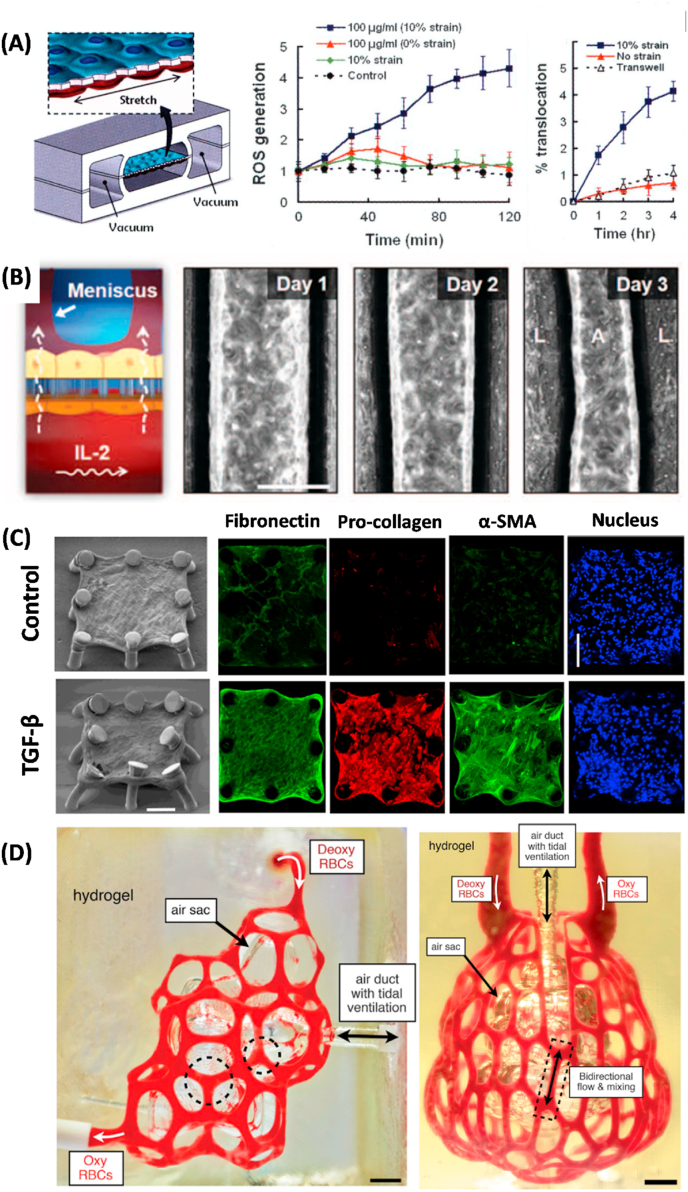
Fig. 3.
Representative lung-on-a-chip systems that can be used to understand nanoparticle induced toxicity. (A) Schematic of a human breathing lung-on-a-chip to study the effect of cyclic mechanical strain on nanoparticle translocation and phenotypic ROS generation. Reproduced with permission [29]. Copyright 2010, The American Association for the Advancement of Science. (B) Schematic of lung-on-a-chip system to study the onset of pulmonary edema (interstitial fluid buildup) upon IL-2 stimulation.
Reproduced with permission [181]. Copyright 2012, The American Association for the Advancement of Science.
(C) Recapitulation of fibrogenesis in lung microtissue. Continuous TGF-β treatment induced increase in the expression of α-SMA, pro-collagen, and EDA-Fibronectin compared to untreated microtissue [185]. Copyright 2018, Springer Nature. (D) Photograph of vascularized alveolar model printed by 3D stereolithography technique. Red blood cells are perfused to demonstrate oxygenation and deoxygenation. Reproduced with permission [187]. Copyright 2019, The American Association for the Advancement of Science.
The ability to capture multiple aspects of the native human environment in in vitro settings has gained significant attention from the scientific communities and has established a strong foundation to study disease-specific responses of human lungs. In Huh's follow up study, the authors investigated how the stimulation of endothelial cells may be implicated in pulmonary edema, which is characterized by abnormal accumulation of intravascular fluid in the alveolar spaces. By applying IL-2 into the endothelialized channel, this platform captures intra-alveolar fluid accumulation caused by the disruption of endothelial cell VE-cadherin junction and epithelial occuludin junction, which results in fibrin deposition and impaired gas exchange [181] (Fig. 3B). As exposure to air pollution is implicated in the systemic upregulation of pro-inflammatory cytokines which are known to cause significant leakage in endothelial cells, this system may provide an exciting opportunity to delineate complex intercellular crosstalk between endothelial cells and epithelial cells in inducing pulmonary complications upon nanoparticle exposure.
It is important to note that majority of lung-on-a-chip models have been focused on the study of nanoparticle toxicity or disease pathophysiology for alveoli regions which only represents tiny fraction of the entire respiratory system. Human respiratory tract is a highly complex biological system with different anatomy and physiology at each region. Unlike epithelial cells in the alveoli, the human airway is mainly lined by ciliated epithelial cells and mucin-secreting goblet cells, which act in concert to drive mucuociliated clearance. Emerging evidence reveals that particulate matter is implicated in the impairment of airway cilia, yet the underlying mechanisms of how these particles affect the overall function of airway remain largely unknown [182]. To understand the biological response of airway epithelial cells in vitro, Benam et al. studied the effect of smoking in the development of chronic obstructive pulmonary disease (COPD) using a microengineered breathing lung-on-a-chip featuring ciliated epithelial cells [183]. Transcriptomic analysis identified key 10 genes in COPD epithelial cells which are selectively activated in the controlled smoking apparatus. Additionally, increased cilia beating irregularity was observed in the smoke-exposed epithelial cells, potentially explaining reduced mucociliary clearance observed in smokers’ lung. Given that cigarette smoke, even in the diluted form, contains a large number of potentially toxic nanoparticles, this platform can be readily translated to predict and understand the pulmonary complications caused by UFP. There are other engineered airway-on-a-chip models that contain differentiated, mucuociliary epithelium for the analysis of organ-level lung physiology [184]. However, the roles of other cells, such as pulmonary fibroblasts and macrophages, are largely neglected in the study given the strong link of air pollutants in driving fibrotic responses and local inflammation. As such, we envision that the synergistic integration of both airway-on-a-chip and alveoli-on-a-chip with all relevant cells, connected through a vascular network, will provide a powerful way to unveil the complex nanoparticle interactions with cells and subsequent molecular mechanisms that are associated with pulmonary dysfunction, including nanoparticle clearance, nanoparticle translocation, and pulmonary inflammatory responses.
The study of the progression of pulmonary disease is not limited to the use of lung-on-a-chip system lined by human alveolar epithelium with endothelial cells. As exposure to air pollutants is implicated in the pathogenesis of pulmonary fibrosis, Asmani et al. developed a novel system to study pulmonary dysfunction which leverages the use of microtissues composed of human lung fibroblasts and human small airway epithelial cells that are suspended on micro-pillars to recapitulate the key aspect of fibrogenesis in lung interstitial tissues. By applying TGF-β1 to induce fibrosis, the authors were able to demonstrate collagen deposition and the subsequent increase in tractional forced generated by individual microtissues based on the displacement of micro-pillar structures [185] (Fig. 3C). Adapting a similar system featuring microtissue array fabricated in a PDMS substrate, Chen et al. showed even low concentration (50 ng/ml) of carboxylate-modified multi-wall carbon nanotubes to BEAS-2B normal lung epithelial cell-populated microtissues lead to the onset of pulmonary fibrosis within 72 h of exposure, as illustrated by the increase of tractional force generated by the microtissue and upregulation of fibrogenic marker miR-21 expression [186].
Another innovative approach employs the use of stereolithography, a 3D printing technique which creates 3D constructs layer-by-layer through the solidification of pre-polymer solution by applying light. Using this technique, Grigoryan et al. developed a complex breathable alveolus intravascular network, which is capable of showing oxygenation and deoxygenation of human red blood cells [187](Fig. 3D). While this system does not permit the transport of macromolecules, the ability to demonstrate such a sophisticated organ-level vascular network in parallel with the ability to mimic breathing motion and gas exchange may allow one to study true in vivo like biological mechanisms with integrated endothelium/epithelium to recapitulate pulmonary interface.
3.3. Heart-on-a-chip
While ambient air-pollution derived PM and UFPs have been closely linked with pulmonary dysfunction, there are mounting clinical and epidemiological data suggesting exposure to UFP is positively correlated with cardiovascular diseases. Both acute and chronic exposure to air pollution has been implicated with exacerbation of cardiovascular disease including ischemic heart disease [188], vascular dysfunction, thrombosis [63], hypertension [64,188], myocardial infarction [2,3], and arrhythmia [189], yet the mechanisms that drive cardiovascular disease and dysfunction remain controversial. One of the hypotheses is that inhalation of nanoparticles into the lungs can instigate the inflammatory responses within the alveolae, and subsequent systemic inflammation results in cardiovascular damage. Animal studies have reported that increased cellular and inflammatory cytokines such as IL-6 [69,126,190,191], IL-1β [191], TNF-α [126], IL-8 [126], and MCP-1 [126] are of importance in the pathogenesis of acute and chronic heart failure contributing to cardiac damage. Inflammatory cytokines modulate myocardial functions by a variety of mechanisms including stimulation of hypertrophy and fibrosis through direct effect on cardiomyocytes and fibroblasts [192,193], impairment of myocardial contractile functions and intracellular calcium transport [194], and stimulation of genes involved in the remodeling process [195].
The second hypothesis proposes that the inhaled nanoparticles cross the air-blood barrier into the blood circulation and accumulate in the secondary organs, including the heart [60,196]. The translocated nanoparticles then trigger decreased cardiac output by oxidative stress, neutrophil mediated cardiac inflammation, and inhibit calcium signaling pathway and cardiac muscle contraction pathway [114], that play a central role in maintaining cardiac function [114]. In another study, treatment of SiO2 nanoparticles exerted cardiac toxicity through downregulation of Connexin-43 expression, a gap junction protein that is responsible for intercellular communication between cardiomyocytes [116]. Inhibition of gap junctions is correlated with the increase in protein expression involved in the mitochondrial pathway related apoptosis such as caspase-3, caspase-9, and cytochrome-C.
Despite advances made in tissue engineering, recreating human heart in vitro has long been a challenging task for scientists. A major limitation of cardiac research is the scarce availability of human adult cardiomyocytes as they have limited potential for regeneration and proliferation, which explains heavy dependence on the use of animal models or animal derived cells for studying nanoparticle induced toxicity. Animal models still remain the gold standard to understand the modulation of heart function upon nanoparticle treatment, yet the discrepancy in physiological responses and lack of functional readout data preclude the detailed delineation of molecular mechanisms. As an alternative to animal models, the Langendorff heart has been proposed for evaluation the toxicity of nanoparticles, an ex vivo technique used to examine the cardiac force of contraction and heart rate without the complications of an intact animal or human. Using this model, Stampft et al. have demonstrated TiO2 and SiO2 nanoparticles induce arrhythmia and increase the heart rate, while monodispersed PS nanoparticles exhibited no effects, elucidating nanoparticle-induced cardiac toxicity is dependent on nanoparticle material composition [118]. However, as Langendorff heart is constantly deteriorating in ex vivo settings, it can be only used for several hours in studying nanoparticle induced toxicity, which is not ideal for long-term exposure experiments.
To address these key limitations of the pre-existing models, it is becoming evident that new models such as engineered cardiac tissues and organs-on-a-chip are needed (Table 2). Despite the lack of cell sources available for research, the convergence of induced pluripotent stem cells (iPSC) and directed cell differentiation greatly transformed the conventional research paradigm. The ability to differentiate human pluripotent stem cells into ventricular- [197], atrial- [198], and nodal-like cells and ability to generate large number of cardiomyocytes for research allows the construction of functional cardiac tissues around supporting structures, such as cantilevers [199], micro-posts [200,201], wires [25,202], or thin films [203,204]. These supporting structures not only provide a mechanical cue for cardiac tissue remodeling process, but also the mechanical displacement of supporting structures by cardiac tissue can be turned into contraction force using mathematical modelling or calibration curves. For instance, Wang et al. utilized Biowire II system, in which cardiac tissues were suspended between a pair of elastomers to demonstrate progression of cardiac fibrosis and evaluate potential therapeutic efficacy of anti-fibrotic compounds [205]. Based on displacement of elastomeric wires, the authors were able to demonstrate that the fibrotic tissue exhibits inferior contractile properties as indicated by the increase in passive tension and reduction in active force (Fig. 4A). While several novel approaches have been developed to study different aspects of the heart disease such as cardiac fibrosis [205,206] and dilated cardiomyopathy [207], evaluation of nanoparticle toxicity on heart-on-a-chip system has been scarce given the strong link of UPFs in inducing cardiovascular disease. Utilizing cell-sheet based culture system integrated with bioelectronics to measure real-time cardiac tissue contractions, Ahn et al. demonstrated TiO2 nanoparticle treatment lowered contractile force and impaired calcium transient propagation through disruption of sarcomere structures, which is the basic contractile unit that is responsible for contraction [208] (Fig. 4B).
Fig. 4.
Representative heart-on-a-chip systems for understanding nanoparticle induced toxicity. (A) Matured cardiac tissue for investigation of cardiac fibrosis (left). Fibrotic tissue exhibited higher expression of vimentin (green) as illustrated in the immunostaining image of cardiac and fibrotic tissue (right). Reproduced with permission [205]. Copyright 2019, American Chemical Society. (B) Fibrin-coated cardiac microphysiological device for contractility assessment. Exposure to TiO2 nanoparticles decreased contractile function of cardiac tissue through disruption of sarcomeric structure. Reproduced with permission [208]. Copyright 2018, Springer Nature. (C) Confocal image of perfusable vascularized scaffold that supports the self-assembly of cardiac tissue (red: CD31, green: sarcomeric α-actinin). This system allows the investigation of complex interaction between endothelial cells and nanoparticles, and their subsequent cardiac toxicity under electrical stimulation in 96-well format. Reproduced with permission [131]. Copyright 2017, Advanced Functional Materials. (D) 3D bioprinted neonatal-scale human heart with multi-scale vasculature composed of collagen material (left). Perfusion of glycerol (red) through coronary artery also permits the perfusion down to vessels ~100 μm in diameter (right). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Reproduced with permission [208]. Copyright 2018, Springer Nature.
Currently, majority of the heart-on-a-chip systems have focused on the treatment of drugs using only ventricular cardiomyocytes, yet the integration of cardiomyocytes from different chambers and all relevant non-myocytes, such as macrophages, that are important for particle clearance has not been achieved. This is partly due to the fact that the origin of heart failure is mostly due to the ventricular region of the heart [209]. Ventricular cardiomyocytes [197,210] were the first chamber specific cells cardiomyocytes for which the high purity differentiation protocols have been developed. Nevertheless, as nanoparticles have ability to reach all regions of the heart through microvasculature, other compartments of the heart, including atrium, sinoatrial node, and Purkinje fibers are as important for the investigation of nanoparticle-related toxicity [211]. Recent animal study revealed that upon inhalation of TiO2 nanoparticles, atrial activation time (P wave duration) was significantly reduced based on electrocardiographic wave analysis, indicating the effect of nanoparticle was not limited to the ventricular region. The integration of multiple cardiomyocytes from different origin has been demonstrated by Zhao et al., in which heteropolar cardiac tissue featuring both ventricular and atrial cells was developed [25]. The authors demonstrated a chamber specific drug response when only atrial section of the cardiac tissue responded to atrial-selective drug. To date, it is still not clearly understood how nanoparticles interact with the heart, but we envision a functional heteropolar cardiac tissue that incorporates all aspect of chamber specific cells could provide a powerful tool to systemically dissected the mechanisms of action of nanoparticle toxicity.
Aside from chamber specific cardiomyocytes, endothelial cells, in particular, are the most abundant non-myocyte cells consisting of 43% of the total heart cell population by number [212], providing vasculature which functions as both conduit for blood flow and the barrier. Yet the integration of stable perfusable vasculature in the pre-existing heart-on-a-chip system is a challenging task. Several platforms have attempted to incorporate vasculature into cardiac tissues without endothelial cells. For example, Xiao et al. establishes a perfusion using a poly-tetrafluoroethylene (PTFE) tubing to demonstrate the role of NO in modulating cardiac tissue beating and cytoskeletal changes [202]. Mathur et al. have developed a novel cardiac micro-physiological system featuring perfusable channel and a cardiac tissue loading channel separated with permeable walls to study the effect of pharmacological agents on cardiac tissue viability. Yet, these systems are not able to directly measure contraction forces, one of the key parameters that is implicated in heart failure [213].
To further increase the complexity of the system, Zhang et al. used a novel 3D stamping technique in which UV cross-linkable polymer layers with precise features are layered onto each other to form highly complex vascularized channel and the support of cardiac tissue [130]. Medium perfusion in the system prevented the development of necrotic core and mitigated cellular death, highlighting the importance of perfusion in maintaining cardiac tissue function. In a follow up study, Lai et al. simplified the platform into a single channel with the cantilever structures for contractile force detection and carbon electrodes, integrated into 96-well format to analyze the cardiac tissue functional changes upon applying epinephrine inside the vasculature [131] (Fig. 4C). As the endothelial channel provides a protective barrier against nanoparticles, these platforms may enable the study of nanoparticle induced toxicity on cardiac tissue that closely mimic native human heart microenvironment. Most recently, Lu et al. utilized the same system to study the toxic effects of air pollution relevant nanoparticles (50 nm CuO and SiO2) on the cardiovascular system under physiologically relevant perfusion. Authors demonstrated CuO nanoparticles are highly toxic, as they were able to translocate from the endothelial cell compartment into the cardiac tissue, which led to electrical and contractile dysfunction through generation of ROS, disruption of cardiac troponin T, and secretion of biomarkers associated with cardiac injury including brain type natriuretic peptide (BNP), N-terminated BNP, and troponin I [214]. More recently, Feinberg and colleagues developed 3D FRESH bioprinting technique made of collagen material to engineer components of the human heart [215]. This system not only promotes in vivo like microvascularization in micro-porous collagen scaffolds which permit the perfusion, but also it allows the construction of a human-scale ventricle that responds to electrical stimulation, integrated with the multiscale vasculature (Fig. 4D). Although there are still many challenges yet to overcome, the ability to construct human heart with high degree of vascularization along with physiologically relevant mechanical properties of native tissue offer an exciting opportunity for nanotoxicological research (Fig. 4D).
3.4. Gut-on-a-chip
The unwanted health consequences of nanoparticles are not limited to the cardiovascular system and lungs. Once controversial, the theory that exposure to nanoparticles modulates the gut function is gaining major traction in the research community. Ingestion is thought to be the primary port of entry, as significant portion of particle deposition in the gastrointestinal tract (GIT) is from drinking water or unintended ingestion of food containing nanoparticles. In the study published by Lomer et al., it is estimated that 1012-1014 particle are ingested per day by a typical Western diet, with roughly ~1% of particles uptake by the mucosal membrane [216]. These ingested dietary nanoparticles, such as titanium oxide, aluminosilicates, or plastic particles in drinking water, are absorbed by the intestinal epithelial lymphocytes that release pro-inflammatory cytokines including IL-1β, IL-6, IL-13, IL-17, and TNF-α [67,217], which are implicated in the pathogenesis of intestinal bowl disease [218,219] and gastric carcinogenesis [87]. The small intestinal epithelial barrier is also shown to be diminished via oxidant-mediated pathways as indicated by disruption of tight junction proteins, increased permeability, and inflammation [119]. More recently, a number of studies demonstrated the rising concentration of air pollution is also implicated in the modulation of the microbiome population in the intestine. Ingestion of UFP is implicated in the reduced diversity in microbiome [220,221] and changes in short chain fatty acid production [67]. As intestinal microbiota provides important symbiotic role in metabolism, maintaining intestinal immunity and homeostasis, imbalance in the gut microbiota may modulate host metabolism and inflammatory responses resulting in the progression of pathological conditions.
While the toxicity of nanoparticles on GIT has been extensively studied in vivo, given the complex structure and cellular composition of GIT accompanied with unique symbiotic interaction between cells and microbiome, the study of nanoparticle interaction with GIT in vitro is somewhat limited in terms of physiological relevance. Majority of in vitro systems recapitulate intestinal interface by culturing enterocytes (Caco-2) or co-culture of Caco-2 and mucin producing cells (HT20-MTX) to represent epithelial barrier. Exposure of 30 nm silver nanoparticles to Caco-2 cells induced the changes in proteins related to protein folding, cell morphology, and metabolic activity based on proteomics analysis [222]. The nanoparticle transport has also been studied in the transwells with Caco-2/HT29-MTX cells, yet the added iron oxide nanoparticle and silicon quantum dots failed to diffuse across the intestinal barrier [223]. A 2-fold increase in the number of 200 nm PS nanoparticles associated with HT29-MTX monolayer was observed after N-acetyl cysteine removal of the mucus layer, indicating that the secreted mucus functions as a protective barrier to particle diffusion.
While 2D in vitro systems shed new insights in the investigation of nanoparticle toxicity, they fail to effectively emulate many key features of human intestine including villus differentiation, production of mucus, and long-term culture of microbiome, all of which play a significant role in modulating nanoparticle toxicity. The most established in vitro GIT models adopt similar chip design as lung-on-a-chip system, in which two parallel perfusable microfluid channels are separated by ECM-coated porous membrane to recapitulate epithelial-endothelial interface. Under the mechanically active environment of small intestine that is simulated by applying cyclic strain, Caco-2 cells are able to polarize rapidly and spontaneously form the structure that resembles intestinal villi [224]. The genome-wide gene profiling analysis revealed that the Caco-2 cells cultured under fluid flow and cyclic stretch also expresseed significantly different gene expression profile compared to the cells cultured in the transwell system [225]. The ability to recapitulate key structure and function of the intestine is particularly important for the assessment of nanotoxicity, as intestinal mucus that is secreted from the goblet cells has been shown to act as the protective barrier in the epithelium, hindering the overall toxicity of nanoparticles by reducing the nanoparticle cellular uptake.
The intestine is also the major organ at which commensal microbes interact with the host cells to maintain intestinal homeostasis. Emerging evidence suggests that air particulate matter and plastic particles can induce gut microbiota dysbiosis and reduction in gut mucin secretion, suggesting the potential role of nanoparticles in disrupting intestinal homeostasis. The analysis of gut microbiome interaction with human intestinal cells in conventional in vitro settings is a challenging task, as bacterial overgrowth occurs rapidly thereby compromising the integrity of the epithelium.The relevance of gut-intestinal model has been further improved by the work of Kasendra et al., in which the primary epithelial cells are expanded as 3D organoids and seeded on PDMS-based microfluidic device. Based on transcriptomic analysis, epithelial cells cultured within 3D organoids closely resemble many important functions of the living intestine, including defense responses, cell proliferation, digestion, and response to nutrients compared to the gut microfluidic system featuring Caco-2 [31] (Fig. 5). The improved barrier function accompanied with villi differentiation allows the co-culture of living human intestinal epithelium with intestinal microbiome (E. coli), allowing both Caco-2 and microbiome to stay fully viable for over 96 h [225]. Another aspect the authors demonstrated was the ability to mimic the tissue inflammatory diseases, such as inflammatory bowel disease (IBD). Upon applying isolated human peripheral blood mononuclear cells (PBMS) into the endothelial channel, the stimulation of lipopolysaccharide (LPS) in the epithelial compartment not only activated the adjacent ICAM-1 endothelium, but also elevated proinflammatory cytokines, including IL-6, IL-1β, IL-8, and TNF-α. In another study, Zhang et al. developed a Gut-Microbiome physiome platform featuring primary human colon epithelial cells and oxygen-sensitive commensal anaerobe (Faecalibacterium prausnizii) to investigate mucosal barrier-bacterial interactions. Under flow condition, it has been demonstrated that F. parausnizzi exerted anti-inflammatory effects through the production of butyrate and subsequently downregulated toll-like receptor (TLR)-3 and TLR-4, responsible for activation of nuclear factor-light-chain-enhancer of activated B cells (NF-KB) signaling and inflammation [226]. In another organ-on-a-chip model featuring liver (human hepatocytes and Kupfer cells) and gut (ulcerative colitis epithelium, dendritic cells, and macrophages), Trapecar et al. showed the involvement of effector CD4 T cells in modulating cellular metabolism and inflammatory response [227]. When microbiome-derived short-chain fatty acids were added into the system, multiomics analysis revealed the increase in metabolic enzymatic activities involving glycolysis and lipogenesis, while immune responses decreased in the gut. However, when T cells (Treg and Th17) were integrated into the system, short-chain fatty acids induced gut barrier disruption and hepatic injury highlighting the that the presence of autoimmune cells greatly improves the relevance of this model in the study of complex diseases. While these devices allow us to monitor the functional interaction between host cells and its microbiomes, it is important to note that the gut microbiota is very diverse, hosting approximately 300–500 different species of bacteria in the healthy human intestine [228]. The Firmicutes, for example, promotes nutrient absorption and correlate with the development of obesity [229]. Therefore, the integration of multiple types of microbiomes is necessary to understand how nanoparticles modulate microbiome population and subsequent effect on gut functions. However, given the symbiotic role of gut microbiome with the host and the lack of in vitro models to reconstitute complex interaction among the epithelium, the endothelium, the microbiome, and the macrophages, these systems offer an unprecedented opportunity to explore the role of nanoparticles in modulating overall function of the gut and understand the progression of disease as well as nanoparticle transport across the endothelial-epithelial interface.
Fig. 5.
Representative gut-on-a-chip systems for understanding nanoparticle induced toxicity. (A) A primary human small intestine-on-a-chip using biopsy-derived organoid. Mucine producing cells (Muc5AC: green) are primarily found along the apical regions of the villi-like structure [31]. Copyright 2018, Springer Nature. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Liver-on-a-Chip
The liver is the largest solid organ in the body, serving more than 500 different functions. One of the critical roles of the liver is the filtration of blood, removing and detoxifying waste products from the body such as alcohols, drugs, and nanoparticles. Based on in vivo biodistribution studies of nanoparticles, the liver filters 30–90% of administered nanoparticles from the blood stream. These nanoparticles can accumulate in the liver resulting in the change in the liver function and cellular morphology. It has been shown that the long-term exposure to air pollution in humans increases the blood serum alanine transaminase (ALT) and alkaline phosphate (AKP) levels, biomarkers that are secreted upon liver injury [37,230]. These factors are implicated in oxidative damage to the liver, leading to non-alcoholic fatty liver disease and liver cancer. Exposure to fine airborne particulate matter also induces hepatic fibrosis in a mouse model through the activation of TGF-β/SMAD-3 signaling pathway, altering hepatic lipid glucose homeostasis and inducing hepatic steatosis [120]. Additionally, nanoparticles are shown to interact with cytochrome-450 (CYP), an enzyme that is responsible for oxidizing steroids, fatty acids, and xenobiotics [122,231]. For instance, intravenous injection of 30 nm SiO2 nanoparticles has been shown to inhibitCYP3A activities [231]. CYP3A represents around 60% of the total hepatic CYP isoforms and is involved in the metabolism of more than 50% drugs. Therefore, modulation of its activities can lead to severe adverse drug-drug interactions and increases the likelihood of further liver malfunction. While the use of in vivo models provides mechanistic insight toward nanoparticle translocation and subsequent impact on the liver, it is important to question the accuracy of animal models as humans differ significantly from animal models with regards to cellular composition and metabolic activities. In fact, when human CYP activities are compared to those of animals, CYP1A, CYP2C, CYP2D and CYP3A show appreciable differences in terms of enzymatic activities [232], therefore the extrapolation of nanoparticle toxicity based of animal models should be made with caution. Based on these facts, the development of reliable in vitro models that recapitulate human physiology and metabolism is of paramount importance in gaining in depth understanding of the interaction between nanoparticles and the liver, and their subsequent effects on the liver function.
Currently, majority of the liver models for nanotoxicological studies are still limited to the use on 2D systems based of primary human hepatocytes. Mounting scientific evidence suggests that the exposure to nanoparticles induces oxidative mediated genotoxicity and cytotoxicity, which then leads to cellular apoptosis. For example, Shuka et al. demonstrated that the exposure to 30–70 nm TiO2 nanoparticles to the hepatic-derived cell line, HepG2, grown in 2D induces oxidative DNA damage and cellular apoptosis through caspase-dependent pathway [233]. Similar findings were observed when silver nanoparticles were applied to BRL-3A rat liver cells in the well-plate, specifically the cells underwent apoptosis through mitochondrial malfunction [234]. The use of well-plates to study nanoparticle toxicity is well established with good repeatability, yet the functional assessment of the liver upon nanoparticle treatment is challenging, as liver cells cultured in 2D exhibit inadequate hepatocyte functions due to the loss of hepatic marker expression and phenotype.
One way to improve hepatic phenotypes is to introduce the perfusion in the system, thus improving the nutrition and waste exchange (Table 2). Microfluidic platforms featuring hepatocytes that are subjected to flow display increased production of albumin/urea, improved drug metabolisms, and prolonged lifespan of the cells compared to hepatocytes cultured under static conditions. For instance, Li et al. cultured primary rat hepatocytes in the microfluidic system and elucidated higher hepatocyte functionality as represented by higher albumin and urea production after 72 h of culture [235]. When superparamagnetic iron oxide nanoparticles were perfused in the microfluidic channel, they resulted in the significant reduction of albumin and urea production, indication of potential liver injury. In another study, Esch et al. developed liver-gut model in which gut was represented by co-culture of Caco-2/HT29-MTX cells and the liver represented by HepG2/C3A cells to simulate toxicity of nanoparticles to liver under ingestion settings (Table 2). Treatment with 50 nm carboxylated PS nanoparticles resulted in the significantly higher AST release in the gut-liver chip compared to the gut-chip alone, demonstrating compounding effects of inter-organ crosstalk between gut and the liver in facilitating nanoparticle-mediated toxicity (Fig. 6A) [236].
Fig. 6.
Representative liver-on-a-chip systems for understanding nanoparticle induced toxicity. (A) Schematic of the body-on-a-chip system to study inter-organ level crosstalk. Treatment of 50 nm carboxylated PS nanoparticles induced AST in the medium. Reproduced with permission [236]. Copyright 2014, Royal Society of Chemistry. (B) Integration of 3D vascularized hepatic organoid in a microfluidic system (top left). Co-culture of hepatocytes and endothelial cells under flow stimulates drug metabolic process, xenobiotic metabolic process, and the response to drugs (top right), as well as albumin expression (bottom). Reproduced with permission [237]. Copyright 2018, John Wiley and Sons. (C) Schematic of liver-on-a-chip featuring complex liver cytoarchitecture (top). Addition of experimental drug (FIAU) induced the appearance of significant lipid droplets in a human liver-chip but no droplets were observed in rat liver-chip, illustrating species differences in steatosis after FIAU treatment (bottom).
Reproduced with permission [239]. Copyright 2019, The American Association for the Advancement Science.
The 3D model of organoids has also been integrated into the microfluidic system to enhance liver function. The intrinsic advantage of using organoids is the fact that they naturally exhibit enhanced liver function compared to cells cultured in 2D, as they naturally resemble avascular like environment with gradients of nutrients, O2 and CO2, and waste exchanges. When the perfusion is introduced into the microfluidic system containing and 3D hepatic organoids with endothelial cells, a gene ontology analysis revealed that genes involved in drug responses, lipopolysaccharide response, drug metabolic process and xenobiotic metabolic process were significantly upregulated under flow conditions (Fig. 6B), emphasizing the importance of fluid flow to enhance hepatic functions for toxicological studies [237].
To further extend the relevance of liver-on-a-chip systems, multiple cell types have been integrated in the microfluidic device to replicate organ-level functions. While hepatocytes comprise approximately 60% of the total population in the liver, non-parenchymal cells including hepatic stellate cells, Kupffer cells, and liver sinusoid endothelial cells act in concert with hepatocytes to form enhanced metabolic environment and establish complex cell-cell communication, which further prolong the duration of cell culture. Co-culture of endothelial cells (EA.hy926 cells)/Kupffer cells (U937 cells), and hepatocytes/hepatic stellate cells (LX-2) separated by a porous membrane, continuously secreted albumin and maintained CYP3A4 activities for 28 days. These findings highlight the importance of crosstalk among different cell types in maintaining hepatocyte morphology and liver functions [238].
More recently, Jang et al. developed multi-species liver-on-a-chip system, in which species-specific primary hepatocytes (rat, dog, and human), liver sinusoidal endothelial cells, Kupffer cells, and hepatic stellate cells were cultured under physiological flow to demonstrate how interspecies differences contribute to differences in drug metabolism and toxicity (Fig. 6C) [239]. The prediction of species-specific liver responses focused on an experimental drug (Fialuridine), which exhibited no toxicity in rat but was discontinued in phase 2 human clinical trial because of liver failure and death. Addition of Fialuridine displayed dose-dependent increase in lipid accumulation and decline in albumin secretion, whereas rat liver-chip displayed no changes, which highlights the interspecies variation in drug toxicity. The liver-chips also detected liver injury phenotype including hepatocellular injury, steatosis, cholestasis, and fibrosis. While most liver-on-a-chip models are focusing on recapitulating drug metabolism, the ability to differentiate species-specific drug responses and subsequent mode of hepatic injury may potentiate the prediction of nanoparticle-induced liver injury and functional changes which provides a new and exciting opportunity to investigate complex nanoparticle interaction under physiologically relevant conditions.
4. Challenges and outlook
Similar to how the field ecotoxicology arose out of toxicology, the current understanding of nanotoxicology in the context of environmental pollution is emerging. Since the early days of toxicological research, the use of well-plate systems complemented with animal models has been the key tools to systematically evaluate the toxicity of nanoparticles, yet the lack of predicative capability and the translation to human health outcome are questioned. Animal models, on the other hand, play a crucial role in formulating and testing the hypotheses under the current research paradigm. We need to further refine methods in order to identify targets that build on samples from human patients. As a matter of fact, the US environmental protection agency has announced plans to reduce its request for mammal studies by 30% by 2025, and completely eliminate the use of mammals by 2035. This motivates urgent need to develop human cell-based platforms which replace animal models in order to support the studies of nanoparticle toxicity [240].
With the discovery of more physiologically relevant cell sources in parallel with the recent advancement in fabricating sophisticated micro-scale structures, the field of organ-on-a-chip engineering has been exponentially growing aiming to capture spatial and temporal microenvironment similar to that of the in vivo settings. While many organ-on-a-chip systems have emerged as new predictive platforms to predict drug efficacy, the scientific adaptation for nanotoxicity assessment has been lagging. One of the primary concerns hindering the adaptation underlies the complexity of studying nanoparticles due to the lack of standardization in nanoparticle preparation and dosage control. Due to the heterogenous amalgam of nanoparticles of varying size, chemical composition, surface charge, and hydrophilicity, controlling nanoparticle aggregation under high salt concentration in the cell culture medium is a challenging task. As cellular uptake of nanoparticles is highly dependent on the size of nanoparticles, nanoparticle aggregation generates significant uncertainty about the nanoparticle toxicity. Various stabilization method such as PEG surface functionalization are known to prevent the aggregation of nanoparticles and enable the study of size effect of nanomaterials, yet the surface coating may alter the toxicity of the native nanoparticles and their subsequent biological interactions with cells.
Nanoparticle dose metric is another topic which has been greatly debated. In traditional well-plate systems, the volume of the cell culture media is standardized based on the type of well-plate and thus the mass or number of nanoparticles can be calculated based on the nanoparticle concentrations. However, as all organ-on-a-chip system differ by design, number of factors including shear stress, volume of the cell culture media, and number of cells may cause significant discrepancies in nanoparticle uptake. Additionally, unrealistically high nanoparticle concentration that is used in majority of studies preclude the understanding of long-term cumulative effects of nanoparticles. With recent development of organ-on-a-chip systems, the inability to keep 2D cell culture for prolonged duration can be resolved by the use of 3D cell culture system with improved and stable microenvironment for the long-tern cultivation of cells, which allows for the repeated-dosage of nanoparticles at the environmentally relevant concentrations to measure the chronic effects.
Regardless of short- or long-term toxicological assessment of nanoparticles, it is important to note that nanoparticle absorption to the substrate material may lead to significant error in the experiment. PDMS is still the predominant material of choice for organ-on-a-chip devices, yet recent literature suggests that the absorption of small hydrophobic molecules by PDMS severely hinders the application of these systems in the investigation of cellular responses to drugs or small molecules [241,242]. Similar to how drugs interact with PDMS substrate, nanoparticles can also interact with the substrate material through van der Waals, electrostatic, and hydrophobic interactions [243,244]. Most commonly used substrates, including PDMS and glass substrate, show that gold nanoparticles preferentially adhere to a hydrophobic surface rather than to a hydrophilic one [245], which raises a concern in such a device that only one surface of the channel is lined with cells such as endothelium and gut-on-a-chip devices. Nanoparticle absorption to the device can leads to misrepresentation of the nanoparticle toxicity due to the reduction of nanoparticle concentration and subsequent concentration-response interpretation, thus the usefulness of the device may be greatly hindered due to the lack of accuracy, requiring a significant amount of additional work. To prevent nanoparticle absorption, there is a gradual shift toward the use of alternative materials [246,247] or chemical modification of the substrate surface to minimize the molecular absorption [248,249]. However such new methods must maintain their stability over the course of experiment while maintaining the proper cellular function.
Modeling changes of micro-engineered tissue function is another important hallmark in the investigation of the progression of nanoparticle-induced toxicity. The assessment of cellular function should go beyond considerations of live/dead viabilities or change of protein expression based on immunohistochemistry techniques. The ability to quantify the overall tissue in real-time fashion will enhance the utility of organ-on-a-chip system. Indeed, several highly innovative electronic components have been integrated to enable the analysis of biological molecules and detection of cellular functional changes. This includes implementation of TEER sensors to non-invasively assess endothelial/epithelial interface upon nanoparticle treatment and integration of conductive polymer materials to measure cardiac tissue contraction. Given the high level of precision and small size, chip-integrated biosensors offers the potential to accurately quantify additional biological readouts, such as soluble factors and generation of reactive oxygen species in the future. We envision future organ-on-a-chip systems will have the capacity to investigate the complex inter-organ level crosstalk to further extend the knowledge on nanoparticle induced toxicity. Indeed, there has been significant push toward developing body-on-a-chip models that consists of multiple engineered tissues to analyze potential indirect effects of nanoparticles. The body on-a-chip systems, that combine human epithelium and the liver within one microfluidic device were able to demonstrate the inter-organ level crosstalk as demonstrated by the release of AST upon PS nanoparticle exposure [236]. With the ability to connect multiple organs in a single platform, it is possible to analyze complex disease which involves multiple organs and which is influenced by the immune cells. For example, the development of lung-heart duo-organ system allows us to study direct and indirect coupling of lungs with the heart in series and understand the effect of pulmonary pro-inflammatory cytokines on cardiovascular system upon nanoparticle exposure. It is important to note that a fine balance between model complexity and model throughput needs to be considered for the final scientific adaption of the nanotoxicological research.
CRediT authorship contribution statement
Rick Xing Ze Lu: Conceptualization, Writing - original draft. Milica Radisic: Writing - review & editing.
Declaration of competing interest
M.R. is a co-founder of TARA Biosystems and holds euiqty in this company.
Acknowledgements
Our work funded by the Canadian Institutes of Health Research Foundation Grant FDN-167274, Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN 326982-10), NSERC-CIHR Collaborative Health Research Grant (CHRP 493737-16). MR is supported by Killam Fellowship and Canada Research Chair. R. X. Z. L. was supported by Alexander Graham Bell Canada Graduate Scholarship-Doctoral Award (PGS-D).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Rick Xing Ze Lu, Email: ricklu0625@gmail.com.
Milica Radisic, Email: m.radisic@utoronto.ca.
References
- 1.Traboulsi H., Guerrina N., Iu M., Maysinger D., Ariya P., Baglole C.J. Int. J. Mol. Sci. 2017;18:1. doi: 10.3390/ijms18020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K., Schneider A., Cyrys J., Wolf K., Meisinger C., Heier M., Von Scheidt W., Kuch B., Pitz M., Peters A. Environ. Health Perspect. 2020;128:1. doi: 10.1289/EHP5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downward G.S., Van Nunen E.J.H.M., Kerckhoffs J., Vineis P., Brunekreef B., Boer J.M.A., Messier K.P., Roy A., Verschuren W.M.M., Van Der Schouw Y.T. Environ. Health Perspect. 2018;126:1. doi: 10.1289/EHP3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei Y., Wang Y., Di Q., Choirat C., Wang Y., Koutrakis P., Zanobetti A., Dominici F., Schwartz J.D. BMJ. 2019;367:1. doi: 10.1136/bmj.l6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lelieveld J., Evans J.S., Fnais M., Giannadaki D., Pozzer A. Nature. 2015;525:367. doi: 10.1038/nature15371. [DOI] [PubMed] [Google Scholar]
- 6.Ambient Air Pollution: A Global Assessment of Exposure and Burden of Disease. 2016. [Google Scholar]
- 7.Lehner R., Weder C., Petri-fink A., Rothen-rutishauser B. Environ. Sci. Technol. 2019;53:1748. doi: 10.1021/acs.est.8b05512. [DOI] [PubMed] [Google Scholar]
- 8.Revel M., Châtel A., Mouneyrac C. Curr. Opin. Environ. Sci. Heal. 2018;1:17. [Google Scholar]
- 9.Kahru A., Ivask A. Acc. Chem. Res. 2013;46:823. doi: 10.1021/ar3000212. [DOI] [PubMed] [Google Scholar]
- 10.Semmler-Behnke M., Kreyling W.G., Lipka J., Fertsch S., Wenk A., Takenaka S., Schmid G., Brandau W. Small. 2008;4:2108. doi: 10.1002/smll.200800922. [DOI] [PubMed] [Google Scholar]
- 11.Kreyling W.G., Hirn S., Möller W., Schleh C., Wenk A., Celik G., Lipka J., Schäffler M., Haberl N., Johnston B.D. ACS Nano. 2014;8:222. doi: 10.1021/nn403256v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takenaka S., Karg E., Roth C., Schulz H., Ziesenis A., Heinzmann U., Schramel P., Heyder J. Environ. Health Perspect. 2001;109:547. doi: 10.1289/ehp.01109s4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decuzzi P., Godin B., Tanaka T., Lee S.Y., Chiappini C., Liu X., Ferrari M. J. Contr. Release. 2010;141:320. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Karlsson H.L., Cronholm P., Gustafsson J., Moeller L. Chem. Res. Toxicol. 2008;21:1726. doi: 10.1021/tx800064j. [DOI] [PubMed] [Google Scholar]
- 15.Karlsson H.L., Gustafsson J., Cronholm P., Möller L. Toxicol. Lett. 2009;188:112. doi: 10.1016/j.toxlet.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Wen T., Yang A., Piao L., Hao S., Du L., Meng J., Liu J., Xu H. Int. J. Nanomed. 2019;14:4475. doi: 10.2147/IJN.S208225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unger R.E., Krump-Konvalinkova V., Peters K., James Kirkpatrick C. Microvasc. Res. 2002;64:384. doi: 10.1006/mvre.2002.2434. [DOI] [PubMed] [Google Scholar]
- 18.Kruepunga N., Hakvoort T.B.M., Hikspoors J.P.J.M., Köhler S.E., Lamers W.H. Biochim. Biophys. Acta - Mol. Basis Dis. 2019;1865:869. doi: 10.1016/j.bbadis.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Richard P. Elsevier Science Publishing Co Inc; San Diego: 2015. Comparative Biology of the Normal Lung. [Google Scholar]
- 20.N N.M., Janssen P.M.L. Pharmacol. Ther. 2010;141:1. [Google Scholar]
- 21.Han S., Shin Y., Jeong H.E., Jeon J.S., Kamm R.D., Huh D., Sohn L.L., Chung S. Sci. Rep. 2015;5:1. doi: 10.1038/srep18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sfriso R., Zhang S., Bichsel C.A., Steck O., Despont A., Guenat O.T., Rieben R. Sci. Rep. 2018;8:1. doi: 10.1038/s41598-018-24273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mannino R.G., Myers D.R., Ahn B., Wang Y., Timmins L.H., Lam W.A. Nat. Publ. Gr. 2015;1 [Google Scholar]
- 24.Huyer L.D., Zhang B., Korolj A., Montgomery M., Drecun S., Conant G., Zhao Y., Reis L., Radisic M. ACS Biomater. Sci. Eng. 2016;2:780. doi: 10.1021/acsbiomaterials.5b00525. [DOI] [PubMed] [Google Scholar]
- 25.Yimu Z., AschZhao R., Yimuar-Sobbi, Radisic M., Backx P.H., Jekic D., Zhang B., Pahnke A., Naguib H.E., Aggarwal P., Wang E.Y. Cell. 2019;176:913. doi: 10.1016/j.cell.2018.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fede C., Fortunati I., Weber V., Rossetto N., Bertasi F., Petrelli L., Guidolin D., Signorini R., De Caro R., Albertin G. Microvasc. Res. 2015;97:147. doi: 10.1016/j.mvr.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y.Y., Syed A.M., Macmillan P., V Rocheleau J., Chan W.C.W. Adv. Mater. 2020;32:1. doi: 10.1002/adma.201906274. [DOI] [PubMed] [Google Scholar]
- 28.Qiu Y., Ahn B., Sakurai Y., Hansen C.E., Tran R., Mimche P.N., Mannino R.G., Ciciliano J.C., Lamb T.J., Joiner C.H. Nat. Biomed. Eng. 2018;2:453. doi: 10.1038/s41551-018-0224-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huh D., Matthews B.D., Mammoto A., Montoya-zavala M., Hsin H.Y., Ingber D.E., Huh D., Matthews B.D., Mammoto A., Montoya-zavala M. Science. 2016;328:1662. doi: 10.1126/science.1188302. (80-. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H.J., Ingber D.E. Integr. Biol. 2013;5:1130. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 31.Kasendra M., Tovaglieri A., Sontheimer-phelps A., Jalili-firoozinezhad S., Bein A., Cha A., Scholl W., Zhang C., Rickner H., Richmond C.A. Sci. Rep. 2018;8:1. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herland A., Van Der Meer A.D., Fitzgerald E.A., Park T., Jelle J. PloS One. 2016;11:1. doi: 10.1371/journal.pone.0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Partyka P.P., Godsey G.A., Galie J.R., Kosciuk M.C., Acharya N.K., Nagele R.G., Galie P.A. Biomaterials. 2017;115:30. doi: 10.1016/j.biomaterials.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Brook R.D. Clin. Sci. 2008;115:175. doi: 10.1042/CS20070444. [DOI] [PubMed] [Google Scholar]
- 35.Breitner S., Peters A., Zareba W., Hampel R., Oakes D., Wiltshire J., Frampton M.W., Hopke P.K., Cyrys J., Utell M.J. Sci. Rep. 2019;1 doi: 10.1038/s41598-019-38531-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajagopalan S., Al-Kindi S.G., Brook R.D. J. Am. Coll. Cardiol. 2018;72:2054. doi: 10.1016/j.jacc.2018.07.099. [DOI] [PubMed] [Google Scholar]
- 37.Pedersen M., Andersen Z.J., Stafoggia M., Weinmayr G., Galassi C., Sørensen M., Eriksen K.T., Tjønneland A., Jaensch A., Nagel G. Environ. Res. 2017;154:226. doi: 10.1016/j.envres.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Perera F. Int. J. Environ. Res. Publ. Health. 2018;15:1. doi: 10.3390/ijerph15010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Liati, D. Schreiber, P. D. Eggenschwiler, Y. Arroyo, R. Dasilva, 2013, DOI 10.1021/es403121y.
- 40.Brauer M., Reynolds C., Hystad P. CMAJ (Can. Med. Assoc. J.) 2013;185:1557. doi: 10.1503/cmaj.121568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saikia B.K., Saikia J., Rabha S., Silva L.F.O., Finkelman R. Geosci. Front. 2018;9:863. [Google Scholar]
- 42.Van Ryswyk K., Anastasopolos A.T., Evans G., Sun L., Sabaliauskas K., Kulka R., Wallace L., Weichenthal S. Environ. Sci. Technol. 2017;51:5713. doi: 10.1021/acs.est.6b05775. [DOI] [PubMed] [Google Scholar]
- 43.Laney A.S., Weissman D.N. J. Occup. Environ. Med. 2015;56:1. doi: 10.1097/JOM.0000000000000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia J., Muralikrishnan S., Ng C., Yung L.L., Bay B. Environ. Biol. Med. 2010;235:1025. doi: 10.1258/ebm.2010.010021. [DOI] [PubMed] [Google Scholar]
- 45.Occupational Exposure to Carbon Nanotubes and Nanofibers. 2014. [Google Scholar]
- 46.C. Lam, J. T. James, R. Mccluskey, R. L. Hunter, 2004, 134, 126. [DOI] [PubMed]
- 47.Muller J., Moreau N., Misson P., Delos M., Arras M., Fonseca A., Nagy J.B., Lison D. Toxicol. Appl. Pharmacol. 2005;207:221. doi: 10.1016/j.taap.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Sukhanova A., Bozrova S., Sokolov P., Berestovoy M., Karaulov A., Nabiev I. Nanoscale Res. Lett. 2018;13:1. doi: 10.1186/s11671-018-2457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho Y.M., Mizuta Y., Akagi J.I., Toyoda T., Sone M., Ogawa K. J. Toxicol. Pathol. 2018;31:73. doi: 10.1293/tox.2017-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirowsky J., Hickey C., Horton L., Blaustein M., Galdanes K., Peltier R.E., Chillrud S., Chen L.C., Ross J., Nadas A. Inhal. Toxicol. 2013;25:747. doi: 10.3109/08958378.2013.846443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.WHO Air Quality Guidelines for Particulate Matter, Ozone, Nitrogen Dioxide and Sulfur Dioxide. 2005. [PubMed] [Google Scholar]
- 52.Li Y., Lane K.J., Corlin L., Patton A.P., Durant J.L., Thanikachalam M., Woodin M., Wang M., Brugge D. Int. J. Environ. Res. Publ. Health. 2017;14:1. doi: 10.3390/ijerph14050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsen Y., Karottki D.G., Jensen D.M., Bekö G., Kjeldsen B.U., Clausen G., Hersoug L., Holst G.J., Wierzbicka A., Sigsgaard T. Environ. Health (Lond.) 2014;13:1. doi: 10.1186/1476-069X-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geiser M., Schu S., Gehr P., Schu S., Gehr P. J. Appl. Physiol. 2003;94:1793. doi: 10.1152/japplphysiol.00514.2002. [DOI] [PubMed] [Google Scholar]
- 55.Park E., Yoon J., Choi K., Yi J., Park K. Toxicology. 2009;260:37. doi: 10.1016/j.tox.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Chung K.F. Eur. Respir. J. 2001;18:50. doi: 10.1183/09031936.01.00040701. [DOI] [PubMed] [Google Scholar]
- 57.Kumar P., Robins A., Vardoulakis S., Britter R. Atmos. Environ. 2010;44:5035. [Google Scholar]
- 58.Oberdörster G., Oberdörster E., Oberdörster J. Environ. Health Perspect. 2005;113:823. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller M.R., Raftis J.B., Langrish J.P., McLean S.G., Samutrtai P., Connell S.P., Wilson S., Vesey A.T., Fokkens P.H.B., Boere A.J.F. ACS Nano. 2017;11:4542. doi: 10.1021/acsnano.6b08551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Jong W.H., Hagens W.I., Krystek P., Burger M.C., Sips A.J.A.M., Geertsma R.E. Biomaterials. 2008;29:1912. doi: 10.1016/j.biomaterials.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 61.Landen D.D., Wassell J.T., Mcwilliams L., Patel A. Am. J. Ind. Med. 2011;54:727. doi: 10.1002/ajim.20986. [DOI] [PubMed] [Google Scholar]
- 62.Du Y., Xu X., Chu M., Guo Y., Wang J. J. Thorac. Dis. 2016;8:8. doi: 10.3978/j.issn.2072-1439.2015.11.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robertson S., Miller M.R. Part. Fibre Toxicol. 2018;15:1. doi: 10.1186/s12989-017-0237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun Q., Yue P., Ying Z., Cardounel A.J., Brook R.D., Devlin R., Hwang J., Zweier J.L., Chen L.C. Arterioscler. Thromb. Vasc. Biol. 2009;28:1760. doi: 10.1161/ATVBAHA.108.166967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossi S., Savi M., Mazzola M., Pinelli S., Alinovi R., Gennaccaro L., Pagliaro A., Meraviglia V., Galetti M., Lozano-garcia O. Part. Fibre Toxicol. 2019;1 doi: 10.1186/s12989-019-0311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mutlu E.A., Meliton A.Y., Soberanes S., Hamanaka R.B., Ni R., Ghio A.J., Budinger G.R.S. Environ. Pollut. 2018;240:817. doi: 10.1016/j.envpol.2018.04.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li R., Yang J., Saffa A., Jacobs J., Baek K.I., Hough G., Larauche M.H., Ma J., Jen N., Moussaoui N. Sci. Tech. Rep. 2017;1 doi: 10.1038/srep42906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcideunas L., Azzerelli B., Acuna H., Garcia R., Gambling T., Osnaya N., Monroy S., Tizapantzi M., Carson J., Calderon A. Toxicol. Pathol. 2002;30:373. doi: 10.1080/01926230252929954. [DOI] [PubMed] [Google Scholar]
- 69.Guo J., Shi T., Cui X., Rong Y., Zhou T., Zhang Z., Liu Y., Shen Y., Chen W. Arch. Toxicol. 2016;90:247. doi: 10.1007/s00204-014-1405-5. [DOI] [PubMed] [Google Scholar]
- 70.Afsar B., Afsar R.E., Kanbay A., Covic A., Ortiz A., Kanbay M. Clin. Kidney J. 2019;12:19. doi: 10.1093/ckj/sfy111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.J. W. Kim, S. Park, C. W. Lim, K. Lee, B. Kim, 2014, 30, 65.
- 72.Andrady A.L., Neal M.A. Philos. Trans. R. Soc. B. 2009;364:1977. doi: 10.1098/rstb.2008.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geyer R., Jambeck J.R., Law K.L. Sci. Adv. 2017;3:25. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Obbard R.W. Curr. Opin. Environ. Sci. Heal. 2015;1:24. [Google Scholar]
- 75.Lusher A.L., Tirelli V., Connor I.O., Officer R. Nat. Publ. Gr. 2015;1 [Google Scholar]
- 76.Brahney J., Hallerud M., Heim E., Hahnenberger M., Sukumaran S. Science. 2020;368:1257. doi: 10.1126/science.aaz5819. (80-. ) [DOI] [PubMed] [Google Scholar]
- 77.Wetherbee B.G., Baldwin A., Ranville J. 2019. It Is Raining Plastic. [Google Scholar]
- 78.Van Cauwenberghe L., Vanreusel A., Mees J., Janssen C.R. Environ. Pollut. 2013;182:495. doi: 10.1016/j.envpol.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 79.Mason S.A., Welch V.G., Neratko J. Front. Chem. 2018;6:1. doi: 10.3389/fchem.2018.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lambert S., Wagner M. Chemosphere. 2016;145:265. doi: 10.1016/j.chemosphere.2015.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jani P., Halbert G.W., Florence A.T. J. Pharm. Pharmacol. 1990;42:821. doi: 10.1111/j.2042-7158.1990.tb07033.x. [DOI] [PubMed] [Google Scholar]
- 82.D. Li, Y. Li, G. Li, Y. Zhang, J. Li, H. Chen, 2019, 1.
- 83.Lu L., Wan Z., Luo T., Fu Z., Jin Y. Sci. Total Environ. 2018;631–632:449. doi: 10.1016/j.scitotenv.2018.03.051. [DOI] [PubMed] [Google Scholar]
- 84.Jin Y., Lu L., Tu W., Luo T., Fu Z. Sci. Total Environ. 2019;649:308. doi: 10.1016/j.scitotenv.2018.08.353. [DOI] [PubMed] [Google Scholar]
- 85.Li B., Ding Y., Cheng X., Sheng D., Xu Z., Rong Q., Wu Y., Zhao H., Ji X., Zhang Y. Chemosphere. 2020;244:125492. doi: 10.1016/j.chemosphere.2019.125492. [DOI] [PubMed] [Google Scholar]
- 86.Mahler G.J., Esch M.B., Tako E., Southard T.L., Archer S.D., Glahn R.P., Shuler M.L. Nat. Nanotechnol. 2012;7:264. doi: 10.1038/nnano.2012.3. [DOI] [PubMed] [Google Scholar]
- 87.Forte M., Iachetta G., Tussellino M., Carotenuto R., Prisco M., De Falco M., Laforgia V., Valiante S. Toxicol. Vitro. 2016;31:126. doi: 10.1016/j.tiv.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 88.Cortés C., Domenech J., Salazar M., Pastor S., Marcos R., Hernández A. Environ. Sci. Nano. 2020;7:272. [Google Scholar]
- 89.Bucci K., Tulio M., Rochman C. Ecol. Appl. 2020;30:1. doi: 10.1002/eap.2044. [DOI] [PubMed] [Google Scholar]
- 90.Schwaferts C., Niessner R., Elsner M., Ivleva N.P. Trends Anal. Chem. 2019;112:52. doi: 10.1021/acs.analchem.9b05336. [DOI] [PubMed] [Google Scholar]
- 91.Coppock R.L., Cole M., Lindeque P.K., Queir A.M., Galloway T.S. Environ. Pollut. 2017;230:829. doi: 10.1016/j.envpol.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 92.Cole M., Webb H., Lindeque P.K., Fileman E.S., Halsband C., Galloway T.S. Sci. Rep. 2014;4:1. doi: 10.1038/srep04528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lusher A.L., Welden N.A., Sobral P., Cole M. Anal. Methods. 2017;9:1346. [Google Scholar]
- 94.Mintenig S.M., Bäuerlein P.S., Koelmans A.A., Dekker S.C., Van Wezel A.P. Environ. Sci. Nano. 2018;5:1640. [Google Scholar]
- 95.Hernandez L.M., Youse N., Tufenkji N. Environ. Sci. Technol. 2017;4:280. [Google Scholar]
- 96.Murray A., Örmeci B. Water. 2020;12:1. [Google Scholar]
- 97.Hildebrandt L., Mitrano D.M., Zimmermann T., Pröfrock D. Front. Environ. Sci. 2020;8:1. [Google Scholar]
- 98.Wang L., Wu W., Bolan N.S., Tsang D.C.W., Li Y., Qin M. J. Hazard Mater. 2020;401:1. doi: 10.1016/j.jhazmat.2020.123415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hurley R.R., Nizzetto L. Curr. Opin. Environ. Sci. Heal. 2018;1:6. [Google Scholar]
- 100.Elder A., Gelein R., Lunts A., Kreyling W. J. Toxicol. Environ. Health. 2002;65:1532. doi: 10.1080/00984100290071658. [DOI] [PubMed] [Google Scholar]
- 101.Li D., Li Y., Li G., Zhang Y., Li J., Chen H. Proc. Natl. Acad. Sci. Unit. States Am. 2018;116:1. [Google Scholar]
- 102.Jani P., Halbert G.W., Langridge J., Florence A.T. J. Pharm. Pharmacol. 1989;41:809. doi: 10.1111/j.2042-7158.1989.tb06377.x. [DOI] [PubMed] [Google Scholar]
- 103.Saleh Y., Antherieu S., Dusautoir R., Alleman L.Y., Sotty J., De Sousa C., Platel A., Perdrix E., Riffault V. Int. J. Environ. Res. Publ. Health. 2019;16:1. doi: 10.3390/ijerph16071210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morimoto Y., Ogami A., Todoroki M., Yamamoto M., Murakami M., Hirohashi M., Oyabu T., Myojo T., Nishi K., Kadoya C. Nanotoxicology. 2010;4:161. [Google Scholar]
- 105.Lai X., Zhao H., Zhang Y., Guo K., Xu Y., Chen S., Zhang J. Sci. Rep. 2018;1 doi: 10.1038/s41598-018-22556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song Y., Li X., Du X. Eur. Respir. J. 2009;34:559. doi: 10.1183/09031936.00178308. [DOI] [PubMed] [Google Scholar]
- 107.Lin W., Huang Y., Zhou X., Ma Y. Toxicol. Appl. Pharmacol. 2006;217:252. doi: 10.1016/j.taap.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 108.Skuland T., Godymchuk A., Nguyen T.H.T., Pham H.L.T., Refsnes M. Hybrid. Mater. 2018:620. doi: 10.1111/bcpt.12963. [DOI] [PubMed] [Google Scholar]
- 109.Vesterdal L.K., Mikkelsen L., Folkmann J.K., Sheykhzade M., Cao Y., Roursgaard M., Loft S., Møller P. Toxicol. Lett. 2012;214:19. doi: 10.1016/j.toxlet.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 110.Nurkiewicz T.R., Porter D.W., Hubbs A.F., Cumpston J.L., Chen B.T., Frazer D.G., Castranova V. Part. Fibre Toxicol. 2008;12:1. doi: 10.1186/1743-8977-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Araujo J.A., Barajas B., Kleinman M., Wang X., Bennett B.J., Gong W., Navab M., Harkema J., Sioutas C., Lusis A.J. Circ. Res. 2011;102:589. doi: 10.1161/CIRCRESAHA.107.164970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhu M., Wang B., Wang Y., Yuan L., Wang H., Wang M., Ouyang H., Chai Z., Feng W., Zhao Y. Toxicol. Lett. 2011;203:162. doi: 10.1016/j.toxlet.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 113.Ng K.W., Xie J.P., Ong C.N., Tan N.S., Leong D.T. Nat. Commun. 2013;4:1. doi: 10.1038/ncomms2655. [DOI] [PubMed] [Google Scholar]
- 114.Duan J., Yu Y., Li Y., Li Y., Liu H., Jing L., Yang M., Wang J., Li C., Sun Z. Nanotoxicology. 2016;10:575. doi: 10.3109/17435390.2015.1102981. [DOI] [PubMed] [Google Scholar]
- 115.Bernal-Ramírez J., Garza J.R., García N., Ortega E., García-García A., Torre-Amione G., Ornelas-Soto N., Lozano O., Castillo E.C., Vela J. Am. J. Physiol. Cell Physiol. 2017;312:H645. doi: 10.1152/ajpheart.00564.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Du Z.J., Cui G.Q., Zhang J., Liu X.M., Zhang Z.H., Jia Q., Ng J.C., Peng C., Bo C.X., Shao H. Int. J. Nanomed. 2017;12:2179. doi: 10.2147/IJN.S127904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Savi M., Rossi S., Bocchi L., Gennaccaro L., Cacciani F., Perotti A., Amidani D., Alinovi R., Goldoni M., Aliatis I. Part. Fibre Toxicol. 2014;11:1. doi: 10.1186/s12989-014-0063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stampfl A., Maier M., Radykewicz R., Reitmeir P., Göttlicher M., Niessner R. ACS Nano. 2011;5:5345. doi: 10.1021/nn200801c. [DOI] [PubMed] [Google Scholar]
- 119.E. A. Mutlu, P. A. Engen, S. Soberanes, D. Urich, C. B. Forsyth, R. Nigdelioglu, S. E. Chiarella, K. A. Radigan, A. Gonzalez, S. Jakate, et al., 2011, 1. [DOI] [PMC free article] [PubMed]
- 120.Zheng Z., Zhang X., Wang J., Dandeka A., Kim H., Qiu Y., Xu X., Cui Y., Wang A., Chen L.C. J. Hepatol. 2016;63:1397. doi: 10.1016/j.jhep.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu M., Ge C., Qin Y., Gu T., Lou D., Li Q. Free Radic. Biol. Med. 2019;130:542. doi: 10.1016/j.freeradbiomed.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 122.Tang H., Xu M., Shi F., Ye G., Lv C., Luo J., Zhao L. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cui Y., Liu H., Zhou M., Duan Y., Li N., Gong X., Hu R. J. Biomed. Mater. Res. 2010;96:221. doi: 10.1002/jbm.a.32976. [DOI] [PubMed] [Google Scholar]
- 124.Brook R.D., Rajagopalan S., Iii C.A.P., Brook J.R., Bhatnagar A., V Diez-roux A., Holguin F., Hong Y., V Luepker R., Mittleman M.A. Circulation. 2010:2331. doi: 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- 125.Zhong J., Brook R.D. Antioxidantas Redox Signal. 2018;28:797. doi: 10.1089/ars.2017.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.C. A. P. Iii, A. Bhatnagar, J. P. Mccracken, W. Abplanalp, D. J. Conklin, T. O. Toole, 2016, 1204. [DOI] [PMC free article] [PubMed]
- 127.Apopa P.L., Qian Y., Shao R., Guo N.L., Schwegler-berry D., Pacurari M., Porter D., Shi X., Vallyathan V., Castranova V. Part. Fibre Toxicol. 2008;14:1. doi: 10.1186/1743-8977-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eweje F., Ardoña A.M., Zimmerman J.F., Connor B.B.O., Ahn S., Grevesse T., Rivera K.N., Bitounis D., Kit K. Nanoscale. 2019;11:17878. doi: 10.1039/c9nr04981a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Savoji H., Huyer L.D., Mohammadi M.H., Fook B., Lai L., Rafatian N., Bannerman D., Shoaib M., Bobicki E.R., Ramachandran A. ACS Biomater. Sci. Eng. 2020;6:1333. doi: 10.1021/acsbiomaterials.9b00676. [DOI] [PubMed] [Google Scholar]
- 130.Zhang B., Montgomery M., Chamberlain M.D., Ogawa S., Korolj A., Pahnke A., Wells L.A., Massé S., Kim J., Reis L. Nat. Mater. 2016;15:669. doi: 10.1038/nmat4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fook B., Lai L., Huyer L.D., Xing R., Lu Z., Drecun S. Adv. Funct. Mater. 2017:1. 1703524. [Google Scholar]
- 132.Buchanan C.F., Voigt E.E., Szot C.S., Freeman J.W., Vlachos P.P., Rylander M.N. Tissue Eng. Part C. 2014;20:16. doi: 10.1089/ten.tec.2012.0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mannino R.G., Myers D.R., Ahn B., Wang Y., Timmins L.H., Lam W.A. Sci. Rep. 2015;5:1. doi: 10.1038/srep12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thomas A., Ou-yang H.D., Lowe-krentz L. Biomicrofluidics. 2016;10:1. doi: 10.1063/1.4936672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cho E.C., Zhang Q., Xia Y. Nat. Nanotechnol. 2011;6:385. doi: 10.1038/nnano.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Park M.S., Yoon T.H. Bull. Kor. Chem. Soc. 2014;35 doi: 10.5012/bkcs.2014.35.1.123. [DOI] [Google Scholar]
- 137.Ballerman B., Dardik A., Eng E., Liu A. Kidney Int. 1998;54:100. doi: 10.1046/j.1523-1755.1998.06720.x. [DOI] [PubMed] [Google Scholar]
- 138.Fede C., Albertin G., Petrelli L., De Caro R. J. Nanopaprt. Res. 2017;19:1. doi: 10.1007/s11051-017-3993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.D. Kim, Y. Lin, C. L. Haynes, 2012, 83, 8377.
- 140.Destefano J.G., Xu Z.S., Williams A.J., Yimam N., Searson P.C. Fluids Barriers CNS. 2017;1 doi: 10.1186/s12987-017-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tovar-lopez F., Thurgood P., Gilliam C., Nguyen N. Front. Bioeng. Biotechnol. 2019;7:1. doi: 10.3389/fbioe.2019.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rodin A., Jr T.H.M., Clark A.G., Sing C.F., Boerwinkle E. Arterioscler. Thromb. Vasc. Biol. 2005;12:1. [Google Scholar]
- 143.Ishibazawa A., Nagaoka T., Yokota H., Ono S. Exp. Eye Res. 2013;116:308. doi: 10.1016/j.exer.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 144.Poduri A., Chang A.H., Raftrey B., Rhee S., Van M., Red-horse K. Development. 2017;144:3241. doi: 10.1242/dev.150904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zheng Y., Chen J. Nat. Commun. 2015;6:1. doi: 10.1038/ncomms8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Thomas A., Tan J., Liu Y. Microvasc. Res. 2014;94:17. doi: 10.1016/j.mvr.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chi J., Chang H.Y., Haraldsen G., Jahnsen F.L., Troyanskaya O.G., Chang D.S., Wang Z., Rockson S.G., Van De Rijn M., Botstein D. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:10623. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Setyawati M.I., Tay C.Y., Bay B.H., Leong D.T. ACS Nano. 2017;11:5020. doi: 10.1021/acsnano.7b01744. [DOI] [PubMed] [Google Scholar]
- 149.Pauty J., Usuba R., Takahashi H., Suehiro J., Fujisawa K., Yano K. Nanother. 2017;1:103. doi: 10.7150/ntno.18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ho Y.T., Adriani G., Beyer S., Nhan P., Kamm R.D., Chen J., Kah Y. Sci. Rep. 2017;1 doi: 10.1038/s41598-017-00750-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim Y., Lobatto M.E., Kawahara T., Lee B., Mieszawska A.J. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:10798. doi: 10.1073/pnas.1322725111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.D. Kim, S. Finkenstaedt-quinn, K. R. Hurley, J. T. Buchman, C. L. Haynes, 2015, DOI 10.1039/c3an01679j.
- 153.Ryu H., Oh S., Lee H.J., Lee J.Y., Lee H.K., Jeon N.L. J. Lab. Autom. 2015;20:296. doi: 10.1177/2211068214562831. [DOI] [PubMed] [Google Scholar]
- 154.Zeinali S., Bichsel C.A., Hobi N., Funke M., Marti T.M., Schmid R.A., Guenat O.T., Geiser T. Angiogenesis. 2018;21:861. doi: 10.1007/s10456-018-9631-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim S., Lee H., Chung M., Jeon N.L. Lab Chip. 2013;13:1489. doi: 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- 156.Kendall R.T., Feghali-bostwick C.A., Rauch B.H. Front. Pharmacol. 2014;5:1. doi: 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tracy L.E., Minasian R.A., Caterson E.J. Adv. Would Care. 2016;5:119. doi: 10.1089/wound.2014.0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rajasekar S., Lin D.S.Y., Abdul L., Liu A., Sotra A., Zhang F., Zhang B. Adv. Mater. 2020;32:1. doi: 10.1002/adma.202002974. [DOI] [PubMed] [Google Scholar]
- 159.Li Y., Wu Y., Liu Y., Deng Q., Mak M., Yang X. Nanoscale. 2019;11:15537. doi: 10.1039/c9nr03622a. [DOI] [PubMed] [Google Scholar]
- 160.Mutlu M., Ghio A., Bundinger G.R.S., Soberanes S., Panduri V., Kamp D.W., Respir Am J. Crit. Care Med. 2006;174:1229. doi: 10.1164/rccm.200602-203OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mutlu M., Snyder C., Bellmeyer A., Wang H., Hawkins K., Soberanes S., Welch L.C., Ghio A.J., Chandel N.S., Kamp D. Am. J. Cell Mol. Biol. 2006;34:670. doi: 10.1165/rcmb.2005-0329OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fischa G., Roder C., Wichman G., Nieber K., Lehmann I. Toxicol. Vitro. 2008;22:359. doi: 10.1016/j.tiv.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 163.Xu X., Jiang S.Y., Wang T., Bai Y., Zhong M., Wang A., Lippmann M., Chen L., Rajagopalan S., Sun Q. PloS One. 2013;8:1. doi: 10.1371/journal.pone.0071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shadie A.M., Herbert C., Kumar R.K. Clin. Exp. Immunol. 2014;177:491. doi: 10.1111/cei.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Soukup J.M., Becker S., Particulate I., Soukup E. Toxicol. Appl. Pharmacol. 2001;26:20. doi: 10.1006/taap.2000.9096. [DOI] [PubMed] [Google Scholar]
- 166.Dagher Z., Garc G., Courcot D., Aboukais A., Shirali P. Toxicology. 2006;225:12. doi: 10.1016/j.tox.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 167.Provoost S., Maes T., Pauwels N.S., Berghe V., Vandenabeele P., Lambrecht B.N., Joos G.F., Tournoy K.G. J. Immunol. 2011;187:3331. doi: 10.4049/jimmunol.1004062. [DOI] [PubMed] [Google Scholar]
- 168.Agostini C., Gurrieri C. Proc. Am. Thorac. Soc. 2006;3:357. doi: 10.1513/pats.200601-010TK. [DOI] [PubMed] [Google Scholar]
- 169.Barnes P.J. J. Clin. Invest. 2008;118:3546. doi: 10.1172/JCI36130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Aukrust P., Ueland T., Lien E., Bendtzen K., Müller F., Andreassen a K., Nordøy I., Aass H., Espevik T., Simonsen S. Am. J. Cardiol. 1999;83:376. doi: 10.1016/s0002-9149(98)00872-8. [DOI] [PubMed] [Google Scholar]
- 171.J. C. Kips, 2001, 24.
- 172.Schütte H., Lohmeyer J., Rosseau S., Ziegler S., Siebert C., Kielisch H., Pralle H., Grimminger F., Morr H., Seeger W. Eur. Respir. J. 1996;9:1858. doi: 10.1183/09031936.96.09091858. [DOI] [PubMed] [Google Scholar]
- 173.Oberdorster G., Ferin J., Lehnert B.E. Environ. Health Perspect. 1994:173. doi: 10.1289/ehp.102-1567252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Müller L., Brighton L.E., Carson J.L., Fischer W.A., Jaspers I. J. Vis. Exp. 2013;1 doi: 10.3791/50646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cozens D., Sutherland E., Marchesi F., Taylor G., Berry C.C., Davies R.L. Sci. Rep. 2018;8:1. doi: 10.1038/s41598-018-33180-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pezzulo A.A., Starner T.D., Scheetz T.E., Traver G.L., Tilley A.E., Harvey B.G., Crystal R.G., McCray P.B., Zabner J. Am. J. Physiol. Lung Cell Mol. Physiol. 2011;300:25. doi: 10.1152/ajplung.00256.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Panas A., Comouth A., Saathoff H., Leisner T., Al-rawi M., Simon M., Seemann G., Dössel O., Mülhopt S., Paur H. J. Nanotechnol. 2014;5:1590. doi: 10.3762/bjnano.5.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lenz A.G., Karg E., Brendel E., Hinze-Heyn H., Maier K.L., Eickelberg O., Stoeger T., Schmid O. BioMed Res. Int. 2013 doi: 10.1155/2013/652632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Loret T., Peyret E., Dubreuil M., Aguerre-Chariol O., Bressot C., le Bihan O., Amodeo T., Trouiller B., Braun A., Egles C. Part. Fibre Toxicol. 2016;13 doi: 10.1186/s12989-016-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bitterle E., Karg E., Schroeppel A., Kreyling W.G., Tippe A., Ferron G.A., Schmid O., Heyder J., Maier K.L., Hofer T. Chemosphere. 2006;65:1784. doi: 10.1016/j.chemosphere.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 181.Huh D., Leslie D.C., Matthews B.D., Fraser J.P., Jurek S., Hamilton G.A., Thorneloe K.S., Mcalexander M.A., Ingber D.E. Sci. Transl. Med. 2018;4:1. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cao Y., Chen M., Dong D., Xie S., Liu M. Thorac. Canc. 2020;11:505. doi: 10.1111/1759-7714.13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Breathing M., Chip L., Benam K.H., Novak R., Nawroth J., Bahinski A., Parker K.K., Ingber D.E., Benam K.H., Novak R. Cell Syst. 2016;3:456. doi: 10.1016/j.cels.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 184.Humayun M., Chow C., Young E.W.K. Lab Chip. 2018;18:1298. doi: 10.1039/c7lc01357d. [DOI] [PubMed] [Google Scholar]
- 185.M. Asmani, S. Velumani, Y. Li, N. Wawrzyniak, Z. Chen, B. Hinz, R. Zhao, Nat. Commun.. n.d., 1. [DOI] [PMC free article] [PubMed]
- 186.Chen Z., Wang Q., Asmani M., Li Y., Liu C., Li C., Lippmann J.M., Wu Y., Zhao R. Nat. Publ. Gr. 2016;1 [Google Scholar]
- 187.Grigoryan B., Paulsen S.J., Corbett D.C., Sazer D.W., Fortin C.L., Zaita A.J., Greenfield P.T., Calafat N.J., Gounley J.P., Ta A.H. Science. 2019;364:458. doi: 10.1126/science.aav9750. (80-. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xie W., Li G., Zhao D., Xie X., Wei Z., Wang W., Wang M., Li G., Liu W., Sun J. Heart. 2015;101:257. doi: 10.1136/heartjnl-2014-306165. [DOI] [PubMed] [Google Scholar]
- 189.Bartell S.M., Longhurst J., Tjoa T., Sioutas C., Delfino R.J. Environ. Health Perspect. 2013;121:1135. doi: 10.1289/ehp.1205914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rückerl R., Greven S., Ljungman P., Aalto P., Antoniades C., Bellander T., Berglind N., Chrysohoou C., Forastiere F., Jacquemin B. Environ. Health Perspect. 2007;1072:1072. doi: 10.1289/ehp.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Van Eeden S.F., Tan W.A.N.C., Suwa T., Mukae H., Terashima T., Fujii T., Qui D., Vincent R., Hogg J.C. Am. J. Respir. Crit. Care Med. 2001;164:826. doi: 10.1164/ajrccm.164.5.2010160. [DOI] [PubMed] [Google Scholar]
- 192.Kong P., Christina P., Frangogiannis N. Cell. Mol. Life Sci. 2014;71:549. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Frangogiannis N.G., Dewald O., Xia Y., Ren G., Haudek S., Leucker T., Kraemer D., Taffet G., Rollins B.J., Entman M.L. Circulation. 2007;115:584. doi: 10.1161/CIRCULATIONAHA.106.646091. [DOI] [PubMed] [Google Scholar]
- 194.Tanaka T., Kanda T., Takahashi T., Saegusa S., Moriya J., Kurabayashi M. J. Int. Med. Res. 2004;32:57. doi: 10.1177/147323000403200109. [DOI] [PubMed] [Google Scholar]
- 195.Liu Y., Goodson J.M., Zhang B., Chin M.T. Front. Physiol. 2015;6:1. doi: 10.3389/fphys.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Takenaka S., Karg E., Kreyling W.G., Lentner B. Inhal. Toxicol. 2006;18:733. doi: 10.1080/08958370600748281. [DOI] [PubMed] [Google Scholar]
- 197.Hsiao C., Azarin S.M., Hazeltine L.B., Lian X., Kamp T.J., Zhang J., Raval K.K., Zhu K., Palecek S.P., Wilson G. Proc. Natl. Acad. Sci. Unit. States Am. 2012;109:E1848. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva S.M., Lopes D.S. EMBO Mol. Med. 2015;7:394. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Boudou T., Legant W.R., Mu A., Borochin M.A., Thavandiran N., Radisic M., Zandstra P.W., Epstein J.A., Margulies K.B., Chen C.S. Tissue Eng. Part A. 2012;18:910. doi: 10.1089/ten.tea.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vandenburgh H., Shansky J., Benesch-Lee F., Barbata V., Reid J., Thorrez L., Valentini R., Crawford G. Muscle Nerve. 2008;37:438. doi: 10.1002/mus.20931. [DOI] [PubMed] [Google Scholar]
- 201.Goldfracht I., Protze S., Shiti A., Setter N., Nartiss Y., Keller G., Gepstein L., Gruber A., Shaheen N. Nat. Commun. 2020:1. doi: 10.1038/s41467-019-13868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xiao Y., Zhang B., Liu H., Miklas J.W., Gagliardi M., Pahnke A., Thavandiran N., Sun Y., Simmons C., Keller G. Lab Chip. 2014;14:869. doi: 10.1039/c3lc51123e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Feinberg A.W., Feigel A., Shevkoplyas S.S., Sheehy S., Whitesides G.M., Parker K.K. Science. 2007;317:1366. doi: 10.1126/science.1146885. (80-. ) [DOI] [PubMed] [Google Scholar]
- 204.Grosberg A., Alford P.W., McCain M.L., Parker K.K. Lab Chip. 2011;11:4165. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang E.Y., Rafatian N., Zhao Y., Lee A., Fook B., Lai L., Lu R.X., Jekic D., Huyer L.D., Knee-walden E.J. ACS Cent. Sci. 2019;5:1146. doi: 10.1021/acscentsci.9b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mastikhina O., Moon B., Williams K., Hatkar R., Gustafson D., Mourad O., Sun X., Koo M., Lam A.Y.L., Sun Y. Biomaterials. 2020;233:119741. doi: 10.1016/j.biomaterials.2019.119741. [DOI] [PubMed] [Google Scholar]
- 207.Ma Z., Huebsch N., Koo S., Mandegar M.A., Healy K.E. Nat. Biomed. Eng. 2019;2:955. doi: 10.1038/s41551-018-0280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.S. Ahn, H. A. M. Ardoña, J. U. Lind, F. Eweje, S. L. Kim, G. M. Gonzalez, 2018, DOI 10.1007/s00216-018-1106-7.
- 209.Mosterd A., Hoes A.W. Heart. 2007;93:1137. doi: 10.1136/hrt.2003.025270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bhattacharya S., Burridge P.W., Kropp E.M., Chuppa S.L., Kwok W., Wu J.C., Boheler K.R., Gundry R.L. J. Vis. Exp. 2014;91:1. doi: 10.3791/52010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rossi S., Savi M., Mazzola M., Pinelli S., Alinovi R., Gennaccaro L., Pagliaro A., Meraviglia V., Galetti M., Lozano-garcia O. Part. Fibre Toxicol. 2019;16:1. doi: 10.1186/s12989-019-0311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pinto A.R., Ilinykh A., Ivey M.J., Kuwabara J.T., Antoni M.L.D., Debuque R., Chandran A., Wang L., Arora K., Rosenthal N.A. Circ. Res. 2016;118:400. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mathur A., Loskill P., Shao K., Huebsch N., Hong S., Marcus S.G., Marks N., Mandegar M., Conklin B.R., Lee L.P. Sci. Rep. 2015;5:1. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lu R.X.Z., Lai B.F.L., Benge T., Wang E.Y., Davenport Huyer L., Rafatian N., Radisic M. Adv. Mater. Technol. 2020:2000726. n/a. [Google Scholar]
- 215.Lee A., Hudson A.R., Shiwarski D.J., Tashman J.W., Hinton T.J., Yerneni S., Bliley J.M., Campbell P.G., Feinberg A.W. Science. 2019:482. doi: 10.1126/science.aav9051. (80-. ) [DOI] [PubMed] [Google Scholar]
- 216.Lomer M.C.E., Hutchinson C., Volkert S., Greenfield S.M., Catterall A., Thompson R.P.H., Powell J.J. Br. J. Nutr. 2004;92:947. doi: 10.1079/bjn20041276. [DOI] [PubMed] [Google Scholar]
- 217.Nogueira C.M., De Azevedo W.M., Lucia M., Dagli Z., Toma S.H., Leite Z.D.A., Lordello M.L., Nishitokukado I., Ortiz-agostinho C.L., Irma M. World J. Gastroenterol. 2012;18:4729. doi: 10.3748/wjg.v18.i34.4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bamias G., Cominelli F. Curr. Opin. Gastroenterol. 2016;32:437. doi: 10.1097/MOG.0000000000000315. [DOI] [PubMed] [Google Scholar]
- 219.Neurath M.F. Nat. Rev. 2014;14:329. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 220.Kish L., Hotte N., Kaplan G.G., Vincent R., Tso R., Ga M., Rioux K.P., Thiesen A., Barkema H.W., Wine E. PloS One. 2013;8:1. doi: 10.1371/journal.pone.0062220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Fouladi F., Bailey M.J., Patterson W.B., Sioda M., Blakley I.C., Fodor A.A., Jones R.B., Chen Z., Kim J.S., Lurmann F. Environ. Int. 2020;138:105604. doi: 10.1016/j.envint.2020.105604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gioria S., Urbán P., Hajduch M., Barboro P., Cabaleiro N., La R., Chassaigne H. Toxicol. Vitro. 2018;50:347. doi: 10.1016/j.tiv.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.A. F. G. Strugari, M. S. Stan, S. Gharbia, A. Hermenean, 2018, DOI 10.3390/nano9010005.
- 224.Kim H., Huh D., Hamilton G., Ingber D. Lab Chip. 2012;12:2165. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 225.Jung H., Li H., Collins J.J., Ingber D.E. Proc. Natl. Acad. Sci. Unit. States Am. 2015;113:7. [Google Scholar]
- 226.Zhang J., Huang Y.-J., Yoon J.Y., Kemmitt J., Wright C., Schneider K., Sphabmixay P., Hernandez-Gordillo V., Holcomb S.J., Bhushan B. Med. 2020 doi: 10.1016/j.medj.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Trapecar M., Communal C., Velazquez J., Yilmaz O., Trumper D., Griffith L.G., Trapecar M., Communal C., Velazquez J., Maass C.A. Cell Syst. 2020;10:223. doi: 10.1016/j.cels.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Guarner F., Malagelada J. Lancet. 2003;360:512. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 229.Ottman N., Smidt H., De Vos W.M., Belzer C. Front. Cell. Infect. Microbiol. 2012;2:1. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pan W., Wu C., Chen M., Huang Y., Chen C., Su H., Yang H. J. Natl. Cancer Inst. 2016;108:1. doi: 10.1093/jnci/djv341. [DOI] [PubMed] [Google Scholar]
- 231.Cornu R., Rougier N., Pellequer Y., Lamprecht A., Hamon P. Nanoscale. 2018;10:5171. doi: 10.1039/c8nr00226f. [DOI] [PubMed] [Google Scholar]
- 232.Martignoni M., Groothuis G.M.M., Kanter R. Expet Opin. Drug Metabol. Toxicol. 2007;2:875. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- 233.Shukla R.K., Kumar A., Gurbani D., Pandey A.K., Dhawan A., Shukla R.K., Kumar A., Gurbani D., Pandey A.K., Singh S. Nanotoxicology. 2013;5390:48. doi: 10.3109/17435390.2011.629747. [DOI] [PubMed] [Google Scholar]
- 234.Hussain S.M., Hess K.L., Gearhart J.M., Geiss K.T., Schlager J.J. Toxicol. Vitro. 2005;19:975. doi: 10.1016/j.tiv.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 235.Li L., Gokduman K., Gokaltun A., Yarmush M.L., Berk O. Nanomedicine. 2019;14:2209. doi: 10.2217/nnm-2019-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Esch M.B., Mahler G.J., Stokol T., Shuler M.L., States U., States U., Road T., States U., Sciences D., States U. Lab Chip. 2015;14:3081. doi: 10.1039/c4lc00371c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Jin Y., Kim J., Lee J.S., Min S., Kim S., Ahn D., Kim Y., Cho S. Adv. Funct. Mater. 2018:1. 1801954. [Google Scholar]
- 238.Prodanov L., Jindal R., Bale S.S., Hedge M., McCarty W., Golberg I., Bhushan A., Yarmush M.L., Usta O.B. Curr. Opin. Pulm. Med. 2017;22:138. [Google Scholar]
- 239.Jang K., Otieno M.A., Ronxhi J., Lim H., Ewart L., Kodella K.R., Petropolis D.B., Kulkarni G., Rubins J.E., Conegliano D. Sci. Transl. Med. 2019;11:1. doi: 10.1126/scitranslmed.aax5516. [DOI] [PubMed] [Google Scholar]
- 240.A. R. Wheeler, 2019.
- 241.van Meer B.J., de Vries H., Firth K.S.A., van Weerd J., Tertoolen L.G.J., Karperien H.B.J., Jonkheijm P., Denning C., IJzerman A.P., Mummery C.L. Biochem. Biophys. Res. Commun. 2017;482:323. doi: 10.1016/j.bbrc.2016.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Toepke M.W., Beebe D.J. Lab Chip. 2006;6:1484. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 243.Luo Y., Zhao R., Pendry J.B. Proc. Natl. Acad. Sci. U.S.A. 2014;111:18422. doi: 10.1073/pnas.1420551111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Auría-Soro C., Nesma T., Juanes-Velasco P., Landeira-Viñuela A., Fidalgo-Gomez H., Acebes-Fernandez V., Gongora R., Parra M.J.A., Manzano-Roman R., Fuentes M. Nanomaterials. 2019;9 doi: 10.3390/nano9101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ahmed S.R., Kim J., Tran V.T., Suzuki T., Neethirajan S., Lee J., Park E.Y. Sci. Rep. 2017;7:1. doi: 10.1038/srep44495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Campbell S.B., Wu Q., Yazbeck J., Liu C., Okhovatian S., Radisic M. ACS Biomater. Sci. Eng. 2020 doi: 10.1021/acsbiomaterials.0c00640. [DOI] [PubMed] [Google Scholar]
- 247.Domansky K., Sliz J.D., Wen N., Hinojosa C., Thompson G., Fraser J.P., Hamkins-Indik T., Hamilton G.A., Levner D., Ingber D.E. Microfluid. Nanofluidics. 2017;21 doi: 10.1007/s10404-017-1941-4. [DOI] [Google Scholar]
- 248.Gökaltun A., Kang Y. B. (Abraham), Yarmush M.L., Usta O.B., Asatekin A. Sci. Rep. 2019;9:1. doi: 10.1038/s41598-019-43625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Gokaltun A., Yarmush M.L., Asatekin A., Usta O.B. Technology. 2017;5:1. doi: 10.1142/S2339547817300013. [DOI] [PMC free article] [PubMed] [Google Scholar]