Abstract
Background & Aims
p21-activated kinase-1 (PAK1) belongs to a family of serine-threonine kinases and contributes to cellular pathways such as nuclear factor-κB (NF-κB), mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT), and Wingless-related integration site(Wnt)/β-catenin, all of which are involved in intestinal homeostasis. Overexpression of PAK1 is linked to inflammatory bowel disease as well as colitis-associated cancer (CAC), and similarly was observed in interleukin (IL)10 knockout (KO) mice, a model of colitis and CAC. Here, we tested the effects of PAK1 deletion on intestinal inflammation and carcinogenesis in IL10 KO mice.
Methods
IL10/PAK1 double-knockout (DKO) mice were generated and development of colitis and CAC was analyzed. Large intestines were measured and prepared for histology or RNA isolation. Swiss rolls were stained with H&E and periodic acid-Schiff. Co-immunoprecipitation and immunofluorescence were performed using intestinal organoids, SW480, and normal human colon epithelial cells 1CT.
Results
When compared with IL10 KO mice, DKOs showed longer colons and prolonged crypts, despite having higher inflammation and numbers of dysplasia. Crypt hyperproliferation was associated with Notch1 activation and diminished crypt differentiation, indicated by a reduction of goblet cells. Gene expression analysis indicated up-regulation of the Notch1 target hairy and enhancer of split-1 and the stem cell receptor leucin-rich repeat-containing G-protein–coupled receptor 5 in DKO mice. Interestingly, the stem cell marker olfactomedin-4 was present in colonic tissue. Increased β-catenin messenger RNA and cytoplasmic accumulation indicated aberrant Wnt signaling. Co-localization and direct interaction of Notch1 and PAK1 was found in colon epithelial cells. Notch1 activation abrogated this effect whereas silencing of PAK1 led to Notch1 activation.
Conclusions
PAK1 contributes to the regulation of crypt homeostasis under inflammatory conditions by controlling Notch1. This identifies a novel PAK1–Notch1 axis in intestinal pathophysiology of inflammatory bowel disease and CAC.
Keywords: PAK1, Notch1, Stem Cell, Inflammation, Colitis-Associated Cancer, Inflammatory Bowel Disease
Abbreviations used in this paper: AKT, protein kinase B; AOM, azoxymethane; CAC, colitis-associated cancer; CRC, colorectal cancer; DAI, disease activity index; DKO, double-knockout; DSS, dextran sodium sulfate; HCEC-1CT, human colon epithelial cells 1CT; HES1, hairy and enhancer of split-1; IBD, inflammatory bowel disease; IL, interleukin; JNK, c-Jun N-terminal kinase; KO, knockout; LBO, large bowel organoid; LGR5, leucin-rich repeat-containing G-protein–coupled receptor 5; MAPK, mitogen-activated protein kinase; MMP, matrix metalloproteinase; mRNA, messenger RNA; NF-κB, nuclear factor-κB; NICD, Notch1 intracellular domain; OLFM4, olfactomedin-4; PAK1, p21-activated kinase-1; PPAR, Peroxisome proliferator-activated receptor; ROS, reactive oxygen species; UC, ulcerative colitis; Wnt, Wingless-related integration site; WT, wild-type
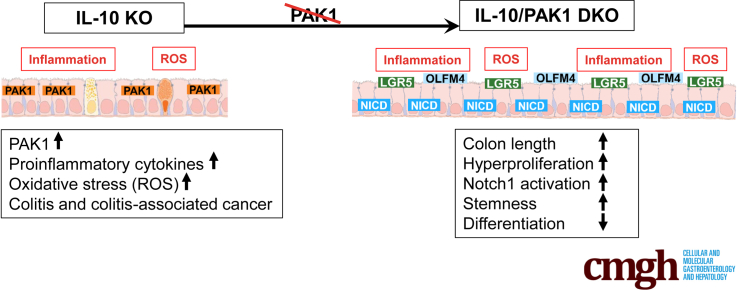
Graphical abstract
Summary.
We identified a novel p21-activated kinase-1 (PAK1)–Notch1 axis, in which PAK1 deletion in the context of intestinal inflammation results in Notch pathway activation, diminished differentiation, and increased crypt stemness. This interplay of PAK1 and Notch1 is fundamental in gut homeostasis.
Current pathogenetic concepts in inflammatory bowel disease (IBD) with its 2 main forms, ulcerative colitis (UC) and Crohn’s disease, include food and smoke-born environmental factors, changes in gut microbiota, disturbance of the epithelial barrier, intestinal immune response, and genetic predisposition.1 Overt oxidative stress is a consequence of chronic inflammation of the gastrointestinal tract, with disturbances of cellular homeostasis resulting in DNA damage as well as changes in differentiation and proliferation.2,3 Such changes ultimately can result in neoplastic transformation and further progress to colitis-associated cancer (CAC).4 Since its generation in the early 1990s, interleukin (IL)10 knockout (KO) mice have served as murine model of chronic enterocolitis and CAC development.5
Our laboratory showed that the p21-activated kinase-1 (PAK1) is overexpressed in IBD and CAC and is a target of mesalamine, the mainstay treatment in mild to moderate colitis.6 In a recent integrated multi-omics analysis, PAK signaling was identified as a driver of colitis.7 PAK1 has effects on cell adhesion, mitogen-activated protein kinase (MAPK), Wingless-related integration site (Wnt)/β-catenin, and nuclear factor-κB (NF-κB) signaling in human beings and in mouse models of colitis and colorectal cancer (CRC), as well as in various other cancers.8, 9, 10, 11, 12 Its multifaceted activation through binding of the small Rho guanosine triphosphatases Ras-related C3 botulinum toxin substrate 1/Cell division control protein 42 homolog (Rac1/CDC42), growth factors, phosphoinositides/membranous lipids, and adaptor proteins underlines its involvement in cell motility and structural organization.13 Further effects on cell migration, proliferation, and survival are attributed to its kinase activity and initiation of transcription pathways.10 For instance, via phosphorylation of Raf murine sarcoma viral oncogene homolog (RAF), PAK1 enhances MAPK signaling as well as phosphatidylinositol 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) and Cellular myelocytomatosis oncogene (c-myc).11 Recent findings also have suggested involvement of PAK1 in nuclear translocation of β-catenin via phosphorylation.14,15
Intestinal development and homeostasis rely on highly conserved pathways such as the Notch and Wnt pathway. Notch signaling is implicated in the regulation of stem cells, intestinal cell fate decision, and proliferation.16 Notch consists of an extracellular, a transmembranous, and an intracellular domain (NICD), and is activated upon binding of its transmembranous ligands such as delta-like 1/3/4 and Jagged1/2 (JAG1, JAG2), which are expressed by neighboring cells. Binding results in cleavage of the NICD by the presenilin-γ-secretase complex, followed by nuclear translocation of NICD where it binds to Recombining binding protein suppressor of hairless (RBPJ), resulting in transcriptional activation of hairy and enhancer of split-1 (HES1),17 thereby initiating intestinal differentiation into the absorptive cell type. In addition, implications have been made that Notch signaling has an important role in CRC and chronic inflammatory conditions such as IBD.3,18, 19, 20, 21, 22, 23
To better understand the role of PAK1 in colitis and colitis-associated carcinogenesis we crossed PAK1-depleted mice into the IL10 KO background. In these double-knockout (DKO) mice, PAK1 deficiency resulted in significant changes in crypt architecture, more severe inflammation, and, ultimately, more dysplastic lesions.
Results
PAK1 Deficiency Abrogates Crypt Differentiation in IL10 KO Mice
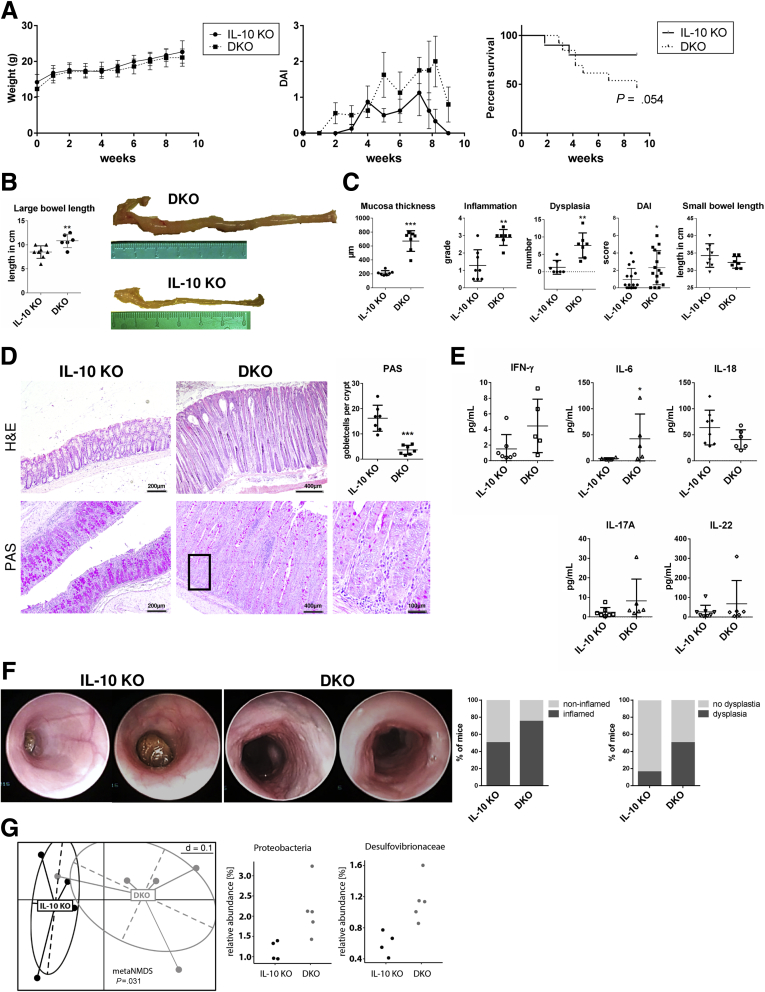
PAK1 is considered a central driver of colitis. Here, we hypothesized that PAK1 deletion will ameliorate colitis and tumorigenesis in IL10 KO mice. IL10 KO mice were crossed with PAK1 KO mice to generate PAK1/IL10 DKO mice. Six- to 8-week-old mice received piroxicam (200 ppm) mixed into their chow for 7 days to enhance and synchronize intestinal inflammation. Total weight gain of IL10 KO and DKO mice was similar over the course of the experiment (Figure 1A). DKOs showed higher average disease activity over time (P < .02) and a trend toward higher mortality (Figure 1A and C). Macroscopically, length measurement of the intestine showed significantly longer colons in DKO compared with IL10 KO mice (10.83 ± 0.57 vs 8.50 ± 0.46 cm; P = .009) (Figure 1B), a phenotype that is not present in PAK1 KO mice.24 The length of the small intestine showed no differences (Figure 1C). Histologic analysis showed hyperproliferative colonic crypts with higher grades of inflammation and more dysplastic lesions in DKO compared with IL10 KO (Figure 1C and D). The crypts of DKO mice showed reduced numbers of goblet cells (as indicated by periodic acid–Schiff staining), reflecting a severe disturbance of crypt differentiation (Figure 1D). To measure systemic inflammation, cytokine multiplexing was used to measure serum cytokines. IL6 was increased 10-fold in DKO mice, whereas interferon-γ, IL18, IL22, and IL17A showed no difference (Figure 1E). In addition, changes in gut microbiota were indicative of a proinflammatory state (Figure 1G).25
Figure 1.
Exacerbated enterocolitis and colitis-associated tumorigenesis upon PAK1 deletion results in loss of goblet cells. (A) Weight curve, DAI, and survival of IL10 KO and DKO mice. Weight gain was similar in IL10 KO and DKO. DKO mice showed higher DAI and a trend toward higher mortality (80% vs 46.15%; P = .054) over the course of the experiment. (B) Length measurement of IL10 KO and DKO colons, DKOs had significantly longer colons (P < .01) compared with IL10 KO mice. (C) Histologic analysis showed significantly higher grades of inflammation (P < .05), more dysplastic lesions (P < .01), and a thicker mucosal layer (P < .05) in DKO compared with IL10 KO mouse colons. The average DAI over time indicated a significant increase in DKO compared with IL10 KO mice (P < .05). Length measurement of the small intestine of IL10 KO and DKO mice did not show significant difference as observed in the DKO colon. (D) Exemplary H&E stains of IL10 KO and DKO colons and periodic acid–Schiff (PAS) staining. DKOs showed a reduction in goblet cells throughout the colon indicating an impaired differentiation. (E) Serum cytokines of IL10 KO and DKO mice were analyzed. IL6 was increased significantly (P < .05) in the serum of DKO mice compared with IL10 KO mice. Other cytokines showed no significant differences. (F) Exemplary colonoscopy pictures of 2 IL10 KO mice with macroscopically normal mucosa and 2 DKO mice with hyperproliferative mucosa. DKO mice showed more signs of inflammation and dysplasia in the distal colon (IL10 KO, n = 12; DKO, n = 8). (G) Nonmetric multidimensional scaling of stool samples indicating different bacterial composition in DKO compared with IL10 KO mice. Composition of the bacterial phylum Proteobacteria showed an increase in the Desulfovibrionaceae family. Independent samples t test, ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
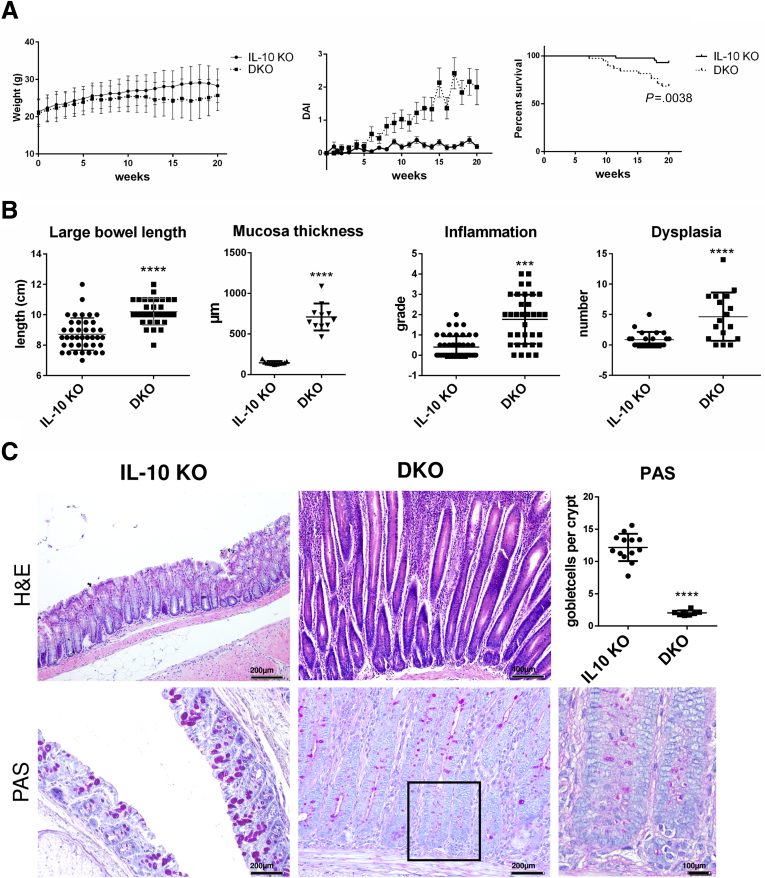
A separate experiment was conducted to exclude any effect of piroxicam on this phenotype. Six- to 8-week-old mice were kept for 20 weeks and disease activity was scored. Colonoscopy was performed after 12 weeks to evaluate inflammation and dysplastic progression. Colonoscopy showed more signs of inflammation and more dysplastic lesions in DKO mice (Figure 1F). Without piroxicam, weight gain was lower in DKO compared with IL10 KO mice (Figure 2A). The disease activity index (DAI) and mortality were significantly higher in DKO compared with IL10 KO mice (Figure 2A). The mice were sacrificed after 20 weeks and the DKO mice showed longer colons (10.2 ± 0.16 vs 8.7 ± 0.17 cm; P < .0001) despite histologically higher grades of inflammation (1.77 ± 0.2 vs 0.4 ± 0.1; P < .0001) and a higher number of dysplastic lesions (4.6 ± 0.96 vs 0.9 ± 0.2; P = .0001) (Figure 2B). Similarly, increased mucosal thickness and goblet cell depletion was observed (Figure 2C). Interestingly, PAK1 KO mice have a macroscopically and histologically normal-appearing colon and small intestine and show no particular signs of hyperproliferation or dysregulation of differentiation.24
Figure 2.
PAK1 deficiency results in exacerbated spontaneous enterocolitis and colitis-associated tumorigenesis with concomitant reduction of goblet cells. (A) Weight curve, DAI, and survival curve of spontaneous CAC model. DKO mice had significantly lower body weight (P < .05), higher DAI (P < .001), and higher mortality (P < .0041) compared with IL10 KO mice. (B) Macroscopic and microscopic analysis of IL10 KO and DKO colons. DKO mice had significantly longer colons (P < .001), higher grades of inflammation (P < .001), and longer crypts (P < .001). (C) Exemplary H&E stains of IL10 KO and DKO colons and periodic acid–Schiff (PAS) staining. Independent samples t test, ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, ∗∗∗∗P < .0001. DAI, Disease activity index.
These data reject our hypothesis that PAK1 deletion ameliorates colitis and carcinogenesis in IL10 KO mice, but rather alters crypt differentiation with loss of protective secretory cells, which results in inflammation and neoplastic growth.
DKO Mice Show Notch1 Pathway Activation and Increased Stemness
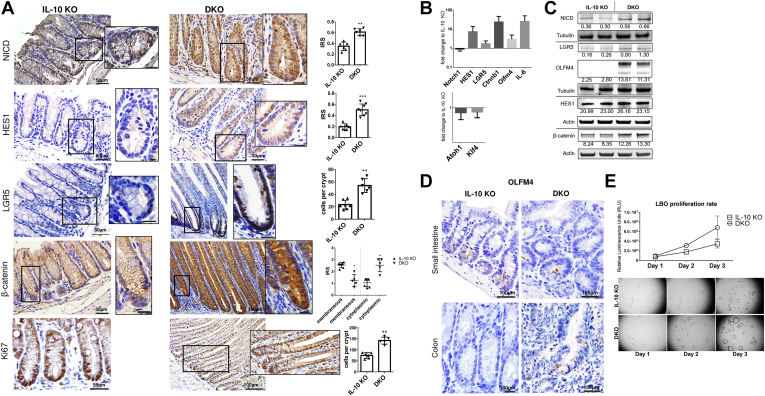
The earlier-described findings show that DKO mice have hyperplastic colonic crypts with increased proliferation and abrogated crypt differentiation into secretory cells. Such changes are indicative of alterations in the Notch pathway. DKO mouse colons showed nuclear accumulation of activated Notch1 (NICD) compared with IL10 KO mice (Figure 3A). In addition, the downstream target of NICD, HES1, the stem cell marker leucin-rich repeat-containing G-protein–coupled receptor 5 (LGR5), and the Wnt/β-catenin pathway, showed higher expression (Figure 3A). HES1 indicated enhanced nuclear staining in DKO compared with IL10 KO mice, suggesting Notch-pathway activation. LGR5 showed expanded positive staining at the colonic crypt base in DKO colons, showing expansion of the stem cell niche. Interestingly, and likely not related to Notch activation and LGR5 expansion, a loss in membranous and an increase in cytoplasmic β-catenin also was observed in DKO mice (Figure 3A). Furthermore, Ki67 staining showed vast expansion of the proliferative crypt zone. Hes1 as well as Olfm4, but also Lgr5, β-catenin, and IL6, also showed increased messenger RNA (mRNA) level while Notch1 mRNA itself remained unchanged. The opposing transcription factor Atoh1 and differentiation marker Klf4 mRNA levels were reduced (Figure 3B), reflecting Notch-pathway activation, increased stemness, and inflammation. NICD and its target olfactomedin-4 (OLFM4) also were increased using immunoblotting from colonic tissue (Figure 3C), and so was β-catenin. HES1 showed no difference in total levels. Colonic OLFM4 expression was confirmed in colonic crypts of DKO mice (Figure 3D). OLFM4, a secreted glycoprotein, usually is not expressed in mouse colon, although it is associated with colitis in human beings.26,27
Figure 3.
PAK1 deficiency results in Notch1 activation and increased stemness in colonic crypts of IL10 KO mice. (A) Immunohistochemistry (IHC) showed increased Notch1 pathway activation and expansion of the stem cell compartment, marked by NICD, Hes1, Olfm4, and Lgr5. In addition, β-catenin showed a cytoplasmic accumulation in DKO compared with IL10 KO. Ki67 indicated expansion of the proliferative zone at the crypt base of DKO mice. (B) mRNA analysis from IL10 KO and DKO colons indicated an increase in the Notch1 downstream target Hes1, stem cell markers Lgr5 and Olfm4, and Wnt target β-catenin, while Atoh1 and Klf4 were down-regulated. In addition, mRNA of the proinflammatory cytokine IL6 was highly up-regulated in DKO. (C) Immunoblotting of protein extracts from IL10 KO and DKO colonic samples corroborated IHC findings for NICD, LGR5, and β-catenin. (D) IHC for OLFM4 marking the stem cell zone at the base of the crypt in the small intestine of IL10 and DKO mice. OLFM4-positive colon crypts were found in DKO mice while IL10 KO colonic crypts showed no OLFM4 in any sample. (E) Proliferation assay indicating a higher rate of proliferation in DKO LBOs. Exemplary pictures of LBOs from IL10 KO and DKO mice over the course of 3 days underlining hyperproliferative phenotype. Graph shows means + SD of 4 wells from 2 independent experiments. For statistical data, a 2-tailed Student t test or the Kruskal-Wallis ANOVA was used. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001, ∗∗∗∗P < .0001.
Overall, these findings indicate the expansion of the stem cell niche (LGR5) and absorptive enterocyte lineage by activation of Notch1 paralleled by overexpression and cytoplasmic accumulation of β-catenin resulting in hyperproliferation and enlargement of colonic crypts and colon length. To further verify the increase in stemness, large-bowel organoids from IL10 KO and DKO mice were used to analyze organoid proliferation. Large-bowel organoids (LBOs) from DKO mice had a higher proliferation rate and were larger in size (Figure 3E), further supporting the in vivo phenotype.
PAK1 and Notch1 Are Associated in Intestinal Epithelial Cells
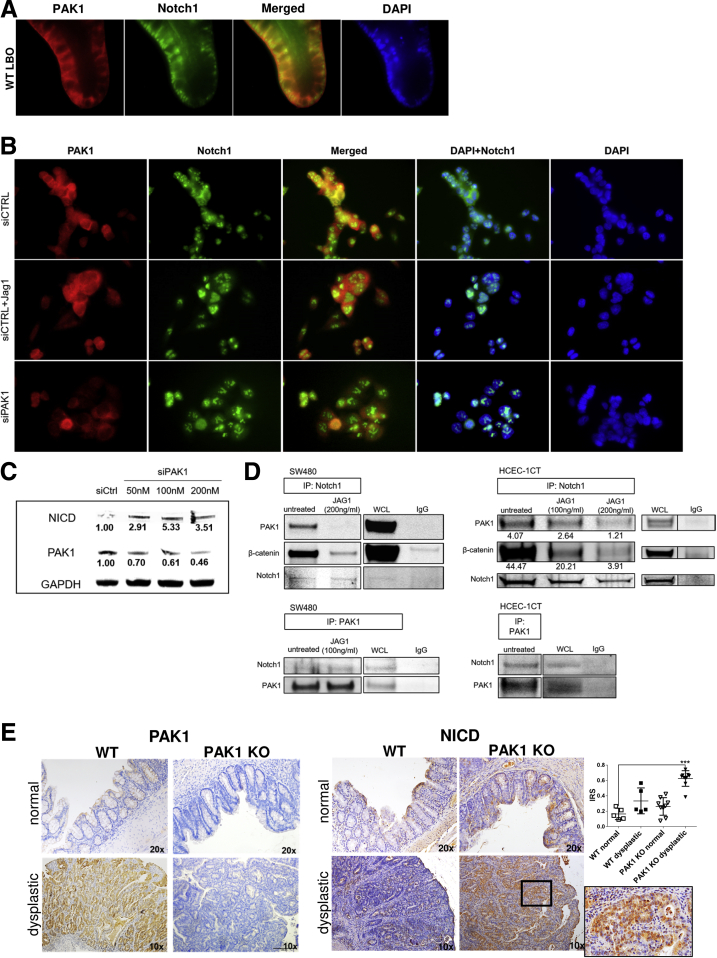
The earlier-described findings imply direct interaction between PAK1 and Notch1 in the intestinal epithelium. In LBOs from wild-type (WT) mice, staining for PAK1 and Notch1 indicated a co-localization in the basal membrane region (Figure 4A). For further analysis we used SW480 cells and normal diploid human colon epithelial cells 1CT (HCEC-1CT). Immunofluorescence in SW480 similarly indicated cytoplasmic co-localization of Notch1 and PAK1, while upon Notch-pathway activation using Jagged1, the co-localization was lost and nuclear Notch1 signal increased (Figure 4B). PAK1 small interfering RNA (siRNA) resulted in a similar strong nuclear Notch1 signal (Figure 4B) and led to an increase of activated Notch1 (Figure 4C)
Figure 4.
PAK1 associates and co-localizes with Notch1. (A) Immunofluorescence with staining for PAK1 (red) and Notch1 (green) in LBOs from WT mice showed co-localization in the cytoplasm. (B) Immunofluorescence analysis of SW480 cells transiently transfected with siPAK1 or siCTRL. Upper panel: siCTRL; middle panel: siCTRL + Jagged1 (100 ng/mL); and lower panel: siPAK1. In siCTRL SW480 cells, PAK1 and Notch1 co-localized in the cytoplasm. Treatment of SW480 cells with Jagged1 (Jag1) led to dissociation of PAK1 and Notch1 with increased nuclear Notch1. Silencing of PAK1 in SW480 resulted in similar observations, with nuclear translocation of Notch1. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Exemplary pictures of 2 independent experiments. (C) Western blot of SW480 cells after transient silencing of PAK1, showing up-regulation of activated Notch1. (D) Co-immunoprecipitation of SW480 and HCEC 1CT cells, using Notch1 or PAK1 as pull-down. Blotting for PAK1 and Notch1, respectively, indicated direct association of PAK1-Notch1, and β-catenin and Notch1. Associations were abrogated upon treatment with Jagged1. (E) Immunohistochemistry of NICD and PAK1 in AOM/DSS-treated WT and PAK1 KO mice. In PAK1 KO dysplastic lesions, NICD is highly up-regulated. PAK1 is up-regulated in inflamed-dysplastic lesions of WT mice. Results were analyzed for differences between the multiple independent groups by the Kruskal-Wallis ANOVA. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001, ∗∗∗∗P < .0001; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IP, immunoprecipitation; IRS, immune-reactivity score; siPAK1, small interfering PAK1; siCTRL, small interfering Control; WCL, whole-cell lysate.
We next conducted co-immunoprecipitation experiments, evaluating a direct interaction between PAK1 and Notch1. Upon Notch1 pull-down, we detected a direct association of Notch1 with PAK1 both in SW480 and HCEC-1CT. This interaction was lost upon Notch1 activation using Jagged1, similar to the observation made in the immunofluorescence assay. A previously reported association between β-catenin and Notch1 also was confirmed in HCEC-1CT and SW480.28 An attenuation of this interaction also was seen after Jagged1 treatment (Figure 4D).
To corroborate the in vitro findings of the PAK1–Notch1 interaction, we used intestinal tissue sections from WT and PAK1 KO mice that were treated with azoxymethane (AOM)/dextran sodium sulfate (DSS).24 Staining for PAK1 showed low expression in noninflamed mucosa, while inflamed dysplastic lesions showed up-regulation of PAK1 (Figure 4E). In contrast to this, NICD was increased in PAK1 KO dysplasia compared with WT dysplastic lesions, indicating the ability of PAK1 to control activation of Notch1 (Figure 4E). The noninflamed mucosa showed no differences in NICD expression between WT and PAK1 KO. These results suggest the possibility of a PAK1/β-catenin/Notch1 complex, which is disrupted upon Notch activation, allowing Notch to translocate to the nucleus.
Oxidative Stress Results in Notch Activation
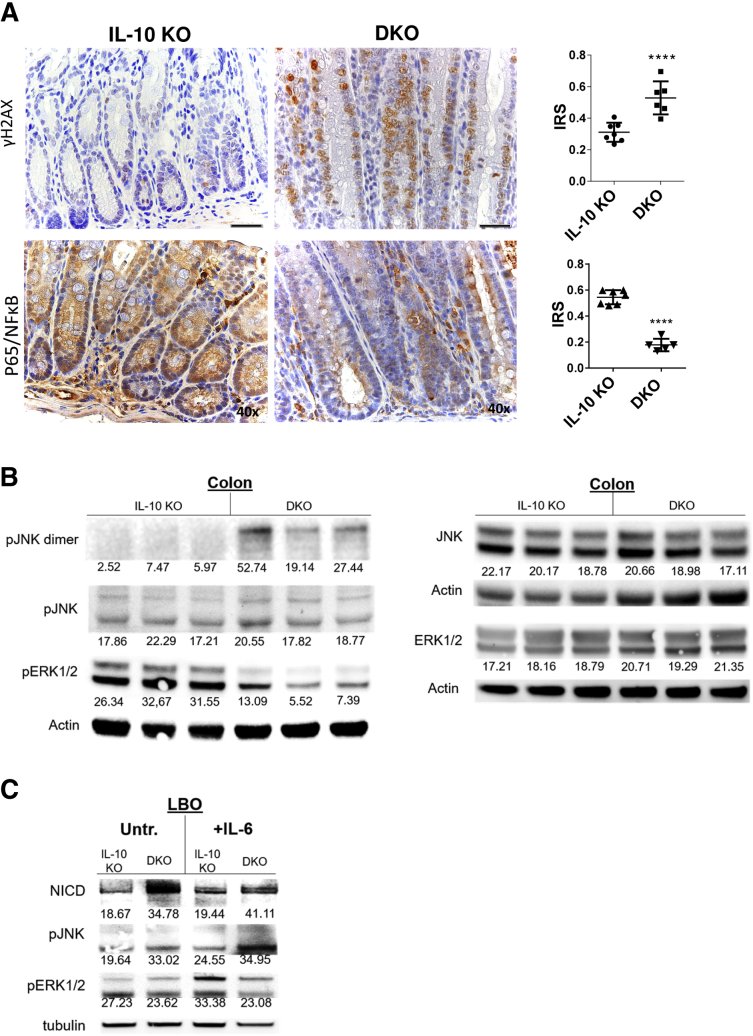
IL10 KO mice are known to show the overt presence of oxidative stress, resulting in colitis-induced DNA damage as measured by phosphorylation of H2A histone family member X (γH2AX).4,29 Staining for γH2AX showed even more double-strand breaks in DKOs, which likely was a consequence of exacerbated oxidative stress (Figure 5A). P65/NF-κB was reduced in DKO, as was the canonical NF-κB activation (Figure 5A).9
Figure 5.
PAK1 deficiency exacerbates oxidative stress–related DNA damage and stress kinases, and drives Notch1 activation. (A) Immunohistochemistry (IHC) of γH2AX marking double-strand breaks. PAK1 deletion resulted in increased double-strand breaks. IHC of p65 indicating NF-κB activation and corresponding IRS. PAK1 deletion attenuated NF-κB signaling. (B) Western blot of colon samples from IL10 KO and DKO mice. In DKO mice, pERK signaling was abrogated, while blotting for pJNK showed increased dimerization, indicating pJNK activation. (C) Western blot of LBOs, which were treated with IL6 (10 ng/mL). Treatment resulted in activation of Notch1 and JNK in DKO, while ERK/AKT activation was impeded upon PAK1 loss. AKT, protein kinase B; γH2AX, phosphorylated H2A histone family member X; IRS, immune-reactivity score; pERK, phosphorylated extracellular signal-regulated kinase1/2; pJNK, phosphorylated c-Jun N-terminal kinase.
Stress kinases in IL10 KO and DKO were analyzed using Western blot. DKO mice showed increased phosphorylated c-Jun N-terminal kinase (JNK) dimers, a state that indicates Stress-activated protein kinases (SAPK)/JNK activation.30 Phosphorylation of extracellular signal-regulated kinase1/2 (ERK1/2) showed a reduction in DKO colons, an expected result of the PAK1 KO (Figure 5B).11 After IL6 treatment, DKO LBOs showed higher levels of NICD in an untreated state and after IL6 treatment. In addition, in line with the findings in colon samples, activation of JNK and impeded ERK signaling in DKO were detected as well (Figure 5C).
Altogether, our data indicate involvement of PAK1 to maintain intestinal homeostasis under conditions of chronic inflammation via a novel interaction between Notch1 and PAK1. Absence of PAK1 results in increased Notch1 activation accompanied with hyperproliferation and loss of the secretory crypt cells. This hyperproliferative state is associated with increased DNA double-strand breaks and neoplastic transformation. Thus, active PAK1 protects from inflammation-driven colon carcinogenesis.
Discussion
Previous findings from our laboratory and other investigators have shown involvement of PAK1 in CRC as well as in IBD.7,11,24 In this study, we sought to investigate the role of PAK1 further in intestinal inflammation and colitis-associated carcinogenesis using the IL10 KO mouse model. Surprisingly, PAK1 loss in IL10 KO mice exacerbated colitis and tumorigenesis and led to longer colons and crypts. This is in contrast to our previous findings, in which PAK1 was modulating tumor initiation, however, a specific hyperproliferative phenotype was not observed upon PAK1 deletion. A hallmark of the changes in our model was the activation of Notch1, accompanied by a loss in differentiated crypts and increased stemness. Here, we identified a novel Notch1–PAK1 axis, showing a direct association and interaction between PAK1 and Notch1, essential in gut homeostasis and immune response.
IL10 KO mice are a well-established model of colitis and colitis-associated carcinogenesis. We previously showed overexpression of PAK1 in neoplasia of IL10 KO mice and in IBD and CAC.10,11 We expected less disease activity and cancer upon PAK1 deletion in these mice, however, surprisingly, we observed the opposite, marked by more inflammation and dysplasia. Moreover, DKO mice showed a paradox phenotype; albeit having generally higher grades of inflammation, these mice also had longer and thicker hyperproliferative colons. This observation was consistent in the piroxicam-induced as well as in the spontaneous CAC model. Our IL10 KO mice in general showed quite a low grade of inflammation and dysplasia, a condition that also depends on the microbial composition of the gut.31 We found a shift in the fecal microbiome, with an increased abundance of bacteria belonging to the phylum Proteobacteria and Desulfovibrionaceae family in our DKO mice. Increased numbers of Proteobacteria also have been observed in stool samples from human beings with sporadic adenocarcinomas.32 Desulfovibrionaceae can produce H2S, which has direct genotoxic properties and can induce hyperproliferation.33 However, if changes in the microbiome are a cause or consequence of carcinogenesis in our model needs to be determined with further experiments. Alongside those changes, the colonic crypts showed a loss of differentiation indicated by a loss of goblet cells together with a marked increase in activated Notch1 and its downstream targets such as HES1. This loss of goblet cells was seen in different models investigating Notch signaling, but also is relevant in UC.3,22,34, 35, 36, 37 Moreover, impaired mucus production owing to a lack of goblet cells results in an impaired mucus layer and may explain the increased inflammation of DKO mice.38
Notch signaling facilitates crypt cell proliferation and amplifies the pool of progenitor cells,39,40 thereby regenerating the intestinal epithelium. In addition, it seems to have tumor-suppressor functions, especially in the setting of chronic inflammation and CAC. This is facilitated via various effectors such as matrix metalloproteinase (MMP)9, p53, reactive oxygen species (ROS) production via NADPH oxidase 1 (NOX1), or regulation of apoptosis and cell-cycle arrest.21,41, 42, 43 Interestingly, Notch inhibition at an early stage of chemically induced colitis led to reduced colitis severity.44 This underlines the necessity of tightly controlled Notch signaling in the setting of rapid proliferation, as seen in epithelial regeneration in the context of colitis and concomitant cell fate decision. With regard to our model, PAK1 might be the important regulator in a proinflammatory environment and increased oxidative stress. PAK1 deletion had a synergistic effect on Notch1, resulting in a Notch signaling disbalance, and thereby Notch loses its protective role and is associated with more dysplasia.
In a similar fashion, Notch downstream target HES1 expresses a dichotomous role in which HES1 deficiency led to dysbiosis and increased susceptibility to DSS colitis.45 On the other hand, ectopic HES1 expression, as seen in UC patients, is associated with goblet cell depletion.22 In line with this, in DKO mice we saw an expansion of the proliferative zone and the stem cell compartment (Ki67 and LGR5), marked by increased nuclear expression of NICD and HES1, alongside increased Hes1 and Olfm4 and down-regulated Atoh1 and Klf4 mRNA levels. Klf4, usually a marker of differentiated epithelial cells, was shown to be associated with colonic inflammation because intestinal-specific Klf4 knockout mice showed increased proliferation and were less sensitive to DSS-induced inflammation. This effect was mediated via NF-κB, which might also explain the hyperproliferation in our model, despite the higher inflammatory burden.46 However, further research is required to clearly characterize the proliferative cells in DKO crypts.
The Wnt/β-catenin pathway is not only crucial for intestinal homeostasis, but also is a driver of tumorigenesis. PAK1 directly interacts with and phosphorylates β-catenin, resulting in its nuclear translocation.14,15 In our model, we detected an increase in β-catenin mRNA and protein level upon PAK1 deletion. Immunohistochemistry showed cytoplasmic accumulation of β-catenin. In line with absent phosphorylation by PAK1, nuclei seemed to lack β-catenin in DKO. In addition, cross-talk between the Notch and Wnt pathway was shown at various signaling levels and intestinal diseases, resulting in either repression or signal enhancement of each other.28,47, 48, 49, 50 Wnt inhibition by transgenic Dickkopf1 expression was shown to disrupt intestinal homeostasis, accompanied by a lack of nuclear β-catenin and an absence of secretory cell lineages, but in contrast to our findings, abrogated proliferation.51 Disruption of β-catenin signaling using Tcf7l2 KO mice results in perinatal death and in loss of the proliferative compartment of the intestine.52 How redundant Wnt ligands produced by stromal stem cells can bypass the loss of nuclear β-catenin in DKO mice remains elusive. The generation of cell-specific KO models is needed to elucidate the role of stromal vs epithelial PAK1 and β-catenin signaling.
Another interesting finding was the detection of OLFM4 in colons of DKO mice, which usually is not expressed in the murine large intestine. OLFM4 is not only a downstream target of the Notch1 pathway, but is itself involved in negatively controlling Wnt/β-catenin, NF-κB, and epithelial renewal.16,26,53 This also might explain the observed down-regulation of nuclear p65 observed in DKO colons. In IBD, OLFM4 also might play an important role because it is overexpressed in active IBD and triggered by IL22.27 However, in our mice, we did not detect increased levels of IL22 but rather IL6 as a general biomarker of systemic inflammation.
We thought of oxidative stress and the presence of ROS as a trigger for Notch1 activation. IL10 KO mice show an overt presence of oxidative stress, which might explain initial Notch1 activation.4,54 One potential mechanism is the up-regulation of Notch-activating MMPs such as Disintegrin and metalloproteinase domain-containing protein 10 (ADAM10), owing to inflammation and increased ROS.55 Remarkably, it also was shown that other MMPs can work as a substrate to activate Notch (ie, MMP7 was shown to directly cleave Notch1, and in pancreatic acinar cells MMP7 was necessary to maintain Notch1 activation to induce transdifferentiation).56,57 On the other hand, PAKs also are known to induce various MMPs via JNK.58,59 IL6 itself is able to induce epithelial regeneration via Transcriptional coactivator YAP and Notch1 activation.60, 61, 62, 63 Nevertheless, the increase of IL6 and oxidative stress is a hallmark for CAC initiation and progression because we also detected more dysplastic lesions in DKO mice.61,64 We further examined samples from WT and PAK1 KO mice from a previous study, in which inflammation and consecutive CAC was triggered using AOM/DSS. Surprisingly, the normal-appearing mucosa, but especially the dysplastic lesions in PAK1 KO mice, showed a strong up-regulation of nuclear NICD compared with WT mice.
PAK1 might regulate Notch1 activation using the activated intracellular form for Notch1 feedback inhibition. However, further research is needed regarding whether the scaffolding or the kinase function of PAK1 is involved in Notch-pathway activation. The direct association of PAK1 and Notch1 already has been shown, interestingly indicating regulation of PAK1 activity via NICD.65 Furthermore, Vadlamudi et al66 showed that phosphorylation of SMRT/HDAC1 Associated Repressor Protein (SHARP) via PAK1 resulted in repression of Notch target genes. In addition, implications for PAKs inducing a stem cell phenotype already have been made in renal cell carcinoma and in vitro using CRC and pancreatic cancer cell lines.67, 68, 69 However, in our model, changes were prominent only in the colons of DKO mice under inflammatory conditions, whereas changes in the small intestine and its stem cell niche were only scarce.
The main treatment for remission in UC is mesalamine. We have shown that mesalamine reduces PAK1 overexpression in a dose-dependent manner at both mRNA and protein levels.6 However, mesalamine does not specifically inhibit phospho-PAK1. A reduction in PAK also could affect MAPK, NF-κB, and AKT signaling, contributing to IBD. Another aspect of mesalamine efficacy is through activation of Peroxisome proliferator-activated receptor gamma (PPARγ),70 and we previously have shown that PAK1 overexpression down-regulates PPARγ.9 Nevertheless, mesalamine also shows PAK1 independent activity such as scavenging ROS.71 Overall, these lines of evidence suggest that PAK1 overexpression and activation in inflammation and CAC promote proinflammatory and oncogenic signaling such as NF-κB, MAPK, AKT, and Wnt β-catenin, and suppression of PPARγ. Mesalamine could interfere with all of these pathways in both a PAK1-dependent and independent manner.
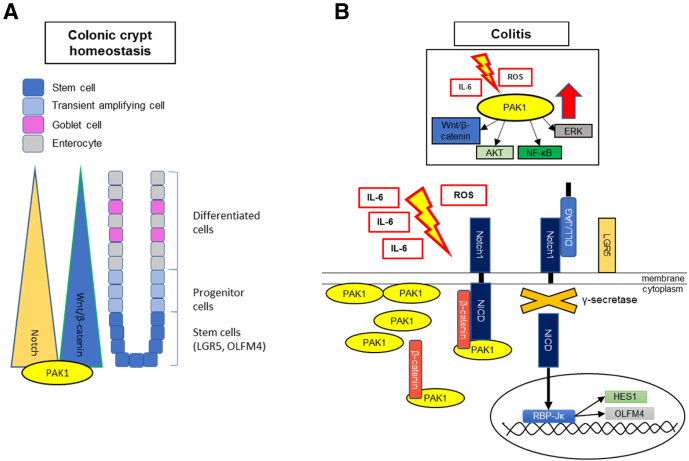
Our findings suggest a novel PAK1–Notch1 axis. Under homeostasis in the intestine, PAK1 seems to be associated with Notch1 and β-catenin. We showed how activation of Notch1 resulted in dissociation of PAK1 from Notch1. Intestinal inflammation results in PAK1 overexpression, thereby tightly controlling the release of Notch1 and β-catenin upon activation and maintaining proper stem cell proliferation and differentiation.15,65 Loss of PAK1 in this setting leads to diminished differentiation and a more stem cell–like intestinal phenotype as shown by Notch1, OLFM4, LGR5, and Wnt/β-catenin activation, and overexpression with an increase in tumorigenesis (Figure 6).
Figure 6.
A novel PAK1–Notch1 axis in colonic inflammation. (A) In colonic crypt homeostasis Wnt/β-catenin and Notch signaling at the crypt base are hallmarks of stem cell–associated proliferation and consecutive differentiation. PAK1 is associated directly with such signaling pathways. (B) In colitis, characterized by exacerbated oxidative stress and proinflammatory cytokines such as IL6, PAK1 is overexpressed. PAK1 regulates downstream targets such as Wnt/β-catenin, AKT, ERK, and NF-κB. Our data suggest a complex of PAK1, Notch1, and β-catenin. In our study, PAK1 deficiency in colitis results in activation of Notch and an increased stemness marked by stem cell markers OLFM4 and LGR5. AKT, protein kinase B; ERK, extracellular signal-regulated kinase
Materials and Methods
Animal Experiments
C57BL/6 PAK1 KO mice were obtained from the Mutant Mouse Regional Resource Center, University of North Carolina, and bred at the Division of Laboratory Animal Science and Genetics (Himberg, Austria) under specific pathogen-free conditions with C57BL/6 IL10 KO mice to generate PAK1/IL10 DKO mice. At the age of 5 weeks, mice were transferred to our animal facility (Institute of Biomedical Research, Medical University of Vienna, Vienna, Austria) and kept under 12-hour light/dark cycles with food and water ad libitum. For experiments, mice (N = 16) received, after 1 week of adaptation, 200 ppm piroxicam in their diet for 7 days to trigger and synchronize intestinal inflammation. Mice were weighed on a daily to weekly basis, and their health status was checked. If weight loss of more than 20% or severe inflammation including rectal prolapse occurred, mice were killed. All animal studies were in accordance with the Austrian Animal Experiments Regulation and approved by the animal experiment committee in accordance with the institutional good scientific practice guidelines (BMWFW-66.009/0268-WF/V/3b/2014). In a second experiment, 6- to 8-week-old IL10 KO (N = 43) and DKO mice (N = 38) were kept for 20 weeks. After 12 weeks, inflammation and dysplasia number were assessed via colonoscopy. The bowel preparation and colonoscopy was performed as described elsewhere.24
Six- to 8-week-old male and female C57BL/6 PAK1 KO (N = 48) or C57BL/6 WT (N = 54) mice received AOM treatment after a 2-week adaption phase on regular chow. All mice received 1 intraperitoneal injection of 10 mg/kg AOM and 4 intermittent cycles of 1.7% DSS in drinking water for 4 days, followed by 14 days of regular water.24
Disease Activity and Histologic Analysis
Over the course of the experiment, the DAI was assessed as previously described.72 The DAI consisted of body weight loss (scores were as follows: 0, no weight loss; 1, 1%–5% weight loss; 2, 6%–10% weight loss; 3, 11%–20% weight loss; and 4, >20% weight loss), stool consistency (scores were as follows: 0, normal; 2, loose stool; and 4, diarrhea), and bloody feces (scores were as follows: 0, none; 2, occult bleeding; and 4, gross bleeding). Intestines were dissected, flushed with phosphate-buffered saline, and coiled up as a Swiss roll followed by 10% formalin fixation or stored in RNAlater (Thermo Fisher, Waltham, MA) for further downstream analysis. H&E-stained slides of paraffin-embedded intestinal tissue sections were analyzed by an independent pathologist, blinded for the genotype, scoring numbers of dysplastic lesions, and grade of inflammation as previously described.4
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections were used for immunohistochemistry as described previously.4 The primary antibodies used are listed in Supplementary Table 1. Briefly, slides were dried and dewaxed with 2 cycles of xylol, followed by rehydration and blocking of endogenous peroxidase. Antigen retrieval was performed by boiling slides in citrate buffer followed by overnight incubation with primary antibody. Staining was performed using a biotinylated secondary antibody with avidin–biotin–horseradish-peroxidase complex (Vectastain, PK-6100, Vector Laboratories, Burlingame, CA) and diaminobenzidine. Nuclei then were counterstained using hematoxylin (5174; Merck, Darmstadt, Germany), and slides were visualized and recorded on an Olympus BX51 microscope (Olympus, Tokyo, Japan). Scoring of immunohistochemistry was, if not stated otherwise, performed as a ratio of positive cells or nuclei per crypt. At least 8 crypts per high-power field were counted in at least 4 high-power fields per sample slide in a 40× magnification.
Intestinal Organoids
LBOs were obtained from 3 different IL10 KO and DKO mice each, as described elsewhere.73 Briefly, after isolation, organoids were pooled and cultured in 24-well plates in 50 μL Matrigel (BD Biosciences, Franklin Lakes, NJ) droplets and covered in 50% conditioned medium with Wnt, R-spondin1, and Noggin. Organoids were passaged every 2–4 days in a 1:3 ratio. Wnt-conditioned medium was obtained from L-WRN cells (ATCC CRL-3276).74
Proliferation Assay
Organoids were split into single-cell suspensions using gentle cell dissociation reagent (Stemcell Technologies, Vancouver, Canada) and Trypsin EDTA (Gibco, Carlsbad, CA). Cells were resuspended to a concentration of 20,000 cells/mL in 50% Matrigel/50% Complete 50% WRN medium. A total of 10 μL of this solution was pipetted per well of a 96-well plate. Each Matrigel droplet was covered with 100 μL Complete 50% WRN medium. At 24 (day 1), 48 (day 2), and 72 (day 3) hours later, wells were imaged on an EVOS XL (Thermo Fisher Scientific, Waltham, MA) and 100 μL of CellTiter-Glo (Promega, Madison, WI) was added to each well and pipetted repeatedly for homogenization. The plate was placed on a shaker at room temperature for 25 minutes, after which luminescence was measured on a Plate Chameleon V (Hidex, Turku, Finland). Medium was replaced on day 2. Relative luminescence unit values were plotted.
Immunofluorescence
Immunofluorescence was performed on LBOs and SW480 cells as described previously.9 Briefly, after fixation, immunostaining was performed with primary antibodies (Supplementary Table 1). Samples then were washed, dried, mounted in medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA), and imaged on an Olympus BX51 microscope.
Immunoblotting and Co-immunoprecipitation
Protein isolation from colonic samples, LBOs, and whole-cells lysates was performed as described elsewhere.73 Co-immunoprecipitation experiments and immunoblotting were conducted as described previously, if not stated otherwise. Data presented are from 3 independent experiments.9
Quantitative Real-Time Polymerase Chain Reaction
RNA was isolated using the Aurum Total RNA Mini Kit (Bio-Rad, Basel, Switzerland) and complementary DNA was synthesized using the High-Capacity complementary DNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA) according to the manufacturer’s protocol. Quantitative reverse-transcription polymerase chain reaction was performed in triplicate or duplicate on custom or predesigned polymerase chain reaction arrays (Bio-Rad) (see Supplementary Table 2 for targets) on an ABI Prism 7500 fast (Applied Biosystems). Data were normalized to endogenous control glyceraldehyde-3-phosphate dehydrogenase and acidic ribosomal phosphoprotein P0 (36B4).
16S Ribosomal RNA Gene Amplicon Sequencing
DNA from stool was extracted using the standard QIAamp DNA stool mini kit protocol (Qiagen, Hilden, Germany) modified by an initial bead-beating step (Lysing Matrix E; MP Biomedicals, Santa Ana, CA). Amplicon sequencing of the V3V4 16S region was performed using an established barcoding approach and Illumina MiSeq technology 10 (Illumina Inc, San Diego, CA). Reads were processed using DADA211 and SINA12.
Cell Lines and Reagents
Primary HCEC-1CTs were a generous gift from Dr Jerry W. Shay and were cultured as described previously.75 The SW480 cell line was obtained from ATCC (Manassas, VA) and cultured in Dulbecco’s modified Eagle medium (Gibco), supplemented with 10% fetal bovine serum (Biochrom, Cambridge, UK). Cell lines were maintained in culture with regular passaging every 2–3 days. Cells were tested for Mycoplasma every 6 months (Mycoplasma detection kit; Lonza, Basel, Switzerland). The cytokine IL6 (Preprotech, Rocky Hill, NJ) and the ligand Jagged1 (Abcam, Cambridge, UK) were used.
RNA Interference and PAK1 Silencing
For silencing RNA (siRNA) experiments, SW480 cell were plated at a density of 1 × 106 per well in a 6-well plate. Transient transfection was performed through electroporation with 100 nmol/L of small interfering PAK1 (siPAK1) on an Amaxa nucleofactor 2b (Lonza, Basel, Switzerland). Oligonucleotides were purchased from Dharmacon (catalog number: D-003521-03; accession number: NM_002576; target sequence: CAUCAAAUAUCACUAAGUC, Lafayette, CO). Control siRNA-A (sc-37007, Santa Cruz Biotechnology, Santa Cruz, TX) was prepared based on the manufacturer’s instructions.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6.01 software (GraphPad Software, La Jolla, CA). Means of normally distributed data were compared using an unpaired Student t test; a nonparametric Mann–Whitney U test was used for not normally distributed data. For the analysis of sample similarity, modified Rhea scripts were used.76 Generalized unique fraction metric (UniFrac) distances were visualized using nonmetric multidimensional scaling. Cluster significance was assessed using permutational multivariate analysis of variance. Testing for significant differences in diversity and bacterial abundances was performed using the Kruskal–Wallis rank sum test. If P values were .05 or less they were considered statistically significant.
Acknowledgments
IL10 KO mice were gratefully provided by Dr Terrence A. Barrett (University of Kentucky, Lexington, KY) and Jeffrey B. Brown (Northwestern University, Feinberg School of Medicine, Chicago, IL).
CRediT Authorship Contributions
Victoria Neumeyer (Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Supporting)
Anna Brutau-Abia (Investigation: Supporting)
Michael Allgäuer (Investigation: Supporting)
Nicole Pfarr (Formal analysis: Supporting; Methodology: Supporting)
Wilko Weichert (Resources: Supporting)
Christina Falkeis-Veits (Formal analysis: Supporting)
Elisabeth Kremmer (Methodology: Supporting)
Michael Vieth (Formal analysis: Supporting)
Markus Gerhard (Supervision: Supporting)
Raquel Mejías-Luque, PhD (Conceptualization: Lead; Formal analysis: Lead; Supervision: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by the Austrian Federal Ministry of Education, Sciene and Research and by the Austrian Sciene Fund (P32302 to C.G.)
Supplementary Material
Supplementary Table 1.
Antibodies
| Target | Name | Supplier | Catalog number |
|---|---|---|---|
| Actin | Anti–β-actin antibody | Santa Cruz Biotechnology, Santa Cruz, TX | sc-47778 |
| Anti-mouse | IRDye 680CW donkey anti-mouse IgG | LICOR (Lincoln, NE) | 926-68072 |
| Anti-rabbit | IRDye 800CW goat anti-rabbit IgG | LICOR | 926-32211 |
| Anti-rabbit | IRDye 680CW goat anti-rabbit IgG | LICOR | 926-68073 |
| CC3 | Cleaved caspase 3 antibody | Cell Signaling Technology (Leiden, Netherlands) | 9664 |
| HES1 | HES1 (D6P2U) rabbit monoclonal antibody | Cell Signaling Technology | 11988 |
| Ki67 | Ki-67 antibody | Abcam (Cambridge, UK) | ab15580 |
| LGR5 | Rabbit polyclonal to LGR5 | Abcam | ab219107 |
| NICD | Anti-activated Notchl | Abcam | ab8925 |
| NOTCH1 | Notch1 antibody (E4) | Santa Cruz | sc-373944 |
| OLFM4 | Olfm4 (D6Y5A) rabbit monoclonal antibody | Cell Signaling Technology | 39141 |
| P65 | NF-κB p65 (D14E12) XP rabbit monoclonal antibody #8242 | Cell Signaling Technology | 8242S |
| PAK1 | Anti-PAK1 antibody | Cell Signaling Technology | 2602 |
| p-AKT | Phospho-AKT (Serine 473) rabbit | Santa Cruz | sc-101629 |
| p-ERK1/2 | Phospho-MAPK antibody phospho (Erk1/2) (mouse) | Cell Signaling Technology | 9106S |
| p-JNK | Phospho-SAPK/JNK (Thr183/Tyr185) | Cell Signaling Technology | 4668S |
| p-P38 | p-p38 (D-8) | Santa Cruz | sc-7973 |
| Tubulin | NF-κB p65 (D14E12) XP rabbit monoclonal antibody #8242 | Abcam | ab7291 |
| β-catenin | Purified Mouse Anti-β-Catenin Clone 14/Beta-Catenin | BD Transduction Laboratories (Franklin Lakes, NJ) | 610153 |
| γ-H2AX | Histone H2AXpSer139: rabbit polyclonal antibody | Cell Signaling Technology | 9718S |
Supplementary Table 2.
PCR targets (BioRad prime PCR, Bio-Rad, Basel, Switzerland)
| Atoh1 | |
| Jak1 | |
| Pparg | |
| Bax | |
| Lgr5 | |
| Stat3 | |
| Bcl2 | |
| Muc2 | |
| Yap1 | |
| Cdh1 | |
| Nfe2I2 | |
| Actb | |
| Nfkb1 | |
| Gapdh | |
| Ctnnb1 | |
| Nfkb1 | |
| Gapdh | |
| Hes1 | |
| Notch1 | |
| Ikbkb | |
| Nox1 | |
| II6 | |
| Olfm4 |
References
- 1.Ananthakrishnan A.N. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 2.Koch S., Nusrat A. The life and death of epithelia during inflammation: lessons learned from the gut. Annu Rev Pathol. 2012;7:35–60. doi: 10.1146/annurev-pathol-011811-120905. [DOI] [PubMed] [Google Scholar]
- 3.Gersemann M., Becker S., Kubler I., Koslowski M., Wang G., Herrlinger K.R., Griger J., Fritz P., Fellermann K., Schwab M., Wehkamp J., Stange E.F. Differences in goblet cell differentiation between Crohn's disease and ulcerative colitis. Differentiation. 2009;77:84–94. doi: 10.1016/j.diff.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Frick A., Khare V., Paul G., Lang M., Ferk F., Knasmuller S., Beer A., Oberhuber G., Gasche C. Overt increase of oxidative stress and DNA damage in murine and human colitis and colitis-associated neoplasia. Mol Cancer Res. 2018;16:634–642. doi: 10.1158/1541-7786.MCR-17-0451. [DOI] [PubMed] [Google Scholar]
- 5.Kühn R., Löhler J., Rennick D., Rajewsky K., Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 6.Khare V., Lyakhovich A., Dammann K., Lang M., Borgmann M., Tichy B., Pospisilova S., Luciani G., Campregher C., Evstatiev R., Pflueger M., Hundsberger H., Gasche C. Mesalamine modulates intercellular adhesion through inhibition of p-21 activated kinase-1. Biochem Pharmacol. 2013;85:234–244. doi: 10.1016/j.bcp.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyons J., Brubaker D.K., Ghazi P.C., Baldwin K.R., Edwards A., Boukhali M., Strasser S.D., Suarez-Lopez L., Lin Y.J., Yajnik V., Kissil J.L., Haas W., Lauffenburger D.A., Haigis K.M. Integrated in vivo multiomics analysis identifies p21-activated kinase signaling as a driver of colitis. Sci Signal. 2018;11 doi: 10.1126/scisignal.aan3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J.B., Lee G., Managlia E., Grimm G.R., Dirisina R., Goretsky T., Cheresh P., Blatner N.R., Khazaie K., Yang G.Y., Li L., Barrett T.A. Mesalamine inhibits epithelial beta-catenin activation in chronic ulcerative colitis. Gastroenterology. 2010;138 doi: 10.1053/j.gastro.2009.10.038. 595–605, e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dammann K., Khare V., Lang M., Claudel T., Harpain F., Granofszky N., Evstatiev R., Williams J.M., Pritchard D.M., Watson A., Gasche C. PAK1 modulates a PPAR gamma/NF-kappa B cascade in intestinal inflammation. Biochim Biophys Acta. 2015;1853:2349–2360. doi: 10.1016/j.bbamcr.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dammann K., Khare V., Gasche C. Tracing PAKs from GI inflammation to cancer. Gut. 2014;63:1173–1184. doi: 10.1136/gutjnl-2014-306768. [DOI] [PubMed] [Google Scholar]
- 11.Khare V., Dammann K., Asboth M., Krnjic A., Jambrich M., Gasche C. Overexpression of PAK1 promotes cell survival in inflammatory bowel diseases and colitis-associated cancer. Inflamm Bowel Dis. 2015;21:287–296. doi: 10.1097/MIB.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar R., Gururaj A.E., Barnes C.J. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 13.Guo D.G., Tan Y.C., Wang D.W., Madhusoodanan K.S., Zheng Y., Maack T., Zhang J.J., Huang X.Y. A Rac-cGMP signaling pathway. Cell. 2007;128:341–355. doi: 10.1016/j.cell.2006.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park M.H., Kim D.J., You S.T., Lee C.S., Kim H.K., Park S.M., Shin E.Y., Kim E.G. Phosphorylation of beta-catenin at serine 663 regulates its transcriptional activity. Biochem Biophys Res Commun. 2012;419:543–549. doi: 10.1016/j.bbrc.2012.02.056. [DOI] [PubMed] [Google Scholar]
- 15.Zhu G., Wang Y., Huang B., Liang J., Ding Y., Xu A., Wu W. A Rac1/PAK1 cascade controls beta-catenin activation in colon cancer cells. Oncogene. 2012;31:1001–1012. doi: 10.1038/onc.2011.294. [DOI] [PubMed] [Google Scholar]
- 16.VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud A., Grosse A.S., Gumucio D.L., Ernst S.A., Tsai Y.H., Dempsey P.J., Samuelson L.C. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noah T.K., Shroyer N.F. Notch in the intestine: regulation of homeostasis and pathogenesis. Ann Rev Physiol. 2013;75:263–288. doi: 10.1146/annurev-physiol-030212-183741. [DOI] [PubMed] [Google Scholar]
- 18.Dahan S., Rabinowitz K.M., Martin A.P., Berin M.C., Unkeless J.C., Mayer L. Notch-1 signaling regulates intestinal epithelial barrier function, through interaction with CD4+ T cells, in mice and humans. Gastroenterology. 2011;140:550–559. doi: 10.1053/j.gastro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahan S., Roda G., Pinn D., Roth-Walter F., Kamalu O., Martin A.P., Mayer L. Epithelial: lamina propria lymphocyte interactions promote epithelial cell differentiation. Gastroenterology. 2008;134:192–203. doi: 10.1053/j.gastro.2007.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reedijk M., Odorcic S., Zhang H., Chetty R., Tennert C., Dickson B.C., Lockwood G., Gallinger S., Egan S.E. Activation of Notch signaling in human colon adenocarcinoma. Int J Oncol. 2008;33:1223–1229. doi: 10.3892/ijo_00000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto R., Tsuchiya K., Nemoto Y., Akiyama J., Nakamura T., Kanai T., Watanabe M. Requirement of Notch activation during regeneration of the intestinal epithelia. Am J Physiol Gastrointest Liver Physiol. 2009;296:G23–G35. doi: 10.1152/ajpgi.90225.2008. [DOI] [PubMed] [Google Scholar]
- 22.Zheng X., Tsuchiya K., Okamoto R., Iwasaki M., Kano Y., Sakamoto N., Nakamura T., Watanabe M. Suppression of hath1 gene expression directly regulated by hes1 via notch signaling is associated with goblet cell depletion in ulcerative colitis. Inflamm Bowel Dis. 2011;17:2251–2260. doi: 10.1002/ibd.21611. [DOI] [PubMed] [Google Scholar]
- 23.Wehkamp J., Salzman N.H., Porter E., Nuding S., Weichenthal M., Petras R.E., Shen B., Schaeffeler E., Schwab M., Linzmeier R., Feathers R.W., Chu H., Lima H., Jr., Fellermann K., Ganz T., Stange E.F., Bevins C.L. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci U S A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dammann K., Khare V., Harpain F., Lang M., Kurtovic A., Mesteri I., Evstatiev R., Gasche C. PAK1 promotes intestinal tumor initiation. Cancer Prev Res. 2015;8:1093–1101. doi: 10.1158/1940-6207.CAPR-15-0205-T. [DOI] [PubMed] [Google Scholar]
- 25.Carvalho F.A., Koren O., Goodrich J.K., Johansson M.E., Nalbantoglu I., Aitken J.D., Su Y., Chassaing B., Walters W.A., Gonzalez A., Clemente J.C., Cullender T.C., Barnich N., Darfeuille-Michaud A., Vijay-Kumar M., Knight R., Ley R.E., Gewirtz A.T. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W., Li H., Hong S.H., Piszczek G.P., Chen W., Rodgers G.P. Olfactomedin 4 deletion induces colon adenocarcinoma in Apc(Min/+) mice. Oncogene. 2016;35:5237–5247. doi: 10.1038/onc.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gersemann M., Becker S., Nuding S., Antoni L., Ott G., Fritz P., Oue N., Yasui W., Wehkamp J., Stange E.F. Olfactomedin-4 is a glycoprotein secreted into mucus in active IBD. J Crohns Colitis. 2012;6:425–434. doi: 10.1016/j.crohns.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Kwon C., Cheng P., King I.N., Andersen P., Shenje L., Nigam V., Srivastava D. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arthur J.C., Perez-Chanona E., Muhlbauer M., Tomkovich S., Uronis J.M., Fan T.J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., Rhodes J.M., Stintzi A., Simpson K.W., Hansen J.J., Keku T.O., Fodor A.A., Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nitta R.T., Badal S.S., Wong A.J. Measuring the constitutive activation of c-Jun N-terminal kinase isoforms. Methods Enzymol. 2010;484:531–548. doi: 10.1016/B978-0-12-381298-8.00026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sellon R.K., Tonkonogy S., Schultz M., Dieleman La, Grenther W., Balish E., Rennick D.M., Sartor R.B. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori G., Rampelli S., Orena B.S., Rengucci C., De Maio G., Barbieri G., Passardi A., Casadei Gardini A., Frassineti G.L., Gaiarsa S., Albertini A.M., Ranzani G.N., Calistri D., Pasca M.R. Shifts of faecal microbiota during sporadic colorectal carcinogenesis. Sci Rep. 2018;8:10329. doi: 10.1038/s41598-018-28671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christl S.U., Eisner H.D., Dusel G., Kasper H., Scheppach W. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: a potential role for these agents in the pathogenesis of ulcerative colitis. Dig Dis Sci. 1996;41:2477–2481. doi: 10.1007/BF02100146. [DOI] [PubMed] [Google Scholar]
- 34.Tsai Y.H., VanDussen K.L., Sawey E.T., Wade A.W., Kasper C., Rakshit S., Bhatt R.G., Stoeck A., Maillard I., Crawford H.C., Samuelson L.C., Dempsey P.J. ADAM10 regulates Notch function in intestinal stem cells of mice. Gastroenterology. 2014;147:822–834 e13. doi: 10.1053/j.gastro.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Es J.H., de Geest N., van de Born M., Clevers H., Hassan B.A. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1:18. doi: 10.1038/ncomms1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Q., Bermingham N.A., Finegold M.J., Zoghbi H.Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 37.Katz J.P., Perreault N., Goldstein B.G., Lee C.S., Labosky P.A., Yang V.W., Kaestner K.H. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCormick D.A., Horton L.W., Mee A.S. Mucin depletion in inflammatory bowel disease. J Clin Pathol. 1990;43:143–146. doi: 10.1136/jcp.43.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed I., Chandrakesan P., Tawfik O., Xia L., Anant S., Umar S. Critical roles of Notch and Wnt/beta-catenin pathways in the regulation of hyperplasia and/or colitis in response to bacterial infection. Infect Immun. 2012;80:3107–3121. doi: 10.1128/IAI.00236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garg P., Jeppsson S., Dalmasso G., Ghaleb A.M., McConnell B.B., Yang V.W., Gewirtz A.T., Merlin D., Sitaraman S.V. Notch1 regulates the effects of matrix metalloproteinase-9 on colitis-associated cancer in mice. Gastroenterology. 2011;141:1381–1392. doi: 10.1053/j.gastro.2011.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pujada A., Bui T.A., Walter L., Denning T.L., Garg P. Notch1 mediates protection in colitis associated cancer. Gastroenterology. 2018;154 S42-S. [Google Scholar]
- 43.Walter L., Canup B., Pujada A., Bui T.A., Arbasi B., Laroui H., Merlin D., Garg P. Matrix metalloproteinase 9 (MMP9) limits reactive oxygen species (ROS) accumulation and DNA damage in colitis-associated cancer. Cell Death Dis. 2020;11:767. doi: 10.1038/s41419-020-02959-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinoda M., Shin-Ya M., Naito Y., Kishida T., Ito R., Suzuki N., Yasuda H., Sakagami J., Imanishi J., Kataoka K., Mazda O., Yoshikawa T. Early-stage blocking of Notch signaling inhibits the depletion of goblet cells in dextran sodium sulfate-induced colitis in mice. J Gastroenterology. 2010;45:608–617. doi: 10.1007/s00535-010-0210-z. [DOI] [PubMed] [Google Scholar]
- 45.Guo X.K., Ou J., Liang S., Zhou X., Hu X. Epithelial Hes1 maintains gut homeostasis by preventing microbial dysbiosis. Mucosal Immunol. 2018;11:716–726. doi: 10.1038/mi.2017.111. [DOI] [PubMed] [Google Scholar]
- 46.Ghaleb A.M., Laroui H., Merlin D., Yang V.W. Genetic deletion of Klf4 in the mouse intestinal epithelium ameliorates dextran sodium sulfate-induced colitis by modulating the NF-kappaB pathway inflammatory response. Inflamm Bowel Dis. 2014;20:811–820. doi: 10.1097/MIB.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishiguro H., Okubo T., Kuwabara Y., Kimura M., Mitsui A., Sugito N., Ogawa R., Katada T., Tanaka T., Shiozaki M., Mizoguchi K., Samoto Y., Matsuo Y., Takahashi H., Takiguchi S. NOTCH1 activates the Wnt/beta-catenin signaling pathway in colon cancer. Oncotarget. 2017;8:60378–60389. doi: 10.18632/oncotarget.19534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H.A., Koo B.K., Cho J.H., Kim Y.Y., Seong J., Chang H.J., Oh Y.M., Stange D.E., Park J.G., Hwang D., Kong Y.Y. Notch1 counteracts WNT/beta-catenin signaling through chromatin modification in colorectal cancer. J Clin Invest. 2012;122:3248–3259. doi: 10.1172/JCI61216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin Y.H., Kim H., Ki H., Yang I., Yang N., Lee K.Y., Kim N., Park H.S., Kim K. Beta-catenin modulates the level and transcriptional activity of Notch1/NICD through its direct interaction. Biochim Biophys Acta. 2009;1793:290–299. doi: 10.1016/j.bbamcr.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 50.Peignon G., Durand A., Cacheux W., Ayrault O., Terris B., Laurent-Puig P., Shroyer N.F., Van Seuningen I., Honjo T., Perret C., Romagnolo B. Complex interplay between beta-catenin signalling and Notch effectors in intestinal tumorigenesis. Gut. 2011;60:166–176. doi: 10.1136/gut.2009.204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinto D., Gregorieff A., Begthel H., Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P.J., Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 53.Tian H., Biehs B., Chiu C., Siebel C.W., Wu Y., Costa M., de Sauvage F.J., Klein O.D. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11:33–42. doi: 10.1016/j.celrep.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boopathy A.V., Pendergrass K.D., Che P.L., Yoon Y.S., Davis M.E. Oxidative stress-induced Notch1 signaling promotes cardiogenic gene expression in mesenchymal stem cells. Stem cell Res Ther. 2013;4:43. doi: 10.1186/scrt190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones J.C., Rustagi S., Dempsey P.J. ADAM proteases and gastrointestinal function. Ann Rev Physiol. 2016;78:243–276. doi: 10.1146/annurev-physiol-021014-071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawey E.T., Crawford H.C. Metalloproteinases and cell fate: Notch just ADAMs anymore. Cell Cycle. 2008;7:566–569. doi: 10.4161/cc.7.5.5531. [DOI] [PubMed] [Google Scholar]
- 57.Sawey E.T., Johnson J.A., Crawford H.C. Matrix metalloproteinase 7 controls pancreatic acinar cell transdifferentiation by activating the Notch signaling pathway. Proc Natl Acad Sci U S A. 2007;104:19327–19332. doi: 10.1073/pnas.0705953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou L., Yan C., Gieling R.G., Kida Y., Garner W., Li W., Han Y.P. Tumor necrosis factor-alpha induced expression of matrix metalloproteinase-9 through p21-activated kinase-1. BMC Immunol. 2009;10:15. doi: 10.1186/1471-2172-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rider L., Oladimeji P., Diakonova M. PAK1 regulates breast cancer cell invasion through secretion of matrix metalloproteinases in response to prolactin and three-dimensional collagen IV. Mol Endocrinol. 2013;27:1048–1064. doi: 10.1210/me.2012-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi K., Wu L.W., Grivennikov S.I., de Jong P.R., Lian I., Yu F.X., Wang K., Ho S.B., Boland B.S., Chang J.T., Sandborn W.J., Hardiman G., Raz E., Maehara Y., Yoshimura A., Zucman-Rossi J., Guan K.L., Karin M. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grivennikov S., Karin E., Terzic J., Mucida D. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong S., Wang R., Chen Q., Luo J., Wang J., Zhao Z., Li Y., Wang Y., Wang X., Cheng B. Cancer-associated fibroblasts promote stem cell-like properties of hepatocellular carcinoma cells through IL-6/STAT3/Notch signaling. Am J Cancer Res. 2018;8:302–316. [PMC free article] [PubMed] [Google Scholar]
- 63.Taniguchi K., Moroishi T., de Jong P.R., Krawczyk M., Grebbin B.M., Luo H., Xu R.H., Golob-Schwarzl N., Schweiger C., Wang K., Di Caro G., Feng Y., Fearon E.R., Raz E., Kenner L., Farin H.F., Guan K.L., Haybaeck J., Datz C., Zhang K., Karin M. YAP-IL-6ST autoregulatory loop activated on APC loss controls colonic tumorigenesis. Proc Natl Acad Sci U S A. 2017;114:1643–1648. doi: 10.1073/pnas.1620290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bollrath J., Phesse T.J., von Burstin V.A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., Matthews V., Schmid R.M., Kirchner T., Arkan M.C., Ernst M., Greten F.R. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Yoon J.H., Mo J.S., Ann E.J., Ahn J.S., Jo E.H., Lee H.J., Hong S.H., Kim M.Y., Kim E.G., Lee K., Park H.S. NOTCH1 intracellular domain negatively regulates PAK1 signaling pathway through direct interaction. Biochim Biophys Acta. 2016;1863:179–188. doi: 10.1016/j.bbamcr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Vadlamudi R.K., Manavathi B., Singh R.R., Nguyen D., Li F., Kumar R. An essential role of Pak1 phosphorylation of SHARP in Notch signaling. Oncogene. 2005;24:4591–4596. doi: 10.1038/sj.onc.1208672. [DOI] [PubMed] [Google Scholar]
- 67.Huynh N., Shulkes A., Baldwin G., He H. Up-regulation of stem cell markers by P21-activated kinase 1 contributes to 5-fluorouracil resistance of colorectal cancer. Cancer Biol Ther. 2016;17:813–823. doi: 10.1080/15384047.2016.1195045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu Y., Liu H., Xu L., An H., Liu W., Liu Y., Lin Z., Xu J. p21-activated kinase 1 determines stem-like phenotype and sunitinib resistance via NF-kappaB/IL-6 activation in renal cell carcinoma. Cell Death Dis. 2015;6:e1637. doi: 10.1038/cddis.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu H.Y., Yang M.C., Ding L.Y., Chen C.S., Chu P.C. p21-Activated kinase 3 promotes cancer stem cell phenotypes through activating the Akt-GSK3beta-beta-catenin signaling pathway in pancreatic cancer cells. Cancer Lett. 2019;456:13–22. doi: 10.1016/j.canlet.2019.04.026. [DOI] [PubMed] [Google Scholar]
- 70.Rousseaux C., Lefebvre B., Dubuquoy L., Lefebvre P., Romano O., Auwerx J., Metzger D., Wahli W., Desvergne B., Naccari G.C., Chavatte P., Farce A., Bulois P., Cortot A., Colombel J.F., Desreumaux P. Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-gamma. J Exp Med. 2005;201:1205–1215. doi: 10.1084/jem.20041948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campregher C., Gasche C. Aminosalicylates. Best Pract Res Clin Gastroenterol. 2011;25:535–546. doi: 10.1016/j.bpg.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 72.Cooper H.S., Murthy S., Kido K., Yoshitake H., Flanigan a. Dysplasia and cancer in the dextran sulfate sodium mouse colitis model. Relevance to colitis-associated neoplasia in the human: a study of histopathology, B-catenin and p53 expression and the role of inflammation. Carcinogenesis. 2000;21:757–768. doi: 10.1093/carcin/21.4.757. [DOI] [PubMed] [Google Scholar]
- 73.Khare V., Krnjic A., Frick A., Gmainer C., Asboth M., Jimenez K., Lang M., Baumgartner M., Evstatiev R., Gasche C. Mesalamine and azathioprine modulate junctional complexes and restore epithelial barrier function in intestinal inflammation. Sci Rep. 2019;9:2842. doi: 10.1038/s41598-019-39401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roig A.I., Eskiocak U., Hight S.K., Kim S.B., Delgado O., Souza R.F., Spechler S.J., Wright W.E., Shay J.W. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138 doi: 10.1053/j.gastro.2009.11.052. 1012–1021 e1-5. [DOI] [PubMed] [Google Scholar]
- 76.Lagkouvardos I., Fischer S., Kumar N., Clavel T. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ. 2017;5 doi: 10.7717/peerj.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]