Abstract
Purpose:
Multiparametric MRI (mpMRI) improves the detection of clinically significant prostate cancer, but is limited by interobserver variation. The second version of theProstate Imaging Reporting and Data System (PIRADSv2) was recently proposed as a standard for interpreting mpMRI. To assess the performance and interobserver agreement of PIRADSv2 we performed a multi-reader study with five radiologists of varying experience.
Materials and Methods:
Five radiologists (n = 2 prostate dedicated, n = 3 general body) blinded to clinicopathologic results detected and scored lesions on prostate mpMRI using PIRADSv2. The endorectal coil 3 Tesla MRI included T2W, diffusion-weighted imaging (apparent diffusion coefficient, b2000), and dynamic contrast enhancement. Thirty-four consecutive patients were included. Results were correlated with radical prostatectomy whole-mount histopathology produced with patient-specific three-dimensional molds. An index lesion was defined on pathology as the lesion with highest Gleason score or largest volume if equivalent grades. Average sensitivity and positive predictive values (PPVs) for all lesions and index lesions were determined using generalized estimating equations. Interobserver agreement was evaluated using index of specific agreement.
Results:
Average sensitivity was 91% for detecting index lesions and 63% for all lesions across all readers. PPV was 85% for PIRADS ≥ 3 and 90% for PIRADS ≥ 4. Specialists performed better only for PIRADS ≥ 4 with sensitivity 90% versus 79% (P = 0.01) for index lesions. Index of specific agreement among readers was 93% for the detection of index lesions, 74% for the detection of all lesions, and 85% for scoring index lesions, and 58% for scoring all lesions.
Conclusion:
By using PIRADSv2, general body radiologists and prostate specialists can detect high-grade index prostate cancer lesions with high sensitivity and agreement.
Level of Evidence:
1
Multi-parametric MRI (mpMRI) and targeted biopsies have improved the ability to detect clinically significant prostate cancer, while reducing the diagnosis of low-grade tumors.1-5 Nevertheless, prostate mpMRI requires a high level of expertise to interpret and has been limited by notable interobserver variability.6,7 This is partly due to nonstandardized criteria for diagnosing an abnormality on mpMRI.
Recently, the second version of the Prostate Imaging Reporting and Data System (PIRADSv2) was published.8-10 The PIRADSv2 guidelines are a joint effort of the American College of Radiology, European Society for Uroradiology, and the AdMeTech Foundation to improve the standardization of prostate mpMRI interpretation.10 PIRADSv2 provides guidance regarding the acquisition and interpretation of prostate mpMRI and proposes a simplified 5-point scale with a score of 5 indicating the highest likelihood of a clinically significant cancer.8,9 This system was created to improve the accuracy and inter-observer agreement of the original PIRADS, which was limited by variable interpretations and no clear threshold for identifying clinically significant cancer.11-13
Initial studies evaluating reader performance using PIRADSv2 have shown good accuracy and moderate agreement between readers, but are limited by study design. Vargas et al14 found a sensitivity of 95% for large predetermined index lesions with PIRADSv2. Muller et al6 evaluated the inter-observer agreement of PIRADSv2 for 5 readers and found a kappa score of 0.46 for 5 readers. However, neither study allowed readers to detect and score lesions as would be done in a typical clinical workflow, but evaluated the ability of readers to correctly score predetermined lesions. Furthermore, in the study by Muller et al,6 they used multiple readers who were not trained radiologists.15
To evaluate PIRADSv2 in a clinically relevant scenario, we evaluated the ability of multiple readers with previous experience in PIRADSv2 to prospectively detect and score prostate cancer lesions on mpMRI. The objective of this study was to assess the accuracy and inter-observer agreement of PIRADSv2 for detecting prostate cancer on mpMRI using whole mount pathology as the reference standard.
Materials and Methods
Sample Size Calculation
A sample size calculation was done to determine the number of patients that must be evaluated by five readers such that the sensitivity for the detection of all lesions across all readers is estimated with a desired precision. Under the assumption that two lesions on average were identified on histopathology per patient, the total number of lesions used in calculating reader-specific sensitivity is two times the number of patients. Because multiple lesions may be identified by multiple readers for each patient, several factors contribute to the sample size calculation, including the variability of reader scores at the patient level, the interlesion correlation of multiple lesions detected by the same reader, and the intralesion correlation between different readers. From in-house data, the inter-reader correlation was defined as 0.1 and intralesion correlation was defined as 0.3. As sample size increases with these two correlations, the inter- and intralesion correlation were conservatively set at 0.2 and 0.4, respectively. To achieve 10% precision with a sensitivity of 70%, 25 patients are required. For a study size achievable while maintaining high precision, we chose a sample size of 35 patients, with an estimated precision of 8%. Multiple lesions, low interlesion correlation and multiple readers contribute to sample size reduction.
Study Population
This single-institution retrospective interpretation of prospectively acquired data was approved by the local institutional review board and was compliant with the Health Insurance Portability and Accountability Act. Written informed consent was obtained from all patients for future use of imaging and pathologic data. The study was comprised of 35 consecutive patients who underwent radical prostatectomy after a preoperative prostate mpMRI from February 2013 to April 2014. Inclusion criteria for this study were: (a) an endorectal coil prostate mpMRI at 3 Tesla (T), including T2-weighted (T2W) images, diffusion-weighted imaging (DWI) with both apparent diffusion coefficient (ADC) maps and high b-value (b2000) DWI scans, and dynamic contrast enhancement (DCE) MRI; and (b) a radical prostatectomy with whole mount histopathology specimen. Exclusion criteria were (a) whole-mount specimen unavailable or (b) one or more MRI sequence not acquired. The final cohort of patients in this study consisted of 34 patients. For the final 34 patient cohort, the median age was 59 years (range, 48–71 yeaars) and median PSA was 6.65 ng/mL (range, 2.38–54.1 ng/mL).
MRI
The prostate mpMRI scans were acquired on a 3T scanner (Achieva 3.0T-TX, Philips Healthcare, Best, Netherlands) using an endorectal coil (BPX-30, Medrad, Pittsburgh, PA) filled with 45 mL fluorinert (3M, Maplewood, MN) and the anterior half of a 32-channel cardiac SENSE coil (InVivo, Gainesville, FL). Table 1 contains the sequences and MRI acquisition parameters used in this study.
TABLE 1.
Multiparametric MR Imaging Sequence Parameters at 3T
| Parameter | T2 Weighted | DWIa | High b-Value DWIb |
DCE MRI |
|---|---|---|---|---|
| Field of view (mm) | 140 × 140 | 140 × 140 | 140 × 140 | 262 × 262 |
| Acquisition matrix | 304 × 234 | 112 × 109 | 76 × 78 | 188 × 96 |
| Repetition time (ms) | 4434 | 4986 | 6987 | 3.7 |
| Echo time (ms) | 120 | 54 | 52 | 2.3 |
| Flip angle (degrees) | 90 | 90 | 90 | 8.5 |
| Section thickness (mm), no gaps | 3 | 3 | 3 | 3 |
| Image reconstruction matrix (pixels) | 512 × 512 | 256 × 256 | 256 × 256 | 256 × 256 |
| Reconstruction voxel imaging resolution (mm/pixel) | 0.27 × 0.27 × 3.00 | 0.55 × 0.55 × 2.73 | 0.55 × 0.55 × 2.73 | 1.02 × 1.02 × 3.00 |
| Time for acquisition (min:s) | 2:48 | 4:54 | 3:50 | 5:16 |
For ADC map calculation. Five evenly spaced b values (0-750 s/mm2) were used.
b = 2000 s/mm2.
Evaluation of Reader Performance Using PIRADSv2 to Detect Prostate Cancer
Five body radiologists evaluated each prostate mpMRI according to PIRADSv2 guidelines.9 All readers were blinded to clinical and pathological outcomes. Two readers were highly experienced with >2000 mpMRI cases read (B.T., P.L.C. with experience of 8, 15 years on prostate imaging, respectively) cumulatively and three readers were general body radiologists with 300–500 mpMRI cases read (J.M., Y.M.L., R.S.; all with 2 years of experience on prostate imaging) cumulatively. Three readers were based at the same institution and two readers were based at other institutions. All readers had prior experience with PIRADSv2 before starting the study. Readers identified and scored up to three suspicious lesions per patient by PIRADSv2 and mapped those lesions on a 12-sector template. This map was a modification of the 36 sector map proposed by PIRADSv2 to simplify correlation between readers. Readers were instructed to detect and score lesions they would include in a clinical report and give an overall PIRADSv2 score for each lesion. Lesions were correlated between readers based on sector location and lesion morphology.
Histopathology Reference Standard
Prostatectomy specimens were used as the reference standard. Each prostate specimen was formalin fixed and processed in a customized 3D-printed in vivo MRI-based specimen mold.16 Lesions on histopathology were visually assessed by an experienced genitourinary pathologist and outlined on whole mount specimens (Fig. 1). Gleason (Gl) scores were assigned an overall primary and secondary pattern for the prostate specimen. Mapped lesion volumes were planimetrically measured on digital pathology images using Aperio ImageScope.17 Discrepancies among sectors were permitted for lesions interpreted to be the same based on pathological morphology and T2W MRI slice location. Index lesion was defined on pathology as the highest grade lesion on pathology, or if of equal grade the largest volume lesion on pathology. Up to two index lesions were permitted per patient if grade and volume were equivalent.
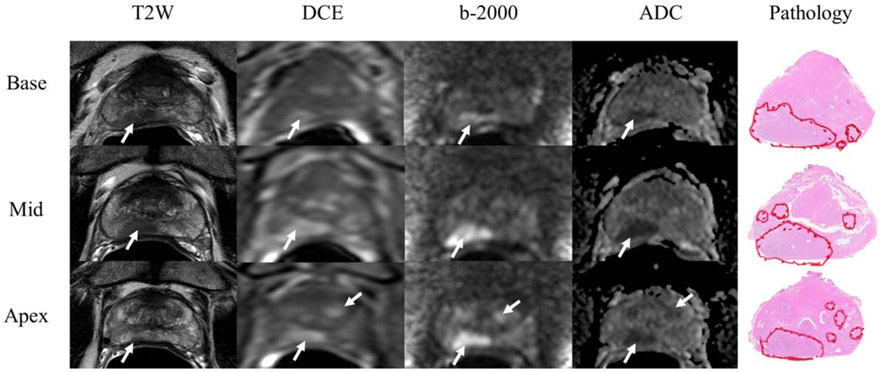
FIGURE 1:
Prostate cancer lesion detection and scoring on mpMRI. Images are representative of one patient in study. Five body radiologists blinded to clinical and pathological outcomes, including lesion location, detected cancer lesions on mpMRI and scored the lesion according to PIRADSv2. Lesions are marked with white arrows of each parameter: T2W, DCE, high-b value DWI (b2000), and ADC maps. Detected lesions were visually registered to whole mount prostate specimens with prostate lesions defined by a pathologist. The index lesion in this case in the right peripheral zone had Gleason 4 + 4 = 8 disease.
Statistical Analysis
Generalized estimating equations (GEE) with a logit link function and working independence correlation structure were used to estimate sensitivity and positive predictive value (PPV) of each reader. In each GEE model, the predictors included readers, covariates, and reader-covariate interaction terms. The averaged PPV and sensitivity values were obtained by taking the average of reader-specific sensitivities and PPVs estimated from each GEE model. Robust variance estimates under the working independence assumption were used to calculate the standard errors of the estimators. Sensitivity and PPV were determined for PIRADS ≥ 1, PIRADS ≥ 3 and PIRADS ≥ 4 for all lesions and index lesions for all readers, highly experienced readers, and moderately experienced readers. PPV for all lesions was defined as the proportion of true positive lesions among all detected lesions. Specificity was not determined because readers detected clinically suspicious lesions, but did not detect “negative” lesions or sectors.
Interobserver agreement was examined with respect to (1) overall scoring lesions with PIRADSv2, (2) detecting lesions (i.e., PIRADS ≥ 3) and (3) scoring lesions PIRADS ≥ 4. Agreement was calculated by the index of specific agreement (ISA), which is defined as the conditional probability given that one of the raters, randomly selected, makes a specific rating, that the other rater will also do so.18-20 To compare with traditional kappa statistic, Cohen’s kappa statistic was also used to examine the pairwise agreement. The bootstrap resampling procedure was used to calculate the standard error of ISA and kappa where the bootstrap sampling unit is patient.
Results
Histopathologic Characteristics
Ninety-four total lesions were identified on histopathology, of which 50 were Gl=3 + 4, 7 were Gl = 4 + 3, 26 were Gl = 4 + 4, and 11 were Gl = 4 + 5. There were 35 index lesions, of which 17 were Gleason 3 + 4 and 18 were Gleason ≥ 4 + 3. The average lesion size of all lesions was 1.36 mL (range, 0.05–8.70 mL). The average size of all index lesions was 2.33 mL (range, 0.57–8.70 mL).
Accuracy of PIRADSv2
On mpMRI, each reader identified an average of 2.1 lesions and 1.7 true positive lesions per patient. There were 7, 20, 58, 122, and 153 lesions at PIRADS thresholds of 1, 2, 3, 4, and 5, respectively, for all readers. Of the PIRADS<3 lesions, 10/27 were true positives. Moderately experienced readers were more likely to score a detected lesion as PIRADS = 3 rather than PIRADS > 3 compared with highly experienced readers, odds ratio (OR) 2.25 (95% confidence interval [CI]: 1.27–3.99; P = 0.005).
The average sensitivity of PIRADSv2 with a threshold of PIRADS≥3 for all readers to detect prostate cancer was 61% (Table 2). The experience level of the reader did not affect the performance of lesion detection. Highly and moderately experienced readers demonstrated similar sensitivities for all lesions with a sensitivity of 63% for experienced readers and 59% for moderately experienced readers (P = 0.30). Experience was more influential for PIRADS ≥ 4, where average sensitivity for all lesions was 60% versus 49% for highly and moderately experienced readers, respectively (P < 0.001).
TABLE 2.
Average Sensitivity and SE of PIRADSv2 Scores for Five Observers for All Lesions
| Experience | All | High | Moderate | P-Valuea |
|---|---|---|---|---|
| PIRADS ≥ 1 | 0.63 (0.04) | 0.66 (0.04) | 0.61 (0.04) | 0.21 |
| PIRADS ≥ 3 | 0.61 (0.04) | 0.63 (0.05) | 0.59 (0.04) | 0.30 |
| PIRADS ≥ 4 | 0.53 (0.05) | 0.60 (0.05) | 0.49 (0.05) | <0.001 |
P-Values are between sensitivities of highly and moderately experienced readers
The average sensitivity for detecting index lesions was 91% at PIRADS ≥ 3 and 83% at PIRADS ≥ 4 (Table 3). The average sensitivity for detecting index lesions with threshold PIRADS ≥ 4 was higher for experienced readers, 90% versus 79% for moderately experienced readers (P = 0.01). Highly and moderately experienced readers did not demonstrate a significantly different sensitivity at PIRADS≥3 (P = 0.30).
TABLE 3.
Average Sensitivity and SE of PIRADSv2 Thresholds for 5 Observers for Index Lesions
| Experience | All | High | Moderate | P-Valuea |
|---|---|---|---|---|
| PIRADS ≥ 1 | 0.91 (0.03) | 0.93 (0.03) | 0.90 (0.04) | 0.30 |
| PIRADS ≥ 3 | 0.91 (0.03) | 0.93 (0.03) | 0.90 (0.04) | 0.30 |
| PIRADS ≥ 4 | 0.83 (0.05) | 0.90 (0.04) | 0.79 (0.05) | 0.01 |
P-Values are between sensitivities of highly and moderately experienced readers.
The PPV improved at each PIRADS cutoff for detecting all lesions (Table 4). Using a cutoff of PIRADS ≥ 3, readers had an average PPV of 85%, with improvement to 90% for PIRADS ≥ 4. Experience was not a significant factor for PPV at all PIRADS thresholds, with highly experienced readers reaching a PPV of 93% for PIRADS ≥ 4 lesions versus 88% for moderately experienced readers (P = 0.26).
TABLE 4.
Average PPV and SE for All Lesions for Five Readers
| Experience | All | High | Moderate | P-Valuea |
|---|---|---|---|---|
| PIRADS ≥ 1 | 0.82 (0.03) | 0.84 (0.03) | 0.80 (0.03) | 0.11 |
| PIRADS ≥ 3 | 0.85 (0.03) | 0.87 (0.03) | 0.84 (0.03) | 0.45 |
| PIRADS ≥ 4 | 0.90 (0.02) | 0.93 (0.03) | 0.88 (0.03) | 0.26 |
P-Values are between sensitivities of highly and moderately experienced readers.
Interobserver Agreement
The average index of specific agreement in assigning PIRADSv2 scores for all detected lesions was 58%, whereas the average agreement in detecting lesions (PIRADS ≥ 3) was 74% (Table 5). The average agreement was 72% for scoring a detected lesion at PIRADS ≥ 4 when all lesions were considered. Interobserver agreement for index lesions was 85% for score assignment, 93% for lesion detection, and 95% for PIRADS ≥ 4.
TABLE 5.
Index of Specific Agreement and SE for Five Readers for Detecting and Scoring Lesions with PIRADSv2
| Overall | H-H | H-M | M-M | ||
|---|---|---|---|---|---|
| All Lesions | PIRADS scoring | 0.58 (0.04) | 0.70 (0.04) | 0.58 (0.04) | 0.53 (0.04) |
| Lesion detection | 0.74 (0.03) | 0.75 (0.04) | 0.74 (0.03) | 0.75 (0.03) | |
| PIRADS≥4 | 0.72 (0.03) | 0.81 (0.04) | 0.72 (0.04) | 0.68 (0.04) | |
| Index Lesions | PIRADS scoring | 0.85 (0.04) | 0.92 (0.03) | 0.86 (0.04) | 0.79 (0.05) |
| Lesion detection | 0.93 (0.02) | 0.92 (0.03) | 0.93 (0.02) | 0.92 (0.04) | |
| PIRADS≥4 | 0.90 (0.03) | 0.95 (0.03) | 0.91 (0.03) | 0.88 (0.04) |
H-H = agreement between highly experienced observers; H-M = agreement between highly and moderately experienced observers; M-M = agreement between moderately experienced observers.
Highly experienced readers agreed more frequently than moderately experienced readers especially for high risk disease: highly experienced readers demonstrated an agreement of 95% for PIRADS ≥ 4 index lesions compared with 88% for agreement between moderately experienced readers and 91% for agreement between highly experienced and moderately experienced readers. Concordance was most divergent among moderately experienced readers scoring PIRADSv2 with an agreement of 53% for all lesions compared with 70% for highly experienced readers.
As readers detected lesions independently, the value of assessing agreement by chance with the kappa statistic is uncertain. Table 6 demonstrates the calculated kappa statistic between two readers. Reader 1 and Reader 2 are the two highly experienced readers with results of index lesions detection tabulated in a 2×2 table. The detection rate was 32/35 for reader 1 and 33/35 for reader 2, with agreement on 30/35 total index lesions. The kappa value calculates the “observed agreement” (po) and standardizes that against “expected agreement” (pe) by chance.21 In this scenario, po is 0.86 and pe is 0.87 yielding a kappa value of −0.07, which defies common sense. Conversely, the ISA for these two readers was 95%.
TABLE 6.
Kappa Statistic Calculations for Two Highly Experienced Readers Detecting Index Lesions
| Reader 2 lesion detected? | ||||||
|---|---|---|---|---|---|---|
| Yes | No | Total | ||||
| Reader 1 lesion detected? | Yes | 30 | 2 | 32 | pe | 0.87 |
| No | 3 | 0 | 3 | po | 0.86 | |
| Total | 33 | 2 | 35 | kappa | −0.07 | |
Discussion
We evaluated the agreement among five radiologists in using PIRADSv2 to detect and score lesions on mpMRI using whole mount histopathology as the reference standard. Average sensitivity for detecting index lesions was 91% for PIRADS ≥ 3 and 83% for PIRADS ≥ 4, demonstrating excellent accuracy for PIRADSv2 to detect index lesions. When secondary and tertiary lesions were included, sensitivity was 63% for all lesions. Highly experienced readers performed better than moderately experienced readers only for index lesions scored as PIRADS ≥ 4 (90% versus 79%; P = 0.01), but did not perform better when PIRADS = 3 lesions were included. The average PPV of PIRADSv2 scores improved for higher PIRADSv2 scores, with PIRADS ≥ 4 demonstrating a PPV of 90%. There was substantial (93%) agreement among readers for detecting index lesions, and good agreement for scoring index lesions (85%). The results of our study indicate that readers with varying levels of experience can use PIRADSv2 to detect index lesions with high sensitivity and agreement.
Our results are concordant with previous studies on PIRADSv2, and our testing is more representative than these studies of evaluating prostate mpMRI in the clinical setting. Zhao et al22 found a sensitivity and specificity over 80% for two readers, but was limited as TRUS-guided biopsy alone was used as the reference standard, where we used prostatectomy as a standard. Vargas et al14 evaluated a “best achievable” performance of PIRADSv2 by identifying high-grade lesions on whole-mount prostatectomy specimens and scoring the correlated mpMRI lesions with PIRADSv2. By preselecting lesions to be assessed, a favorable bias is introduced. Thus, readers correctly identified 95% of large tumors (≥0.5 mL), but only 20–26% of small high-grade tumors. Unlike the previous study, however, readers in our study were asked to detect and score the scans as they would in a routine clinical scenario, not to score predetermined lesions. Our results for reader-defined rather than preselected lesions indicated a sensitivity of 91% for index lesions and 63% for all lesions. Our sensitivity was similar to that of Muller et al6 who evaluated PIRADSv2 in biopsy naïve patients and found a sensitivity of 85–88% for detecting clinically significant tumors using PIRADSv2, with a kappa score of 0.46 for 5 readers.
However, this study has been criticized for its inclusion of nonradiologists as readers.15 In the current study, all readers were trained body radiologists and had prior familiarity with PIRADSv2. Much higher agreement for detecting and scoring lesions was observed in these conditions with agreement reaching 93% for index lesions. This underscores the importance of proper training in interpreting mpMRI. Additionally, we used whole mount prostate specimens whereas Muller et al used biopsy specimens as the reference standard, which are more likely to miss clinically significant lesions. Although this study cannot be directly compared with studies on PIRADSv1, Hamoen et al12 performed a meta-analysis of all previously published PIRADSv1 studies and found a collective sensitivity of 78% among 14 qualifying studies. Our study suggests similar sensitivity of PIRADSv2 to PIRADSv1 but with high reader agreement. This is similar to other studies that directly compared PIRADSv1 to PIRADSv2, such as the study of Kasel-Seibert et al23 with AUC values improved from 0.70–0.79 to 0.83 for PI-RADSv1 compared with PI-RADSv2, respectively.
Prior reader experience was most evident in scoring lesions using a PIRADS ≥ 4 threshold. The sensitivity was not different between highly and moderately experienced readers at a threshold of PIRADS ≥ 3 for index lesions (P = 0.30), but highly experienced readers performed better at a threshold of PIRADS ≥ 4 (P = 0.01). While presumably all PIRADS≥3 lesions would be indicated for biopsy, the question remains why moderately experienced readers were less accurate at PIRADS ≥ 4 and more likely to score a lesion as PIRADS = 3. One possible explanation is the criteria on PIRADSv2 for distinguishing PIRADS = 3 from PIRADS = 4 is not adequate for readers without experience to make the distinction between intermediate (PIRADS = 3) and high (PIRADS = 4) suspicion lesions.24 The discordance in agreement for lesion detection (74%) and scoring (58%) may also be due to the ambiguity of scoring these lesions. The ambiguity for inexperienced readers between PIRADS 3 and PIRADS 4 lesions is an important area for improvement in the next version of PIRADS.
The tool by which reproducibility is measured greatly influences the result. We elected to use the index of specific agreement over the kappa statistic. The kappa statistic has been widely applied in medical research as a measure of reliability for nominal classification procedures. It corrects for the proportion of agreement by chance. Kappa statistics can give rise to paradoxical results caused by uneven distribution of classification rates. As readers independently detected lesions, the probability that readers found the same lesions in the same location purely by chance is low. The kappa statistic is more appropriate in study designs where lesions are predetermined and then are shown to readers to score as a dichotomized decision, not where readers prospectively detect and score lesions.
An alternative measure of agreement is the index of specific agreement. Analogous to sensitivity and specificity, which are used together to evaluate the accuracy of a test procedure, the index of specific agreement is used to evaluate inter-reader agreement at each score category. This makes it well suited to the task of comparing PIRADSv2 scoring, and as such, the source of agreement and disagreement can be identified.19,20,25
There were several limitations in our study. One of the major challenges was histopathological correlation, a common difficulty for studies involving prostate cancer rad-path interpretation. Despite the use of customized patient molds allowing for patient-specific axial histological sections corresponding to axial MRI, there were artifacts introduced from differences in morphology between the formalin-fixed histologic sections and MRI images. We attempted to correct for any mis-registration between prostate sections to correlate reader-defined lesions with each other and with pathology, but errors were still possible due to the thick histological sections (6 mm) compared with the MRI slices (≤3 mm). We were also limited by the cohort size (n = 34). Additionally, all patients underwent radical prostatectomy. This could introduce a bias toward intermediate–high risk prostate cancer patients and a detection bias among readers; however, this reflects a patient population in which PIRADSv2 would be commonly used. Further research is needed to validate PIRADv2 in a prospective patient population with a larger risk distribution.
In conclusion, by using PIRADSv2, general body radiologists and prostate dedicated radiologists can detect high grade index lesion prostate cancers on mpMRI with high sensitivity and high agreement. Thus, PIRADSv2 appears to be a major step forward in standardizing interpretation of prostate mpMRI across readers of varying experience.
Acknowledgments
This research was funded in part by the Intramural Research Program, National Institutes of Health, National Cancer Institute and Clinical Center. This research was also made possible through the National Institutes of Health (NIH) Medical Research Scholars Program, a public-private partnership supported jointly by the NIH and generous contributions to the Foundation for the NIH from Pfizer Inc., The Doris Duke Charitable Foundation, The Newport Foundation, The American Association for Dental Research, The Howard Hughes Medical Institute, and the Colgate-Palmolive Company, as well as other private donors. For a complete list, please visit the Foundation website at: http://fnih.org/work/education-training-0/medical-research-scholars-program. Authors have no conflicts of interest to disclose.
References
- 1.Menard C, Smith IC, Somorjai RL, et al. Magnetic resonance spectros-copy of the malignant prostate gland after radiotherapy: a histopathologic study of diagnostic validity. Int J Radiat Oncol Biol Phys 2001;50:317–323. [DOI] [PubMed] [Google Scholar]
- 2.Pinto PA, Chung PH, Rastinehad AR, et al. Magnetic resonance imaging/ultrasound fusion guided prostate biopsy improves cancer detection following transrectal ultrasound biopsy and correlates with multiparametric magnetic resonance imaging. J Urol 2011;186:1281–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyagawa T, Ishikawa S, Kimura T, et al. Real-time virtual sonography for navigation during targeted prostate biopsy using magnetic resonance imaging data. Int J Urol 2010;17:855–860. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015;313:390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddiqui MM, Rais-Bahrami S, Truong H, et al. Magnetic resonance imaging/ultrasound-fusion biopsy significantly upgrades prostate cancer versus systematic 12-core transrectal ultrasound biopsy. Eur Urol 2013;64:713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller BG, Shih JH, Sankineni S, et al. Prostate cancer: interobserver agreement and accuracy with the revised prostate imaging reporting and data system at multiparametric MR imaging. Radiology 2015;277:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghai S, Haider MA. Multiparametric-MRI in diagnosis of prostate cancer. Indian J Urol 2015;31:194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett T, Turkbey B, Choyke PL. PI-RADS version 2: what you need to know. Clin Radiol 2015;70:1165–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate imaging - reporting and data system: 2015, version 2. Eur Urol 2016;69:16–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radiology ACo. MR prostate imaging reporting and data system version 2.0. 2015. Available at: http://www.acr.org/~/media/ACR/Documents/PDF/QualitySafety/Resources/PIRADS/PIRADS%20V2.pdf.
- 11.Tewes S, Hueper K, Hartung D, et al. Targeted MRI/TRUS fusion-guided biopsy in men with previous prostate biopsies using a novel registration software and multiparametric MRI PI-RADS scores: first results. World J Urol 2015;33:1707–1714. [DOI] [PubMed] [Google Scholar]
- 12.Hamoen EH, de Rooij M, Witjes JA, Barentsz JO, Rovers MM. Use of the Prostate Imaging Reporting and Data System (PI-RADS) for prostate cancer detection with multiparametric magnetic resonance imaging: a diagnostic meta-analysis. Eur Urol 2015;67:1112–1121. [DOI] [PubMed] [Google Scholar]
- 13.Renard-Penna R, Mozer P, Cornud F, et al. Prostate imaging reporting and data system and Likert scoring system: multiparametric MR imaging validation study to screen patients for initial biopsy. Radiology 2015;275:458–468. [DOI] [PubMed] [Google Scholar]
- 14.Vargas HA, Hotker AM, Goldman DA, et al. Updated prostate imaging reporting and data system (PIRADS v2) recommendations for the detection of clinically significant prostate cancer using multiparametric MRI: critical evaluation using whole-mount pathology as standard of reference. Eur Radiol 2016;26:1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barentsz JO, Weinreb JC, Verma S, et al. Synopsis of the PI-RADS v2 guidelines for multiparametric prostate magnetic resonance imaging and recommendations for use. Eur Urol 2016;69:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah V, Pohida T, Turkbey B, et al. A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum 2009;80:104301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aperio Technologies I. ImageScope. 12.2.2.5015 ed; 2016. Available at: http://www.leicabiosystems.com/digital-pathology/digital-pathology-management/imagescope/. Accessed 4 April, 2016.
- 18.Guggenmoos-Holzmann I How reliable are chance-corrected measures of agreement? Stat Med 1993;12:2191–2205. [DOI] [PubMed] [Google Scholar]
- 19.Feinstein AR, Cicchetti DV. High agreement but low kappa: I. The problems of two paradoxes. J Clin Epidemiol 1990;43:543–549. [DOI] [PubMed] [Google Scholar]
- 20.Cicchetti DV, Feinstein AR. High agreement but low kappa: II. Resolving the paradoxes. J Clin Epidemiol 1990;43:551–558. [DOI] [PubMed] [Google Scholar]
- 21.Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Fam Med 2005;37:360–363. [PubMed] [Google Scholar]
- 22.Zhao C, Gao G, Fang D, et al. The efficiency of multiparametric magnetic resonance imaging (mpMRI) using PI-RADS Version 2 in the diagnosis of clinically significant prostate cancer. Clin Imaging 2016;40:885–888. [DOI] [PubMed] [Google Scholar]
- 23.Kasel-Seibert M, Lehmann T, Aschenbach R, et al. Assessment of PI-RADS v2 for the detection of prostate cancer. Eur J Radiol 2016;85:726–731. [DOI] [PubMed] [Google Scholar]
- 24.Rosenkrantz AB, Oto A, Turkbey B, Westphalen AC. Prostate Imaging Reporting and Data System (PI-RADS), version 2: a critical look. AJR Am J Roentgenol 2016;206:1179–1183. [DOI] [PubMed] [Google Scholar]
- 25.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159–174. [PubMed] [Google Scholar]