Abstract
Structural details of protein interactions are invaluable to the understanding of cellular processes. However, the identification of interactions at atomic resolution is a continuing challenge in the systems biology era. Although the number of structurally resolved complexes in the Protein Databank increases exponentially, the complexes only cover a small portion of the known structural interactome. In this chapter, we review the PRISM system that is a protein–protein interaction (PPI) prediction too—its rationale, principles, and applications. We further discuss its extensions to discover the effect of single residue mutations, to model large protein assemblies, to improve its performance by exploiting conformational protein ensembles, and to reconstruct large PPI networks or pathway maps.
Keywords: Structural matching, PPI prediction, Mutation mapping, PPI network, Structural pathway modeling
1. Introduction
1.1. Review of Template-Based Approaches
In the cell biological processes are realized through interactions among proteins and between proteins and other molecules. Protein–protein interactions (PPIs) orchestrate complex processes including signaling and catalysis. With the recent advances in experimental techniques, the number of identified PPIs keeps increasing. However, we are still far from complete knowledge of PPIs and their characterization at the atomistic level. Template-based computational methods to predict PPIs have recently become popular due to the significant increase in protein sequence and structural data. In this chapter, we review template-based approaches, not only to predict PPIs but also to provide structural models of the interacting proteins. Here, we describe PRISM, one of the earliest template-based algorithms that aimed to build PPI networks (called structural PPI networks) based on augmented sets of structural models of the interactions. The integration of protein structures into the PPI network allows mapping of single nucleotide variations (SNVs) in protein-coding areas to interactions and predicting the effect of the mutations in larger settings. The fundamental concept behind PRISM is that there are favorable structural motifs at protein–protein interfaces and that these architectural motifs resemble those found in protein cores [1–5].
1.2. Template-Based Docking
Template-based computational approaches for predicting PPIs and their structural models utilize the accumulated sequence and structure data of known PPIs [2]. Given two unbound proteins, the task of the template-based method is to find a similar complex (template) in the database of known interactions, aligning the unbound proteins to the template. One of the earliest approaches is the homology-based prediction of PPIs [6], which is based on the observation that homologous pairs of proteins tend to interact in a similar manner. The homology-based method requires significant sequence similarity (at least 30 %) and uses the whole sequence. Interactome3D is among the homology-based methods. Additionally, the domain–domain preferences of protein interactions are considered as partial templates in Interactome3D [7]. The Instruct method also uses domain-mediated interaction templates and searches for domain availability in target proteins [8]. In the case of low sequence similarity, a protein threading approach can be used [9, 10]. This method was initially used for predicting the structure of single proteins, and later the methodology has been extended to model PPIs [9, 10]. In both cases, similarity between the unbound proteins and templates as a whole is sought. This limits the applicability at large scale due to many factors such as low coverage of available data, conformational changes of the proteins, and so on.
Another template-based approach is to use partial structures as templates, where the most representative case is the use of protein interfaces. PRISM is one of the first methods that uses protein interfaces to predict PPIs and their structural models [11, 12]. In this chapter, we outline the basics of PRISM and its extensions for PPI prediction. We include in silico analysis of functional variations, constructing large assemblies (see Note 1) and modeling PPI networks in atomic detail. We also detail the functionalities of the PRISM web server for online prediction.
2. Methods
2.1. Principles of PPI Prediction by PRISM
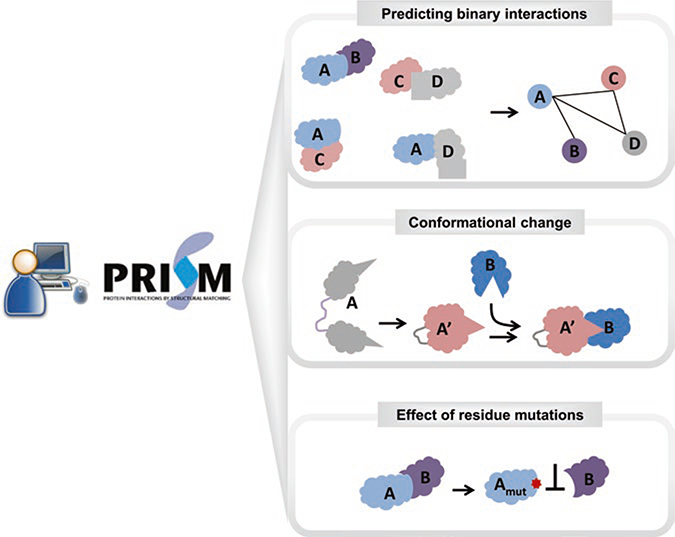
PRISM combines sequence and structural information about known protein interfaces to discover not only potential novel interactions but also the binding modes of known protein interactions. The method is well established, accurate, and computationally efficient. PRISM serves both as an online resource [13] and a downloadable protocol [12] (Fig. 1). To run the PRISM system, two datasets are necessary: (1) a template set and (2) a target set. The idea is that if partner chains of a template interface are spatially similar to any region on the surfaces of the two targets and share some evolutionarily important residues, then these target proteins can interact with each other with an architecture resembling that of the template interface.
Fig. 1.
A schematic representation of PRISM functionalities. PRISM can be run on local computers as well as interactively on the web server. PRISM predictions can be used for multiple purposes. Predicting the interaction between two proteins and PPI networks is possible with PRISM. In addition, with an accurate design of the template set, conformational changes can be handled in the prediction. The effect of residue mutations can be analyzed by comparing the wild-type and mutated predictions
2.1.1. Template Set
The template set is composed of known protein interfaces. Protein complexes deposited in the Protein Data Bank (PDB) are the main resource to extract protein interfaces. The performance of PRISM depends on the diversity of the interfaces in the template dataset. The ideal set should cover all available architectures of protein interfaces. An approximate template set can be formed by clustering structurally similar interfaces and choosing one member from each cluster. Details on how to construct an interface dataset using this approach is discussed in [14–16]. PRISM provides a built-in (default) template set prepared in this way, which is a subset of the protein interfaces in the PDB. Depending on the system under investigation, the template set can be modified—e.g., using only oncogenic interfaces to model cancer-specific interactions. The latest version of the interface dataset used in PRISM was constructed from the all PDB complexes deposited before January 2012. This version includes 22,604 unique interface architectures.
In addition to structural similarity, PRISM uses matching of some critical residues. The residues in protein interfaces do not equally contribute to the binding energy of interactions. Some interface residues are more important; these are called “hot spots”. Experimentally, they can be determined by alanine-scanning mutagenesis experiments, but these data are available only for a limited number of complexes. Therefore, computational approaches emerged to accurately and efficiently predict binding hot spots. Hotpoint, which considers solvent accessibility and contact potentials of residues [17, 18], is one of these approaches. If a residue is highly packed and buried in the interface, it has a higher tendency to be a hot spot. Hot spots predicted by the Hotpoint server are used in the built-in template set of PRISM. However, the user is free to label hot spots with other available methods.
2.1.2. Target Set
The target set is composed of all proteins under consideration. It should contain at least two proteins. The atomic coordinates of the targets are retrieved from the PDB (for details see Note 2). For proteins not available in the PDB, homology modeling–based techniques can be used to increase the structural coverage of the target set.
The target set can be shaped based on the purpose of the analysis. If the aim is to discover a specific interaction between two proteins, the target set should contain all available structures of the two proteins. To structurally model a known pathway, all proteins functioning in that pathway form the target set. Another possibility is to construct an organism-specific structural interactome with PRISM. In this case, all proteins having structural information or homology models in that organism are included in the target set. If the aim is to construct a tissue-specific structural interactome, the target set should cover all proteins expressed in the selected tissue.
Recent efforts using PRISM have shown that considering all available 3D conformations of proteins improves the accuracy of prediction [19] (see Note 3). Proteins are flexible and prone to conformational changes. Binding of an activator to the extracellular portion of a membrane protein or binding a small molecule to a region of a protein that is distant from the functional region can lead to allosteric changes in global structure. Domain motions including hinge motions also reflect conformational changes, typically on a larger scale. For example, when a small molecule (trifluoperazine) binds to calmodulin, calmodulin adopts a new conformation that opens up two helices and changes its binding preferences. Even binding of an ion (Ca2+) shifts the equilibrium between the open and closed calmodulin states. The PDB is a rich source for multiple protein conformations.
2.1.3. Prediction
The prediction procedure follows four consecutive steps: (1) extraction of the surface regions of target proteins, (2) structural alignment of the templates to the targets, (3) transformation of the targets onto the templates and filtering unrealistic cases, and (4) flexible refinement and energy calculation.
Proteins interact using their surfaces. Therefore, the first step is extracting the atomic coordinates of target protein surface residues. The NACCESS [20] tool, which calculates the solvent accessibility of protein residues, is used for this purpose. Residues are defined as being on the surface if the ratio of their solvent accessible surface area in the protein state and in an extended tripeptide state is greater than 15 %. To conserve the secondary structure, residues within 6 Å of the surface are also extracted. These are called “nearby” residues. The output of this step is the atomic coordinates of the surface and nearby residues of each of the target proteins in PDB format.
The next stage is rigid body structural alignment, which is sequence order independent where only geometric similarity is sought. PRISM uses Multiprot [21] for structural alignment. At the alignment stage, conservation of the template interface hot spots on the target surface is also checked to limit the search space. If the surfaces of two targets have spatially similar regions to complementary partners of a template interface and at least one hot spot in each partner chain is conserved on the target surfaces, the global structures of the targets are superimposed onto the template. In this way, the first putative complex is modeled. The super-imposition of the targets may cause atomic clashes. At the filtering stage, predicted complexes having many atomic clashes are removed from the final list. The last stage entails refinement of predicted complexes and ranking them according to their binding affinities. Fiberdock [22] is employed for refinement and energy calculation purposes. First, the side chains in the interaction inter face and the backbone of the predicted complex are optimized. The binding energy score is also calculated. In this way, PRISM considers not only geometric complementarity but also the flexibility of targets and their chemical complementarity. The final list gives a set of predicted complexes refined with docking protocols.
After completion of all intermediate stages, the final result includes the list of putative binary interactions, their binding energy scores, and their 3D modeled complexes. The output is a rich resource for further analysis and modeling. PRISM is multifunctional. It can be used for predicting binary interactions, discovering the effect of residue mutations, constructing protein interaction networks, and structural modeling of known pathways (Fig. 1). Each of these functionalities is reviewed in the following sections.
2.2. PRISM2.0 Web Server
PRISM can be used in two ways: (1) PRISM stand-alone version [12] or (2) PRISM web server [13]. Advanced users may prefer to install the PRISM protocol and use it on their local computers. This allows adjustment of parameters for prediction or getting output for intermediate steps. The stand-alone version runs only in the Linux environment. PRISM is continuously upgraded to improve its performance and user-friendliness as well as its computational effectiveness. The current stand-alone protocol (version 1.0) can be accessed at http://prism.ccbb.ku.edu.tr/prism_protocol/. An upgraded stand-alone version will be released soon. The PRISM web server, on the other hand, has been developed to provide a simple user interface to perform all the steps of the PRISM algorithm with default settings. The web server (PRISM2.0) [13] implements a slightly modified version of the stand-alone protocol [12]. In addition, the web server aims to provide a repository (database) of structural models of all predicted interactions. All the predicted interactions between proteins having PDB IDs are stored in its database. As the community uses the server, it is expected that the structural models in the database will grow and become an invaluable resource.
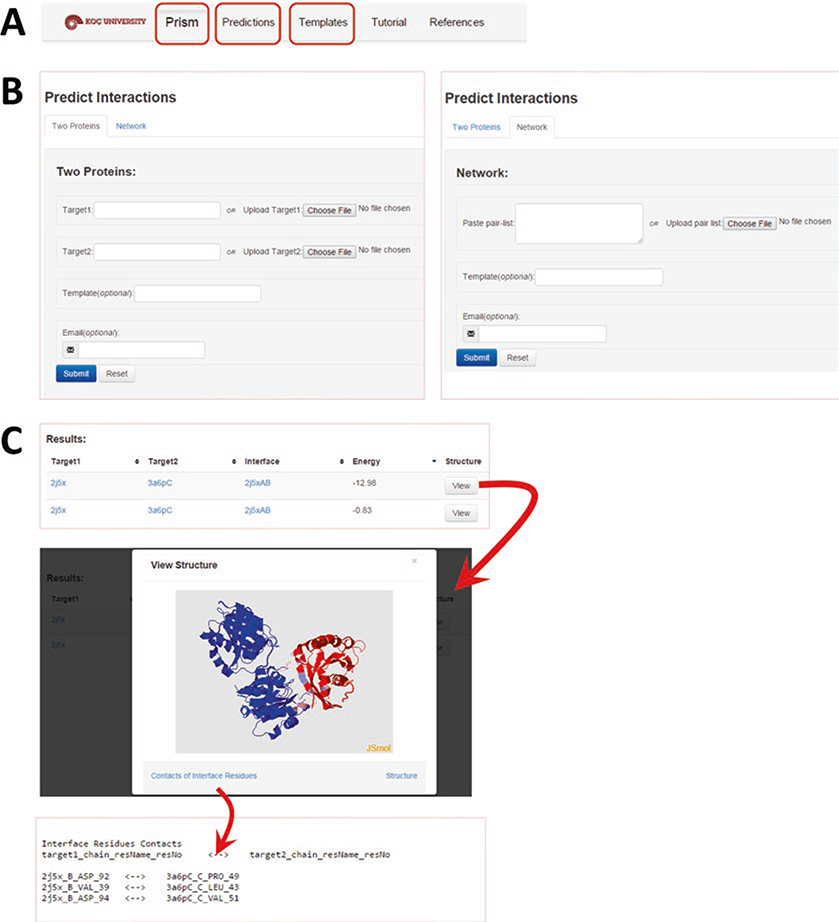
The web server is multifunctional, which allows for online prediction as well as browsing the accumulated data. We provide an overview of the web server in Fig. 2. The step-by-step procedure to run the prediction algorithm on the PRISM server is provided in Subheading 2.2.1.
Fig. 2.
Overview of the PRISM web server. (a) The “Prism” tab is designed for online prediction. The “Predictions” tab is for browsing the accumulated data in the web server. The “Templates” tab is for browsing detailed information about the built-in template set. (b) For online prediction, two options are available: (1) predicting the interaction between two proteins (left panel) or (2) predicting interactions in a network (right panel). For the former option, the input is the PDB codes or PDB files of “Target 1” and “Target 2.” For the latter option, the input is the list of protein pairs with their PDB codes. The template code and e-mail address are optional inputs for both. (c) The “Results” page lists the predicted interactions with the target codes, the template (the column labeled “Interface”), and the calculated binding energy. Each interaction has a “View” button where an interactive visualization of the predicted complex is possible. Target 1 is colored blue and Target 2 is red. Their interface regions are in pink and light blue, respectively. The contacts in the interface region and the PDB file of the predicted complex are downloadable using the “Contact of Interface Residues” and “Structure” links, respectively
2.2.1. Online Prediction
The user first needs to prepare the target pairs and determine the templates to be used in the prediction.
Because PRISM requires the structures of target proteins, protein names or any other identifiers must be cross-referenced to the PDB identifiers. As mentioned in Note 2, one protein may have many PDB identifiers; each may represent partial structures of different fragments or structures at different resolutions. For example, human RAF protein has many structures in the PDB (e.g., chain B of 1c1y covers positions 55–131, chain A of 1faq covers positions 136–187, and chain A of 3omv covers 323–618). All structures of the corresponding target protein should be included for a better accuracy. As input, the user can provide the PDB codes of these proteins with or without chain identifiers or upload the PDB formatted files of the target structures. If chain identifiers are included in the PDB id, then only those chains will be used. For example, the input 1a0hAE will use only the A and E chains (skipping the other chain in 1a0h) to form the protein structure.
If the protein does not have information in PDB, homology modeling techniques can be used to predict the structure. Some useful resources for homology modeling are MODELLER [23], SWISS-MODEL [24], and ModBase [25]. The input for online prediction in PRISM would then be the PDB formatted files of the homology models.
With its default settings, PRISM uses the built-in template set. However, the user can enter a template name to run PRISM only for one interface. This option is useful if the user is interested in predicting an interaction using a specific template (much faster than using the whole template set) or in using a template interface that is not available in the built-in set.
There are two prediction options (Fig. 2b): the first is to search for the interaction between two proteins and the second is to predict a network of interactions for up to 10 pairs of proteins. If the target set contains a structure prepared by the user with homology modeling, then only the “predict between two proteins” option can be used.
After preparing the target pairs and determining the templates to be used, the job can be submitted. The user can wait on the page, bookmark the page to browse later, or enter an email address to receive a notification after the job is completed.
For each prediction, the PDB codes and chain identifiers of the targets, the template interface used in the prediction and the calculated binding energy are listed (Fig. 2c). Also, a “View” button is available for interactive visualization of the predicted complex in JMol where user can examine the predicted interface as well as noninterface regions in the overall complex. The PDB formatted file of the predicted complex and contacting residues in its interface are downloadable on the results page.
2.2.2. Browsing Accumulated PPI Predictions in PRISM
The PRISM web server also provides a search option and a download option to retrieve predictions from the accumulated data.
On the upper panel (shown in Fig. 2a), the user can select the “Predictions” tab to browse the PPIs already predicted and deposited at PRISM. Only the predictions for structures available in PDB are stored in the database. User-supplied protein structures (i.e., from homology modeling) are not accumulated. All the functionalities including visualization and download are also available for the accumulated data.
In addition, the user can search the data either with the target code or with the template interface code. For example, the target 3g9wD has five predicted interactions in the accumulated data and these interactions are predicted from the templates 3g9wAD and 1jfiBC. When we search the data with a template interface code, e.g., 3fxdBD, we find that there are 38 predicted interactions based on this interface.
2.2.3. Analysis of Template Interfaces
The built-in template dataset can be also browsed and analyzed in detail.
On the upper panel (shown in Fig. 2a), the user can select the “Templates” tab to browse the built-in template set. PRISM provides a table listing the representative of each cluster of interfaces (“Representative” column) as well as the number of interfaces in the cluster (“Members” column).
Clicking on an interface in the “Representative” column displays a page in the HotRegion database where the user can find the computational hot spots and hot regions.
-
Clicking on a number in the “Members” column opens a new window listing the interfaces structurally similar to the representative interface. Each member interface has a web link to the HotRegion database.
The PRISM web server has also a “Tutorial” page where the definitions of all terms are mentioned and the principles of prediction procedure can be found in detail (Fig. 2a).
2.3. Mapping Residue Level Mutations onto Predicted Binding Sites
The reconstructed structural PPI networks of signaling pathways help complete missing parts of pathways and provide PPI details. For example, they reveal the details of how signals coming from different upstream pathways merge and propagate downstream, how parallel pathways compensate for each other, and how multi-subunit signaling complexes form. These networks can also offer the possibility of observing the effects of SNPs and mutations on signal propagation.
There has been a tremendous increase in genomic variations data as well as in PPI data. Genome-wide association studies, whole-genome sequencing, and exome sequencing have shown that each personal genome contains millions of variants, thousands of which are nonsynonymous single nucleotide variants (nsSNVs) causing changes at the residue level. Some of these variations are neutral, whereas some are disease associated. For example, cancer results from somatically acquired variations in the DNA of cancer cells. However, not all the somatic changes present in a cancer genome are involved in development of the cancer. Indeed, it is likely that some have no contribution at all. These are termed as passenger mutations, whereas the ones that do contribute to disease are driver mutations. The identification and characterization of disease-associated, driver variations are important tasks in personalized medicine and genomic sequencing [26]. Genetic variation and mutation data are publicly available in databases such as the Online Database of Mendelian Inheritance in Man (OMIM) [27], the Human Gene Mutation Database (HGMD) [28], Humsavar (http://www.uniprot.org/docs/humsavar) of Uniprot, ClinVar [29], COSMIC (Catalogue of Somatic Mutations in Cancer) database [30], and cBioPortal for Cancer Genomics (which contains data from the Cancer Genome Atlas (TCGA) project) [31]. These databases capture variants at the level of the gene nucleotide sequence as well as the corresponding amino acid changes in the protein (gene product). These variations can be polymorphisms, variations between strains, isolates, or cultivars, disease-associated mutations, or RNA editing events. Most databases provide detailed information about the mutations, including the mutated residue numbers.
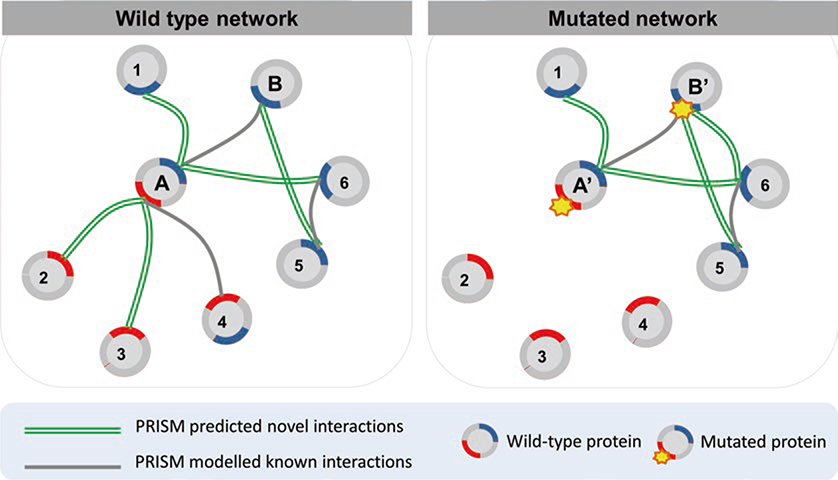
In our previous studies [32, 33], we extracted available point mutations (and data related to these mutations, e.g., phenotypic effects) from different sources and modeled the effects of oncogenic mutations. First, we identified mutations that mapped to the interface region of our modeled protein–protein complexes. Next, we performed in silico mutagenesis to observe the contributions of these specific mutations to the interaction (see Subheading 2.5). We re-ran PRISM on the mutant structures [32, 33] and modeled the new interaction between the mutant target and its partner. PRISM results for the wild-type and mutated cases give insight into the edgetic effects of the analyzed mutation. A mutation may cause a loss of an interaction, cause a gain, or be neutral. In Fig. 3, we show a conceptual representation of edgetic effects in a PRISM predicted network. A hypothetical protein “A” having two binding sites loses three interactions when a mutation occurs in one of its binding sites. However, a mutation in the binding site of hypothetical protein “B” is neutral for its wild-type interactions. However, a new interaction is gained when this mutation occurs in B. This type of analysis has been applied to the interleukin 1 (IL-1)-initiated signaling pathway [32]. Mapping residue mutations onto the reconstructed structural IL-1-initiated signaling pathway improved our understanding of the activation/inhibition mechanism.
Fig. 3.
A representation of the concept of a mutation effect in a PRISM predicted network. Protein A has two binding sites (red and blue colored regions) and the mutation occurs in the red binding site in A’ (mutated protein A). Known interactions are gray and novel interactions are green double lines. A mutation in protein A leads to loss of interactions between A and P2, P3, P4. On the other hand, when the mutation occurs in protein B, it gains a new interaction (with P6) while conserving its wild-type interactions
2.4. From PRISM Predictions to Interaction Networks and Pathway Modeling
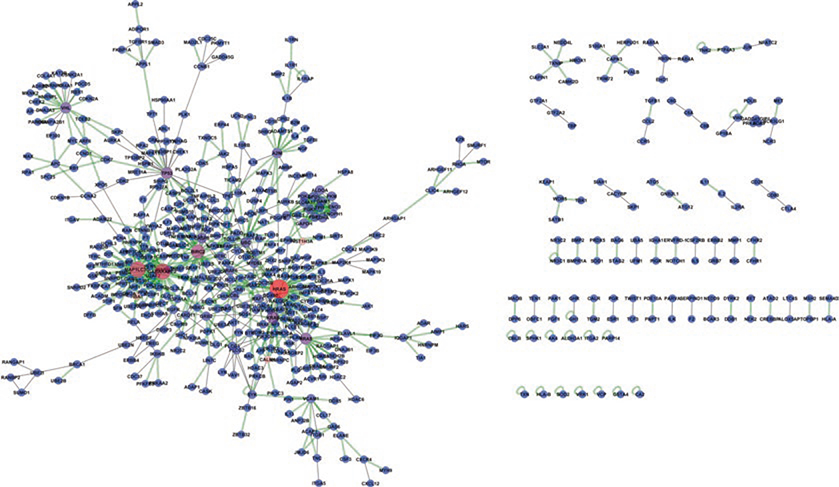
PRISM predicts PPIs at the proteome scale in a computationally efficient way, which enables us to construct large PPI networks. In these networks, nodes represent proteins and edges represent the interaction between them. The output of PRISM is the PDB codes of the chains of predicted interacting pairs, their binding energy, the structural model of the predicted complex, and the name of the template interface. To convert the PRISM output to a classical PPI network, each structural state is mapped to its corresponding protein name (see Note 4). The Uniprot database can be used as the central cross-referencing resource to map PDB chains to their unique protein identifiers. To illustrate a sample network predicted by PRISM, we compiled all the interactions accumulated in the web server. Since 2014, 542 unique human protein targets from 1193 PDB chains have been tested by external users. In total, 749 unique interactions have been predicted (Fig. 4). Among them, 246 are known interactions supported by experimental or database evidence in the STRING database [34] with a confidence score greater than 0.80. The remaining 503 interactions are novel—discovered by PRISM. The sizes of the nodes are scaled according to their centrality in the network. The node colors change from blue to red based on their centrality.
Fig. 4.
A network constructed with the interactions that have been predicted by users of the PRISM web server. Node sizes are scaled based on their centrality in the network. Gray edges are PRISM predicted and STRING validated interactions. Green two-line edges are PRISM predicted novel interactions
Additionally, a known pathway map can be structurally reconstructed by PRISM predictions. Adding the structures of protein interactions is necessary for rational pathway modeling and improves the understanding of how a function is exerted or a signal is transduced in a pathway. In previous work, PRISM has been applied to structurally model several pathways. Among them, the reconstructed p53-mediated pathway has been used to illustrate the multipartner interaction preferences of hub proteins including p53 and Mdm2 [35].
In general, pathway maps are incomplete. Predicted novel interactions help in reconstructing the pathway and in elucidating unknown functionalities. The human ubiquitination pathway, IL-1-initiated signaling pathway, MAPK pathway, and Toll-like receptor signaling pathway have also been structurally modeled with PRISM. For this type of analysis, the initial target set is all PDB chains of the proteins present in the pathway of interest. The output is structural models of known interactions and predicted novel interactions. By superimposing all partners of a protein, overlapping and distinct binding sites can be identified. This enables discriminating simultaneous or mutually exclusive interactions.
2.5. Case Study
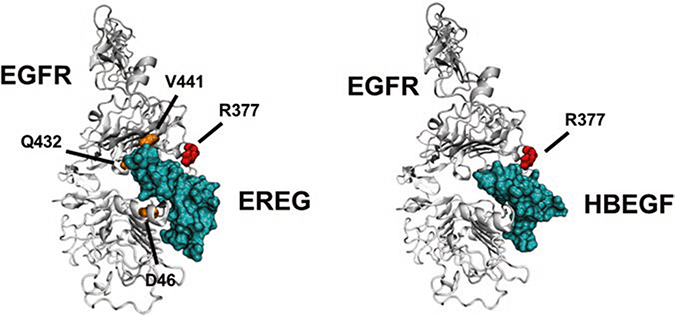
PRISM can also be utilized to reconstruct phenotype-specific subnetworks of PPIs. Previously, we formed subnetworks using the critical seed genes for two different phenotypes: lung and brain metastasis from primary breast tumors [33]. The seed genes were obtained from the literature [36, 37]. We extended the networks around these genes by using GUILD [38]. Finally, we modeled the structures of protein–protein complexes in these subnetworks and mapped the mutations from COSMIC [30] to the protein interfaces. This permitted the evaluation of the effect of mutations on the interactions. For example, EGFR was observed to make different interactions in the two reconstructed subnetworks. In brain metastasis, EGFR was found to interact with HBEGF, whereas the same protein was found to interact with EREG in lung metastasis. Indeed, HBEGF and EREG were known to have roles in brain [36] and lung [39] metastasis of breast cancer, respectively. When we examined the modeled structures of EGFR–HBEGF and EGFR–EREG complexes (Fig. 5), we realized that both complexes’ interfaces included different genetic variations obtained from the COSMIC database [30]. The residue R377S mutation affects both EGFR–EREG and EGFR–HBEGF interactions. However, the D46N, Q432H, and V441I mutations were found only in the EGFR–EREG interface (see Note 5 about how to determine residue positions from sequence to structure). Consequently, one can speculate that the latter three mutations might have important roles in lung metastases initiating from breast tumors. These mutations may contribute in different ways to the stability of the complexes and thus to their signaling pathways. Some mutations can make the complex active all the time (a gain-of-function mutation), whereas others can make the interaction weaker (a loss-of-function mutation). Calculation of the effect of these mutations on the stability of the complex is needed to determine whether a mutation is likely to cause a loss- or gain-of-function.
Fig. 5.
The structures of EGFR-EREG and EGFR-HBEGF complexes modeled by PRISM
Acknowledgments
N.T. thanks to the TUBITAK-Marie Curie Co-funded Brain Circulation Scheme (114C026) and the Young Scientist Award Program of the Science Academy (Turkey) for the support. O.K and A.G. are members of the Science Academy (Turkey). We acknowledge the partial funding from TUBITAK projects (114M196 and 113E164). This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research.
Footnotes
The following are some points to be kept in mind:
Proteins often form large assemblies. Besides pairwise prediction of PPIs, discovering the organization of proteins in large assemblies is crucial to accurately model functional pathways. PRISM can to be used to build three-dimensional models of protein assemblies from predicted binary interactions. PRISM has also been successfully applied to model symmetric cyclic structures and asymmetric complexes. Electron microscopy (EM) maps have been used for the validation of constructed assemblies. In a recent study, the TIR domain signalosome in the Toll-like receptor-4 signaling pathway has been constructed with PRISM predictions [40].
If the complete structure of the targeted protein is not available, all partial structures can be added to the target set (partial crystallization).
Using multiple conformations from the PDB increases the accuracy. Rather than using a single structure, structures captured under different conditions help to increase the conformational space.
A gene product might have several PDB codes. When preparing the target set and/or constructing PPI networks, mapping from gene codes to the corresponding protein codes needs to be done (possibly using UniProt).
The positions of mutations in the protein sequence may not match the residue position in the protein structure. To solve this problem, the fasta sequence of the protein structure can be aligned with the complete protein sequence and the residue positions can be relabeled. Another problem is that if a structure includes only a fragment of a protein, a particular position might not be present in the structure at all.
References
- 1.Keskin O, Gursoy A, Ma B, Nussinov R (2008) Principles of protein–protein interactions: what are the preferred ways for proteins to interact? Chem Rev 108(4):1225–1244 [DOI] [PubMed] [Google Scholar]
- 2.Muratcioglu S, Guven-Maiorov E, Keskin O, Gursoy A (2015) Advances in template-based protein docking by utilizing interfaces towards completing structural interactome. Curr Opin Struct Biol 35:87–92 [DOI] [PubMed] [Google Scholar]
- 3.Keskin O, Nussinov R (2007) Similar binding sites and different partners: implications to shared proteins in cellular pathways. Structure 15(3):341–354 [DOI] [PubMed] [Google Scholar]
- 4.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R (1996) Protein–protein interfaces: architectures and interactions in protein–protein interfaces and in protein cores. Their similarities and differences. Crit Rev Biochem Mol Biol 31(2):127–152 [DOI] [PubMed] [Google Scholar]
- 5.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R (1996) A dataset of protein–protein interfaces generated with a sequence-order-independent comparison technique. J Mol Biol 260(4):604–620 [DOI] [PubMed] [Google Scholar]
- 6.Aloy P, Russell RB (2002) Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci U S A 99(9):5896–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosca R, Ceol A, Aloy P (2013) Interactome3D: adding structural details to protein networks. Nat Methods 10(1):47–53 [DOI] [PubMed] [Google Scholar]
- 8.Meyer MJ, Das J, Wang X, Yu H (2013) INstruct: a database of high-quality 3D structurally resolved protein interactome networks. Bioinformatics 29(12):1577–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosur R, Xu J, Bienkowska J, Berger B (2011) iWRAP: an interface threading approach with application to prediction of cancer-related protein–protein interactions. J Mol Biol 405(5):1295–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L, Lu H, Skolnick J (2002) MULTIPROSPECTOR: an algorithm for the prediction of protein–protein interactions by multimeric threading. Proteins 49(3):350–364 [DOI] [PubMed] [Google Scholar]
- 11.Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A (2005) PRISM: protein interactions by structural matching. Nucleic Acids Res 33(Web Server issue):W331–W336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuncbag N, Gursoy A, Nussinov R, Keskin O (2011) Predicting protein–protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc 6(9):1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baspinar A, Cukuroglu E, Nussinov R, Keskin O, Gursoy A (2014) PRISM: a web server and repository for prediction of protein–protein interactions and modeling their 3D complexes. Nucleic Acids Res 42(Web Server issue):W285–W289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cukuroglu E, Gursoy A, Nussinov R, Keskin O (2014) Non-redundant unique interface structures as templates for modeling protein interactions. PLoS One 9(1):e86738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keskin O, Tsai CJ, Wolfson H, Nussinov R (2004) A new, structurally nonredundant, diverse data set of protein–protein interfaces and its implications. Protein Sci 13(4):1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuncbag N, Gursoy A, Guney E, Nussinov R, Keskin O (2008) Architectures and functional coverage of protein–protein interfaces. J Mol Biol 381(3):785–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuncbag N, Gursoy A, Keskin O (2009) Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics 25(12):1513–1520 [DOI] [PubMed] [Google Scholar]
- 18.Tuncbag N, Keskin O, Gursoy A (2010) HotPoint: hot spot prediction server for protein interfaces. Nucleic Acids Res 38(Web Server issue):W402–W406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuzu G, Gursoy A, Nussinov R, Keskin O (2013) Exploiting conformational ensembles in modeling protein–protein interactions on the proteome scale. J Proteome Res 12(6):2641–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbard SJT, JM (1993) Naccess, Department of biochemistry and molecular biology University College, London. [Google Scholar]
- 21.Shatsky M, Nussinov R, Wolfson HJ (2004) A method for simultaneous alignment of multiple protein structures. Proteins 56(1):143–156 [DOI] [PubMed] [Google Scholar]
- 22.Mashiach E, Nussinov R, Wolfson HJ (2010) FiberDock: Flexible induced-fit backbone refinement in molecular docking. Proteins 78(6):1503–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiser A, Sali A (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374:461–491 [DOI] [PubMed] [Google Scholar]
- 24.Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31(13):3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pieper U et al. (2014) ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 42(Database issue):D336–D346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nussinov R, Tsai CJ (2015) ‘Latent drivers’ expand the cancer mutational landscape. Curr Opin Struct Biol 32:25–32 [DOI] [PubMed] [Google Scholar]
- 27.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A (2015) OMIM. org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43(Database issue):D789–D798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenson PD et al. (2014) The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 133(1):1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum MJ et al. (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42(Database issue):D980–D985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes SA et al. (2015) COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43(Database issue):D805–D811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerami E et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5):401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acuner Ozbabacan SE, Gursoy A, Nussinov R, Keskin O (2014) The structural pathway of interleukin 1 (IL-1) initiated signaling reveals mechanisms of oncogenic mutations and SNPs in inflammation and cancer. PLoS Comput Biol 10(2):e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engin HB, Guney E, Keskin O, Oliva B, Gursoy A (2013) Integrating structure to protein–protein interaction networks that drive metastasis to brain and lung in breast cancer. PLoS One 8(11):e81035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franceschini A et al. (2013) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41(Database issue):D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuncbag N, Kar G, Gursoy A, Keskin O, Nussinov R (2009) Towards inferring time dimensionality in protein–protein interaction networks by integrating structures: the p53 example. Mol Biosyst 5(12):1770–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bos PD et al. (2009) Genes that mediate breast cancer metastasis to the brain. Nature 459(7249):1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minn AJ et al. (2005) Genes that mediate breast cancer metastasis to lung. Nature 436(7050):518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guney E, Oliva B (2012) Exploiting protein–protein interaction networks for genome-wide disease-gene prioritization. PLoS One 7(9):e43557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Heyningen V, Yeyati PL (2004) Mechanisms of non-Mendelian inheritance in genetic disease. Human molecular genetics 13 Spec No 2:R225–233. [DOI] [PubMed] [Google Scholar]
- 40.Guven-Maiorov E et al. (2015) The architecture of the TIR domain signalosome in the toll-like receptor-4 signaling pathway. Sci Rep 5:13128. [DOI] [PMC free article] [PubMed] [Google Scholar]