Abstract
Conjugation of Nedd8 (neddylation) to Cullins (Cul) in Cul-RING E3 ligases (CRLs) stimulates ubiquitination and polyubiquitination of protein substrates. CRL is made up of two Cul-flanked arms: one consists of the substrate-binding and adaptor proteins and the other consists of E2 and Ring-box protein (Rbx). Polyubiquitin chain length and topology determine the substrate fate. Here, we ask how polyubiquitin chains are accommodated in the limited space available between the two arms and what determines the polyubiquitin linkage topology. We focus on Cul5 and Rbx1 in three states: before Cul5 neddylation (closed state), after neddylation (open state), and after deneddylation, exploiting molecular dynamics simulations and the Gaussian Network Model. We observe that regulation of substrate ubiquitination and polyubiquitination takes place through Rbx1 rotations, which are controlled by Nedd8–Rbx1 allosteric communication. Allosteric propagation proceeds from Nedd8 via Cul5 dynamic hinges and hydrogen bonds between the C-terminal domain of Cul5 (Cul5CTD) and Rbx1 (Cul5CTD residues R538/R569 and Rbx1 residue E67, or Cul5CTD E474/E478/N491 and Rbx1 K105). Importantly, at each ubiquitination step (homogeneous or heterogeneous, linear or branched), the polyubiquitin linkages fit into the distances between the two arms, and these match the inherent CRL conformational tendencies. Hinge sites may constitute drug targets.
Introduction
Covalent ligation of ubiquitin to substrates and ubiquitins is achieved via a three-enzyme cascade: activation of ubiquitin by E1 ubiquitin-activating enzymes, conjugation of ubiquitin to E2-conjugating enzymes, and finally transfer of ubiquitin by E2 bound-E3 ubiquitin ligases to the substrate [1–4]. Covalent attachment of the first ubiquitin to the substrate is followed by polyubiquitin chain elongation where the Gly76 carboxy terminus of each ubiquitin is connected to one of the seven ubiquitin Lys residues [5]. The number of ubiquitins in the polyubiquitin chain and the ubiquitination site determine the fate of the tagged substrate protein [6]. Tagging proteins with a homogeneous polyubiquitin chain of at least four ubiquitins linked via isopeptide bonds between K48 ε-amide group and G76 carboxyl end marks the protein for proteasomal degradation [7,8]. In addition to the homogeneous K48-linked ubiquitins [9], homogeneous [10–13] and heterogeneous linkages [14–16] are observed. These can be in a linear organization, where an amino group of one ubiquitin is linked to a following ubiquitin [17] or in a branched (forked) arrangement [9,16]. The number of ubiquitins and their alternating linkages provide immensely diversified polyubiquitin chain topologies [18–20]. Here, we ask how these diverse polyubiquitin chains and topologies are accommodated in the limited space between the two Cullin (Cul)-RING E3 ligase (CRL) arms. The hypothesis we set out to test is that the answer may lie in CRL conformational dynamics. We reason that dynamics can link these specific polyubiquitin chains with the energy landscape of CRL. Proteins are not rigid rocks; thus, the CRL ensemble may already include conformations that fit each of those distinct polyubiquitin topologies. Even though the population of those conformations may be low, an allosteric event is capable of enhancing them through a ‘population shift’. Below, we illustrate this to be the case, with the allosteric regulator being a covalently linked Nedd8 molecule. Post-translational modifications are known to act allosterically [21].
Based on their core domains, E3 ubiquitin ligases can be divided into two major classes: HECT (homologous to the E6-AP carboxyl terminus) and RING (really interesting new gene) [22,23]. Cul-RING ligases are the largest E3 ubiquitin ligase family. The amino terminal domain (NTD) of Culs recruits adaptor proteins that bind to specific substrate receptors (SRs) (Figure 1) [24,25]. The carboxy terminal domain (CTD) of Culs binds Ring-box proteins Rbx1 or Rbx2, which present docking sites for specific E2 enzymes. CRL is activated by conjugation of Nedd8 to Cul [26,27]. Nedd8 conjugates to a conserved Lys residue (K724 in Cul5) in CulCTD and leads to a significant conformational change in the CRLCTD (CulCTD and Rbx1) (Figure 1) [28,29]. Neddylation is a reversible process. Deneddylation of CRLs is regulated by the COP9 signalosome (or CSN) and/or DEN1 [30]. After deneddylation, CAND1, a CRL exchange factor, binds to the deneddylated CRL complexes to block the binding sites of adaptor proteins and Nedd8, and keeps CRLs in a rigidoff-state [31,32]. Inhibition of CRL catalytic activity and interaction with substrate proteins is a promising anticancer [33–40] and antiretroviral [41–43] therapeutic strategy.
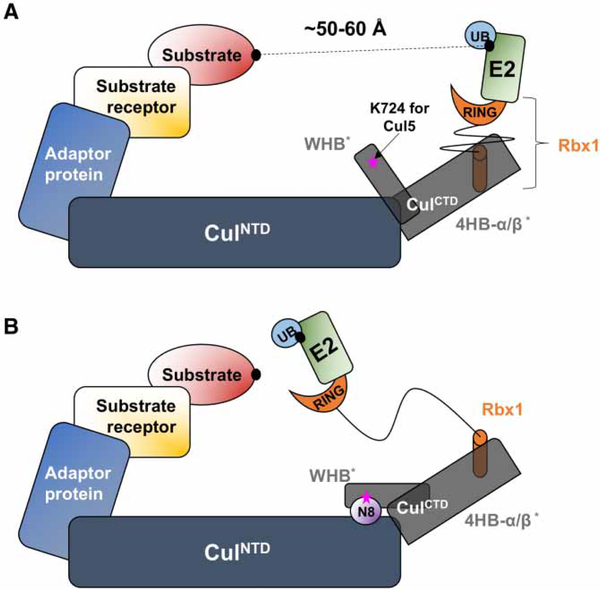
Figure 1. Schematic representation of closed (A) and open (B) state Cul-RING Ligase (CRL)s.
A sketch to present the effect of Cul neddylation in CRLs. Cul have two domains: CulNTD (dark blue) and CulCTD (dark gray) where CulCTD consists of WHB domain, 4HB, and α/β-domains (shown with asterisks). Adaptor and SR proteins bind to CulNTD. Rbx1 (orange) binds to CulCTD. Ubiquitin-carrier E2 enzyme (green) binds to Rbx1 for substrate ubiquitination. Nedd8 (N8, shown in purple) conjugates to the CulCTD H29 of the WHB domain (K724 on Cul5, shown with a pink star) and leads to significant conformational change in CulCTD. Crystal structures report the distance between E2 Cys and the substrate in the closed state as ∼50–60 Å [26,57].
Earlier studies of E3 ubiquitin ligase Rbx1 RING domains did not present the active state [26,29]. A new Rbx1 conformation showed that Rbx1 RING domain rotations are required to place Rbx1-associated E2 (UBC12) in proximity to Cul’s Nedd8-acceptor lysine for an effective ubiquitin transfer [44]. More recently, cryo-EM and biochemical studies showed that E3 anaphase-promoting complex/cyclosome, a RING E3 enzyme, adopts two distinct conformations with its two E2 partners for branched and linear polyubiquitination [45]. These findings reveal a RING domain rotation in the absence of Cul neddylation. RING domain rotations and polyubiquitin topologies in the neddylated state are still elusive. Although allostery has been suggested as the mechanism for CRL function [46–49], how it relates to RING domain rotations and how this rotation contributes to polyubiquitination in CRLs have not been resolved.
To obtain a mechanistic view of Cul neddylation and polyubiquitination, we performed molecular dynamics (MD) simulations combined with Gaussian Network Model (GNM) analysis [50,51] on the crystal structures of Cul5CTD–Rbx1 (closed state), Cul5CTD–Rbx1–Nedd8 (open state) complexes, and the complete closed-state Cul5 model. We observe abrogation of rigid CRL dynamics and capture multiple Rbx1 RING domain confor-mations/ configurations emerging via alternating RING domain rotations in the neddylated Cul state. Intrinsic flexible segments in Cul5 are the main elements of strongly coupled dynamics between Nedd8 and Rbx1. Nedd8 dynamizes the Cul5–Rbx1 complex while allosterically controlling Rbx1 RING domain conformations through a dynamic hinge plane mechanism. We find that distinct Rbx1 RING domain rotations together with the flexible Cul dynamics can provide the basis for efficient ubiquitin transfer during polyubiquitination in neddylated CRLs. This not only allows varied distances and paths between the ubiquitin on the substrate and E2 to be overcome, but also provides additional conformational space for the growing polyubiquitin chain. In closed-state CRLs, the gap adjustment between ubiquitin on the substrate and E2 is predominantly provided by Cul dynamics. In particular, the dynamic structure of CRL complexes that we observe suggests (1) how RING domain conformations are controlled and regulated by Nedd8 for effective ubiquitination/polyubiquitination;(2) the role of alternating RING domain rotations during ubiquitination/polyubiquitination in the neddylated state; and in particular, (3) coupling between Nedd8-controlled CRL functional dynamic states and polyubiqui-tin chain topologies.
Materials and methods
Structures and system set-up
CRLs include a Cul bound to Rbx RING protein. Unmodified Cul–Rbx RING E3 ligase is referred to as closed-state CRL, whereas Nedd8-conjugated Cul–Rbx RING E3 ligase as open state. In the present study, we use Cul5CTD bound to Rbx1 RING protein. Cul5CTD–Rbx1 RING E3 ligase (CRLCTD) crystal structures in the closed and open states are extracted from the Protein Data Bank (PDB ID: 3DPL for the closed and 3DQV chains A, C and R for the open CRL state). The pseudo-deneddylated state CRL is obtained by removal of the Nedd8 protein from the open state CRL (3DQV chain C and R only). Missing residues in Rbx1 in the closed state (T64-E66) and in Cul5 in the open and deneddylated state (N516-K518) are homology-modeled using MODELLER [52].
The complete closed state Cul5 model is built by MODELLER with the crystal structures of human Cul5NTD (PDB code: 4JGH, chain: D) and human Cul5CTD (PDB code: 3DPL, chain: C) by using the complete Cul1 (PDB code: 1U6G, chain: A) as a template.
CRLCTD crystal structures and the complete closed state Cul5 model are protonated at 310 K and the system pH is kept at 7 by the Molecular Operating Environment (MOE). The protonation states of the histidine residues are then modified in the PDB files. The final structures are placed in a TIP3P rectangular water box with 10 Å padding and neutralized with Cl− and Na+ ions. Subsequently, the PSFs (protein structure files) of the prepared PDBs are generated by VMD 1.9.2 [53] PSF-Builder plug-in to be used in the MD simulations.
CRL models set-up
The closed/open state CRL models are built based on multiple superpositions: (1) the 4-helix bundle (4HB) and α/β-domains of the closed/open states of MD-sampled Cul5CTDs and the 4HB and α/β-domains from the complete closed state of the Cul5 model; (2) the RING domain of E2 UbcH7 (PDB code: 1FBV chain: A) with MD-sampled closed/open state Rbx1 RING domain; (3) Cul1NTD (PDB code: 1LDK chain: A) with the NTD from the complete closed state of the Cul5 model; and (4) Skp1 of β-TrCP1–Skp1–β-catenin complex (PDB code: 1P22 chain: B) with the F-box of the Cul1–Rbx1–Skp1–Fbox Skp2 complex (PDB code: 1LDK chain: D).
Simulation protocol
Explicit MD simulations are performed with the CHARMM 27 force field with CMAP correction [54,55] with the NAMD program [56]. Initially, the prepared CRLCTD crystal structures are energetically minimized for 20 000 steps by MOE. For the closed state and the pseudo-deneddylated state, two parallel MD simulation trajectories, for the open state three parallel MD simulation trajectories, and for the complete closed Cul5 model single MD simulation trajectory are performed with a 2 fs integration time step within the NPT ensemble. Langevin dynamics is used to keep the temperature and pressure constant at 310 K and 101.3 kPa, respectively. Periodic boundary conditions are applied. All covalent bonds involving hydrogen atoms are kept at their equilibrium distances by the SHAKE constraint. During the simulations, particle mesh Ewald summation is used to treat the long range electrostatic interactions and the distances between the Zn atoms in the Rbx1 RING domain Zn fingers. VMD 1.9.2 [53] is used for structural alignments. MD simulation time details are given in Supplementary Table S1.
Analysis of MD simulations and structures
The details about the MD simulation analysis (RMSD, MSF, cross-correlations between residue fluctuations; Supplementary Figure S1 and Tables S1 and S2) and the identification of the main conformers and their analysis by the GNM [50,51] are given in Supplementary Text File 1.
Results and discussion
X-ray crystal structures of the Cul5CTD–Rbx1 (CRLCTD) complex describe the closed and open states. The compact structure of CulCTD WHB (winged-helix B) domain with the Rbx1 RING domain is the unmodified closed state, whereas the open state is the modified, neddylated state obtained upon Nedd8 conjugation to CulCTD (Figure 1). We also looked at a third pseudo-state from the open conformation where Nedd8 is removed to observe the deneddylated state of the CRLCTD machinery (Supplementary Text File 2).
Below, we first introduce the closed and open state CRLCTD conformations, which relate to the unneddylated and neddylated Cul states. MD simulations and GNM analysis reveal that Nedd8 controls RING domain conformations. We introduce the hinge residues coordinating the allosteric communication between Nedd8 and Rbx1 RING domains. Next, we describe distinct Rbx1 RING domain positions based on the dynamic hinge coordination in CRLs. Finally, we model complete CRLs, where the substrate is on one CRL arm and ubiquitin-bound E2 on the other, to measure distances between the substrate and E2. This aims to understand the role of Cul flexibility and Rbx RING domain rotations (collectively referred to as neddylated CRL functional dynamic states). We conclude by proposing that polyubiquitin linkage topologies may be coupled to the neddylated CRL functional dynamic states that are modulated by the hinge residues.
Rbx1 RING domain rotations provide multiple conformations irrespective of Cul’s neddylation state
Crystallography reports that the initial gap between the substrate-carrying arm and the E2 catalytic Cys on the other arm is ∼50–60 Å in the closed state CRL (Figure 1) [26,57]. The gap decreases in the open state as Rbx1 gains flexibility after Cul neddylation [58]. Rbx1 is not the only flexible CRL component adjusting the target distances. The role of flexibility of Cul [59], the substrate-binding protein–substrate complexes [60,61], and even ubiquitin [62], has previously been elucidated in shortening the distance between the arms. In particular, for Rbx1, flexibility is exerted via RING domain rotations [44,45]. A recent study has discovered an intermediate Rbx1 RING E3 conformation during the Nedd8 transfer from Ubc12 to Cul1 acceptor Lys [44]. This structure has demonstrated that RING domain rotations are inherent, present before Nedd8 transfer to Cul. They were observed when the Nedd8–E2 complex binds to RING E3 and positions the RING-E2-Nedd8 catalytic center close to the Cul acceptor Lys. Although Rbx1 RING domain rotations were suggested as a general mechanism for ubiquitin transfer before Cul neddylation, the mechanism following CulCTD neddylation has been elusive.
We present Rbx1 RING domain conformations with rotations about the Rbx1 N-terminal β-strand (K20-W35) in the closed and open CRLCTD complexes. MD simulations have shown that Rbx1 W35, with a pivot role before Cul neddylation [44], regulates RING domain rotations after Cul5CTD neddylation, resulting in conformers with distinct rotations (Figure 2A–C). The first two open state CRLCTD conformers (Figure 2A, B) show strong Nedd8–Rbx1 RING domain coupling (Supplementary Figure S2A,B), indicating that Rbx1 H1 and H2 helices are almost vertical to the Cul5CTD WHB domain. The Rbx1 RING domain rotation mechanism exposes Rbx1-binding site residues (A43, C45, I54, Q57, C83, R86, and P95) to Nedd8-specific E2 enzyme-UBE2M or UBE2F [63] (Supplementary Figure S3). Such arrangement reflects the H-bonding between E67 (Rbx1 RING domain)-R569 (Figure 2A) or R538 (Figure 2B) of the Cul5CTD α/β-domain. On the other hand, the Rbx1 RING domain rotation in the open state with lower Nedd8–Rbx1 RING domain coupling (Supplementary Figure S2C) placed the RING domain horizontal to the Cul5CTD WHB domain (Figure 2C). Three open state conformations (Figure 2A–C) may correspond to Rbx1 conformations in ubiquitination/polyubiquitination stages. The initiation of substrate ubiquitination can be achieved via a RING domain rotation that favors closure of the gap between the E2 active Cys and the substrate. However, the Rbx1 RING domain may adopt different rotations for further ubiquitin transfers to accommodate the growing polyubiquitin chain. The preferred Rbx1 states are expected to change with the growing polyubiquitin chain and this may affect polyubiquitin chain topology.
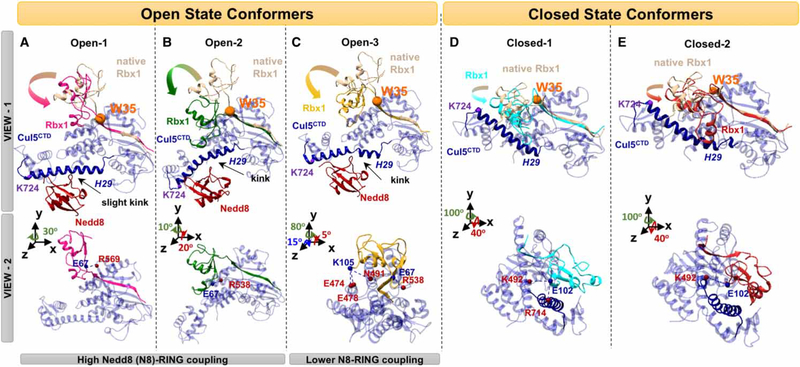
Figure 2. RBX RING domain rotations.
(A–C) Nedd8 (red) conjugated CRLCTD (open state) conformers that have three distinct Rbx1 RING domain rotations, and (D and E) unmodified CRLCTD (closed state) conformers are shown in two different views. The H29 of Cul (blue) are highlighted by making the rest of Cul elements transparent. Distinct Rbx1 conformations are shown with different colors (pink, green, and yellow for open; cyan and red for closed). Rotations from native — crystal structure — Rbx1 (beige) are highlighted with arrows. The Rbx1 W35 residue shown with an orange sphere has a pivotal role in RING domain rotations after (A–C) and before (D and E) Cul neddylation. Nedd8-binding site on CulCTD, K724, is shown with a purple sphere in (D and E). In view 2, the H-bonds between Cul residues (red spheres) and Rbx1 (blue spheres) stabilizing the Rbx1 RING domain conformation are shown for both states.
The dynamics of the closed state complex also suggest that the RING domain rotates (Figure 2D,E), however differing from the open state (Figure 2A–C). In the closed (before Cul neddylation) state, we observed a rigid body rotation in the Rbx1 RING domain around Rbx1 W35 (Figure 2D,E). The rotations shifted the Rbx1 RING domain closer to K724, which is the Nedd8-acceptor residue in the helix 29 (H29), while preserving the Rbx1 helix configuration with respect to Cul5CTD. Such rotation reflects the H-bonding between E102 (Rbx1 RING domain)-K492 of the Cul5CTD 4HB domain and R714 of the Cul5CTD H29 (Figure 2D,E).
A challenging question is how RING domain rotations are dynamically established and fine-tuned upon Cul neddylation. The answer may lie in the arrangement of CRL hinge residues after neddylation. Our results indicate that Rbx1 and Nedd8 are dynamically connected by neddylated Cul. The control of Rbx1 motions, leading to the RING domain rotations, positions the domain such that it is poised for effective ubiquitination.
Nedd8 controls RING domain conformations via Cul5 flexible elements
Nedd8 conjugation to Cul needs to be communicated to Rbx1. Fluctuation dynamics of the Cul5CTD –Rbx1–Nedd8 complex suggests strong couplings between Nedd8 and Rbx1 V38-C42 and the entire RING domain (C42-K105; Supplementary Figure S2D). These indicate the cooperativity of Cul5CTD with Rbx1 and Nedd8 through the following Cul5CTD segments: V477-M496 in the 4HB domain, N532-S539 and H566-K570 in the α/β-domain and E703-G706 in the WHB domain (Supplementary Figure S2D). These Cul5CTD segments lie in-between hinge residues where the dynamic domain continuity within the Cul5CTD is broken. These segments are able to dynamically associate with Rbx1 and Nedd8. We call these fragments Cul5CTD flexible elements. Similarly, fluctuation dynamics of the closed state complex display strong couplings between Cul5CTD and Rbx1, where the following Cul5CTD flexible segments are in play: P405-K501 in the 4HB domain, L531-R538, I555-N579, S615-L634, and E651-L665 in the α/β-domain, and R691-R714 in the WHB domain (Supplementary Figure S2E).
We analyzed the global dynamics of CRLCTD of the MD-sampled closed and open states by the GNM that delineate the hinges of flexible segments (Supplementary Text File 1). The slowest GNM mode reveals hinge residues at the Cul5CTD linker between the α/β and WHB domains that mainly contribute to the most global conformational changes in the neddylated Cul, while the second slowest mode discloses unique hinge residues in allosteric communication between Cul5CTD, Rbx1 and Nedd8 (Figure 3A,B and Supplementary Table S3). The open state Cul5CTD hinges that define the Cul5 flexible elements and contribute to the Nedd8–Cul5–Rbx1 allosteric communication are R495, F497, L506 of the 4HB domain, I555, L571 of the α/β-domain and E701, N704, E705, I707 of H29 in the WHB domain (Figure 3A). Similarly, closed state hinge residues plausible for Cul5–Rbx1 allosteric communication are I500, V502-E504 of the 4HB domain, I528, K529, S540, E541, V543, D553, P556, Y562, H574, T696 of the α/β-domain and at I712, L713, R714, L742, F746 of the WHB domain (Figure 3B). The similarity between hinge residues in the closed and open states, particularly in the Cul5CTD α/β and 4HB domain, reveals the key role of global dynamics in the regulation of CRLCTD. The intrinsic Cul5CTD hinge behavior can be further observed by analyzing crystal structures in both states (Supplementary Table S4). Closed and open Cul5CTDs expose similar hinge residues alone or in complex with Rbx1 and Nedd8 (Supplementary Table S4).
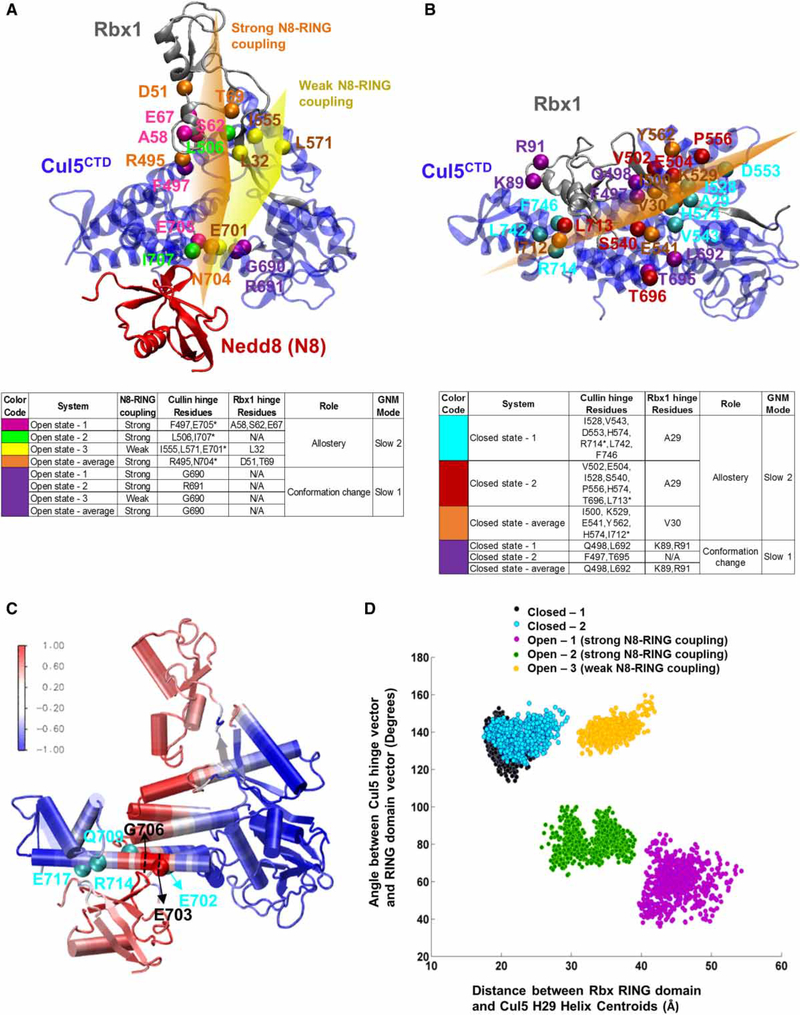
Figure 3. Hinge sites and coupled Cul regions on CRLCTD provide a dynamic measure for RING domain rotation.
Open-1 and closed-1 CRLCTD conformers are shown in (A and C) and (B), respectively, to illustrate the hinge sites. The open and closed state CRLCTD (Cul5CTD–Rbx1) hinge residues of the slowest mode of motion during Cul neddylation are shown in purple. Hinge residues coordinating the allosteric communication are colored differently for each conformer. Open state-average and closed state-average yield the average conformers for both states. Side tables are provided for guidelines. The Cul5 H29 hinge residues are marked with asterisk in the side tables. Approximate imaginary planes are drawn for illustration of hinge coordination. In (A), the orange and yellow hinge planes represent the hinge coordination for Nedd8–Cul5–Rbx1 allosteric communication when Nedd8–RING coupling is high and weak, respectively. In (B), the orange plane represents the hinge coordination for Cul5–Rbx1 allosteric communication in closed state conformers. (C) Open state CRLCTD conformer is colored according to the Cul5 flexible element showing highest correlation with Nedd8, Glu703–Gly706, to highlight the allosteric communication pathway from Nedd8 through CRLCTD. The normalized correlation values are in the range of [−1,1]. The Cul5 residues that H-bond with Nedd8 are shown in cyan. (D) Dynamic RING domain positions throughout each MD simulation are depicted; x-axis: distance between the RING domain (Cys42–Lys105) centroid and the Nedd8-binding Cul5 WHB domain’s H29 (Trp695–Met725) centroid, y-axis: angle between the state-specific hinge vector and the RING domain vector from Rbx1 Cys42 to the RING domain centroid.
Nedd8 conjugates to the Cul5CTD H29, where flexible elements display coupled dynamics with most flexible elements in the closed and open states, providing an allosteric path (Figure 3C and Supplementary Figure S2D). Upon neddylation, the H29 hinge (I712-R714) shifts toward the N-terminus of the helix closer to the α/ β-domain (E701-I707; Figure 3A,B). In the open state, a kink (toward Nedd8) forms in H29 (Figure 2A–C). A kink at E686-Q693 of the H29 in Cul4A disturbs the CulCTD WHB and Rbx1 RING domains [64]. These residues correspond to E705-R711 in Cul5CTD. Shifts in hinge positions together with kink formations point to the extended flexible nature of Cul5CTD H29. Nedd8 covalently attaches to the conserved K724 of the H29 and also interacts with the H29 hinge residue region (specifically G706, Q709, L710, L713, and R714). Furthermore, Nedd8 H-bonds with E702, Q709, R714, and E717 within the CRLCTD WHB domain (Figure 3C) and forms non-bonded contacts with the highly conserved second half of the H29 toward the CTD. The RING domain rotations place the Rbx1 Q57–E67 loop as the closest Rbx1 fragment to the H29 in both closed and open states; however, in the open state, the loop is stabilized by neddylation and displays high correlations with the coupled Cul5CTD flexible elements (Supplementary Figure S2D).
The cooperativity among the Cul5CTD flexible elements either with each other or with Rbx1 RING (in both closed and open states) and with Nedd8 (open state) suggests that Nedd8 conjugation to Cul5CTD H29 reorganizes CRL. Nedd8 fine-tunes and maintains the conformation and motion of Rbx1 for selective positioning with respect to Cul5CTD for effective ubiquitin transfer and polyubiquitination. Here, we reveal strong allosteric dynamics between Nedd8 and Rbx1 RING domains that adjust and control Rbx1 RING domain conformations during polyubiquitination.
Dynamic hinge coordination through the flexible CulCTD underlies Rbx1 RING domain motions
In the allosteric communication of the closed state, the coordination of the Cul5CTD and Rbx1 hinges divides the structure into two dynamic domains with an imaginary hinge plane passing through the Cul5CTD WHB domain H29 hinge (I712-R714 on the second half of the helix) up to the Cul5CTD α/β-domain-Rbx1 N-terminal β-strand K20-W35 (Figure 3B). In the closed state conformers, the coordination of the hinges does not change dramatically, showing the restrictive dynamics of CRLs before Cul neddylation (Figure 3B). H-bond formations between the Rbx1 E102 with K492 of the Cul5CTD 4HB domain and R714 of the Cul5CTD H29 (Figure 2D,E) accompany the coupled dynamics among the Cul5CTD flexible elements (Supplementary Figure S2E) along this imaginary hinge plane (Figure 3B, cyan, red, and orange hinges). Such coordination leads the Rbx1 RING domain to stay parallel to the Cul5CTD WHB domain, but shifts toward Nedd8-acceptor residue K724 in the H29 (Figure 2D,E).
On the other hand, a different arrangement of the Cul5CTD and Rbx1 hinges elicits allostery in the open state leading to different Rbx1 RING domain rotations (Figure 3A). Within the open state conformers, the hinge plane coordination varies with the strength of Nedd8–Rbx1 coupling (Figure 3A). In the strong Nedd8–Rbx1 coupling (Supplementary Figure S2A,B), the Cul5CTD hinges on the H29 shift toward the Cul5CTD WHB domain (N704, E705, and I707), close to the middle of the helix, and additional hinges appear in the Cul5CTD 4HB domain (R495, F497, and L506) and in the Rbx1 RING domain (D51, A58-S62, E67, and T69) upon Cul neddylation (Figure 3A, pink, green, and orange hinges). H-bond formations between the Rbx1 E67 — either with R569 or with R538 of the Cul5CTD α/β-domain — accompany the coupled dynamics among the Cul5CTD flexible elements along this imaginary hinge plane (Figures 2A,B and 3A). These H-bonds place the Rbx1 RING domain helices vertical to the Cul5CTD WHB domain (Figure 2A,B) and support a strong allosteric communication between Nedd8 and Rbx1 RING domains as a whole (Supplementary Figure S2A,B).
On the other hand, in the weaker Nedd8–Rbx1 coupling compared with the other conformers (Supplementary Figure S2C), the coordination among the Cul5CTD and Rbx1 hinges deviates (Figure 3A, yellow hinges). The H29 hinge position shifts to E701. The hinges align along a plane from the H29 hinge (E701) through the Cul5CTD α/β-domain hinges (I555, L571) and the Rbx1 RING domain hinge (L32). In addition to the H-bonding of Rbx1 E67 and R538 (Cul5CTD α/β-domain) as observed in the other conformers, H-bond formation between the Rbx1 RING and the Cul5CTD 4HB domains (Rbx1 K105-Cul5CTD E474, E478, and N491) is observed (Figure 2C — view 2). H-bond formation between the Rbx1 RING and the Cul5CTD 4HB domains is a typical closed state behavior; we thus suggest that the additional H-bonds do not allow RING domain rotation as in other conformers and subsequently leads the RING domain to adopt a conformation similar to the one in the closed state (Figure 2D,E). In this state of open CRLCTD, Nedd8 is coupled with Rbx1 A29-C53 (from the second half of the RING domain N-terminus to the Rbx1 H1 helix) and S65-K89 (which includes the linker between the Rbx1 H1 and H2 helices, and the H2 helix) regions instead of the RING domain as a whole (Supplementary Figure S2C).
Consequently, we deduce that, depending on the Cul5CTD hinge coordination in the CRLCTD complex, the strength of Nedd8–Rbx1 cooperativity and accordingly the nature of Nedd8 allosteric conformational control of Rbx1 RING domain varies.
Dynamic description of Rbx1 RING domain positioning based on hinge coordination in CRLs
We consider a metric to reflect the RING domain conformation positions observed in the closed and open state conformers (Figure 3D): the distance between the Rbx1 RING domain (Rbx1 C42-K105) center of mass (centroid) and the centroid of the Cul5CTD conformer WHB domain H29 (Trp695–Met725) (x-axis) versus the angle between the Cul5 hinge vector and the RING domain vector (tying the Rbx1 C42 and the Rbx1 RING domain centroids) (y-axis). The Cul5 hinge vector from each conformer (for y-axis) is state-specific. It connects the centroids of Cul5CTD 4HB-α/β and WHB domain (H29) hinges (F497 with E705 and L506 with I707 for the open state conformers where Nedd8–Rbx1 RING domain coupling is high, and I555 with E701 for the open state conformer with lower Nedd8–Rbx1 RING domain coupling; D553 with R714 and P556 with L713 for the closed state conformers). The hinge vector reflects the direction of the imaginary hinge plane passing through the CRLCTD complex. These measurements reflect two points: (1) the distances indicate the proximity of the Rbx1 RING domain to the Cul5CTD H29 and (2) the angle between the RING domain vector and the hinge plane classifies the RING domain positions by rotations and identifies distinct arrangements of Rbx1 helices with respect to the Cul5CTD. The most significant classification of the positions of the RING domain is achieved by the CRLCTD state-specific hinge plane information. This classifier segregated the open CRLCTD conformers into two classes: those in which strong Nedd8–Rbx1-coupled dynamics are at play in distinct RING domain configurations with respect to Cul5CTD and those with weaker Nedd8–Rbx1 coupling along different RING domain rotations (Figure 3D).
To understand the functional effect of these RING domain rotations within the open-state CRLs during ubiquitination/polyubiquitination, we constructed complete CRL models and measured the distances between the substrate located on one CRL arm and the E2 on the other. To discover the putative Cul flexibility contribution in CRLs during ubiquitination/polyubiquitination in the open state, we performed a single MD simulation with the complete closed state Cul5 model to include a set of flexible Cul5 conformations in the distance measurements.
The critical role of large hinge-bending conformational changes in Cul
The MD simulation of the complete closed state Cul5 model demonstrates large conformational changes via contraction motion in the NTD and CTD, indicating flexibility in the elongated CRL models (Figure 4A). Of particular interest, the Cul5CTD H29 hinge region, where Nedd8 binds Cul5CTD, is highly coupled with the Cul5NTD center, Cul5CTD 4HB, and WHB domains (Figure 4B,C and Supplementary Figure S4). Not surpris-ingly, the hinges observed in the CTD of the complete Cul5 comprise the Cul5CTD hinges observed in the dynamics of the closed/open-state CRLCTD (Supplementary Table S5, GNM second slowest mode). We thus speculate that Nedd8 attachment onto the Cul5CTD not only induces conformational change in the CTD, but also in the NTD. Hence, it would be anticipated to observe a change in the configuration at the NTD–CTD junction upon neddylation.
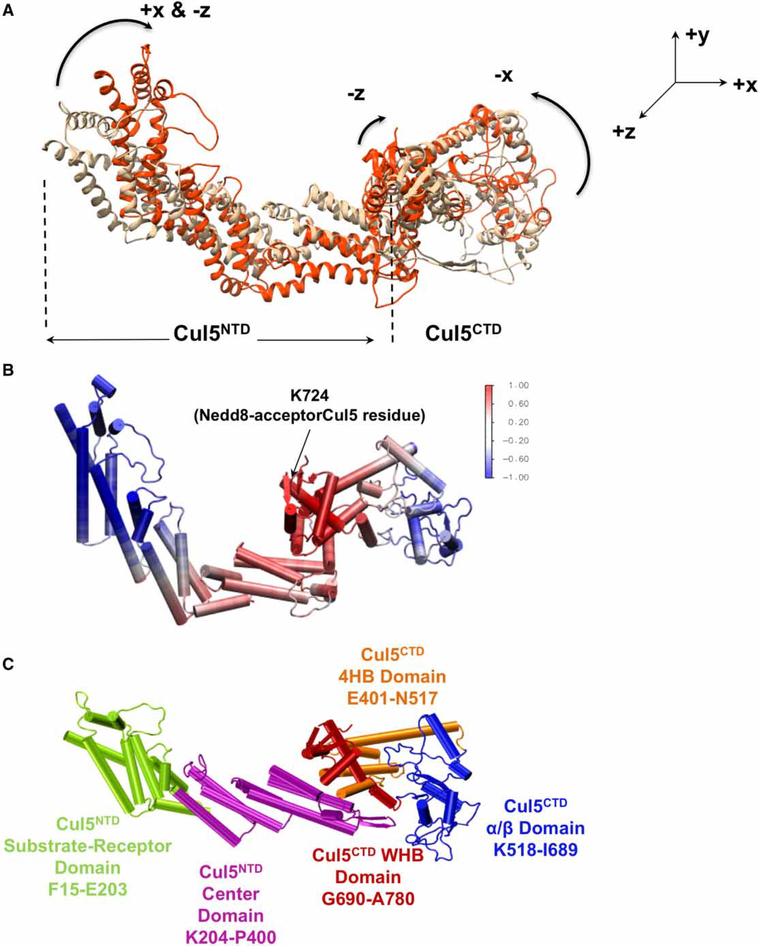
Figure 4. Cul coupled dynamics and flexibility.
(A) Contraction motions observed in complete closed state Cul5 MD simulation are shown by aligning the most contracted Cul conformation (Frame 547 — used in Figure 5; orange) on the initial complete Cul5 model (beige). (B) Cul5 conformer is colored according to the Cul5 correlations with Nedd8-acceptor Cul5 residue K724. Color code is done based on the normalized correlation values in the range of [−1,1], where −1 reflects highest negative correlation and 1 shows the highest positive correlations in fluctuation dynamics during the MD simulation. (C) The Cul5 domains (including NTD dynamic domains determined from the network of correlated motions) are labeled.
The simultaneous role of Cul flexibility and RING domain rotation in polyubiquitination in CRLs
To elucidate the effects of RING domain rotations and Cul5 flexibility on ubiquitination/polyubiquitination in CRLs, we modeled complete CRLs with diverse sets of Cul5 NTD and Cul5CTD–Rbx1 RING (CRLCTD) domain conformations. We used six distinct Cul5 NTDs from complete closed-state Cul5 MD simulation in combination with closed and open state CRLCTD MD conformations. We measured the distances between the tip of the substrate (PDB: 1P22 chain: C residue: T40) held by the CRLNTD and the active Cys of E2 (PDB: 1FBV chain: C residue: 1086) held by the CRLCTD arms. The measured distances are a function of RING domain rotations (x-axis) and Cul5 flexibility (y-axis) (Figure 5A–C). Of note, we do not know the relationship between Cul flexibility and the mechanism of the RING domain rotation, nor their explicit order. This representation enables us to probe whether the distance decrease between the CRL arms during the ubiquitination/polyubiquitination stages is dependent solely on the RING domain rotations, Cul flexibility or both. Not surprisingly, Cul contraction motions have decreased the gap between the arms of the CRL in both closed and open state CRLs. In contrast, the RING domain rotations show diverse effects on the gap between the CRL arms in the open state.
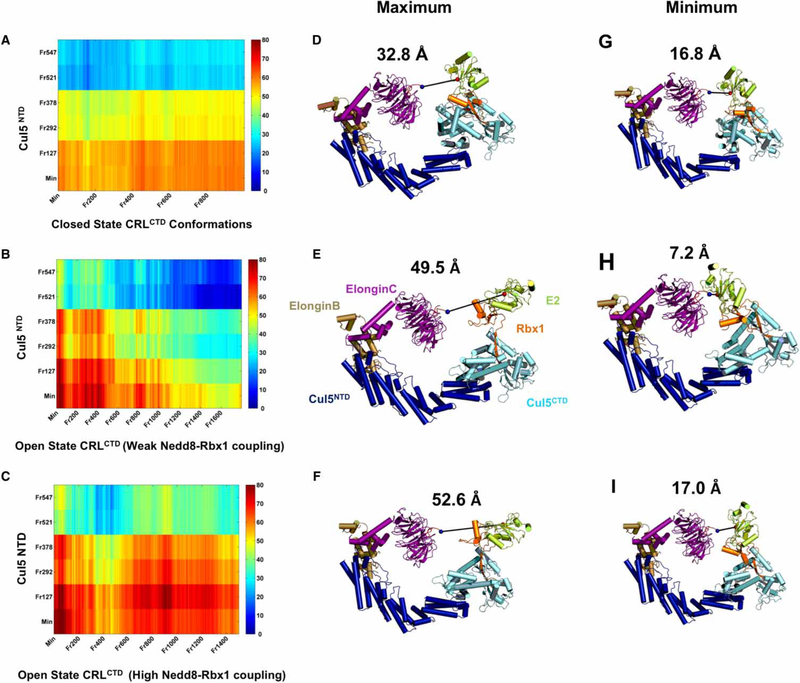
Figure 5. Distance measurements between the CRL arms.
(A–C) show the distance maps for closed (A), and open (B and C) state CRLs. CRLCTD conformations (frames from MD simulations; x-axes) are modeled with the NTD of selected Cul5 conformations (frames from the complete closed state Cul5 MD simulation) (y-axes) to obtain complete CRL models and measure the distance between the substrate tip and active E2 Cys on CRLs. The color bar provides the measured distances in Ångstrom. (D–F) show the CRL models with the most contracted Cul5 conformation (Frame 547) having maximum distance between the arms according to the distance maps shown in (A–C), whereas (G–I) show the minimum. (D and G), (E and H), and (F and I) comparisons highlight the effect of RING domain rotations on the distance reduction between the CRL arms in each CRL functional state.
The open state conformer with lower Nedd8–Rbx1 cooperativity — compared with the other two conformers — whose Rbx1 is rotated such that the RING domain lies parallel to the Cul5CTD WHB domain, contributed the most to the reduction in the gap between the CRL arms (Figure 5B). In this open state, with the most contracted Cul5 conformation, the RING domain rotations dramatically reduce the distance between the arms of the CRLs from a maximum of 49.5 Å to a minimum of 7.2 Å (Figure 5E,H). We speculate that this RING domain position is favored for the initial ubiquitin transfer alone or in coordination with another RING-between-RING E3 ligase ARIH [65]. In two other open state conformers, we observe higher cooperativity between Nedd8 and Rbx1. There Nedd8 restricts the RING domain, which restrains the distance between the CRL arms (Figure 5C and Supplementary Table S6) from a maximum of 52.6 Å to a minimum of 17 Å (Figure 5F,I and Supplementary Table S6). The contracted Cul conformations are compatible with reduction in the gap. The coupled dynamics results in CRLCTD conformations in which the position of the RING domain may be in line with certain polyubiquitination topologies, which would prevent hindrance of the growing polyubiquitin chain (Figure 6). This points to different Rbx1 configurations with respect to the Cul5CTD WHB domain between the first ubiquitin transfer and subsequent polyubiquitination events (homogeneous or heterogeneous). Taken together, polyubiquitin chain topologies in different CRLs may reflect different Rbx RING domain rotations, with Rbx flexibility stimulated by Nedd8’s allosteric action. Therefore, we hypothesize that polyubiquitin linkage topologies may be coupled to neddylated CRL functional dynamic states that are modulated by hinges.
Figure 6. Schematic representation for the role of neddylated CRL dynamics during polyubiquitination.
Flexibility in Cul (A) and diverse Rbx1 conformations due to diverse RING domain rotations with varying level of Nedd8 allosteric control (B) are the key determinants of neddylated CRL dynamics. Heterogeneous (C and D) and homogeneous (E and F) polyubiquitin chains on the substrates held by neddylated CRLs are depicted with different neddylated CRL conformational tendencies (different combinations of Cul flexibility and RING domain rotations). The ubiquitin of the polyubiquitin chain which is going to be ubiquitinated and the ubiquitin on the E2 which is going to be transferred are shown by red in (C–F). It is proposed that neddylated CRL conformational tendencies may adapt/effect different polyubiquitin chain topologies.
Comparison of the closed and open states suggests that the closed state Rbx1 does not have sufficient motional degrees of freedom. In the distance reduction between the closed state CRL arms, Rbx1 RING domain rotations do not play a major role. The gap reduction from a maximum of 32.8 Å to a minimum of 16.8 Å is achieved only via Cul5 contraction motions (Figure 5A). This shows that the open state places the E2-ubiquitin complex in proximity to the substrate for polyubiquitination, whereas the RING domain rotations in the closed state play a major role in bringing the active E2 Cys closer to the Nedd8-acceptor K724 on Cul5 (Figure 2D,E). These results indicate that the flexibility of the Cul and presumably the substrate and substrate-binding protein are the major, or the only factors facilitating the reduction in the distance between the arms in the closed state. Apparently, the closed state Rbx1 does not have an active role in the ubiquitination when compared with the open state. Thus, facilitating the initiation of ubiquitination is not sufficient for substrate tagging for degradation; coordinating the polyubiquitin chain elongation with RING domain rotations is also essential. Low Rbx1 flexibility may impede the coordination chore. At this stage, Nedd8 serves as a catalyst for further activation of Rbx1 and controls Rbx1 configurations for efficient regulation of polyubiquitination. This may be the reason why closed state CRLs are in the off or inactive state.
Conclusions
Here, we reveal the effect of Nedd8 on CRLs, specifically Rbx1 RING domain conformations/configurations. We hypothesize that because these involve changes in the distances between the two CRL arms, they may also relate to polyubiquitin chain topologies. Our results show that selective Rbx1 RING domain conformations/configurations are maintained by Nedd8 allosteric control via dynamically connected Cul5 hinges. With limited Rbx1 motional degrees of freedom in the absence of a Nedd8 action, the closed state may not satisfy the required dynamics for polyubiquitination; in contrast, in the open state, Nedd8 dynamizes the CRL complex. The Nedd8–Rbx1 coupled dynamics promote efficient ubiquitin transfer in polyubiquitination through distinct Rbx1 RING domain rotations. Importantly, not only the Rbx1 RING domain, but also Cul5 flexibility, plays a significant role in controlling the distance between the CRL arms. Taken together, we hypothesize that there may be coupling between neddylated CRL functional dynamic states that are differentially modulated by hinges and polyubiquitin linkage topologies. The cooperativity of the hinge positions and allosteric interactions suggest that the open state Cul5 hinges (Figure 3A) can serve as drug targets in cancer and in retroviral therapeutics. Allosteric control of interactions via hinge sites can be tested by biochemical, crystallographic, and single-molecule studies of the mutant and wild-type Cul5.
Supplementary Material
Acknowledgments
Funding
This research was funded by Bogazici University (BAP 6512). T.H. acknowledges Turkish State Planning Organization grant 2009K120520 and Betil Fund. This project has also been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. It was also supported by research grants from the Hungarian National Science Foundation (OTKA K83314) and by the EU (TÁMOP-4.2.2/B-10/1-2010-0013). This research was also supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Abbreviations
- 4HB
4-helix bundle
- H29
helix 29
- CAND1
Cullin-associated NEDD8-dissociated protein 1
- CMAP correction
effect of phi, psi cross-term map correction
- CRLs
Cullin-RING E3 ligases
- CSN
COP9 signalosome
- CTD
carboxy terminal domain
- Cul5
Cullin 5
- Den1
deneddylase 1
- EM
electron microscopy
- GNM
Gaussian Network Model
- HECT
homologous to the E6-AP carboxyl terminus
- MD
molecular dynamics
- MOE
Molecular Operating Environment
- MSF
mean square fluctuation
- NTD
amino terminal domain
- PSFs
protein structure files
- Rbx
Ring-box protein
- RING
really interesting new gene
- RMSD
root mean square deviation
- SRs
substrate receptors
- UBE2F
Nedd8-conjugating enzyme Ube2f
- UBE2M
Nedd8-conjugating enzyme Ubc12
- WHB
winged-helix B
Footnotes
Competing Interests
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Streich FC Jr and Lima CD (2014) Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 43, 357–379 doi: 10.1146/annurev-biophys-051013-022958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershko A (2005) The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 12, 1191–1197 doi: 10.1038/sj.cdd.4401702 [DOI] [PubMed] [Google Scholar]
- 3.Komander D and Rape M (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 doi: 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 4.Buetow L and Huang DT (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642 doi: 10.1038/nrm.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciechanover A and Stanhill A (2014) The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta, Mol. Cell Res. 1843, 86–96 doi: 10.1016/j.bbamcr.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 6.Sadowski M and Sarcevic B (2010) Mechanisms of mono- and poly-ubiquitination: ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine. Cell Div. 5, 19 doi: 10.1186/1747-1028-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK et al. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 doi: 10.1126/science.2538923 [DOI] [PubMed] [Google Scholar]
- 8.Thrower JS, Hoffman L, Rechsteiner M and Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 doi: 10.1093/emboj/19.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kravtsova-Ivantsiv Y, Sommer T and Ciechanover A (2013) The lysine48-based polyubiquitin chain proteasomal signal: not a single child anymore. Angew. Chem. Int. Ed. Engl. 52, 192–198 doi: 10.1002/anie.201205656 [DOI] [PubMed] [Google Scholar]
- 10.Jin L, Williamson A, Banerjee S, Philipp I and Rape M (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 doi: 10.1016/j.cell.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bedford L, Layfield R, Mayer RJ, Peng J and Xu P (2011) Diverse polyubiquitin chains accumulate following 26S proteasomal dysfunction in mammalian neurones. Neurosci. Lett. 491, 44–47 doi: 10.1016/j.neulet.2010.12.064 [DOI] [PubMed] [Google Scholar]
- 12.Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A et al. (2009) Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 28, 359–371 doi: 10.1038/emboj.2008.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J et al. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 doi: 10.1016/j.cell.2009.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D et al. (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8, 700–710 doi: 10.1038/ncb1436 [DOI] [PubMed] [Google Scholar]
- 15.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D et al. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 doi: 10.1074/jbc.M609659200 [DOI] [PubMed] [Google Scholar]
- 16.Meyer H-J and Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 doi: 10.1016/j.cell.2014.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fushman D and Wilkinson KD (2011) Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 Biol. Rep. 3, 26 doi: 10.3410/B3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suryadinata R, Roesley SN, Yang G and Šarčević B (2014) Mechanisms of generating polyubiquitin chains of different topology. Cells 3, 674–689 doi: 10.3390/cells3030674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Tang C, Wang E and Wang J (2014) Polyubiquitin chain linkage topology selects the functions from the underlying binding landscape. PLoS Comput. Biol. 10, e1003691 doi: 10.1371/journal.pcbi.1003691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadowski M, Suryadinata R, Tan AR, Roesley SNA and Sarcevic B (2012) Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 64, 136–142 doi: 10.1002/iub.589 [DOI] [PubMed] [Google Scholar]
- 21.Nussinov R, Tsai C-J, Xin F and Radivojac P (2012) Allosteric post-translational modification codes. Trends Biochem. Sci. 37, 447–455 doi: 10.1016/j.tibs.2012.07.001 [DOI] [PubMed] [Google Scholar]
- 22.Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N and Schulman BA (2011) Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 21, 257–264 doi: 10.1016/j.sbi.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshaies RJ and Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 doi: 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 24.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P et al. (2002) Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 doi: 10.1038/416703a [DOI] [PubMed] [Google Scholar]
- 25.Lydeard JR, Schulman BA and Harper JW (2013) Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 14, 1050–1061 doi: 10.1038/embor.2013.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M and Schulman BA (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 doi: 10.1016/j.cell.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enchev RI, Schulman BA and Peter M (2015) Protein neddylation: beyond cullin–RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 doi: 10.1038/nrm3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabut G and Peter M (2008) Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9, 969–976 doi: 10.1038/embor.2008.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calabrese MF, Scott DC, Duda DM, Grace CRR, Kurinov I, Kriwacki RW et al. (2011) A RING E3–substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nat. Struct. Mol. Biol. 18, 947–949 doi: 10.1038/nsmb.2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei N, Serino G and Deng X-W (2008) The COP9 signalosome: more than a protease. Trends Biochem. Sci. 33, 592–600 doi: 10.1016/j.tibs.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 31.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y et al. (2004) Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528 doi: 10.1016/j.cell.2004.10.019 [DOI] [PubMed] [Google Scholar]
- 32.Helmstaedt K, Schwier EU, Christmann M, Nahlik K, Westermann M, Harting R et al. (2011) Recruitment of the inhibitor Cand1 to the cullin substrate adaptor site mediates interaction to the neddylation site. Mol. Biol. Cell. 22 153–164 doi: 10.1091/mbc.E10-08-0732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Morgan MA and Sun Y (2014) Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid. Redox Signal. 21, 2383–2400 doi: 10.1089/ars.2013.5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Li L, Liang Y, Li C, Zhao H, Ye D et al. (2014) Targeting the neddylation pathway to suppress the growth of prostate cancer cells: therapeutic implication for the men’s cancer. Biomed. Res. Int. 2014, 974309 PMID:25093192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Q, Yu G-Y, Shi J-Y, Li L-H, Zhang W-J, Wang Z-C et al. (2014) Neddylation pathway is up-regulated in human intrahepatic cholangiocarcinoma and serves as a potential therapeutic target. Oncotarget 5, 7820–7832 doi: 10.18632/oncotarget.2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S and Yu L (2015) Targeting cullin-RING ligases for cancer treatment: rationales, advances and therapeutic implications. Cytotechnology 68, 1–8 PMID:25899169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan H, Tang Z, Jin H and Sun Y (2016) Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci. Rep. 6, 24218 doi: 10.1038/srep24218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benamar M, Guessous F, Du K, Corbett P, Obeid J, Gioeli D et al. (2016) Inactivation of the CRL4-CDT2-SET8/p21 ubiquitylation and degradation axis underlies the therapeutic efficacy of pevonedistat in melanoma. EBioMedicine 10, 85–100 PMID:27333051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulatov E and Ciulli A (2015) Targeting Cullin–RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation. Biochem. J. 467, 365–386 doi: 10.1042/BJ20141450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y and Sun Y (2013) Cullin-RING Ligases as attractive anti-cancer targets. Curr. Pharm. Des. 19, 3215–3225 doi: 10.2174/13816128113199990300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanley DJ, Bartholomeeusen K, Crosby DC, Kim DY, Kwon E, Yen L et al. (2012) Inhibition of a NEDD8 cascade restores restriction of HIV by APOBEC3G. PLoS Pathog. 8, e1003085 doi: 10.1371/journal.ppat.1003085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann H, Norton TD, Schultz ML, Polsky SB, Sunseri N and Landau NR (2013) Inhibition of CUL4A Neddylation causes a reversible block to SAMHD1-mediated restriction of HIV-1. J. Virol. 87, 11741–11750 doi: 10.1128/JVI.02002-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nekorchuk MD, Sharifi HJ, Furuya AKM, Jellinger R and de Noronha CMC (2013) HIV relies on neddylation for ubiquitin ligase-mediated functions. Retrovirology 10, 138 doi: 10.1186/1742-4690-10-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW et al. (2014) Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157, 1671–1684 doi: 10.1016/j.cell.2014.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown NG, VanderLinden R, Watson ER, Weissmann F, Ordureau A, Wu K-P et al. (2016) Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell 165, 1440–1453 doi: 10.1016/j.cell.2016.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma B, Tsai C-J, Haliloğlu T and Nussinov R (2011) Dynamic allostery: linkers are not merely flexible. Structure 19, 907–917 doi: 10.1016/j.str.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J and Nussinov R (2013) The role of allostery in the ubiquitin–proteasome system. Crit. Rev. Biochem. Mol. Biol. 48, 89–97 doi: 10.3109/10409238.2012.742856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nussinov R, Tsai C-J and Liu J (2014) Principles of allosteric interactions in cell signaling. J. Am. Chem. Soc. 136, 17692–17701 doi: 10.1021/ja510028c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papaleo E, Saladino G, Lambrughi M, Lindorff-Larsen K, Gervasio FL and Nussinov R (2016) The role of protein loops and linkers in conformational dynamics and allostery. Chem. Rev. 116, 6391–6423 doi: 10.1021/acs.chemrev.5b00623 [DOI] [PubMed] [Google Scholar]
- 50.Erman B (2006) The Gaussian network model: precise predictions of residue fluctuations and application to binding problems. Biophys. J. 91, 3589–3599 doi: 10.1529/biophysj.106.090803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haliloglu T and Erman B (2009) Analysis of correlations between energy and residue fluctuations in native proteins and determination of specific sites for binding. Phys. Rev. Lett. 102, 088103 doi: 10.1103/PhysRevLett.102.088103 [DOI] [PubMed] [Google Scholar]
- 52.Sánchez R and Sali A (2000) Comparative protein structure modeling. Introduction and practical examples with modeller. Methods Mol. Biol. 143, 97–129 PMID:11084904 [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W, Dalke A and Schulten K (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–8 doi: 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 54.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 doi: 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- 55.Mackerell AD Jr, Feig M and Brooks CL III (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 doi: 10.1002/jcc.20065 [DOI] [PubMed] [Google Scholar]
- 56.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E et al. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 doi: 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saifee NH and Zheng N (2008) A ubiquitin-like protein unleashes ubiquitin ligases. Cell 135, 209–211 doi: 10.1016/j.cell.2008.09.049 [DOI] [PubMed] [Google Scholar]
- 58.Liu J and Nussinov R (2010) Rbx1 flexible linker facilitates Cullin-RING ligase function before neddylation and after deneddylation. Biophys. J. 99, 736–744 doi: 10.1016/j.bpj.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J and Nussinov R (2011) Flexible cullins in cullin-RING E3 ligases allosterically regulate ubiquitination. J. Biol. Chem. 286, 40934–40942 doi: 10.1074/jbc.M111.277236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J and Nussinov R (2009) The mechanism of ubiquitination in the cullin-RING E3 ligase machinery: conformational control of substrate orientation. PLoS Comput. Biol. 5, e1000527 PMID:19798438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J and Nussinov R (2010) Molecular dynamics reveal the essential role of linker motions in the function of Cullin–RING E3 ligases. J. Mol. Biol. 396, 1508–1523 doi: 10.1016/j.jmb.2010.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chong RA, Wu K, Spratt DE, Yang YY, Lee C, Nayak J et al. (2014) Pivotal role for the ubiquitin Y59-E51 loop in lysine 48 polyubiquitination. Proc. Natl. Acad. Sci. U.S.A. 111, 8434–8439 doi: 10.1073/pnas.1407849111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC et al. (2009) E2-RING expansion of the NEDD8 cascade confers specificity to Cullin modification. Mol. Cell 33, 483–495 doi: 10.1016/j.molcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Angers S, Li T, Yi XH, MacCoss MJ, Moon RT and Zheng N (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593 PMID:16964240 [DOI] [PubMed] [Google Scholar]
- 65.Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA et al. (2016) Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166, 1198–1214.e24 doi: 10.1016/j.cell.2016.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.