Abstract
Self-assembly of peptides is closely related to many diseases, including Alzheimer’s, Parkinson’s, and prion diseases. Understanding the basic mechanism of this assembly is essential for designing ultimate cure and preventive measures. Template-assisted self-assembly (TASA) of peptides on inorganic substrates can provide fundamental understanding of substrate-dependent peptides assemble, including the role of hydrophobic interface on the peptide fibrillization. Here, we have studied the self-assembly process of a potential pentapeptide inhibitor on the surface of highly oriented pyrolytic graphite (HOPG) using real time atomic force microscopy (RT-AFM) as well as molecular dynamics (MD) simulation. Experimental and simulation results show nanofilament formation consisting of β-sheet structures and epitaxial growth on HOPG. Height analysis of the nanofilaments and MD simulation indicate that the peptides adopt a lying down configuration of double-layered antiparallel β-sheets for its epitaxial growth, and the number of nanofilament layers is concentration-dependent. These findings provide new perspective for the mechanism of peptide-based fibrillization in amyloid diseases as well as for designing well-ordered micrometrical and nanometrical structures.
Graphical Abstract
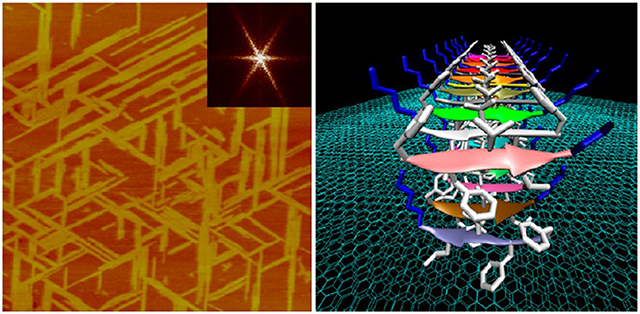
INTRODUCTION
Fibril formation by self-assembly of peptides or proteins under pathogenic conditions is relevant to amyloidosis.1 As an example, Alzheimer’s disease (AD) is linked to the accumulation of amyloid β (Aβ), a 39–43 amino acids long peptide deposited in the brain parenchyma and cerebrovasculature.2,3 Understanding fibrillization of such peptide in terms of their structure and intermolecular interactions is therefore of great interest in finding cure designing preventive measures.4–6
Amyloidosis often initiates at biomembranes.7–9 For example, small aggregates of Aβ peptides disrupt cellular calcium homeostasis by forming channel-like pores in the membrane, suggesting that Aβ-lipid interaction at the cell membrane initiates the neurotoxic cascade in AD.9,10 The template-assisted self-assembly (TASA) technology originally developed for inorganic particles11 has quickly extended to biomolecule-inorganic interactions, especially inorganic substrates (like mica and graphite)-directed self-assembly of proteins/peptides.12–14 Inorganic substrates are commonly used in TASA as a mimic of biomembrane surfaces and lipoprotein particles in vivo.15–18
A pentapeptide with the KLVFF sequence corresponding to Aβ(16–20) has been considered a core sequence of Aβ that drives the formation of fibrils.19 Previous studies using in situ cryo-transmission electron microscopy (TEM), small-angle Xray scattering, Fourier transform infrared (FTIR) spectroscopy, and fiber X-ray diffraction have shown that KLVFF peptide itself can form β-sheet fibrils and even hydrogels in a higher concentration.6,20 Because KLVFF is the core sequence that drives the Aβ aggregation, ligands capable of binding this sequence have been investigated as possible drug candidates to inhibit Aβ fibrillization.21,22
Other peptides using KLVFF sequence as a core have also been studied. For examples, a peptide with AALKVFF sequence was reported to form distinct structures of twisted fibrils in water or on nanotubes in methanol.23 KLVFFAE24 and AcK(Me)LV(Me)FF-NH2 25 were also shown to form fibrils in aqueous solutions. Interestingly, a retro-inverso (RI) peptide of KLVFF, ffvlk, has been reported to bind artificial fibrils of Aβ with moderate affinity.26 The written form “ffvlk” without using capital letters indicates D-amino acids, and the corresponding technology in combination with reversing the primary sequence of a peptide called retro-inversion has provided an opportunity for designing better peptide-based therapeutics.27 However, the binding mechanisms as well as the fibrillization properties are not well studied.
In order to understand potential drawbacks for clinic use of these peptides for treatment of AD, it is important to examine the potential of RI peptide self-assembly into fibrils as well as the interaction mechanism at a hydrophobic interface. Here we synthesized a pentapeptide NH2-FFVLK-CONH2 (in short “P5”), in which all the amino acids are just of L-enantiomer for convenience (D-enantiomer is most prone to digestion by natural enzymes in vivo28). We introduced an amine group to its C-terminal to remove the terminal charge of carboxylic acid. The modified peptides more closely mimic the native protein, increases its ability to enter cells and prolongs its shelf life.29 We used both real time atomic force microscopy (RT-AFM) and MD simulation to understand the self-assembly of P5 on a highly oriented pyrolytic graphite (HOPG). Our results reveal that P5 self-assembles on HOPG following the atomic lattice of HOPG. MD simulation results provide more details for this epitaxial growth in which the double-layered antiparallel β-sheets are the most stable nanostructures.
EXPERIMENTAL SECTION
Peptide Sample Preparation.
The peptide P5 (NH2-FFVLKCONH2) was custom-synthesized from Chinapeptides Company (Shanghai, China). The final purity of the peptide is more than 97% as verified by high-performance liquid chromatography (HPLC) and was further characterized with mass spectrum. The lyophilized powder was stored at −22 °C. The peptide powder was dissolved in Milli-Q water to a certain peptide concentration. Then the solutions were centrifuged at 10 000 rpm for 10 min to remove the possible aggregates and the supernatant was immediately used or stored in aliquots to different PE tubes in the freezer (−22 °C). Other chemicals were purchased from Sigma and used without further purification.
Thioflavin T Assay.
The fibrosis of P5 was studied by monitoring the thioflavin T (ThT) fluorescence with a fluorescence spectrometer (Fluorolog-MAX 4, Horiba) equipped with a 1.0 cm quartz cell at 25 °C. The P5 lyophosized powder was directly dissolved in 10 μM ThT water solution to get a concentration of 0.8 mg/mL (1.23 mM). The mixture was agitated at 900 rpm with a magnetic stirring bar. The fluorescence signal was recorded every 10 min within the first 200 min, and afterward the signal was recorded every 100 min.
RT-AFM.
A commercial AFM instrument (Nanoscope IIId, Bruker) equipped with either a J-scanner (125 μm × 125 μm) or a E-scanner (12 μm × 12 μm) and a liquid cell was used. All images were captured with a scan rate at 1–2 Hz. Experiments were performed in tapping mode under liquid phase. Silicon nitride cantilevers with a nominal spring constant of 0.35 N/m (SNL-10, Bruker) were used. HOPG (ZYB grade, 12 mm × 12 mm × 2 mm, Bruker) were freshly cleaved by adhesive tape before each experiment. All the real time imaging process was conducted as the following: (1) the peptide solution was slowly added into the liquid chamber through a connected tube; (2) the final concentration of the peptide in the liquid chamber was calculated by considering the previous volume of the buffer solution. AFM imaging started before adding the peptides into the liquid chamber in order to capture the whole dynamics of the peptide assembly. AFM images were processed by using NanoScope Analysis (version 1.40) that was supplied by the AFM manufacture.
Atomistic Molecular Dynamics Simulations.
Two Aβ monomer conformations were extracted from small Aβ1–40 protofibrils (PDB codes: 2LMN and 2LMO)30 and Aβ1–42 fibril (PDB code: 2MXU)31 where both fibril structures were based on the ssNMR models. In both structures, the N-terminal coordinates, residues 1–8 for the former and 1–10 for the latter structures, are missing due to disorder. To generate P5 peptide, the coordinates of the central region of Aβ, residues 16–20, were taken from both monomer conformations. The sequence of these Aβ fragments, KLVFF, was inverted to FFVLK, generating two P5 conformers. In conformer 1 of P5 peptide, two Phe side chains point in the opposite direction, while they point in the same direction in conformer 2. The monomeric P5 conformers were used to construct the 16-mer single-layered, 32-mer double-layered, and 48-mer triple-layered β-sheet fibrils with a linear shape. The initial P5 fibrils were minimized with a rigid body motion for the peptides, which enhanced the formation of intermolecular backbone hydrogen bonds (H-bonds) within a β-sheet. Using the same protocol for the HOPG simulation as in our previous work,32 three layers of graphite in a hexagonal phase were created to mimic HOPG. The minimized P5 fibrils were deposited onto the HOPG surface. To avoid the unfavorable interaction between the positively charged Lys residue and the hydrophobic graphite surface, we aligned the P5 fibrils with the hydrophobic residues, Phe2 and Leu3, pointing to the HOPG surface.
The CHARMM program33 with version 3634 was used to construct the set of initial conformations and to relax the systems for the atomistic MD simulations. The same MD simulation techniques as used in the amyloid studies 10'35–37 were applied to the P5/HOPG simulations. In the preequilibrium stages, a series of combined minimization and dynamics cycles with electrostatic cutoffs (12 Å) were performed for the initial configurations to gradually relax the solvents around the harmonically restrained P5 peptides in the fibrils. The harmonic restraints were gradually diminished with the full Ewald electrostatics calculation and constant temperature (Nosé—Hoover) thermostat/barostat at 300 K. Following preequilibrium, a 200 ns production run was performed with the NAMD 2.10 parallel computing code38 on a Biowulf cluster at the National Institute of Health.
RESULTS AND DISCUSSION
We used ThT molecules to check whether the synthesized P5 can form fibrils. ThT fluorescent molecules have been widely used for the kinetic study of amyloid fibrillization because the molecule can specifically bind to β-sheet-containing amyloid fibrils and show an increase in fluorescence peak at around 482 nm.14 A typical ThT fluorescent emission spectrum of P5 showed an increasing fluorescence peak at around 470 nm (Figure 1A), indicating that P5 formed amyloid-like β-sheet structures. The blue shift of ThT emission from 482 to 470 nm could arise from small changes in the local hydrophobicity.14 By monitoring the fluorescence intensity (IF) at 470 nm, the kinetics of P5’s fibrillization was examined in detail. As expected, the intensity of ThT fluorescence signals of P5 solutions increased remarkably with the increasing incubation time (Figure 1B). The fluorescence spectrum in Figure 1B can be roughly divided into two parts: (i) After about 80 min lag phase, the fluorescence intensity (IF) increases rapidly from 80 to 200 min, indicating a fast elongation which is believed to be the critical stage for amyloid fibrillization; (ii) after that, the fluorescence intensity tends to be stationary for more than 10 h, suggesting that P5 can assemble into robust β-sheet structures.
Figure 1.

(Left panel) The ThT fluorescence spectrum of P5. ThT fluorescence shows a maximum emission peak at about 470 nm when mixed in P5 solutions. The fluorescence signal was measured after 5 h incubation at room temperature. (Right panel) Fibrillization of P5 monitored by ThT fluorescence. ThT fluorescence was monitored at 470 nm every 10 min within the first 200 min, and afterward the signal was recorded every 100 min. The excitation wavelength was 440 nm.
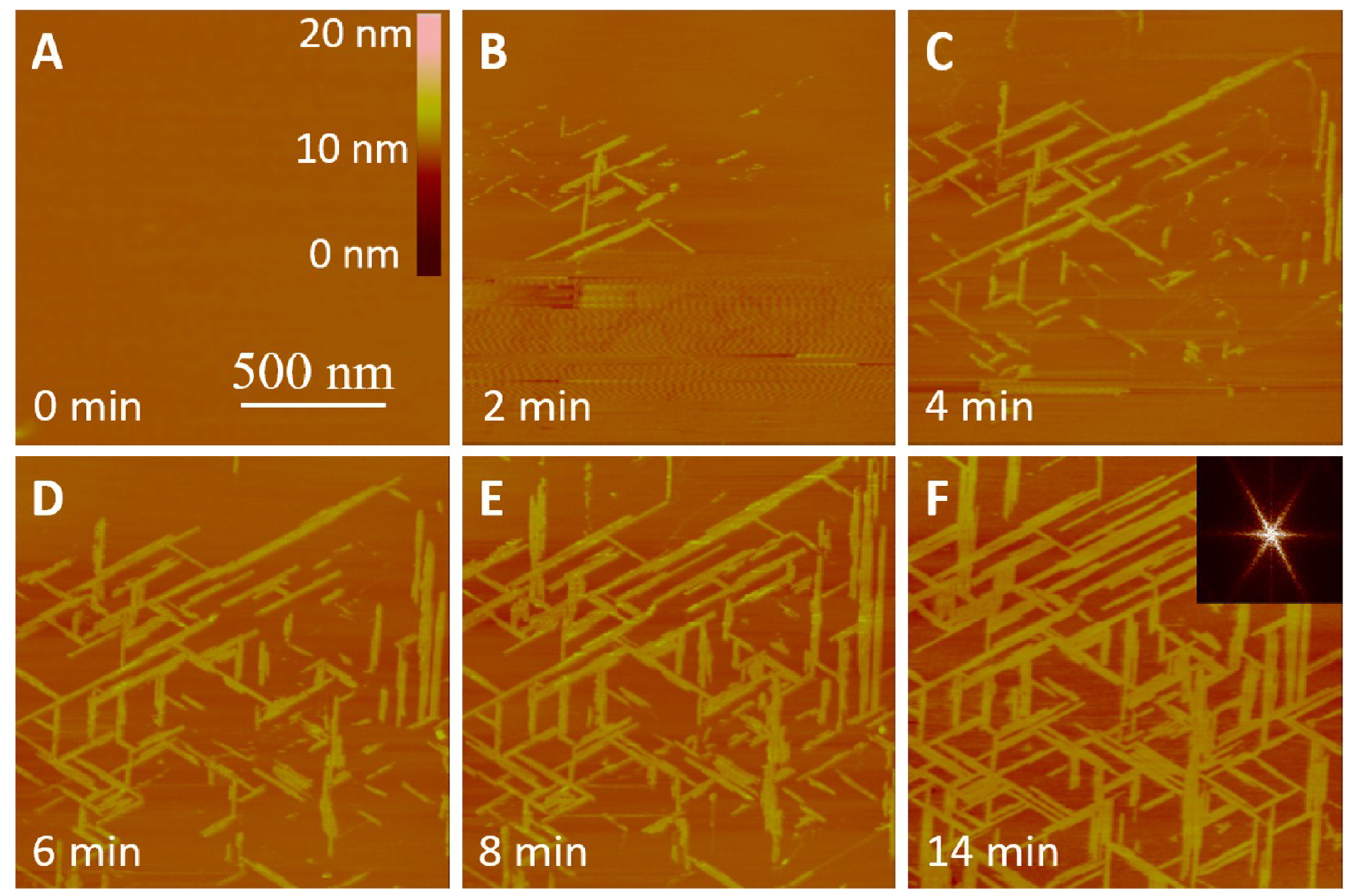
We explored the self-assembly dynamics of P5 on HOPG using RT-AFM.39,40 At the concentration of 0.15 mg/mL, P5 adsorbed onto HOPG surface at a very high rate (Figure 2). Interestingly, most of the freshly adsorbed P5 molecules formed unregular structures on HOPG. However, with the time going on we can see the loose unregular structures in Figure 2B became denser and denser from Figure 2 panel C to panel D, and meanwhile the structures became more and more regular until densely aligned fibrils in Figure 2E,F. This phenomenon indicated that the adsorbed P5 molecule can be still capable to reorganize their self-assembly style on HOPG within about 1 h. We think this is due to the too concentrated P5 solution made the rapid adsorption without enough time to arrange the P5 molecule into an ordered nanostructure (nanofilaments), and also leads to a very loose aggregate style on HOPG (Figure 2B) and later due to the stability (lowest energy) driving, these molecules found their relatively better positions and orientations in the limited space since the rapidly adsorbed P5 molecules took most of it.
Figure 2.

Time-lapse monitoring of P5 fibrillization on HOPG at 0.15 mg/mL in Milli-Q water. (A-F) Tapping-mode RT-AFM images of P5 self-assembling at room temperature for different time periods as denoted in each image. The scale bars in (A) apply to all images.
From the measurement of the heights of nanofilaments formed by P5 molecules, we observed that the formed nanofilaments are mostly about 2 nm in height (Figure 3). When the nanofilaments were densely packed, the precise height measurements were only limited at several marginal regions where the AFM tips can touch the surface of HOPG. Most of nanofilaments show homogeneous in height (Figure 2E,F). The calculation of the orientations of nanofilaments show that several angles, 30°, 60°, and 90° coexist between those connected nanofilaments (Figure 2D–F, Figure 3C,D). To slow down the growth rate of fibrils to obtain more detailed information, one can reduce either temperature or concentration. Because reducing temperature requires us to pay additional expensive accessories, we prepared low concentrations of P5 solutions to observe their self-assembly processes on HOPG.
Figure 3.

Height measurements of P5 nanofilaments on HOPG. (A-D) Representation of the cross-section analysis of the nanofilament structures (insets) from Figure 2F. Different colors indicated the cutting lines and the corresponding vertical distances (heights) nearby two arrows with same color. The axis units are all nanometers.
The first diluted concentration was 10 μg/mL, however, this concentration was still too high to obtain more intermediate growth states of nanofilaments (Figure S1). So we again diluted to 1 μg/mL. With the RT-AFM observation, we found that almost no unregular adsorption happens at this concentration. P5 grew bidirectionally and formed nanofilaments with an extending rate of ~25 μm/min. The lengths of these filaments were up to several micrometers (Figure 4). Notably and obviously, the nanofilaments never overlapped with each other during their growth, indicating that the growth of the nanofilaments is anisotropic. They arranged themselves into nanofilaments with dimensional selectivity and specificity (binding sites). The fast Fourier transform image (inset in Figure 4F) revealed three preferred orientations with an angle of 60° to each other, similar to the hexagonal atomic lattice of the substrate. This indicates an “epitaxial” growth or templateassisted effect.12–14,18,41 Similar images of nanofilaments growth are included in Figure S1.
Figure 4.
Time lapse monitoring of P5 fibrillization on HOPG at 1 μg/mL in Milli-Q water. (A-F) Tapping-mode RT-AFM images of P5 self-assembling at room temperature for different time periods as denoted in each image. The scale bars in (A) apply to all images. The inset in (F) shows the periodic spatial distribution in (F) formed by using the two-dimensional Fourier transfer function. The wave-like signal in (B,C) was resonance noise during liquid phase AFM imaging.
The height of the P5 filaments was monitored during the self-assembling from different growth regions. It is ranged from 1.0 to 2.2 nm for different nanofilaments (Figure 5). The difference in heights on a single filament was about 0.6 nm (Figure 5B). Previously, several different self-assembling peptides showed the multilayered nanostructures by monitoring the assembly in a concentration-dependent manner.12,14,42 To observe more single layered nanofilaments, we further diluted the concentration of P5 from 1 μg/mL to 1 ng/mL because the nanofilament growth rate was still high enough. The nanofilaments covered almost 50% of the HOPG surface within only 14 min, suggesting that the concentration can be more diluted.
Figure 5.

Height measurements of P5 nanofilaments on HOPG. (A-D) Represention of the cross-section analysis of the nanofilament structures (insets) from Figure 4F. Different colors indicated the cutting lines and the corresponding vertical distances (heights) nearby two arrows with same color. The axis units are all nanometers.
The dilution of P5 concentration resulted not only a slow self-assembly progress of P5 on HOPG but also more and lower nanofilaments formed. We observed that all nanofilaments grew with a hexagonal symmetry on HOPG as seen at higher concentration (Figure 6). The growth rate of nanofilaments decreased to ~1 μm/min, but the length of nanofilaments increased as compared with those grown under higher concentrations. This indicates that nonisotropic 2D growth was derived from the chiral property of P5 molecule itself. Because of the single nanofilament, the widening rate was much less than the lengthening rate, which further indicates that the synergistic effect from both peptide-to-peptide and peptide-to-HOPG played a dominant role in the fibrillization process. The body-to-body interactions should be much stronger than head/ tail-to-head/tail interactions between peptides, which had also discussed by other MD researches.43,44 Without exception, none of the nanofilaments appears to cross the edges of HOPG, which further show the necessity of a precise match for the epitaxial growth. We found that one of the preferred growth directions was parallel to the edges of HOPG, suggesting the correlation between the atomic lattice and the easiest cleavage plane of HOPG.
Figure 6.

A dynamic epitaxial growth procedure of P5 on HOPG at 1 ng/mL in Milli-Q water. (A-F) A series of top-view tapping-mode AFM images at room temperature for different time periods as denoted in each image. The scale bars in (A) applied to all images.
The cross-section analysis showed that the initial heights of nanofilaments were less than 1 nm (Figure 7), which was comparable to P5’s theoretical diameter (0.8 nm), and much less than its length (1.7 nm), indicating that P5 adopts a “lying down” manner on HOPG for its fibrillization. Small variations from the height measurements were also observed. Such variations could be attributed to different amino acid residues. For an example, the lysine residue can be more flexible than both leucine and valine residues, because it is on the terminus with more freedom to fluctuate than those residues located in the middle of P5.
Figure 7.

Height measurements of P5 nanofilaments on HOPG at a concentration of 1 ng/mL. (A-D) Representation of the cross-section analysis of Figure 6B,D-F. The black lines in the inset AFM images indicated the section cutting lines. The measured heights indicated by two red arrows were 0.65, 0.71, 0.95, and 0.98 nm, respectively.
To systematically compare the properties of nanofilaments at different conditions, we have also made the statistical analysis of both the height and orientation of nanofilaments on HOPG at different concentrations (Figure 8). As shown in Figure 8A, the height distributions of nanofilaments changed significantly from 0.8–1.1 nm at 1 ng/mL to 1.6–2.2 nm at 1 μg/mL. At the higher concentration of 0.15 mg/mL, the heights below 1 nm were hardly to found, and the dominant height distribution was also around 2 nm. Because the theoretical diameter and length of P5 are about 0.8 and 1.7 nm, respectively, the similar concentration-dependent discrete height subpopulations of nanostructures could be formed by either “lying down” peptides on HOPG or “standing up/upright” peptides on mica.12 Moreover, the estimated heights (more than 3 nm) of double-layered nanostructures formed by upright P5 were not found in all concentrations. Thus, our results suggest that the observed individual height subpopulations are likely formed by P5 with a lying down orientation rather than the standing up/ upright orientation. More detailed discussions will be shown in MD simulation results. In addition, the orientations of nanofilaments formed on HOPG at different concentrations (Figure 8B) also protruded the differences between lower concentrations and higher concentrations. The histogram showed that at concentrations lower than 1 μg/mL (even 10 μg/mL as shown in Figure S1), only 60° presented in the images. Strictly three-folded symmetric orientations were formed, which resembled the atomic lattice orientations of HOPG. However, at concentration of 0.15 mg/mL, three different angles coexited in the self-assembling process. The rapid adsorption of P5 molecules experienced a rearrangement to form dense nanofilaments on HOPG (Figure 2). We hypothesized that the angles at 30° and 90° could be the secondary favorable orientations for P5 epitaxial growth, which could be also significantly influenced by the steric hindrance of the crowed molecules adsorbed in vicinity and thus the limited rearrangement space. From this histogram, we can also conclude that the strict epitxial growth requires a slow process that has enough time to allow molecules for tuning their orientations to fit into the right positions in the nanostructures.
Figure 8.

Statistical data for the heights (A) and orientations (B) of P5 nanofilaments on HOPG at different concentrations. For the angle measurements, only the acute angles were recorded. There were at least 40 measurements for each condition.
In order to verify the “epitaxial growth”, we investigated the atomic lattice of HOPG using contact mode AFM instead of STM12 to minimize the disturbance of orientations. Both contact and tapping modes can be performed in the same set up. We first imaged the HOPG before adding any P5 peptide in the solution. Figure 9A shows the atomic lattice of HOPG with a regular hexagonal symmetry under contact AFM mode. Then we changed to tapping mode and added P5 solution to image the real-time self-assembly of P5 on the same region of HOPG. When we compared the orientations of the nanofilaments growths (Figure 9B) to the underneath atomic lattice of HOPG (Figure 9A), we found that both orientations perfectly matched each other. Our results indicate that the self-assembly of P5 peptides is template-assisted/epitaxial procedure which follows the similar manner found previously for other peptides on substrates.12–14
Figure 9.
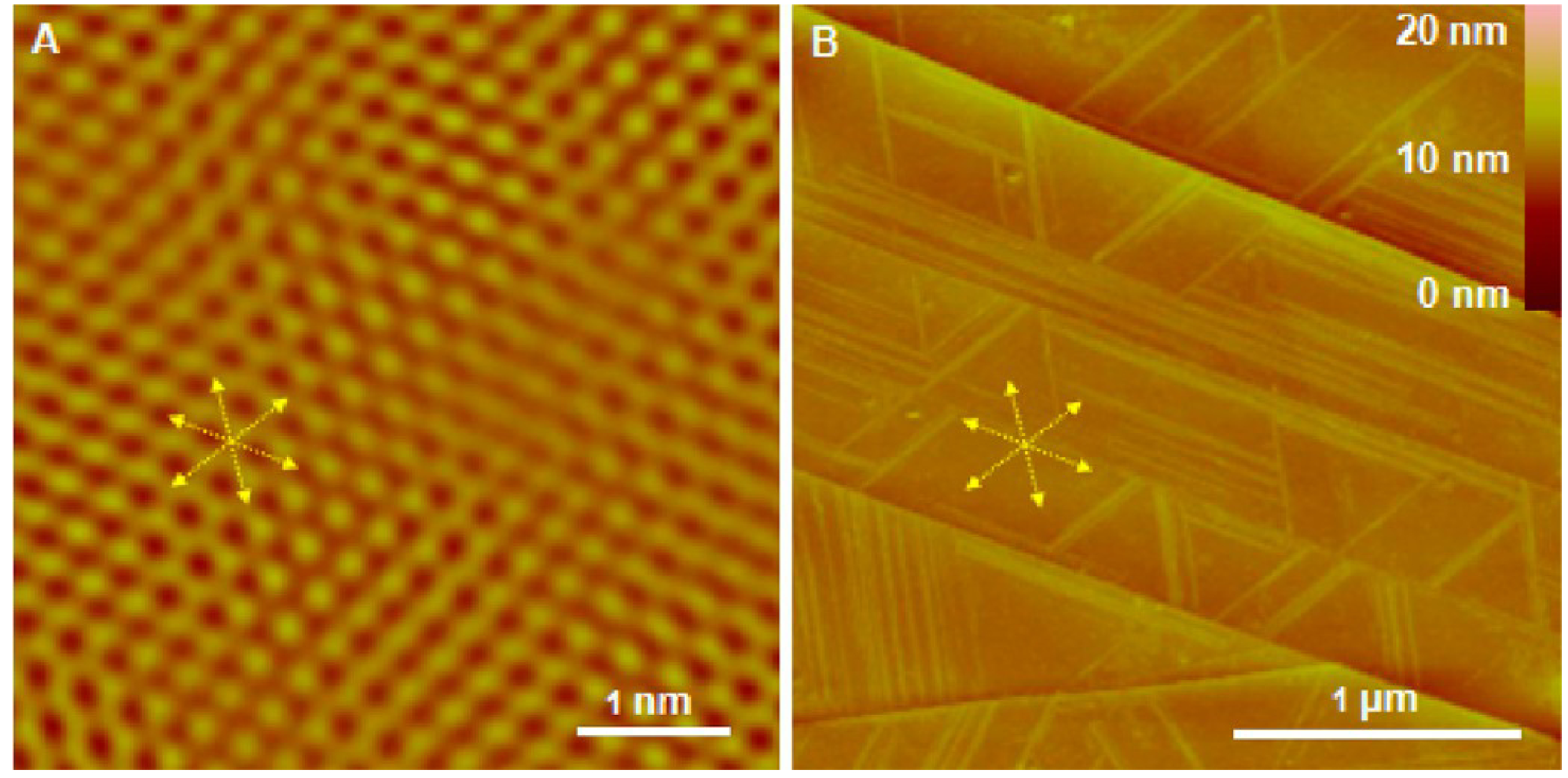
Comparison between the atomic lattice of underneath HOPG and the growth orientations of the P5 nanofilaments. (A) The height image of the HOPG’s atomic lattice obtained prior to P5 self-assembly using friction channel of contact mode AFM. (B) The height image of P5 nanofilaments grown on the surface of HOPG from (A). The yellow arrows and lines denote the consistence of their orientations.
To understand the arrangement of P5 peptides in the nanofilaments at the atomic level, we used MD simulations of eight P5/HOPG systems with different combinations of conformers (Figure S2), layers, and parallel/antiparallel arrangements (Table 1). During the simulations, we found that the double-layered antiparallel β-sheet morphology in the out-of-phase is the most stable architecture (Figure S8). Out-of-phase stacking refers to the β-strand stacking within an antiparallel fashion between two adjacent β-sheets, whereas in-phase stacking denotes the parallel β-strand stacking.23 All parallel arrangements (Figures S5, S6, S7, and S9) including the upright orientation (Figure S7) are not stable enough to preserve the nanostructures within the given period. The phenyl groups in Phe residues have stronffi affinity to the graphite surface due to the π-π interactions. In term of the β-sheet content from the initial structure, double-layered conformation preserves the β-sheet much better than single-layered ones (Figure S11), indicating that the intermolecular affinity of P5 plays an important role in stabilizing the nanofilaments (Figure S12). The modeled single-layered nanofilaments are correlated with those observed in a super diluted solution (Figures 6, 7, and 9), whereas the nanofila-ments observed in Figures 2-5 are related to the double-layered structures. We speculate that both the intermolecular affinity of P5 and the attraction between P5 molecules and HOPG surface are the driving force for the current TASA system.
Table 1.
Summarized Results from the MD Simulations of P5 Fibrils
| systems | conformers | β-strand arrangements | layers | molecular numbers | % of β-sheet content from the initial structure | final morphologies |
|---|---|---|---|---|---|---|
| S1 | 1 | antiparallel | single | 16 | 56 | collapsed aggregate with partial β-sheets |
| S2 | 2 | antiparallel | single | 16 | 0 | disassembled |
| S3 | 1 | parallel | single | 16 | 0 | disassembled |
| S4 | 2 | parallel | single | 16 | 0 | disassembled |
| S5 | 2 | parallel (upright) | single | 16 | 31 | collapsed aggregate with partial β-sheets |
| D1 | 1 | antiparallel, out of phase | double | 32 | 97 | double-layered antiparallel β-sheet preserved |
| D2 | 1 | parallel, out of phase | double | 32 | 63 | reduced to broken β-sheets due to twist |
| TL | 1 | antiparallel, out of phase | triple | 48 | 85 | unstable 1st and 3rd layers, starting to dissociate |
From the well-conserved nanostructures on HOPG (Figure 9A), the growth axis of nanofilaments well matches the atomic lattice of HOPG (Figures 9, 10), which is also consistent with experimental results (Figure 9). It is interesting to note that only one of the phenyl ring from the Phe residue of each P5 molecule interacts with the HOPG surface, while the other one at the N-terminal Phe residue is oriented opposite to the HOPG surface due to the steric hindrance between two phenyl rings and the HOPG surface. To stabilize β-sheet, the P5 peptide is arranged as a β-strand adjacent to other strands forming an extensive intermolecular backbone H-bond network. As a result, the phenyl ring at the second Phe residue and the Leu side chain are directed in the same direction toward the HOPG surface, satisfying the β-sheet secondary structure. Because of the steric hindrance and the electrostatic interaction, the Lys side chain is pointing toward solution instead of the HOPG surface (Figure 10B,E). In the P5 nanofilament with the double-layered antiparallel β-sheet arrangement, the second β-sheet layer is stabilized through the hydrophobic interactions with the aliphatic side chains in the first layer. The Phe and Val side chains in the first layer interact with the Leu and Phe side chains in the second β-sheet layer, respectively. No π-π interaction between phenyl rings can be observed because the π-stacking requires an orientation of the backbone of Phe residue, which can disrupt the β-sheet network. Our results suggest that the dominated hydrophobic interaction between the β-sheet layers can serve as a driving force for the epitaxial growth. This explains why P5 can form free-standing β-sheets even in solutions at a relative higher concentration (Figure 1).
Figure 10.

Representative nanostructures from MD simulations. (A-C) The top view, angle, and side views for single- layered antiparallel β-sheet formed by conformer 1, respectively. (D-F) The top view, angle, and side views for double-layered antiparallel β-sheets formed by conformer 1, respectively. Both panels A and D showed that the epitaxial growth directions were consistent with the atomic lattice of HOPG, and the antiparallel β-strands were indicated by arrows (the tail of arrows denote the N terminal of P5). Both panels B and E showed the close look at the interaction between amino acid residues and HOPG surface. Both panels C and F showed the height estimates of antiparallel β-sheets.
The calculated average heights of both modeled singlelayered and double-layered β-sheet structures are 0.8 and 1.7 nm, respectively. Considering the flexibility and side chain’s length variation of different amino acid residues, the actual measured heights can be varied within a certain range. Because the AFM imaging could exert a certain pressure on the layered structures even though the force is minimized during the imaging, the measured heights can be underestimated. In fact, the measured heights of β-sheet structures show various values in the ranges of 0.65—0.98 nm at a lower concentrated solution and 1.05—2.23 nm at a higher concentrated P5 solution, respectively. The large value of the height corresponds to that of triple-layered β-sheet structures on HOPG. From the simulations, such triple-layered β-sheet structures can be formed, although they finally started to dissociate at the first and third layers (Table 1). However, compared with the conservation rate of single-layered β-sheet structures, the triplelayered score is much higher, suggesting that the highest measured heights of nanofilaments corresponds to the triplelayered β-sheet structure (Figure S10).
CONCLUSION
We studied the graphite-assisted self-assembly of a de novo pentapeptide P5 using both experimental and computational methods. Experimentally, the concentration-dependent epitaxial growth of P5 peptides on HOPG was observed in real time. At different concentrations, P5 adopted a lying down manner to grow into nanofilaments with different heights and growth rates. The MD simulation results showed that P5 molecules follow the epitaxial principle by registering themselves exactly along with the atomic lattice of HOPG and forming nanofilaments. Double-layered antiparallel β-sheets were the most stable conformation in the nanofilaments. Both experimental and simulation results agree well with each other. In combination with the biocompatibility of graphite and the special designing of P5 molecules, these results might provide useful information in understanding the aggregation of amyloid peptides in diseases and the fast-developing researches on the bionano interface.
Supplementary Material
ACKNOWLEDGMENTS
We are grated to Inner Mongolia Autonomous Region Natural Science Foundation (No. 2015MS0806, 2016MS0211). The support provided by China Scholarship Council (CSC) during a visit of “Feng Zhang” to UCSD is acknowledged. This project was supported in part by the National Institute on Aging of National Institutes of Health (Grant AG028709). This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. All simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.lang-muir.7b00414.
More real-time AFM observation of P5 self-assembly at different concentrations on HOPG, and the two different initial conformations of monomeric P5 and their different simulation results (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Kagan BL Amyloidosis and protein folding. Science 2005, 307 (5706), 42b–43b. [DOI] [PubMed] [Google Scholar]
- (2).DeToma AS; Salamekh S; Ramamoorthy A; Lim MH Misfolded proteins in Alzheimer’s disease and type II diabetes. Chem. Soc. Rev. 2012, 41 (2), 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Goedert M; Spillantini MG A century of Alzheimer’s disease. Science 2006, 314 (5800), 777–781. [DOI] [PubMed] [Google Scholar]
- (4).Choi JS; Braymer JJ; Nanga RP; Ramamoorthy A; Lim MH Design of small molecules that target metal-Aβ species and regulate metal-induced Aβ aggregation and neurotoxicity. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (51), 21990–21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Yoo SI; Yang M; Brender JR; Subramanian V; Sun K; Joo NE; Jeong SH; Ramamoorthy A; Kotov NA Inhibition of Amyloid Peptide Fibrillation by Inorganic Nanoparticles: Functional Similarities with Proteins. Angew. Chem., Int. Ed. 2011, 50, 5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Eisenberg DS; Sawaya MR Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 10.1146/annurev-biochem-061516-045104 [DOI] [PubMed] [Google Scholar]
- (7).Brender JR; Lee EL; Cavitt MA; Gafni A; Steel DG; Ramamoorthy A Amyloid fiber formation and membrane disruption are separate processes localized in two distinct regions of IAPP, the type-2-diabetes-related peptide. J. Am. Chem. Soc. 2008, 130 (20), 6424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Arispe N; Rojas E; Pollard HB Alzheimer-disease amyloid beta-protein forms calcium channels in bilayer-membranes - blockade by tromethamine and aluminum. Proc. Natl. Acad. Sci. U. S. A. 1993, 90 (2), 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Jang H; Arce FT; Ramachandran S; Kagan BL; Lal R; Nussinov R Disordered amyloidogenic peptides may insert into the membrane and assemble into common cyclic structural motifs. Chem. Soc. Rev. 2014, 43 (19), 6750–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jang H; Arce FT; Ramachandran S; Capone R; Azimova R; Kagan BL; Nussinov R; Lal R Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer’s Disease and Down syndrome. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (14), 6538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yin Y; Lu Y; Gates B; Xia Y Template-Assisted SelfAssembly: A Practical Route to Complex Aggregates of Monodispersed Colloids with Well-Defined Sizes, Shapes, and Structures. J. Am. Chem. Soc. 2001, 123 (36), 8718–8729. [DOI] [PubMed] [Google Scholar]
- (12).Zhang F; Du HN; Zhang ZX; Ji LN; Li HT; Tang L; Wang HB; Fan CH; Xu HJ; Zhang Y; Hu J; Hu HY; He JH Epitaxial growth of peptide nanofilaments on inorganic surfaces: Effects of interfacial hydrophobicity/hydrophilicity. Angew. Chem., Int. Ed. 2006, 45 (22), 3611–3613. [DOI] [PubMed] [Google Scholar]
- (13).Hou JH; Du QQG; Zhong RB; Zhang P; Zhang F Temperature manipulating peptide self-assembly in water nanofilm. Nucl. Sci. Tech 2014, 25 (6), 060502. [Google Scholar]
- (14).Du Q; Dai B; Hou J; Hu J; Zhang F; Zhang Y A comparative study on the self-assembly of an amyloid-like peptide at water-solid interfaces and in bulk solutions. Microsc. Res. Tech. 2015, 78 (5), 375–381. [DOI] [PubMed] [Google Scholar]
- (15).Kowalewski T; Holtzman DM In situ atomic force microscopy study of Alzheimer’s beta-amyloid peptide on different substrates: New insights into mechanism of beta-sheet formation. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (7), 3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Brown CL; Aksay IA; Saville DA; Hecht MH Template-directed assembly of a de novo designed protein. J. Am. Chem. Soc. 2002, 124 (24), 6846–6848. [DOI] [PubMed] [Google Scholar]
- (17).Hoyer WG; Cherny D; Subramaniam V; Jovin TM Rapid self-assembly of alpha-synuclein observed by in situ atomic force microscopy. J. Mol. Biol. 2004, 340 (1), 127–139. [DOI] [PubMed] [Google Scholar]
- (18).Kellermayer MS; Karsai A; Benke M; Soos K; Penke B Stepwise dynamics of epitaxially growing single amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (1), 141–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hamley IW Peptide Fibrillization. Angew. Chem., Int. Ed. 2007, 46 (43), 8128–8147. [DOI] [PubMed] [Google Scholar]
- (20).Krysmann MJ; Castelletto V; Kelarakis A; Hamley IW; Hule RA; Pochan DJ Self-Assembly and Hydrogelation of an Amyloid Peptide Fragment. Biochemistry 2008, 47 (16), 4597–4605. [DOI] [PubMed] [Google Scholar]
- (21).Watanabe K; Nakamura K; Akikusa S; Okada T; Kodaka M; Konakahara T; Okuno H Inhibitors of fibril formation and cytotoxicity of beta-amyloid peptide composed of KLVFF recognition element and flexible hydrophilic disrupting element. Biochem. Biophys. Res. Commun. 2002, 290 (1), 121–4. [DOI] [PubMed] [Google Scholar]
- (22).Arai T; Sasaki D; Araya T; Sato T; Sohma Y; Kanai M A cyclic KLVFF-derived peptide aggregation inhibitor induces the formation of less-toxic off-pathway amyloid-beta oligomers. ChemBioChem 2014, 15 (17), 2577–83. [DOI] [PubMed] [Google Scholar]
- (23).Hamley IW; Nutt DR; Brown GD; Miravet JF; Escuder B; Rodriguez-Llansola F Influence of the solvent on the selfassembly of a modified amyloid beta peptide fragment. II. NMR and computer simulation investigation. J. Phys. Chem. B 2010, 114 (2), 940–51. [DOI] [PubMed] [Google Scholar]
- (24).Gordon DJ; Sciarretta KL; Meredith SC Inhibition of βAmyloid(40) Fibrillogenesis and Disassembly of β-Amyloid(40) Fibrils by Short β-Amyloid Congeners Containing N-Methyl Amino Acids at Alternate Residues†. Biochemistry 2001, 40 (28), 8237–8245. [DOI] [PubMed] [Google Scholar]
- (25).Gordon DJ; Tappe R; Meredith SC Design and characterization of a membrane permeable N-methyl amino acidcontaining peptide that inhibits Aβ1–40 fibrillogenesis. J. Pept. Res. 2002, 60 (1), 37–55. [DOI] [PubMed] [Google Scholar]
- (26).Zhang G; Leibowitz MJ; Sinko PJ; Stein S Multiplepeptide conjugates for binding beta-amyloid plaques of Alzheimer’s disease. Bioconjugate Chem. 2003, 14 (1), 86–92. [DOI] [PubMed] [Google Scholar]
- (27).Sundaram RK; Kasinathan C; Stein S; Sundaram P Novel Detox Gel Depot sequesters beta-Amyloid Peptides in a mouse model of Alzheimer’s Disease. Int. J. Pept. Res. Ther. 2012, 18 (2), 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kokkoni N; Stott K; Amijee H; Mason JM; Doig AJ NMethylated peptide inhibitors of beta-amyloid aggregation and toxicity. Optimization of the inhibitor structure. Biochemistry 2006, 45 (32), 9906–18. [DOI] [PubMed] [Google Scholar]
- (29).Arispe N; Diaz JC; Flora M Efficiency of HistidineAssociating Compounds for Blocking the Alzheimer’s A beta Channel Activity and Cytotoxicity. Biophys. J. 2008, 95 (10), 4879–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Petkova AT; Yau WM; Tycko R Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry 2006, 45 (2), 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Xiao Y; Ma B; McElheny D; Parthasarathy S; Long F; Hoshi M; Nussinov R; Ishii Y Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015, 22 (6), 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Arce FT; Jang H; Ramachandran S; Landon PB; Nussinov R; Lal R Polymorphism of amyloid beta peptide in different environments: implications for membrane insertion and pore formation. Soft Matter 2011, 7 (11), 5267–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Brooks BR; Brooks CL 3rd; Mackerell AD Jr.; Nilsson L; Petrella RJ; Roux B; Won Y; Archontis G; Bartels C; Boresch S; Caflisch A; Caves L; Cui Q; Dinner AR; Feig M; Fischer S; Gao J; Hodoscek M; Im W; Kuczera K; Lazaridis T; Ma J; Ovchinnikov V; Paci E; Pastor RW; Post CB; Pu JZ; Schaefer M; Tidor B; Venable RM; Woodcock HL; Wu X; Yang W; York DM; Karplus M CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009, 30 (10), 1545–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Klauda JB; Venable RM; Freites JA; O’Connor JW; Tobias DJ; Mondragon-Ramirez C; Vorobyov I; MacKerell AD Jr.; Pastor RW Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 2010, 114 (23), 7830–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Jang D; Na W; Kang M; Kim N; Shin S Ultrasensitive Detection of Single-Walled Carbon Nanotubes Using Surface Plasmon Resonance. Anal. Chem. 2016, 88 (1), 968–973. [DOI] [PubMed] [Google Scholar]
- (36).Jang H; Connelly L; Arce FT; Ramachandran S; Kagan BL; Lal R; Nussinov R Mechanisms for the Insertion of Toxic, Fibril-like beta-Amyloid Oligomers into the Membrane. J. Chem. Theory Comput. 2013, 9 (1), 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Jang H; Arce FT; Ramachandran S; Kagan BL; Lal R; Nussinov R Familial Alzheimer’s disease Osaka mutant (▽E22) βbarrels suggest an explanation for the different Aβ1–40/42 preferred conformational states observed by experiment. J. Phys. Chem. B 2013, 117 (39), 11518–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kale L; Schulten K Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26 (16), 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hillner P; Manne S; Hansma P; Gratz t. l. A. Atomic force microscope: a new tool for imaging crystal growth processes. Faraday Discuss. 1993, 95, 191–197. [Google Scholar]
- (40).Hansma P; Elings V; Marti O; Bracker C Scanning tunneling microscopy and atomic force microscopy: application to biology and technology. Science 1988, 242 (4876), 209–216. [DOI] [PubMed] [Google Scholar]
- (41).Zhang F; Zhang P; Hou J; Yun X; Li W; Du Q; Chen Y Large scale anomalous patterns of muscovite mica discovered by atomic force microscopy. ACS Appl. Mater. Interfaces 2015, 7 (16), 8699–705. [DOI] [PubMed] [Google Scholar]
- (42).Parbhu A; Lin H; Thimm J; Lal R Imaging real-time aggregation of amyloid beta protein (1–42) by atomic force microscopy. Peptides 2002, 23 (7), 1265–70. [DOI] [PubMed] [Google Scholar]
- (43).Kang S. g.; Li H; Tien H; Zhang F; Xia Z; Zhang Y; Zhou R Molecular Mechanism of Surface-Assisted Epitaxial Self-Assembly of Amyloid-like Peptides. ACS Nano 2012, 6 (10), 9276–9282. [DOI] [PubMed] [Google Scholar]
- (44).Kang SG; Huynh T; Xia Z; Zhang Y; Fang HP; Wei GH; Zhou RH Hydrophobic Interaction Drives Surface-Assisted Epitaxial Assembly of Amyloid-like Peptides. J. Am. Chem. Soc. 2013, 135 (8), 3150–3157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


