Abstract
The CD274 (PD-L1)/PDCD1 (PD-1) pathway is crucial for the modulation of immune responses and self-tolerance. Aberrantly expressed CD274 allows tumor cells to evade host immune system and is considered to be a mechanism of adaptive immune resistance. Inhibition of the CD274/PDCD1 immune checkpoint offers a promising new therapeutic strategy. Although CD274-expressing tumor cells have been identified in different types of tumors including colorectal cancer, clinicopathologic profile of these CD274-positive tumors has not been extensively studied. In this study, 454 primary colorectal carcinomas were analyzed histologically and immunohistochemically for CD274, mismatch repair (MMR) proteins, intestinal differentiation marker (CDX2), and stem cell markers (ALCAM, ALDH1A1, and SALL4). CD274-positive colorectal carcinomas (54/454 (12%)) usually (83%) involved the right or transverse colon with poorly differentiated and solid/medullary histology. On the basis of multivariate logistic regression analysis, CD274 positivity was significantly associated with poorly differentiated histotype (OR: 3.32; 95% CI: 1.46–7.51; P = 0.004), MMR deficiency (OR: 10.0; 95% CI: 4.66–21.5; P<0.001), and ‘stem-like’ immunophenotype defined by the loss or weak expression of CDX2 and ALCAM-positivity (OR: 5.51; 95% CI: 1.66–18.3; P = 0.005). Mutation analysis of 66 arbitrary selected colorectal carcinomas revealed that CD274-positive tumors usually (88%) carried the BRAF V600E mutation. Thus, colorectal carcinomas defined by CD274 positivity displayed features associated with tumors arising via the serrated neoplasia pathway. Moreover, colorectal carcinomas characterized by lack of CDX2 and prominent expression of ALCAM frequently (71%) showed CD274 positivity. This might suggest association of CD274 expression with ‘stem-like’ phenotype. Further evaluation of a larger cohort or experimental analyses would be needed to confirm this notion.
Introduction
The PD-Ls (programmed cell death ligands)/PDCD1 (programmed cell death 1, PD-1) axis are crucial for the modulation of the immune system to reduce collateral tissue damage from the inflammatory response in peripheral tissues as well as for T-cell responses and the maintenance of self-tolerance to avoid autoimmune diseases.1,2
CD274 (B7-H1, PD-L1) and PDCD1LG2 (B7-DC, CD273, PD-L2), two physiological ligands for PDCD1, have been identified as cell-surface glycoproteins belonging to the B7 family. In peripheral tissues, CD274 is primarily induced by interferon-γ (IFNγ), from T helper 1 (TH1) cells under inflammatory conditions.3–6
PDCD1 is a cell-surface receptor that belongs to the immunoglobulin superfamily and is mainly expressed on activated T cells, whereas other non-T lymphocytes such as B cells and natural killer (NK) cells express PDCD1 only on induction.7,8 PDCD1, once engaged by its ligands, inhibits kinases that are involved in T-cell activation through the phosphatase SHP2.4 When T cells were exposed to chronic antigen stimulation such as chronic viral infection or cancer, high levels of persistent PDCD1 expression is induced and leads to T-cell exhaustion or anergy.9
Variable CD274 expression has been reported in different types of tumors, including esophageal, gastric, and lung cancers, and linked to tumor aggressiveness and a poor prognosis. 10–13 In colorectal cancer, CD274 expression was reported to associate positively with mismatch repair (MMR)-deficient phenotype, BRAF mutation, or cytotoxic tumor-infiltrating lymphocytes, and inversely with regulatory T lymphocytes.13–15 However, the impact of CD274 expression on clinical outcome of colorectal cancer patients is controversial.15–17 Furthermore, the biological characteristics, such as stem cell features, of CD274-expressing colorectal cancers have not been fully identified.
A majority of colorectal carcinomas arise from conventional adenomas via a classical adenomacarcinoma pathway leading to microsatellite stable carcinomas. However, a subset of adenocarcinomas arises along the serrated neoplasia pathway from the sessile serrated adenomas/polyps. These tumors are characterized by mutational BRAF activation, CpG island methylator and MMR-deficient phenotypes, and loss of CDX2 (Caudal-type homeobox transcription factor 2), a marker for intestinal differentiation.18,19
Stem cell markers such as ALCAM (activated leukocyte cell adhesion molecule, CD166), ALDH1A1 (aldehyde dehydrogenase 1 family member A1, ALDH1), and SALL4 (spalt like transcription factor 4), are variably expressed in colorectal carcinomas and linked to unfavorable clinical outcome in some studies.20–23 More recently, expression of CDX2 was found to be inversely correlated with expression of ALCAM. Furthermore, loss of CDX2 expression was a poor prognostic indicator in early stage (stage II and stage III) colon carcinoma. 24
In this study, comprehensive immunohistochemistry of MMR molecules, intestinal differentiation (CDX2), and stem cell markers (ALCAM, ALDH1A1, and SALL4) and mutation analyses for BRAF, KRAS, NRAS, PIK3CA, and GNAS, were performed to characterize CD274-positive colorectal carcinomas.
Material and Methods
Primary 454 colorectal carcinomas and 46 normal colonic and small intestinal mucosa derived from colon carcinoma specimens were assembled to multitumor blocks containing up to 50 rectangular tissue samples as previously described.25 The size of tumor tissue samples was estimated to exceed the size of a single 0.6 mm2 core by a factor of 10–15. All tumors selected for this study were extensively characterized histologically. Also, colonic (n = 43) and terminal ileal (n = 3) mucosa adjacent to the tumor was analyzed for expression of the studied markers: CD274, MMR proteins, CDX2, ALCAM, ALDH1A1, and SALL4.
The antibody source, dilution, and staining protocols are summarized in Supplementary Table S1. All immunostaining was performed with the Leica Bond-Max automation and Leica Refine detection kit (Leica Biosystems, Bannockburn, IL, USA). The protocol included in situ deparaffinization and epitope retrieval for 25 min, incubation with primary antibody for 30 min, polymer for 15 min, postpolymer for 15 min, and DAB chromogen for 10 min, followed by 5- min hematoxylin counterstain. For the sequential double staining, second signal was visualized by Fast Red chromogen. Immunohistochemistry of MLH1, MSH2, MSH6, and PMS2 were performed as previously reported.26 The immunostains were independently evaluated by two pathologists (SI and MM). The concordance rates of initial immunohistochemical evaluation are shown in Supplementary Table S2. For the discordant cases, the results were confirmed by the discussion of the two pathologists. Immunoreactivity of CD274 (membrane and/or cytoplasm), ALCAM (membrane), ALDH1A1 (cytoplasm), and SALL4 (nucleus) was evaluated with a detection cut-off of 5%, whereas CDX2 immune reactivity (nucleus) was classified into two categories; strongly-positive (same or stronger than the normal colonic mucosa) and weekly-positive/negative expression (weaker than the normal colonic mucosa or loss of expression). Representative images of immunohistochemistry are shown in Figure 1 and Supplementary Figure S1.
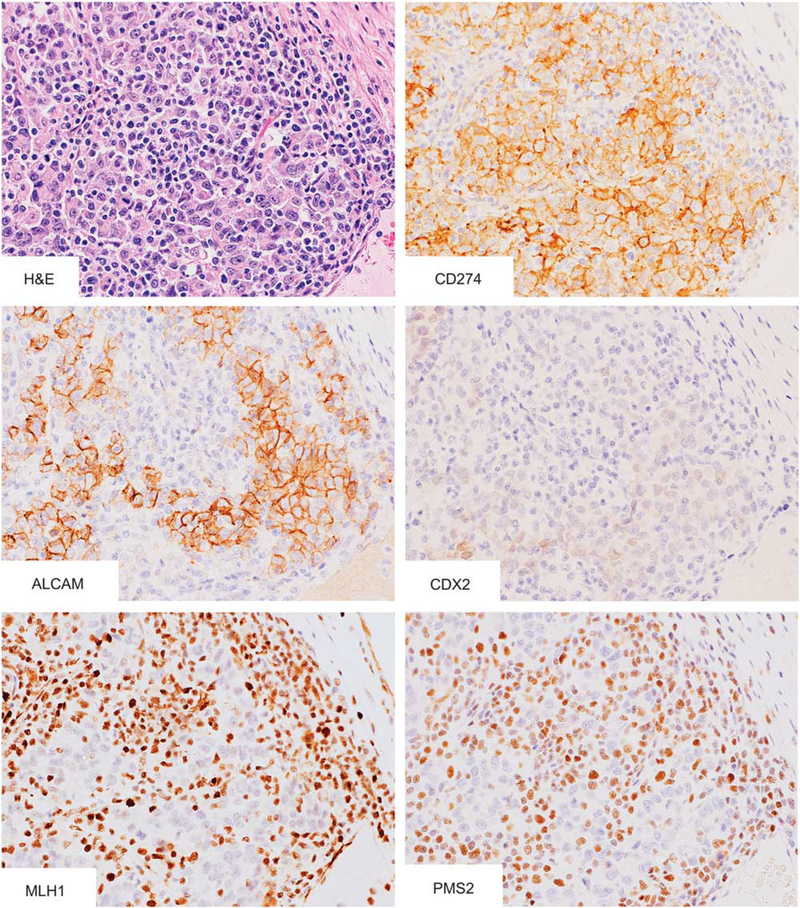
Figure 1.
Histology and immunophenotypes of colon carcinomas with diffuse CD274 expression. A case of poorly differentiated adenocarcinoma with massive lymphoplasmacytic infiltration showed diffuse CD274 and ALCAM expression on the cell membrane. Nuclear CDX2, MLH1, and PMS2 were expressed under detectable levels in tumor cells.
BRAF, KRAS, NRAS, PIK3CA, and GNAS mutation analyses were done as previously reported.26 Primer sequences, PCR conditions and size of amplicons are provided in Supplementary Table S3.
Chi-square test or Fisher’s exact test was performed with EZR version 1.32. software27 to analyze the statistical correlation between categorical data. Simple Boneferroni correction for multiple hypothesis testing were applied to adjusted two-sided alpha level at 0.0041 (=0.05/12). ‘Stem-like’ immunophenotype was defined by CDX2-negativ-ity/weakly-positivity and ALCAM-positivity. Multivariate logistic regression analysis was also performed with EZR version 1.32. software to analyze the association of CD274 expression (dependent variable) and other factors (independent variables). Variables with P-value < 0.05 on univariate analysis, such as age ( <69 vs >470 years old), tumor location (cecum and ascending colon vs transverse colon including hepatic and splenic flexures vs descending to sigmoid colon vs rectum), tumor differentiation (well to moderate vs poor including signet ring cell-like feature), solid/medullary histology with a threshold of 50% of area (present vs absent), MMR status (preserved vs deficient), and ‘stem-like’ immunophenotype (CDX2-weakly-positive/negative and ALCAM-positive vs CDX2-strongly-positive and/or ALCAM-negative), were included in the initial multivariable logistic regression analysis model. A backward elimination with a threshold of P = 0.05 was used to select variables in the final model. Cases with missing information were eliminated from the statistical analysis of that parameter.
Results
CD274 Expression in Colorectal Cancers
Fifty-four (30 on the cell membrane and 24 in the cytoplasm) of 454 colorectal carcinomas showed variable CD274 expression (12% of cases, with 5–100% of positive cells, median 60%). Clinical, pathological, and immunohistochemical features of analyzed tumors have been summarized in Tables 1A, 1B, and Table 2. Representative images of the colon carcinomas with diffuse CD274 expression on the cell membrane are shown in Figure 1. CD274 positivity tended to occur in older patients (mean age: 72.6±13.1 years). CD274-expressing tumors were found more frequently in the right or transverse colon with poorly differentiated and/or solid/medullary histology. Weak or negative expression of CDX2 was seen in 54% of CD274-positive colorectal tumors. ‘Stem-like’ markers, ALCAM, ALDH1A1, and SALL4 were expressed in 48, 33, and 5%, respectively, in CD274-positive tumors. However, there was no significant difference in expression of ALDH1A1 and SALL4 between CD274 positive and negative tumors. Supplementary Table S4 presents comparison of CDX2 expression and all three stem cell markers. Multivariate logistic regression analysis revealed significant association between CD274 expression and poorly differentiated histotype (OR: 3.32; 95% CI: 1.46–7.51; P = 0.004), MMR deficiency (OR: 10.0; 95% CI: 4.66–21.5; P <0.001) and ‘stem-like’ immunophenotype (OR: 5.51; 95% CI: 1.66–18.3; P =0.005).
Table 1A.
Clinicopathological characteristics of 454 colorectal carcinomas with or without CD274 expression
|
CD274 expression |
||||
|---|---|---|---|---|
|
Total no. 454 (100%) [100%] |
Positive 54 (12%) [100%] |
Negative 400 (88%) [100%] |
P-value | |
| Sex | 0.24a | |||
| Male | 202 (100%) [46%] |
20 (10%) [38%] |
182 (90%) [47%] |
|
| Female | 235 (100%) [54%] |
32 (14%) [62%] |
203 (86%) [53%] |
|
| Age, years (mean ± s.d.) | 68.1 ± 13.3 | 72.6 ± 13.1 | 67.4 ± 13.2 | 0.0080b |
| Tumor size, cm (mean ± s.d.) | 5.6 ± 2.6 | 6.1 ± 2.4 | 5.5 ± 2.6 | 0.19b |
| Tumor location | < 0.001a | |||
| Cecum and ascending colon | 191 (100%) [46%] |
33 (17%) [66%] |
158 (83%) [44%] |
|
| Transverse colon | 60 (100%) [15%] |
10 (17%) [20%] |
50 (83%) [14%] |
|
| Descending and sigmoid colon | 126 (100%) [31%] |
6 (5%) [12%] |
120 (95%) [33%] |
|
| Rectum | 36 (100%) [9%] |
1 (3%) [2%] |
35 (97%) [10%] |
|
| Tumor differentiation | < 0.001a | |||
| Well to moderate | 388 (100%) [85%] |
26(7%) [48%] |
362 (93%) [91%] |
|
| Poor | 66 (100%) [15%] |
28 (42%) [52%] |
38 (58%) [10%] |
|
| Mucinous histology | 0.058a | |||
| 0–49% | 418 (100%) [92%] |
46 (11%) [85%] |
372 (89%) [93%] |
|
| 50%– | 36 (100%) [8%] |
8 (22%) [15%] |
28 (78%) [7%] |
|
| Solid/medullary histology | < 0.001a | |||
| 0–49% | 417 (100%) [92%] |
34 (8%) [63%] |
383 (92%) [96%] |
|
| 50%– | 37 (100%) [8%] |
20 (54%) [37%] |
17 (46%) [4%] |
|
P-values were calculated by the χ2 test for CD274 expression.
T-test was used to compare the means of age and tumor size. The Bonferroni-corrected P-value for significance was P = 0.0041 (0.05/12).
Table 1B.
Immunohistochemical characteristics of 454 colorectal carcinomas with or without CD274 expression
|
CD274 expression |
||||
|---|---|---|---|---|
|
Total No. 454 (100%) [100%] |
Positive 54 (12%) [100%] |
Negative 400 (88%) [100%] |
P-value | |
| MMR status | < 0.001 | |||
| Deficient | 88 (100%) [19%] |
40 (45%) [74%] |
48 (55%) [12%] |
|
| Preserved | 366 (100%) [81%] |
14 (4%) [26%] |
352 (96%) [88%] |
|
| CDX2 | < 0.001 | |||
| Negative/weakly-positive | 53 (100%) [12%] |
29 (55%) [54%] |
24 (45%) [6%] |
|
| Strongly-positive | 401 (100%) [88%] |
25 (6%) [46%] |
376 (94%) [94%] |
|
| ALCAM | < 0.001 | |||
| Positive | 107 (100%) [24%] |
26 (24%) [48%] |
81 (76%) [20%] |
|
| Negative | 347 (100%) [77%] |
28 (8%) [52%] |
319 (92%) [80%] |
|
| ALDH1A1 | 0.52 | |||
| Positive | 132 (100%) [29%] |
18 (14%) [33%] |
114 (86%) [29%] |
|
| Negative | 322 (100%) [71%] |
36 (11%) [67%] |
286 (89%) [72%] |
|
| SALL4 | 1 | |||
| Positive | 22 (100%) [5%] |
2 (9%) [5%] |
20 (91%) [5%] |
|
| Negative | 432 (100%) [95%] |
52 (12%) [96%] |
380 (88%) [95%] |
|
| ‘Stem-like’ immune phenotype | < 0.001 | |||
| CDX2-negative/-weak and ALCAM-positive | 24 (100%) [5%] |
17 (71%) [31%] |
7 (29%) [2%] |
|
| CDX2-strong and/or ALCAM-negative | 428 (100%) [95%] |
37 (9%) [69%] |
391 (91%) [98%] |
|
P-values were calculated by the χ2 test for CD274 expression. The Bonferroni-corrected P-value for significance was P = 0.0041 (0.05/12).
Table 2.
Multivariate logistic regression analysis to assess association between CD274 expression and other factors
|
95% CI |
||||
|---|---|---|---|---|
| Odds ratio | Min | Max | P-value | |
| Poorly differentiated histotype | 3.32 | 1.46 | 7.51 | 0.004 |
| MMR deficiency | 10.0 | 4.66 | 21.5 | < 0.001 |
| ‘Stem-like’ immune phenotype | 5.51 | 1.66 | 18.3 | 0.005 |
The multivariable logistic regression analysis model initially included age, tumor location, tumor differentiation, solid/medullary histology, MMR status and ‘stem-like’ immune phenotype. A backward elimination with a threshold of P = 0.05 was used to select variables in the final model.
Mutation Analysis in Colorectal Cancers
The results of pathological, immunohistochemical, and mutation analyses of 66 arbitrary selected colorectal carcinomas from the study group are summarized in Table 3 and Supplementary Table S5. Most CD274-expressing tumors (14/16, (88%)) were BRAF mutants with a MMR-deficient phenotype. Only one CD274-positive case (1/16 (6%)) carried KRAS c.35G>A (G12D) mutation. In contrast, CD274-negative cases were frequently KRAS (26/50 (52%)) and rarely BRAF (5/50 (10%)) mutants with or without PIK3CA mutation. Statistical analysis showed significant correlation between CD274 expression and BRAF mutation (Table 4). BRAF and KRAS mutations were observed in a mutually exclusive manner. No GNAS or NRAS mutations were detected in CD274-positive colorectal carcinomas.
Table 3.
Characterization of 16 CD274-positive colorectal carcinomas
| Age | Sex | Location | Size (cm) | Tumor differentiation | Mucinous histology | Solid/medullary histology | CD274 (%) Localization | CDX2 | ALCAM | ALDH1A1 | SALL4 | MLH1 | PMS2 | MSH2 | MSH6 | BRAF | KRAS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 66 | F | HF | 8 | Poor | − | + | 100 | Membrane | − | + | − | − | − | − | + | − | MUT | WT |
| 62 | F | Colon NOS | — | Poor | − | − | 100 | Membrane | − | + | − | − | − | − | + | + | MUT | WT |
| 78 | M | T | 7 | Poor | − | + | 100 | Membrane | − | + | − | − | − | − | + | + | MUT | WT |
| 65 | F | S | 5 | Poor | − | + | 100 | Membrane | − | − | − | − | − | − | + | + | MUT | WT |
| 85 | M | Colon NOS | 6 | Well | − | − | 100 | Cytoplasm | + | + | − | − | − | − | + | + | MUT | WT |
| 86 | F | C | 4 | Well | + | − | 100 | Cytoplasm | + | − | + | − | − | − | + | + | MUT | WT |
| 77 | F | A | 6 | Poor | − | + | 80 | Cytoplasm | + | − | − | − | − | − | + | + | MUT | WT |
| 70 | F | A | 10 | Poor | − | + | 80 | Cytoplasm | + | + | + | − | − | − | + | + | MUT | WT |
| 81 | M | A | 8 | Moderate | − | − | 80 | Cytoplasm | + | + | + | − | − | − | + | + | MUT | WT |
| 80 | F | C | 9 | Moderate | − | − | 40 | Membrane | + | − | − | − | + | + | + | + | WT | WT |
| 65 | M | C | 6.5 | Well | − | − | 40 | Cytoplasm | + | − | − | − | − | − | + | + | MUT | WT |
| 61 | F | C | 4 | Well | − | − | 30 | Cytoplasm | + | − | − | − | − | − | + | + | MUT | WT |
| 66 | F | C | 3.5 | Well | − | − | 20 | Membrane | + | − | + | − | + | + | + | + | WT | Codon12 GGT>GAT |
| 71 | F | C | 5 | Well | − | − | 20 | Cytoplasm | + | + | + | − | − | − | + | + | MUT | WT |
| 82 | F | C | 7 | Moderate | − | − | 10 | Cytoplasm | + | − | + | − | − | − | + | + | MUT | WT |
| 77 | F | A | 4 | Moderate | − | − | 10 | Cytoplasm | + | − | + | − | − | − | + | + | MUT | WT |
Abbreviations: A, ascending colon; C, cecum; HF, hepatic flexure; T, transverse colon. WT, wild type. MUT, c.1799T > A (BRAF V600E) mutant.
No mutation was found in NRAS, PIK3CA, and GNAS.
Table 4.
BRAF and KRAS mutants in 66 colorectal carcinomas
|
CD274 expression |
||||
|---|---|---|---|---|
|
Total No. 66 (100%) [100%] |
Positive 16 (24%) [100%] |
Negative 50 (76%) [100%] |
P-value | |
| Gene mutation | P < 0.001a | |||
| BRAF mutant | 19 (100%) [29%] |
14 (74%) [88%] |
5 (25%) [10%] |
|
| KRAS mutant | 27 (100%) [41%] |
1 (4%) [6%] |
26 (96%) [52%] |
|
| Wild type | 20 (100%) [30%] |
1 (5%) [6%] |
19 (95%) [38%] |
|
P-value was calculated by the Fisher’s exact test for CD274 expression. The Bonferroni-corrected P-value for significance was P = 0.017(0.05/3). P < 0.001 (BRAF mutant vs KRAS mutant), P < 0.001 (BRAF mutant vs wild type).
CD274 Expression in Normal Intestinal Epithelium
No CD274 expression was detected in 46 normal intestinal mucosal tissues. Also, double staining for CD274 (red) and CDX2 (brown) showed no CD274 expression in CDX2-negative epithelial cells (Figures 2a and b). A case of CDX2-negative and CD274-positive tumor was studied as a control (Figure 2c). Representative images of the ALDH1A1 and ALCAM staining in normal epithelia are shown in Supplementary Figure S2. No SALL4 expression was detected in the normal intestinal epithelium (data not shown).
Figure 2.
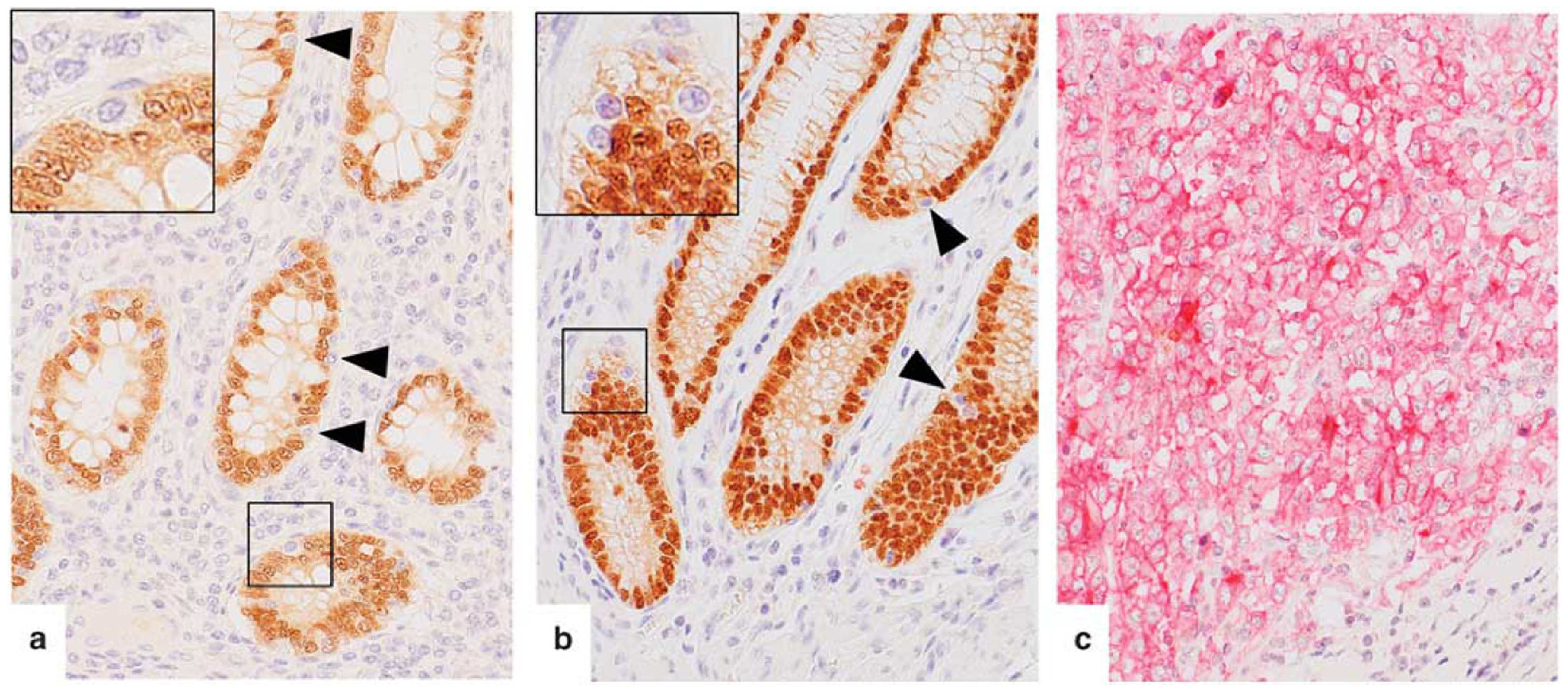
CDX2 and CD274 expression in normal intestinal epithelium and colon carcinoma. (a–c) Sequential immunostaining of CDX2 (brown) and CD274 (red). (a,b) Normal intestinal epithelium including CDX2-negative epithelial cells of the terminal ileum (a) and colon (b) showed no CD274 expression. (c) A case of poorly differentiated adenocarcinoma of the colon with diffuse CD274 expression on the cell membrane without nuclear CDX2.
Discussion
In this study, 454 well characterized colorectal carcinomas were screened immunohistochemically for CD274, CDX2 (intestinal differentiation marker), and stem cell markers (ALCAM, ALDH1A1, and SALL4) expression. In addition, mutational status of BRAF, KRAS, NRAS, PIK3CA, and GNAS was evaluated in arbitrary selected cases.
CD274-positive colorectal carcinomas, represented 12% of analyzed cases, tend to occur in older patients and frequently involve the right or transverse colon. Significant association between CD274 expression and poorly differentiated histology, solid/medullary histology, MMR deficiency, ‘stem-like’ characteristics, and BRAF c.1799T>A (BRAF V600E) mutation was detected.
Approximately 15% of colorectal carcinomas are believed to develop through the serrated neoplasia pathway, which is an alternative pathway to the classical adenoma-carcinoma pathway characterized by mutations in APC, KRAS, and TP53. Colorectal carcinomas of serrated neoplasia pathway frequently show mucinous and/or poorly differentiated histology, CpG island methylator phenotype and MMR deficiency, mutational BRAF activation, and in some cases, CDX2-negativity.18 These features correspond to the characteristics of the CD274-positive colorectal cancers.
CD274 is variably expressed in subsets of many tumor types and allows tumors to evade host immune system. CD274/PDCD1 immune checkpoint inhibitors were introduced to cancer treatment and, in some cases, showed significant anti-cancer effects. Thus, CD274 immunohistochemistry has been used as a potential biomarker to predict clinical response to these drugs.28–31 However, in some cases, CD274/PDCD1 immunotherapy has shown anti-cancer effects in spite of low CD274 expression (1% or less positive cells).29 This has led to the hypothesis that CD274 might be expressed on cancer cells harboring specific characteristics, such as tumor initiating cells or cancer stem cells.
Colorectal cancer stem cells characterized by the expression of markers such as ALCAM, ALDH1A1, and SALL4 display tumor initiating and highly proliferative potentials in tumor xenograft models.21,32 Furthermore, high content of colorectal cancer cells with stem cell marker-expression in surgically resected specimens was reported to worsen the clinical outcome.20–23 In this study, an inverse correlation between ALCAM and CDX2 expression, which was recently pointed out by gene expression screening,24 was confirmed by immunohistochemistry. Furthermore, it was uncovered that colorectal carcinomas with ‘stem-like’ immunophenotype frequently (71%) showed CD274 positivity. This observation might suggest association of CD274 expression with ‘stem-like’ phenotype such as highly proliferative and tumor initiating capability, however, further evaluation by a large cohort or experimental analyses would be necessary for the confirmation. In the present study, prognostic analysis by the expression status of CD274 and other molecules was not done because of the lack of clinical outcome data. Although several recent studies showed the association between CD274 expression and poor clinical outcome in the patients with MMR-deficient colorectal cancer,15,17 further study would also be needed to clarify this point.
In the present study, no CD274 expression was detected immunohistochemically in normal intestinal epithelial tissue including CDX2-negative epithelial cells. Thus, similar to other tumors,33,34 aberrantly activated oncogenic signaling might be responsible for CD274 expression in colorectal carcinoma cells. Also, loss of CDX2 or activation of other pathways including genes encoding ‘stem-like’ markers might accelerate CD274 expression. Further studies are needed to better understand the biological mechanism underlying CD274 expression in colorectal carcinomas.
In the present study, according to the past report,13 immunohistochemistry using monoclonal anti-CD274 antibody clone E1L3N with a detection cut-off 5% was applied to colorectal carcinomas. However, there are many clinicopathological studies showing variable CD274 positivity (5–89%) using different antibodies and/or cut-off values for the evaluation of CD274.14–17 Furthermore, as a ‘complementary diagnostics’, independent immunohis tochemical staining methods using different antibodies and/or cut-off value were applied to four different anti-PD-Ls/PDCD1 immune checkpoint inhibitors (eg, nivolumab and pembrolizumab against PDCD1 or MPDL3280A and MEDI-4736 against CD274). In the near future, standardized CD274 immunohistochemistry should be established for anti-PD-Ls/PDCD1 immune checkpoint therapy.
In summary, this study characterized colorectal carcinomas expressing CD274. The tumors defined by CD274 positivity were associated with poorly differentiated and solid/medullary histology, MMR deficiency and mutational BRAF activation, features typically seen in adenocarcinomas arising via the serrated neoplasia pathway. Moreover, significant numbers of colorectal carcinomas with ‘stem-like’ immunophenotype expressed CD274. Further evaluation of a large cohort of cases, or experimental analyses, could confirm the ‘stem-like’ phenotypes of CD274-positve colorectal cancers.
Supplementary Material
Acknowledgments
We thank Dr Yasuyuki Arai (National Institutes of Health) for advice on statistical analyses. This work is supported as a part of National Cancer Institute’s intramural research program.
Footnotes
Disclosure/conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on Modern Pathology website (http://www.nature.com/modpathol)
References
- 1.Nishimura H, Honjo T. PD-1: an inhibitory immunoreceptor involved in peripheral tolerance. Trends Immunol 2001;22:265–268. [DOI] [PubMed] [Google Scholar]
- 2.Nishimura H, Nose M, Hiai H, et al. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 1999;11:141–151. [DOI] [PubMed] [Google Scholar]
- 3.Dong H, Zhu G, Tamada K, et al. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med 1999; 5:1365–1369. [DOI] [PubMed] [Google Scholar]
- 4.Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000;192:1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latchman Y, Wood CR, Chernova T, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol 2001;2:261–268. [DOI] [PubMed] [Google Scholar]
- 6.Tseng SY, Otsuji M, Gorski K, et al. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp Med 2001;193:839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanoni D, Tavecchio S, Recalcati S, et al. New monoclonal antibodies against B-cell antigens: possible new strategies for diagnosis of primary cutaneous B-cell lymphomas. Immunol Lett 2011;134:157–160. [DOI] [PubMed] [Google Scholar]
- 8.Terme M, Ullrich E, Aymeric L, et al. IL-18 induces PD-1-dependent immunosuppression in cancer. Cancer Res 2011;71:5393–5399. [DOI] [PubMed] [Google Scholar]
- 9.Fife BT, Bluestone JA. Control of peripheral T-cell tolerance and autoimmunity via the CTLA-4 and PD-1 pathways. Immunol Rev 2008;224:166–182. [DOI] [PubMed] [Google Scholar]
- 10.Gao Q, Wang XY, Qiu SJ, et al. Overexpression of PD-L1 significantly associates with tumor aggressiveness and postoperative recurrence in human hepatocellular carcinoma. Clin Cancer Res 2009;15: 971–979. [DOI] [PubMed] [Google Scholar]
- 11.Ohigashi Y, Sho M, Yamada Y, et al. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res 2005;11: 2947–2953. [DOI] [PubMed] [Google Scholar]
- 12.Wu C, Zhu Y, Jiang J, et al. Immunohistochemical localization of programmed death-1 ligand-1 (PD-L1) in gastric carcinoma and its clinical significance. Acta Histochem 2006;108:19–24. [DOI] [PubMed] [Google Scholar]
- 13.Inaguma S, Wang Z, Lasota J, et al. Comprehensive immunohistochemical study of programmed cell death ligand 1 (PD-L1): analysis in 5536 cases revealed consistent expression in trophoblastic tumors. Am J Surg Pathol 2016;40:1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masugi Y, Nishihara R, Yang J, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut 2016; doi: 10.1136/gutjnl-2016-311421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbaum MW, Bledsoe JR, Morales-Oyarvide V, et al. PD-L1 expression in colorectal cancer is associated with microsatellite instability, BRAF mutation, medullary morphology and cytotoxic tumor-infiltrating lymphocytes. Mod Pathol 2016;29: 1104–1112. [DOI] [PubMed] [Google Scholar]
- 16.Droeser RA, Hirt C, Viehl CT, et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer 2013;49:2233–2242. [DOI] [PubMed] [Google Scholar]
- 17.Lee LH, Cavalcanti MS, Segal NH, et al. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod Pathol 2016; doi: 10.1038/modpathol.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.JE IJ, Vermeulen L, Meijer GA, et al. Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat Rev Gastroenterol Hepatol 2015;12:401–409. [DOI] [PubMed] [Google Scholar]
- 19.Muto T, Bussey HJ, Morson BC. The evolution of cancer of the colon and rectum. Cancer 1975;36:2251–2270. [DOI] [PubMed] [Google Scholar]
- 20.Levin TG, Powell AE, Davies PS, et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 2010;139:2072–2082 e2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007;1:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 2009;69:3382–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng J, Deng R, Zhang P, et al. miR-219–5p plays a tumor suppressive role in colon cancer by targeting oncogene Sall4. Oncol Rep 2015;34:1923–1932. [DOI] [PubMed] [Google Scholar]
- 24.Dalerba P, Sahoo D, Paik S, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med 2016;374:211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miettinen M. A simple method for generating multi-tissue blocks without special equipment. Appl Immu-nohistochem Mol Morphol 2012;20:410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasota J, Kowalik A, Wasag B, et al. Detection of the BRAF V600E mutation in colon carcinoma: critical evaluation of the imunohistochemical approach. Am J Surg Pathol 2014;38:1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant 2013;48:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med 2015;372:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015;373:1803–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang WL, Yang MH, Tsai ML, et al. SNAIL regulates interleukin-8 expression, stem cell-like activity, and tumorigenicity of human colorectal carcinoma cells. Gastroenterology 2011;141:279–291. [DOI] [PubMed] [Google Scholar]
- 33.Marzec M, Zhang Q, Goradia A, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc Natl Acad Sci USA 2008;105:20852–20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsa AT, Waldron JS, Panner A, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med 2007;13: 84–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.