Abstract
Major histocompatibility complex class I (MHC-I) molecules play a crucial role in immunity by capturing peptides for presentation to T cells and natural killer (NK) cells. The peptide termini are tethered within the MHC-I antigen-binding groove, but it is unknown whether other presentation modes occur. Here we show that 20% of the HLA-B*57:01 peptide repertoire comprises N-terminally extended sets characterized by a common motif at position 1 (P1) to P2. Structures of HLA-B*57:01 presenting N-terminally extended peptides, including the immunodominant HIV-1 Gag epitope TW10 (TSTLQEQIGW), showed that the N terminus protrudes from the peptide-binding groove. The common escape mutant TSNLQEQIGW bound HLA-B*57:01 canonically, adopting a dramatically different conformation than the TW10 peptide. This affected recognition by killer cell immunoglobulin-like receptor (KIR) 3DL1 expressed on NK cells. We thus define a previously uncharacterized feature of the human leukocyte antigen class I (HLA-I) immunopeptidome that has implications for viral immune escape. We further suggest that recognition of the HLA-B*57:01-TW10 epitope is governed by a ‘molecular tension’ between the adaptive and innate immune systems.
HLA-I molecules serve as a vital component of cellular immunity by presenting endogenous peptide epitopes on the cell surface for scrutiny by CD8+ T cells and NK cells. These peptides are derived from intracellular proteins that have been proteolytically degraded, and during the course of a viral infection they will include epitopes originating from the invading pathogen. HLA-I molecules bind compatible peptides via a specialized groove capped by a conserved network of residues that form hydrogen bonds with the N termini (P1) and C termini (PΩ) of the peptide1. The peptide is further anchored at specific pockets spaced along the groove; for many HLA allotypes this anchoring typically occurs at the P2 and PΩ positions2–5. The nature of these ‘anchoring pockets’ differs between HLA-I molecules, and thus defines their unique peptide repertoires5.
The closed-end nature of the HLA-I antigen-binding cleft generally constrains the repertoire of peptides to 9–11 amino acids in length. Some longer HLA-I-restricted peptides have been described, but because of the conserved polar interaction network with the N and C termini in the P1 and PΩ pockets, longer peptides are canonically accommodated in the groove only after they adopt a ‘bulged’ conformation in which the central portion of the peptide epitope projects from the groove6,7. The T cell receptor (TCR) binds these bulged regions of the peptide while simultaneously contacting the HLA-I molecule. Although the KIR family of receptors on NK cells demonstrate broader peptide recognition than TCRs, they also bind HLA in a peptide-dependent manner8–10. Particularly important are residues at positions PΩ−1 and PΩ−2 that often directly contact the KIR11,12. Isolated reports have described peptides that protrude at the C terminus from the peptide-binding groove, with the PΩ−1 residue acting as an alternate C-terminal anchor, although the extent to which this represents a common occurrence remains unclear13–15. Such noncanonical HLA-I-peptide landscapes are important because they potentially form unique contact surfaces for TCR and KIR recognition, with attendant implications for disease-relevant immune responses14,16,17. In contrast, it is not known whether peptides can protrude from the N terminus of the HLA-I antigen-binding cleft.
The HLA-B*57:01 allele is associated with delayed disease progression in HIV-1-infected individuals18–21 and hypersensitivity to the antiviral drug abacavir22–24. In HLA-B*57:01+ HIV-1-infected individuals, robust CD8+ T cell responses specific for four epitopes within the p24 Gag protein (TW10, IW9, QW9 and KF11) are associated with such protective immunity19,25–27. Cellular immune responses against these epitopes drive the selection of amino acid mutations that allow viral escape from the immune system28–30. A common escape mutation in the p24 Gag epitope TW10 (TSTLQEQIGW; residues 240–249) is Thr242Asn, which lies at P3 of the epitope (T3N)31–33. The emergence of this variant coincides with loss of the original CD8+ T cell response in many patients27,32 and with impaired viremic control, especially after the acquisition of compensatory mutations30,34. In addition to acting as a restriction element for CD8+ T cell responses, HLA-B*57:01 is also a potent ligand for KIR3DL1, which regulates NK cell function. Furthermore, genetic association studies have implicated the interaction between HLA-B*57 and certain allelic variants of KIR3DL1 with delayed progression to AIDS35–37.
Here we show that the repertoire of endogenous and viral peptides presented by HLA-B*57:01 contains sets of both N- and C-terminally extended epitopes. We identified a characteristic motif as a signature of N-terminally extended peptides, and our structural studies revealed a noncanonical binding mode whereby the N-terminal residue over-hangs the peptide-binding groove of HLA-B*57:01. This novel mode of peptide binding enables HIV-1 to escape immune recognition via a register shift within the TW10 epitope that alters the presented antigenic structure upon mutation to T3N. In addition to its consequences for the adaptive immune response, this mutation also had an effect on KIR3DL1 recognition, which highlights the differences and ‘molecular tension’ in the adaptive and innate immune systems’ recognition of this common HIV-1 epitope.
RESULTS
Structure of HLA-B*57:01–TW10 in complex with KIR3DL1
To extend our previous analysis of KIR3DL1 bound to self-peptide-presenting HLA-B*57:01 (ref. 12), we determined the crystal structure of KIR3DL1*001–HLA-B*57:01 in complex with the HIV-1 p24 Gag epitope TW10 (TSTLQEQIGW; residues 240–249) (Table 1).
Table 1.
Data collection and refinement statistics for HLA-B*57:01 complexes
| TSTFEDVKILAF (PDB 5T6Y) | TSTTSVASSW (PDB 5T6X) | SSTRGISQLW (PDB 5T6W) | KIR3DL1-TSTLQEQIGW (PDB 5T6Z) | KIR3DL1-TSNLQEQIGW (PDB 5T70) | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P212121 | P212121 | P212121 | P1 | P1 |
| Cell dimensions | |||||
| a, b, c (Å) | 50.7, 82.0, 110.1 | 50.5, 81.7, 109.6 | 50.8, 81.2, 109.8 | 51.8, 61.4, 65.4 | 51.7, 61.2, 65.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 95.3, 98.0, 109.2 | 96.0, 97.5, 108.7 |
| Resolution (Å) | 46.06–1.76 (1.85–1.76)a | 45.89–1.69 (1.78–1.69) | 54.89–1.90 (2.00–1.90) | 57.32–2.00 (2.11–2.00) | 63.84–2.10 (2.21–2.10) |
| Rmerge | 0.122 (0.682) | 0.077 (0.581) | 0.197 (0.553) | 0.082 (0.218) | 0.083 (0.579) |
| I /σ I | 8.7 (2.3) | 15.0 (3.1) | 4.7 (2.3) | 11.2 (5.1) | 12.7 (2.4) |
| Completeness (%) | 98.5 (90.6) | 95.7 (92.6) | 99.6 (99.8) | 94.4 (94.2) | 91.4 (91.7) |
| Redundancy | 6.4 (5.5) | 4.9 (4.8) | 3.9 (3.9) | 4.9 (4.9) | 3.8 (3.9) |
| Refinement | |||||
| Resolution (Å) | 1.76 | 1.69 | 1.90 | 2.0 | 2.1 |
| No. reflections | 291,141 (33,508) | 243,607 (33,315) | 141,190 (20,620) | 232,052 (34,355) | 150,929 (22,413) |
| Rwork /Rfree | 0.1778/0.2189 | 0.2085/0.2443 | 0.2114/0.2630 | 0.2030/0.2395 | 0.1791/0.2310 |
| No. atoms | |||||
| Protein | 3,174 | 3,332 | 3,139 | 5,361 | 5,377 |
| Ligand/ion | 22 | 0 | 6 | 42 | 42 |
| Water | 302 | 180 | 207 | 651 | 334 |
| B factors | |||||
| Protein | 26.0 | 21.4 | 27.6 | 27.7 | 36.1 |
| Ligand/ion | 50.3 | 0 | 44.8 | 51.8 | 82.0 |
| Water | 34.7 | 29.4 | 30.3 | 33.6 | 40.1 |
| R.m.s. deviation | |||||
| Bond length (Å) | 0.0069 | 0.0070 | 0.0073 | 0.0070 | 0.0197 |
| Bond angle (°) | 1.0296 | 1.0480 | 1.0448 | 1.2082 | 1.8504 |
All data sets are collected from a single crystal.
Values in parentheses are for the highest-resolution shell.
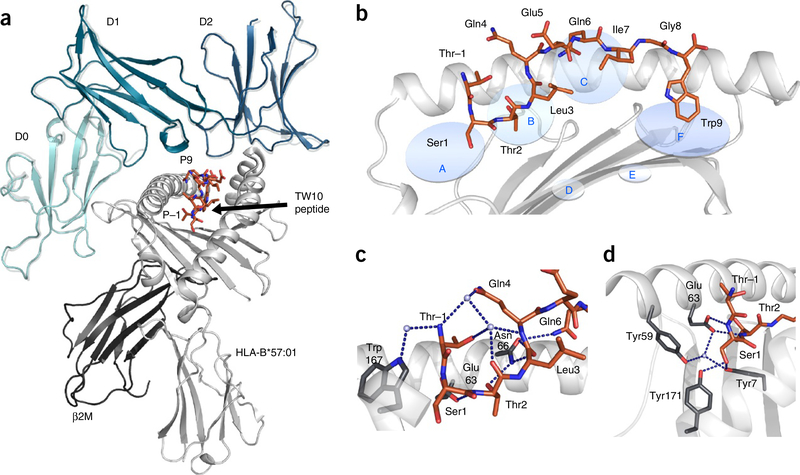
The mode of KIR3DL1 recognition of the viral epitope was essentially identical to that of the self-peptide bound to HLA-B*57:01 (ref. 12) (Fig. 1a). The three KIR3DL1 domains engaged HLA-B*57:01 in a conserved orientation, with the D0 domain docking at the side and the D1 and D2 domains docking at the C-terminal end of the peptide binding cleft, above the α1 and α2 helices, respectively (Fig. 1a). The D1 domain interacted with the Bw4 motif on the α1 helix. In the course of our analysis, we noted that the TW10 epitope adopted a unique conformation not previously observed for HLA-I–peptide complexes. Namely, the N-terminal region of the TW10 epitope protruded from the antigen-binding cleft and thus did not occupy the A pocket (Fig. 1b). Instead, the second and third residues of the TW10 epitope were located in the A and B pockets, respectively (Fig. 1b). Comparison with available HLA-B*57:01–peptide structures12,23,38 showed that, with the exception of a 0.7-Å shift of Trp167 (not shown), there were minimal structural distortions to the peptide-binding cleft that accompanied this previously unobserved peptide conformation (r.m.s. deviation < 0.25 Å over Cα positions 1–180). Because of the closed nature of the peptide-binding groove, the P–1 residue (one residue N-terminal of P1) of the peptide projected at a right angle from the plane of the floor of the groove, with the side chain pointing back along the length of the peptide (Fig. 1c). Contacts between P–1-Thr and HLA-B*57:01 were limited to water-mediated H-bonds between Trp167 and the P–1-Thr peptide backbone, and van der Waals interactions between Glu63 and Leu163 and the P–1-Thr side chain (Fig. 1c). Contacts between P–1-Thr and the rest of the peptide included water-mediated H-bonds with P2-Thr and P4-Gln (Fig. 1c).
Figure 1.
HLA-B*57:01 in complex with the TW10 peptide. (a) The overall structure of KIR3DL1 (green) in complex with HLA-B*57:01–TW10 (orange). The structure of KIR3DL1 when bound to HLA B*57:01–LF9 is overlaid12 (gray) for comparison of the binding modes. (b) Cartoon representation of the crystal structure of HLA-B*57:01 (light gray) in complex with the TW10 peptide TSTLQEQIGW (orange) shown against the α1 helix of HLA and oriented N-C-terminal from left. Anchor pockets of the HLA are indicated along the binding groove at P1 (A) to PΩ (F). (c,d) Orientation of the protruding Thr residue of the peptide at P–1 (orange) and its interaction with residues of the HLA (dark gray sticks) (c), and the conserved hydrogen-bonding network (blue dashed lines) at the N-terminal end of HLA-B*57:01 (dark gray sticks) maintained to Ser at P1 (orange) (d). Hydrogen bonds are shown as blue dashed lines.
The A pocket is characterized by a highly conserved set of residues including Tyr7 and Tyr171 that contribute a network of H-bonds, thus stabilizing the N terminus of the presented peptide. In the HLA-B*57:01–TW10 complex, the P1-Ser residue of the TW10 epitope occupied the A pocket of HLA-B*57:01 and thus maintained a network of H-bonds with Tyr7 and Tyr171 (Fig. 1d). As a result, the P1-Ser side chain was rotated approximately 180° from the canonical positioning of a P1 anchored residue. Accordingly, the slippage of TW10 from the groove of HLA-B*57:01 was accompanied by distortion of the peptide at the P1 residue and not by structural distortion of the peptide-binding groove. As peptide repertoire data for HLA-B*57:01 had previously demonstrated the P2 anchor preference for Ser or Thr23, it was conceivable that either P2-Ser or P3-Thr could act as an N-terminal anchor for TW10. However, the high-resolution structure of HLA-B*57:01–TW10 showed unambiguous electron density across the peptide, thus indicating that only one conformation of TW10 was present in the crystal lattice. Further, we confirmed by the use of hydrogen-deuterium exchange with mass spectrometry that, in contrast to the peptide free in solution, TW10 adopts a single conformation while in complex with B*57:01 (Supplementary Fig. 1). Thus, the P–1–P2 residues enable the peptide to extend from the peptide-binding cleft of HLA-B*57:01.
N-terminal extensions within the HLA-B*57:01 immunopeptidome
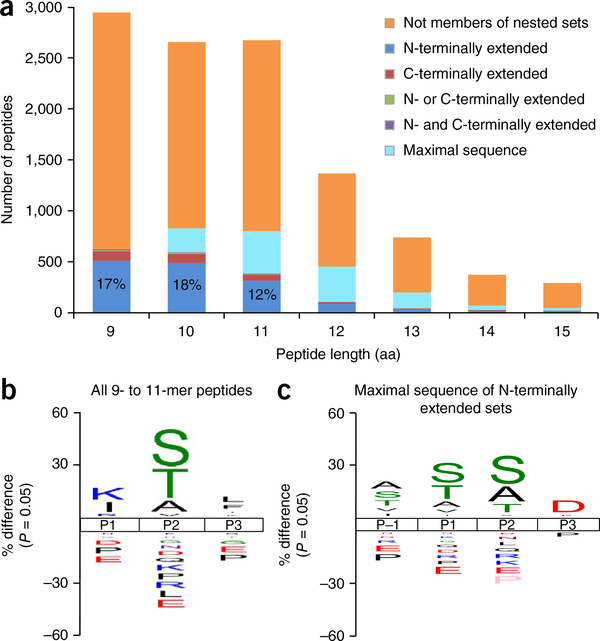
To determine whether N-terminally extended peptides are a general feature of the HLA-B*57:01 restricted immunopeptidome, we sought to define the range of self-peptides and HIV-Gag epitopes presented by HLA-B*57:01. We isolated peptides from C1R cells transfected with expression constructs for HLA-B*57:01 and HIV-1-Gag and sequenced them using tandem mass spectrometry. The resulting data set was (i) filtered for peptides known to bind the endogenous HLA-I and HLA-II of the parental cell line, or bearing the peptide-binding motifs of HLA-Cw4 and HLA-B*35:03 (endogenous HLA-I of C1R cells)17, and (ii) further augmented with previously defined peptides from the immunopeptidome of HLA-B*57:01 (ref. 23), which yielded 11,954 peptides in total (Supplementary Table 1). As described previously23, HLA-B*57:01 ligands were predominantly 9–11 amino acids in length and showed enrichment of S/T/A at the P2 anchor, and aromatic residues at the C terminus (Fig. 2). In addition, a thorough bioinformatic interrogation of this expanded HLA-B57 peptidome revealed a number of peptides that contained these shorter peptides (9–11 amino acids) with an N-terminal extension and were isolated from both HIV-1-Gag+ and HIV-1-Gag− cells.
Figure 2.
Characterization of N-terminally extended peptides. (a) Length distribution of HLA-B*57:01 ligands, showing those classified within nested sets. Definitions of nested-set subcategories can be found in Supplementary Note 1. The values in the first three bars respectively represent the percentage of peptides 9, 10 and 11 amino acids in length that were part of extended sets and extended at the N terminus only. (b,c) Sequence logos showing the percent difference in the abundance of amino acids in comparison to the human proteome at each location in the N-terminal portion of (b) all 9–11-residue HLA-B*57:01 ligands (n = 8268), and in (c) the maximal sequences of N-terminal-extended sets possessing C-terminal aromatic anchors (n = 972). Sequence logos were generated using the iceLogo stand-alone version39. Source data are available in Supplementary Tables 1 and 2.
Extended sets of peptides were defined on the basis of the following criteria: (i) minimal core sequence of 7–11 amino acids and (ii) maximal sequence of >9 amino acids. A total of 1,275 sets of peptides met these criteria; among these, 17%, 18% and 12% of 9-, 10- and 11-residue peptides possessed only N-terminal extensions, whereas far fewer were purely C-terminally extended (3%, 3% and 2%, respectively) (Fig. 2a). To define the sequence features that were predisposed to N-terminal extensions, we aligned peptides exhibiting N-terminal heterogeneity on the basis of their minimal-sequence P1 (Supplementary Table 2). We visualized the motif as a sequence logo encompassing P–1 to P3 with iceLogo39. We also generated a similar motif from P1 to P3 of all 9–11 residue peptides within the HLA-B*57:01 peptide data set and aligned them at P1 for comparison (Fig. 2b,c). Whereas the global HLA-B*57:01 9–11-amino-acid peptide motif showed enrichment of Lys, Ile and Arg at P1, the N-terminally extended peptides were enriched for Ser, Thr and Ala at P1 and P–1. Given that Ser, Thr and Ala are preferred P2 anchors for HLA-B*57:01, these residues could potentially be acting as alternative anchor sites in longer peptides, thus resulting in bulged conformations within the peptide-binding groove. However, given the appearance of numerous sets in which a minimal core sequence was found with several extension lengths (for example, set 233: STTSVASSW, TSTTSVASSW, DTSTTSVASSW, HDTSTTSVASSW, SHDTSTTSVASSW, SSHDTSTTSVASSW and TASSHDTSTTSVASSW), not all of which contained Ser, Thr or Ala at the second residue, it seemed likely that the excess residues might overhang the cleft. Thus, we defined the N-terminal protrusion motif as Ser/Thr/Ala at P–1 to P2.
TW10 is part of an extended set restricted by HLA-B*57:01
Of the 11,954 peptides used in this analysis, 9 were derived from the Gag polypeptide. Among the Gag-derived epitopes, we observed two N-terminally extended epitope sets: QW9 and TW10 (set 33 and set 500; Fig. 3a and Supplementary Table 2). The TW10 epitope was the minimal peptide observed within its N-terminally extended set that contained multiple extension lengths (Fig. 3a). To determine the relative abundance of each peptide in the set, we measured the extracted ion chromatogram specific for each of the peptides (Fig. 3b). The N-terminally extended AGTTSTLQEQIGW peptide was the most intense ion and by inference the most abundant, followed by TTSTLQEQIGW, GTTSTLQEQIGW and TW10 (Fig. 3b). We also identified two longer N-terminally extended peptides (DIAGTTSTLQEQIGW and SDIAGTTSTLQEQIGW), at significantly lower levels (Fig. 3b). Taken together, our data suggest that a common motif characterizes N-terminally protruding HLA-B*57:01-restricted peptides regardless of origin (i.e., self or viral).
Figure 3.
The HIV-1 Gag repertoire of HLA-B*57:01. (a) HIV-1 Gag epitopes presented by HLA-B*57:01. The QW9 and TW10 epitopes form part of N-terminally extended sets. (b) Relative levels of N-terminally extended variants compared with the HIV Gag TW10 epitope. Values are mean and s.d.; n = 3; data represent three independent experiments. Source data are available in Supplementary Table 3.
Structure of HLA-B*57:01 presenting extended self-peptides
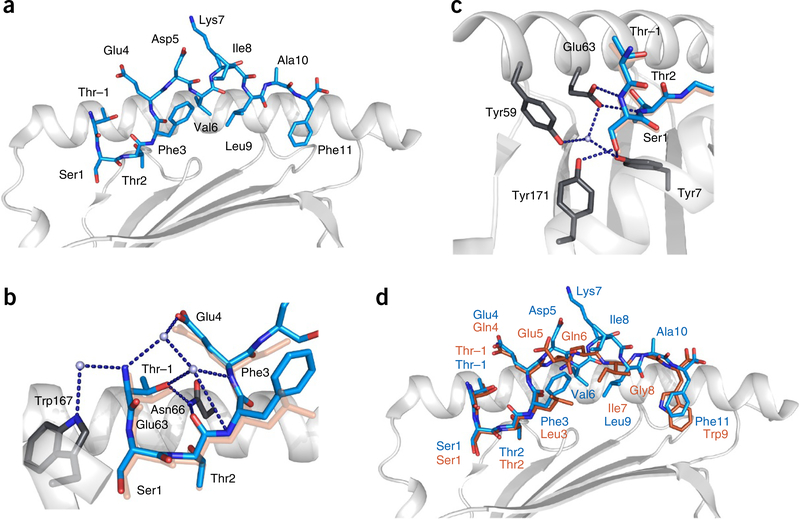
To further probe the structural nature of the N-terminal extension motif, we investigated three epitopes from the self-repertoire of HLA-B*57:01. We selected the peptides TSTTSVASSW (UniProt Q14679), TSTFEDVKILAF (UniProt Q6YHU6) and SSTRGISQLW (UniProt Q96HA1), each of which formed part of a nested set (set 233, set 926 and set 736, respectively; Supplementary Table 2). Crystals of the refolded HLA-B*57:01–peptide complexes formed in the space group P212121 with unit cell dimensions of ~a = 50, b = 82 and c = 110 Å, and diffracted to high resolution (Table 1).
All of these HLA-B*57:01–peptide complexes adopted the noncanonical conformation observed for HLA-B*57:01–TW10. Namely, the Ser residue in the second position of the TSTFEDVKILAF (Fig. 4a), TSTTSVASSW and SSTRGISQLW peptides occupied the A pocket, which normally accommodates the N-terminal amide. As these self-peptide complexes were highly similar, we restricted analysis to the HLA-B*57:01–TSTFEDVKILAF structure. The water-mediated H-bonding network to the Thr at position P–1 to residues Asn66 and Trp167 of HLA-B*57:01 was highly conserved (Figs. 1c and 4b). Similarly, the H-bonding network at the N terminus of the peptide-binding groove was also conserved, with the hydroxyl of P1-Ser maintaining the contacting Tyr7, Tyr59, Glu63 and Tyr171 (Figs. 1d and 4c,d). Accordingly, the P–1-to-P2 motif identified in the repertoire analysis is a common motif that enables the peptide to protrude from the N terminus of HLA-B*57:01.
Figure 4.
Cartoon representations of the crystal structure of HLA-B*57:01 (light gray) in complex with the TSTFEDVKILAF peptide (cyan). (a) HLA-B*57:01 in complex with the TSTFEDVKILAF peptide, shown against the α1 helix of HLA and oriented N-C-terminal from the left. (b) The network of direct and water-mediated hydrogen bonds (dark blue dashed lines) around the protruding P–1 residue, showing the interaction of P–1-Thr with Trp167 and Asn66 of HLA. The TW10 peptide is underlaid (orange) for comparison. (c) The network of conserved hydrogen bonds at the N terminus of the HLA-B*57:01 peptide-binding groove, showing the P1-Ser1 side chain replacing contacts normally mediated by the N terminus of the peptide. The TW10 peptide is underlaid (orange) for comparison. (d) Overlay representation of the TSTFEDVKILAF (blue) and TSTLQEQIGW (orange) structures.
The structural basis of viral escape via the T3N mutation
The TW10 epitope undergoes a common and rapid substitution at the N terminus, namely, T3N (TSNLQEQIGW), to facilitate HIV-1 immune escape in HLA-B*57:01+ individuals. To address this phenomenon at the molecular level, we determined the crystal structure of HLA-B*57:01 in complex with the T3N peptide and KIR3DL1 (Table 1). Overall, the contacts between KIR3DL1 and HLA-B*57:01 were conserved between the TW10 and T3N structures. However, contacts between KIR3DL1 and the HLA-bound peptide were markedly different for TW10 and T3N. KIR3DL1 formed two contacts with the T3N peptide. The D1 Gly116 bound P7-Gln, whereas Tyr200 at the D1/D2 hinge loop bound P8-Ile (Fig. 5a). In contrast, KIR3DL1 did not directly contact the TW10 peptide. Instead, water-mediated contacts were formed between Tyr200 and the backbones of P8-Gly and P9-Trp (Fig. 5b). KIR3DL1 therefore interacted differently with TW10 and T3N as a result of changes in the bound peptide conformation (detailed below).
Figure 5.
Comparison of the HLA-B*57:01–TW10 and HLA-B*57:01–T3N ternary complex structures with KIR3DL1*001. (a) Cartoon representation of the interactions between KIR3DL1 (teal) and the T3N peptide (blue) presented by HLA-B*57:01 (light gray). (b) The interactions between KIR3DL1 (teal) and the TW10 peptide (orange). (c) Cartoon representation of the crystal structure of HLA-B*57:01 (light gray) in complex with the T3N peptide TSNLQEQIGW (blue) shown against the α1 helix of the HLA and oriented N-C-terminal from left. Hydrogen bonds are shown as blue dashed lines. (d) Overlay of the TW10 (orange) and T3N (blue) peptide conformations.
The Thr3Asn substitution caused the peptide to adopt a conventional anchoring mode in which it bound to HLA-B*57:01 with the A and B pockets occupied by the Thr and Ser in the first two peptide positions, respectively (Fig. 5c). Notably, the HLA-B*57:01–TW10 complex was more stable than the HLA-B*57:01–T3N complex, with a melting temperature of 61.8 °C, compared with 55.0 °C in circular dichroism assays (Supplementary Fig. 2). The inability of Asn3 to occupy the B pocket is consistent with a previous HLA-B*57:01 peptide-elution study in which Asn was not observed as an anchor residue at P2 (ref. 23). The change in anchoring at P2 shifted the peptide register across all positions except the C-terminal anchor and P9-Gly (Fig. 5d). Accordingly, the secondary anchor positions also differed between TW10 and T3N. Specifically, the TW10 secondary anchors at P3-Leu, P6-Gln and P7-Ile were reassigned to P3-Asn, P5-Gln and P6-Glu, respectively, in T3N. Consequently, the P–1-Thr, P4-Gln and P5-Glu side chains were considerably solvent exposed in TW10, whereas P4-Leu, P5-Gln and P8-Ile were considerably solvent exposed in T3N (Fig. 5d). Thus, the Thr3Asn mutation shifts the peptide register within the peptide-binding groove, thereby resulting in the presentation of an altered peptide conformation to immune cell receptors.
Escape mutants in TW10 attenuate KIR3DL1 recognition
To determine whether the T3N escape mutation affects binding to KIR3DL1, we used surface plasmon resonance to measure the affinity of KIR3DL1*001 for HLA-B*57:01–TW10 and HLA-B*57:01–T3N. The TW10 complex bound more strongly to KIR3DL1 (KD ~ 60 μM) than the T3N complex did (KD ~ 100 μM) (Fig. 6a). We then used fluorochrome-labeled tetrameric forms of HLA-B*5701–TW10 and HLA-B*5701–T3N to stain HEK293T cells transfected with prevalent KIR3DL1 allotypes (*001, *005 and *015) (portions of the data for TW10 have been published previously40). The T3N mutation almost entirely abrogated HLA-B*57:01 tetramer binding to all three surface-expressed allotypic variants of KIR3DL1 (Fig. 6b). However, we did observe some differences between allotypes, and found KIR3DL1*015 to be the most sensitive to changes in the epitope.
Figure 6.
HLA-B*57:01 TW10 and T3N binding to KIR3DL1. (a) (i) Representative surface plasmon resonance (SPR) injection series from two independent experiments for KIR3DL1 binding to the TW10 (top) and T3N (bottom) HLA-B*57:01 complexes. (ii,iii) SPR-based affinity measurements of the interaction between KIR3DL1*001 and B57:01 T3N (ii) and B*57:01 TW10 (iii) complexes; results represent two independent experiments. Data are shown as mean ± range of two values. Concentration values represent the equilibrium binding constants of KIR3DL1 for the HLA-peptide complexes. RU, response units. (b) Staining of KIR3DL1 allotypes with HLA-B*57:01 TW10 (orange) and T3N (blue) tetramers (0.2 μg each), normalized to TW10 binding. Results represent two (015), three (005) and four (001) independent replicates. Data are shown as mean ± range. (c,d) HLA-B*57:01 TW10 and T3N tetramer staining of HEK293 cells transfected with KIR3DL1*001 and a panel of KIR3DL1*001 interface residue mutants, normalized to TW10 tetramer binding to KIR3DL1*001 (c) or to the respective tetramer binding of KIR3DL1*001 transfectants (d). All results for c and d represent three independent experiments and are shown as mean and s.e.m. WT, wild-type.
We probed the relative contribution of specific KIR3DL1*001 residues to the HLA-B*57:01-TW10 and HLA-B*57:01-T3N interfaces further by mutational analysis. Overall, there were distinct differences in the recognition of these HLA-B*57:01–peptide complexes (Fig. 6c,d). Starting with the D0 domain, the T3N complex was less sensitive than the TW10 complex to substitution at Phe9, but more sensitive to substitution at Phe34 (Fig. 6c,d). In the D1 domain, substitutions at Gly138 and Ser140 dramatically altered recognition of the TW10 complex yet had a minimal effect on recognition of the T3N complex, whereas the Leu166Ala mutation was sufficient to restore T3N binding to wild-type (TW10) levels (Fig. 6c,d). Substitutions across the D2 domain indicated similar contributions from Tyr200, Glu201, Ser227, Ser228, Phe276, His278 and Ser279. In contrast, the Arg277Ala substitution abrogated binding to the T3N complex while improving recognition of the TW10 complex.
Taken together, these findings demonstrate that the viral escape mutant T3N attenuates KIR3DL1 recognition in an allotype-dependent manner. Furthermore, the KIR3DL1 residue Leu166, which sat proximal to the interface with the peptide in the ternary structure, has a central role in this effect. The altered peptide conformation of T3N therefore limits the interaction despite the presence of additional direct contacts with KIR3DL1. Accordingly, while the HIV escape mutants within the TW10 epitope evade recognition by T cells, these same substitutions can impair their interaction with the prototypical inhibitory receptor KIR3DL1.
DISCUSSION
HLA-II molecules possess open-ended antigen-binding clefts that allow peptides to extend from the groove at both N and C termini. In contrast, the N- and C-terminal pockets of HLA-I molecules are closed, which restricts the bound peptides to a typical length of 9–11 amino acids. Nevertheless, longer MHC-I-restricted epitopes have been described that adopt a centrally bulged conformation while maintaining the N- and C-terminal anchor residues. In addition, there have been isolated reports of peptides with residues overhanging the C-terminal pocket of HLA-A*02:01 (refs. 13,14) and H2-M3 (ref. 41). It has been proposed that N-terminally extended peptides could be presented42,43, yet whether such peptides could overhang the more buried N-terminal pocket or whether they are centrally bulged remained unclear.
Our comprehensive analysis of the repertoire of peptides naturally presented by HLA-B*57:01 shows that approximately 27% are clustered into extended sets, most of which are solely N-terminally extended (20%), whereas the others are either C-terminally extended or both N- and C-terminally extended. We have also shown that N-terminally extended peptides are accommodated via protrusion from the groove. From the amino acid enrichment data for N-terminally extended HLA-B*57:01 epitopes and our structure-based insights, we can devise the following rules that predict the P–1 overhang: (i) the peptide should be at least one residue longer than the minimal epitope length that can be presented, (ii) the P2 residue should be a residue that favors binding in the B pocket and (iii) the P1 residue should be smaller than leucine to enable a 180° rotation within the A pocket. Notably, none of these criteria are specific to HLA-B*57:01, and thus we speculate that N-terminally protruded peptides will be a common feature of immunopeptidomes. Accordingly, we define a new mechanism that allows MHC-I molecules to present lengthy epitopes.
Viruses have developed numerous strategies to evade HLA-mediated immune detection, including mechanisms that directly interfere with the presentation of viral epitopes and HLA molecules on the cell surface44,45. For example, HIV-1 can increase cellular endocytosis of HLA molecules via nef46, limit HLA transcription and peptide processing via tat47, and suppress TAP-mediated peptide transport into the endoplasmic reticulum48. Under selection pressure from the immune system, HIV-1 also mutates rapidly to evade cytotoxic T lymphocyte responses. These epitope-centric escape mutations typically alter the conformation of exposed residues that interact with the TCR49–51 or abrogate peptide presentation via effects on antigen processing or HLA-I binding52,53. Here we report a novel mode of escape whereby the common T3N mutant exploits the P–1 overhang of TW10 and the anchor-residue preferences in the A and B pockets to shift the register of the peptide within HLA-B*57:01. This structural rearrangement alters the conformation of exposed peptide residues to enable immune escape31–33,54.
There has been considerable interest in the idea that KIR-dependent recognition of HLA-I can shape the nature of viral escape mutations, with some studies suggesting that inhibitory receptors expressed by NK cells may exert some selective pressure during HIV infection55,56. Although immune recognition and the subsequent immune escape observed for the TW10 epitope is likely to be a predominantly T-cell-mediated phenomenon, it is plausible that this epitope also affects NK cell recognition. Such effects would be accentuated by the viral modulation of the peptide repertoire through the inclusion of viral peptides, which have been observed to constitute up to 50% of the immunopeptidome in the early stages of vaccinia infection57, and alterations to self-peptide presentation. However, it is unclear whether the T3N escape variant actually affects NK cell inhibition via engagement of KIR3DL1. A previous study using surface plasmon resonance reported reduced interaction with T3N presented by HLA-B*57:03 (ref. 33). In contrast, another study based on tetramer staining of KIR3DL1-transfected cell lines reported no interaction58. Although this discrepancy is probably a result of the different experimental approaches, it is clear from our structural, surface plasmon resonance and tetramer-staining data that KIR3DL1 recognizes T3N, albeit with lower affinity than for TW10 in the context of HLA-B*57:01. There are also well-described C-terminal escape mutations within the TW10 epitope, such as G9D, that abrogate KIR3DL1 recognition40. Notably, these effects are KIR3DL1-allotype-dependent, in agreement with the findings of a recent study59. This is in line with recent reports that demonstrated allotypic variation in the capacity of KIR3DL1 to bind defined HLA-I–peptide complexes40,59. Accordingly, there appears to be a ‘molecular tension’ between the inhibitory germline-encoded and activating rearranging receptors of the innate and adaptive arms of the immune system as they converge to target a common HIV-1 determinant and its escape variants presented by HLA-B*57:01. In summary, our structural and proteomics analyses reveal a novel mechanism of viral immune escape whereby HIV-1 mutates to exploit a previously undescribed mode of peptide presentation by MHC-I.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
ONLINE METHODS
Isolation of MHC-I peptide ligands.
C1R HLA-B*57:01 transfectants generated as described in ref. 38 were grown to high density in RPMI 1640 media (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FCS (Bovogen Biologicals Pty. Ltd., Melbourne, Australia), 7.5 mM HEPES (MP Biomedicals, Eschwege, Germany), 150 μg/mL streptomycin (Sigma-Aldrich, St. Louis, MO), 150 U/mL benzylpenicillin (CSL, Parkville, Australia), 2 mM L-glutamine (MP Biomedicals), 76 μM β-mercaptoethylamine (Sigma-Aldrich) and 150 μM non-essential amino acids (Life Technologies, Carlsbad, CA). The Gag plasmid, a kind gift from Johnson Mak (Deakin University, Melbourne, Australia), was introduced into cell lines by electroporation. We assayed the maintenance of Gag expression in these transfectants by western blotting using an antibody to Gag that was generated by Johnson Mak (Deakin University, Melbourne, Australia). Transfectants were maintained under hygromycin (0.3 mg/mL, HLA-B*57:01) and G418 (0.5 mg/mL Gag) selection. Cells were tested for mycoplasma contamination in-house at regular intervals and for continued expression of HLA-B*57:01 with the 3E12 antibody60. Cells were harvested by centrifugation (1,200g, 20 min, 4 °C) and snap-frozen in liquid nitrogen. Clarified lysates were generated from 5 × 109 cells with a combination of cryogenic milling and detergent-based lysis. HLA–peptide complexes were immunoaffinity purified from cell lysates using the W6/32 monoclonal antibody in solid phase as described previously61,62. Bound complexes were eluted by acidification with 10% acetic acid and fractionated in a 4.6-mm (internal diameter) × 100-mm (length) monolithic reversed-phase C18 high-performance liquid chromatography (HPLC) column (Chromolith SpeedROD; Merck Millipore, Darmstadt, Germany) with the ÄKTAmicro HPLC system (GE Healthcare, Little Chalfont, UK). The mobile phase consisted of buffer A (0.1% trifluoroacetic acid; Thermo Fisher Scientific) and buffer B (80% acetonitrile, 0.1% trifluoroacetic acid; Thermo Fisher Scientific). HLA-peptide mixtures were loaded onto the column at a flow rate of 1 mL/min with separation based on a B gradient of 2–40% for 4 min, 40–45% for 4 min and a final rapid 2-min increase to 100%. Fractions (500 μL) were collected, vacuum-concentrated to 10 μL and diluted in 0.1% formic acid to reduce the acetonitrile concentration.
Liquid chromatography–tandem mass spectrometry (LC-MS/MS) sequencing of MHC-I-bound peptides.
For LC-MS/MS acquisition, peptide-containing fractions were loaded onto a microfluidic trap column packed with ChromXP C18-CL 3-μm particles (300-Å nominal pore size; equilibrated in 0.1% formic acid, 2% acetonitrile) at 5 μL/min with a NanoUltra cHiPLC system (Eksigent). An analytical (75 μm × 15 cm ChromXP C18-CL, 3 μm, 120 Å, Eksigent) microfluidic column was switched in line, and peptides were separated by linear gradient elution with 0–30% buffer B (80% acetonitrile, 0.1% formic acid) over 50 min and 30–80% over 5 min flowing at 300 nL/min. Separated peptides were analyzed with an AB SCIEX 5600+ TripleTOF mass spectrometer equipped with a Nanospray III ion source and accumulating up to 20 MS/MS spectra per second. The following instrument parameters were used: ion spray voltage, 2,400 V; curtain gas, 25 l/min; ion source gas, 10 l/min; and interface heater temperature, 150 °C. MS/MS switch criteria included the following: ions of m/z >200 amu; charge state, +2 to +5; and intensity, >40 counts per second. The top 20 ions meeting these criteria were selected for MS/MS per cycle. We calibrated the instrument every four LC runs using [Glu1]-Fibrinopeptide B standard as per the manufacturer’s instructions.
LC-MS/MS data were searched against the human proteome (UniProt/Swiss-Prot v2014_10) and the HIV NL4–3-gag and AD8-env sequence with ProteinPilot software (version 4.5, SCIEX). A 5% false discovery rate cutoff was applied. Peptides known to bind the endogenous MHC-I of C1R cells (HLA-C*04:01 and HLA-B*35:03)17 and those identified as binders of MHC-II in similar MHC-II isolations in the laboratory and commonly observed contaminants were removed from the data set before subsequent analysis of HLA-B*57:01 peptide ligands.
Analysis of the HLA peptidome for extended sets.
The final combined data set used to estimate the prevalence of N-terminally extended HLA-B*57:01 ligands and characterize the extension motif (Supplementary Table 1) contained 11,954 unique peptides. We identified 2,746 of those as constitutive HLA-B*57:01 ligands in studies that examined the impact of abacavir on the HLA-B*57:01 peptidome23. In this study, 8,432 peptides were eluted from same parental CIR cell line in the absence of the HIV Gag antigen, and 8,233 from HIV Gag transfected cells (Supplementary Table 1). We identified extended sets through an iterative process of examining longer peptides to see whether they encompassed shorter peptides. Sets containing minimal core sequences of 7–11 amino acids consistent with canonical class I ligands and maximal sequences greater than 9 amino acids were defined as extended sets. Motifs showing the enrichment (compared to the human proteome) of specific amino acids at defined locations within the peptide ligands were generated using the iceLogo stand-alone version with the static reference method39. Motifs were generated from the N-terminal portion of all 9–11-residue peptides in the HLA-B*57:01 data set (n = 8,268) aligned on the basis of the first residue assignment of P1, and from P–1 to P3 of the maximal sequences of nested sets containing purely N-terminal extensions and possessing C-terminal aromatic anchors (n = 972) (Supplementary Table 2) aligned on the basis of the minimal sequences of the nested set. Letter height in sequence logos corresponds to the difference in frequency of the amino acid compared to the human proteome. Only significantly regulated amino acids—those for which the z score falls outside the confidence interval for a P value of 0.05—are shown in the figures. Extended sets containing C-terminal or N- and C-terminal extensions are shown in Supplementary Table 3.
Protein expression and purification.
The HLA-B*57:01 and β2-microglobulin genes were subcloned into the pET-30 expression vector and expressed separately into inclusion bodies in Escherichia coli before refolding and purification as described previously63. Briefly, HLA-B*57:01 was refolded by rapid dilution in a solution containing 100 mM Tris-HCl, pH 8.0, 400 mM l-arginine-HCl, 5 mM reduced glutathione, and 0.5 mM oxidized glutathione in the presence of β2-microglobulin the appropriate peptide. The refolded HLA-B*57:01 complex was purified by anion exchange and size-exclusion chromatography.
KIR3DL1*001 (residues 1–299) was subcloned into the pHLSec mammalian expression vector with N-terminal 6×His and secretion tags. KIR3DL1 was expressed and secreted from transiently transfected HEK293S cells and harvested from the culture media after 3 d by nickel affinity and gel filtration chromatography with an S200 16/60 column (GE Healthcare) in 10 mM Tris, pH 8.0, 300 mM NaCl. Purified KIR3DL1 was then concentrated to 15 mg/mL and deglycosylated with endoglycosidase H (New England BioLabs, Ipswich, MA). The extent of deglycosylation was monitored by SDS–PAGE before crystallization trials. For surface plasmon resonance studies, a similar construct of KIR3DL1*001 was prepared in the pFastBac vector and expressed from Hi-5 insect cells (Invitrogen, Carlsbad, CA). KIR3DL1 was purified as described above, but excluding the endoglycosidase H deglycosylation step.
Hydrogen-deuterium exchange and analysis by LC-MS.
A sample containing 5 μg (0.1 nmol) of peptide or protein complex containing peptide was diluted 24-fold with 50 mM Tris and 50 mM NaCl dissolved in D2O (Cambridge Isotope Laboratories) at 25 °C to label the sample. The deuteration reaction was quenched at 10 s by the addition of an equal volume of 100 mM NaH2PO4, pH 2.4, and quickly frozen in a dry ice–ethanol bath. The frozen sample was quickly thawed and immediately injected onto a micropeptide trap column connected to a C18 HPLC column coupled to a Bruker Micro quadrupole time-of-flight mass spectrometer. The HLA-bound peptide was separated using a 12-min gradient of 10–45% acetonitrile at a flow rate of 50 μL/min. The micropeptide trap and C18 HPLC column were immersed in ice to minimize back exchange. Because the mass of a peptide increases by one for every amide hydrogen atom exchanged with deuterium, the amount of deuterium can be determined by comparison of the mass of the labeled peptide with the mass of the same peptide without the label. The centroid mass of each peptide was determined using the software package MagTran64. The data presented in the main text are representative of three independent experiments.
Crystallization, data collection, structure determination and refinement.
HLA-B*57:01 binary and ternary complexes with KIR3DL1*001 were concentrated to ~10 mg/mL and crystallized at 294K via the hanging-drop vapor-diffusion method. Binary complexes (TSTTSVASSW, TSTFEDVKILAF and SSTRGISQLW) crystallized from a solution comprising 12–20% PEG 4000, 0.2 M ammonium acetate and 0.1 M tri-sodium citrate, pH 5.4–5.6. Ternary complexes (KIR3DL1–TW10 and KIR3DL1–T3N) crystallized from a solution comprising 16% PEG 3350, 2% tacsimate, pH 5.0, and 0.1 M tri-sodium citrate, pH 5.6. Before data collection, crystals were equilibrated in reservoir solution with 10% glycerol added as a cryoprotectant and then flash-cooled in a stream of liquid nitrogen at 100 K. Data sets were collected at the MX2 beamline (Australian Synchrotron, Clayton, Victoria, Australia). The data were recorded on a Quantum-315 CCD (charge-coupled device) detector and were integrated and scaled using MOSFLM and SCALA from the CCP4 program suite65–67. Details of the data processing statistics are summarized in Table 1. The crystal structures were solved by molecular replacement, as implemented in PHASER68, with HLA-B*57:01-LF9 used as the search model (PDB 2RFX). Refinement of the models proceeded with iterative rounds of manual building in COOT69 and refinement in PHENIX70. The structures were validated with MOLPROBITY71. Refinement statistics are summarized in Table 1.
Surface plasmon resonance.
Surface plasmon resonance experiments were conducted at 298 K on a Biacore 3000 instrument using HBS buffer (10 mM HEPES-HCl, pH 7.4, 150 mM NaCl and 0.005% surfactant P20). The W6/32 antibody was immobilized on a CM5 chip via amine coupling to capture HLA–peptide complexes, creating a surface density of approximately 700 response units (RU). KIR3DL1*001 (2.37–300 μM) was injected over the chip at a flow rate of 5 μL/min. The response to W6/32 alone was subtracted from the response to KIR3DL1*001-HLA-B*5701–peptide. Equilibrium data were analyzed with GraphPad Prism. Data in the main text are representative of two independent experiments. Equilibrium binding constants were generated with nonlinear regression analysis using the following equation: Y = BmaxX/(Kd + X), where Bmax is the extrapolated binding at saturation.
Transfection studies.
FLAG-tagged KIR3DL1*001 was cloned into the pEF6 vector. Specific nucleotide residues were mutated with the QuikChange II site-directed mutagenesis kit (Stratagene), and constructs were introduced into HEK293T cells with FuGene 6 Transfection Reagent (Roche). After 48 h, the cells were harvested and stained with anti-Flag (clone M2, Sigma-Aldrich) or tetramer for 30 min at 4 °C. The cells were then washed and analyzed on a Fortessa flow cytometer (BD Biosciences). Staining with anti-Flag showed that none of the introduced mutations substantially affected cell surface expression of KIR3DL1*001 (data not shown). KIR3DL1 mutant data in the main text are representative of three independent experiments. KIR3DL1 allotype data in the main text are representative of four independent experiments; data were not analyzed if Flag expression was present on >5% of cells, which resulted in a minimum of two replicates in this assay. HEK293T cells were regularly tested and maintained as mycoplasma free for the duration of these studies.
Data availability.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the following accession codes: TSTTSVASSW, 5T6X; TSTFEDVKILAF, 5T6Y; SSTRGISQLW, 5T6W; KIR3DL1–TW10, 5T6Z; and KIR3DL1-T3N, 5T70. The source data for Figures 2 and 3 are available in Supplementary Tables 1–3. Other data from this study are available from the corresponding authors upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by project grants from the National Health and Medical Research Council of Australia (NH&MRC; APP1063829 to A.W.P., and APP1099814 to J.P.V. and D.A.P.) and the Australian Research Council (ARC; DP150104503 to J.R. and A.W.P.). A.W.P. is an NH&MRC Senior Research Fellow. P.T.I. is an NH&MRC Early Career Fellow. S.H.R. is the recipient of an Australian Postgraduate Award. D.A.P. is supported by a Wellcome Trust Senior Investigator Award. J.R. is supported by an ARC Laureate Fellowship. This work was funded in part by the intramural program of the National Institutes of Health, National Cancer Institute. This research was carried out in part on the MX2 beamline at the Australian Synchrotron, Victoria, Australia. J. Mak (Deakin University, Melbourne, Victoria, Australia) provided the Gag plasmid and generated the antibody used to assay Gag expression in transfectants.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Saper MA, Bjorkman PJ & Wiley DC Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 Å resolution. J. Mol. Biol. 219, 277–319 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Rammensee HG, Friede T & Stevanoviíc S MHC ligands and peptide motifs: first listing. Immunogenetics 41, 178–228 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Deres K, Beck W, Faath S, Jung G & Rammensee HG MHC/peptide binding studies indicate hierarchy of anchor residues. Cell. Immunol. 151, 158–167 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Wilson IA & Fremont DH Structural analysis of MHC class I molecules with bound peptide antigens. Semin. Immunol. 5, 75–80 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Garrett TP, Saper MA, Bjorkman PJ, Strominger JL & Wiley DC Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature 342, 692–696 (1989). [DOI] [PubMed] [Google Scholar]
- 6.Speir JA, Stevens J, Joly E, Butcher GW & Wilson IA Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa. Immunity 14, 81–92 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Tynan FE et al. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J. Biol. Chem. 280, 23900–23909 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Malnati MS et al. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science 267, 1016–1018 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Peruzzi M, Parker KC, Long EO & Malnati MS Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J. Immunol. 157, 3350–3356 (1996). [PubMed] [Google Scholar]
- 10.Stewart-Jones GB et al. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur. J. Immunol. 35, 341–351 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Fan QR, Long EO & Wiley DC Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat. Immunol. 2, 452–460 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Vivian JP et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 479, 401–405 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins EJ, Garboczi DN & Wiley DC Three-dimensional structure of a peptide extending from one end of a class I MHC binding site. Nature 371, 626–629 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Tenzer S et al. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat. Immunol. 10, 636–646 (2009). [DOI] [PubMed] [Google Scholar]
- 15.McMurtrey C et al. Toxoplasma gondii peptide ligands open the gate of the HLA class I binding groove. eLife 5, e12556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stryhn A, Pedersen LO, Holm A & Buus S Longer peptide can be accommodated in the MHC class I binding site by a protrusion mechanism. Eur. J. Immunol. 30, 3089–3099 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Schittenhelm RB, Dudek NL, Croft NP, Ramarathinam SH & Purcell AW A comprehensive analysis of constitutive naturally processed and presented HLA-C*04:01 (Cw4)-specific peptides. Tissue Antigens 83, 174–179 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Carrington M & O’Brien SJ The influence of HLA genotype on AIDS. Annu. Rev. Med. 54, 535–551 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Migueles SA et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97, 2709–2714 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 11, 1290–1292 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Kaslow RA et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2, 405–411 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Mallal S et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 359, 727–732 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Illing PT et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486, 554–558 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Hetherington S et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet 359, 1121–1122 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Goulder PJ et al. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retroviruses 12, 1691–1698 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Klein MR et al. Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gag- and RT-specific cytotoxic T lymphocyte responses. J. Gen. Virol. 79, 2191–2201 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Bailey JR, Williams TM, Siliciano RF & Blankson JN Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203, 1357–1369 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernardin F, Kong D, Peddada L, Baxter-Lowe LA & Delwart E Human immunodeficiency virus mutations during the first month of infection are preferentially found in known cytotoxic T-lymphocyte epitopes. J. Virol. 79, 11523–11528 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganusov VV et al. Fitness costs and diversity of the cytotoxic T lymphocyte (CTL) response determine the rate of CTL escape during acute and chronic phases of HIV infection. J. Virol. 85, 10518–10528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Picado J et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80, 3617–3623 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novitsky V et al. Dynamics and timing of in vivo mutations at Gag residue 242 during primary HIV-1 subtype C infection. Virology 403, 37–46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura T et al. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J. Virol. 83, 2743–2755 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brackenridge S et al. An early HIV mutation within an HLA-B*57-restricted T cell epitope abrogates binding to the killer inhibitory receptor 3DL1. J. Virol. 85, 5415–5422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crawford H et al. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 81, 8346–8351 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alter G et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 204, 3027–3036 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin MP et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31, 429–434 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Qi Y et al. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2, e79 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chessman D et al. Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity 28, 822–832 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Colaert N, Helsens K, Martens L, Vandekerckhove J & Gevaert K Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 (2009). [DOI] [PubMed] [Google Scholar]
- 40.O’Connor GM et al. Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity. J. Immunol. 192, 2875–2884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang CR et al. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell 82, 655–664 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Escobar H et al. Large scale mass spectrometric profiling of peptides eluted from HLA molecules reveals N-terminal-extended peptide motifs. J. Immunol. 181, 4874–4882 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Samino Y et al. A long N-terminal-extended nested set of abundant and antigenic major histocompatibility complex class I natural ligands from HIV envelope protein. J. Biol. Chem. 281, 6358–6365 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Petersen JL, Morris CR & Solheim JC Virus evasion of MHC class I molecule presentation. J. Immunol. 171, 4473–4478 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Jost S & Altfeld M Evasion from NK cell-mediated immune responses by HIV-1. Microbes Infect. 14, 904–915 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz O, Maréchal V, Le Gall S, Lemonnier F & Heard JM Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2, 338–342 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Seeger M, Ferrell K, Frank R & Dubiel W HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J. Biol. Chem. 272, 8145–8148 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Kutsch O, Vey T, Kerkau T, Hünig T & Schimpl A HIV type 1 abrogates TAP-mediated transport of antigenic peptides presented by MHC class I. Transporter associated with antigen presentation. AIDS Res. Hum. Retroviruses 18, 1319–1325 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Iglesias MC et al. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood 118, 2138–2149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu YC et al. A molecular basis for the interplay between T cells, viral mutants, and human leukocyte antigen micropolymorphism. J. Biol. Chem. 289, 16688–16698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ladell K et al. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity 38, 425–436 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Goulder PJ et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412, 334–338 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Schneidewind A et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81, 12382–12393 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crawford H et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206, 909–921 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alter G & Altfeld M NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J. Intern. Med. 265, 29–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lichterfeld M et al. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 204, 2813–2824 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croft NP et al. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog. 9, e1003129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fadda L et al. Common HIV-1 peptide variants mediate differential binding of KIR3DL1 to HLA-Bw4 molecules. J. Virol. 85, 5970–5974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saunders PM et al. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J. Exp. Med. 213, 791–807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kostenko L et al. Rapid screening for the detection of HLA-B57 and HLA-B58 in prevention of drug hypersensitivity. Tissue Antigens 78, 11–20 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Dudek NL et al. Constitutive and inflammatory immunopeptidome of pancreatic β-cells. Diabetes 61, 3018–3025 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purcell AW & Gorman JJ The use of post-source decay in matrix-assisted laser desorption/ionisation mass spectrometry to delineate T cell determinants. J. Immunol. Methods 249, 17–31 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Clements CS et al. The production, purification and crystallization of a soluble heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta Crystallogr. D Biol. Crystallogr 58, 2131–2134 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z & Marshall AG A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 9, 225–233 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 66.Evans P Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Leslie AGW Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography No. 26 (1992). [Google Scholar]
- 68.McCoy AJ et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the following accession codes: TSTTSVASSW, 5T6X; TSTFEDVKILAF, 5T6Y; SSTRGISQLW, 5T6W; KIR3DL1–TW10, 5T6Z; and KIR3DL1-T3N, 5T70. The source data for Figures 2 and 3 are available in Supplementary Tables 1–3. Other data from this study are available from the corresponding authors upon reasonable request.