Staphylococcus aureus SdrC forms a unique dimer through homophilic interactions, and a sandwich-like clamp at the C-terminus is formed by the two monomers in the stabilized SdrC dimer. Comparison with other Sdr proteins led to the proposal of a model for the confomational change of SdrC upon ligand binding.
Keywords: Staphylococcus aureus, adhesion, SdrC, dimer, homophilic interaction
Abstract
Staphylococcus aureus is an opportunistic disease-causing pathogen that is widely found in the community and on medical equipment. A series of virulence factors secreted by S. aureus can trigger severe diseases such as sepsis, endocarditis and toxic shock, and thus have a great impact on human health. The transformation of S. aureus from a colonization state to a pathogenic state during its life cycle is intimately associated with the initiation of bacterial aggregation and biofilm accumulation. SdrC, an S. aureus surface protein, can act as an adhesin to promote cell attachment and aggregation by an unknown mechanism. Here, structural studies demonstrate that SdrC forms a unique dimer through intermolecular interaction. It is proposed that the dimerization of SdrC enhances the efficiency of bacteria–host attachment and therefore contributes to the pathogenicity of S. aureus.
1. Introduction
The Gram-positive bacterium Staphylococcus aureus is a commensal opportunistic pathogen that colonizes human tissues, increasing the risk of developing severe infections or diseases such as endocarditis, bacteremia, sepsis and osteomyelitis (Kluytmans et al., 1997 ▸; von Eiff et al., 2001 ▸; Lowy, 1998 ▸). Virulence factors secreted by S. aureus play significant roles in the initial contact with host cells, and are also responsible for the stepwise progression of S. aureus from an asymptomatic colonizer to an invasive pathogen (Jenkins et al., 2015 ▸).
In S. aureus, there are numerous cell-wall-anchored (CWA) virulence factors that are involved in colonization or infection (Foster et al., 2014 ▸). Among these CWA proteins, a family called MSCRAMM (microbial surface components recognizing adhesive matrix molecules) mediate the adherence of S. aureus to host tissues. Effective initiation of adherence is fundamental for subsequent infection by and pathogenicity of S. aureus. However, the adhesion process that is involved in bacteria–host interactions is highly sophisticated.
Three prototype MSCRAMMs (FnBPA, CNA and ClfA) are similar in structure, with a long (∼40-amino-acid) signal sequence at the N-terminus that is required for protein secretion and an extracellular A-region that can be divided into three subdomains called N1–N3 for ligand binding (McDevitt et al., 1997 ▸; Savage et al., 1995 ▸). At the C-terminus, there is a proline- or glycine-rich wall-spanning region, an LPXTG motif called the sorting signal that is cleavable by sortase, and a hydrophobic membrane-spanning domain (Patti et al., 1994 ▸; Schneewind et al., 1995 ▸). A unique R-domain with a Ser–Asp dipeptide repeat (SD-repeat) follows the A-region.
The gene loci of sdrC, sdrD and sdrE were identified by Southern blotting using DNA encoding the R-domain of ClfA as a probe, and the three genes are closely linked and are tandemly arrayed (McDevitt et al., 1994 ▸; Josefsson, McCrea et al., 1998 ▸). Sdr proteins are similar to Clf proteins in structural organization, but have additional B-repeats of 110–113 residues located between the A-region and the SD-repeat with unknown function (Speziale et al., 2014 ▸; Josefsson, McCrea et al., 1998 ▸). Among members of the Clf–Sdr subfamily, the A-region is similar in size but has a low sequence identity of less than 30% (Ni Eidhin et al., 1998 ▸). TYTFTDYVD, a consensus motif in the A-region that overlaps the C-terminus of the EF-hand-like motif in ClfA, is shared by all proteins in the subfamily. An EF-hand loop with a high affinity for Ca2+ was found in each B-repeat and was indispensable for the structural integrity of the protein (Josefsson, O’Connell et al., 1998 ▸; Josefsson, McCrea et al., 1998 ▸). Whether the B-repeats have ligand-binding activity or only act in projecting the A-region away from the cell surface for better ligand interaction remains unresolved.
It has been reported that gene expression of sdrC is upregulated during the shift of S. aureus from colonization to invasion, suggesting a potential role for SdrC in staphylococcal pathogenesis (Jenkins et al., 2015 ▸). Moreover, the N2 domain of SdrC has been implicated to be involved in biofilm formation through homophilic interactions in previous research (Barbu et al., 2014 ▸), but the binding details were not fully clarified (Pi et al., 2020 ▸). Here, we determined the crystal structure of the N2 and N3 domains of SdrC (SdrC-N2N3) by X-ray diffraction. The dimer-form structure provided solid evidence for homophilic binding in SdrC. From the structural analysis, we further identified crucial residues in the dimer interface that are involved in intermolecular interactions. These results provide insights into the molecular mechanism of the adhesin SdrC that promotes colonization by S. aureus and also establish a model for the enhanced pathogenicity of the bacterium.
2. Materials and methods
2.1. Gene cloning, protein expression and purification
The cDNA sequence encoding the fragment 178–496 of SdrC (UniProt entry ID Q99W48) from S. aureus strain Mu50 was cloned into pET-28a-C6His vector and then transformed into Escherichia coli Rosetta2 (DE3) cells for protein expression. The cells were grown in Luria broth containing 50 mg l−1 ampicillin and 20 mg l−1 chloramphenicol at 37°C until the OD600 reached 0.4. Expression of the recombinant protein was induced by adding isopropyl β-d-1-thiogalactopyranoside to the medium to a concentration of 0.5 mM and the cells were further grown overnight at 16°C. The cells were harvested by centrifugation at 5000g for 15 min, and the cell pellet was resuspended in lysis buffer [50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5%(v/v) glycerol, 1 mM tris(2-carboxyethyl)phosphine] and lysed using a sonicator. The cell lysate was centrifuged at 10 000g for 1 h at 4°C and the supernatant was incubated with pre-equilibrated Ni–NTA resin at 4°C for 30 min, followed by reloading onto a nickel-affinity column to remove unbound components. The immobilized proteins were washed with a gradient of imidazole (5–100 mM) and finally eluted as 5 ml fractions. The fractions containing the desired protein SdrC (178–496) were further purified by size-exclusion chromatography on a Superdex 200 (16/60) prep-grade gel-filtration column in SEC buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl) using an ÄKTA purification system (GE Healthcare). The purified protein was assessed by SDS–PAGE, and its concentration was roughly measured by the Bradford assay using a spectrophotometer at 595 nm. Information on protein production is shown in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | S. aureus strain Mu50 |
| DNA source | Amplified from S. aureus strain Mu50 genome |
| Forward primer† | 5′-CATGCCATGGCAGCTCCACAACAAGGAAC-3′ |
| Reverse primer‡ | 5′-CCGCTCGAGTTTCTTTTGGTCGCCATTTGCAG-3′ |
| Cloning vector | pET-28a-C6His |
| Expression vector | pET-28a-C6His |
| Expression host | E. coli Rosetta2 (DE3) |
| Complete amino-acid sequence of the construct produced | MAAPQQGTNVNDKVHFTNIDIAIDKGHVNKTTGNTEFWATSSDVLKLKANYTIDDSVKEGDTFTFKYGQYFRPGSVRLPSQTQNLYNAQGNIIAKGIYDSKTNTTTYTFTNYVDQYTNVSGSFEQVAFAKRENATTDKTAYKMEVTLGNDTYSKDVIVDYGNQKGQQLISSTNYINNEDLSRNMTVYVNQPKKTYTKETFVTNLTGYKFNPDAKNFKIYEVTDQNQFVDSFTPDTSKLKDVTGQFDVIYSNDNKTATVDLLNGQSSSDKQYIIQQVAYPDNSSTDNGKIDYTLETQNGKSSWSNSYSNVNGSSTANGDQKKLEHHHHHH |
The NcoI site is underlined.
The XhoI site is underlined.
2.2. Crystallization
The purified SdrC (178–496) with a C-terminal His6 tag was concentrated to 10 mg ml−1 as determined by a OneDrop spectrophotometer using a molar extinction coefficient of 42 290 M −1 cm−1 and an Abs 0.1% (= 1 g l−1) of 1.141 (estimated using the ProtParam tool from ExPASy). The protein was crystallized at 16°C by hanging-drop vapor diffusion. Optimal crystals for data collection were obtained using a well solution consisting of 0.08 M magnesium formate dihydrate, 25%(w/v) polyethylene glycol 3350. The crystals were cryoprotected in a solution consisting of 80% well solution and 20%(v/v) glycerol and were immediately flash-cooled in liquid nitrogen. Crystallization information is given in Table 2 ▸.
Table 2. Crystallization conditions.
| Method | Sitting-drop vapor diffusion |
| Plate type | 48-well plates |
| Temperature (K) | 277 |
| Protein concentration (mg ml−1) | 18 |
| Buffer composition of protein solution | 20 mM Tris–HCl pH 8.0, 150 mM NaCl |
| Composition of reservoir solution | 0.08 M magnesium formate dihydrate, 25%(w/v) polyethylene glycol 3350 |
| Volume ratio of drop | 1:1 |
| Volume of reservoir (µl) | 0.5 |
2.3. Data collection and processing
X-ray diffraction data were collected at a wavelength of 0.9793 Å on beamline BL18U1 at the National Center for Protein Science Shanghai (NCPSS) under gaseous nitrogen (100 K) using a Dectris PILATUS 6M detector. The best data were collected to 1.54 Å resolution and were indexed, integrated and scaled using HKL-3000 (Minor et al., 2006 ▸). Data-collection and processing statistics are shown in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | Beamline BL18U1, NCPSS |
| Wavelength (Å) | 0.9793 |
| Temperature (K) | 100 |
| Detector | PILATUS 6M |
| Crystal-to-detector distance (mm) | 450 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 0.2 |
| Space group | P1211 |
| a, b, c (Å) | 63.73, 87.13, 63.97 |
| α, β, γ (°) | 90, 97.99, 90 |
| Mosaicity (°) | 0.1–0.2 |
| Resolution range (Å) | 100–1.49 (1.52–1.49) |
| Total No. of reflections | 678848 (100843) |
| No. of unique reflections | 57973 (2869) |
| Completeness (%) | 99.9 (100.0) |
| Multiplicity | 6.7 (6.8) |
| 〈I/σ(I)〉 | 20.2 (3.6) |
| R p.i.m. (%) | 17.5 (60.4) |
| Overall B factor from Wilson plot (Å2) | 19.5 |
2.4. Structure refinement
The structure was solved by molecular replacement using MOLREP (Vagin & Teplyakov, 2010 ▸) as implemented in the CCP4 suite (Winn et al., 2011 ▸) using the homologous protein UafA (PDB entry 3irp) as a search model (Matsuoka et al., 2011 ▸). The structural model of SdrC was automatically rebuilt using PHENIX AutoBuild (Terwilliger et al., 2008 ▸) and refinement was carried out using REFMAC5 (Murshudov et al., 2011 ▸) and Coot (Emsley et al., 2010 ▸) iteratively. Structure-refinement statistics are summarized in Table 4 ▸. Structural analysis and figure generation were performed using PyMOL (http://www.pymol.org).
Table 4. Structure refinement.
Values in parentheses are for the highest resolution shell. The structure was refined using REFMAC 5.8.0103 (Murshudov et al., 2011 ▸); MolProbity (Chen et al., 2010 ▸) was used for Ramachandran analysis.
| Resolution range (Å) | 63.35–1.54 (1.58–1.54) |
| Completeness (%) | 99.1 (89.8) |
| No. of reflections, working set | 95689 (6424) |
| No. of reflections, test set | 5023 (323) |
| Final R cryst | 0.126 (0.188) |
| Final R free | 0.177 (0.242) |
| No. of non-H atoms | |
| Protein | 5078 |
| Mg | 2 |
| Glycerol | 30 |
| Water | 1138 |
| Total | 6248 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.027 |
| Angles (°) | 2.255 |
| Average B factors (Å2) | |
| Protein | 17.6 |
| Mg | 18.6 |
| Glycerol | 41.8 |
| Water | 35.6 |
| Ramachandran plot | |
| Favored regions (%) | 98.9 |
| Additionally allowed (%) | 1.1 |
3. Results and discussion
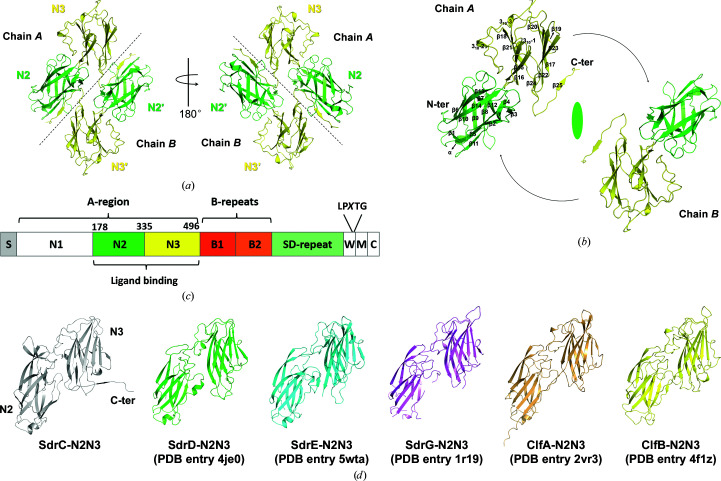
Since SdrC promotes biofilm accumulation by homophilic binding (Barbu et al., 2014 ▸; Feuillie et al., 2017 ▸), the structure of SdrC-N2N3 (residues 178–496) was determined to verify self-interactions. Inadvertently, a crystal of the single-point mutation P366H was obtained with reasonable diffraction (Table 3 ▸). In this structure, there are two molecules in the asymmetric unit and they are highly identical, with an r.m.s.d. of 0.2 Å (DALI pairwise structure comparison; Holm, 2019 ▸). Each molecule is composed of the N2 and N3 subdomains consisting of residues 181–334 and 335–495, respectively, including one short α-helix, three 310-helices, 25 β-strands and several connecting loops. Facing one another, the two molecules have a relative rotation of ∼180°, so that the C-terminus of N3 from one chain (chain A) contacts N2′ from the other chain (chain B) (Figs. 1 ▸ a and 1 ▸ b).
Figure 1.
Overall structure of SdrC-N2N3. (a) Crystal structure of SdrC. The N2 and N3 subdomains are colored green and yellow, respectively. Black dotted lines roughly separate the two chains. (b) Relationship of the two chains in the dimer. Secondary-structure elements are indicated on chain A. The dyad axis is shown as an ellipsoid and black arrows show the rotation between the two monomers. (c) Structural organization of SdrC. S, signal sequence, The A-region includes N1–N3. The ligand-binding region, N2N3, is located within the A-region. The B-repeats include B1 and B2. SD-repeat, Ser-Asp dipeptide repeats. W, short wall-spanning region. M, membrane-spanning segment. C, intracellular region. The LPXTG motif is located between the W and M domains. (d) Structural comparison of SdrC with other Clf–Sdr family proteins. One copy (chain A) was extracted from the apo structures of SdrC-N2N3 (gray), SdrD-N2N3 (green), SdrE-N2N3 (cyan) and SdrG-N2N3 (violet) and the structures of the ClfA–Fg (light orange) and ClfB–keratin (yellow) complexes. The overall structure of the six proteins was composed of two Ig-like subdomains with a similar arrangement.
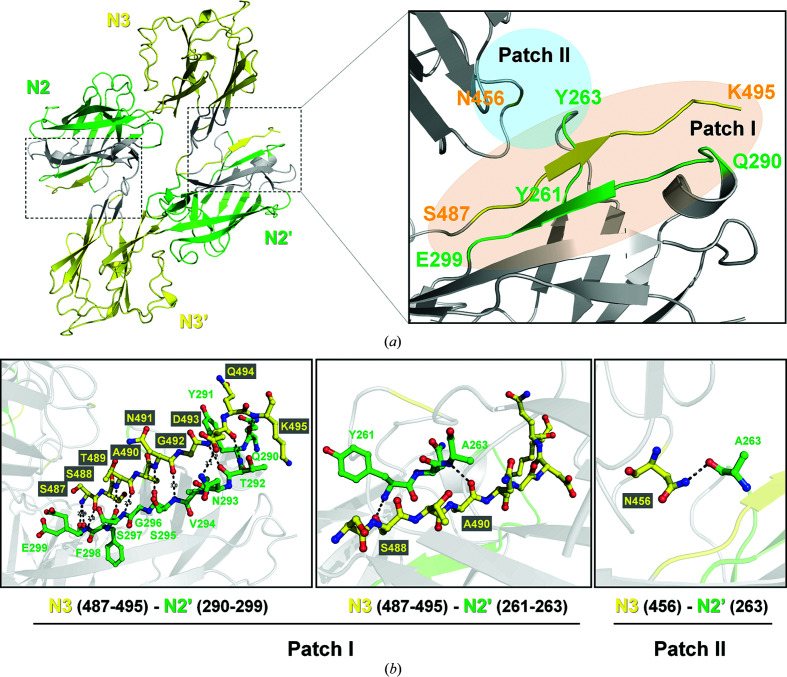
The domain organization of Sdr proteins is highly conserved (Fig. 1 ▸ c), and the overall structure of SdrC-N2N3 is similar to those of other members of the Clf–Sdr subfamily (Figs. 1 ▸ d and 3a; Supplementary Table S1; Wang et al., 2013 ▸; Zhang et al., 2017 ▸; Ponnuraj et al., 2003 ▸; Ganesh et al., 2008 ▸; Xiang et al., 2012 ▸). All six proteins are composed of two Ig-like subdomains with a similar arrangement, but SdrC forms a homodimer through intermolecular interactions. The dimer interface contains two major interaction regions (Fig. 2 ▸ a): (i) between the C-terminus of N3 (yellow, residues 487–495) and N2′ (green, residues 290–299 and 261–263) and (ii) between a loop from N3 (yellow, residue 456) and a loop from N2′ (green, residue 263), and vice versa. The major interactions in the SdrC-N2N3 dimer are between the C-terminal tail (residues 487–495) of N3 and residues 290–299 of N2′, and the contact area as estimated by PISA (Proteins, Interfaces, Structures and Assemblies; http://pdbe.org/pisa/) is ∼1750 Å2 (Fig. 2 ▸ a and Supplementary Fig. S5).
Figure 2.
Intermolecular interactions within the SdrC-N2N3 dimer. (a) Dimer interface of SdrC. Two interaction regions, patch I, where residues 487–495 of N3 (yellow) interact with residues 290–299 and 261–263 of N2′ (green), and patch II, where residue 456 of N3 (yellow) interacts with residue 263 of N2′ (green), are highlighted using light orange and aquamarine, respectively. (b) Residues involved in homophilic binding of SdrC. Interacting residues at the dimer interface are indicated. Hydrogen bonds are shown as black dashed lines.
Based on the crystal structure, the potential interactions between two molecules predicted by PDBsum (Laskowski et al., 2018 ▸) include 35 hydrogen bonds and 298 nonbonded contacts, which are provided by 39 residues (Supplementary Fig, S5). Structural analysis indicated that 16 interacting residues form 13 hydrogen bonds, and most of the bonds are mediated by backbone interactions (Fig. 2 ▸ b). Ser487, Thr489, Asn491 and Asp493 of one chain form eight main-chain hydrogen-bond pairs with Glu299, Ser297, Ser295 and Thr292 from the opposite chain, respectively. The residue pairs Ala490–Ala263 and Lys495–Gln290 also provide two main-chain interactions. In addition, three side-chain interactions are provided by Ser488, Asp493 and Asn456 of one molecule and Asn293 from the other molecule (Fig. 2 ▸ b). Thus, the C-terminal tail of N3 is clamped and fixed by two fragments from N2′ to form a sandwich-like architecture (Fig. 2 ▸ a). The hydrogen bond provided by Asn456 might also contribute to this. This ingenious sandwich-like arrangement performed well for the dimerization of SdrC.
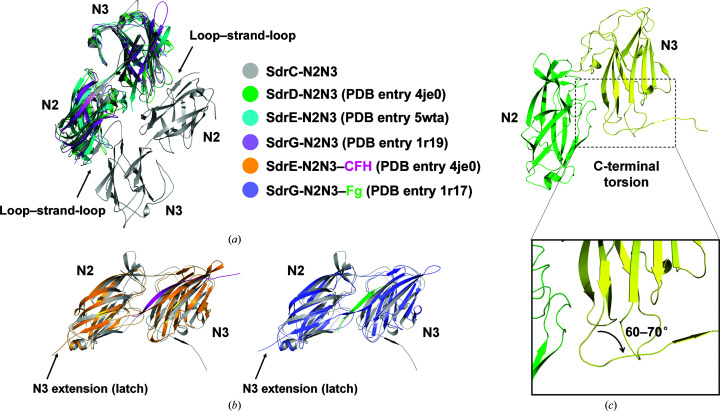
When compared with the structures of other Sdr proteins and ligand-bound Sdr, an extra loop–strand–loop (residues 487–495) was found in the C-terminus of SdrC-N2N3 (Fig. 3 ▸ a). In apo structures of SdrD-N2N3 (PDB entry 4je0) and SdrE-N2N3 (PDB entry 5wta), electron density for the coordinate region was not visible due to flexibility (Wang et al., 2013 ▸). However, in the SdrE-N2N3–CFH complex (PDB entry 5wtb), the C-terminal region (residues 587–598) of N3 extended towards N2 and formed an antiparallel β-sheet to latch the ligand (Fig. 3 ▸ b; Zhang et al., 2017 ▸). Moreover, the structures of the apo form of S. epidermidis SdrG (PDB entry 1r19) and of its complex with a fibrinogen-derived peptide (PDB entry 1r17) showed the same features as SdrE (Fig. 3 ▸ b; Ponnuraj et al., 2003 ▸; Bowden et al., 2008 ▸).
Figure 3.
Structural comparison of Sdr family proteins. (a) Superimposition of SdrC with other Sdr proteins (apo form). SdrD, SdrE and SdrG are colored as in Fig. 1 ▸. The SdrC dimer is shown in gray. Compared with other Sdr proteins, an extra loop–strand–loop (residues 487–495) is found in the C-terminus of SdrC-N2N3 (black arrow). (b) Superimposition of SdrC with the SdrE–CFH and SdrG–Fg complexes. SdrC is compared with SdrE (bright orange) and SdrG (slate) in their complexes with CFH (light magenta) and Fg (green), respectively. The latch structure in SdrE–CFH and SdrG–Fg formed by the C-terminal extension of N3 is indicated with a black arrow. (c) The deviation at the C-terminus in SdrC-N2N3. The local shift at the C-terminus of SdrC-N2N3 triggered a 60–70° deviation.
For some MSCRAMM proteins, such as the fibronectin-binding protein FnBP, the collagen-binding protein CNA and the fibrinogen-binding protein Clf, their adhesion functions are achieved by recognition of the host extracellular matrix (ECM) as a ligand. However, for other adhesins homophilic binding is the major contribution to bacterial adhesion and colonization. In the case of SraP, cadherin-like domains in the BR-region promote biofilm formation by homophilic interactions between pairs of CDHL domains (Yang et al., 2014 ▸). In Aap from S. epidermidis, the repeated domains G5-E can undergo a Zn2+-dependent homophilic interaction to form an antiparallel twisted cable in an end-to-end manner (Conrady et al., 2013 ▸). Like SraP, the SdrC protein can form a homophilic dimer through intermolecular interactions, with the potential to promote adhesion, colonization and subsequent biofilm accumulation of S. aureus.
In previous studies, Barbu and coworkers identified two fragments (RPGSV247–251 and VDQYT288–292) that are potentially involved in the homophilic interaction of SdrC through phage display. They also generated a hypothetical model of SdrC-N2N3 using in silico methods based on the crystal structure of ClfA (Barbu et al., 2014 ▸; Feuillie et al., 2017 ▸). In this predicted model, the RPGSV247–251 fragment located in the cleft between N2 and N3 was related to ligand binding. Recently, Pi and coworkers solved a dimer structure of Ca2+-bound SdrC and found that the VDQYT288–292 fragment was adjacent to the Ca2+-binding site so that the dimer could be destroyed by Ca2+ deficiency. Moreover, they also reported another SdrC dimer (without Ca2+) that was formed in the same way as our structure (Pi et al., 2020 ▸). Although both of the research groups mentioned above reported that N2 was the only subdomain that was involved in SdrC self-association, we assumed that in addition to Ca2+-mediated dimerization the homophilic interaction between N2 and N3 may be another mechanism for SdrC biofilm formation (Supplementary Figs. S1–S5). Three structures of SdrC-N2N3 and the structure of the complex of SdrE-N2N3 with its ligand peptide CFH were compared to show that the dimerization reported for our SdrC-N2N3 structure was complementary to the N2–N2′ self-association mediated by Ca2+ (Supplementary Figs. S1 and S2), and most of the SdrC-N2N3 obtained from size-exclusion chromatography during purification was shown to be dimeric in solution (Supplementary Figs. S3 and S4). Moreover, analysis of the interactions at the dimer interface also supported our assumption (Supplementary Fig. S5).
Since the C-terminus of N3 of SdrE and SdrG became visible in the map upon ligand binding, we speculated that the C-terminal extension of SdrC was probably induced by dimer formation. Notably, in the ‘Dock, Lock, Latch’ (DLL) model of SdrE or SdrG, the ligand was latched tightly within the binding groove between the N2 and N3 subdomains, making the C-terminus stretch to form an antiparallel β-sheet with N2 (Zhang et al., 2017 ▸; Ponnuraj et al., 2003 ▸). However, in the case of our SdrC structure a local shift starting from the end of β12 in N3 changed the track of the C-terminus and triggered a 60–70° deviation (Fig. 3 ▸ c). This large torsion made it easier for SdrC to contact the neighboring molecule and form a dimeric structure.
In contrast to the apo form of SdrE, in which the intrinsic loopA–B closed the ligand-binding cleft between N2 and N3 (Zhang et al., 2017 ▸), the putative binding pocket was exposed with no shelter in our SdrC dimer, similar to as in SdrG. With the C-terminus clamped, SdrC-N2N3 could not finish the subsequent process of ‘lock and latch’ upon ligand binding (Fig. 2 ▸ a) unless the dimer were to disassemble. Therefore, we assumed that ligand binding is followed by breakage of the SdrC dimer so as to release the C-terminal extension for latch formation.
SdrC has been reported to bind the ligand neurexin-1β (Nrx1β) with high affinity and specificity (Barbu et al., 2010 ▸), and the potential binding site in SdrC overlapped with the RPGSV247–251 fragment (Barbu et al., 2014 ▸; Feuillie et al., 2017 ▸). Thus, it was likely that binding of Nrx1β may interfere with SdrC dimerization; however, solid structural evidence was lacking, although inhibition of the interaction of SdrC was demonstrated by competition assays. However, in our SdrC structure the residues associated with the dimer interface were distinct from those hypothesized by modeling (Fig. 2 ▸; Feuillie et al., 2017 ▸). We assumed that breakage of the SdrC dimer was not the reason for the hypothesized breakage of homophilic bonds by Nrx1β, but that it is necessary for the subsequent ‘lock and latch’ steps upon ligand binding. Therefore, we proposed a model (Dock, Break, Lock, Latch; DBLL) to show how SdrC undergoes homophilic binding and dimer breakage through conformational change (Supplementary Fig. S6). In this model, the SdrC dimer was dynamically regulated by ligand binding (Dock), which exerts a disturbance of the overall structure, which subsequently separates into monomers (Break). The released monomer rotates its free C-terminal tail to form a latch structure extending to N2, locking the ligand in the binding cleft between N2 and N3 (Lock, Latch). Therefore, the conformational rearrangement induced by binding of a ligand (such as Nrx1β) might disrupt homophilic interactions of SdrC and attenuate bacterial adhesion as well as biofilm accumulation, thus greatly affecting the pathogenicity of S. aureus (Supplementary Fig. S6).
In conclusion, the adhesin SdrC may use two different mechanisms of dimerization to accelerate biofilm formation. Compared with Ca2+-bound SdrC, our structure implied that the SdrC dimer may play a role in favoring biofilm dynamics as a regulatory mechanism to flexibly adjust the environment or change the status of S. aureus, such as the transformation from the initial adhesion state to the subsequent invasion stage. These results indicated the possibility that pathogenic adhesins in antibiosis can serve as potential therapeutic targets. Further studies are required to explore the mechanism of SdrC–ligand binding and the regulation of other physiological processes by Sdr family proteins.
Supplementary Material
PDB reference: N2 and N3 domains of SdrC, 6leb
Supplementary Table and Figures. DOI: 10.1107/S2053230X21000741/ft5112sup1.pdf
Acknowledgments
We thank the staff of beamline BL18U1 of NCPSS (National Center for Protein Science Shanghai) for assistance with data collection.
Funding Statement
This work was funded by Anhui Department of Education grant KJ2019ZD02 to Min Zhang. Natural Science Foundation of Anhui Province grant 2008085QC161 to Tianrong Hang.
References
- Barbu, E. M., Ganesh, V. K., Gurusiddappa, S., Mackenzie, R. C., Foster, T. J., Sudhof, T. C. & Höök, M. (2010). PLoS Pathog. 6, e1000726. [DOI] [PMC free article] [PubMed]
- Barbu, E. M., Mackenzie, C., Foster, T. J. & Höök, M. (2014). Mol. Microbiol. 94, 172–185. [DOI] [PMC free article] [PubMed]
- Bowden, M. G., Heuck, A. P., Ponnuraj, K., Kolosova, E., Choe, D., Gurusiddappa, S., Narayana, S. V. L., Johnson, A. E. & Höök, M. (2008). J. Biol. Chem. 283, 638–647. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Conrady, D. G., Wilson, J. J. & Herr, A. B. (2013). Proc. Natl Acad. Sci. USA, 110, E202–E211. [DOI] [PMC free article] [PubMed]
- Eiff, C. von, Becker, K., Machka, K., Stammer, H. & Peters, G. (2001). N. Engl. J. Med. 344, 11–16. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Feuillie, C., Formosa-Dague, C., Hays, L. M. C., Vervaeck, O., Derclaye, S., Brennan, M. P., Foster, T. J., Geoghegan, J. A. & Dufrêne, Y. F. (2017). Proc. Natl Acad. Sci. USA, 114, 3738–3743. [DOI] [PMC free article] [PubMed]
- Foster, T. J., Geoghegan, J. A., Ganesh, V. K. & Höök, M. (2014). Nat. Rev. Microbiol. 12, 49–62. [DOI] [PMC free article] [PubMed]
- Ganesh, V. K., Rivera, J. J., Smeds, E., Ko, Y.-P., Bowden, M. G., Wann, E. R., Gurusiddappa, S., Fitzgerald, J. R. & Höök, M. (2008). PLoS Pathog. 4, e1000226. [DOI] [PMC free article] [PubMed]
- Holm, L. (2019). Bioinformatics, 35, 5326–5327. [DOI] [PubMed]
- Jenkins, A., Diep, B. A., Mai, T. T., Vo, N. H., Warrener, P., Suzich, J., Stover, C. K. & Sellman, B. R. (2015). mBio, 6, e02272–14. [DOI] [PMC free article] [PubMed]
- Josefsson, E., McCrea, K. W., Ní Eidhin, D., O’Connell, D., Cox, J., Höök, M. & Foster, T. J. (1998). Microbiology, 144, 3387–3395. [DOI] [PubMed]
- Josefsson, E., O’Connell, D., Foster, T. J., Durussel, I. & Cox, J. A. (1998). J. Biol. Chem. 273, 31145–31152. [DOI] [PubMed]
- Kluytmans, J., van Belkum, A. & Verbrugh, H. (1997). Clin. Microbiol. Rev. 10, 505–520. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A., Jabłońska, J., Pravda, L., Vařeková, R. S. & Thornton, J. M. (2018). Protein Sci. 27, 129–134. [DOI] [PMC free article] [PubMed]
- Lowy, F. D. (1998). N. Engl. J. Med. 339, 520–532. [DOI] [PubMed]
- Matsuoka, E., Tanaka, Y., Kuroda, M., Shouji, Y., Ohta, T., Tanaka, I. & Yao, M. (2011). Protein Sci. 20, 406–416. [DOI] [PMC free article] [PubMed]
- McDevitt, D., Francois, P., Vaudaux, P. & Foster, T. J. (1994). Mol. Microbiol. 11, 237–248. [DOI] [PubMed]
- McDevitt, D., Nanavaty, T., House-Pompeo, K., Bell, E., Turner, N., McIntire, L., Foster, T. & Höök, M. (1997). Eur. J. Biochem. 247, 416–424. [DOI] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ní Eidhin, D., Perkins, S., Francois, P., Vaudaux, P., Höök, M. & Foster, T. J. (1998). Mol. Microbiol. 30, 245–257. [DOI] [PubMed]
- Patti, J. M., Allen, B. L., McGavin, M. J. & Höök, M. (1994). Annu. Rev. Microbiol. 48, 585–617. [DOI] [PubMed]
- Pi, Y., Chen, W. & Ji, Q. (2020). Biochemistry, 59, 1465–1469. [DOI] [PubMed]
- Ponnuraj, K., Bowden, M. G., Davis, S., Gurusiddappa, S., Moore, D., Choe, D., Xu, Y., Hook, M. & Narayana, S. V. L. (2003). Cell, 115, 217–228. [DOI] [PubMed]
- Savage, B., Bottini, E. & Ruggeri, Z. M. (1995). J. Biol. Chem. 270, 28812–28817. [DOI] [PubMed]
- Schneewind, O., Fowler, A. & Faull, K. F. (1995). Science, 268, 103–106. [DOI] [PubMed]
- Speziale, P., Pietrocola, G., Foster, T. J. & Geoghegan, J. A. (2014). Front. Cell. Infect. Microbiol. 4, 171. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C., Grosse-Kunstleve, R. W., Afonine, P. V., Moriarty, N. W., Zwart, P. H., Hung, L.-W., Read, R. J. & Adams, P. D. (2008). Acta Cryst. D64, 61–69. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wang, X., Ge, J., Liu, B., Hu, Y. & Yang, M. (2013). Protein Cell, 4, 277–285. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Xiang, H., Feng, Y., Wang, J., Liu, B., Chen, Y., Liu, L., Deng, X. & Yang, M. (2012). PLoS Pathog. 8, e1002751. [DOI] [PMC free article] [PubMed]
- Yang, Y.-H., Jiang, Y.-L., Zhang, J., Wang, L., Bai, X.-H., Zhang, S.-J., Ren, Y.-M., Li, N., Zhang, Y.-H., Zhang, Z., Gong, Q., Mei, Y., Xue, T., Zhang, J.-R., Chen, Y. & Zhou, C.-Z. (2014). PLoS Pathog. 10, e1004169. [DOI] [PMC free article] [PubMed]
- Zhang, Y. J., Wu, M. H., Hang, T. R., Wang, C. L., Yang, Y., Pan, W. M., Zang, J. Y., Zhang, M. & Zhang, X. (2017). Biochem. J. 474, 1619–1631. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: N2 and N3 domains of SdrC, 6leb
Supplementary Table and Figures. DOI: 10.1107/S2053230X21000741/ft5112sup1.pdf