Abstract
Pioneer transcription factors have the intrinsic biochemical ability to scan partial DNA sequence motifs that are exposed on the surface of a nucleosome and thus access silent genes that are inaccessible to other transcription factors. Pioneer factors subsequently enable other transcription factors, nucleosome remodeling complexes, and histone modifiers to engage chromatin, thereby initiating the formation of an activating or repressive regulatory sequence. Thus, pioneer factors endow the competence for fate changes in embryonic development, are essential for cellular reprogramming, and rewire gene networks in cancer cells. Recent studies with reconstituted nucleosomes in vitro and chromatin binding in vivo reveal that pioneer factors can directly perturb nucleosome structure and chromatin accessibility in different ways. This review focuses on our current understanding of the mechanisms by which pioneer factors initiate gene network changes and will ultimately contribute to our ability to control cell fates at will.
Keywords: pioneer factor, nucleosome, chromatin, differentiation, reprogramming, gene networks
INTRODUCTION AND BACKGROUND
Transcription factors are adaptor proteins that recognize particular DNA sequences and cooperate with other factors to regulate target genes. Such regulation involves the recruitment of transcriptional coactivators or corepressors, histone modifiers, and nucleosome remodeling proteins to a local domain in chromatin. Consequently, functional gene regulatory regions, including promoters and enhancers, gene repressing elements, and genome architectural sites, assemble large complexes. The complexes often assemble in a cell-type-specific manner, with many bound proteins that can span hundreds of base pairs of DNA. Transcription factors that can act as pioneer factors were initially discovered while researchers were investigating how enhancer regulatory complexes are first formed in silent chromatin during embryonic development, leading to the induction of a tissue-specific gene network. Pioneer transcription factors were subsequently found to enable hormone-responsive gene networks in human cancers and artificially change the fate of cells in culture. The diverse roles of pioneer factors, particularly in cancer, have been reviewed (56, 61, 84, 94). This review focuses on the latest studies revealing the molecular processes by which pioneer factors bind to nucleosomes, perturb nucleosome structure to enable regulatory complex assembly, and reset genetic networks during cell fate transitions.
Diverse Types of Transcriptionally Silent Chromatin
Transcriptionally silent genes in a given cell type or state may be neutral to a cell’s function, necessary for a future function, or antithetical to a cell’s current and future functions. Thus, gene silencing involves mechanisms that range from allowing the competence of a gene to respond to stimuli to ensuring gene silence under most or all physiologic conditions.
An understanding of the diverse ways that chromatin structure silences gene activity is needed to determine how transcription factors overcome such barriers during gene network and cell fate changes. The DNA in eukaryotic cells winds nearly twice around an octamer of the four core histones, thereby forming arrays of nucleosomes (64). Within a nucleosome, part of the surface of DNA, along its long axis, faces the globular domains of the histones, causing the DNA sequence on that side to be hidden sterically. Thus, nucleosomal DNA can be inherently restrictive to transcription factor access and transcription itself (80). Furthermore, the N-terminal tail of histone H4, which extends from the nucleosome core, can interact with an acidic patch on the globular histone domain of an adjacent nucleosome, helping to stabilize internucleosome interactions that promote local chromatin condensation (4, 32, 102). The condensation of a nucleosome array can be further stabilized by linker histone, which binds at the entry and exit points of DNA on the nucleosome (40) and restricts the action of nucleosome remodelers (46, 50, 112). Chromatin compaction can be further modified by histone variants within the nucleosome core (34). Histone H3 can be covalently methylated on its N-terminal tail at lysine 9 (H3K9me2 or H3K9me3) (113) or lysine 27 (H3K27me3) (26, 96), which promotes the binding of heterochromatin protein 1 (HP1) or canonical Polycomb repressive complex 1 (PRC1), respectively, to further compact the chromatin (71, 107). The nuclear periphery is lined with lamin proteins (69) that associate with H3K9me2 heterochromatin to silence genes (110), while H3K9me3 heterochromatin occurs in apparent phase-separated globules throughout the nucleus (117, 132). Consistent with the many mechanisms to silence gene activity, imaging of nuclei by electron microscopy and optical super-resolution methods reveals clusters of nucleosomes in diverse compaction states (97, 99, 115). Thus, there are many mechanisms of repression that build upon the nucleosome repeats in chromatin.
Discovery of Pioneer Factors
Pioneer transcription factors were originally discovered by in vivo footprinting (88, 104) to determine which transcription factors bound first to a liver-specific enhancer of the alb1 gene in mouse embryo development (44). At the time, it was assumed that the earliest occupancy of the liver enhancer would be when liver genes are first expressed in hepatoblasts, as they differentiate from the embryonic endoderm (15); the endoderm itself was considered to be a negative control. The endoderm outside the prehepatic region is multipotent, and although liver genes are silent in this tissue, they remain competent to be activated for several days after the period of liver induction (44).
Unexpectedly, the extrahepatic endoderm exhibited occupancy of adjacent binding sites for FoxA and GATA transcription factors (11, 44), and this occupancy was retained throughout the embryonic period, during which the endoderm remained competent to induce liver genes (12). In hepatoblasts, multiple other factors bind the liver-specific enhancer as alb1 is activated (44). Notably, FoxA and GATA factors bind to the enhancer in silent chromatin. The factors were then tested for ability to bind the same enhancer target sequence on nucleosomes assembled in vitro. Recombinant FoxA1, but not other transcription factors, bound to its sites on mono- and dinucleosomes (21, 124) and helped GATA4 bind to an adjacent sequence (22). Finally, recombinant FoxA1 and, to a lesser extent, GATA4 could create a local nuclease-sensitive region beneath their binding sites in the middle of a 13x nucleosome array compacted with linker histone (20). Taken together, the studies led to the proposal that FoxA and, to a lesser extent, GATA4 are pioneer transcription factors that are able to initiate the formation of regulatory complexes in chromatin by targeting nucleosomes and endowing competence to induce new developmental networks (20) (Figure 1).
Figure 1.
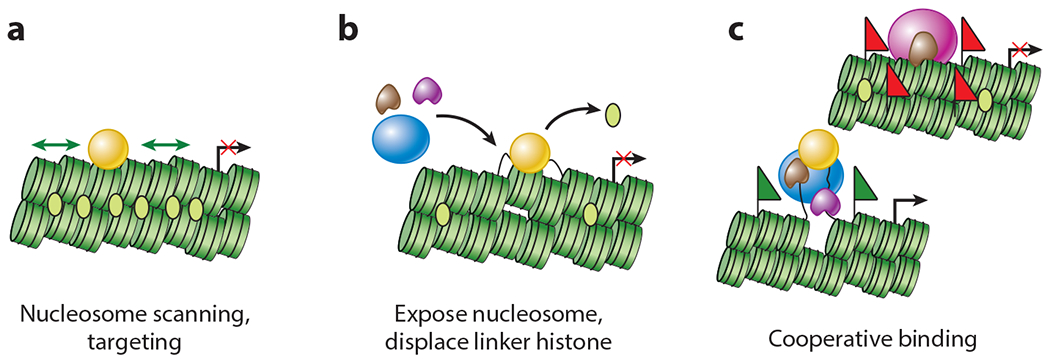
Actions of pioneer transcription factors. (a) The pioneer transcription factor (gold sphere) scans laterally across chromatin and targets a nucleosome. (b) The pioneer transcription factor exposes an underlying nucleosome in chromatin, displacing linker histone. (c) The pioneer transcription factor enables the binding of other transcription factors, coactivators or corepressors, and nucleosome remodelers. Green flags represent activating histone modifications; red flags represent repressive histone modifications.
The early experiments underscored cooperativity between FoxA and GATA pioneer factors in engaging silent chromatin sites in vivo. Subsequent genetic studies revealed that both FoxA1 and FoxA2 genes (68) and both Gata4 and Gata6 genes (49, 145, 152) are necessary for hepatic induction from the embryonic endoderm. Other transcription factors with similar biochemical and genetic properties have since been found in diverse animal and plant developmental contexts (1, 37, 42, 98), including zygotic gene activation (87, 134), the conferral of hormone responsiveness and invasiveness in human cancers (2, 28, 61, 101, 116), the expression of circadian rhythm genes (90), and nongene regulatory processes in chromatin, including DNA recombination (129). Together, these observations indicate that pioneer transcription factors are fundamental to gene network changes and have led to interest in their mechanism of action.
MODES OF TRANSCRIPTION FACTOR INTERACTIONS WITH NUCLEOSOMES
Assessing the Ability of a Transcription Factor to Bind Target Sites on Nucleosomes
The central feature of pioneer factors is their ability to target DNA on nucleosomes. The most direct ways to assess nucleosome targeting are testing whether a purified transcription factor binds to motif-bearing mononucleosomes assembled in vitro (21, 138); whether transcription factors detected by chromatin immunoprecipitation (ChIP) are cooccupied, as assayed by re-ChIP, for core histones (17, 55); or whether an ectopically expressed transcription factor in cells targets preexisting, micrococcal nuclease (MNase)-resistant, nucleosome-sized DNA (127). Nucleosome targeting can also be inferred from the local resistance to the assay for transposase accessible chromatin using sequencing (ATAC-seq) (14), particularly if fragment sizes are taken into account (120), though such inferences need to be confirmed with defined nucleosome and chromatin substrates in vitro. Most important for in vivo assays is comparing the chromatin state prior to transcription factor expression with where the factor binds once it is newly expressed. The cleavage under targets and release using nuclease (CUT&RUN) assay can reveal nucleosome-sized DNA fragments that are released from chromatin by a protein A-MNase fusion that binds to an antibody, which in turn binds to a transcription factor in situ (89, 125). As with ATAC-seq, it will be important to compare the method with untethered MNase on defined chromatin templates to define exactly how the protein A-MNase/antibody complex maps nucleosomal target sites.
Transcription Factors Bind to Nucleosomes in Different Ways
A recent study assessed transcription factor binding to nucleosomes with a highly parallel approach, using randomized DNA sequence libraries assembled into nucleosomes in vitro (153). Over 200 transcription factor DNA-binding domains (DBDs) and selected full-length proteins were tested for binding to nucleosomes and their corresponding free DNA sequences in cycles of PCR-based selection of histone H2A-tagged complexes and tagged transcription factors. Enriched sequences were assessed for specific sequence motifs to serve as evidence for where the factors bind on the DNA and nucleosomes. The study concluded that, while nucleosomes generally inhibit transcription factor binding, a subset of factors are capable of binding nucleosomal DNA in vitro (153). Significantly, the same factors target nucleosome-sized, MNase-resistant chromatin fragments in vivo. After binding nucleosomes in vitro, transcription factors often, but not always, facilitate the dissociation of the histones. Notably, nucleosome binding could occur at different positions throughout the nucleosome, with target motifs, as expected, oriented outside of the histone core. The results contradict a model wherein transcription factors would only access DNA after its spontaneous dissociation from the histones (72, 109).
Different classes of pioneer transcription factors exhibit distinct nucleosome-binding characteristics. Certain basic leucine zipper (bZIP) and basic helix-loop-helix (bHLH) factors, particularly those that surround more than 180° of the DNA circumference, bind to sites at the edge of the nucleosome (153) (Figure 2a). Yet CREB, another bZIP factor, and ETS factors bind asymmetrically near the dyad axis (i.e., center) of the nucleosome (Figure 2b). Zinc-finger, homeodomain, and forkhead box (FOX) transcription factors bind to DNA periodically around the nucleosome (Figure 2c), while Sox (HMG box-containing) and RFX5 factors bind close to the nucleosome dyad axis (Figure 2d), with the Sox2 result confirmed in an independent study (74). HMG box-containing factors like Sox2, which bend DNA (114), can target nucleosomes because the DNA is prebent when wrapped around histones (127). Interestingly, T-box (TBX) factors bind simultaneously to the two double-stranded DNA gyres as they pass around the histone octamer, selecting for motifs separated by 80 bp in the linear DNA and thereby stabilizing nucleosomes (Figure 2e) (153). These studies and more limited parallel comparisons in vitro (36, 127) indicate that nucleosome binding exhibits a spectrum of affinities and mechanisms, rather than being a binary characteristic.
Figure 2.
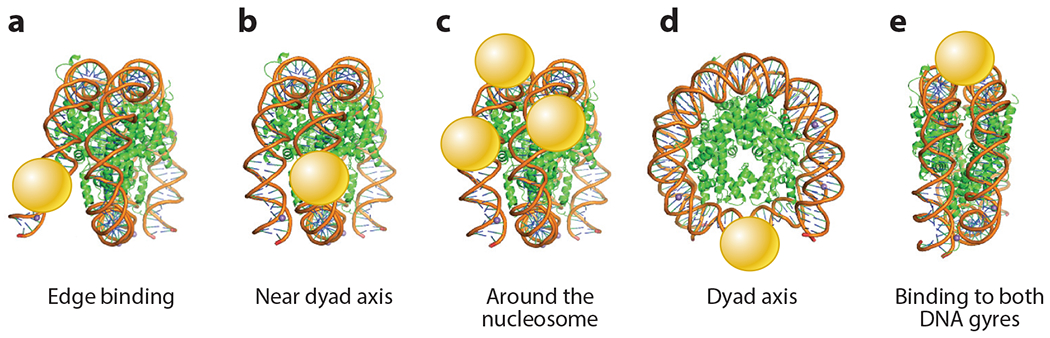
Transcription factor DNA-binding domains (spheres) bind to nucleosomes in diverse methods, such as (a) edge binding, (b) near dyad axis, (c) around the nucleosome, (d) dyad axis, and (e) binding to both DNA gyres (153).
DNA-Binding Domain Structure Can Predict Nucleosome Binding
To investigate how structural features of DBDs relate to nucleosome binding, over 1,000 full-length transcription factors were attached to a glass slide and assessed for binding to three different end-labeled nucleosome DNA sequences and their corresponding free DNAs (36). The nucleosome sequences were curated from natural genomic sites of enriched MNase-resistant segments that are commonly targeted by groups of fate-changing transcription factors. While follow-up studies demonstrated that binding characteristics on the slides were not recapitulated for many of the proteins in solution, trends in DBD structure consistent with nucleosome binding could be discerned (36).
Transcription factors that recognize their DNA motifs via a short anchoring α-helix protruding into the major groove, exemplified by FOX, ETS, homeodomain, and zinc finger factors, were generally observed to target nucleosomal DNA (36) (Figure 3a). The use of natural sequence nucleosomes enabled the ETS factor to bind with an ~5-nM dissociation constant (36), whereas using artificial 601 sequence nucleosomes (108) prevented PU.1 binding (93). The latter result is consistent with another study showing that natural nucleosome-forming sequences are more accommodating of transcription factor binding than the 601 model (52). Indeed, the PU.1 factor targets genomic DNA sites in vitro and in vivo that preferentially form nucleosomes (5). bHLH and bZIP proteins, which have scissors-like pairs of α-helices that reach across the major groove orthogonally to the long axis of DNA, were variable in nucleosome binding (36). Various bHLH factors with potent reprogramming ability, such as Ascl1 (144), use relatively short α-helices to bind DNA and target sequences on nucleosomes, whereas bHLH factors such as c-Myc, with extended α-helices that could sterically interfere with the histone octamer, bind nucleosomes poorly (Figure 3b). Based on its structure, the TALE-homeodomain protein PBX1 was predicted to bind nucleosomes well and thereby could cooperatively enable potent muscle programming by myoblast determination protein 1 (MyoD) (8), a bHLH factor with long α-helices (81). TBX factors were not found to bind natural sequence nucleosomes that lack a pair of TBX motifs (36), unlike the binding seen on artificial sequence nucleosomes containing such a pair (153). Notably, various factors that were tested bound free DNA well but did not bind nucleosomes (36). Thus, strong DNA binding does not necessarily predict nucleosome binding or histone displacement.
Figure 3.
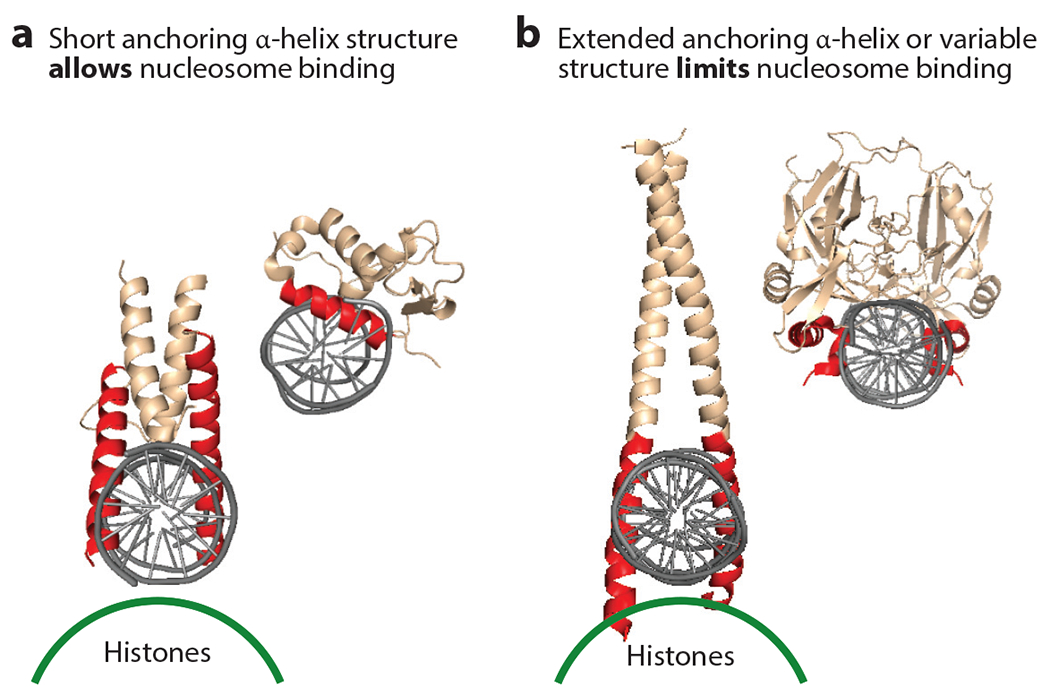
DNA-binding domain structures that enable or limit binding to nucleosomes. (a) Two different DNA-binding domain structures are shown interacting with DNA orthogonal to the long axis of the double helix. The DNA-binding domain on the left illustrates how short α-helices enable interactions that would not sterically collide with the underlying histone surface, depicted by a green line, on a nucleosome (36, 127). (b) By contrast, extended α-helices or other structures can sterically interfere with nucleosome binding. Figure adapted from Reference 36 with permission.
How does a transcription factor recognize a target motif on the surface of the nucleosome, where the histone octamer partially blocks access? Curation of motifs at nucleosome-targeted sites in vivo, compared to nucleosome-free DNA sites, revealed that various transcription factors target a partial DNA motif on the nucleosome (89, 127). This mode of binding may be particularly relevant to factors that bind DNA with closely apposed but structurally separate globular domains, such as octamer-binding transcription factor 4 (Oct4), which uses a linked POU and homeodomain (33, 52, 91, 114), and PAX factors, which use paired domains and homeodomains (13). Similarly, zinc-finger factors with three or more DNA recognition modules can target nucleosomal sequences that employ only two of the modules engaging adjacent motifs exposed on the nucleosome surface (127). Molecular dynamics simulations illustrate that even when binding free DNA–harboring complete motifs, transcription factors could initially engage a partial motif driven by the binding of a subset of their DBD modules (127). Clusters of suboptimal motifs are now understood to increase the cell-type specificity of enhancers in embryonic development (24, 35). These findings illustrate the challenges in deciphering transcription factors associated with chromatin features by examining local sequences for consensus or optimal DNA-binding motifs.
Scanning Chromatin via Nonspecific Nucleosome Binding
The interaction of transcription factors with nucleosomes raises the question of how that property is used to scan chromatin for specific DNA sequences. The issue of chromatin scanning was initially studied by comparing diverse green fluorescent protein-tagged transcription factors in living cells by fluorescence recovery after photobleaching. The experiments revealed a wide range of nuclear protein mobilities (105, 121). However, interpreting such data with regard to chromatin binding in vivo can be complicated by the extent to which the tagged factors are overexpressed, issues regarding diffusion rates and photobleaching, and whether such factors have nonchromatin components to which they bind (95, 128). A way to address DNA target specificity is to compare transcription factors that are point-mutated in their DBDs and expressed equally in cells. For example, FoxA1 has exceptionally low nuclear mobility, yet two amino acid point mutations that diminish FoxA1’s target sequence recognition had a small effect on FoxA1’s nuclear mobility and minimally affected nonspecific DNA and nucleosome binding in vitro (121). Thus, the factor’s slow nuclear mobility and low diffusion in the nucleus are not driven by stable binding site occupancy. By contrast, two amino acid point mutations that diminish FoxA1’s nonspecific DNA binding, and virtually abolish nucleosome binding, markedly increased the factor’s diffusion through the nucleus, while allowing target sequence recognition on free DNA (111, 121). Two major implications can be drawn from these and related studies (6, 45, 62). First, even pioneer factors interact transiently with chromatin and are mobile in the nucleus. Second, given the abundance of nucleosomes, the dependency of the slow movement of FoxA1 on nonspecific binding could reflect its lateral on-and-off scanning across nucleosomes in chromatin, rather than nucleoplasm diffusion as seen for fast-moving nuclear proteins (23, 141).
Transient Localization to Sites in Nuclei
Recent advances in single-molecule tracking (SMT) reveal how individual molecules move in the nucleus (57, 79, 147). Presently, the most sensitive SMT involves a transcription factor that is genetically fused to HALO protein, which can covalently bind to a bright fluorophore (18). Long exposure times (slow-SMT), with caveats regarding photobleaching, allow an estimation of the residence time of a factor at a single nuclear location, whereas short exposure times (fast-SMT) allow motion tracking. Transcription factors, regardless of their nucleosome-binding capacity, have apparent average residence times in mammalian nuclei within a range of several to 100 s (18, 57, 77, 100, 136, 139), with some factors, such as CTCF, residing at sites in nuclei for over 200 s (77). A common interpretation of long residence time measurements of transcription factors is that they represent site-specific binding in chromatin. However, for all factors tested, mutations that eliminate DNA motif binding in vitro lead to only a partial reduction in apparent long residence times in vivo (18, 86, 100, 133). Without understanding whether a HALO-tagged factor is bound to free DNA or nucleosomes at the time of measurement, and whether such behaviors are different during cell fate transitions versus the maintenance of gene activity, residence times are not particularly informative about pioneer factor activity. Cooperating transcription factors, coactivators, and nucleosome remodelers can affect apparent residence times of HALO-tagged transcription factors in nuclei (18, 47, 136). Recent SMT studies indicate that pioneer factors can explore some of the most mobility-restricted domains in the nucleus, where non-pioneer factors cannot (70). In summary, the results of live-cell imaging methods agree that transcription factor occupancy in chromatin is transient and that nonspecific chromatin binding is a significant feature.
Active gene transcription occurs in bursts of initiation events on a time scale of minutes (39, 131, 135). Transcription factors may stochastically bind and release, enabling cyclic rebinding of other factors into functional regulatory complexes to control transcription (142). Alternatively, multicomponent regulatory complexes may undergo wholesale loss from the chromatin and need to be reestablished in a particular order for functional activity. Along these lines, MNase titration studies, along with core histone immunoprecipitation, reveal that nucleosomes are detectable at active enhancers (55, 92). Furthermore, FoxA2 can be coimmunoprecipitated with core histones at MNase-sensitive enhancer nucleosomes in liver cells, while its cooperating transcription factors can be detected at the same sequences but not cobound to core histones; this is consistent with the enhancers cycling through different factor- and nucleosome-occupied states (55, 75).
Separate from considerations of how ongoing gene activity is maintained, the concept of pioneer factors emerged from an effort to understand how new genetic networks are established in naive chromatin or in repressed, silent chromatin. That is, transient and nonspecific binding allows cell fate–controlling factors to scan chromatin, as discussed above, and to enable secondary and cooperative events at nucleosomal sites that are targeted by the factors, as discussed below.
DIRECT NUCLEOSOME PERTURBATIONS BY PIONEER FACTORS
Pioneer Factor Binding Can Perturb Mononucleosome Structure
Just as pioneer factors have different mechanisms for binding to their target sequences on nucleosomes, they also perturb chromatin structure in different ways. For example, p53 seems to access nucleosomal DNA that is transiently dissociated at the edge of a nucleosome and facilitates histone eviction (67, 151), fitting a site exposure model (72) (Figure 4a). However, it may have transient affinity for the histone octamer or bent nucleosomal DNA (25, 76). By contrast, biophysical studies employing fluorescent resonance energy transfer have revealed that the yeast pioneer factors Reb1 and Cbf1 can elicit partial DNA unwrapping from the nucleosome without histone eviction (31) (Figure 4b). While Reb1 binds nucleosomes at the entry-exit point of DNA on the nucleosome (31, 63), the lack of histone loss and a lower dissociation rate than from free DNA suggest that Reb1 may interact directly with the histones themselves. The yeast Cbf1 (31) and mammalian FoxA1 (22) factors also exhibit lower dissociation rates from nucleosomes than from free DNA, which would compensate for a slower apparent on-rate for nucleosomes. These details once again underscore the importance of understanding the nucleosomal versus free DNA status in nuclear residence time data.
Figure 4.
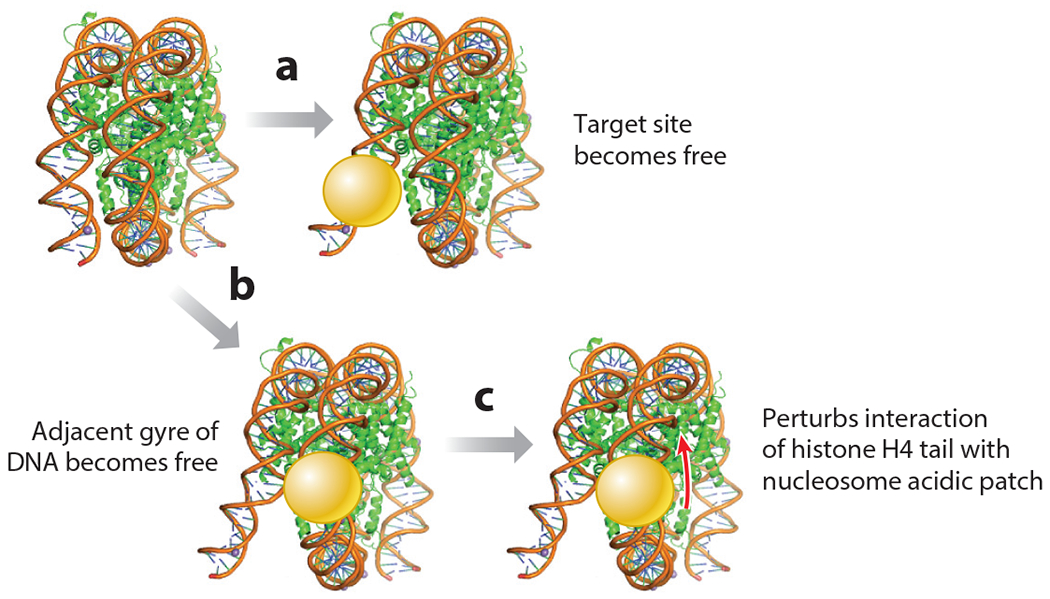
Pioneer transcription factors perturb nucleosome structure in different ways. (a) Pioneer factor binding can facilitate the freeing of its target site from the nucleosome. (b) Binding can cause the adjacent gyre of DNA to be released from the nucleosome. (c) Binding can perturb the interaction between the histone H4 N-terminal tail and the acidic patch of the nucleosome (target of the red arrow), which normally stabilizes internucleosome interactions.
A different mechanism for dissociating part of the DNA from the nucleosome has been revealed by combining cryo-electron microscopy and crystallography of complexes of the pioneer factors Sox2 and Sox11 with selected-sequence nucleosomes (29, 153). The Sox factors bound to nucleosomes that were two DNA helical turns from the dyad axis and not to DNA released at the edge, and they induced perturbations of the underlying DNA contacts with histones. Also, the protein mass bound to one DNA gyre clashed with the adjacent DNA gyre on the nucleosome, eliciting the partial release of the latter (29) (Figure 4b). Such dynamics were not observed on artificial 601 sequence nucleosomes with engineered Sox2 target sites, where Sox2 bound strongly only at the entry and exit point of the DNA (91). Single-molecule assays showed that, in certain binding site configurations, Oct4 can increase the dwell time of Sox2 when both are bound to nucleosomes, but not when they are bound to the same free DNA sequence (74). This result illustrates how the nucleosome itself can promote binding factor cooperativity. Interestingly, Sox11 DBD binding to mononucleosomes repositions the histone H4 tail (29) (Figure 4c), the latter of which, as noted above, normally promotes internucleosomal interactions that locally stabilize the chromatin fiber. Thus, Sox factors may act as pioneer factors by binding to nucleosomes, partially releasing DNA to allow other factors to bind, and destabilizing local nucleosome interactions in chromatin.
Correspondence of Nucleosome Perturbations In Vitro and In Vivo
Single-molecule, total internal reflection fluorescence microscopy studies showed that the full-length yeast pioneer factor Rap1 binds mononucleosomes in vitro with similar on-rates to free DNA, but with lower residence times and a lack of DNA-unwrapping from the histones (83). However, when Rap1 targets the middle nucleosome of a 13x nucleosome array that is compacted with high salt or divalent cations, it destabilizes internucleosomal interactions to make the targeted nucleosome accessible. Furthermore, Rap1 binds to its sites on mononucleosomes of a natural DNA sequence and modulates the activity of the complex called remodeling the structure of chromatin (RSC) to move an underlying nucleosome in a manner similar to that seen at the position of a Rap1-dependent, nucleosome-free region, as observed in vivo (83). The Rap1 effect was marginal on 601 sequence nucleosomes harboring the same Rap1-binding sites (83). These experiments illustrate how biochemical reconstitution studies can address questions that are difficult to address in cells, in this case showing that the pioneer factor is sufficient to provide directionality to a nucleosome remodeler.
FoxA1 and FoxA2 use a core histone-interacting domain, outside of the DBD, to promote the accessibility of a targeted nucleosome in a linker histone-compacted nucleosome array (20). Recent work showed that the core histone interactions involve a conserved, α-helical region in FoxA1 and FoxA2 (54). Early mouse embryos homozygous for FoxA2-RFP or FoxA2-RFP-Δ-helix, the latter having a 10-amino acid deletion of the α-helical domain, exhibited similar RFP nuclear distributions and expression patterns (54). Yet the FoxA2-RFP-Δ-helix homozygous embryos exhibited about 14,000 fewer sites of open chromatin compared to FOXA2-RFP controls, as assessed by ATAC-seq in endoderm cells, along with perturbed RNA-seq patterns and developmental defects leading to embryonic or perinatal lethality (54). Taken together, the FoxA and Rap1 studies with artificial chromatin substrates in vitro have been useful models to reveal different mechanisms by which pioneer factors perturb chromatin in vivo.
TARGETING AND OPENING SILENT CHROMATIN IN VIVO
Targeting Silent Chromatin During Reprogramming to Pluripotency
How much silent, closed chromatin is there in a somatic cell? Studies of 111 human tissues and cell lines by the Roadmap Epigenomics Consortium (66) have found that, on average, enhancers, promoters, and actively transcribed regions constitute up to 10% of a cell’s genome, and another 12% is weakly transcribed, constituting open, active chromatin. The remainder, constituting closed, DNase-resistant, and transcriptionally silent chromatin, contains H3K27me3- and H3K9me3-repressed domains as well as about 40% of the genome in a quiescent or low signal state, not enriched for known chromatin modifications as seen with super-resolution imaging observations (10). Strikingly, low signal state, DNase-resistant chromatin is most frequently targeted by the pluripotency factors Oct4, Sox2, and Klf4 (OSK) induced in human fibroblasts for 48 h (126). Most of the sites targeted independently by the factors are nucleosomal and exhibit incomplete DNA motifs compared to the minority of free DNA sites targeted that exhibit complete motifs (127). c-Myc, by contrast, is necessary for human induced pluripotent stem cell (iPSC) reprogramming (137) but preferentially targets preexisting, DNase-accessible sites at promoters that possess activating histone modifications (126). While the distinctions between OSK initially binding to enhancers and c-Myc to promoters were also observed at 48 h of OSKM induction in mouse embryonic fibroblasts (MEFs), OSKM in MEFs primarily targets open enhancer sites (19). OSKM was induced in secondary MEFs derived from an iPSC-generated mouse embryo (130), and thus they could exhibit low-level priming by the factors rather than in the de novo OSKM lentiviral induction system used in human cells. Regardless, the mouse studies revealed that OSKM perturbed resident fibroblast transcription factors, causing fibroblast enhancer decommissioning for pluripotency conversion.
Pioneer Factor Targeting of Closed Chromatin Can Precede Chromatin Opening In Vivo
Careful time course studies of chromatin opening by pioneer factor binding in vivo have revealed a surprising difference from in vitro studies. Shortly after inducing expression in factor-naive cells, EBF1, a B cell differentiation factor; Pax7, an intermediary pituitary factor; or C/EBPa, a macrophage factor, exhibits binding hours prior to chromatin opening, as assessed by the ATAC-seq assay (73, 85, 118). At later time points, additional transcription factors are recruited, DNA demethylation can occur, and the sites become accessible by ATAC-seq. FoxA1 targets closed chromatin sites in the G1 phase of the cell cycle, thus not requiring the chromatin opening that occurs in DNA replication (30). The Caenorhabditis elegans FoxA homolog PHA-4, which binds nucleosomes in vitro as well as FoxA1 does, targets closed chromatin sites in C. elegans embryos and recruits RNA polymerase II, which forms a transcriptionally poised complex prior to chromatin opening (51). c-Myb recruits the histone acetyltransferase p300 to elicit chromatin opening (38). The activated progesterone receptor preferentially targets MNase-resistant nucleosomal sites, with consequent loss of linker histone H1 and core histones H2A and H2B (3). In summary, time course studies reveal that there are intervening steps between pioneer factor binding and overt chromatin opening in vivo. The nucleosome perturbations elicited by pioneer factors thus seem restricted to the underlying nucleosome, while secondary events enabled by the factors in vivo result in the observed open chromatin state that typically spans several nucleosomes.
Pioneer Factor Recruitment of Nucleosome Remodelers to Open Local Chromatin
Quantitative analysis of many pioneer factors found directionality in local DNase hypersensitivity, with regard to the orientation of their targeted motifs in chromatin (123). As noted by the authors (123), detecting and characterizing pioneer factors solely by eliciting open chromatin will fail to map binding events that recruit corepressors and create more inaccessible chromatin (101, 122, 146). For example, in embryonic stem cells, FOXD3 first recruits the BRG1 nucleosome remodeler to promote nucleosome removal but then recruits histone deacetylates to repress its targeted enhancers (65). A systematic study of nucleosome-binding transcription factors in yeast observed a correlation between ATP-dependent nucleosome remodelers recruited by the factors and the consequent patterns of open chromatin (150). Nucleosome-binding transcription factors, including the glucocorticoid receptor (48, 103), BZLF1 (119), Isl1 (41), and PU.1 (36, 93), all recruit nucleosome remodelers to chromatin sites that become open. Remodelers interact with the nucleosome acidic patch (27, 140), which, as noted above, could be exposed in compacted chromatin by the initial binding of pioneer factors. In summary, pioneer factor recruitment of ubiquitous nucleosome remodeling complexes to new target sites seems crucial for resetting gene networks in cells.
Cooperative Priming of Chromatin Endows Competence for Gene Expression
Chromatin opening elicited by pioneer factors can prime subsequent developmental events rather than resulting in immediate gene activation (9, 44, 58, 143). Indeed, pioneer factors themselves, during differentiation of embryonic stem cells in culture, can target sites that are often primed by marginal but detectable open chromatin features, or prior binding of pluripotency (and pioneering) transcription factors or repressors (16, 65, 82, 89, 149), and recruit additional factors. For such analyses, it is crucial to assess the individual targeted sites, e.g., with heat maps, for the extent to which the factors target closed versus open chromatin sites, rather than using metaplot averages. In another example of establishing competence, the prior binding of FoxA1 can enable hormonally activated estrogen and androgen receptors to bind many sites in breast and prostate cancer cells, respectively, with consequent gene activation (2, 7, 53, 61, 101, 106). Conversely, in a small percentage of the FoxA1-binding events that lack FoxA1 target motifs and are detected by some laboratories (60, 136) but not others (43), FoxA1 may be recruited by hormone-activated receptors (136). De novo FoxA1 expression in human fibroblasts induces DNA demethylation at FoxA1 target sites, providing an epigenetic mechanism for inheriting expression competence (30). In another example, in embryonic stem cells, 10% of Sox2-binding events at nucleosomes require cobinding with PARP1, independent of poly-ADP ribosylation (78).
These studies and others raise the question of how pioneer factors exhibit cell-type-binding specificity, as opposed to binding all target motifs in all cell types. A comparison of binding signals for FoxA and GATA4 at all sites targeted among three different cell lines showed that, as for other transcription factors, peaks were most frequently seen at different sites in different cells, depending upon cooperative interactions with other factors specific to each cell (30). However, FoxA and GATA4 also exhibited low but detectable binding at most of the sites targeted in all cells, which the authors (30) referred to as sampling. Analogously, when peaks for the initial binding of OSKM in human fibroblasts were called at increasingly low thresholds, the peak counts for c-Myc plateaued, whereas peak counts for Oct4, Klf4, and especially Sox2 kept rising (127). To summarize, pioneer factors may transiently sample many motifs and create functional complexes at a subset of nucleosomal sites, where they enable cooperativity with other factors.
FUTURE DIRECTIONS OF INTEREST
What determines the ability of a pioneer factor to target a particular type of silenced chromatin? TCF1, the nucleosome-targeting T cell factor, efficiently targets its motifs and opens sites marked by H3K27me3 (59). Pax7 targets and opens heterochromatic sites with high levels of H3K9me2 and intermediate levels of H3K9me3 but is resistant to sites with high levels of both (85). FoxA2 can target lamin-enriched sites and decrease underlying levels of H3K9me3 (148). Given that the DBD of a pioneer factor must bind nucleosomes, presumably a non-DBD portion of the protein may confer heterochromatin binding. Can hybrid pioneer factors expand chromatin-targeting features?
Biophysical and structural studies on a few pioneer factors indicate distinct ways to perturb the underlying nucleosome. Comparisons of additional pioneer factors seem likely to reveal more nucleosome-perturbative mechanisms. The adaptability of natural sequence DNA on the octamer is more revealing about pioneer mechanisms than artificial 601 DNA engineered to contain a binding motif. HP1 binding to nucleosomes reveals orientation changes within the histone octamer itself (117). Thus, future assessments of nucleosome dynamics on pioneer factor binding will be enabled with natural sequence DNA and direct assessments of histone organization. For SMT studies, the development of additional highly fluorescent tags will allow a careful dissection of the consequences of pioneer factor binding to nucleosome target sequences in vivo.
There is a gap in understanding how nucleosome perturbations that occur upon pioneer factor binding lead to the cooperative engagement of other transcription factors, chromatin- and DNA-modifying proteins, and nucleosome remodelers. Are pioneer factors sufficient to enable nucleosome-remodeling complexes to access targeted sites on H1-compacted nucleosome arrays? Does such access require a direct interaction between the pioneer factor and the remodeler, or is the locally exposed nucleosome generated by the pioneer factor sufficient to allow remodeler engagement? Answers will be found with complex chromatin templates in vitro, including nucleosome arrays with natural sequence targets.
With regard to understanding how pioneer factors target different types of silent chromatin, it will be informative to employ nucleosome array templates that are not only marked with H3K9me3 or H3K27me3 but also bound by the respective protein complexes that elicit chromatin compaction. Are additional cofactors or remodelers required for pioneer factor engagement? In all cases for the in vitro studies, it is essential to identify protein domains on the pioneer factors that elicit a chromatin change and test their function in a natural, cell fate–changing context (54). In summary, understanding the mechanisms of action of pioneer factors continues to reveal how genetic networks are rewired during cell fate changes. Ultimately, the insights can be used to control cell fate at will.
ACKNOWLEDGMENTS
The author acknowledges valuable comments on the manuscript from Roberto Bonasio, Alyssa Miller, Jonathan Lerner, Kelsey Kaeding, Jingchao Zhang, Megan Frederick, and Golnaz Vahedi and support from National Institutes of Health (NIH) grant R01GM36477.
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adachi K, Kopp W, Wu G, Heising S, Greber B, et al. 2018. Esrrb unlocks silenced enhancers for reprogramming to naive pluripotency. Cell Stem Cell 23:266–75.e6 [DOI] [PubMed] [Google Scholar]
- 2.Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, et al. 2019. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 571:408–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballare C, Castellano G, Gaveglia L, Althammer S, Gonzalez-Vallinas J, et al. 2013Nucleosome-driven transcription factor binding and gene regulation. Mol. Cell 49:67–79 [DOI] [PubMed] [Google Scholar]
- 4.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, et al. 2006. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science 311:856–61 [DOI] [PubMed] [Google Scholar]
- 5.Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, et al. 2014. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell 54:844–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker M, Baumann C, John S, Walker DA, Vigneron M, et al. 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3:1188–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belikov S, Berg OG, Wrange O. 2016. Quantification of transcription factor-DNA binding affinity in a living cell. Nucleic Acids Res. 44:3045–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465–77 [DOI] [PubMed] [Google Scholar]
- 9.Bevington SL, Cauchy P, Piper J, Bertrand E, Lalli N, et al. 2016. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J. 35:515–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, et al. 2016. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529:418–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossard P, Zaret KS. 1998. GATA transcription factors as potentiators of gut endoderm differentiation. Development 125:4909–17 [DOI] [PubMed] [Google Scholar]
- 12.Bossard P, Zaret KS. 2000. Repressive and restrictive mesodermal interactions with gut endoderm: possible relation to Meckel’s Diverticulum. Development 127:4915–23 [DOI] [PubMed] [Google Scholar]
- 13.Budry L, Balsalobre A, Gauthier Y, Khetchoumian K, L’Honore A, et al. 2012. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 26:2299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buenrostro JD, Giresi PG,Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10:1213–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cascio S, Zaret KS. 1991. Hepatocyte differentiation initiates during endodermal-mesenchymal interactions prior to liver formation. Development 113:217–25 [DOI] [PubMed] [Google Scholar]
- 16.Cernilogar FM, Hasenöder S, Wang Z, Scheibner K, Burtscher I, et al. 2019. Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 47:9069–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaya D, Hayamizu T, Bustin M, Zaret KS. 2001. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J. Biol. Chem. 276:44385–89 [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Zhang Z, Li L, Chen BC, Revyakin A, et al. 2014. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156:1274–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chronis C, Fiziev P, Papp B, Butz S, Bonora G, et al. 2017. Cooperative binding of transcription factors orchestrates reprogramming. Cell 168:442–59.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9:279–89 [DOI] [PubMed] [Google Scholar]
- 21.Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, et al. 1998. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 17:244–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirillo LA, Zaret KS. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4:961–69 [DOI] [PubMed] [Google Scholar]
- 23.Cortini R, Filion GJ. 2018. Theoretical principles of transcription factor traffic on folded chromatin. Nat. Commun. 9:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, et al. 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160:191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui F, Zhurkin VB. 2014. Rotational positioning of nucleosomes facilitates selective binding of p53 to response elements associated with cell cycle arrest. Nucleic Acids Res. 42:836–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185–96 [DOI] [PubMed] [Google Scholar]
- 27.Dann GP, Liszczak GP, Bagert JD, Muller MM, Nguyen UTT, et al. 2017. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548:607–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, et al. 2016. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell 166:328–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dodonova SO,Zhu F, Dienemann C, Taipale J, Cramer P 2020. Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 580:669–72 [DOI] [PubMed] [Google Scholar]
- 30.Donaghey J, Thakurela S, Charlton J, Chen JS, Smith ZD, et al. 2018. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 50:250–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donovan BT, Chen H, Jipa C, Bai L, Poirier MG. 2019. Dissociation rate compensation mechanism for budding yeast pioneer transcription factors. eLife 8:e43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorigo B, Schalch T, Bystricky K, Richmond TJ. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327:85–96 [DOI] [PubMed] [Google Scholar]
- 33.Echigoya K, Koyama M, Negishi L, Takizawa Y, Mizukami Y, et al. 2020. Nucleosome binding by the pioneer transcription factor OCT4. Sci. Rep. 10:11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan JY, Rangasamy D, Luger K, Tremethick DJ. 2004. H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell 16:655–61 [DOI] [PubMed] [Google Scholar]
- 35.Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. 2015. Suboptimization of developmental enhancers. Science 350:325–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez Garcia M, Moore CD, Schulz KN, Alberto O, Donague G, et al. 2019. Structural features of transcription factors associating with nucleosome binding. Mol. Cell 75:921–32.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiedler M, Graeb M, Mieszczanek J, Rutherford TJ, Johnson CM, Bienz M. 2015. An ancient Pygodependent Wnt enhanceosome integrated by Chip/LDB-SSDP. eLife 4:e09073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuglerud BM, Ledsaak M, Rogne M, Eskeland R, Gabrielsen OS. 2018. The pioneer factor activity of c-Myb involves recruitment of p300 and induction of histone acetylation followed by acetylation-induced chromatin dissociation. Epigenet. Chromatin 11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukaya T, Lim B, Levine M. 2016. Enhancer control of transcriptional bursting. Cell 166:358–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y. 2018. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19:192–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao R, Liang X, Cheedipudi S, Cordero J,Jiang X, et al. 2019. Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell Res. 29:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gehrke AR, Neverett E, Luo YJ, Brandt A, Ricci L, et al. 2019. Acoel genome reveals the regulatory landscape of whole-body regeneration. Science 363:eaau6173. [DOI] [PubMed] [Google Scholar]
- 43.Glont SE, Chernukhin I, Carroll JS. 2019. Comprehensive genomic analysis reveals that the pioneering function of FOXA1 is independent of hormonal signaling. Cell Rep. 26:2558–65.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 10:1670–82 [DOI] [PubMed] [Google Scholar]
- 45.Hager GL,McNally JG,Misteli T. 2009Transcription dynamics. Mol. Cell 35:741–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill DA, Imbalzano AN. 2000. Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry 39:11649–56 [DOI] [PubMed] [Google Scholar]
- 47.Hipp L, Beer J, Kuchler O, Reisser M, Sinske D, et al. 2019. Single-molecule imaging ofthe transcription factor SRF reveals prolonged chromatin-binding kinetics upon cell stimulation. PNAS 116:880–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman JA, Trotter KW, Ward JM, Archer TK. 2018. BRG1 governs glucocorticoid receptor interactions with chromatin and pioneer factors across the genome. eLife 7:e35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holtzinger A, Evans T. 2005. Gata4 regulates the formation of multiple organs. Development 132:400514. [DOI] [PubMed] [Google Scholar]
- 50.Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, et al. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 9:263–67 [DOI] [PubMed] [Google Scholar]
- 51.Hsu HT, Chen HM, Yang Z, Wang J, Lee NK, et al. 2015. Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science 348:1372–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huertas J, MacCarthy CM, Scholer HR, Cojocaru V. 2020. Nucleosomal DNA dynamics mediate Oct4 pioneer factor binding. Biophys. J. 118:2280–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwafuchi M, Cuesta I, Donahue G, Takenaka N, Osipovich AB, et al. 2020. Gene network transitions in embryos depend upon interactions between a pioneer transcription factor and core histones. Nat. Genet. 52:418–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, et al. 2016. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62:79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwafuchi-Doi M, Zaret KS. 2016. Cell fate control by pioneer transcription factors. Development 143:1833–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Izeddin I, Recamier V, Bosanac L, Cisse II, Boudarene L, et al. 2014. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife 3:e02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobs J, Atkins M, Davie K, Imrichova H, Romanelli L, et al. 2018. The transcription factor Grainy head primes epithelial enhancers for spatiotemporal activation by displacing nucleosomes. Nat. Genet. 50:1011–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson JL, Georgakilas G, Petrovic J, Kurachi M, Cai S, et al. 2018. Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T cells. Immunity 48:243–57.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson TA, Chereji RV, Stavreva DA, Morris SA, Hager GL, Clark DJ. 2018. Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo. Nucleic Acids Res. 46:20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jozwik KM Carroll JS. 2012. Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer 12:381–85 [DOI] [PubMed] [Google Scholar]
- 62.Karpova TS, Chen TY, Sprague BL, McNally JG. 2004. Dynamic interactions of a transcription factor with DNA are accelerated by a chromatin remodeller. EMBO Rep. 5:1064–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koerber RT, Rhee HS, Jiang C, Pugh BF. 2009. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol. Cell 35:889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kornberg RD, Lorch Y. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285–94 [DOI] [PubMed] [Google Scholar]
- 65.Krishnakumar R, Chen AF, Pantovich MG, Danial M, Parchem RJ, et al. 2016. FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell 18:104–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kundaje A, Meuleman W, Ernst J, Bilenky M, Meuleman W, et al. (Roadmap Epigenomics Consort.). 2015. Integrative analysis of 111 reference human epigenomes. Nature 518:317–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laptenko O, Beckerman R, Freulich E, Prives C. 2011. p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. PNAS 108:10385–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. 2005. The initiation of liver development is dependent on Foxa transcription factors. Nature 435:944–47 [DOI] [PubMed] [Google Scholar]
- 69.Leemans C, van der Zwalm MCH, Brueckner L, Comoglio F, van Schaik T, et al. 2019. Promoter-intrinsic and local chromatin features determine gene repression in LADs. Cell 177:852–64.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lerner J, Gomez-Garcia PA, McCarthy RL, Liu Z, Lakadamyali M, Zaret KS. 2020. Two-parameter mobility assessments discriminate diverse regulatory factor behaviors in chromatin. Mol. Cell 79:677–88.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li G, Reinberg D. 2011. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 21:175–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li G, Widom J. 2004. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11:763–69 [DOI] [PubMed] [Google Scholar]
- 73.Li R, Cauchy P, Ramamoorthy S, Boller S, Chavez L, Grosschedl R. 2018. Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev. 32:96–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li S, Zheng EB, Zhao L, Liu S. 2019. Nonreciprocal and conditional cooperativity directs the pioneer activity of pluripotency transcription factors. Cell Rep. 28:2689–703.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, et al. 2012. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151:1608–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lidor Nili E, Field Y, Lubling Y, Widom J, Oren M, Segal E. 2010. p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy. Genome Res. 20:1361–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu H, Dong P, Ioannou MS, Li L, Shea J, et al. 2018. Visualizing long-term single-molecule dynamics in vivo by stochastic protein labeling. PNAS 115:343–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Z, Kraus WL. 2017. Catalytic-independent functions ofPARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol. Cell 65:589–603.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Z, Tjian R.2018. Visualizing transcription factor dynamics in living cells. J. Cell Biol. 217:1181–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251–55 [DOI] [PubMed] [Google Scholar]
- 81.Ma PC, Rould MA, Weintraub H, Pabo CO. 1994. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77:451–59 [DOI] [PubMed] [Google Scholar]
- 82.Mahony S, Edwards MD, Mazzoni EO, Sherwood RI, Kakumanu A, et al. 2014. An integrated model of multiple-condition ChIP-seq data reveals predeterminants of Cdx2 binding. PLOS Comput. Biol. 10:e1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mivelaz M, Cao A-M, Kubik S,Zencir S, Hovius R, et al. 2020. Chromatin fiber invasion and nucleosome displacement by the Rap1 transcription factor. Mol. Cell 77:488–500.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mayran A, Drouin J. 2018. Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 293:13795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mayran A, Khetchoumian K, Hariri F, Pastinen T, Gauthier Y, et al. 2018. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat. Genet. 50:259–69 [DOI] [PubMed] [Google Scholar]
- 86.Mazza D, Abernathy A, Golob N, Morisaki T, McNally JG. 2012. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McDaniel SL, Gibson TJ, Schulz KN, Fernandez Garcia M, Nevil M, et al. 2019. Continued activity of the pioneer factor Zelda is required to drive zygotic genome activation. Mol. Cell 74:185–95.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McPherson CE, Shim EY, Friedman DS, Zaret KS. 1993. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75:387–98 [DOI] [PubMed] [Google Scholar]
- 89.Meers MP, Janssens DH, Henikoff S. 2019. Pioneer factor-nucleosome binding events during differentiation are motif encoded. Mol. Cell 75:562–75.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Menet JS, Pescatore S, Rosbash M. 2014. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 28:8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Michael AK, Grand RS, Isbel L, Cavadini S, Kozicka Z, et al. 2020. Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 368(6498):1460–65 [DOI] [PubMed] [Google Scholar]
- 92.Mieczkowski J, Cook A, Bowman SK, Mueller B, Alver BH, et al. 2016. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7:11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Minderjahn J, Schmidt A, Fuchs A, Schill R, Raithel J, et al. 2020. Mechanisms governing the pioneering and redistribution capabilities of the non-classical pioneer PU.1. Nat. Commun. 11:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morris SA. 2016. Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks. Development 143:2696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mueller F, Wach P, McNally JG. 2008. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys. J. 94:3323–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, et al. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197–208 [DOI] [PubMed] [Google Scholar]
- 97.Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, et al. 2017. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell 67:282–93.e7 [DOI] [PubMed] [Google Scholar]
- 98.Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, et al. 2014. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 55:708–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC. 2017. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357:eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paakinaho V, Presman DM, Ball DA, Johnson TA, Schiltz RL, et al. 2017. Single-molecule analysis of steroid receptor and cofactor action in living cells. Nat. Commun. 8:15896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parolia A, Cieslik M, Chu SC, Xiao L, Ouchi T, et al. 2019. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 571:413–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pepenella S, Murphy KJ, Hayes JJ. 2014. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma 123:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perlmann T, Wrange O. 1988. Specific glucocorticoid receptor binding to DNA reconstituted in nucleosome. EMBOJ. 7:3073–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pfeifer GP, Steigerwald SD, Mueller PR, Wold B, Riggs AD. 1989. Genomic sequencing and methylation analysis by ligation medicated PCR. Science 246:810–13. Erratum. 1990. Science 248:802 [DOI] [PubMed] [Google Scholar]
- 105.Phair RD, Scaffidi P,Elbi C, Vecerova J, Dey A, et al. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pihlajamaa P, Sahu B, Lyly L, Aittomaki V, Hautaniemi S, Janne OA. 2014. Tissue-specific pioneer factors associate with androgen receptor cistromes and transcription programs. EMBO J. 33:312–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, et al. 2019. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 33:799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Polach KJ, Widom J. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130–49 [DOI] [PubMed] [Google Scholar]
- 109.Polach KJ, Widom J. 1996. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 258:800–12 [DOI] [PubMed] [Google Scholar]
- 110.Poleshko A Smith CL, Nguyen SC Sivaramakrishnan P, Wong KG, et al. 2019. H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis. eLife 8:e49278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raccaud M, Friman ET, Alber AB, Agarwal H, Deluz C, et al. 2019. Mitotic chromosome binding predicts transcription factor properties in interphase. Nat. Commun. 10:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ramachandran A, Omar M Cheslock P Schnitzler GR. 2003Linkerhistone H1 modulates nucleosome remodeling by human SWI/SNF. J. Biol. Chem. 278:48590–601 [DOI] [PubMed] [Google Scholar]
- 113.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–99 [DOI] [PubMed] [Google Scholar]
- 114.Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. 2003. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17:2048–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ricci MA, Manzo C, Garcia-Parajo MF, Lakadamyali M, Cosma MP. 2015. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160:1145–58 [DOI] [PubMed] [Google Scholar]
- 116.Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, et al. 2017. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 170:875–88.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sanulli S,Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, et al. 2019. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575:390–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sardina JL, Collombet S, Tian TV, Gomez A, Di Stefano B, et al. 2018. Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell 23:727–41.e9 [DOI] [PubMed] [Google Scholar]
- 119.Schaeffner M,Mrozek-Gorska P, Buschle A,Woellmer A, Tagawa T, et al. 2019. BZLF1 interacts with chromatin remodelers promoting escape from latent infections with EBV. Life Sci. Alliance 2:e201800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, Greenleaf WJ. 2015. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25:1757–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. 2009. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 23:804–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sekiya T, Zaret KS. 2007. Repression by Groucho/TLE/Grg proteins: Genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol. Cell 28:291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, et al. 2014. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 32:171–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shim EY, Woodcock C, Zaret KS. 1998. Nucleosome positioning by the winged-helix transcription factor HNF3. Genes Dev. 12:5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping ofDNA binding sites. eLife 6:e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151:994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161:555–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sprague BL, Pego RL, Stavreva DA, McNally JG. 2004. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86:3473–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Spruce C, Dlamini S,Ananda G, Bronkema N, Tian H, et al. 2020. HELLS and PRDM9 form a pioneer complex to open chromatin at meiotic recombination hot spots. Genes Dev. 34:398–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, et al. 2013. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 15:872–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stavreva DA, Garcia DA, Fettweis G, Gudla PR, Zaki GF, et al. 2019. Transcriptional bursting and co-bursting regulation by steroid hormone release pattern and transcription factor mobility. Mol. Cell 75:1161–77.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547:241–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sugo N, Morimatsu M, Arai Y, Kousoku Y, Ohkuni A, et al. 2015. Single-molecule imaging reveals dynamics of CREB transcription factor bound to its target sequence. Sci. Rep. 5:10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sun Y,Nien CY, Chen K,Liu HY, Johnston J, et al. 2015Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25:1703–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. 2011. Mammalian genes are transcribed with widely different bursting kinetics. Science 332:472–74 [DOI] [PubMed] [Google Scholar]
- 136.Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, et al. 2016. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165:593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–72 [DOI] [PubMed] [Google Scholar]
- 138.Taylor IC, Workman JL, Schuetz TJ, Kingston RE. 1991. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 5:1285–98 [DOI] [PubMed] [Google Scholar]
- 139.Teves SS, An L, Bhargava-Shah A, Xie L, Darzacq X, Tjian R. 2018. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7:e35621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Valencia AM, Collings CK, Dao HT, St Pierre R, Cheng YC, et al. 2019. Recurrent SMARCB1 mutations reveal a nucleosome acidic patch interaction site that potentiates mSWI/SNF complex chromatin remodeling. Cell 179:1342–56.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.von Hippel PH, Berg OG. 1989. Facilitated target location in biological systems. J. Biol. Chem. 264:675–78 [PubMed] [Google Scholar]
- 142.Voss TC, Hager GL. 2014. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 15:69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang A, Yue F, Li Y, Xie R, Harper T, et al. 2015. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16:386–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, et al. 2013. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155:621–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Watt AJ, Zhao R, Li J, Duncan SA. 2007. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev. Biol. 7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Watts JA, Zhang C,Klein-Szanto AJ, Kormish JD, Fu J, et al. 2011. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLOS Genet. 7:e1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, et al. 2016. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165:75–87 [DOI] [PubMed] [Google Scholar]
- 148.Whitton H, Singh LN, Patrick MA, Price AJ, Osorio FG, et al. 2018. Changes at the nuclear lamina alter binding of pioneer factor Foxa2 in aged liver. Aging Cell 17:e12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, et al. 2009. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 23:2824–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yan C, Chen H, Bai L. 2018. Systematic study of nucleosome-displacing factors in budding yeast. Mol. Cell 71:294–305.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yu X, Buck MJ. 2019. Defining TP53 pioneering capabilities with competitive nucleosome binding assays. Genome Res. 29:107–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. 2005. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell. Biol. 25:2622–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, et al. 2018. The interaction landscape between transcription factors and the nucleosome. Nature 562:76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]


