Abstract
Background
The antibody‐dependent enhancement (ADE) of dengue virus (DENV) has critically restricted vaccine development. Prior research suggested pr4 as the probable ADE epitope of DENV.
Methods
Chimeric DENV was constructed by replacing the DENV pr4 gene with the corresponding Japanese encephalitis virus (JEV) gene to determine whether it can reduce ADE activities. An alanine scanning method and bioinformatics analysis were utilized to identify the amino acid of pr4 that was crucial as an ADE epitope.
Results
Chimeric virus reduced ADE and virulence. The amino acids at the following locations on the mutant peptides showed significantly reduced binding ability to prM antibody: pr4.5 (position 5 – leucine), pr4.6 (position 6 – leucine), pr4.7 (position 7 – phenyalanine) and pr4.16 (position 16 – cysteine). The four amino acids had formed a pocket‐like structure, which could increase the possibility of binding to an antibody.
Conclusions
ADE activities could be reduced by replacing the DENV pr4 gene with the corresponding JEV gene. Leucine at position 5, leucine at position 6, phenyalanine at position 7 and cysteine at position 16 were the key amino acid sites in the ADE response of DENV. The occurrence of ADE can potentially be reduced by the replacement of key amino acids, hence highlighting its possible contribution to dengue vaccine design, paving a way for future vaccine research.
Keywords: ADE, dengue virus, epitope, prM antibodies, vaccine
The pr4 gene was replaced with the JEV pr4 gene. The chimeric virus reduced virulence and the anti‐sera could reduce the ADE of DENV infection significantly. An alanine scanning method was used to confirm which amino acid of pr4 was the key ADE epitope. We found that the binding ability of pr4.5, pr4.6, pr4.7 and pr4.16 in the mutant peptides to prM antibody was significantly reduced, indicating that amino acids at positions 5 (leucine), 6 (leucine), 7 (phenylalanine) and 16 (cysteine) played an important role in the binding process of pr4 epitope and prM antibody. The tertiary structure of pr peptide was analyzed by Swiss‐Model homology modeling. We found that the leucine, phenylalanine and cysteine of pr4 could form a pocket‐like structure in space, which could increase the possibility of combining the antibody. The key amino acid site mutant peptide has a reduced ADE effect on DENV2.
1. INTRODUCTION
Dengue virus (DENV) is an arbovirus transmitted mainly by Aedes aegypti and Aedes albopictus. It belongs to the genus Flavivirus of the Flaviviridae family and includes four serotypes (DENV 1–4). It causes asymptomatic infection, symptoms such as mild dengue fever, and even severe dengue hemorrhagic fever and dengue shock syndrome (DHF/DSS). The fatality rate is 15%–50%. 1 , 2 , 3 DHF/DSS is associated with infection by multiple serotypes. 4 Being infected with one serotype stimulates an effective immunity against reinfection of the same serotype; however, it does not provide protection against the other serotypes. 5 Primary infection of infants to dengue‐immune mothers may pose greater risk. 6 , 7 So far, the pathogenic and immune mechanisms of DHF/DSS have not been fully elucidated, although the antibody‐dependent enhancement (ADE) of DENV has been identified as a crucial factor. 8 , 9 , 10 The difficulties in balancing immunity to the four serotypes and the risk of ADE are major hurdles in the development of an effective vaccine against DENV infection. 11 , 12 Although DENV infection imposes one of the largest economic burdens to the world, approved vaccine remains unavailable despite decades of effort. 13
DENV is single‐stranded RNA virus with a total genome length of approximately 11 kb, encoding three structural proteins (C, prM/M and E) and seven non‐structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b and NS5). 14 , 15 prM acts as a chaperone of protein E during virus assembly and maturation. 16 prM contains a furin cleavage site that is cleaved into C‐terminal M protein and N‐terminal pr protein, resulting in the formation of a mature infectious virus. 17 , 18 Cells infected with DENV secrete a heterogeneous mixture of virus particles that vary as a result of the inefficient cleavage of prM to M by furin during DENV maturation, ranging from fully mature (containing M) and partially mature (containing prM and M) to fully immature (containing prM) virions. 19 , 20
Recent studies have indicated that the prM protein is related to ADE of DENV, suggesting the potential role of prM‐specific monoclonal antibodies in enhancing DENV infection. prM protein is a major target of the humoral immune response to DENV. Balakrishnan et al. 21 reported that DENV infection could cause B cell activation, whereas epitopes are the focus of research involving DENV pathogenesis, vaccine development and diagnostic reagent. 22 , 23 Accordingly, our research group had screened the prM protein‐specific B cell epitopes using peptide scanning and bioinformatics. Although it was found that the pr4 epitope greatly influences ADE, 24 , 25 , 26 no further study was carried out to confirm its accuracy.
Based on the full‐length infectious cDNA clone of DENV2 ZS01/01 constructed in our laboratory, 27 , 28 the sequence of DENV prM protein ADE epitope pr4 was replaced with the corresponding gene sequence of Japanese encephalitis virus (JEV) without the ADE effect through infusion technology. The DENV/JEV chimeric virus was analyzed for its effect on ADE. It is confirmed that the ADE effect was indeed reduced after pr4 replacement. The immunological effects of key amino acid sites of pr4 were analyzed and it can be inferred that leucine at position 5, leucine at position 6, phenyalanine at position 7 and cysteine at position 16 were the key amino acid sites in the ADE response of DENV. The results of the present study provide a scientific basis for the ADE effect of the DENV prM protein, enlightening novel approaches in the development of dengue vaccines.
2. MATERIALS AND METHODS
2.1. Cells and viruses
C6/36 cells were cultured in modified essential medium (MEM) supplemented with 5% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, 25 mm Hepes and 2 mm l‐glutamine at 28°C. BHK‐21 cells and LoVo cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 25 mm Hepes and 2 mm l‐glutamine at 37°C with 5% CO2. K562 cells were cultured in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 25 mm Hepes and 2 mm l‐glutamine at 37°C with 5% CO2. DENV1 Hawaii strain (GenBank No. EU848545), DENV2 New Guinea strain (GenBank No. AF038403), DENV2 ZS01/01 strain (GenBank No. EF051521.1), DENV3 H87 strain (GenBank No. M93130), DENV4 H241 strain (GenBank No. AY947539) and JEV (GenBank No. JN711458.1) were maintained in the Key Laboratory for Tropic Diseases Control (Sun Yat‐sen University, Guangzhou, Guangdong, China). C6/36 cells were infected with the viruses at a multiplicity of infection (MOI) of 1 for 1.5 hours at 37°C. Cells were observed every day. When the cell lesion was obvious, the supernatants were harvested and stored at −80°C. Immature DENV2 particles were produced on LoVo cells. LoVo cells were cultured in Ham's medium supplemented with 20% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 25 mm Hepes and 2 mm l‐glutamine, at 37°C with 5% CO2. LoVo cells were infected with DENV at a MOI of 10 for 1.5 hours at 37°C. Virus titers were determined by standard plaque assay on C6/36 cells.
2.2. Construction of chimeric JEVpr4/DENV2 cDNA in pACYC177
pACYC177 (GenBank No. X06402) was used as the plasmid vector (Figure 1A). Based on the published nucleotide sequence of DENV2 ZS01/01, the full‐length cDNA was divided into four fragments and amplified by a polymerase chain reaction (PCR) using Phusion® High Fidelity DNA Polymerases (New England Biolabs, Beverly, MA, USA). Four pairs of primers are listed in Table 1. A 15‐bp overlap sequence was designed to be included in the four pairs of primers to enable ligation between the PCR products and the pACYC177 plasmid using an In‐Fusion™ HD Cloning Kit (Clontech, Palo Alto, CA, USA). To clone both DENV2S1 and DENV2S2 into the pACYC177 vector, the SacII endonuclease site was added to two pairs of the primers for DENV2S1F, and the ClaI endonuclease site was added to DENV2S2R. Both DENV2S1R and DENV2S2F have a 15‐bp reverse‐complement sequence. Therefore, there was 15 bp of overlapping regions at the junctions of the same vectors for DENV2S1 and DENV2S2 (Figure 1B, C). The ligation reaction was performed with a Clontech Infusion HD cloning kit. The restriction sites XhoI and ClaI were used to double digest pACYC177‐DENV2 (1–5505) plasmid and create ends that could be connected to two fragments of DENV 5426–10,723 (DENV2S3, DENV2S4). The above method was repeated to clone the 5426–10,723‐bp section of the gene into the vector, so that a complete cDNA clone was formed (Figure 1D, E).
FIGURE 1.
The steps of constructing of pACYC177‐JEVpr4DENV2 plasmid. (A) The structure of pACYC177 plasmid. (B) The pACYC177 plasmid was digested by SacII and ClaI and purified; specific PCR products were purified. (C) Construction of a part of pACYC177‐DENV2(1–5505) cDNA infectious clone using an In‐Fusion™ HD cloning kit. (D) The pACYC177‐DENV2(1–5505) plasmid was digested by XhoI and ClaI and purified; specific PCR products were purified. (E) Construction of a full‐length pACYC177‐DENV2(10723) cDNA infectious clone using an In‐Fusion™ HD cloning kit. (F) pACYC177‐DENV2(10723) plasmid was digested by SacII and SpeI. The construction of pACYC177‐JEVpr4DENV2 plasmid is shown
TABLE 1.
Oligonucleotide primers used to construct the full‐length cDNA clone of DENV‐2 and JEVpr4/DENV‐2
| Primers | Primer sequence (5'‐ to 3') | Introduced sequence/restriction site |
|---|---|---|
| NV2S1F |
GAAAAACGGCTTTG CCGCGG ATTTAGGTGACACTATAG AGTTGTTAGTCTACGTGGACCGACA |
Linker sequence between the vector pACYC177 and the dengue 2 virus genome (italic); SacII endonuclease recognition sequences (underlined); the core sequence of the SP6 RNA polymerase promoter (bold); |
| DENV2S1R | GGTAAATACTCCAAAGCCATAGTCTTCAAC | |
| DENV2S2F | TTTGGAGTATTTACCACCAACATATGGC | |
| DENV2S2R | CTTCCCATACAATCGATGCTCTGAGGAAATGGGTCTCTGCTT | ClaI endonuclease recognition sequences (underlined) |
| DENV2S3F | TACATTTCAACTCGAGTTGAGATGG | XhoI endonuclease recognition sequences (underlined) |
| DENV2S3R | TGATGGTCCGTTTCTCCTCTTT | |
| DENV2S4F | GAGAAACGGACCATCACGCTGT | |
| DENV2S4R | CTTCCCATACAATCGATAGAACCTGTTGATTCAACAGCAC | ClaI endonuclease recognition sequences (underlined) |
| JEV/pr4 S1F |
GAAAAACGGCTTTG CCGCGG ATTTAGGTGACACTATAG AGTTGTTAGTCTACGTGGACCGACA |
Linker sequence between the vector pACYC177 and the dengue 2 virus genome (italic); the core sequence of the SP6 RNA polymerase promoter (bold) SacII endonuclease recognition sequences (underlined) |
| JEV/pr4 S1R |
GTTCTCTCCTTTTGAGGTGGGAATCACGATAACGTCTGC AATCTCTTGTCTACTG |
|
| JEV/pr4 S2F | TCAAAAGGAGAGAACAGATGCACCCTCATGGCCATG | |
| JEV/pr4 S2F | TCAAAAGGAGAGAACAGATGCACCCTCATGGCCATG | |
| JEV/pr4 S2R | CCCCACTAATACTAGTGACACAGACA | Linker sequence between the vector pACYC177 and the dengue 2 virus genome (italic); Spe I endonuclease recognition sequences (underlined) |
Similarly, to construct the pr4 mutant plasmid, PCR amplification primers were designed. A 5‐bp upstream SP6 RNA polymerase promoter core sequence was added upstream of the primer. According to the methods above, the pr4 sequence of DENV: AAGGGAAAAGTCTTCTGTTTAAAACAGAGAACGGTGTGAACATGTGT would be changed into the JEV pr4 sequence TTGCAGACGTTATCGTGATTCCCACCTCAAAAGGAGAGAACAGATGC (Figure 1F). The pACYC177‐JEVpr4DENV2 plasmid was verified by enzyme digestion and sequencing.
2.3. RNA transcripts and recovery virus acquisition
The pACYC177‐JEVpr4/DENV2 plasmid was linearized with ClaI to acquire the template for in vitro transcription. To ensure the infectivity of the transcript, 2.5 μg of linearized plasmid DNA was added to the SP6 RNA in vitro transcription reaction system (Ribo MAXTM Large Scale RNA Production Systems; Promega, Madison, WI, USA). The 100‐μl reaction system comprised: 20 μl of SP6 Transcription 5 × Buffer, 20 μl of rNTPs (25 mm ATP, CTP, UTP and 3 mm GTP), 7.5 μl of Ribo m7G Cap Analog (40 mm), 10 μl of Enzyme Mix (SP6), 5 μg of linear DNA template and 42.5 μl of Nuclease‐Free Water. RNase‐Free DNase was added according to the ratio of 1 U/1 μg DNA and the mixture was maintained at 37°C for 15 minutes. BHK‐21 cells were used for transfection. On day 5 after transfection, the virus was harvested from BHK21 and transferred into monolayered C6/36 cells and incubated at 37°C for 2 hours. Next, the culture solution was discarded, 2 ml of MEM virus maintenance solution was added and then incubated at 37°C with 5% CO2 for 5–7 days. When the cytopathy reached 3+, the virus supernatant was collected after centrifugation, the pH value was adjusted with 9.6% NaHCO3 to weakly alkaline, the chimeric virus was titrated with the plaque formation test and compared with the wild‐type virus.
2.4. Titration of JEVpr4/DENV2 chimeric virus by plaque‐forming assay
C6/36 cells were diluted to 5 × 104/500 μl and added to a 24‐well cell culture plate to grow into a single layer within 24 hours. The virus suspension (DENV1–4, DENV2ZS01/01, JEVpr4/DENV2) was serially diluted in a 10‐fold manner from 10−1 to 10−8. Then, 250 μl of the diluted virus suspension was added to each well for 2 hours. The virus suspension in the well was discarded and washed twice with 250 μl of phopshate‐buffered saline (PBS). Next, 850 μl of DMEM medium containing 2% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mm l‐glutamine and 1.2% (w/v) carboxymethylcellulose (Sigma, St Louis, MO, USA) was added, and the mixture was incubated for 5–7 days. The cells were observed every day for plaque formation. Once plaque formation was noticed, the methylcellulose in the wells was discarded, 500 μl of 4% (v/v) formaldehyde was added and the cells were maintained at room temperature for 10 minutes. The formaldehyde was then discarded and the cells were washed with running water. Subsequently, 250 μl of 0.8% (w/v) crystal violet was added to each well and maintained at room temperature for 10 minutes. Finally, the crystal violet was discarded, the number of plaques was counted and the virus concentrations were expressed as plaque forming units per milliliter (PFU/ml).
2.5. Indirect immunofluorescence assay of infectivity of chimeric virus on BHK21 and C6/36 cells
BHK‐21 cells and C6/36 cells were cultured in 24‐well plates containing a coverslip inside at a density of 5 × 104 cells/500 μl per well overnight. The cells were infected with viruses at a MOI of 10 for 2 hours. The plates were maintained at 37°C with 5% CO2. At 48 hours post‐transfection, the cells were washed with PBS. The coverslips were fixed in 4% paraformaldehyde for 15 minutes and then washed with PBS. The cells were permeabilized with 0.5% Triton X‐100/PBS at room temperature for 20 minutes. The coverslips were subsequently blocked with 5% bovine serum albumin. Each coverslip was incubated with 300 μl of 1:100 diluted DENV monoclonal antibody and placed in a wet box at 4°C overnight. Samples were incubated with a 500‐fold dilution of Alexa Fluor 488 conjugated goat anti‐mouse immunoglobulin (Ig)G (Invitrogen, Carlsbad, CA, USA) in a wet box, avoiding light at 37°C for 45 minutes. The coverslips were fixed to glass slides and the cell nuclei were stained with 4',6‐diamidino‐2‐phenylindole (Sigma) and anti‐fluorescence quencher. Images were observed and collected using an upright fluorescence microscope.
2.6. Immunogenicity analysis of JEVpr4/DENV2 chimeric virus
The JEVpr4/DENV2 chimeric virus was inactivated at 60°C for 30 minutes, fully emulsified with an equal volume of Freund's complete adjuvant, and then used to immunize 4–6‐week‐old Balb/C female mice. The negative control group was injected with the same volume of PBS plus adjuvant. The mouse was immunized once every 2 weeks. The immunization was emulsified with Freund's incomplete adjuvant plus inactivated chimeric virus. Two weeks after the last immunization of the mice, blood was taken from the eyeball and the serum was separated. Serum titer was detected by an enzyme‐linked immunosorbent assay (ELISA), with JEVpr4/DENV2 as antigens coated on the ELISA plate, and the mouse immune serum of JEVpr4/DENV2 was diluted 10‐fold, starting from 1:10 as the primary antibody, Horseradish peroxidase‐labeled goat anti‐mouse IgG antibody was added as a secondary antibody to the reaction system. Sulfuric acid was added to terminate the reaction. Finally, absorbance at 450 nm was detected using a spectrophotometer.
2.7. ADE of JEVpr4/DENV2 immune serum
Real‐time quantitative PCR and flow cytometry were used to detect the effect of JEVpr4/DENV2 immune serum on the ADE of DENV1–4 and ImDENV2. Relative quantitative analysis was used to determine the expression of DENV. Real‐time PCR primer sequences were designed according to the DENV E protein gene and β‐actin gene. The primers were : DENV, forward: AGCATCTCGCTGCCCAACACAAGG; DENV, reverse: TAATCCGCATCCATTTCCCCA TCC; β‐actin, forward: AAGGAGAAGCTGTGCTACGTCGC; β‐actin, reverse: AGACAGCACTGTGTT GGCGTACA. The reaction system comprised: 10 μl of SYBR FAST qPCR Master Mix (2 ×) Universal, 0.4 μl of upstream primer, 0.4 μl of downstream primer, 1 μl of the sample DNA and 20 μl of ddH2O. The reaction conditions were: 95°C for 3 min; 95°C for 1–3 seconds; 60°C for 20 seconds; and 72°C for 5 seconds; with 40 cycles in total. ADE can also be detected by flow cytometry. JEVpr4/DENV2 immune serum was inactivated at 56°C for 30 minutes, and was sterilized with a 0.22‐μm filter. Two‐fold serially diluted serum (10−1–10−8) was incubated with an equal volume of DENV1–4 and ImDENV2 for 2 hours. Mouse negative serum was used as the negative control. The virus‐serum complexes were transferred to K562 cell suspension (2 × 105 per sample) at a MOI of 1 and incubated for 2 hours, then K562 cells were collected by centrifugation at 1500 rpm (200 g) for 5 minutes. The supernatant was discarded and 700 μl of IMDM virus maintenance solution was added to resuspend the cells. The mixture was then transferred to a 24‐well plate to be incubated at 37°C with 5% CO2 for 72 hours. Infected K562 cells were fixed and permeabilized with Cytofix/Cytoperm™ Fixation/Permeabilization kit (Becton‐Dickinson Biosciences, Franklin Lakes, NJ, USA). DENV antigens were stained with specific anti‐E antibody, followed by fluorescein isothiocyanate‐conjugated goat anti‐mouse IgG antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The samples were washed twice and the percentage of K562 cells infected by DENV was detected by flow cytometry.
2.8. Neutralization assay of JEVpr4/DENV2 immune serum
The neutralizing activity of JEVpr4/DENV2 immune serum was measured via a 50% plaque reduction neutralization test (PRNT50). C6/36 cells were diluted to 5 × 104/500 μl and JEVpr4/DENV2 immune serum was inactivated at 56°C for 30 minutes. Serial two‐fold dilutions of inactivated sera (10−1 to 10−8) were mixed with an equal volume of approximately 30 PFU of DENV1–4 and ImDENV2 at 37°C with 5% CO2 for 2 hours. Mouse negative serum was used as negative control, whereas MEM maintenance solution without virus was used as a blank control. The virus‐serum mixtures were then transferred to cells in 24‐well plates followed by the plaque forming assay as described previously. The percentage of plaque reduction was calculated. Percentage reduction of plaque = [(number of plaques in negative serum group – number of plaques in antibody group)/number of plaques in negative serum group] × 100%. The serum titer required to reduce dengue viral plaques by 50% (50% neutralization titer) was calculated from three experiments.
2.9. Screening to determine the key amino acid sites of ADE epitope pr4
The “alanine scanning method” was used for epitope screening. Sixteen amino acids of epitope peptide pr4 were mutated to alanine, with pr4 as a positive control and PM10 as an irrelevant control. The mutant peptide was synthesized by Jill Biochemical (Shanghai) Co., Ltd (Shaghai, China). The pr4 mutant peptides pr4.1–pr4.16 were coated, respectively. pr4 rabbit polyclonal antibody was diluted to 100 μl per well. Horseradish peroxidase‐labeled goat anti‐rabbit IgG secondary antibody was added and incubated at 37°C for 1 hour, and washed five times with PBS‐Tween 20. After maintaining for 20 minutes at room temperature for color rendering, an equal volume of H2SO4 was added to terminate the reaction. The absorbance at 450 nm was measured.
2.10. Bioinformatics analysis of key amino acid sites
The amino acid sequences of DENV1–4, flavivirus (YFV), JEV, West Nile virus and tick‐borne encephalitis virus were downloaded from NCBI, and the pr peptides were sequenced with Meg Align Compare (USA). The Swiss‐Model (Basel / Switzerland) homology modeling method was used to predict the tertiary structure of the pr peptide of dengue type 2 virus. The key amino acid positions in the resulting tertiary structure were analyzed using Swiss‐Pdb Viewer (Basel / Switzerland), version 4.1.0.
2.11. Preparation of pr4 key site mutation peptide antibody
The mutant peptides, positive control pr4 and irrelevant control PM10 were used to immunize 4–6‐week‐old female Balb/C mice. The peptides were coupled with keyhole limpet hemocyanin. Samples were emulsified with Freund's complete adjuvant for the first immunization and immunized by a multi‐point method. The animal experiments were carried out as previously described. The epitope peptides pr4, pr4.5, pr4.6, pr4.7 and pr4.16 were diluted to 10 μg/ml. The immune serum of each peptide was diluted by two‐fold ratios (1:50, 1:100, 1:200, 1:400, 1:800, 1:1600, 1:3200 and 1:6400). An ELISA was performed to detect the titer of each mutant peptide mouse immune serum.
2.12. The ADE effect of mutant peptide immune serum on DENV
The mutant peptide mouse immune serum was inactive according to the method oulined above and was sterilized by filtration with a 0.22‐μm filter. Two‐fold dilution was carried out from 1:2 to 1:1024, and the enhanced effect of DENV2 on K562 cells mediated by mutant peptide mouse antiserum was analyzed by flow cytometry.
2.13. Neutralizing effect of mutant peptide immune serum on DENV
The mutant peptide mouse immune serum was inactive according to the method outlined above and was sterilized by filtration with a 0.22‐μm filter. Two‐fold ratio dilution was carried out from 1:2 to 1:1024, and the neutralization effect of mutant peptide mouse antiserum on DENV2 was detected by the plaque reduction neutralization test.
2.14. Statistical analysis
Statistical significance of differences between experimental groups was determined using Student's t test with Prism, version 6.0 (GraphPad Software Inc., San Diego, CA, USA). Analysis of variance and Tukey's post‐hoc statistical test was used make comparisons between the experimental groups and the control group for the ADE assay. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. Construction of DENV pr4 epitope to JEV corresponding sequences
The pACYC177‐JEVpr4DENV2 plasmid was successfully constructed according to the procedure described. pACYC177‐DENV2 was double‐digested with SacII and SpeI to obtain a large linearized vector fragment that was connected to two small fragments. The larger fragment was the target fragment, which was used to connect with the small fragment to construct the chimeric plasmid. The two gene fragments used to construct pr4 gene replacement cDNA were amplified with Phusion high‐fidelity enzymes (Thermo Fisher, Waltham, MA, USA). The results are shown in the Supporting information (Figure S1). The products from the PCR amplification were 525 bp and 1834 bp. 10 pairs of primer sequences were designed covering the entire genome (see Supporting information, Table S1). The successful construction of pACYC177‐JEVpr4DENV2 plasmid was detected by PCR products of 10 fragments (see Supporting information, Figure S1A). The construction of the plasmid was also identified by enzyme digestion. The full‐length plasmid had a ClaI, a SacII and a XhoI restriction site, which could be digested to form a single linear plasmid with a size of approximately 14,000 bp; two fragments could be formed by SacII and ClaI double enzymes: the larger fragment was approximately 11,000 bp and the smaller fragment was approximately 3,200 bp. Two fragments were formed after ClaI and XhoI double enzyme digestion: the large fragment was 8,500 bp and the small fragment was approximately 5,500 bp. The plasmid contained three BamHI restriction sites, and it could be divided into three fragments: 9798 bp, 3659 bp and 508 bp, respectively. It also contained four HindIII restriction sites, which could be divided into 4752 bp, 4165 bp, 3186 bp and 1862 bp, respectively. It contained six EcoRI restriction sites, which could be divided into 6521 bp, 3213 bp, 1284 bp, 1122 bp, 793 bp and 109 bp. The specific results of restriction digestion are shown in the Supporting information (Figure S1B) and the digestion sizes were consistent with expectations. The sequencing results showed that the chimeric plasmids of pACYC177‐JEVpr4/DENV2 were successfully constructed. The pr4 fragment was replaced by the corresponding fragments of JEV (see Supporting information, Figure S1C; the box in the figure is the sequence of DENV2pr4 that was replaced by JEV). BLAST (https://blast.ncbi.nlm.nih.gov) was used to align the pACYC177/DENV2 and pACYC177‐JEVpr4/DENV2 sequencing, and the corresponding gene of JEV can be seen to have replaced the DENV2 pr4 sequence (see Supporting information, Figure S1D).
3.2. Infectivity and immunogenicity of JEVpr4/DENV2
BHK‐21 and C6/36 cells were infected with the JEVpr4/DENV2 chimeric virus at a MOI of 10. After 48 hours, fluorescence could be seen through a fluorescence microscope, indicating that the chimeric virus was proliferating in BHK‐21 and C6/36. The fluorescence results of JEVpr4/DENV2‐infected BHK21 and C6/36 cells are shown in Figure 2A. The infectivity was detected using plaque formation test in C6/36. The results are shown in Figure 2B. The plaques formed by the JEVpr4/DENV2 chimeric virus were smaller than DENV2‐ZS01/01. From the growth curve, the replication of the JEVpr4/DENV2 chimeric virus in C6/36 cells was significantly lower than that of DENV2‐ZS01/01, and the highest titers appeared at 48 hours post‐infection, whereas it took only 24 hours for the DENV2‐ZS01/01 strain to achieve the same (Figure 2C). The results suggested that the chimeric virus had a reduced replication capacity compared to wild‐type DENV2 strain. After injection of the JEVpr4/DENV2 chimeric virus and the immune adjuvant, Balb/C mice were immunized to obtain JEVpr4/DENV2 immune serum. The titer of the immune serum was 1:6400.
FIGURE 2.
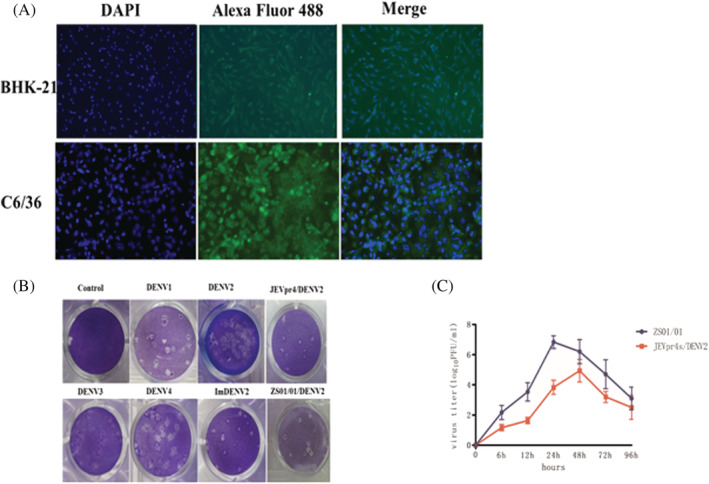
Verification of JEVpr4/DENV2 chimeric virus and production of chimeric virus antibody. (A) Indirect immunofluorescence detection of JEVpr4/DENV2 infectivity to BHK21 and C6/36 cells. (B) Plaque morphology of DENV2 in C6/36 plaque. The plaques formed by the JEVpr4/DENV2 chimeric virus were smaller than DENV2‐ZS01/01. (C) The titers of JEVpr4/DENV2 and DENV2‐ZS01/01 at different time points in C6/36 cells. The replication of the JEVpr4/DENV2 chimeric virus in C6/36 cells was significantly lower than that of DENV2‐ZS01/01, where the highest titers appeared 48 hours after infection for JEVpr4/DENV2, whereas DENV2‐ZS01/01 took only 24 hours
3.3. ADE of JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2
Real‐time PCR was used to detect the viral load in K562 cells that were infected with DENV1–4 and ImDENV2 in the presence of JEVpr4/DENV2 immune serum at different dilutions in 72 hours. The group without antibody was used as a control. JEVpr4/DENV2 immune serum had different degree of infection enhancement effect on the different types of DENV, and the infection enhancement effect was different under different dilutions. Figure 3 shows the ADE effect of JEVpr4/DENV2 immune serum on DENV1, DENV2, DENV3, DENV4 and ImDENV2. The JEVpr4/DENV2 immune serum had the greatest enhancement effect on immature DENV infection, and had an obvious effect on DENV1. It had the lowest enhancement effect on the same type of DENV2. The enhancement effect on the other types of mature DENV had a greater enhancement effect range, from 1:40 to 1:1280, reaching its peak at 1:80 to 1:320.
FIGURE 3.
Detection of the ADE effect of JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2 by real‐time PCR. The ADE of JEVpr4/DENV2 antibody on (A) DENV1, (B) DENV2, (C) DENV3, (D) DENV4 and (E) ImDENV2. The JEVpr4/DENV2 immune serum had the greatest enhancement effect on immature DENV infection, and had an obvious effect on DENV1. It had the lowest enhancement effect on the same type of DENV2. The enhancement effect on other types of mature DENV had a greater enhancement effect range, from 1:40 to 1:1280, reaching a peak at 1:80 to 1:320
FCM was used to compare the ADE effects of JEVpr4/DENV2 immune serum with DENV2 wild‐type strain immune serum. The results are shown in Figure 4. Compared with the negative control without immune serum, the different dilutions of DENV2 wild‐type strain immune serum and JEVpr4/DENV2 immune serum had different degrees of enhancement effect on DENV infection of K562 cells. The ADE of DENV2 wild‐type strain immune serum and JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2 was obvious over a wide range of dilution. The enhancement effect of DENV2 wild‐type strain immune serum and JEVpr4/DENV2 immune serum on Im‐DENV2 infection was the strongest. This reached a peak at 1:320 dilution, and the enhancement effect reached 75.6% and 38.7%, respectively. The results were consistent with the qPCR results. Compared to the ADE of DENV2 wild‐type strain immune serum, the ADE effects of JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2 were significantly weakened, with the difference being significant (p < 0.05).
FIGURE 4.
Detection of the ADE effect of JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2 by FCM. The ADE of JEVpr4/DENV2 antibody on (A) DENV1, (B) DENV2, (C) DENV3, (D) DENV4 and (E) ImDENV2. The ADE of DENV2 wild‐type strain immune serum and JEVpr4/DENV2 immune serum on Im‐DENV2 infection was the strongest, reaching a peak at 1:320 dilution. The enhancement effect reached 75.6% and 38.7%, respectively. Compared to the ADE of DENV2 wild‐type strain immune serum, the ADE effects of JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2 were significantly weakened, with the difference being significant (p < 0.05)
3.4. Neutralization effect of JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2
The neutralizing effects of DENV2 wild‐type strain immune serum and JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2 were detected by a plaque reduction test. The results showed that the DENV2 wild‐type strain immune serum had a high neutralizing activity against DENV1–4 and ImDENV2 in the serum dilution range 1:10 to 1:40, whereas it had a neutralizing activity against DENV2 in the dilution range 1:10 to 1:160. The activity was higher than 50%. The chimeric virus immune serum exhibited a neutralizing activity similar to DENV1, DENV3, DENV4 and ImDENV2. Meanwhile, the JEVpr4/DENV2 immune serum had a high neutralizing activity against DENV1–4 and ImDENV2 within the dilution range 1:10 to 1:80, and had the highest neutralizing effect on DENV2. In addition, the neutralization effect of ImDENV2 was stronger than that of the wild‐type strains. The neutralizing activity of JEVpr4/DENV2 immune serum to DENV2 and ImDENV2 at 1:160 dilution was still close to 80%. The results showed that JEVpr4/DENV2 immune serum is a type of cross‐antibody that can react with different types of DENV. Both the JEVpr4/DENV2 immune serum and the DENV2 immune serum exhibited a similar neutralizing effect on DENV1–4. The PRNT results are shown in Figure 5.
FIGURE 5.
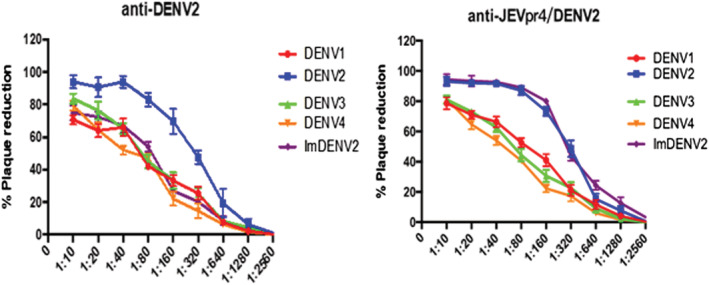
Detection of neutralization effect of JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2. The DENV2 wild‐type strain immune serum had a high neutralizing activity against DENV1–4 and ImDENV2 in the serum dilution range 1:10 to 1:40, whereas it had a neutralizing activity against DENV2 in the dilution range 1:10 to 1:160. The chimeric virus immune serum had similar neutralizing activity on DENV1, DENV3, DENV4 and ImDENV2. The JEVpr4/DENV2 immune serum had a high neutralizing activity against DENV1–4 and ImDENV2 within the dilution range 1:10 to 1:80, and had the highest neutralizing effect on DENV2
3.5. Screening of key amino acid of pr4 epitope
The 16 amino acids of pr4 had been mutated to alanine in sequence (Table 2). Each mutant peptide was coated with 5 μg per well and prM antibody diluted at 1:200, 1:1000 and 1:5000. The results are shown in Figure 6A‐C. It was observed that the reactivity of antigen and antibody was greatly reduced when amino acids 5, 6, 7 and 16 of pr4 were mutated to alanine. The four amino acids identified were leucine at position 5, leucine at position 6, phenyalanine at position 7 and cysteine at position 16. More details are provided in Figure 6D.
TABLE 2.
The amino acid sequence of pr4 and its mutant peptide
| Polypeptide | Amino acid sequence | Polypeptide | Amino acid sequence |
|---|---|---|---|
| pr4 | KGKSLLFKTENGVNMC | pr4.9 | KGKSLLFKAENGVNMC |
| pr4.1 | AGKSLLFKTENGVNMC | pr4.10 | KGKSLLFKTANGVNMC |
| pr4.2 | KAKSLLFKTENGVNMC | pr4.11 | KGKSLLFKTEAGVNMC |
| pr4.3 | KGASLLFKTENGVNMC | pr4.12 | KGKSLLFKTENAVNMC |
| pr4.4 | KGKALLFKTENGVNMC | pr4.13 | KGKSLLFKTENGANMC |
| pr4.5 | KGKSALFKTENGVNMC | pr4.14 | KGKSLLFKTENGVAMC |
| pr4.6 | KGKSLAFKTENGVNMC | pr4.15 | KGKSLLFKTENGVNAC |
| pr4.7 | KGKSLLAKTENGVNMC | pr4.16 | KGKSLLFKTENGVNMA |
| pr4.8 | KGKSLLFATENGVNMC | PM10 | SQNPPHRHQS |
Note: Bold emphasis the mutant peptide site of the pr4 amino acid sequence.
FIGURE 6.
Screening and identification of the key amino acid of ADE in pr4. (A–C) Screening of the key amino acids of pr4 by prM. The mutation of amino acids 5, 6, 7 and 16 of pr4 to alanine greatly reduced the reactivity of both antigen and antibody. (D) The key amino acid on ADE epitope pr4. The four amino acids were leucine at position 5, leucine at position 6, phenyalanine at position 7 and cysteine at position 16
3.6. Bioinformatics analysis of key amino acid sites
The results of amino acid sequence alignment between DENV1–4 and the other members of the family Flaviviridae are shown in Figure 7A. Amino acids 5, 6 and 7 of the pr4 peptide were conserved among the different DENV serotypes, which were different from the other members of the genus Flavivirus. However, cysteine at position 16 was conserved among members of the genus Flavivirus.
FIGURE 7.
Bioinformatics analysis of key amino acid sites. (A) Alignment of amino acids of the pr protein of flaviviruses with DENV1–4. Amino acids 5, 6, and 7 of the pr4 peptide are conserved among different DENV serotypes, which are different from other members of the genus Flavivirus. However, cysteine at position 16 is conserved among members of the genus Flavivirus. (B) The tertiary structure of pr peptide was obtained by Swiss‐Model homology modeling. Amino acids 5, 6, 7 and 16 of pr4 formed a pocket‐like structure, which increased the possibility for these four sites being the key amino acid sites in antibody‐binding epitopes
The tertiary structure of pr peptide was obtained by Swiss‐Model homology modeling. The results are shown in Figure 7B. The amino acids 5, 6, 7 and 16 of pr4 formed a pocket‐like structure, which increased the possibility for these four sites to be the key amino acid sites in antibody binding epitopes.
3.7. Detection of mutant peptide immune serum titer
pr4, pr4.5, pr4.6, pr4.7 and pr4.16 were conjugated with hemocyanin and emulsified with immune adjuvant to immunize Balb/C female mice. The results are shown in Figure 8. The titer of antiserum in most mice could reach more than 1:6400, although the pr4.16 mutant peptide antiserum had no titer at all.
FIGURE 8.
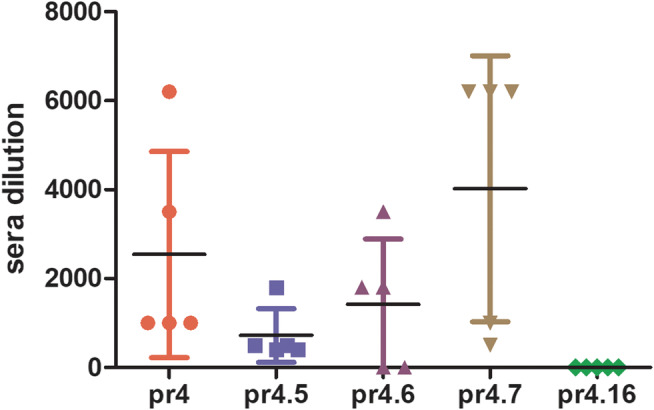
The titer of each mutant peptide immune serum detected by an ELISA. The titer of immune serum could reach more than 1:6400. The pr4.16 mutant peptide immune serum had no titer at all
3.8. ADE of mutant peptide antiserum
The ADE results are shown in Figure 9. It can be seen that the key amino acid site mutant peptide reduced the enhancement effect on DENV2, and the reduction level was more apparent when the antibody dilution was higher. The difference was statistically significant (p < 0.05).
FIGURE 9.
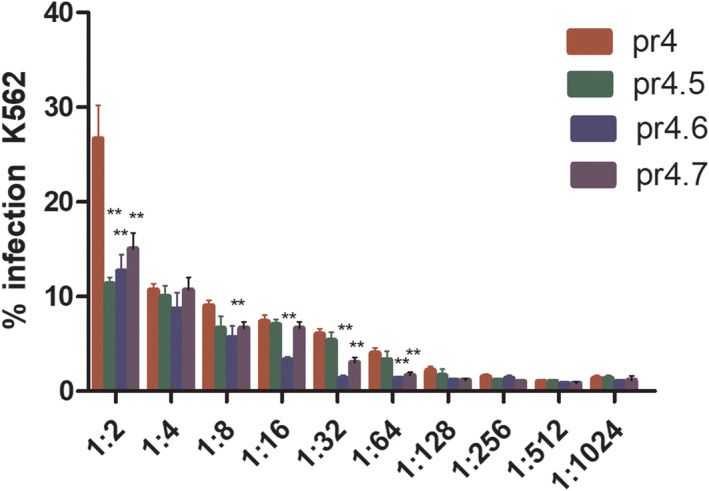
ADE of DENV2 infection on K562 cells mediated by anti‐pr4.5, pr4.6 and pr4.7 pAb. The key amino acid site mutant peptide reduced the enhancement effect on DENV2, and the reduction level was more obvious when the antibody dilution was higher. The difference was significant (p < 0.05)
3.9. Neutralizing effect of mutant peptide antiserum on DENV2
The neutralizing effects of mutant peptide antiserum on DENV2 are shown in Figure 10. The neutralization effects of pr4.5 and pr4.7 antiserum reach more than 80%, whereas the antiserum neutralization effect of the positive control pr4 reached only 42.8%. The difference was statistically significant (p < 0.05).
FIGURE 10.
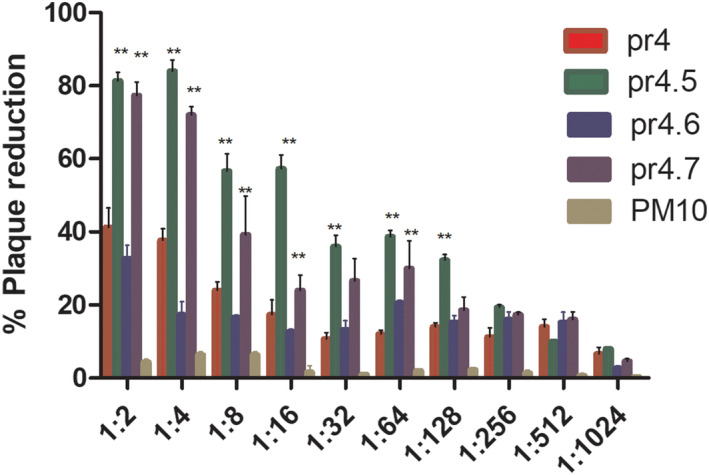
Neutralizing activities of anti‐pr4.5, pr4.6 and pr4.7 pAb with respect to DENV infection. The neutralization effects of pr4.5 and pr4.7 antiserum reach more than 80.0%, whereas the antiserum neutralization effect of the positive control pr4 could only reach 42.8%. The difference was significant (p < 0.05)
4. DISCUSSION
DENV is spread by mosquito vectors in tropical and subtropical regions around the world. It is estimated that 390 million people worldwide are infected every year, with 96 million of them showing obvious clinical symptoms, and the serious development into life‐threatening DHF/DSS, causing serious public health concerns. In addition, the DENV is inextricably linked to the Zika virus that has become a pandemic in the past 2 years, driving the need for profound research on DENV vaccine.
ADE is an important cause of DHF/DSS, which poses as the main hurdle in the development of vaccines for DENV. 29 , 30 The DENV vaccine mainly contains C‐prM‐E protein, which results in the presence of prM antibody in the host after immunization, subsequently leading to the ADE effect during secondary infection of atypical dengue. 31 , 32 Therefore, ways of reducing the occurrence of ADE must be considered when developing vaccines. Because anti‐prM antibodies could enhance DENV infection, 33 , 34 and the epitopes of these infection‐enhancing anti‐prM antibodies are mainly located in the amino acid residues of the pr peptide, 24 , 26 strategies at the molecular level can be developed to replace the ADE epitope, thus reducing ADE. This may eventually provide new insights and novel ideas for vaccine development.
Luo et al. 26 used peptide scanning to screen the linear epitope pr4 of the DENV pr peptide that played a crucial role in ADE. To replace, or better still, eliminate the effect of ADE, the epitope must be considered when designing the vaccine. A plasmid was constructed with pr4 replaced. The chimeric virus JEVpr4/DENV2 showed reduced virulence and good immunogenicity. The enhancement effect of JEVpr4/DENV2 immune serum on DENV1–4 and ImDENV2 was weaker than that of DENV2 wild‐type strain immune serum. These experimental results showed that the ADE effect of JEVpr4/DENV2 immune serum on DENV could be significantly reduced in terms of range and intensity compared to the immune serum of the wild‐type strain. We also constructed a gene deleted plasmid pACYC177‐pr4△/DENV2 (see Supporting information, Figure S2) and found that the pr4 gene deleted plasmid could not be transformed to rescued virus. As a result, it was hypothesized that the pr4 gene played a key role in the proliferation of the virus in mammalian cells. Therefore, we cannot evaluate the ADE by deleting the pr4 gene. These data verify pr4 as the ADE epitope.
The screening of key amino acid sites includes peptide scanning, bioinformatics analysis, crystal structure analysis and other methods. However, the key amino acid sites obtained by preliminary screening require further biological identification to determine the role of these key sites. The commonly used identification method is to replace the key amino acid sites by site‐directed mutagenesis, investigate the change of the mutant peptide or the biological function of the virus, and then finally identify the key sites. Alanine is a neutral aliphatic amino acid and the chiral amino acid with the shortest side chain. Using it to replace other amino acids can keep the overall configuration of the protein unchanged. Therefore, when performing site‐directed mutagenesis, the target site is generally selected to be alanine.
The present study used the alanine mutation scanning method to identify the amino acids at positions 5, 6, 7 and 16 of pr4 as being significant in ADE. Bioinformatics analysis shows that the amino acids at positions 5, 6 and 7 of pr4 were conserved among different serotypes of DENV, whereas the amino acid at position 16 was conserved in flavivirus. Conserved amino acid positions were beneficial with respect to inducing cross‐reacting antibodies. The tertiary structure of the pr peptide was then predicted using Swiss‐Model homology modeling. It was observed that the four sites obtained from the screening were very close. They formed a pocket‐like spatial structure on the outside of the protein, which was conducive to the induction and binding of antibodies. In addition, Smith et al. 29 prepared a series of human prM monoclonal antibodies and screened the corresponding key amino acid positions of their corresponding epitopes. Among them, there were several key positions of monoclonal antibodies, including leucine at position 6 (L).
The present study has identified the immunological effects of key amino acids at the proteomic and genetic levels. It was found that the immune serum had no titer at all after the mice were immunized with the amino acid mutated peptide at position 16. The amino acid at this position was highly conserved in the genus Flavivirus, and mutation of this position might lead to a substantial change in the structure of pr4, thereby causing the immunogenicity of the mutant peptide to disappear. The overall titer of the antiserum was not very high. This might be because the mutant peptide was a hapten. Although hemocyanin was coupled, the main antibody produced after immunization of the mice was against hemocyanin (the titer of anti‐hemocyanin in the serum of immunized mice was more than 1:100,000), resulting in a lower specific antibody titer of the mutant peptide antiserum (Figure 8). In addition, the low titer of the mutant peptide antiserum was also one of the reasons for the low neutralization effect of pr4. The neutralization effect of mutant peptides has indeed improved, whereas the effect of ADE was reduced, although the overall decrease in the effect of ADE was not obvious. It might be because the effect of ADE depends on multiple amino acids, which are independent of one another. Mutations in the chosen amino acid sites did not have a greater impact on the immune functions of epitope pr4 (Figure 9).
To identify which one of the four key amino acids of pr4 was the most important one in ADE, we manually mutated four infectious cDNA clones of DENV type 2 to ensure that one key amino acid was replaced with alanine in each cDNA clone. However, only one strain of mutant virus was successfully rescued with abilities of infectivity and proliferation. We will continue our aim of confirming that this comprises the most critical amino acid site through further experiments (see Supporting information, Figure S3).
In the face of increasingly severe dengue epidemic, the development of vaccines that are safe and effective is still the best strategy for combatomg viral infections. The key to success for an effective dengue vaccine is having balanced and long‐lasting antiviral protection against all four serotypes of DENVes at the same time. More importantly, it must be able to avoid safety issues, such as the severe dengue‐related immune responses that are brought upon by vaccination. Nevertheless, creating a dengue vaccine that meets all these requirements is not easy. Besides being based on an exploration of the pathogenic mechanism of DENV and host antiviral immune regulation, it must also be supplemented by breakthroughs in new biotechnology and vaccine adjuvants. The present study provides important ideas for the development of attenuated vaccines. It should be followed up by more profound research, in the hope of promoting the development of DENV vaccines, and paving a way for the translational research of other vaccines.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
GHC and LFJ conceived and designed the experiments. GHC and LLS performed most of the experiments. YW helped to construct the plasmid. JMZ and HJY analyzed the data. LFJ revised the manuscript. All authors read and approved the final manuscript submitted for publication.
Supporting information
Table S1. Primer designed to verify plasmid sequence integrity.
Figure S1. The verification of pACYC177‐JEVpr4/DENV2 construction. (A) Detection of pACYC177‐JEVpr4/DENV2 using the primers in Table S1. (B) pACYC177‐JEVpr4/DENV2 was digested by 1 ClaI, 2 XhoI, 3 ClaI‐ XhoI, 4 BamHI, 5 EcoRI, 6 Hind III, 7 SacII and 8 ClaI‐SacII. (C) Part of the results from pACYC177‐JEVpr4/DENV2 sequencing. (D) BLAST alignment of pACYC177‐JEVpr4/DENV2 and the sequence of the DENV2 prototype.
Figure S2. The sequence of virus plasmid (pACYC177‐pr4△/DENV2). (A) Sequence of pACYC177‐pr4△/DENV2. (B) BLAST of sequences of pACYC177‐pr4△/DENV2 and DENV2‐ZS01/01.
Figure S3. The sixth position amino acid of pr4 might be the key ADE site. (A) Alignment of nucleotides and amino acids on pr4 of the full‐length plasmid mutations. (B) Identification of the specificity of the mutant viruses by real‐time PCR. (C) Identification of the infectivity of the mutant viruses pr4.6 by IFA.
Cui G, Si L, Wang Y, Zhou J, Yan H, Jiang L. Antibody‐dependent enhancement (ADE) of dengue virus: Identification of the key amino acid that is vital in DENV vaccine research. J Gene Med. 2021;23:e3297 10.1002/jgm.3297
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Hosseini S, Oliva‐Ramirez J, Vazquez‐Villegas P, et al. Dengue fever: a worldwide threat an overview of the infection process, environmental factors for a global outbreak, diagnostic platforms and vaccine developments. Curr Top Med Chem. 2018;18:1531‐1549. [DOI] [PubMed] [Google Scholar]
- 2. Bhatt S, Gething PW, Brady OJ, et al. The global distribution and burden of dengue. Nature. 2013;496:504‐507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castro MC, Wilson ME, Bloom DE. Disease and economic burdens of dengue. Lancet Infect Dis. 2017;17:e70‐e78. [DOI] [PubMed] [Google Scholar]
- 4. Gubler DJ. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found Symp. 2006;277:3‐16. [DOI] [PubMed] [Google Scholar]
- 5. Rothman AL. Dengue: defining protective versus pathologic immunity. J Clin Invest. 2004;113:946‐951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simmons CP, Chau TN, Thuy TT, et al. Maternal antibody and viral factors in the pathogenesis of dengue virus in infants. J Infect Dis. 2007;196:416‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Halstead SB, Lan NT, Myint TT, et al. Dengue hemorrhagic fever in infants: research opportunities ignored. Emerg Infect Dis. 2002;8:1474‐1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khetarpal N, Khanna I. Dengue fever: causes, complications, and vaccine strategies. J Immunol Res. 2016;2016:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kesorn K, Ongruk P, Chompoosri J, et al. Morbidity rate prediction of dengue hemorrhagic fever (DHF) using the support vector machine and the Aedes aegypti infection rate in similar climates and geographical areas. PLoS ONE. 2015;10(5):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Noisakran S, Perng GC. Alternate hypothesis on the pathogenesis of dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS) in dengue virus infection. Exp Biol Med. 2008;233:401‐408. [DOI] [PubMed] [Google Scholar]
- 11. Murphy BR, Whitehead SS. Immune response to dengue virus and prospects for a vaccine. Annu Rev Immunol. 2011;29:587‐619. [DOI] [PubMed] [Google Scholar]
- 12. Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat Rev Microbiol. 2007;5:518‐528. [DOI] [PubMed] [Google Scholar]
- 13. Halstead SB, Dans LF. Dengue infection and advances in dengue vaccines for children. Lancet Child Adolesc Health. 2019;3:734‐741. [DOI] [PubMed] [Google Scholar]
- 14. Anasir MI, Ramanathan B, Poh CL. Structure‐based Design of Antivirals against envelope glycoprotein of dengue virus. Viruses. 2020;12(4):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klein DE, Choi JL, Harrison SC. Structure of a dengue virus envelope protein late‐stage fusion intermediate. J Virol. 2013;87:2287‐2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li L, Lok SM, Yu IM, et al. The flavivirus precursor membrane‐envelope protein complex: structure and maturation. Science. 2008;319:1830‐1834. [DOI] [PubMed] [Google Scholar]
- 17. Chan AH, Tan HC, Chow AY, et al. A human PrM antibody that recognizes a novel cryptic epitope on dengue E glycoprotein. PLoS ONE. 2012;7(4):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Che P, Tang H, Li Q. The interaction between claudin‐1 and dengue viral prM/M protein for its entry. Virology. 2013;446:303‐313. [DOI] [PubMed] [Google Scholar]
- 19. Putri DH, Sudiro TM, Yunita R, et al. Immunogenicity of a candidate DNA vaccine based on the prM/E genes of a dengue type 2 virus cosmopolitan genotype strain. Jpn J Infect Dis. 2015;68:357‐363. [DOI] [PubMed] [Google Scholar]
- 20. Sangiambut S, Pethrak C, Anupap C, et al. Enhanced production of infectious particles by adaptive modulation of C‐prM processing and C‐C interaction during propagation of dengue pseudoinfectious virus in stable CprME‐expressing cells. J Gen Virol. 2020;101:59‐72. [DOI] [PubMed] [Google Scholar]
- 21. Balakrishnan T, Bela‐Ong DB, Toh YX, et al. Dengue virus activates polyreactive, natural IgG B cells after primary and secondary infection. PLoS ONE. 2011;6(12):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Paula SO, Lima DM, de Oliveira Franca RF, Gomes‐Ruiz AC, da Fonseca BA. A DNA vaccine candidate expressing dengue‐3 virus prM and E proteins elicits neutralizing antibodies and protects mice against lethal challenge. Arch Virol. 2008;153:2215‐2223. [DOI] [PubMed] [Google Scholar]
- 23. Feng K, Zheng X, Wang R, et al. Long‐term protection elicited by a DNA vaccine candidate expressing the prM‐E antigen of dengue virus serotype 3 in mice. Front Cell Infect Microbiol. 2020;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo Y, Guo X, Yan H, et al. Comprehensive mapping infection‐enhancing epitopes of dengue pr protein using polyclonal antibody against prM. Appl Microbiol Biotechnol. 2015;99:5917‐5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hua RH, Chen NS, Qin CF, et al. Identification and characterization of a virus‐specific continuous B‐cell epitope on the PrM/M protein of Japanese encephalitis virus: potential application in the detection of antibodies to distinguish Japanese encephalitis virus infection from West Nile virus and dengue virus infections. Virol J. 2010;7:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo YY, Feng JJ, Zhou JM, et al. Identification of a novel infection‐enhancing epitope on dengue prM using a dengue cross‐reacting monoclonal antibody. BMC Microbiol. 2013;13:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carvalho A, Van Roy R, Andrus J. International dengue vaccine communication and advocacy: challenges and way forward. Expert Rev Vaccines. 2016;15:539‐545. [DOI] [PubMed] [Google Scholar]
- 28. Gao N, Li J, Sheng Z, et al. Japanese encephalitis virus prM‐E antigen immunization conferred protection against challenge by four different serotypes of dengue viruses in mice. Appl Microbiol Biotechnol. 2019;103:4977‐4986. [DOI] [PubMed] [Google Scholar]
- 29. Smith SA, Nivarthi UK, de Alwis R, et al. Dengue virus prM‐specific human monoclonal antibodies with virus replication‐enhancing properties recognize a single Immunodominant antigenic site. J Virol. 2016;90:780‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kalayanarooj S. Clinical manifestations and Management of Dengue/DHF/DSS. Trop Med Health. 2011;39(4 Suppl):83‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ketloy C, Keelapang P, Prompetchara E, et al. Strategies to improve the immunogenicity of prM+E dengue virus type‐2 DNA vaccine. Asian Pac J Allergy Immunol. 2017;35:11‐19. [DOI] [PubMed] [Google Scholar]
- 32. Lima DM, de Paula SO, Franca RF, et al. A DNA vaccine candidate encoding the structural prM/E proteins elicits a strong immune response and protects mice against dengue‐4 virus infection. Vaccine. 2011;29:831‐838. [DOI] [PubMed] [Google Scholar]
- 33. Beltramello M, Williams KL, Simmons CP, Macagno A, et al. The human immune response to dengue virus is dominated by highly cross‐reactive antibodies endowed with neutralizing and enhancing activity. 2010;8:271‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodpothong P, Boonarkart C, Ruangrung K, Onsirisakul N, Kanistanon D, Auewarakul P. Relative contribution of dengue prM‐ and E‐specific polyclonal antibodies to neutralization and enhancement. Acta Virol. 2016;60:249‐259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer designed to verify plasmid sequence integrity.
Figure S1. The verification of pACYC177‐JEVpr4/DENV2 construction. (A) Detection of pACYC177‐JEVpr4/DENV2 using the primers in Table S1. (B) pACYC177‐JEVpr4/DENV2 was digested by 1 ClaI, 2 XhoI, 3 ClaI‐ XhoI, 4 BamHI, 5 EcoRI, 6 Hind III, 7 SacII and 8 ClaI‐SacII. (C) Part of the results from pACYC177‐JEVpr4/DENV2 sequencing. (D) BLAST alignment of pACYC177‐JEVpr4/DENV2 and the sequence of the DENV2 prototype.
Figure S2. The sequence of virus plasmid (pACYC177‐pr4△/DENV2). (A) Sequence of pACYC177‐pr4△/DENV2. (B) BLAST of sequences of pACYC177‐pr4△/DENV2 and DENV2‐ZS01/01.
Figure S3. The sixth position amino acid of pr4 might be the key ADE site. (A) Alignment of nucleotides and amino acids on pr4 of the full‐length plasmid mutations. (B) Identification of the specificity of the mutant viruses by real‐time PCR. (C) Identification of the infectivity of the mutant viruses pr4.6 by IFA.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


