Abstract
N6-methyladenosine (m6A) has emerged as the most abundant mRNA modification that regulates gene expression in many physiological processes. m6A modification in RNA controls cellular proliferation and pluripotency and has been implicated in the progression of multiple disease states, including cancer. RNA m6A methylation is controlled by a multiprotein “writer” complex including the enzymatic factor methyltransferase-like protein 3 (METTL3) that regulates methylation and two “eraser” proteins, RNA demethylase ALKBH5 (ALKBH5) and fat mass- and obesity-associated protein (FTO), that demethylate m6A in transcripts. FTO can also demethylate N6,2′-O-dimethyladenosine (m6Am), which is found adjacent to the m7G cap structure in mRNA. FTO has recently gained interest as a potential cancer target, and small molecule FTO inhibitors such as meclofenamic acid have been shown to prevent tumor progression in both acute myeloid leukemia and glioblastoma in vivo models. However, current FTO inhibitors are unsuitable for clinical applications due to either poor target selectivity or poor pharmacokinetics. In this work, we describe the structure-based design, synthesis, and biochemical evaluation of a new class of FTO inhibitors. Rational design of 20 small molecules with low micromolar IC50’s and specificity toward FTO over ALKBH5 identified two competitive inhibitors FTO-02 and FTO-04. Importantly, FTO-04 prevented neurosphere formation in patient-derived glioblastoma stem cells (GSCs) without inhibiting the growth of healthy neural stem cell-derived neurospheres. Finally, FTO-04 increased m6A and m6Am levels in GSCs consistent with FTO inhibition. These results support FTO-04 as a potential new lead for treatment of glioblastoma.
Introduction
The role of mRNA modifications in regulation of gene expression, stem-cell maintenance, and differentiation has gained significant interest upon transcriptome-wide mapping of the most abundant internal modification, N6-methyladenosine (m6A), which was identified in over 25% of all mRNAs.1−3 m6A methylation is considered a reversible modification, where addition of the methyl group is controlled by a multiprotein “writer” complex requiring a heterodimer comprised of METTL3 and METTL14 proteins and supported by WTAP, KIAA1429, and RBM15.4−7 Demethylation is controlled primarily by two “eraser” Fe(II)- and 2-oxoglutarate-dependent dioxygenases, RNA demethylase ALKBH5 (ALKBH5) and fat mass- and obesity-associated protein (FTO).8−16 FTO has also been shown to demethylate N6,2′-O-dimethyladenosine (m6Am) modified RNA transcripts.15,17−20 An additional host of “reader” proteins is composed primarily of the YTH-domain containing family that binds m6A-containing mRNAs and triggers a variety of downstream fates, including RNA degradation, stabilization, and translation.3,21−28
While the role of m6A modification in stem cell differentiation is well-known, the role of this modification in dedifferentiation and tumor progression is still emerging. Geula et al. have shown that pluripotent stem cells depleted in m6A modifications show resistance to differentiation, suggesting that alterations in m6A can alter differentiation pathways.2 As such pathways are known to be directly linked to the acquisition of stem-like cell properties in solid and hematological tumors, it is suspected that m6A dysregulation may play a role in the generation of tumor-initiating cells and cancer progression.29 Several studies have shown that dysregulation of any part of the adenosine-m6A equilibrium is associated with poor prognosis and tumorigenesis in a wide variety of cancers, including acute myeloid leukemia (AML).30−40 Recent studies have started to illuminate the role of RNA methylation dynamics in regulating the outcomes of cancer immunotherapies41−44 Su et al. have shown that FTO regulates MYC/CEBPA expression, and inhibition of FTO by the α-ketoglutarate mimic R-2-hydroxyglutarate reduces proliferation and viability of leukemia cells both in vitro and in vivo.36 Recently, a new derivative of meclofenamic acid (MA) called FB23-2 was also shown to suppress proliferation and promote differentiation in AML cells and prolong survival in AML mouse models.38
The m6A methylation machinery has also been identified as a potential therapeutic target in glioblastoma. ALKBH5 has been shown to be an oncogene for glioblastoma, where shRNA knockdown of ALKBH5 in patient-derived glioblastoma stem cells (GSCs) decreased tumor cell proliferation and tumorigenesis by reducing the expression of FOXM1.33 Depletion of m6A by knockdown of either METTL3 or METTL14 leads to growth and self-renewal in GSCs both in vitro and in vivo.(35) Reduction of m6A levels in vivo were further correlated with poor survival outcomes in GSC-grafted mice, while increased m6A levels via overexpression of METTL3 impaired tumor proliferation in multiple GSC lines in vitro.35 Furthermore, treatment of orthotopically transplanted GSC tumors with the small molecule FTO inhibitor MA prevented tumor progression in vivo, supporting the role of m6A methylation pathways in GSC growth and self-renewal.35 Conversely, Visvanathan et al. showed that silencing of METTL3 impaired neurosphere formation in GSCs and sensitized neurospheres to γ-irradiation via downregulation of SOX2-mediated DNA repair; the authors further demonstrate that knockdown of METTL3 prolonged lifespan in an intracranial orthotopic mouse model.45 While the role of m6A methylation in glioblastoma is still unclear, these studies illustrate the emerging interest in the m6A methylation machinery and FTO specifically as potential targets for cancer chemotherapy. However, most existing small molecule inhibitors of FTO show poor pharmacokinetic profiles or inadequate selectivity toward FTO and are considered unsuitable for clinical study. Therefore, it is important to identify novel chemical scaffolds for targeting FTO that may offer advantages over existing selectivity and physicochemical properties.
Results and Discussion
Structure-Based Design and Synthesis of Pyrimidine-Based FTO Inhibitors
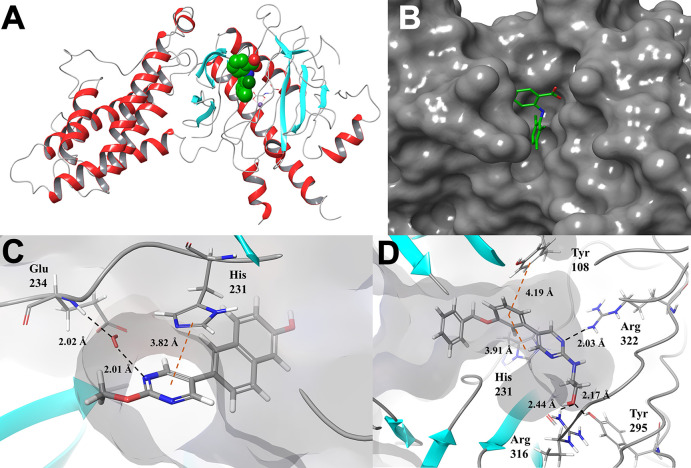
In order to identify chemically distinct inhibitors of FTO, we used a combination of structure-based drug design and molecular docking with the Schrödinger software suite to target the MA binding site of FTO. As MA has previously been shown to preferentially inhibit FTO over ALKBH5, we rationalized that targeting this site would be more likely to identify unique inhibitors that also maintained selectivity against ALKBH5.46 An X-ray crystal structure of the MA-FTO complex (PDB ID: 4QKN) was first prepared using the Prime module, and the docking grid was defined as a 5 × 5 × 5 Å cube centered on MA (Figure 1A and B).46 Docking was performed using Glide XP.47−49 Scaffold hopping of the benzoic acid region identified a pyrimidine scaffold as a promising replacement, and fragment growth was directed toward an unoccupied binding pocket containing residues Glu234, Tyr106, Tyr108, and Arg322. Interactions with these four residues were considered highly favorable. Additional contacts with the nucleotide recognition lid (β3i and β4i, including Val83–Pro93) were considered favorable, as this flexible loop is unique to FTO among homologous α-ketoglutarate dependent dioxygenases and the selectivity of MA toward FTO over ALKBH2, -3, and -5 has been attributed to interactions with this region.46 Representative docking poses for two inhibitors (FTO-02 and FTO-18) are shown in Figure 1C and D. Docking poses for FTO-1–20 are in the Supporting Information (Figures S1–S20). Hits showing promising docking scores (absolute value ≥7) were also analyzed by QikProp to assess their physicochemical properties. As existing FTO inhibitors fail to progress to clinical applications due to poor pharmacokinetic profiles, it was important to filter our screen for compounds with more favorable physicochemical properties. Priority was placed on compounds with high predicted membrane permeability (>500 nm/s), a clogP between 1 and 4, and a low molecular weight (<350 g/mol). These criteria were selected due to multiple studies indicating that compounds with low molecular weight and moderate lipophilicity are more likely to show favorable adsorption and clearance rates and less toxicity due to target promiscuity. As such, controlling the physicochemical properties of inhibitors during the initial screening stages should select for better leads for future optimization and development. On the basis of these criteria, the top 20 inhibitors were selected for synthesis (Table S1). These parameters were also calculated for MA, FB23-2, and its precursor FB23 (Table S2). Of these, only FB23-2 was found to have a clogP value in between 1 and 4 (3.46) and all three are predicted to have limited membrane permeability. In Huang et al., FB23 was shown to have limited cellular efficacy due to poor cellular uptake.38 FB23-2 was designed to overcome this limitation, and the cellular concentration of FB23-2 was found to be ∼3–10× greater than that of FB23 in MONOMAC6 and NB4 cells, although still limited.38 Similarly, our predicted permeability models estimate the rate of passive diffusion for FB23-2 to be ∼2.5× greater than that of FB23. Of the 20 compounds selected for synthesis, 15 were predicted to have improved permeability relative to MA, FB23, and FB23-2 while still adhering to the ideal lipophilicity range (Tables S1 and S2).
Figure 1.
Molecular docking targeting the meclofenamic acid binding site of FTO. (A) X-ray crystal structure of human FTO in complex with meclofenamic acid (MA; PDB ID: 4QKN). The docking site for in silico screening is shown in green spheres. (B) Surface representation of human FTO in complex with MA in green (PDB ID: 4QKN). (C) Predicted binding mode of FTO-02 at the MA binding site. A water mediated hydrogen bond is expected between the pyrimidine ring of FTO-02 and the backbone of Glu 234. A π–π stacking interaction is observed with His 231. (D) Predicted binding pose of FTO-18 at the MA binding site of FTO. A benzene ring of FTO-18 is observed to form π–π stacking interactions with His 231 and Tyr 108, and the pyrimidine ring of FTO-18 is expected to form a hydrogen bond to Arg 322. Tyr 295 and Arg 316 are predicted to form a bifurcated hydrogen bond to the hydroxyl group of FTO-18.
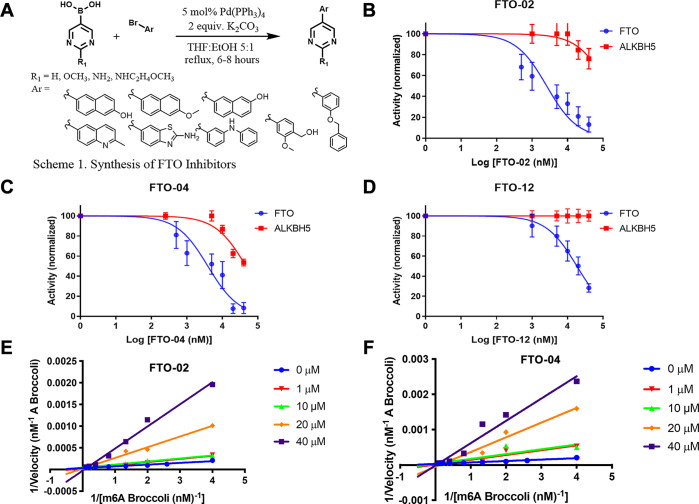
Compounds were synthesized via Suzuki–Miyaura cross-coupling, affording all compounds on a milligram scale in moderate yields (52–75%, Scheme 1 in Figure 2A, general procedure A). Substituted pyrimidine boronic acids were coupled with a variety of commercially available aryl bromides by tetrakis(triphenylphosphine)palladium in tetrahydrofuran and ethanol. While most compounds were synthesized without the use of protecting groups, the amino group of the amino-benzothiazole ring in FTO-04 was protected with a tertbutyloxycarbonyl (Boc) group prior to coupling (SI, procedure B). The Boc group was then removed under acidic conditions to obtain FTO-04 (SI, procedure C). After purification by silica gel column chromatography, a total of 20 potential FTO inhibitors were obtained.
Figure 2.
FTO Inhibitors are selective and competitive. (A) Synthesis of FTO inhibitors by Suzuki coupling. (B) Sigmoidal dose–response curves for FTO-02. Inhibition against FTO is shown in blue, and inhibition of ALKBH5 is shown in red. (C) Sigmoidal dose–response curves for FTO-04. Inhibition against FTO is shown in blue, and inhibition of ALKBH5 is shown in red. (D) Sigmoidal dose–response curves for FTO-12. Inhibition against FTO is shown in blue, and inhibition of ALKBH5 is shown in red. (E) Double reciprocal plot for FTO-02. FTO-02 inhibits FTO by a competitive mechanism. (F) Double reciprocal plot for FTO-04. FTO-04 inhibits FTO by a competitive mechanism.
FTO-02 and FTO-04 Are Potent and Selective Competitive Inhibitors of FTO
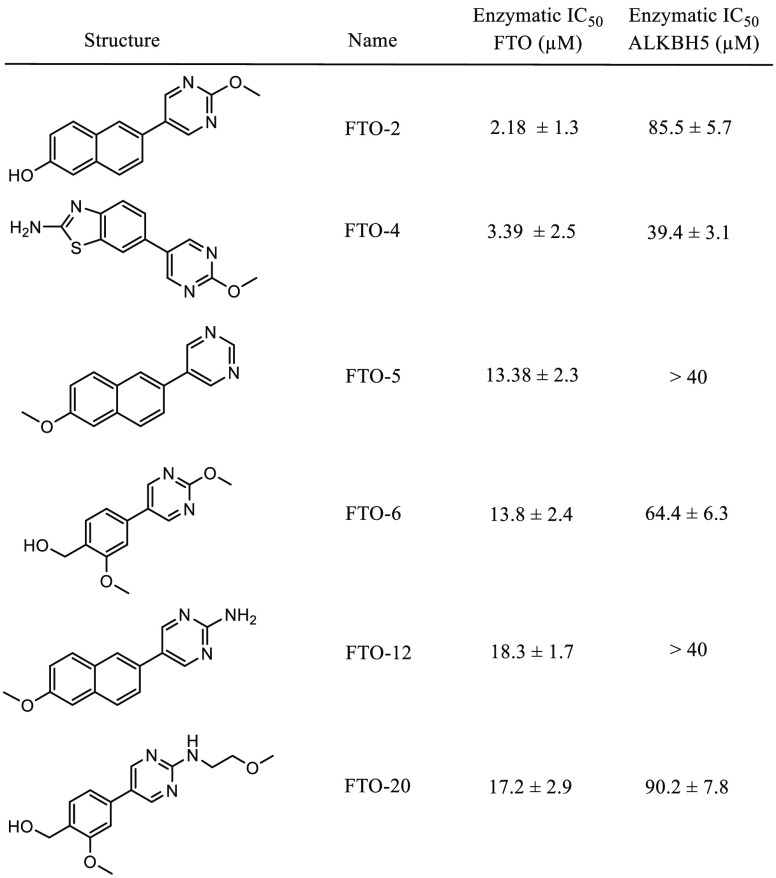
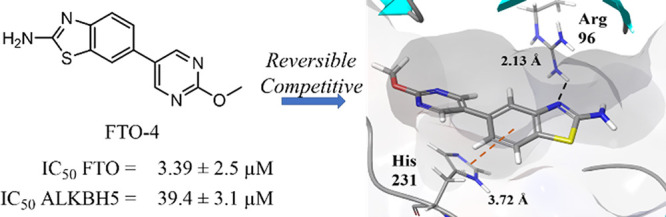
In order to determine their efficacy as FTO inhibitors, the compounds were screened by a fluorescence enzymatic inhibition assay developed previously by the Jaffrey lab.50 Briefly, a nonfluorescent methylated RNA substrate termed “m6A7-Broccoli” is incubated with FTO in the presence of 2-oxoglutarate (300 μM), (NH4)2Fe(SO4)2·6H2O (300 μM), and l-ascorbate (2 mM) for 2 h at RT in reaction buffer (50 mM NaHEPES, pH 6). Read buffer (250 mM NaHEPES, pH 9, 1 M KCl, 40 mM MgCl2) containing the small molecule 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI-1T, 2.2 μM) was added to the reaction mixture, and DFHBI-1T binds preferentially to demethylated Broccoli to produce a fluorescent signal after incubation for 2 h at RT. MA was used as a positive control, and the observed IC50 was in agreement with literature values (IC50 = 12.5 ± 1.8 μM, Figure S21).46,50 The enzymatic activity of FTO was tested at six concentrations of each inhibitor ranging from 0 to 40 μM in triplicate. As a negative control, the assays were repeated with demethylated Broccoli to ensure that any change in fluorescence was not due to interference with the Broccoli-DHBI-1T complex (Figure S22); no compounds were observed to significantly alter the fluorescence signal at concentrations up to 40 μM. To ensure that DMSO did not interfere with the fluorescence signal or enzyme activity, the activity was determined for FTO under concentrations of DMSO ranging from 0 to 10% (Figure S23). DMSO was found to interfere with enzyme activity at concentrations >1%; all inhibitor concentrations were restricted to a final concentration of 0.2% DMSO. Compounds FTO-02 and FTO-04 were also screened against FTO without the presence of cofactor 2-oxoglutarate; under these conditions, no fluorescence was observed (Figure S24). Two compounds, FTO-03 and FTO-15, showed significant precipitation in assay buffer, and the dose response could not be determined. All other compounds showed IC50’s in the micromolar range, with six compounds showing IC50’s below 15 μM and seven showing IC50’s above 40 μM (Table 1, Table S1). Of the four pyrimidine scaffolds tested, 2-methoxypyrimidine appeared to be the most potent against FTO, as all compounds with this moiety had an IC50 below 15 μM. Compounds with the unsubstituted pyrimidine scaffold varied in IC50 from 13 to 41 μM, and both the 2-aminopyrimidine and the pyrimidine-2-aminoethanol scaffolds showed little inhibitory potency. Of the aryl bromides, the 6-methoxynaphthalene and the (2-methoxyphenyl)methanol scaffolds both consistently showed potency toward FTO, where all compounds containing these scaffolds had IC50’s below 20 μM (Table 1, Table S1). The potency of other aryl bromide scaffolds varied widely and appeared dependent on the corresponding pyrimidine scaffold. In general, compounds containing either the 2-methoxypyrimidine or the 6-methoxynaphthalene were the most potent inhibitors of FTO; the two most potent inhibitors, FTO-02 and FTO-04 (IC50 = 2.2 and 3.4 μM respectively), were found to inhibit FTO approximately 4× more potently than MA (IC50 = 12.5 μM) with comparable potency to FB23-2 (reported IC50 = 2.6 μM).38
Table 1. Selective Inhibitors of FTO.
The top two inhibitors were also screened against FTO using an ELISA-based inhibition assay as an orthogonal assay control. Biotinylated m6A-RNA was incubated with FTO for 2 h at RT in reaction buffer (50 mM NaHEPES pH 6, 300 μM 2-oxoglutarate, 300 μM (NH4)2Fe(SO4)2·6H2O, and 2 mM L-ascorbate) with 0–40 μM FTO-02 or FTO-04. The reaction mixture was then incubated with neutravidin coated 96-well plates overnight at 4 °C, washed and blocked, incubated with m6A-specific antibody for 1 h at RT, washed and blocked, and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at RT. After extensive washing, the wells were treated with 3,3′,5,5′-tetramethylbenzidine (TMB) for 30 min at RT, and the absorbance was measured at 390 nm. Absorbance was normalized to control wells for each concentration of inhibitor without cofactor 2-oxoglutarate to control for nonspecific antibody binding, and the data were fit to a sigmoidal dose–response curve in GraphPad Prism 6. These assays reported IC50 values consistent with those observed in the Broccoli assays (1.48 ± 0.7 μM FTO-02, 2.79 ± 1.3 μM FTO-04, Figure S25).
All compounds which did not show precipitation were also screened in the same manner against ALKBH5 to determine if there was any specificity toward FTO (Table 1, Table S1). Of the 18 compounds tested, nine displayed poor activity toward ALKBH5 (IC50 ≥ 40 μM), and five of these showed no measurable inhibition at the highest concentration measured (FTO-01, FTO-05, FTO-07, FTO-12, and FTO-18). This selectivity against ALKBH5 is comparable to that observed for MA and FB23-2, which were reported to show little to no inhibition of FTO at 50 μM.38 Importantly, the two most potent inhibitors FTO-02 and FTO-04 (FTO IC50 = 2.2 and 3.4 μM, respectively) both reported significant selectivity over ALKBH5 (ALKBH5 IC50 = 85.5 and 39.4 μM respectively), with FTO-02 showing ∼40× greater potency toward the target FTO. Compounds FTO-05, FTO-06, FTO-12, and FTO-20 showed a preference for FTO over ALKBH5 of 5-fold or higher (Table 1, Figure 2B–D). Four compounds, FTO-08, FTO-10, FTO-11, and FTO-19, were considered equivalent inhibitors toward both demethylases. Interestingly, two compounds, FTO-09 and FTO-13, showed a distinct preference toward ALKBH5 over FTO, where FTO-09 was almost 10 times more potent toward ALKBH5 (IC50 = 5.2 vs >40 μM). Both FTO-09 and FTO-13 feature the 2-aminopyrimidine ring previously identified as a poor inhibitor of FTO. In general, three of the five compounds which reported IC50’s against ALKBH5 below 40 μM contained the 2-aminopyrimidine ring, suggesting this scaffold preferentially inhibits ALKBH5 over FTO.
Of the six selective inhibitors shown in Table 1, five are predicted to form hydrophobic contacts with residues of the nucleotide recognition lid, specifically residues Val83, Ile85, Leu90, Thr92, Pro93, and Val94. While it has been suggested that the selective inhibition of MA against FTO over ALKBH2, -3, and -5 can be attributed to contacts with this loop, it is unclear if these contacts also control the selectivity of FTO-02, -04, -05, -06, -12, and -20 without crystal structures. As ALKBH2, -3, and -5 do not contain this loop, it is likely that inhibitors selective against ALKBH5 will also be selective against ALKBH2 and -3. However, as the fluorescent inhibition assay is not amenable to the DNA demethylating enzymes ALKBH2 and -3, off-target inhibition of these enzymes cannot be ruled out.
The mechanism of inhibition was established for the two most potent and highly selective inhibitors, FTO-02 and FTO-04, using steady-state inhibition kinetics. The reaction velocity was determined for FTO in the presence of 0, 0.5, 1, 10, and 40 μM of inhibitor with a range of 10 substrate concentrations between 0 and 10 μM. A plot of the reaction velocity versus substrate concentration shows that vmax is reached when substrate concentrations exceed 5 μM, for all concentrations of FTO-02 and FTO-04 (Figure S26A,B). The double-reciprocal plots show that all concentrations of FTO-02 and FTO-04 converge on a common y-intercept, indicating vmax is independent of the concentration of either inhibitor, supporting a competitive mechanism of inhibition (Figure 2E,F). This mechanism is consistent with the initial in silico modeling targeted toward the MA binding site and the competitive mechanism previously reported for MA.46
FTO-04 Impairs Self-Renewal in GSC-Derived Neurospheres
Recent studies have indicated that the m6A methylation machinery mediates tumorigenesis and self-renewal in glioblastoma stem cells. Depletion of m6A methylation promotes tumor growth both in vitro and in vivo, while knockdown of the demethylase ALKBH5 was found to impede tumorigenesis and prolong life span in GSC-derived tumor bearing mice.33 Additionally, the small molecule FTO inhibitor meclofenamic acid was observed to prolong lifespan in intracranial GSC xenograft mice.35 However, other reports suggest that depletion of m6A methylation can impair tumor growth and sensitize GSC neurospheres to γ-irradiation and prolong the lifespan in tumor-bearing mice.45 While the role of m6A methylation in glioblastoma is still emerging, these data suggest that targeting the m6A methylation machinery to alter m6A levels could prove a promising strategy for treating glioblastoma.
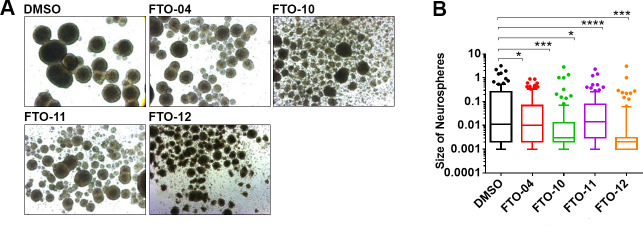
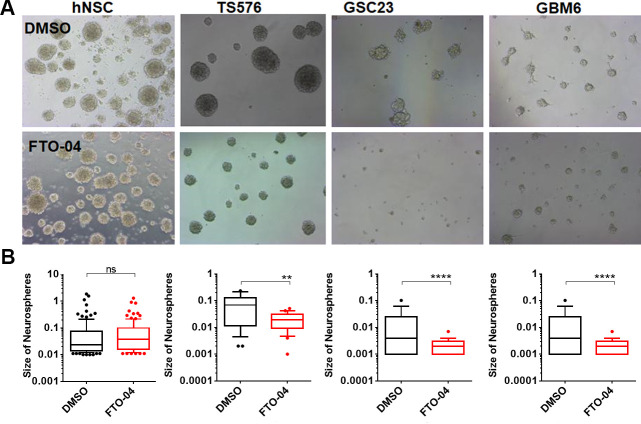
To understand the effects of our FTO inhibitors on the self-renewal properties of GSCs, neurospheres cultured from the patient-derived GSC line TS576 were treated with 30 μM of FTO-04, FTO-10, FTO-11, or FTO-12 (Figure 3A,B; cell line gifted from the Furnari lab).51,52 The GSCs were cultured in sphere-forming assays for 24 h, then treated with either inhibitors or DMSO control for 2 days. The size of the neurospheres was calculated using ImageJ. The neurospheres model was chosen over traditional monolayer cell screening assays as it is known to better replicate the tumor microenvironment.53−57 As dysregulation of m6A methylation processes has been associated with hypoxia, the neurospheres model was considered a more favorable model system.31,32,58 Changes in neurosphere size after treatment with FTO-04 was also compared to lentiviral knockdown of FTO as a positive control (Figure S27). Knockdown of FTO was found to significantly reduce the size of neurospheres relative to shControl, and the reduction in neurosphere size was proportional to the knockdown efficiency (Figure S27B,C). As observed in Figure 3A,B, all four inhibitors showed a significant reduction in size of the neurospheres compared to vehicle control. Furthermore, FTO-04 was also shown to significantly decrease the size of neurospheres cultured from patient-derived TS576, GSC-23, and GBM-6 GSC lines at 20 μM (Figure 4A,B; cell lines gifted from the Furnari lab).51,52 The assay was repeated for neurospheres derived from healthy human neural stem cells (hNSCs), which showed no alteration in neurosphere size after treatment with 20 μM FTO-04, indicating that inhibition of self-renewal is specific to the GSC lines at this dose (Figure 4A,B). Collectively, these data indicate that FTO-04 can significantly impair the self-renewal properties of GSCs to prevent neurospheres formation without significantly impairing the growth of hNSC neurospheres.
Figure 3.
FTO inhibitors impair the self-renewal of GSC neurospheres. (A) Bright field images of neurospheres after 2 days treatment with 30 μM FTO inhibitors in TS576 glioblastoma cells. (B) Size of neurospheres as quantified by ImageJ. Box and whisker plots show 10–90th percentile. N > 50 neurospheres per group. **p < 0.01, ****p < 0.0001, by Student’s t test.
Figure 4.
FTO-04 inhibits GSC neurospheres formation in multiple patient-derived stem cell lines without impairing hNSC neurosphere growth. (A) Bright field images of neurospheres after 2 days of treatment with the FTO-04 inhibitor (20 μM) to normal human neural stem cells (hNSC) and glioblastoma cell lines (TS576, GBM-GSC-23 and GBM-6). (B) Size of neurospheres as quantified by ImageJ. Box and whisker plots show 10–90th percentile. N > 50 neurospheres per group. **p < 0.01, ****p < 0.0001, by Student’s t test.
Next, we sought to determine if FTO-04 was able to alter m6A levels in purified mRNA from GSCs by m6A dot blot assay. TS576 cells were treated with shControl or shFTO to establish the relative change in m6A mRNA levels due to FTO knockdown. As observed in Figure S28A, m6A levels remain high under shFTO treatment relative to shControl. TS576 cells were also treated with either DMSO or FTO-04 (Figure S28B). As observed with FTO knockdown, m6A levels are increased in cells treated with FTO-04 relative to DMSO control. These results indicate that FTO-04 reduces the neurosphere size of GSCs by altering m6A mRNA levels consistent with the inhibition of FTO. However, it is important to note that this assay does not distinguish between m6A and m6Am transcripts; it is possible that the increase in m6A mRNA levels is due at least in part to alterations of m6Am transcripts.
FTO-04 Enhances m6A and m6Am Levels in GSCs
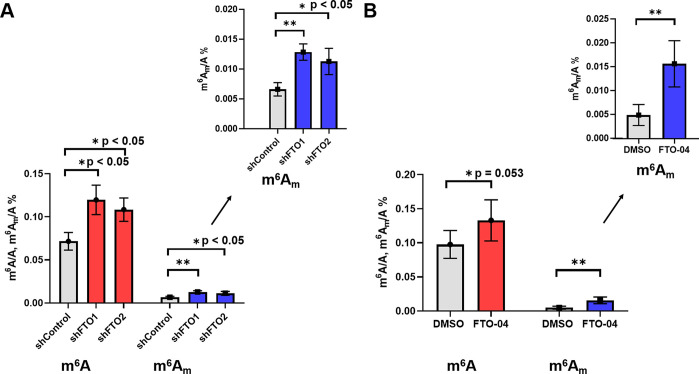
To determine the effects of FTO inhibition on mRNA modification in GSCs, we quantified m6A and m6Am levels after the FTO silencing and FTO-04 treatment. To establish the effects of FTO knockdown on methylated nucleosides, GSCs were treated with either shControl, shFTO1, or shFTO2. The polyadenylated capped RNAs were decapped and digested to single nucleosides, then high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS/MS) was used to quantify the levels of m6A and m6Am as described previously.59−61 As observed in Figure 5A, the levels of m6Am after treatment with shControl are approximately 1/10–1/15 that of m6A, consistent with previous reports in untreated HeLa, HEK293T, and 3T3-L1 cell lines.16,18 FTO knockdown with shFTO1 or shFTO2 resulted in significant increases in levels of both m6A and m6Am. Both shFTO1 and shFTO2 showed an approximately 1.5–2-fold increase in m6A and m6Am levels (Figure 5 A). These results indicate that FTO knockdown is able to alter the levels of both m6A and m6Am in glioblastoma stem cells.
Figure 5.
FTO-04 increases m6A and m6Am levels in GSCs consistent with FTO knockdown. (A) FTO knockdown increases both m6A and m6Am levels relative to shControl. (B) Treatment of GSCs with FTO-04 increases both m6A and m6Am levels. *p < 0.05 unless otherwise noted, **p < 0.01, by Student’s t test.
Next, to determine if FTO-04 was able to impede adenosine demethylation in RNA transcripts consistent with FTO knockdown results, we quantified the m6A and m6Am levels in mRNA samples isolated from GSCs treated with either FTO-04 or DMSO control. Polyadenylated decapped RNAs were again isolated, digested, and quantified by HPLC-MS/MS/MS. As shown in Figure 5B, the m6Am levels in cells treated with the DMSO control remain ∼1/10–1/15 the levels observed for m6A. Treatment with FTO-04 was found to increase the levels of both m6A and m6Am modifications, with m6Am modifications showing the largest fold-change relative to DMSO control (∼3.2× increase, Figure 5B). Treatment with FTO-04 increased m6A levels by ∼1.4×, similar to the increase in m6A observed after FTO knockdown (∼1.5×). However, the increase in m6Am levels was larger after FTO-04 treatment (∼3.2×) than treatment with either shFTO1 (∼2×) or shFTO2 (∼1.7×). These results indicate that FTO-04 is able to alter the levels of both m6A and m6Am in a manner consistent with FTO knockdown, suggesting that FTO is a cellular target of FTO-04.
Conclusions
As interest in characterizing the role of m6A modification in tumor progression and proliferation gains momentum, it will be critical to identify small molecule inhibitors which can be used as high quality chemical probes both in vitro and in vivo. To that end, it is necessary to identify chemical scaffolds which are not only potent and selective inhibitors but also that have physicochemical properties that are favorable for future in vivo proof of concept models and potential pharmacokinetic development. Collectively, this work represents an important step forward by combining structure-based drug design and a high throughput in vitro inhibition assay system to identify a new chemical class of FTO inhibitors with tightly defined physicochemical properties. Many of these compounds were found to inhibit FTO selectively over ALKBH5 with micromolar potency and the most potent and selective inhibitors FTO-02 and FTO-04 were found to inhibit FTO through a competitive mechanism, consistent with the initial in silico screening at the MA-binding site. Importantly, FTO-04 was found to inhibit neurosphere formation in cultures derived from multiple GSC lines without significantly altering hNSC neurosphere formation. A comparison of m6A mRNA levels in GSCs after FTO knockdown or treatment with FTO-04 indicate that FTO-04 increases m6A mRNA levels in a manner consistent with FTO inhibition. Quantification of m6A and m6Am nucleosides in GSCs indicated that FTO knockdown is able to significantly increase both m6A and m6Am levels. Furthermore, treatment of GSCs with FTO-04 resulted in increases of both m6A and m6Am levels consistent with FTO knockdown, suggesting that FTO is a cellular target of FTO-04. These data indicate that targeting the m6A methylation machinery, and the demethylase FTO specifically, could prove an effective mechanism for treating glioblastoma and identify FTO-04 as a new lead for therapeutic development.
Materials and Methods
Detailed procedures for in silico screening and docking of the FTO inhibitors can be found in the SI. Protocols for protein expression and purification, in vitro inhibition assays, and steady-state enzyme kinetics can be found in the SI. Detailed synthetic procedures are presented in the SI. All cell culture procedures can also be found in the SI.
Acknowledgments
We thank F. Furnari for the GBM cell lines and S. Jaffrey for providing ALKBH5 and FTO plasmids for protein expression and purification. The authors thank D. Siegel and B. Duggan of UCSD Skaggs School of Pharmacy for assistance with synthetic chemistry and NMR spectroscopy. We also thank members of the Rana lab for helpful discussions and advice. This work was supported in part by grants from the National Institutes of Health (R35 ES031707 to Y.W. and CA177322, DA049524, and DA046171 to T.M.R) and T32 training grants through the National Cancer Institute and the National Institute of Environmental Health Sciences of the National Institutes of Health (Award Numbers T32 CA121938 to S.H. and T32 ES018827 to G.M.G.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.0c00841.
Detailed experimental procedures for in silico modeling, protein expression and purification, in vitro inhibition assays, steady-state enzyme kinetics, synthesis, and cell culture, Figures S1–S22, and Tables S1 and S2 (PDF)
Author Contributions
S.H. designed and performed the experiments, analyzed the data, and wrote the manuscript. S.K.T. performed experiments, analyzed the data, and prepared figures. G.M.G. performed LC-MS/MS/MS experiments. Y.G. analyzed the data. T.M.R. conceived and planned the project and participated in experimental design, data analysis, data interpretation, and manuscript writing.
Author Contributions
∥ These authors contributed equally to this work
The authors declare the following competing financial interest(s): T.M.R. is a founder of ViRx Pharmaceuticals and has an equity interest in the company. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies.
Supplementary Material
References
- Meyer K. D.; Saletore Y.; Zumbo P.; Elemento O.; Mason C. E.; Jaffrey S. R. (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S.; Moshitch-Moshkovitz S.; Dominissini D.; Mansour A. A.; Kol N.; Salmon-Divon M.; Hershkovitz V.; Peer E.; Mor N.; Manor Y. S.; Ben-Haim M. S.; Eyal E.; Yunger S.; Pinto Y.; Jaitin D. A.; Viukov S.; Rais Y.; Krupalnik V.; Chomsky E.; Zerbib M.; Maza I.; Rechavi Y.; Massarwa R.; Hanna S.; Amit I.; Levanon E. Y.; Amariglio N.; Stern-Ginossar N.; Novershtern N.; Rechavi G.; Hanna J. H. (2015) Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006. 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Dominissini D.; Moshitch-Moshkovitz S.; Schwartz S.; Salmon-Divon M.; Ungar L.; Osenberg S.; Cesarkas K.; Jacob-Hirsch J.; Amariglio N.; Kupiec M.; Sorek R.; Rechavi G. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Liu J.; Yue Y.; Han D.; Wang X.; Fu Y.; Zhang L.; Jia G.; Yu M.; Lu Z.; Deng X.; Dai Q.; Chen W.; He C. (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Feng J.; Xue Y.; Guan Z.; Zhang D.; Liu Z.; Gong Z.; Wang Q.; Huang J.; Tang C.; Zou T.; Yin P. (2016) Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- Meyer K. D.; Jaffrey S. R. (2017) Rethinking m(6)A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H.; Wei J.; He C. (2019) Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 74, 640–650. 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z.; Niu T.; Chang J.; Lei X.; Zhao M.; Wang Q.; Cheng W.; Wang J.; Feng Y.; Chai J. (2010) Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 464, 1205–1209. 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- Jia G.; Fu Y.; Zhao X.; Dai Q.; Zheng G.; Yang Y.; Yi C.; Lindahl T.; Pan T.; Yang Y. G.; He C. (2011) N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A.; Bencokova Z.; Poole R.; Loenarz C.; Adam J.; O’Flaherty L.; Schodel J.; Mole D.; Giaslakiotis K.; Schofield C. J.; Hammond E. M.; Ratcliffe P. J.; Pollard P. J. (2011) Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1alpha (HIF-1alpha). PLoS One 6, e16210 10.1371/journal.pone.0016210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.; Dahl J. A.; Niu Y.; Fedorcsak P.; Huang C. M.; Li C. J.; Vagbo C. B.; Shi Y.; Wang W. L.; Song S. H.; Lu Z.; Bosmans R. P.; Dai Q.; Hao Y. J.; Yang X.; Zhao W. M.; Tong W. M.; Wang X. J.; Bogdan F.; Furu K.; Fu Y.; Jia G.; Zhao X.; Liu J.; Krokan H. E.; Klungland A.; Yang Y. G.; He C. (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aik W.; Scotti J. S.; Choi H.; Gong L.; Demetriades M.; Schofield C. J.; McDonough M. A. (2014) Structure of human RNA N(6)-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 42, 4741–4754. 10.1093/nar/gku085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.; Liu K.; Tempel W.; Demetriades M.; Aik W.; Schofield C. J.; Min J. (2014) Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J. Biol. Chem. 289, 17299–17311. 10.1074/jbc.M114.550350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S.; Toh J. D.; Wong K. H.; Gao Y. G.; Hong W.; Woon E. C. (2016) N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci. Rep. 6, 25677. 10.1038/srep25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J.; Luo X.; Blanjoie A.; Jiao X.; Grozhik A. V.; Patil D. P.; Linder B.; Pickering B. F.; Vasseur J. J.; Chen Q.; Gross S. S.; Elemento O.; Debart F.; Kiledjian M.; Jaffrey S. R. (2017) Reversible methylation of m(6)Am in the 5′ cap controls mRNA stability. Nature 541, 371–375. 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wei L. H.; Wang Y.; Xiao Y.; Liu J.; Zhang W.; Yan N.; Amu G.; Tang X.; Zhang L.; Jia G. (2019) Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. U. S. A. 116, 2919–2924. 10.1073/pnas.1820574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J.; Jaffrey S. R. (2018) FTO, m(6) Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 592, 2012–2022. 10.1002/1873-3468.13092. [DOI] [PubMed] [Google Scholar]
- Wei J.; Liu F.; Lu Z.; Fei Q.; Ai Y.; He P. C.; Shi H.; Cui X.; Su R.; Klungland A.; Jia G.; Chen J.; He C. (2018) Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 71, 973–985. 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J.; Sindelar M.; Despic V.; Guez T.; Hawley B. R.; Vasseur J. J.; Rentmeister A.; Gross S. S.; Pellizzoni L.; Debart F.; Goodarzi H.; Jaffrey S. R. (2019) FTO controls reversible m(6)Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 15, 340–347. 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh C. W. Q.; Goh Y. T.; Goh W. S. S. (2019) Atlas of quantitative single-base-resolution N(6)-methyl-adenine methylomes. Nat. Commun. 10, 5636. 10.1038/s41467-019-13561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.; Wang X.; Liu K.; Roundtree I. A.; Tempel W.; Li Y.; Lu Z.; He C.; Min J. (2014) Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929. 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- Zhu T.; Roundtree I. A.; Wang P.; Wang X.; Wang L.; Sun C.; Tian Y.; Li J.; He C.; Xu Y. (2014) Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 24, 1493–1496. 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Lu Z.; Gomez A.; Hon G. C.; Yue Y.; Han D.; Fu Y.; Parisien M.; Dai Q.; Jia G.; Ren B.; Pan T.; He C. (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Zhao B. S.; Roundtree I. A.; Lu Z.; Han D.; Ma H.; Weng X.; Chen K.; Shi H.; He C. (2015) N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Zhao D.; Wu J.; Shi Y. (2014) Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 24, 1490–1492. 10.1038/cr.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S.; Tong L. (2014) Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U. S. A. 111, 13834–13839. 10.1073/pnas.1412742111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theler D.; Dominguez C.; Blatter M.; Boudet J.; Allain F. H. (2014) Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 42, 13911–13919. 10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D. P.; Pickering B. F.; Jaffrey S. R. (2018) Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 28, 113–127. 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey S. R.; Kharas M. G. (2017) Emerging links between m6A and misregulated mRNA methylation in cancer. Genome Med. 9, 2. 10.1186/s13073-016-0395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriack-Sjodin P. A.; Ribich S.; Copeland R. A. (2018) RNA-modifying proteins as anticancer drug targets. Nat. Rev. Drug Discovery 17, 435–453. 10.1038/nrd.2018.71. [DOI] [PubMed] [Google Scholar]
- Panneerdoss S.; Eedunuri V. K.; Yadav P.; Timilsina S.; Rajamanickam S.; Viswanadhapalli S.; Abdelfattah N.; Onyeagucha B. C.; Cui X.; Lai Z.; Mohammad T. A.; Gupta Y. K.; Huang T. H.; Huang Y.; Chen Y.; Rao M. K. (2018) Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 4, eaar8263 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Zhi W. I.; Lu H.; Samanta D.; Chen I.; Gabrielson E.; Semenza G. L. (2016) Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 7, 64527–64542. 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Zhao B. S.; Zhou A.; Lin K.; Zheng S.; Lu Z.; Chen Y.; Sulman E. P.; Xie K.; Bogler O.; Majumder S.; He C.; Huang S. (2017) m(6)A Demethylase ALKBH5Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31, 591–606. 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I.; Tzelepis K.; Pandolfini L.; Shi J.; Millan-Zambrano G.; Robson S. C.; Aspris D.; Migliori V.; Bannister A. J.; Han N.; De Braekeleer E.; Ponstingl H.; Hendrick A.; Vakoc C. R.; Vassiliou G. S.; Kouzarides T. (2017) Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131. 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q.; Shi H.; Ye P.; Li L.; Qu Q.; Sun G.; Sun G.; Lu Z.; Huang Y.; Yang C. G.; Riggs A. D.; He C.; Shi Y. (2017) m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 18, 2622–2634. 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R.; Dong L.; Li C.; Nachtergaele S.; Wunderlich M.; Qing Y.; Deng X.; Wang Y.; Weng X.; Hu C.; Yu M.; Skibbe J.; Dai Q.; Zou D.; Wu T.; Yu K.; Weng H.; Huang H.; Ferchen K.; Qin X.; Zhang B.; Qi J.; Sasaki A. T.; Plas D. R.; Bradner J. E.; Wei M.; Marcucci G.; Jiang X.; Mulloy J. C.; Jin J.; He C.; Chen J. (2018) R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 172, 90–105. 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu L. P.; Pickering B. F.; Cheng Y.; Zaccara S.; Nguyen D.; Minuesa G.; Chou T.; Chow A.; Saletore Y.; MacKay M.; Schulman J.; Famulare C.; Patel M.; Klimek V. M.; Garrett-Bakelman F. E.; Melnick A.; Carroll M.; Mason C. E.; Jaffrey S. R.; Kharas M. G. (2017) The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376. 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Su R.; Sheng Y.; Dong L.; Dong Z.; Xu H.; Ni T.; Zhang Z. S.; Zhang T.; Li C.; Han L.; Zhu Z.; Lian F.; Wei J.; Deng Q.; Wang Y.; Wunderlich M.; Gao Z.; Pan G.; Zhong D.; Zhou H.; Zhang N.; Gan J.; Jiang H.; Mulloy J. C.; Qian Z.; Chen J.; Yang C. G. (2019) Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 35, 677–691. 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Du B. (2019) Novel positioning from obesity to cancer: FTO, an m(6)A RNA demethylase, regulates tumour progression. J. Cancer Res. Clin. Oncol. 145, 19–29. 10.1007/s00432-018-2796-0. [DOI] [PubMed] [Google Scholar]
- Li Z.; Weng H.; Su R.; Weng X.; Zuo Z.; Li C.; Huang H.; Nachtergaele S.; Dong L.; Hu C.; Qin X.; Tang L.; Wang Y.; Hong G. M.; Huang H.; Wang X.; Chen P.; Gurbuxani S.; Arnovitz S.; Li Y.; Li S.; Strong J.; Neilly M. B.; Larson R. A.; Jiang X.; Zhang P.; Jin J.; He C.; Chen J. (2017) FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.; Liu J.; Chen C.; Dong L.; Liu Y.; Chang R.; Huang X.; Liu Y.; Wang J.; Dougherty U.; Bissonnette M. B.; Shen B.; Weichselbaum R. R.; Xu M. M.; He C. (2019) Anti-tumour immunity controlled through mRNA m 6 A methylation and YTHDF1 in dendritic cells. Nature 566, 270. 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N.; Kang Y.; Wang L.; Huff S.; Tang R.; Hui H.; Agrawal K.; Gonzalez G. M.; Wang Y.; Patel S. P.; Rana T. M. (2020) ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. U. S. A. 117, 20159–20170. 10.1073/pnas.1918986117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Hui H.; Agrawal K.; Kang Y.; Li N.; Tang R.; Yuan J.; Rana T. M. (2020) m6A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. e104514. 10.15252/embj.2020104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Wei J.; Cui Y.-H.; Park G.; Shah P.; Deng Y.; Aplin A. E.; Lu Z.; Hwang S.; He C.; He Y.-Y. (2019) m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10, 2782. 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan A.; Patil V.; Arora A.; Hegde A. S.; Arivazhagan A.; Santosh V.; Somasundaram K. (2018) Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533. 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Yan J.; Li Q.; Li J.; Gong S.; Zhou H.; Gan J.; Jiang H.; Jia G. F.; Luo C.; Yang C. G. (2015) Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43, 373–384. 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Svensen N.; Jaffrey S. R. (2016) Fluorescent RNA Aptamers as a Tool to Study RNA-Modifying Enzymes. Cell Chem. Biol. 23, 415–425. 10.1016/j.chembiol.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inda M. M.; Bonavia R.; Mukasa A.; Narita Y.; Sah D. W.; Vandenberg S.; Brennan C.; Johns T. G.; Bachoo R.; Hadwiger P.; Tan P.; Depinho R. A.; Cavenee W.; Furnari F. (2010) Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 24, 1731–1745. 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez J. A.; Ma J.; D’Antonio M.; Boyer A.; Camargo M. F.; Zanca C.; Kelly S.; Khodadadi-Jamayran A.; Jameson N. M.; Andersen M.; Miletic H.; Saberi S.; Frazer K. A.; Cavenee W. K.; Furnari F. B. (2017) PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nat. Commun. 8, 15223. 10.1038/ncomms15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L.; Huang Z.; Zhou W.; Wu Q.; Donnola S.; Liu J. K.; Fang X.; Sloan A. E.; Mao Y.; Lathia J. D.; Min W.; McLendon R. E.; Rich J. N.; Bao S. (2013) Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell 153, 139–152. 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D.; Hu Z.; Lu L.; Lu H.; Xu X. (2017) Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncol. Lett. 14, 6999–7010. 10.3892/ol.2017.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirkse A.; Golebiewska A.; Buder T.; Nazarov P. V.; Muller A.; Poovathingal S.; Brons N. H. C.; Leite S.; Sauvageot N.; Sarkisjan D.; Seyfrid M.; Fritah S.; Stieber D.; Michelucci A.; Hertel F.; Herold-Mende C.; Azuaje F.; Skupin A.; Bjerkvig R.; Deutsch A.; Voss-Bohme A.; Niclou S. P. (2019) Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 10, 1787. 10.1038/s41467-019-09853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro T.; Ohata H.; Sato A.; Yamawaki K.; Enomoto T.; Okamoto K. (2017) Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications. Cancer Sci. 108, 283–289. 10.1111/cas.13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell N.; Larion M.; Giles A. J.; Seldomridge A. N.; Sizdahkhani S.; Gilbert M. R.; Park D. M. (2017) Hypoxia in the glioblastoma microenvironment: shaping the phenotype of cancer stem-like cells. Neuro Oncol 19, 887–896. 10.1093/neuonc/now258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Samanta D.; Lu H.; Bullen J. W.; Zhang H.; Chen I.; He X.; Semenza G. L. (2016) Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A. 113, E2047 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G.; Rana T. M. (2019) Profiling of N(6)-Methyladenosine in Zika Virus RNA and Host Cellular mRNA. Methods Mol. Biol. 1870, 209–218. 10.1007/978-1-4939-8808-2_16. [DOI] [PubMed] [Google Scholar]
- Lichinchi G.; Zhao B. S.; Wu Y.; Lu Z.; Qin Y.; He C.; Rana T. M. (2016) Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe 20, 666–673. 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G.; Gao S.; Saletore Y.; Gonzalez G. M.; Bansal V.; Wang Y.; Mason C. E.; Rana T. M. (2016) Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol 1, 16011. 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.