Abstract
Ovarian cancer (OC) is a type of cancer with high prevalence and shocking mortality in women around the world. Radioresistance is a major reason for OC relapse. Mounting studies have shown the significant function of dysregulated microRNAs (miRNAs) in cancer progression and the cellular response to irradiation. The present study inquired about the function and mechanism of microRNA (miR)-4478 in regulating radiosensitivity of OC cells. Results showed that miR-4478 was downregulated in OC, and a low miR-4478 level indicated a disappointing prognosis for OC patients. Besides, in OC cells exposed to irradiation, the expression of miR-4478 decreased over time. Functionally, the upregulation of miR-4478 retarded OC cell proliferation and sensitized OC cells to irradiation. Mechanistically, miR-4478 targeted and inhibited fused in sarcoma (Fus). Additionally, Fus was upregulated in OC and its expression further elevated in OC cells under irradiation. Furthermore, miR-4478 targeted Fus to inhibit autophagy, therefore sensitizing OC cells to irradiation. Collectively, our study uncovered miR-4478 as a novel radiosensitizer by targeting Fus in OC cells, which may shed a new light on developing targets for treating patients with OC, particularly those with radioresistance.
Keywords: autophagy, Fus, miR-4478, ovarian cancer, radiosensitivity
Graphical Abstract

Corresponding authors and colleagues discovered that microRNA-4478 sensitizes ovarian cancer cells to irradiation through targeting fused in sarcoma, suggesting a promising target for treating OC patients, especially those with radioresistance.
Introduction
Ovarian cancer (OC), as the fourth most common malignancy in females, is a major contributor to gynecological cancer-related deaths.1,2 The common therapeutic methods for OC include surgical dissection, chemotherapy, and radiotherapy.3 Although progress has been achieved in the treatment of primary OC, most OC patients at advanced stages turn out to suffer from recurrence.4, 5, 6, 7 The development of resistance is a common limit for the treatment efficacy of radiotherapy.8 In spite of accumulating knowledge in this field during the past decades, the mechanism behind radioresistance in OC remains poorly understood. Therefore, the investigation of the molecular mechanism underlying radioresistance of OC can be beneficial for the identification of a new, potential radiosensitizer for OC patients.
MicroRNAs (miRNAs) are recognized as a cluster of endogenous short noncoding RNAs. Functionally, miRNAs are able to directly bind to the 3′ untranslated region (3′ UTR) of target messenger RNAs (mRNAs) to induce translation suppression or mRNA degradation.9, 10, 11 Over the decades, miRNAs have been reported to regulate biological processes in multiple cancers by numerous studies and have also been currently identified to affect the response of cancer cells to irradiation.12, 13, 14, 15, 16 Interestingly, several studies have illustrated that enhanced cell autophagy might contribute to radioresistance in cancers, including OC.17, 18, 19 Also, miRNAs have been documented to sensitize cancer cells to irradiation through inhibiting autophagy.18,20 Previously, microRNA (miR-4478) has been identified as a tumor suppressor in colorectal cancer.21,22 However, its role in OC is still unknown, and whether it modulates autophagy and radioresistance in OC also needs to be revealed.
Fused in sarcoma (Fus) is a gene belonging to the Ten-eleven translocation (TET) protein family, which encodes an RNA binding protein.23,24 Fus has been reported to be involved in the progression of multiple cancers.25, 26, 27, 28 Additionally, the suppression of Fus by miRNAs in cancers has been revealed by a number of studies.27,28 Intriguingly, it has been discovered that Fus can promote DNA damage repair and that the mutation of Fus leads to higher radiosensitivity in mice.29,30 Moreover, researches demonstrated that stress granules containing Fus could be colocalized with autophagosomes and that mutation in Fus led to impaired autophagy compared with cells expressing wild-type Fus.31, 32, 33 All of these findings prompted us to wonder whether Fus could regulate autophagy and therefore control radiosensitivity in cancer cells like OC cells.
The present study explores the role and downstream mechanism of miR-4478 in regulating the radioresistance of OC cells.
Results
Downregulation of miR-4478 was correlated with poor prognosis and radioresistance in OC patients
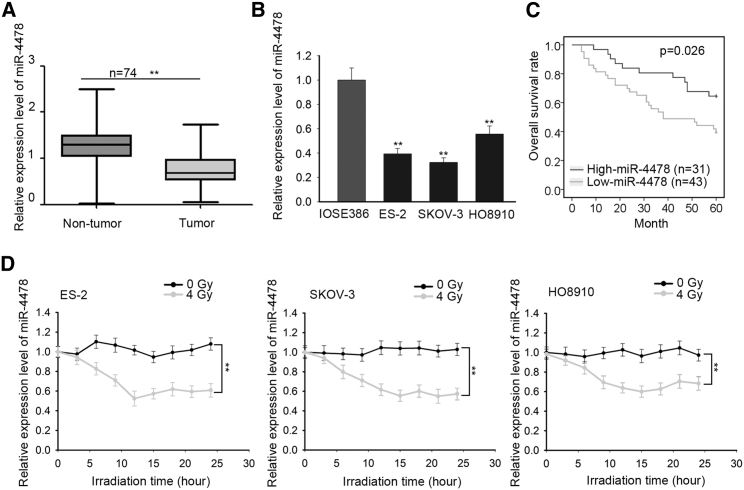
First, to explore the involvement of miR-4478 in OC, we examined the expression of miR-4478 in OC tissues and cell lines. Results of quantitative real-time reverse-transcriptase polymerase chain reaction RT-PCR) displayed that miR-4478 presented a significant low level in OC samples (n = 74) and cell lines (IOSE80, SKOV-3, and HO8910) compared with corresponding noncancerous tissues (n = 74) and a normal ovarian epithelial cell line (IOSE386) (Figures 1A and 1B). Additionally, OC samples were separated into 2 groups by the mean level of miR-4478 (high miR-4478: n = 31; low miR-4478: n = 43). Then, we evaluated the association of miR-4478 expression with clinical features of OC patients. It was uncovered that miR-4478 expression was closely related to tumor size (p = 0.018) and International Federation of Gynecology and Obstetrics (FIGO) stage (p = 0.019) (Table 1). Further, Kaplan-Meier analysis revealed that miR-4478 downregulation was closely associated with poor overall survival of OC patients (p = 0.026) (Figure 1C). Also, multivariate analysis showed that the expression of miR-4478 (p = 0.02) was an independent prognostic factor for OC patients (Table 2). For OC patients, resistance to radiotherapy is correlated with poor prognosis.3, 4, 5, 6, 7 Therefore, we investigated the association between miR-4478 level and radioresistance in OC cells. We treated three OC cell lines with 0, 2, 4, 6, or 8 Gy of irradiation. The results of the Cell Counting Kit 8 (CCK-8) assay showed that irradiation resulted in a significant decrease in cell viability (Figure S1A). Based on above data, we then used the dose of 4 Gy for subsequent assays. Three OC cell lines were treated with 4 Gy radiation, and the expression of miR-4478 was measured every 5 h after treatment. Results manifested that miR-4478 was downregulated in three OC cell lines after radiation (Figure 1D). These results suggested that miR-4478 downregulation was correlated with poor prognosis and radioresistance in OC patients.
Figure 1.
Downregulation of miR-4478 was correlated with poor prognosis and radioresistance in OC patients
(A and B) Quantitative real-time RT-PCR examined the expression level of miR-4478 in OC tissues and cell lines. (C) Kaplan-Meier analysis and log-rank test evaluated the association of miR-4478 expression with overall survival (OS) in OC patients. (D) Quantitative real-time RT-PCR examined the variation of miR-4478 level with 0 or 4 Gy radiation treatments in three OC cell lines. Data presented as mean ± SD (namely error bar). All assays were performed in triplicate. ∗∗p < 0.01.
Table 1.
Correlation between miR-4478 expression and clinical characteristics (n = 74)
| Variable | miR-4478 expression |
p value | |
|---|---|---|---|
| Low | High | ||
| Age | |||
| <60 | 12 | 11 | 0.612 |
| ≥60 | 31 | 20 | |
| Histological subtype | |||
| Serous | 19 | 15 | 0.815 |
| Others | 24 | 16 | |
| Tumor size | |||
| <3 cm | 15 | 20 | 0.018a |
| ≥3 cm | 28 | 11 | |
| FIGO stage | |||
| I−II | 13 | 18 | 0.019a |
| III~IV | 30 | 13 | |
| Histological grade | |||
| G1−G2 | 26 | 20 | 0.810 |
| G3 | 17 | 11 | |
| Lymph node metastasis | |||
| Absent | 17 | 14 | 0.642 |
| Present | 26 | 17 | |
Low/high by sample mean. Pearson χ2 test.
p < 0.05 was viewed as statistically significant.
Table 2.
Multivariate analysis of prognostic parameters of OC patients by Cox regression analysis
| Variable | Category | p value |
|---|---|---|
| Age | ||
| <60 | 0.509 | |
| ≥60 | ||
| Histological subtype | ||
| serous | 0.016a | |
| others | ||
| Tumor size | ||
| <3 cm | 0.321 | |
| ≥3 cm | ||
| FIGO stage | ||
| I−II | 0.026a | |
| III−IV | ||
| Histological grade | ||
| G1−G2 | 0.974 | |
| G3 | ||
| Lymph node metastasis | ||
| absent | 0.118 | |
| present | ||
| miR-4478 level | ||
| low | 0.02a | |
| high | ||
Proportional hazards method analysis showed a positive and independent prognostic importance of miR-4478 expression (p = 0.02).
p < 0.05 was considered statistically significant.
miR-4478 overexpression sensitized OC cells to irradiation
To examine the effect of miR-4478 on radioresistance in OC, we overexpressed it in IOSE80 and SKOV-3 cells (Figure 2A), which presented the lowest miR-4478 level in previous detection. In the meantime, we also silenced miR-4478 expression in HO8190 cells (Figure S1B), which presented the highest miR-4478 level among three OC cell lines. Subsequently, through the CCK-8 assay, we found that cell proliferation was attenuated by the ectopic expression of miR-4478 and that miR-4478 overexpression further enhanced the prohibitive effect of irradiation on proliferation of OC cells (Figure 2B). In HO8190 cells, inhibition of miR-4478 facilitated cell proliferation under normal conditions and also recovered the decreased cell proliferation caused by 4 Gy irradiation (Figure S1C). Later, with accumulated treatment of radiation, miR-4478 overexpression induced a sharp decrease in the survival fraction (SF) of OC cells (Figure 2C), whereas miR-4478 inhibition retarded the decrease in the OC cell SF owing to irradiation (Figure S1D). These data suggested that miR-4478 reduced the resistance of OC cells to irradiation. To further investigate the effect of miR-4478 on radioresistance, western blotting analysis was carried out for detecting the level of poly-(ADP-ribose) polymerase (PARP), a protein contributing to DNA damage repair. Results illustrated that under 4 Gy radiation, cells transfected with a miR-4478 mimic exhibited a lower PARP level than the control group (Figure 2D), whereas cells with a miR-4478 inhibitor exhibited opposite results (Figure S1E). The results indicated that miR-4478 suppressed DNA damage repair in response to irradiation, leading to enhanced radiosensitivity of OC cells. All data above supported that miR-4478 sensitized OC cells to irradiation.
Figure 2.
miR-4478 overexpression enhanced the radiosensitivity in OC cells
(A) Quantitative real-time RT-PCR analyzed the overexpression efficiency of miR-4478. (B) CCK-8 assay tested the proliferation of NC mimic- or miR-4478 mimic-transfected OC cells with or without irradiation. (C) The survival fraction curves of NC mimic- or miR-4478 mimic-transfected OC cells under irradiation were determined by colony-formation assay. (D) Western blot analyzed the protein level of PARP in irradiated OC cells transfected with NC mimic or miR-4478 mimic. Data presented as mean ± SD (error bar). All assays were performed in triplicate. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
miR-4478 targeted and negatively regulated Fus
It has been reported that miRNAs regulate gene expression by directly binding to the 3′ UTR of mRNAs in numerous cancers.9, 10, 11 Hence, we hypothesized that miR-4478 might exert its function in OC through this way. We searched for the potential target genes of miR-4478 by using the TargetScan tool (http://www.targetscan.org/vert_72/). Fus was then identified as one of the targets for miR-4478. Fus has been reported to be targeted by miRNAs in multiple cancers and intriguingly, was identified to promote DNA damage repair.27, 28, 29, 30 Thus, we selected Fus for further investigation. The putative binding sequences between miR-4478 and Fus were shown in Figure 3A. Luciferase reporter assay showed that miR-4478 upregulation reduced the luciferase activity of wild type (WT)-Fus rather than mutant (mut)-Fus (Figure 3B). RNA pulldown assay manifested that Fus mRNA could be pulled down by a bio-WT-miR-4478 probe rather than a bio-mut-miR-4478 or bio-negative control (NC) probe and that knockdown of miR-4478 led to a decrease in the enrichment of Fus mRNA in the bio-WT-miR-4478 group (Figure 3C). Moreover, quantitative real-time RT-PCR and western blotting analyses presented that miR-4478 overexpression resulted in the downregulation of Fus at mRNA and protein levels, whereas miR-4478 inhibition exerted opposite effects (Figure 3D). Additionally, quantitative real-time RT-PCR results showed that Fus was significantly upregulated in OC tissues and cell lines (Figure 3E). Kaplan-Meier analysis revealed that a high level of Fus indicated unsatisfactory overall survival of OC patients (p = 0.019) (Figure 3F). Moreover, Fus was upregulated after a 4-Gy dose of radiation in all the three OC cell lines (Figure 3G), indicating its correlation with radioresistance in OC cells. The negative correlation between Fus and miR-4478 expressions in OC tissues was validated by Spearman’s correlation analysis (Figure 3H). Together, these results elucidated that Fus was positively associated with poor prognosis and radioresistance and negatively regulated by miR-4478 in OC.
Figure 3.
miR-4478 negatively regulated Fus by targeting it
(A) The binding sites between miR-4478 and Fus were identified by searching TargetScan. The sequences of WT-Fus and mut-Fus with WT or mut miR-4478 binding sites were designed for luciferase reporter assay. (B) Luciferase reporter assay detected the impact of miR-4478 overexpression on the luciferase activity of WT-Fus and mut-Fus reporters. (C) RNA pulldown assay tested the enrichment of Fus by indicated probes in OC cells with or without miR-4478 inhibition. (D) Quantitative real-time RT-PCR and western blot analyses detected the effect of miR-4478 on Fus mRNA and protein levels. (E) Quantitative real-time RT-PCR examined the expression level of Fus in OC tissues and cell lines. (F) Kaplan-Meier analysis and log-rank test evaluated the association between Fus expression with OS in OC patients. (G) Quantitative real-time RT-PCR examined the level variation of Fus in three OC cell lines with 0 or 4 Gy radiation treatment. (H) Spearman’s correlation curve revealed the correlation between miR-4478 and Fus expression in OC samples. Data presented as mean ± SD (error bar). All assays were performed in triplicate. ∗p < 0.05, ∗∗p < 0.01.
Cell autophagy was induced by irradiation and contributed to radioresistance in OC cells
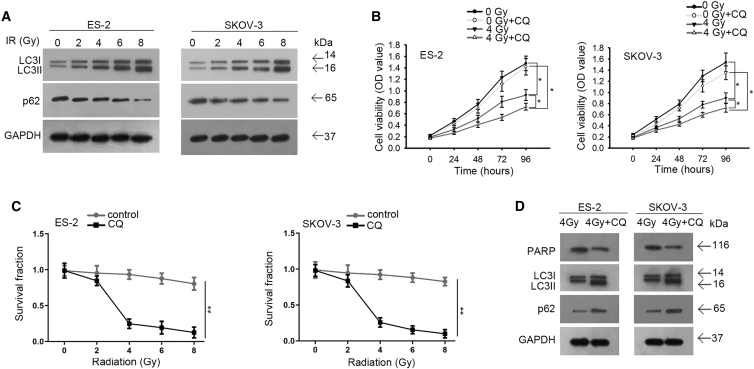
The facilitating effect of autophagy on radioresistance of cancer cells has been uncovered by previous studies, including in OC cells,18,19,34 and miRNAs have been proven to regulate radiosensitivity through modulating autophagy.18,20 Based on our findings that miR-4478 could prohibit the radioresistance of OC cells and target Fus, we interrogated whether miR-4478/Fus could regulate radioresistance through modulating autophagy. Before that, we tested the influence of irradiation on OC cell autophagy. Autophagy markers LC3II and LC3I, which are the isoforms of LC3,35 and the autophagy substrate p62 were examined in OC cells exposed to 0, 2, 4, 6, or 8 Gy of irradiation. We observed from western blot results that LC3II/LC3I was increased, whereas p62 was decreased with the increasing irradiation doses (Figure 4A), indicating enhanced autophagy in OC cells responding to irradiation. To examine whether targeting autophagy could affect radioresistance in OC cells, we treated OC cells with chloroquine (CQ) to inhibit autophagosome generation. It turned out that CQ treatment augmented the inhibitive effect of 4 Gy irradiation on cell proliferation (Figure 4B). Furthermore, the inhibition of autophagy by CQ could sharpen the decrease in the OC cell SF under accumulated irradiation (Figure 4C). Western blot analysis confirmed that CQ treatment decreased the levels of PARP and LC3II/I, whereas increased p62 expression in irradiated OC cells (Figure 4D). These results suggested that cell autophagy was induced by irradiation and contributed to the radioresistance of OC cells.
Figure 4.
Cell autophagy was induced by irradiation and contributed to the radioresistance of OC cells
(A) Western blot detected the levels of LC3II/LC3I and p62 in OC cells under above treatments. (B) CCK-8 assay examined the proliferation of OC cells from indicated groups. (C) Colony-formation assay determined the survival fraction curve of indicated OC cells. (D) Western blot tested the levels of PARP, LC3II/LC3I, and p62 in indicated OC cells. Data presented as mean ± SD (error bar). All assays were performed in triplicate. ∗p < 0.05, ∗∗p < 0.01.
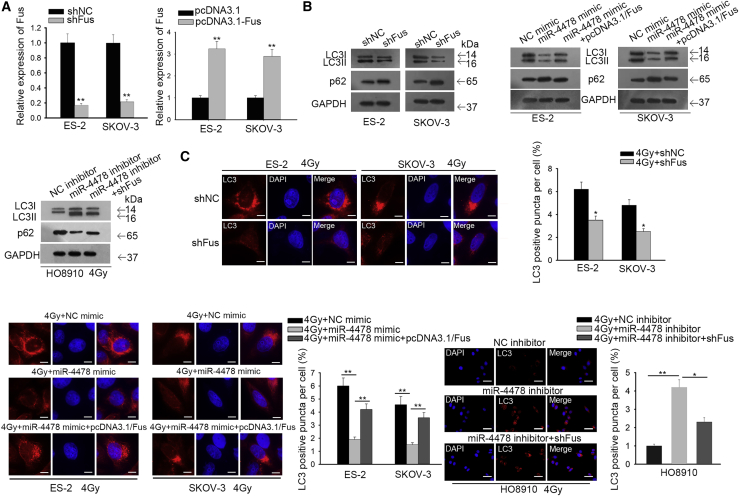
Silencing Fus abrogated OC cell autophagy, and miR-4478 inhibited autophagy through targeting Fus
After confirming that increased autophagy contributed to radioresistance in OC cells, we continued to investigate whether miR-4478 could inhibit autophagy through targeting Fus. Therefore, we silenced Fus to test its function in cell autophagy and overexpressed Fus to examine whether miR-4478 attenuated cell autophagy through Fus (Figure 5A). Western blot results showed that under irradiation, silencing Fus or overexpressing miR-4478 could decrease the level of LC3II/LC3I and increase the level of p62 (Figure 5B). Additionally, overexpression of Fus reversed the effect of miR-4478 overexpression on decreasing LC3II/LC3I and increasing p62, and silencing of Fus reversed the effect of miR-4478 inhibition on increasing LC3II/LC3I and decreasing p62 (Figure 5B). Also, the immunofluorescence assay helped us to observe that silencing Fus reduced the ratio of LC3 dots (Figure 5C). Overexpression of Fus counteracted the repressive effect of miR-4478 overexpression on LC3 positivity in OC cells, and silencing of Fus abrogated the strengthening effect of the miR-4478 inhibitor on LC3 positivity in OC cells (Figure 5C). Together, these results indicated that silencing Fus abrogated OC cell autophagy, and miR-4478 inhibited autophagy through targeting Fus.
Figure 5.
Silencing Fus abrogated OC cell autophagy, and miR-4478 inhibited cell autophagy through targeting Fus
(A) Quantitative real-time RT-PCR verified the knockdown and overexpression efficiency of Fus. (B) Western blot detected the levels of LC3II/LC3I and p62 in OC cells under different transfections. (C) Immunofluorescence assay examined the LC3 dots in OC cells under 4 Gy irradiation and diverse transfections (scale bars, 10 μm or 50 μm). Data presented as mean ± SD (error bar). All assays were performed in triplicate. ∗p < 0.05, ∗∗p < 0.01.
miR-4478 contributed to cell radiosensitivity by targeting Fus in OC
Finally, rescue assays were designed and conducted to validate the effect of the miR-4478/Fus axis on the radioresistance of OC cells. The CCK-8 assay demonstrated that under 4 Gy radiation, the hampered cell proliferation caused by miR-4478 overexpression was partially recovered by the upregulation of Fus (Figure 6A). Colony-formation assay depicted that cotransfection of pcDNA3.1-Fus reversed the suppression of miR-4478 mimic on the SF of OC cells under increased radiation (Figure 6B). Furthermore, western blotting illustrated that ectopic expression of Fus rescued the reduction in PARP level due to miR-4478 upregulation in OC cells under the treatment of 4 Gy radiation (Figure 6C). To conclude, these results indicated that miR-4478 contributed to the radiosensitivity of OC cells via targeting Fus.
Figure 6.
miR-4478 contributed to radiosensitivity by targeting Fus in OC
(A) CCK-8 assay evaluated the proliferation of OC cells with 4 Gy irradiation and different transfections. (B) Colony-formation assay analyzed the survival fraction of indicated OC cells under increasing doses of irradiation. (C) Western blotting analyzed changes in PARP protein level in OC cells transfected with different plasmids under 4 Gy radiation. Data presented as mean ± SD (error bar). All assays were performed in triplicate. ∗p < 0.05.
Discussion
As one of the most common gynecological malignant tumors, OC is life threatening to women all over the world.1,2 Although radiotherapy is one of the most frequent therapeutic methods for OC,3 resistance to radiotherapy leads to the limited treatment effectiveness, thereby inducing the relapse of OC.4, 5, 6, 7, 8 Despite mounting achievements in improving treatment, there is still an urgent need for further exploration of the underlying mechanism underlying OC cell radioresistance.
Currently, miRNAs have been discovered to regulate the effectiveness of radiotherapy in cancers.12, 13, 14, 15, 16 Downregulation of miR-4478 has been identified as a biological marker in colorectal cancer.21,22 However, miR-4478 has neither been explored in OC nor been associated with radioresistance. The present study first identified the low expression level of miR-4478 in OC tissues and cell lines. Additionally, we identified the positive correlation of miR-4478 expression with the prognosis of OC patients. Then, we confirmed that downregulation of miR-4478 was related to the radioresistance of OC cells. Further, the results of gain-of-function assays suggested that miR-4478 had suppressive effect on the radioresistance of OC cells.
Mechanistically, miRNAs have been reported to regulate cancer development via binding to the 3′ UTR of target genes to silence these genes at a post-transcriptional level.9, 10, 11 In this study, Fus was recognized as the target of miR-4478 in OC cells. Previously, Fus has been reported to be an oncogene in human cancers, and it also has been verified to regulate DNA damage repair.27, 28, 29, 30 However, the impact of Fus on cellular resistance to irradiation has not been explored in OC. In the current study, we first confirmed that miR-4478 targeted Fus to inhibit its expression. Furthermore, Fus was found to be upregulated in OC tissues and cell lines. Upregulation of Fus predicted unfavorable prognosis for patients with OC. Importantly, our study first revealed the association between the expression of Fus and the radioresistance of OC cells.
Moreover, former studies have found that increased autophagy could contribute to radioresistance in cancer cells,17,18 including in OC cells.19 In concordance, we confirmed that autophagy was facilitated in OC cells in response to the irradiation and that inhibiting autophagy by CQ sensitized OC cells to irradiation, suggesting that irradiation-elevated autophagy aggravated radioresistance in OC. Previous research has demonstrated that miRNAs could attenuate cancer cell radioresistance through inhibiting autophagy.18,20 Based on these findings, we tried to figure out whether miR-4478/Fus could regulate autophagy to affect radiosensitivity in OC cells. Expectedly, we first found that miR-4478 could inhibit cell autophagy in OC and that overexpressing Fus reversed such effects. These findings suggested that miR-4478 inhibited autophagy through targeting Fus. Meanwhile, since previous studies have shown that mutation of Fus can impair cell autophagy in contrast to WT Fus,31, 32, 33 we sought to probe whether WT Fus could exhibit a promoting effect on autophagy in OC cells. As a result, our study first showed that silencing Fus could retard autophagy in OC cells, suggesting that Fus might contribute to autophagy in OC cells. Further, we demonstrated via rescue assays that upregulation of Fus counteracted the inhibitive effect of miR-4478 overexpression on OC cell radioresistance, indicating that miR-4478 targeted Fus to control the radioresistance of OC cells.
Taken together, these results proved that miR-4478 contributed to cell radiosensitivity by targeting Fus to inhibit cell autophagy in OC. The findings indicated that miR-4478 might be as a novel prognostic marker and a potential radiosensitizer for OC patients.
Materials and methods
Tissue samples
OC tissues and the paired adjacent normal tissues were collected from 74 patients with OC at Harbin Medical University Cancer Hospital. These patients were assessed according to FIGO staging. The clinical characteristics of these patients were summarized in Table 1. None of the patients received radiotherapy or chemotherapy prior to surgical resection. After resection, tissues were immediately stored in liquid nitrogen and then maintained at −80°C until use. The present study was approved by the Ethics Committee of Harbin Medical University Cancer Hospital. Written, informed consents were obtained from all patients.
Cell lines and cell culture
Human OC cell lines (ES-2, SKOV3, and HO8910) and the normal human ovarian epithelial cell line (IOSE386) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) at 37°C in a humidified atmosphere containing 5% CO2.
Quantitative real-time RT-PCR
Total RNA was extracted from OC tissues and cell lines using TRIzol reagent (Thermo Fisher Scientific) under the manufacturer’s protocol. Quantitative real-time RT-PCR was performed to examine the expression levels of miR-4478 and Fus using the Applied Biosystems PowerUp SYBR Green Master Mix kit (Thermo Fisher Scientific) by an ABI 7500 fluorescence qRT-PCR machine (Applied Biosystems; Thermo Fisher Scientific), according to the manufacturer’s protocol. U6 was used as the internal reference for miR-4478, whereas glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference for Fus. The relative expression levels were analyzed using the 2−ΔΔCt method. Primer sequences were designed according to the sequences of miR-4478 and Fus obtained from NCBI (https://www.ncbi.nlm.nih.gov/gene/). The accession numbers of GenBank were the following: NCBI: NC_000009.12 for miR-4478 and NC_000016.10 for Fus. We used the “primer-blast” tool in NCBI to test the specificity of primers. Primers with a melting temperature (Tm) difference less than 2 and the single-strand PCR products were identified to be qualified. The primer sequences used were as follows: 5′-CAGGGCTGCAGGGGATG-3′ (forward) and 5′-CTGAGGAGCCTCCAAACCTG-3′ (reverse) for miR-4478; 5′-AGGGCTAGGTGGAAAGACCT-3′ (forward) and 5′-CCTTCCTGATCGGGACATCG-3′ (reverse) for Fus; 5′-CTCCGGGAAACTGTGGCGTG-3′ (forward) and 5′-ACAAAGTGGTCGTTGAGGGCA-3′ (reserve) for GAPDH; 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reserve) for U6.
Cell transfection
miR-4478-mimic was used to overexpress miR-4478, and pcDNA3.1 vector subcloned with Fus cDNA sequence was used to overexpress Fus. NC-mimic and pcDNA3.1 vector were used as corresponding NCs. The miR-4478 inhibitor was used to inhibit miR-4478, with NC inhibitor as the NC. Specific short hairpin (sh)RNAs targeting Fus (shFus) were used to silence Fus expression, with scramble shRNA as the NC (shNC). All vectors were obtained from Shanghai Integrated Biotech Solutions (Shanghai, China). Based on the manufacturer’s instruction, Lipofectamine 2000 (Invitrogen) was used for cell transfection.
X-ray irradiation exposure
Plastic flasks with OC cells underwent the exposure to the X-ray irradiation with the utilization of a linear accelerator (Primus, Siemens, Germany) at the dose of 4 Gy. The X-ray irradiation was implemented when the concentration of OC cells had reached 80%. Thereafter, OC cells were collected at indicated time points subsequent to irradiation for later experiments.
CCK-8
ES-2 and HO8910 cells were seeded in 96-well plates overnight at concentrations of 2 × 103 cells/well. Then, CCK-8 solution (Dojindo) was added into cells after 0, 24, 48, 72, and 96 h of incubation. After 2 h of incubation at 37°C under humidified air, the absorbance was determined at 450 nm using a SpectraMax M5 microplate reader (Molecular Devices, USA).
Colony-formation assay
After transfection, cells were plated into 6-well plates and treated with irradiation at the dose of 0, 2, 4, 6, or 8 Gy. After 2 weeks of incubation under 37°C, cells were fixed with the use of 95% methanol and then stained by 0.1% crystal violet for 15 min. Observable colonies with above 50 cells were counted manually. SF was evaluated as plating efficiency ratio for nonirradiated and irradiated cells. The definition of plating efficiency was as follow: colony number divided by seeded cell number × 100%. Data were expressed with y = 1 − (1 − e−D/D0)N for the presentation of survival curves. Radiation-associated parameters, including Dq (quasi-threshold dose), D0 (mean lethal dose), and N (extrapolation number), were evaluated in accordance with the curve. The sensitivity enhancement ratio (SER) was expressed as the ratio of the D0 value of the control group to that of treatment group.
Luciferase reporter assay
With bioinformatics predictions, the binding sequences for miR-4478 in the 3′ UTR region of Fus were predicted. Then, the full-length Fus 3′ UTR containing WT or mut of miR-4478 binding sequences was loaded into pmirGLO luciferase reporter vectors (Promega, Madison WI, USA), and the recombinant plasmid was, respectively, termed as WT-Fus or mut-Fus. To study the interaction between Fus and miR-4478, WT-Fus or mut-Fus luciferase reporter plasmid was cotransfected into ES-2 and SKOV-3 cells with miR-4478-mimic (or NC-mimic). Following transfection for 48 h, the luciferase activity was determined using the Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA). The firefly luciferase activities were normalized to Renilla luciferase activity.
RNA pulldown assay
Biotin-labeled WT-miR-4478 or mut-miR-4478 probe (Beijing Dingguo Changsheng Biotech, China) was acquired, with NC-probe as the NC. Cells were collected and lysed using the lysis buffer. Streptavidin magnetic beads were used to capture the biotin-labeled probes following the manufacturer’s protocol. Then the biotinylated compounds were incubated with the extract of cells in a 42°C water bath for 40 min. After elution of the magnetic beads, the levels of Fus mRNA were detected by quantitative real-time RT-PCR analysis.
Autophagy detection by GFP-LC3 fluorescence microscopy
The transfected OC cells were fixed and underwent 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO, USA) nuclei staining. The autophagy marker LC3 was stained utilizing the GFP-LC3 (ab48394; Abcam). The green dots in OC cells were observed and calculated by the utilization of an Olympus BX53 fluorescence microscope (Olympus, Tokyo, Japan). Autophagy positivity of OC cells was defined by the green highlights ≥5. Four horizons were randomly picked in each well for observation, and at least 3 wells were calculated for each group.
Western blotting
OC cells were, respectively, lysed in cold radioimmunoprecipitation assay buffer (Thermo Fisher Scientific). Then, the protein concentration was determined using the Bicinchoninic Acid (BCA) Protein Assay Kit (Thermo Fisher Scientific), according to the manufacturer’s protocol. Proteins were separated using 12% sodium dodecyl sulfate polyacrylamide-gel electrophoresis (SDS-PAGE) and then shifted to a polyvinylidene fluoride (PVDF) membrane (Thermo Fisher Scientific). The PVDF membrane was sealed with 5% nonfat dried milk in phosphate-buffered saline (PBS; Thermo Fisher Scientific) for 2 h under room temperature. Subsequently, the membrane was incubated with primary antibodies for 12 h, followed by the incubation with horseradish peroxidase-conjugated secondary antibodies (Abcam, Cambridge, UK) for 1 h. The protein band was revealed using the Enhanced Chemiluminescence Western Blotting Kit (Thermo Fisher Scientific). Protein expression was determined using Image-Pro Plus software 6.0 (Media Cybernetics, Rockville, MD, USA), and GAPDH was used as the internal reference. The primary antibodies were the following: anti-Fus (ab70381), anti-p62 (ab91526), anti-cleaved PARP (ab32561), and anti-LC3B (ab48394) from Abcam (Cambridge, UK) and anti-LC3II (cat. no. 4108) and anti-GAPDH (cat. no. 5174) from Cell Signaling Technology (Danvers, MA, USA).
Statistical analysis
Data were presented as the mean ± standard deviation (SD). SPSS 19.0 software (IBM, Armonk, NY, USA) was used for statistical analysis. Student’s t test was used for the comparisons between two groups, whereas one-way ANOVA with Turkey’s post hoc test was used for the comparisons among multiple groups. A chi-square test was used to evaluate the association between miR-4478 expression and clinicopathological characteristics of OC patients. The association of miR-4478 or Fus expression with the overall survival of OC patients was analyzed by Kaplan-Meier analysis plus log-rank test. p <0.05 was considered to indicate a statistically significant difference. All assays were triplicated.
Acknowledgments
We are very grateful to all individuals and groups involved in this study. This study was supported by Postdoctoral Foundation of Hei Long Jiang Province of China (no. LRB14-173).
Author contributions
L.W. and Y.L. designed the study. H.L., C.Z., and H.W. conducted the experiments. S.D. and W.C. wrote the paper. Y.S. and X.Z. analyzed the data. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.11.024.
Contributor Information
Ying Liu, Email: liutan6922992@163.com.
Yan Sun, Email: sy_493yansun00@163.com.
Xiulan Zheng, Email: xiu2lan505zheng_99@163.com.
Supplemental information
References
- 1.Cannistra S.A. Cancer of the ovary. N. Engl. J. Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Rai B., Bansal A., Patel F.D., Sharma S.C. Radiotherapy for ovarian cancers - redefining the role. Asian Pac. J. Cancer Prev. 2014;15:4759–4763. doi: 10.7314/apjcp.2014.15.12.4759. [DOI] [PubMed] [Google Scholar]
- 4.Hussein M.A. Nontraditional cytotoxic therapies for relapsed/refractory multiple myeloma. Oncologist. 2002;7(Suppl 1):20–29. doi: 10.1634/theoncologist.7-suppl_1-20. [DOI] [PubMed] [Google Scholar]
- 5.Odunsi K., Jungbluth A.A., Stockert E., Qian F., Gnjatic S., Tammela J., Intengan M., Beck A., Keitz B., Santiago D. NY-ESO-1 and LAGE-1 cancer-testis antigens are potential targets for immunotherapy in epithelial ovarian cancer. Cancer Res. 2003;63:6076–6083. [PubMed] [Google Scholar]
- 6.Gagnon A., Kim J.-H., Schorge J.O., Ye B., Liu B., Hasselblatt K., Welch W.R., Bandera C.A., Mok S.C. Use of a combination of approaches to identify and validate relevant tumor-associated antigens and their corresponding autoantibodies in ovarian cancer patients. Clin. Cancer Res. 2008;14:764–771. doi: 10.1158/1078-0432.CCR-07-0856. [DOI] [PubMed] [Google Scholar]
- 7.Yahara K., Ohguri T., Imada H., Yamaguchi S., Kawagoe T., Matsuura Y., Hachisuga T., Korogi Y. Epithelial ovarian cancer: definitive radiotherapy for limited recurrence after complete remission had been achieved with aggressive front-line therapy. J. Radiat. Res. (Tokyo) 2013;54:322–329. doi: 10.1093/jrr/rrs108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Concin N., Zeillinger C., Stimpfel M., Schiebel I., Tong D., Wolff U., Reiner A., Leodolter S., Zeillinger R. p53-dependent radioresistance in ovarian carcinoma cell lines. Cancer Lett. 2000;150:191–199. doi: 10.1016/s0304-3835(99)00393-6. [DOI] [PubMed] [Google Scholar]
- 9.White N.M.A., Khella H.W.Z., Grigull J., Adzovic S., Youssef Y.M., Honey R.J., Stewart R., Pace K.T., Bjarnason G.A., Jewett M.A. miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br. J. Cancer. 2011;105:1741–1749. doi: 10.1038/bjc.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beresneva E.V., Rykov S.V., Hodyrev D.S., Pronina I.V., Ermilova V.D., Kazubskaia T.P., Braga E.A., Loginov V.I. [Methylation profile of group of miRNA genes in clear cell renal cell carcinoma; involvement in cancer progression] Genetika. 2013;49:366–375. doi: 10.7868/s001667581303003x. [DOI] [PubMed] [Google Scholar]
- 11.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 12.Schickel R., Boyerinas B., Park S.M., Peter M.E. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 13.Li B.-L., Lu W., Qu J.-J., Ye L., Du G.-Q., Wan X.-P. Loss of exosomal miR-148b from cancer-associated fibroblasts promotes endometrial cancer cell invasion and cancer metastasis. J. Cell. Physiol. 2019;234:2943–2953. doi: 10.1002/jcp.27111. [DOI] [PubMed] [Google Scholar]
- 14.Weidhaas J.B., Babar I., Nallur S.M., Trang P., Roush S., Boehm M., Gillespie E., Slack F.J. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemoeller O.M., Niyazi M., Corradini S., Zehentmayr F., Li M., Lauber K., Belka C. MicroRNA expression profiles in human cancer cells after ionizing radiation. Radiat. Oncol. 2011;6:29. doi: 10.1186/1748-717X-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q., Zhang S. miR-214 promotes radioresistance in human ovarian cancer cells by targeting PETN. Biosci. Rep. 2017;37 doi: 10.1042/BSR20170327. BSR20170327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He W.S., Dai X.F., Jin M., Liu C.W., Rent J.H. Hypoxia-induced autophagy confers resistance of breast cancer cells to ionizing radiation. Oncol. Res. 2012;20:251–258. doi: 10.3727/096504013x13589503483012. [DOI] [PubMed] [Google Scholar]
- 18.Hu J.L., He G.Y., Lan X.L., Zeng Z.C., Guan J., Ding Y., Qian X.L., Liao W.T., Ding Y.Q., Liang L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7:16. doi: 10.1038/s41389-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peracchio C., Alabiso O., Valente G., Isidoro C. Involvement of autophagy in ovarian cancer: a working hypothesis. J. Ovarian Res. 2012;5:22. doi: 10.1186/1757-2215-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q., Liu T., Yuan Y., Guo Z., Xie G., Du S., Lin X., Xu Z., Liu M., Wang W. MiR-200c inhibits autophagy and enhances radiosensitivity in breast cancer cells by targeting UBQLN1. Int. J. Cancer. 2015;136:1003–1012. doi: 10.1002/ijc.29065. [DOI] [PubMed] [Google Scholar]
- 21.Ghanbari R., Mosakhani N., Asadi J., Nouraee N., Mowla S.J., Poustchi H., Malekzadeh R., Knuutila S. Decreased expression of fecal miR-4478 and miR-1295b-3p in early-stage colorectal cancer. Cancer Biomark. 2015;15:189–195. doi: 10.3233/CBM-140453. [DOI] [PubMed] [Google Scholar]
- 22.Ghanbari R., Rezasoltani S., Hashemi J., Mohamadkhani A., Tahmasebifar A., Arefian E., Mobarra N., Asadi J., Nazemalhosseini Mojarad E., Yazdani Y. Expression Analysis of Previously Verified Fecal and Plasma Dow-regulated MicroRNAs (miR-4478, 1295-3p, 142-3p and 26a-5p), in FFPE Tissue Samples of CRC Patients. Arch. Iran Med. 2017;20:92–95. [PubMed] [Google Scholar]
- 23.Morohoshi F., Ootsuka Y., Arai K., Ichikawa H., Mitani S., Munakata N., Ohki M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene. 1998;221:191–198. doi: 10.1016/s0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- 24.Bertolotti A., Bell B., Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- 25.Rabbitts T.H., Forster A., Larson R., Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 26.Crozat A., Åman P., Mandahl N., Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 27.Ke H., Zhao L., Feng X., Xu H., Zou L., Yang Q., Su X., Peng L., Jiao B. NEAT1 is Required for Survival of Breast Cancer Cells Through FUS and miR-548. Gene Regul. Syst. Bio. 2016;10(Suppl 1):11–17. doi: 10.4137/GRSB.S29414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Lei H., Sun Q. MicroRNA-141 and its associated gene FUS modulate proliferation, migration and cisplatin chemosensitivity in neuroblastoma cell lines. Oncol. Rep. 2016;35:2943–2951. doi: 10.3892/or.2016.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroda M., Sok J., Webb L., Baechtold H., Urano F., Yin Y., Chung P., de Rooij D.G., Akhmedov A., Ashley T., Ron D. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratti A., Buratti E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 2016;138(Suppl 1):95–111. doi: 10.1111/jnc.13625. [DOI] [PubMed] [Google Scholar]
- 31.Ramesh N., Pandey U.B. Autophagy Dysregulation in ALS: When Protein Aggregates Get Out of Hand. Front. Mol. Neurosci. 2017;10:263. doi: 10.3389/fnmol.2017.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soo K.Y., Sultana J., King A.E., Atkinson R., Warraich S.T., Sundaramoorthy V., Blair I., Farg M.A., Atkin J.D. ALS-associated mutant FUS inhibits macroautophagy which is restored by overexpression of Rab1. Cell Death Discov. 2015;1:15030. doi: 10.1038/cddiscovery.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryu H.-H., Jun M.-H., Min K.-J., Jang D.-J., Lee Y.-S., Kim H.K., Lee J.A. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol. Aging. 2014;35:2822–2831. doi: 10.1016/j.neurobiolaging.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 34.Ma H., Zheng S., Zhang X., Gong T., Lv X., Fu S., Zhang S., Yin X., Hao J., Shan C., Huang S. High mobility group box 1 promotes radioresistance in esophageal squamous cell carcinoma cell lines by modulating autophagy. Cell Death Dis. 2019;10:136. doi: 10.1038/s41419-019-1355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanida I., Ueno T., Kominami E. LC3 and autophagy. In: Deretic V., editor. Autophagosome and Phagosome. Humana Press; 2008. pp. 77–88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.