How protein endocytic recycling changes in response to environmental cues is poorly understood. Mao et al. demonstrated that multiple stresses inhibited endocytic recycling via MAPK11/14-mediated phosphorylation of SNX27 at Ser51. Phosphorylation of SNX27 decreases its binding to cargo proteins and suppresses protein endocytic recycling.
Abstract
Endocytosed proteins can be delivered to lysosomes for degradation or recycled to either the trans-Golgi network or the plasma membrane. It remains poorly understood how the recycling versus degradation of cargoes is determined. Here, we show that multiple extracellular stimuli, including starvation, LPS, IL-6, and EGF treatment, can strongly inhibit endocytic recycling of multiple cargoes through the activation of MAPK11/14. The stress-induced kinases in turn directly phosphorylate SNX27, a key regulator of endocytic recycling, at serine 51 (Ser51). Phosphorylation of SNX27 at Ser51 alters the conformation of its cargo-binding pocket and decreases the interaction between SNX27 and cargo proteins, thereby inhibiting endocytic recycling. Our study indicates that endocytic recycling is highly dynamic and can crosstalk with cellular stress–signaling pathways. Suppression of endocytic recycling and enhancement of receptor lysosomal degradation serve as new mechanisms for cells to cope with stress and save energy.
Introduction
Integral cell surface proteins, together with associated ligands, proteins, and lipids, are internalized in a clathrin-dependent or independent manner. Endocytosed proteins can be delivered to lysosomes for degradation or the TGN or plasma membrane (PM) for recycling (Burd and Cullen, 2014; Cullen and Steinberg, 2018; Wang et al., 2018). As one of the most fundamental cellular processes, endocytic recycling determines the fate of many receptors, thus fine-tuning a large number of signal pathways. Accordingly, it is also tightly regulated by many intracellular and extracellular events, such as gene expression, protein posttranslational modifications, nutritional availability, and bacterial and viral infection (Cullen and Steinberg, 2018). Genetic variations in endocytic recycling components have been linked to a wide range of human diseases, including Alzheimer’s disease, Parkinson’s disease, cancer, and diabetes (Cullen and Steinberg, 2018; McMillan et al., 2017; Tu et al., 2020).
Since the discovery of retromer, a central regulator of endocytic recycling, over 20 yr ago, we have gained significant insights into the process of endocytic recycling. Several highly conserved proteins or protein complexes, including retromer (VPS35/VPS26/VPS29 in higher eukaryotes; Hierro et al., 2007; Kovtun et al., 2018; Seaman et al., 1998), retriever (McNally et al., 2017; VPS35L/VPS26C/VPS29), TBC1D5 (Jia et al., 2016; Seaman et al., 2009), and multiple members of the sorting nexin (SNX) family (Lucas et al., 2016; Steinberg et al., 2013; Temkin et al., 2011), have been shown to mediate the biogenesis of cargo-enriched transport carriers or promote endocytic recycling via other mechanisms. SNX27 is a key regulator of endocytic recycling and often mediates recycling through cooperation with retromer and SNX-Bin/Amphiphysin/Rvs (BAR) proteins (Steinberg et al., 2013; Temkin et al., 2011). The PDZ domain of SNX27 directly binds to the PDZ-binding motifs (PDZbms) located at the C terminus of many proteins, and interaction with the VPS26 subunit of retromer further increases its binding with PDZbms (Clairfeuille et al., 2016; Gallon et al., 2014). Established cargoes of SNX27 include the glucose transporter GLUT1, SEMA4C, G-protein–coupled receptors such as the β2 adrenergic receptor, and parathyroid hormone receptor (PTHR; Chan et al., 2016; McGarvey et al., 2016; Steinberg et al., 2013; Temkin et al., 2011). While the major protein machineries regulating both the recycling and degradation pathways are known, it remains obscure what determines the recycling versus degradation pathways for cargo proteins.
Cells are subjected to different types of stresses, including (but not limited to) starvation, inflammation, DNA damage, and oxidative stress (Hotamisligil and Davis, 2016). One of the best-described pathways directing cellular responses to stresses is the conserved MAPK pathway. Multiple stimuli activate the MAPK through a three-kinase module, and the activation involves the MAP2K-mediated phosphorylation of MAPKs on a threonine and a tyrosine residue of the activation loop. In mammals, the MAPK pathway is organized into several different signaling modules, including p38, JNK, ERK1/2, and ERK5 (Johnson and Lapadat, 2002). Among them, members of the p38 MAPK family, including MAPK14 (p38α) and MAPK11 (p38β), phosphorylate a wide range of intracellular targets that include membrane transporters, cytoskeletal elements, transcription factors, and other protein kinases (Ono and Han, 2000). As a result, p38 MAPK activation contributes to inflammation, apoptosis, cell differentiation, and cell cycle regulation (Zarubin and Han, 2005). However, it remains unclear whether p38 MAPKs also regulate endocytic recycling.
To explore how cellular stress influences endosomal sorting, we investigated endocytic trafficking of multiple cargoes under various treatments. We found that multiple modes of cellular stress inhibit the endocytic recycling of GLUT1 and other cargoes. The inhibition involves MAPK11/14-mediated phosphorylation of SNX27 at serine 51 (Ser51) within the PDZ domain but is independent of TBC1D5 (Roy et al., 2017). This phosphorylation event inhibits the binding of SNX27 to PDZbm-containing proteins and specifically suppresses endocytic recycling of PDZbm-containing cargoes. Thus, inhibition of endocytic recycling could help cells and organisms adapt to changing environmental conditions.
Results
Starvation inhibits endocytic recycling of multiple cargoes
To investigate endocytic trafficking under starvation, we selected the following transmembrane proteins that are known to recycle between endosomes and the PM: GLUT1, a member of the SLC2 family of transmembrane glucose transporters; SEMA4C, a receptor for PLXNB2 that plays an important role in cell–cell signaling; PTHR, a receptor for parathyroid hormone and parathyroid hormone–related peptide; and TRAILR1, a receptor for TNF-related apoptosis-inducing ligand (TRAIL). All of them, except for TRAILR1, contain a PDZbm and are recycled in a SNX27-dependent manner (Steinberg et al., 2013). Although endocytic recycling of TRAILR1 depends on SNX27, TRAILR1 encompasses a SNX-BARs binding motif, but not a PDZbm (Yong et al., 2020).
After internalization from the cell surface, GLUT1 is recycled from endosomes to the cell surface, a process critical for maintaining glucose homeostasis (Steinberg et al., 2013). To address whether starvation alters GLUT1 recycling, we compared the subcellular localization of GLUT1 (Table S1) in HeLa cells stably expressing GFP-LC3 in normal medium or Earle’s balanced salt solution (EBSS; Table S2). Autophagy was clearly induced, as evidenced by the accumulation of GFP-LC3 puncta, when cells were treated with EBSS alone or together with bafilomycin A1 (BafA1; E+B), which inhibits lysosomal functions. Starvation also increased the distribution of GLUT1 to LAMP2-positive organelles (i.e., late endosomes or lysosomes; Fig. 1 A) and reduced the colocalization of GLUT1 and β-catenin, a PM marker (Fig. S1 A), indicating that starvation may perturb the normal recycling of GLUT1 and lead to an accumulation at late endosomes or lysosomes.
Figure 1.
Starvation inhibits endocytic recycling of multiple transmembrane proteins, and autophagy deficiency abolishes the inhibition. (A) Confocal microscopy imaging for GLUT1 (gray) and LAMP2 (red) immunofluorescence staining in HeLa cells stably expressing GFP-LC3 (green). The cells were starved for 4 h in EBSS in the presence or absence of 100 nM BafA1 (E+B). Representative images of three independent experiments are shown. Scale bars: 10 µm. Quantification of GLUT1 immunofluorescence on LAMP2-positive lysosomes. Results for individual cells are plotted, along with the mean and SD for each group (n = 77 cells for control [Con], n = 74 for EBSS, and n = 54 for E+B); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. (B) Confocal microscopy imaging of internalized CD8A-SEMA4C (green) and phalloidin/EEA1/LAMP1 (red) immunofluorescence staining in HeLa cells. HeLa cells were transfected with a plasmid encoding CD8A-SEMA4C for 24 h, followed by incubation with anti-human CD8A antibody on ice for 30 min. Unbound antibodies were removed, and the internalization of antibody-bound CD8A was performed in complete medium, or EBSS in the presence or absence of 100 nM BafA1 (E+B) for 0, 1, or 3 h. Representative images of two independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficients for CD8A-SEMA4C and phalloidin/EEA1/LAMP1. Results for individual cells are plotted, along with the mean and SD for each group: CD8A-SEMA4C versus phalloidin (n = 43 cells for control, n = 29 for EBSS, and n = 54 for E+B), CD8A-SEMA4C versus EEA1 (n = 35 cells for control, n = 31 for EBSS, and n = 39 for E+B), and CD8A-SEMA4C versus LAMP1 (n = 45 cells for control, 48 for EBSS, 44 for E+B); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001;***, P < 0.001; *, P < 0.05. (C) Confocal microscopy imaging of internalized CD8A-PTHR (green) and phalloidin/EEA1/LAMP1 (red) immunofluorescence staining in HeLa cells. Cells were transfected with a plasmid encoding CD8A-PTHR and treated as in B. Representative images of two independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficients for CD8A-PTHR and phalloidin/EEA1/LAMP1. Results for individual cells are plotted, along with the mean and SD for each group: CD8A-PTHR versus phalloidin (n = 41 cells for control, n = 33 for EBSS, and n = 47 for E+B), CD8A-PTHR versus EEA1 (n = 59 cells for control, n = 86 for EBSS, and n = 89 for E+B), and CD8A-PTHR versus LAMP1 (n = 46 cells for control, n = 43 for EBSS, and n = 60 for E+B); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01. (D) Starvation results in specific loss of GLUT1 and SEMA4C at the cell surface. HeLa cells were starved for 4 h in EBSS in the presence or absence of 100 nM BafA1 (E+B), followed by surface biotinylation. One of three independent experiments is shown. Representative immunoblots show the protein levels in the surface (PM) and total fractions (lysis). (E) Confocal imaging for GLUT1 (green) and LAMP2 (red) immunofluorescence staining of WT and ATG7-KO HEK293T cells. The cells were starved for 4 h in EBSS in the presence or absence of 100 nM BafA1 (E+B). Representative images from three independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficients for GLUT1 and LAMP2. Results for individual cells are plotted, along with the mean and SD for each group (293T-WT control, n = 37 cells; 293T-WT EBSS, n = 72 cells; 293T-WT E+B, n = 71 cells; 293T-ATG7-KO control, n = 63 cells; 293T-ATG7-KO EBSS, n = 59 cells; and 293T-ATG7-KO E+B, n = 45 cells); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001; **, P < 0.01. HSD, honestly significant difference; ns, not significant.
Figure S1.
Starvation inhibits endocytic recycling of multiple transmembrane proteins, and autophagy deficiency abolishes the inhibition. (A) Confocal microscopy imaging for GLUT1 (gray) and β-catenin (red) immunofluorescence staining in HeLa cells stably expressing GFP-LC3 (green). The cells were starved for 4 h in EBSS in the presence or absence of 100 nM BafA1 (E+B). Representative images of three independent experiments are shown. Scale bars: 10 µm. Quantification of GLUT1 immunofluorescence on β-catenin-positive cellular membranes. Results for individual cells are plotted, along with the mean and SD for each group (n = 63 cells for control, n = 65 for EBSS, and n = 85 for E+B); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. (B) Confocal imaging for TRAILR1 (gray) and phalloidin (red) immunofluorescence staining in HeLa cells stably expressing GFP-LC3 (green). The cells were treated as in A. Representative images of three independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficients for TRAILR1 and phalloidin. Results for individual cells are plotted, along with the mean and SD for each group (n = 88 cells for control, n = 85 for EBSS, and n = 108 for E+B); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. (C) Confocal imaging for GLUT1 (green) and phalloidin (red) immunofluorescence staining of WT and ATG7 KO HEK293T cells. The cells were starved for 4 h in EBSS in the presence or absence of 100 nM BafA1 (E+B). Representative images from three independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficients for GLUT1 and phalloidin. Results for individual cells are plotted, along with the mean and SD for each group (293T-WT con, n = 45 cells; 293T-WT E+B, n = 28; 293T-ATG7-KO con, n = 34; 293T-ATG7-KO E+B, n = 36); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. (D) Confocal imaging for GLUT1 (green) and LAMP2 (red) immunofluorescence staining of WT and ATG7 KO MEF cells. The cells were starved for 4 h in EBSS in the presence or absence of 100 nM BafA1 (E+B). Representative images are shown. Scale bars: 10 µm. Quantification of GLUT1 immunofluorescence on LAMP2-positive lysosomes. Results for individual cells are plotted, along with the mean and SD for each group (MEF-WT con, n = 10; MEF-WT E+B, n = 19; MEF-ATG7-KO con, n = 19; MEF-ATG7-KO E+B, n = 19); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. (E) Confocal imaging for Venus-GLUT1 (green) and LAMP2 (red) immunofluorescence staining of WT and ATG7 KO HEK293T cells. The cells were transfected with a plasmid encoding Venus-GLUT, and starved for 4 h in EBSS in the presence or absence of 100 nM BafA1 (E+B). Representative images from two independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficients for Venus-GLUT1 and LAMP2. Results for individual cells are plotted, along with the mean and SD for each group (293T-WT con, n = 35; 293T-WT EBSS, n = 34; 293T-WT E+B, n = 32; 293T-ATG7-KO con, n = 27; 293T-ATG7-KO EBSS, n = 47; 293T-ATG7-KO E+B, n = 40); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. HSD, honestly significant difference; ns, not significant.
To determine whether starvation affects protein endocytosis or recycling, we engineered a CD8A-SEMA4C or CD8A-PTHR chimera (Table S3; Tourian et al., 2004; Beenstock et al., 2014; Stefanoska et al., 2018) and performed uptake assays using an antibody against the extracellular region of CD8A (Seaman, 2004; Yong et al., 2020). Treatment of cells with EBSS alone or together with bafA1 (E+B) did not alter internalization of CD8A-SEMA4C or CD8A-PTHR, as indicated by the colocalization of CD8A and phalloidin (staining for F-actin that marks cell boundaries) 30 min after adsorption (Fig. 1 B, top, and Fig. 1 C, top). When evaluated either by immunofluorescence (Fig. 1, B and C) or surface protein biotinylation (Fig. 1 D), endocytic recycling of CD8A-SEMA4C (Fig. 1 B, middle and bottom) and CD8A-PTHR (Fig. 1 C, middle and bottom) was markedly inhibited by starvation. In contrast to SEMA4C and PTHR, the surface level of TRAILR1 was not altered by starvation (Fig. 1 D and Fig. S1 B). Thus, starvation impairs endocytic recycling of some, but not all, proteins that are recycled between endosomes and cell surface.
Since nutrient starvation induces autophagy, we next tested whether autophagy is required for starvation-mediated inhibition of endocytic recycling. WT HEK293T cells displayed an increased colocalization between GLUT1 and LAMP2 upon starvation; in contrast, deletion of ATG7, an essential regulator of autophagy, abolished the increase in colocalization (Fig. 1 E). Similarly, WT HEK293T cells, but not ATG7 knockout (KO) cells, showed a reduction of colocalization between GLUT1 and phalloidin upon starvation (Fig. S1 C). To rule out a cell-type–specific effect, we performed the same experiment in WT and ATG7-KO mouse embryonic fibroblast (MEF) cells and obtained similar results (Fig. S1 D). Finally, when WT and ATG7-KO HEK293T cells exogenously expressing Venus-GLUT1 were starved, similar results were obtained (Fig. S1 E). Taken together, our data indicate that starvation-induced autophagy may impair endocytic recycling either directly or indirectly by compromising signaling pathways that can regulate endocytic recycling.
TBC1D5 is unlikely to mediate starvation-induced inhibition of endocytic recycling
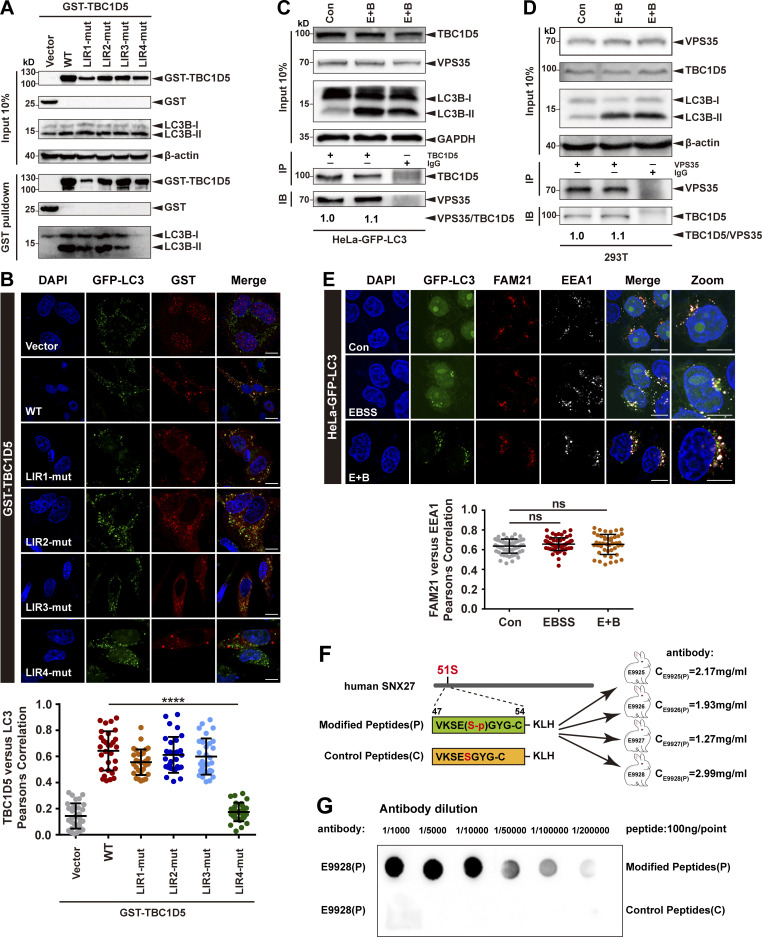
Next, we sought to determine how starvation impairs endocytic recycling of GLUT1. One interesting candidate protein that could mediate the inhibition is TBC1D5, which directly interacts with retromer and functions as a GTP-activating protein for Rab7 (Jia et al., 2016; Jimenez-Orgaz et al., 2018; Seaman et al., 2009; Seaman et al., 2018). TBC1D5 is proposed to shuttle between retromer-positive structures and autophagosomes, depending on cellular metabolic states (Roy et al., 2017). A prerequisite of this model is that TBC1D5 binds to retromer and LC3 competitively (Popovic et al., 2012; Roy et al., 2017). Thus, we next assessed the interactions of TBC1D5 under normal and starvation conditions. Two regions from the TBC domain of TBC1D5 have been shown to be critical for contacting retromer, namely Ins1 (aa 127–153) and Ins2 (aa 263–293; Jia et al., 2016). Using iLIR (http://repeat.biol.ucy.ac.cy/cgi-bin/iLIR/iLIR_cgi; Kalvari et al., 2014), we identified four potential LC3-binding sites in TBC1D5, LIR1 (aa 59–62), LIR2 (aa 566–569), LIR3 (aa 715–718), and LIR4 (aa 787–790; Fig. 2 A). To determine which LIR of TBC1D5 is responsible for contacting LC3, we made a series of plasmids which delete one LIR each time. Similar to TBC1D5 WT, ΔLIR1, ΔLIR2, and ΔLIR3 robustly pulled down endogenous LC3B-II; in contrast, ΔLIR4 almost completely lost the ability to associate with LC3B-II (Fig. 2 B). Next, we determined which LIR of TBC1D5 is required for its colocalization with GFP-LC3 puncta in cells. TBC1D5 WT and variants carrying LIR1, LIR2, or LIR3 deletions colocalized with GFP-LC3 puncta. Quantitative immunofluorescence assays revealed that TBC1D5 WT, ΔLIR1, ΔLIR2, and ΔLIR3, but not ΔLIR4, displayed strong colocalization with GFP-LC3 (Fig. 2 C). These results were further confirmed by experiments using TBC1D5 constructs that carry mutations in each of the LIR motif (Fig. S2, A and B). Thus, the interaction between TBC1D5 and LC3B-II is primarily mediated by the LIR4 motif, which is outside of the TBC domain and distinct from the retromer-binding Ins1 and Ins2 regions. LC3B is unlikely to compete with retromer for the binding to TBC1D5.
Figure 2.
TBC1D5 binds retromer and LC3 independently. (A) Schematic representation of human TBC1D5, with black fonts indicating the predicted LIR motifs and blue fonts indicating the identified Ins motifs. (B) Pull-down assay to assess binding of GST-TBC1D5 and endogenous LC3B-II. HEK293T cells were transfected with GST-tagged TBC1D5-WT, TBC1D5-ΔLIR1, TBC1D5-ΔLIR2, TBC1D5-ΔLIR3, or TBC1D5-ΔLIR4 for 24 h. Cells were starved for 4 h in EBSS in the presence of 100 nM BafA1 (E+B). Control cells (vector) were transfected with empty vector. One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GST resin, followed by immunoblotting using antibodies against GST-TBC1D5 and LC3B. Representative images from one of three independent experiments are shown. (C) Confocal imaging for GST-TBC1D5 (red) and GFP-LC3 (green) immunofluorescence staining in HeLa cells stabling expressing GFP-LC3. TBC1D5 WT, LIR deletions, or empty vector was transiently transfected for 24 h, followed by starvation for 4 h. Representative images from one of two independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficient for TBC1D5 and LC3 colocalization. Results for individual cells are plotted, along with the mean and SD for each group (n = 30); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. (D) Immunoprecipitation of endogenous VPS35 and LC3B from HEK293T cells transfected with HA-YFP tagged TBC1D5. WT, ATG5-KO, or ATG7-KO HEK293T cells were transfected with a HA-YFP-TBC1D5 plasmid, followed by starvation for 4 h (E+B). One tenth of the cell lysate was prepared as input, and the rest was used for immunoprecipitation with anti-HA antibody, followed by immunoblotting with antibodies against VPS35 and LC3B. Representative images from one of three independent experiments are shown. Column chart represents the relative expression of VPS35 to HA-YFP-TBC1D5. Band intensities were quantified with ImageJ and normalized to their respective control group (n = 3). Data are presented as means ± SD, and one-way ANOVA by Tukey’s HSD test or t test was used for data analysis. (E) Immunoprecipitation (IP) of endogenous VPS35 and LC3B from HEK293T cell lysates. WT, ATG5-KO, or ATG7-KO HEK293T cells were starved for 4 h in EBSS in the presence of 100 nM BafA1 (E+B). One tenth of the cell lysate was prepared as input, and the rest was used for immunoprecipitation with anti-TBC1D5 antibodies, followed by immunoblotting (IB) with antibodies against VPS35 and LC3B. Representative images from one of three independent experiments are shown. Column chart represents the relative expression of VPS35 to TBC1D5. Band intensities were quantified with ImageJ and normalized to their respective control group (n = 3). Data are presented as means ± SD, and one-way ANOVA by Tukey’s HSD test or t test was used for data analysis. (F) Model showing the interaction between TBC1D5 and retromer (VPS35/VPS26/VPS29) or LC3-II. HSD, honestly significant difference.
Figure S2.
The interaction between TBC1D5 and retromer is independent of LC3 and is not affected by starvation. (A) Pull-down assay to assess binding of GST-TBC1D5 and endogenous LC3B-II. HEK293T cells were transfected with GST-tagged TBC1D5 WT, LIR mutants, or empty vector (vector) for 24 h. Cells were starved for 4 h in EBSS in the presence of 100 nM BafA1 (E+B). One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with glutathione (GSH) resin, followed by immunoblotting using antibodies against GST-TBC1D5 and LC3B. Representative images from one of three independent experiments are shown. (B) Confocal imaging for GST-TBC1D5 (red) and GFP-LC3 (green) immunofluorescence staining in HeLa cells stabling expressing GFP-LC3. TBC1D5 WT, each LIR mutant, or empty vector was transiently transfected in the cells for 24 h, followed by starvation for 4 h. Representative images from one of two independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficient for TBC1D5 and LC3 colocalization. Results for individual cells are plotted, along with the mean and SD for each group (n = 30); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. (C) Immunoprecipitation of endogenous VPS35 from HeLa cells stably expressing GFP-LC3. The cells were either starved for 4 h (E+B) or not starved. One tenth of the cell lysate was prepared as input, and the rest was used for immunoprecipitation with anti-TBC1D5 antibody, followed by immunoblotting with antibody against VPS35. IgG served as a negative control. Representative images from one of two independent experiments are shown. The numbers under the bands represent the relative expression of VPS35 to TBC1D5. Band intensities were quantified with ImageJ and normalized to the control group. (D) Immunoprecipitation of endogenous TBC1D5 from HeLa cells stably expressing GFP-LC3. The cells were either starved for 4 h (E+B) or not starved. One tenth of the cell lysate was prepared as input, and the rest was used for immunoprecipitation with anti-VPS35 antibody, followed by immunoblotting with antibody against TBC1D5. IgG served as a negative control. Representative images from one of two independent experiments are shown. The numbers under the bands represent the relative expression of TBC1D5 to VPS35. Band intensities were quantified with ImageJ and normalized to control group. (E) Confocal microscopy imaging for FAM21 (red) and EEA1 (gray) immunofluorescence staining in HeLa cells stably expressing GFP-LC3 (green). The cells were starved for 4 h in EBSS in the presence or absence of 100 nM BafA1 (E+B). Representative images of two independent experiments are shown. Scale bars: 10 µm. Quantification of FAM21 immunofluorescence on EEA1-positive early endosomes. Results for individual cells are plotted, along with the mean and SD for each group (n = 59 cells for control, n = 66 for EBSS, and n = 44 for E+B); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. (F) The strategy to raise phospho-SNX27-Ser51-specific antibodies. Sequences of a phosphorylated peptide used for immunization, as well as the control peptide, are shown. (G) The phosphorylated peptide (P) or control nonphosphorylated peptide (C) was spotted on a nitrocellulose membrane, followed by immunoblotting with affinity-purified antibodies at different concentrations. Purified antibody E9928(P) was used in this study. HSD, honestly significant difference; ns, not significant.
It was previously reported that starvation can inhibit the association between TBC1D5 and retromer (Popovic et al., 2012; Roy et al., 2017). To validate this observation, we examined the interaction between TBC1D5 and retromer in autophagy-competent and deficient cells in the presence or absence of starvation (E+B). Immunoprecipitation experiments showed that exogenously expressed HA-YFP-TBC1D5 precipitated similar amount of VPS35 across all three cell lines (WT, ATG5 KO, and ATG7 KO), irrespective of starvation (Fig. 2 D). To confirm this observation, we performed immunoprecipitation experiments for endogenous proteins using antibody against TBC1D5, which also revealed similar results (Fig. 2 E and Fig. S2 C). These observations were further confirmed by reverse immunoprecipitation experiments, which revealed that VPS35 precipitated similar amount of TBC1D5, irrespective of starvation (Fig. S2 D). Given that the evolutionary conserved Wiskott-Aldrich syndrome protein and SCAR homolog (WASH) complex is required for GLUT1 recycling to the cell surface (Gomez and Billadeau, 2009; Jia et al., 2010; Piotrowski et al., 2013; Steinberg et al., 2013), we tested whether starvation alters the cellular distribution of WASH complex subunit FAM21. The colocalization between FAM21 and EEA1 did not display obvious change in HeLa cells stably expressing GFP-LC3, indicating that FAM21 unlikely plays a major role in regulating of GLUT1 recycling upon starvation (Fig. S2 E). Altogether, our results show that TBC1D5 associates with retromer and LC3B through its TBC domain (Jia et al., 2016) and LIR4, respectively (Fig. 2 F), and starvation does not dissociate TBC1D5 from retromer. Thus, TBC1D5 and the WASH complex are unlikely to mediate starvation-induced inhibition of GLUT1 trafficking.
Starvation promotes phosphorylation of SNX27 at Ser51
We next asked whether SNX27, another key modulator of endosomal trafficking, could respond to starvation to regulate GLUT1 trafficking. To assess the protein expression and degradation of SNX27 under autophagy, cells were starved in the presence or absence of the protein biosynthesis inhibitor cycloheximide. The protein level of SNX27 was barely changed upon starvation, suggesting that starvation does not regulate endocytic recycling by controlling the protein amount of SNX27 (Fig. 3 A). Interestingly, phosphorylation of immunoprecipitated SNX27, detected by an anti-phospho-(Ser/Thr) antibody, increased ∼1.4- to 1.5-fold upon starvation, suggesting that starvation might promote the phosphorylation of SNX27 (Fig. 3 B). To identify the putative phosphorylation sites in SNX27, we used a mass spectroscopy (MS)–based approach. HEK293T cells stably expressing GST-tagged human SNX27 were treated with EBSS or E+B, followed by GST-pull-down and quantitative label-free mass spectrometry. Starvation resulted in an ∼1.5× increase in phosphorylation of the peptide IVKSEpSGYGFNVR at Ser51 (Fig. 3 C). Mass spectrometric measurement did not detect changes of other phosphorylation events on SNX27. Interestingly, phosphorylation of Ser51 was also reported when epithelial cells are exposed to cholera toxin, although the identity of the kinases involved in this phosphorylation event is unknown (Singh et al., 2018).
Figure 3.
Starvation promotes SNX27 phosphorylation at Ser51. (A) Expression of SNX27 was determined by immunoblotting in the indicated HEK293T or HeLa cells stably expressing GFP-LC3. The cells were starved for 4 h in the presence or absence of cycloheximide (CHX; 50 µg/ml for 9 h). Representative images from one of three independent experiments are shown. The numbers under the bands represent the relative expression of SNX27 to β-actin. Band intensities were quantified with ImageJ and normalized to their respective control group. (B) Starvation increases phosphorylation of endogenous SNX27, determined by the pan-phospho-Ser/Thr antibody. HEK293T cells were starved for 4 h (E+B) or not. One tenth of the cell lysate was prepared as input, and the rest was used for immunoprecipitation with anti-SNX27 antibody, followed by immunoblotting using antibody against pan-phospho-Ser/Thr. IgG served as a negative control. Representative images from one of three independent experiments are shown. The numbers under the bands represent the relative ratio of pan-phospho-Ser/Thr to SNX27. Band intensities were quantified with ImageJ and normalized to the control group. (C) Full MS/MS spectrum for the identification of Ser51 phosphorylation on SNX27 (b and y ions indicate peptide backbone fragment ions). HEK293T cells stably expressing GST-SNX27 were starved (EBSS or E+B). GST-SNX27 was captured by GST-affinity resin, proteolyzed, and used for LC-MS/MS analysis. Changes in phosphorylation status were determined by label-free quantification, and the relative phosphorylation was normalized to total protein amount. MS/MS spectrum shows the phosphorylation at Ser51, and column chart represents the relative phosphorylation of SNX27 on Ser51, determined from two (EBSS) or three (E+B) independent experiments. Data are presented as means ± SD, and one-way ANOVA by Tukey’s HSD test or t test was used for data analysis. ***, P < 0.001. (D) Starvation increases phosphorylation of endogenous SNX27 on Ser51, determined by a phospho-Ser51-specific antibody. HEK293T cells were transiently transfected with GST-tagged SNX27 for 24 h. Cells expressing SNX27 proteins were starved or not. The cell lysate was used for pull-down with GST-affinity resin, followed by immunoblotting using antibodies against GST-SNX27 and p-SNX27-Ser51. Representative images from one of three independent experiments are shown. The numbers under the bands represent the relative level of phospho-SNX27 to GST-SNX27. Band intensities were quantified with ImageJ and normalized to control group. HSD, honestly significant difference; S51, Ser51.
To facilitate the study of SNX27 phosphorylation, we raised a phospho-specific antibody against Ser51 of SNX27 (Fig. S2 F). This antibody displayed at least a 200,000-fold preference toward a phosphorylated peptide over an unphosphorylated one (Fig. S2 G). We next used this antibody to explore how the phosphorylation of Ser51 changes upon starvation. As our antibody failed to detect phosphorylation of endogenous SNX27, we examined cells transiently expressing GST-SNX27 WT or mutants where Ser51 was replaced with a nonphosphorylatable alanine residue (Ser51A) or a phosphomimetic aspartic acid reside (Ser51D). The phospho-Ser51–specific antibody specifically detected GST-SNX27 pull downed from cells. After starvation treatment with EBSS or E+B, the relative amount of phosphorylated SNX27 increased about twofold compared with the basal level (Fig. 3 D). Taken together, our data indicate that starvation increases phosphorylation of Ser51 in the PDZ domain of SNX27.
Phosphorylation of SNX27 Ser51 is mediated by MAPK11/14 in vitro and in vivo
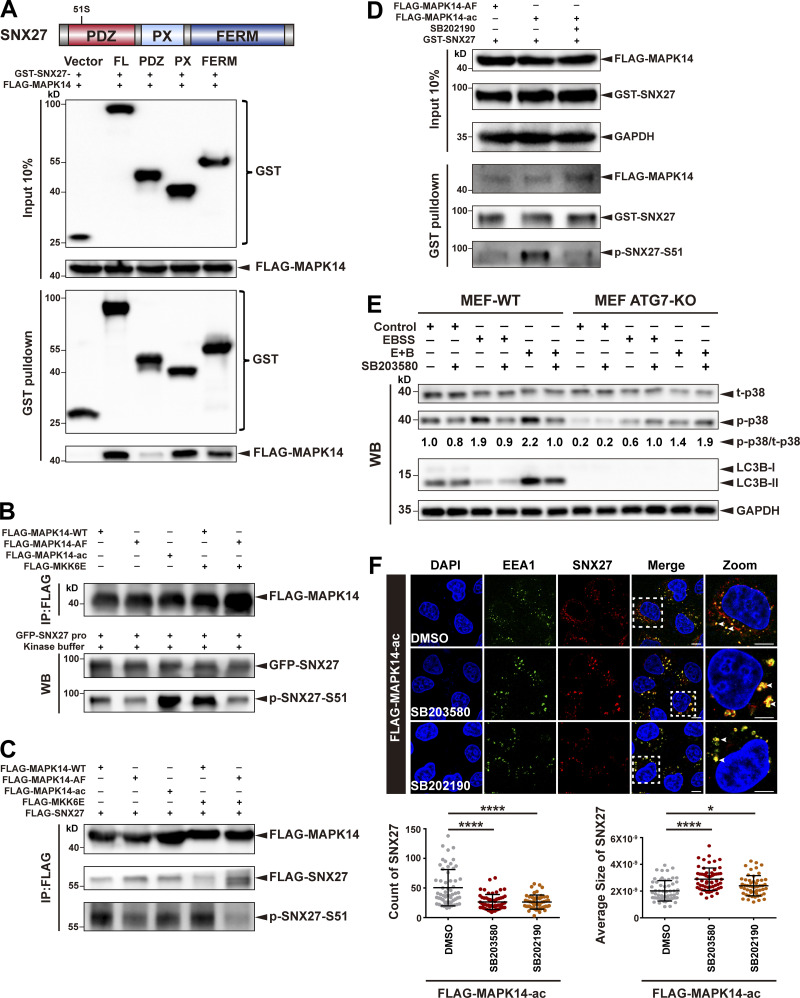
We sought to determine which kinase is responsible for phosphorylation of SNX27-Ser51. The kinase-specific phosphorylation site prediction program iGPS predicts MAPK11/14 as likely candidates (Song et al., 2012). MAPK11 and MAPK14 are two closely related members of the ERK/MAPK family and can be activated by different stimuli, such as growth factors, inflammatory cytokines, and environmental stresses (Ono and Han, 2000). Many MAPK substrates possess a MAPK-binding site (often referred to as docking domains), in addition to the serine/threonine phospho-acceptor sites. We thus determined whether SNX27 can interact with MAPK11/14. GST-SNX27, but not GST alone, robustly pulled down FLAG-MAPK14 from cell lysates. Both the PX and FERM domains, but not the PDZ domain of SNX27, mediated the interaction with MAPK14 (Fig. 4 A). Next, we tested whether MAPK14 phosphorylates SNX27 in an in vitro kinase assay using purified GFP-tagged SNX27 and a series of FLAG-tagged MAPK14 kinases isolated from cells. As shown in Fig. 4 B, SNX27 was barely phosphorylated by MAPK14 WT or kinase-inactive mutant of MAPK14 (MAPK14-AF) but strongly phosphorylated by a kinase-active mutant of MAPK14 (MAPK14-ac; lanes 1 and 2 versus lane 3). Moreover, coexpression of the upstream kinase MKK6E with MAPK14 WT, but not the AF mutant, strongly promoted the phosphorylation (lane 4 versus lane 5).
Figure 4.
MAPK14 phosphorylates SNX27 at Ser51 in vitro and in vivo. (A) Schematic representation of human SNX27, indicating the different domains (top). Pull-down assay to assess interaction between MAPK14 and different domains of SNX27. HEK293T cells were cotransfected with FLAG-tagged MAPK14 and GST-tagged SNX27 FL, PDZ, PX, or FERM domain for 24 h. Control cells (vector) were transfected with empty vector. One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GST resin, followed by immunoblotting using antibodies against GST-tagged SNX27 and FLAG-tagged MAPK14. Representative images from one of three independent experiments are shown. (B) MAPK14 phosphorylates SNX27 at Ser51 in vitro. HEK293T cells were transfected to FLAG-tagged MAPK14 WT, inactive MAPK14 (MAPK14-AF), or active MAPK14 (MAPK14-ac), with or without FLAG-tagged active MKK6 (MKK6E), as indicated. MAPK14 was immunoprecipitated with anti-FLAG beads and mixed with purified GFP-tagged SNX27 and ATP. The assay was performed at 37°C and analyzed using the anti-phospho-SNX27-Ser51 antibody. WB, western blotting. Representative of three independent iterations. (C) MAPK14 phosphorylates SNX27 at Ser51 in vivo. HEK293T cells were cotransfected with FLAG-tagged SNX27 and various versions of FLAG-tagged MAPK14 as in B. Kinase and substrate proteins were immunoprecipitated with anti-FLAG beads, followed by immunoblotting using anti-FLAG and phospho-SNX27-Ser51 antibodies. Representative of three independent iterations. (D) In vivo MAPK14 kinase assay in the presence of kinase inhibitor, B202190. HEK293T cells were cotransfected with GST-tagged SNX27 and FLAG-tagged MAPK14-AF, MAPK14-ac, or MAPK14-ac with or without SB202190 (10 µM for 2 h). One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GST-affinity resin, followed by immunoblotting for FLAG-MAPK14, GST-SNX27, and p-SNX27-Ser51. Representative of three independent iterations. (E) ATG7 deficiency decreases the phosphorylation of p38. WT and ATG7 KO MEF cells were starved in the presence or absence of p38 inhibitor (SB203580, 10 µM for 2 h), followed by immunoblotting using antibodies against total p38, p-p38, LC3B, and GAPDH. Representative of two independent iterations. The numbers under the bands represent the represent the p-p38/t-p38 ratio in each group. Band intensities were quantified with ImageJ and normalized to the MEF-WT control group (first lane). (F) Confocal imaging for EEA1 (green) and SNX27 (red) immunofluorescence staining of HeLa cells. Cells were transfected with FLAG-tagged MAPK14-ac for 24 h and treated with 10 µM SB203580 or 10 µM SB202190 for 2 h. Representative images from two independent experiments are shown. Scale bars: 10 µm. Arrows indicate SNX27 puncta. Statistical analysis was performed based on the numbers and average sizes of the SNX27 puncta in each of the cells. Results for individual cells are plotted, along with the mean and SD for each group (n = 59 cells for DMSO, n = 58 for SB203580, and n = 51 for SB202190); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001; *, P < 0.05. HSD, honestly significant difference.
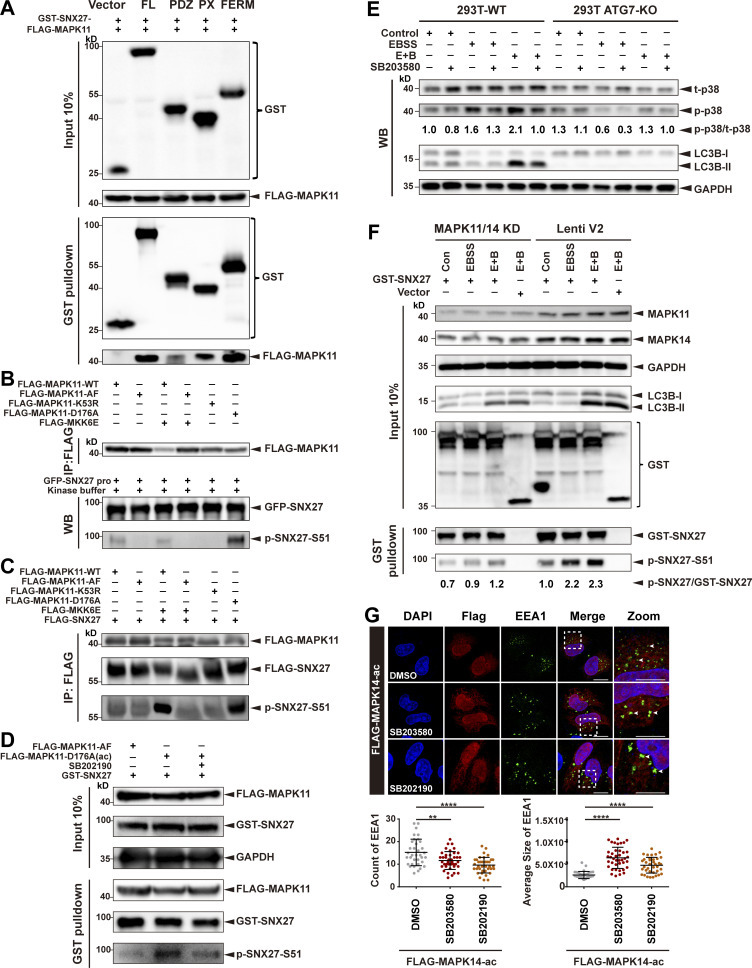
To test whether MAPK14 phosphorylates SNX27 in vivo, cells were cotransfected with FLAG-SNX27 and several MAPK14 variants. MAPK14 and kinase-active MAPK14-ac mutant displayed much stronger phosphorylation activity than the kinase-inactive MAPK14-AF mutant. Analogous to our in vitro results, phosphorylation of SNX27 was strongly stimulated by the coexpression of MKK6E with MAPK14 WT, but not the AF mutant (Fig. 4 C). Similar to MAPK14, MAPK11 phosphorylated SNX27 in vitro and in vivo (Fig. S3, A–C). Further confirming our conclusion that MAPK11/14 are kinases for SNX27, treatment of cells with a MAPK11/14-specific inhibitor, SB202190, abrogated the enhanced phosphorylation of SNX27 by kinase-active MAPK14-ac mutant (Fig. 4 D and Fig. S3 D). Altogether, these data illustrate that SNX27 is specifically phosphorylated by MAPK11 and MAPK14 at Ser51.
Figure S3.
MAPK11 phosphorylates SNX27 at Ser51 in vitro and in vivo. (A) Pull-down assay to assess interaction between MAPK11 and different domains of SNX27. HEK293T cells were cotransfected with FLAG-tagged MAPK11 and GST-tagged SNX27 FL, PDZ, PX, or FERM domain for 24 h. One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GSH affinity resin, followed by immunoblotting for GST-tagged SNX27 and FLAG-tagged MAPK11. Representative of three independent iterations. (B) MAPK11 phosphorylates SNX27 at Ser51 in vitro. HEK293T cells were transfected to FLAG-tagged MAPK11 WT, inactive MAPK11 (MAPK11-AF), inactive MAPK11 (MAPK11-K53R), or active MAPK11 (MAPK11-D176A), with or without FLAG-tagged active MKK6 (MKK6E), as indicated. MAPK11 was immunoprecipitated with anti-FLAG beads and mixed with purified GFP-tagged SNX27 and ATP. The assay was performed at 37°C and analyzed using the anti-phospho-SNX27-Ser51 antibody. Representative of three independent iterations. (C) MAPK11 phosphorylates SNX27 at Ser51 in vivo. HEK293T cells were cotransfected with FLAG-tagged SNX27 and various versions of FLAG-tagged MAPK14 as in B. Kinase and substrate proteins were immunoprecipitated with anti-FLAG beads, followed by immunoblotting using anti-FLAG and phospho-SNX27-Ser51 antibodies. Representative of three independent iterations. (D) In vivo MAPK11 kinase assay in the presence of kinase inhibitor (SB202190). HEK293 cells were cotransfected with GST-tagged SNX27 and FLAG-tagged MAPK11-AF, MAPK11-D176A, or MAPK11-D176A plus SB202190 (10 µM for 2 h). One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GSH resin, followed by immunoblotting for FLAG-MAPK11, GST-SNX27, and p-SNX27-Ser51. Representative of three independent iterations. (E) ATG7 deficiency decreases the phosphorylation of p38. WT and ATG7-KO HEK293T cells were starved in the presence or absence of p38 kinase inhibitor (SB203580, 10 µM for 2 h), followed by immunoblotting using antibodies against total p38, p-p38, LC3B, and GAPDH. Representative of two independent iterations. The numbers under the bands represent the p-p38/t-p38 ratio in each group. Band intensities were quantified with ImageJ and normalized to the 293T-WT control group (first lane). (F) MAPK11/14 depletion reduces phosphorylation of SNX27 at Ser51. HeLa cells were transfected with CRISPR-Cas9 plasmids targeting MAPK11 and MAPK14 (MAPK11/14 knockdown) or empty vector (Lenti V2). The knockdown and control cells were transfected with GST-tagged SNX27 for 24 h and starved for 4 h (E+B). One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GSH affinity resin, followed by immunoblotting for GST-SNX27 and p-SNX27-Ser51. Representative of two independent iterations. The numbers under the bands represent the p-SNX27/GST-SNX27 ratio in each group. Band intensities were quantified with ImageJ and normalized to the Lenti V2 control group. (G) Confocal imaging for EEA1 (green) puncta immunofluorescence staining of HeLa cells. Cells were transfected with FLAG-tagged MAPK14-D176A(ac) for 24 h and treated with 10 µM SB203580 or 10 µM SB 202190 for 2 h. Representative images from two independent experiments are shown. Scale bars: 10 µm (nonzoomed images) and 5 µm (zoomed images). Arrows indicate EEA1 puncta. The statistical analysis was performed based on the numbers and average sizes of the EEA1 puncta in each of the cells. Results for individual cells are plotted, along with the mean and SD for each group (n = 35 cells for DMSO, n = 40 for SB203580, and n = 38 for SB202190); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001; **, P < 0.01. HSD, honestly significant difference.
To understand why the distribution of GLUT1 was not affected by starvation in autophagy-deficient cells, we compared levels of p38 phosphorylation in autophagy-competent and deficient cells under starvation conditions. In normal cells, phosphorylation of p38 was significantly increased following treatment with EBSS or E+B and was also suppressed by the p38 pathway inhibitor SB203580 (Fig. 4 E). In contrast, MEF ATG7-KO cells displayed significantly reduced phosphorylation of p38 (Fig. 4 E). Similarly, deletion of ATG7 in HEK293T cells also dramatically impaired phosphorylation of p38 (Fig. S3 E). Furthermore, knockdown of MAPK11/14 using CRISPR-Cas9 vectors simultaneously targeting MAPK11 and MAPK14 (MAPK11/14 knockdown) dramatically reduced phosphorylation of SNX27 upon starvation stress (Fig. S3 F). Thus, it is likely that autophagy deficiency leads to insensitivity to the p38 pathway in cells, which might in turn impair GLUT1 recycling.
p38 MAPKs can modulate the morphology of early endosomes (Cavalli et al., 2001; Fratti et al., 2003; Pelkmans et al., 2005). Indeed, EEA1 exhibited a characteristic punctate distribution in cells overexpressing MAPK14-ac. Consistent with a previous report, the addition of the p38 inhibitor SB203580 or SB202190 increased the average size of EEA1 puncta and reduced the number of EEA1-positive early endosomes (Fig. S3 G). Interestingly, inhibition of MAPK14 activity by SB203580 and SB202190 also decreased the number of SNX27-positive puncta and increased their average size (Fig. 4 F). Altogether, our study suggests that MAPK11 and MAPK14 mediate the phosphorylation of SNX27 at Ser51 in vitro and in vivo.
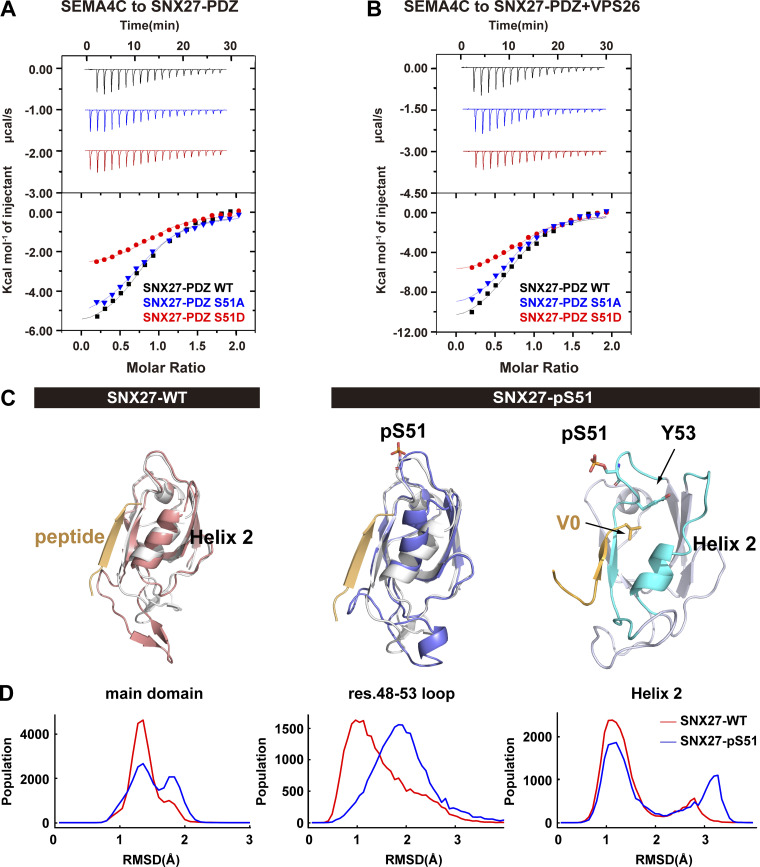
Phosphorylation of Ser51 reduces cargo binding to SNX27
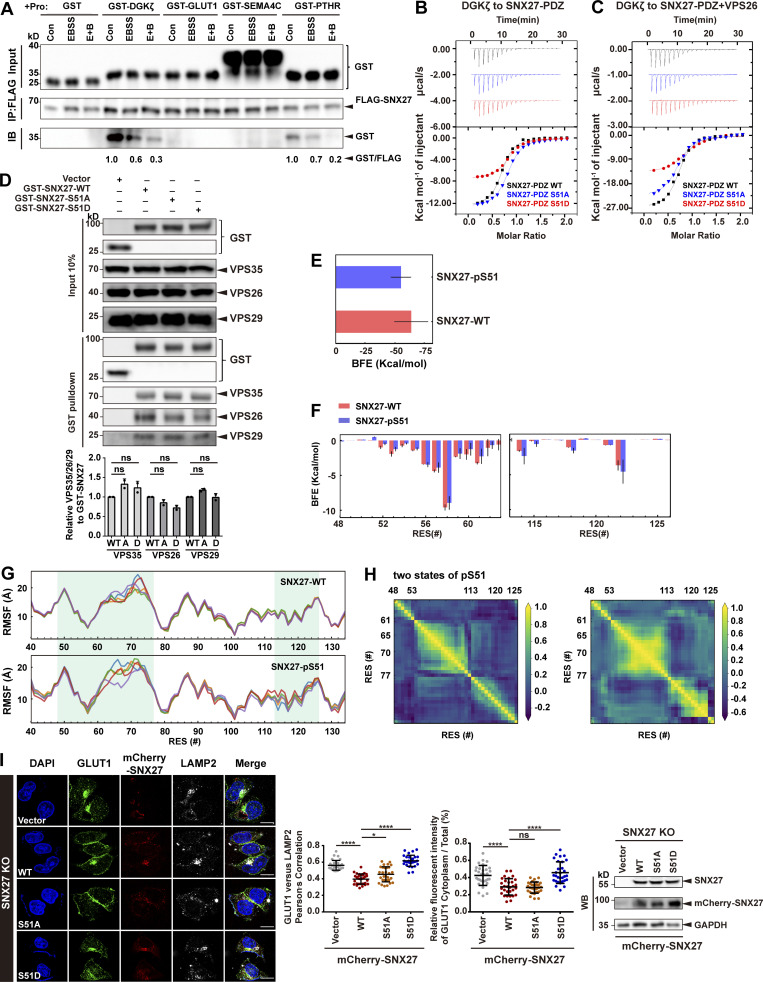
As phosphorylation of Ser51 occurs within the PDZ domain and may reduce SNX27 binding to its cargoes (Singh et al., 2018), we chose four PDZbm-containing proteins and tested their interaction with SNX27. Among them, GLUT1, SEMA4C, and PTHR are transmembrane proteins trafficked by SNX27, whereas diacylglycerol kinase ζ (DGKζ) is a soluble SNX27-binding enzyme (Rincón et al., 2011). Starvation significantly reduced the amount of SNX27 precipitated by DGKζ and PTHR, supporting the notion that phosphorylation of SNX27 Ser51 induced by starvation reduces binding to cargo proteins (Fig. S4 A). Consistent with previous measurements (Clairfeuille et al., 2016), the binding between SNX27 and GLUT1 or SEMA4C was too weak to be detected in our assay (Fig. S4 A). To precisely determine the impact of phosphorylation on cargo binding, we measured the binding between SEMA4C and SNX27 WT, phosphomimetic mutant Ser51D, or nonphosphorylatable mutant Ser51A using isothermal titration calorimetry (ITC). SEMA4C (aa 686–833) bound to SNX27 WT with a dissociation constant value of ∼4.5 M. SNX27 Ser51A displayed a similar affinity, whereas Ser51D mutant reduced the binding ∼2.2-fold (Fig. 5 A). It was reported that VPS26 could cooperate with SNX27 for the binding to PDZbms (Gallon et al., 2014). Thus, we also performed ITC experiments in the presence of VPS26. Indeed, VPS26 increased the affinity of SNX27 for SEMA4C, with SNX27 WT and both mutants being increased similarly (approximately twofold; Fig. 5 B). As a result, SNX27 Ser51D still displayed 2.2-fold weaker affinity relative to SNX27 WT and Ser51A. Similar results were obtained for DGKζ, a high-affinity binder of SNX27-PDZ. DGKζ bound to SNX27 WT, Ser51A, and Ser51D with dissociation constants of ∼1.2, 1.3, and 3 µM, respectively, in the absence VPS26 (Fig. S4 B). VPS26 increased the affinity approximately threefold for all three proteins (Fig. S4 C). Finally, SNX27 WT, Ser51D and Ser51A immunoprecipitated similar amount of retromer subunits, suggesting that phosphorylation at Ser51 (pSer51) unlikely affects the binding between SNX27 and retromer (Fig. S4 D).
Figure S4.
Phosphorylation of Ser51 reduces SNX27 binding to its cargoes. (A) Starvation decreases the interaction between SNX27 and PDZbm-containing proteins. HEK293T cells were transfected with FLAG-tagged SNX27 for 24 h and starved or not for 4 h. The whole-cell lysates were immunoprecipitated with anti-FLAG beads, followed by incubation with four purified GST-tagged PDZbms (DGKζ, GLUT1, SEMA4C, and PTHR) or GST. The bound samples were then washed and analyzed by immunoblotting. Representative of three independent iterations. The bands of GST-tagged DGKζ and PTHR were quantified by ImageJ, normalized to the level of the respective control group, and labeled below the blots. (B) ITC-binding curves for DGKζ peptide (REDQETAV) titrated into SNX27-PDZ WT, Ser51A, or Ser51D at 25°C. Top and bottom panels show raw and integrated heat from injections, respectively. The black curve in the bottom panel represents a fit of the integrated data to a single-site binding model. (C) ITC-binding curves for DGKζ peptide (REDQETAV) titrated into SNX27-PDZ WT, Ser51A, or Ser51D supplemented with equimolar VPS26 at 25°C. Top and bottom panels show raw and integrated heat from injections, respectively. The black curve in the bottom panel represents a fit of the integrated data to a single-site binding model. (D) Pull-down assay to assess binding of GST-SNX27 and endogenous VPS35/26/29. HEK293T cells were transfected with GST-tagged SNX27-WT, Ser51A, or Ser51D for 24 h. Control cells (vector) were transfected with empty vector. One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GST resin, followed by immunoblotting using antibodies against GST-SNX27, VPS35, VPS26, and VPS29. Representative images from one of two independent experiments are shown. Column chart represents the relative expression of VPS35/26/29 to GST-SNX27-WT, Ser51A, or Ser51D. Band intensities were quantified with ImageJ and normalized to their respective WT group (n = 2). Data are presented as means ± SD, and one-way ANOVA by Tukey’s HSD test or t test was used for data analysis. (E) Comparison of binding free energy (BFE) between DGKζ peptide to SNX27 PDZ WT and pSer51. (F) Per-residue free energy decomposition for sequences around the DGKζ-binding regions. (G) Residue fluctuation, measured by root mean square fluctuation (RMSF) of the entire PDZ domain in the WT and pSer51 systems. (H) Dynamic cross-correlation maps for two representative conformations of pSer51, shown in Fig. 5 B. Correlated (positive values) and anticorrelated (negative values) motions varied between residue (RES) 48-53 loop, residue 61–77 loop, or helix 2 (residues 113–126) in the two conformations. (I) Confocal imaging for GLUT1 (green) and LAMP2 (gray) immunofluorescence staining in HeLa cells. SNX27-KO cells were transfected with mCherry-tagged SNX27 WT, Ser51A or Ser51D, or empty vector, for 24 h. Representative images from two independent experiments are shown. Scale bar: 10 µm. Pearson’s correlation coefficients for GLUT1 and LAMP2. Relative fluorescent intensity of GLUT1 in cytoplasm/total ratio in each cell. Results for individual cells are plotted, along with the mean and SD for each group (n = 30); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001; *, P < 0.05. Knockdown and transfection efficiency were determined by immunoblotting with antibodies as indicated (right). HSD, honestly significant difference; ns, not significant.
Figure 5.
Phosphorylation of Ser51 reduces SNX27 binding to PDZbm-containing cargoes via altering the conformation of the cargo-binding pocket. (A) Isothermal titration calorimetry-binding curves for SEMA4C (aa 686–833) titrated into SNX27-PDZ WT, Ser51A, or Ser51D at 25°C. Top and bottom panels show raw and integrated heat from injections, respectively. The black curve in the bottom panel represents a fit of the integrated data to a single-site binding model. (B) ITC-binding curves for SEMA4C (aa 686–833) titrated into SNX27-PDZ WT, Ser51A, or Ser51D supplemented with equimolar VPS26 at 25°C. Top and bottom panels show raw and integrated heat from injections, respectively. The black curve in the bottom panel represents a fit of the integrated data to a single-site binding model. (C) MD-simulated conformations of SNX27 PDZ WT (red) and pSer51(blue), and comparison with the crystal structure (white, PDB accession no. 5ELQ). DGKζ peptide is shown in orange. (D) Distribution of root-mean square deviation (RMSD) between MD-simulated conformations and the crystal structure of SNX27 PDZ (PDB accession no. 5ELQ). Main domain: entire PDZ domain excluding residue (res.) 61–77 loop; residue 48–53 loop; helix 2.
To explore how pSer51 alters SNX27 binding to PDZbm, we performed molecular dynamics (MD) simulations starting from the crystal structure of the SNX27 PDZ domain in complex with the DGKζ peptide (Clairfeuille et al., 2016). Our 8-µs MD simulation reveals that pSer51 induces larger conformational changes compared with the structure with nonphosphorylated Ser51 (WT; Fig. 5, C and D). Specifically, pSer51 increases the dynamics of its in situ loop (residues 48–53) and induces translocation of helix 2 (residues 114–123) via the action of Y53 (Fig. 5 D). Structural clustering indicates that the WT polypeptide has one main conformation that is similar to what is observed in the crystal structure (Fig. 5 C). In contrast, the pSer51 polypeptide has two major conformations; one differs from the crystal structure in the position of helix 2, and the other differs by the decreased helicity of helix 2 (Fig. 5 C). The SNX27–DGKζ complex structure reveals that the DGKζ V0 forms hydrogen bonds with SNX27 Y53 and G54, while the side chain of V0 is cradled in a hydrophobic cavity formed by SNX27 Y53, F55, and I121. Thus, the movement of the in situ loop (residues 48–53) and helix 2 (residues 114–123) could alter the recognition of PDZbm by SNX27. Indeed, the calculated free energy of binding between DGKζ and WT is ∼9 kcal/mol lower than that between DGKζ and pSer51 (Fig. S4, E–H). In agreement with our data, several previous studies showed that conformational flexibility of helix 2 is critical for the PDZ domain’s binding specificity and affinity (Ho and Agard, 2010; Münz et al., 2012). Collectively, our data indicate that pSer51 decreases the binding between SNX27 PDZ and PDZbm, most likely by altering the conformation of the in situ loop and adjacent helix 2.
Phosphorylation of SNX27 Ser51 specifically inhibits endocytic trafficking of PDZbm-containing cargoes
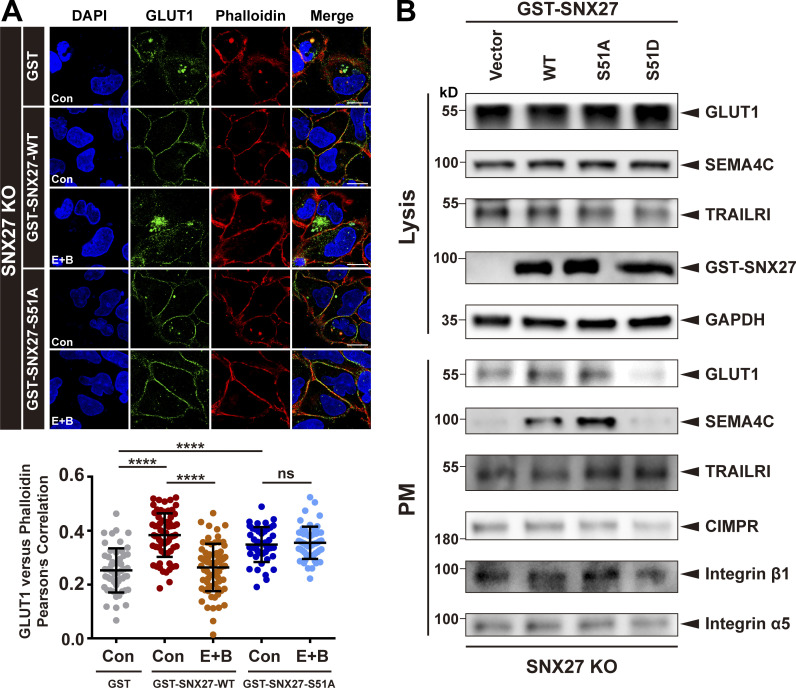
Next, we investigated how phosphorylation of Ser51 affects endocytic trafficking of GLUT1. SNX27-KO cells were generated using CRISPR-Cas9 (Ran et al., 2013; Yong et al., 2020), and SNX27 WT, Ser51A, or Ser51D was then reintroduced into these cells through transient expression. Reexpression of SNX27 WT, but not empty vector, markedly decreased the colocalization of GLUT1 and LAMP2 and reduced the cytoplasmic GLUT1 level, consistent with the notion that SNX27 is necessary for endocytic recycling of GLUT1 (Fig. S4 I). Importantly, SNX27 Ser51A, but not Ser51D, could support normal trafficking of GLUT1, similar to SNX27 WT, suggesting that binding of GLUT1 by the PDZ domain of SNX27 is critical for its normal trafficking (Fig. S4 I). To assess the importance of Ser51 phosphorylation, we performed the experiments under starvation (E+B). In cells transfected with SNX27 WT (KO+SNX27 WT), starvation decreased the colocalization between GLUT1 and phalloidin, consistent with the notion that phosphorylation of Ser51 inhibits endocytic trafficking of GLUT1. In contrast, the colocalization between GLUT1 and phalloidin was not affected by starvation in cells expressing SNX27 Ser51A (KO+SNX27 Ser51A; Fig. 6 A). Hence, phosphorylation of Ser51 acts as a switch to regulate endocytic trafficking of GLUT1.
Figure 6.
Phosphorylation of SNX27 at Ser51 specifically inhibits endocytic recycling of PDZbm-containing cargoes. (A) Starvation decreases endocytic recycling of GLUT1 in cells expressing SNX27, but not Ser51A. Confocal imaging for GLUT1 (green) and phalloidin (red) immunofluorescence staining of SNX27-KO HeLa cells. Cells were transfected with empty vector, GST-tagged SNX27 WT, or Ser51A for 24 h and starved for 4 h (E+B). Representative images from two independent experiments are shown. Scale bars: 10 µm. Pearson’s correlation coefficients for GLUT1 and phalloidin. Results for individual cells are plotted, along with the mean and SD for each group (GST control, n = 52 cells; GST-SNX27-WT control, n = 70; GST-SNX27-WT E+B, n = 74; GST-SNX27-Ser51A control, n = 44; and GST-SNX27-Ser51A E+B, n = 55); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. (B) SNX27 Ser51D specifically inhibits the recycling of SEMA4C and GLUT1, as indicated by surface protein biotinylation. SNX27-KO HeLa cells were transfected with GST-tagged SNX27 WT, Ser51A, or Ser51D for 24 h, followed by surface biotinylation and immunoblotting using indicated antibodies. Representative immunoblots show the protein levels at the PM and total. In contrast with GLUT1 and SEMA4C, which are transported by SNX27, CI-MPR, and TRAILR1, integrin β1 and integrin α5 are cargoes of SNX-BARs and SNX17, respectively. HSD, honestly significant difference; ns, not significant.
Next, we investigated how distribution of cell surface proteins was modulated by phosphorylation of Ser51. A surface protein biotinylation assay (Singla et al., 2019; Yong et al., 2020) revealed that both GLUT1 and SEMA4C could be easily detected in cells reexpressing GST-SNX27 WT (KO+SNX27 WT) or Ser51A mutant (KO+SNX27 Ser51A), but their amounts were significantly reduced in those reexpressing Ser51D (KO+SNX27 Ser51D) or GST control (KO+GST; Fig. 6 B). In contrast, the surface protein level of integrin β1, integrin α5, and CI-MPR remained unchanged in all cells (Fig. 6 B). This observation is consistent with the fact that endocytic recycling of CI-MPR, integrin β1, and integrin α5 depends on SNX-BARs, SNX17, and retriever, respectively (Kvainickas et al., 2017; McNally et al., 2017; Simonetti et al., 2017; Simonetti et al., 2019; Yong et al., 2020). Interestingly, the surface abundance of TRAILR1 also remained constant, consistent with the observation that starvation did not alter recycling of TRAILR1 (Fig. 6 B and Fig. S1 B). One explanation is that although endocytic recycling of TRAILR1 depends on SNX27, recognition of TRAILR1 is mediated by SNX-BARs, but not SNX27 (Yong et al., 2020). Thus, phosphorylation of SNX27 is unlikely to alter the recognition of TRAILR1 by SNX-BARs. Hence, phosphorylation on Ser51 specifically impairs trafficking of cargo proteins that directly bind to the PDZ domain of SNX27.
Phosphorylation of SNX27 Ser51 by MAPK11/14 functions as a switch to regulate endocytic trafficking in response to multiple types of stress
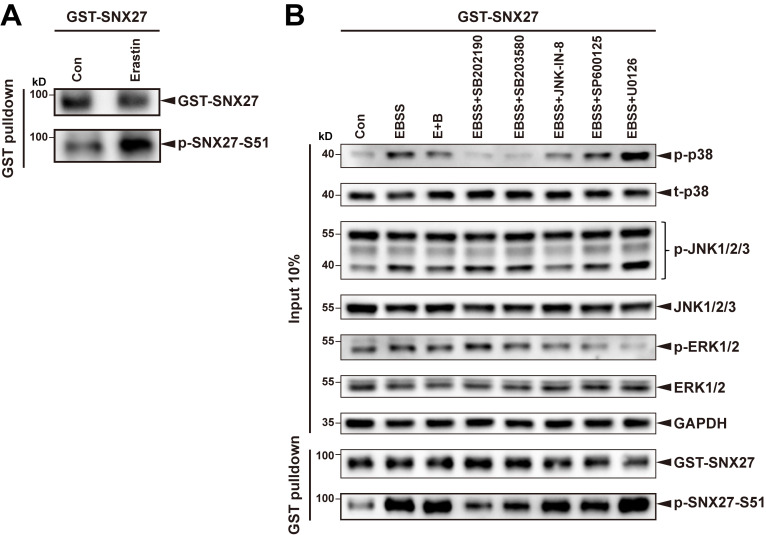
Cells have to deal with distinct types of stress, both intracellular and extracellular, in order to survive and proliferate (Hotamisligil and Davis, 2016). We next investigated which types of stress can modulate endocytic recycling through phosphorylation of SNX27 Ser51. Several classical stress-related stimuli were chosen for analysis, including starvation, lipopolysaccharide (LPS; inflammation inducer), carbonyl cyanide 3-chlorophenylhydrazone (CCCP; mitochondrial depolarizer), thapsigargin (TG; ER stress inducer), cytokine IL-6, EGF, excessive ATP, and erastin (ferroptotic inducer; Dixon et al., 2012; Jimenez-Orgaz et al., 2018; Ono and Han, 2000; Wang and Kaufman, 2016). GST-pull-down experiments revealed that treatment with LPS, IL-6, EGF, excessive ATP, and erastin significantly increased phosphorylation of SNX27, detected by our Ser51-phospho-specific antibody, in addition to EBSS and E+B (Fig. 7 A and Fig. S5 A). On the other hand, treatment with CCCP or TG did not alter the level of SNX27 phosphorylation (Fig. 7 A), although mitophagy was clearly induced when HeLa cells stably expressing GFP-Parkin were treated with the same concentration of CCCP (Trempe et al., 2013; Fig. 7 B). Consistently, quantitative immunofluorescence analysis showed that treatment of EBSS, E+B, LPS, or IL-6 resulted in a significant decrease in the colocalization of GLUT1 and phalloidin compared with untreated cells or cells treated with CCCP (Fig. 7 C). Thus, Ser51 phosphorylation of SNX27 can function as a general switch for cells to cope with multiple types of cellular stress.
Figure 7.
MAPK11/14 transmit multiple types of stress to regulate phosphorylation of SNX27 at Ser51 and endocytic trafficking. (A) Pull-down assay to assess phosphorylation of SNX27 under different stimulation conditions. HEK293T cells were transfected with GST-tagged SNX27 for 24 h and treated with multiple stimulations: EBSS (for 4 h), E+B (100 nM for 4 h), LPS (50 ng/ml for 1 h), CCCP (20 µM for 4 h), TG (1 µM for 4 h), IL-6 (10 ng/ml for 1h), EGF (15 ng/ml for 1h), or ATP (5 mM for 1 h). Complete medium (con) was used as a negative control. The cell lysate was used for pull-down with GST-affinity resin, followed by immunoblotting for GST-SNX27 and p-SNX27-Ser51S. Representative of three independent iterations. (B) Confocal imaging for GLUT1 (gray) and LAMP2 (red) immunofluorescence staining in the stable GFP-Parkin (green) HeLa cells. The cells were treated with CCCP (20 µM for 4 h) or not. Representative images are shown. Scale bars: 10 mm. Pearson’s correlation coefficients for GLUT1 and LAMP2. Results for individual cells are plotted, along with the mean and SD for each group (n = 34); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. (C) Confocal imaging for GLUT1 (gray) and phalloidin (red) immunofluorescence staining in HeLa cells stably expressing GFP-LC3 (green). The cells were treated with classical stimulations as indicated in A. Complete medium (con) was used as a negative control. Representative images are shown. Scale bars: 10 mm. Quantification of GLUT1 immunofluorescence on phalloidin-positive cell membrane. Results for individual cells are plotted, along with the mean and SD for each group (n = 30); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001. (D) Pull-down assay to assess how LPS treatment alters phosphorylation of SNX27 Ser51 in the presence or absence of different MAPK pathway inhibitors. HEK293T cells were transfected with GST-tagged SNX27 for 24 h, pretreated with or without various types of inhibitors: the MAPK11/14-specic inhibitor SB202190 (10 µM for 2 h), the p38 pathway inhibitor SB203580 (10 µM for 2 h), the JNK inhibitor JNK-IN-8 (5 µM for 2 h), and the ERK1/2 inhibitor U0126 (25 µM for 2 h). The cells were then treated or not with 50 ng/ml LPS for 1 h. Control cells (vector) were transfected with empty vector. One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GST-affinity resin, followed by immunoblotting for GST-SNX27 and p-SNX27-Ser51. Representative of three independent iterations. (E) Confocal imaging for Venus-GLUT1 (green) and phalloidin (red) immunofluorescence staining of HeLa cells. Cells were transfected with Venus-GLUT1 for 24 h, treated with or without different pathway inhibitors: the MAPK11/14-specic inhibitor SB202190 (10 µM for 2 h), the p38 pathway inhibitor SB203580 (10 µM for 2 h), the JNK inhibitor JNK-IN-8 (5 µM for 2 h), and the ERK1/2 inhibitor U0126 (25 µM for 2 h). Cells were then starved for 4 h (EBSS or E+B). Representative images are shown. Scale bars: 10 mm. Pearson’s correlation coefficients for Venus-GLUT1 and phalloidin. Results for individual cells are plotted, along with the mean and SD for each group (con, n = 33 cells; EBSS, n = 28; E+B, n = 29; SB202190+E+B, n = 36; SB203580+E+B, n = 40; JNK-IN-8+E+B, n = 41; U0126+E+B, n = 32); P values were calculated using one-way ANOVA by Tukey’s HSD test or t test. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01. HSD, honestly significant difference; ns, not significant.
Figure S5.
Multiple types of stress promote SNX27 phosphorylation via activating MAPK11/14. (A) Pull-down assay to assess Ser51 phosphorylation of SNX27 under erastin treatment. HEK293T cells were transfected with GST-tagged SNX27 for 24 h and treated with erastin (5 µM) for 6 h. Complete medium (con) was used as a negative control. The cell lysate was used for pull-down with GSH affinity resin, followed by immunoblotting for GST-SNX27 and p-SNX27-Ser51S. Representative of three independent iterations. (B) Pull-down assay to assess how starvation alters phosphorylation of SNX27 Ser51 in the presence or absence of different MAPK pathway inhibitors. HEK293T cells were transfected with GST-tagged SNX27 or empty vector (vector) for 24 h, pretreated with or without different pathway inhibitors: the MAPK11/14-specic inhibitor SB202190 (10 µM for 2 h), the p38 pathway inhibitor SB203580 (10 µM for 2 h), the JNK inhibitor JNK-IN-8 (5 µM for 2 h), the JNK inhibitor SP600125 (10 µM for 2 h), and the ERK1/2 inhibitor U0126 (25 µM for 2 h). Cells were then starved for 4 h. One tenth of the cell lysate was prepared as input, and the rest was used for pull-down with GSH affinity resin, followed by immunoblotting for GST-SNX27 and p-SNX27-Ser51. Representative of two independent iterations.
Besides MAPK11/14, other kinases in the MAPK pathway, including ERK1/2 and JNK, are also involved in coping with cell stress (Johnson and Lapadat, 2002). To determine whether these kinases can also phosphorylate SNX27 Ser51, cells expressing SNX27 were pretreated with different inhibitors (SB202190, a MAPK11/14-specific inhibitor; SB203580, an inhibitor of all four p38 kinases, including MAPK11/14; JNK-IN-8, a JNK inhibitor; and U0126, ERK1/2 inhibitor) and then treated with LPS (Fig. 7 D) or starvation (Fig. S5 B). Immunoblotting demonstrated that the inhibition of each kinase-mediated signaling pathway was significant and specific. For instance, among all inhibitors, only U0126 strongly inhibited the phosphorylation of ERK1/2. Two MAPK11/14 inhibitors, SB202190 and SB203580, clearly decreased the phosphorylation of SNX27 upon LPS treatment (lanes 4 and 5 versus lane 3), whereas JNK-IN-8 and U0126 had little effect (lanes 6 and 7 versus lane 3), as shown in Fig. 7 D. Thus, Ser51 phosphorylation of SNX27 is specifically mediated by MAPK11/14, but not by other related kinases in the MAPK pathway.
To fully address the specificity of MAP11/14 in regulating endocytic trafficking in response to cellular stresses, we assessed the effect of different MAPK inhibitors on GLUT1 trafficking (Fig. 7 E). HeLa cells expressing Venus-GLUT1 were pretreated with different inhibitors for 2 h, followed by incubation with E+B. Similar to our previous results, starvation clearly led to a shift of GLUT1 localization from the cell surface to intracellular compartments. The shift was strongly inhibited by the MAPK11/14 inhibitors SB202190 and SB203580, but not by the JNK inhibitor JNK-IN-8 or the ERK1/2 inhibitor U0126 (Fig. 7 E).
Hence, phosphorylation of Ser51 can serve as a switch to regulate endocytic trafficking. Altogether, our study suggests that multiple types of stresses can modulate endocytic recycling via phosphorylation of SNX27 by MAPK11/14 (Fig. 8).
Figure 8.
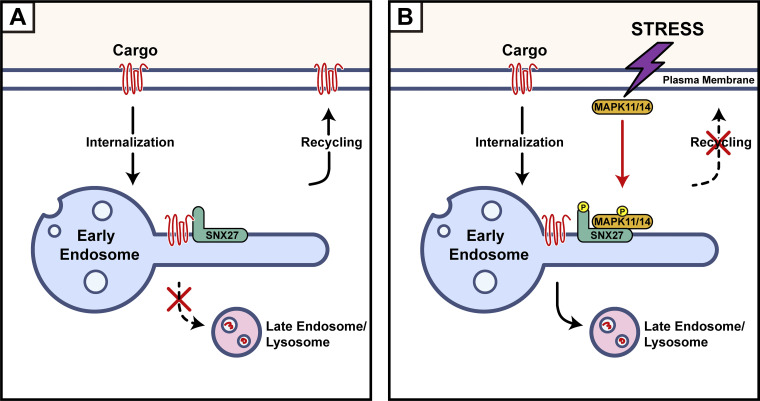
A proposed model for MAPK11/14-mediated phosphorylation of SNX27 inhibits cargo recycling. (A) Under normal conditions, SNX27 recognizes PDZbm-containing cargoes and promotes their recycling from endosomes to the PM. (B) Multiple types of stress activate MAPK11/14, which in turn phosphorylate SNX27 at Ser51. Phosphorylation of SNX27 reduces cargo binding and recycling, leading to enhanced lysosomal degradation.
Discussion
The importance of endocytic trafficking for homeostasis in cells, tissues, and organisms is well established, but the dynamic regulation of endocytic trafficking is poorly understood (Cullen and Steinberg, 2018). In this study, we demonstrate that multiple stresses, including starvation, LPS, or IL-6, lead to an inhibition of endocytic recycling, indicating that endocytic recycling is more highly dynamic than previously appreciated. The inhibition is mediated by crosstalk between endocytic recycling and the MAPK pathway, as we demonstrate that MAPK11/14 specifically phosphorylate SNX27 on Ser51, inducing conformational changes of the PDZbm-binding pocket and resulting in reduced cargo recycling. Our data suggest that endocytic trafficking is influenced by environmental cues that can be dynamically regulated through signaling cascades like many other cellular events.
A previous study indicated that treatment of cells with cholera toxin increases phosphorylation of rat SNX27 at S49 (Singh et al., 2018). Our study greatly extends this observation by showing that multiple stresses lead to phosphorylation of the same site in human SNX27. More importantly, we have determined that MAPK11/14 are specifically responsible for this modification. As cholera toxin exposure could activate the p38 MAPK pathway (Ono and Han, 2000), our study indicates that cholera toxin likely regulates SNX27-mediated trafficking via activation of MAPK11/14. In the present study, we show that starvation decreases GLUT1 accumulation in the PM, which is distinct from a recent study that concluded that autophagy promotes cell surface expression of GLUT1 (Roy et al., 2017). One reason for this difference may be due to the different experimental conditions (starvation versus hypoxia). However, we have demonstrated that retromer and LC3B bind to distinct regions of TBC1D5, suggesting that autophagy is unlikely to promote TBC1D5 shuttling between the retromer complex and LC3+ autophagosomes through a direct competition between retromer and LC3B.
The p38 MAPK pathway has been shown to regulate a wide range of cellular processes, including endocytosis. First, MAPK14 phosphorylates Rab5 effectors (EEA1 and Rabenosyn-5) and regulators (GDI; Cavalli et al., 2001; Macé et al., 2005; Wälchli et al., 2008), thus regulating the endocytosis of multiple membrane receptors. Second, MAPKs can directly phosphorylate multiple transmembrane proteins and regulate their transport, such as EGFR and FGFR1 (Sørensen et al., 2008; Zwang and Yarden, 2006). By demonstrating that MAPK11/14 can phosphorylate SNX27, a key regulator involved in the endosome-to-PM recycling of numerous receptors, we now provide the first evidence that connects p38 MAPK signaling and endocytic recycling. Future studies will be necessary to determine why starvation and other types of stress inhibit endocytic trafficking of some cargoes, whereas these cellular stresses do not impact others, like TRAILR1.
Although phosphorylation of a cargo protein can serve as a mechanism to alter the trafficking route of a specific protein (Cao et al., 1999), our study reveals that phosphorylation of SNX27 within its PDZ domain can act as a general mechanism to regulate the trafficking of many different cargo proteins. It remains to be determined whether phosphorylation of SNX27 at Ser51 or other sites could increase binding to other cargo proteins and enhance their endosome-to-PM recycling. A greater understanding of how SNXs themselves are subject to phosphorylation and other posttranslational modifications may lead to better insights into the mechanisms to control the fate of many distinct proteins. In this regard, it has been noted that many SNXs and other retrieval machinery proteins could be phosphorylated (Cui et al., 2017; Cullen and Steinberg, 2018; Horazdovsky et al., 1997). For instance, phosphorylation of a conserved serine in the PX domain has been shown to regulate the membrane localization of SNX3 (Lenoir et al., 2018). Given the increasing number of modification events, future studies will be necessary to define whether these modifications can regulate cargo retrieval and recycling. These studies will additionally broaden our understanding of endocytic trafficking in the context of development and signaling.
Materials and methods
Cell culture and transfection
HeLa cell lines stably expressing GFP-LC3 or GFP-Parkin were kind gifts from Dr. Quan Chen (Nankai University). The WT and ATG7-KO cell lines, including HEK293T and MEFs, were kind gifts from Dr. Li Yu (Tsinghua University). The SNX27-KO HeLa cell line was previously described (Yong et al., 2020). HEK293T and HeLa cell lines were authenticated by Shanghai Biowing Biotechnology. PCR detection for mycoplasma contamination yielded negative results. All cells were maintained in high-glucose DMEM, supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin–streptomycin. Cells were grown in an incubator at 37°C in humidified air with 5% CO2 and transfected using polyethylenimine or LipoFiter 3.0 Reagent according to the manufacturer’s advising protocol.
Immunofluorescence staining and confocal microscopy
Immunofluorescence experiments were performed as previously described (Singla et al., 2019; Yong et al., 2020). Cells were washed with PBS and then fixed with 4% PFA for 15 min at room temperature. The plates were rinsed with PBS three times and then permeabilized with 0.1% Triton X-100 solution for 15 min, followed by washing with PBS three times. After being blocked with 5% newborn calf serum at 37°C for 1 h, the cells were incubated with 2% newborn calf serum containing primary antibodies overnight at 4°C. The coverslips were washed three times with ice-cold PBST and then incubated with fluorophore-coupled secondary antibody for 1 h at 37°C. Finally, sections were washed gently in PBST three times, stained with DAPI, and mounted in Vectashield fluorescent mounting media. Images were captured using Olympus FV-1000 (100× oil objective, NA = 1.40) and Zeiss LSM 780 (63× oil objective, NA = 1.40) confocal microscopes at 16°C. Minor adjustments of brightness and contrast of digital images were performed with FV10-ASW 4.2 Viewer software. No gamma adjustments were made. Following acquisition, images were rotated, cropped, and sized using Adobe Photoshop and composed using Adobe Illustrator. Images were analyzed by National Institutes of Health ImageJ software. All experiments were repeated two or three times.
Immunoblotting
Whole-cell lysates were prepared in 1X SDS-PAGE sample buffer containing 1% protease and phosphatase inhibitors and boiled at 98°C for 10 min. Next, proteins were separated by SDS-PAGE and then blotted onto a polyvinylidene difluoride membrane. Membrane was blocked with 5% nonfat milk at room temperature for 1 h and incubated with specific primary antibodies overnight at 4°C. Subsequently, the membranes were incubated with appropriate HRP-conjugated secondary antibodies for 1 h at room temperature. Signals were detected using chemiluminescent HRP substrate, performed using QuantityOne (Bio-Rad) and quantified using ImageJ.
GST pull-down
For the GST pull-down assay, HEK293T cells were transfected with various combinations of expression vectors, similar to previous studies (Liu et al., 2020; Qin et al., 2020). The proteins were purified from cells using glutathione Sepharose beads in lysis buffer with protease and phosphatase inhibitors. After extensive washing with lysis buffer, bound proteins were separated by SDS-PAGE and visualized by immunoblotting.
Recombinant protein expression and purification
Recombinant proteins were expressed and purified as previously described (Jia et al., 2012; Jia et al., 2016; Yong et al., 2018). All proteins were expressed in BL21(DE3) cells, and protein expression was induced by the addition of 0.5 mM IPTG at 20°C overnight. Cells were harvested and resuspended with the lysis buffer (20 mM Tris-HCl, 200 mM NaCl, and 1 mM PMSF, pH 8.0). The cells were then lysed with a high-pressure crusher, followed by centrifugation for 30 min at 18,000 rpm. SNX27 PDZ WT and mutants were first purified with GST-affinity resin, and the GST tag was removed by tobacco etch virus (TEV) protease cleavage. Human VPS26A and His6-GFP-tagged SNX27 were purified using Ni-NIA resin (Qiagen) and eluted with buffer A (20 mM Tris-HCl, 200 mM NaCl, and 300 mM imidazole, pH 8.0). GST-tagged DGKζ, GLUT1, SEMA4C, and PTHR were all purified using GST-affinity resin. All proteins were then purified by Superdex 200 increase gel filtration column equilibrated with buffer B (20 mM Tris-HCl, 200 mM NaCl, and 5 mM β-mercaptoethanol, pH 8.0).
ITC
ITC experiments were performed at 25°C using ITC 200 (Microcal) in ITC buffer (20 mM Tris-HCl and 200 mM NaCl, pH 8.0), similar to previous studies (Jia et al., 2016; Yao et al., 2018). DGKζ peptide (sequence: REDQETAV; ∼200–1,000 µM) and SEMA4C (aa 686–833, ∼300–1,000 µM) were titrated into the sample cell containing SNX27 PDZ WT or mutants (∼20–100 µM), supplemented with ∼100–500 µM VPS26 when required. Data were analyzed with the Origin 7.0 software package (OriginLab) by fitting the “one set of sites” model.
MD simulations
Crystal structure of SNX27 PDZ domain bound to a DGKζ peptide (PDB accession no. 5ELQ) was used to prepare systems of WT phosphoserine at Ser51 (pSer51) with or without the DGKζ peptide. The initial aqueous systems were prepared using AMBER (Case et al., 2018), in which a protein or protein–peptide complex was solvated in ∼6,000 TIP3P water molecules, counter ions, and 0.10 M NaCl, totaling ∼19,000 atoms in a periodic box of ∼56 × 62 × 72 Å3. All simulations were performed with the AMBER FF14SB force field (Case et al., 2018). Each system went through energy minimization, 125 ps constant-temperature, constant-volume ensemble (NVT) equilibration, and MD simulations using the AMBER18 package with graphics processing unit acceleration (Case et al., 2018). Production runs were performed in the constant-temperature, constant-pressure ensemble (NPT) ensemble (310K, 1 bar, Langevin dynamics thermostat, and Monte Carlo barostat) with a time step of 2 fs. All of the bond lengths to hydrogen atoms in each system were constrained with SHAKE. The particle mesh Ewald technique was used for the electrostatic calculations. The van der Waals and short-range electrostatics were cut off at 12.0 Å with switch at 10.0 Å. The PDZ-domain–only systems were simulated in five copies with 4 µs each; PDZ domain–DGKζ peptide systems were simulated in 10 copies with 8 µs each. Structural analysis was performed with AMBER built-in tool and Tcl scripts implemented in visual molecular dynamics (Humphrey et al., 1996) and plotted by matplotlib (Hunter, 2007). The Molecular Mechanics Generalized Born Surface Area method (Miller et al., 2012) was used to calculate the binding free energy of DGKζ peptide to the SNX27 PDZ domain.
Immunoprecipitation
Cells were collected with lysis buffer supplemented with 1% protease and phosphatase inhibitors at 4°C for 30 min. After centrifugation, the supernatant was incubated with antibody (1 µg/ml)–coated Protein A+G Agarose overnight at 4°C. Normal IgG was used as the negative control antibody. The beads were washed five times with cold lysis buffer, boiled for 10 min, and analyzed by immunoblotting.
In vitro and In vivo kinase assay
In vitro kinase assays were performed as previously described (Xu and Derynck, 2010). In brief, the bacterially purified recombinant GFP-SNX27 was incubated with immunopurified kinases (including HA-FLAG-tagged MAPK11 and FLAG-tagged MAPK14 or their mutant, MKK-6E) with kinase buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 50 mM MgCl2, 1 mM Na3VO4, 1 mM PMSF, 1 mM DTT, and 5 mM ATP) in 200 µl for 30 min at 37°C. The reaction mixtures were then added 5X SDS-PAGE loading buffer and boiled at 98°C for 10 min. Immunoblotting was performed to analyze the amount of phospho-SNX27(51S). For in vivo kinase assays, SNX27 and p38 MAPK were transfected into HEK293T cells and immunoprecipitated using beads and washed with lysis buffer. Samples were subsequently detected by immunoblotting.
Surface protein biotinylation assay
Biotinylation assay was performed as previously described (Singla et al., 2019). Briefly, cells were washed three times with ice-cold PBS (pH 8.0) and incubated with Sulfo-NHS-SS-Biotin at room temperature for 30 min. Excess biotin reagent was removed by washing with ice-cold PBS three times, and cells were lysed in lysis buffer (50 mM Tris, pH 7.5, 50 mM NaCl, and 0.5% NP-40). The supernatant was incubated with streptavidin resin for 2 h at room temperature. The beads were then washed with PBS, and the bound proteins were used for immunoblotting analysis.
Surface protein internalization and recycling assays
Internalization and recycling assays were performed as described previously (Yong et al., 2020), with a few changes. HeLa cells were cultured and transfected with plasmids encoding CD8A-SEMA4C or CD8A-PTHR fusion proteins for 24 h. Cells were then added anti-CD8A (5 µg/ml in DMEM) for 30 min at low temperature (on ice). Unbound antibodies were removed by washing with ice-cold PBS three times. The medium was replaced with complete medium for an additional 1 or 3 h at 37°C. After treatment, cells were fixed with 4% paraformaldehyde and made permeable with 0.1% Triton X-100. The internalized CD8A–antibody detection was conducted with Alexa Fluor 488 secondary antibody.
Generation of CRISPR-KO cell lines
SNX27-KO cell lines were produced in our previous study (Yong et al., 2020). MAPK11 and MAPK14 knockdown cell lines were generated similarly to the previous study (Huang et al., 2019). Briefly, the target guide RNAs were inserted in the plasmid Lenti-CRISPR-V2. HEK293T cells were transfected with modified Lenti-CRISPR-V2 and lentivirus packaging plasmids. After 2 d of transfection, the supernatant fluid containing virus was used to infect HeLa cells. These cells were selected in puromycin (4 µg/ml) for two generations. Remaining cells were expanded and selected by immunoblotting.
Liquid chromatography (LC)-MS/MS analysis
LC-MS/MS analysis was performed in accordance with a previous report, with minor modifications (Goodman et al., 2018). Prior to MS/MS analysis, all peptide samples were lyophilized and resuspended in buffer A (2% acetonitrile and 0.1% formatic acid), and LC-MS/MS analysis was performed using an EASY-nLC 1000 nano flow LC instrument coupled to a Q Exactive quadrupole-orbitrap mass spectrometer (Thermo Fisher Scientific). Full-scan MS spectra (from m/z 350–1,800) were acquired with resolving power of 70,000 at m/z = 200. For MS/MS scans, the top 20 most intense parent ions were selected with a 1.6-m/z isolation window and fragmented with a normalized collision energy of 25%. All the raw files were searched against the Swiss-Prot human protein sequence database (updated April 2019, 20,431 sequences) in MaxQuant (version 1.6). Phosphorylation was set as variable modifications. The minimum score for modified peptides was set at 40, and label-free quantification was used for relative quantification.
Statistical analysis
All experiments were repeated at least two times, and the representative results are presented. Measurement data are expressed as mean ± SD and were analyzed using a one-way ANOVA test followed by a Tukey’s test using GraphPad Prism 6.0 software. Colocalization analysis of immunofluorescence images was performed with the software FV10-ASW 4.2b (Pearson’s coefficients) or ImageJ (Manders’ coefficients). Statistical significance was established at ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Online supplemental material
Fig. S1 shows that starvation inhibits endocytic recycling of multiple transmembrane proteins and that autophagy deficiency abolishes the inhibition. Fig. S2 shows that the interaction between TBC1D5 and retromer is independent of LC3 and is not affected by starvation. Fig. S3 shows that MAPK11 phosphorylates SNX27 at Ser51 in vitro and in vivo. Fig. S4 shows that phosphorylation of Ser51 reduces SNX27 binding to its cargoes. Fig. S5 shows that multiple types of stress promote SNX27 phosphorylation by activating MAPK11/14. Table S1 lists antibodies used in this study. Table S2 lists reagents used in this study. Table S3 lists plasmids used in this study.
Supplementary Material
lists antibodies used in this study,
lists reagents used in this study.
lists DNA constructs used in this study.
Acknowledgments
We thank Drs. Li Yu (Tsinghua University, Beijing, China), Quan Chen (Nankai University, Tianjin, China), and Wei Liu and Pinglong Xu (Zhejiang University, Hangzhou, China) for materials and Drs. Jizhong Lou and Xiaocheng Wang (Institute of Biophysics, Chinese Academy of Sciences, Beijing, China) for helpful discussion.
This research was supported by the Natural Science Foundation of China (grants 91854121 and 31871429 to D. Jia and grant 21625302 to G. Li), the National Key Research and Development Program of China (grant 2018YFC1005004 to D. Jia), the Sichuan Science and Technology Program (grant 2018RZ0128 to D. Jia), and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB 37000000 to G. Li).
The authors declare no competing financial interests.
Author contributions: D. Jia conceived the project. L. Mao carried out cellular studies with assistance from S. Li. C. Liao and G. Li performed the simulations and data analysis, and Y. Gong and L. Dai completed MS analysis. J. Qin and H. Deng performed ITC assays. Y. Zhou, Z. Liu, W. Deng Q. Sun, X. Mo, Y. Xue, and D.D. Billadeau provided technical assistance. L. Mao and D. Jia prepared the manuscript with input from every author.
References
- Beenstock, J., Ben-Yehuda S., Melamed D., Admon A., Livnah O., Ahn N.G., and Engelberg D.. 2014. The p38β mitogen-activated protein kinase possesses an intrinsic autophosphorylation activity, generated by a short region composed of the α-G helix and MAPK insert. J. Biol. Chem. 289:23546–23556. 10.1074/jbc.M114.578237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C., and Cullen P.J.. 2014. Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6: a016774. 10.1101/cshperspect.a016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, T.T., Deacon H.W., Reczek D., Bretscher A., and von Zastrow M.. 1999. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature. 401:286–290. 10.1038/45816 [DOI] [PubMed] [Google Scholar]
- Case, D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S.. T.E. Chatham III, V.W.D. Cruzeiro, T.A. Darden, R.E. Duke, D. Ghoreishi, M.K. Gilson, et al. 2018. AMBER 2018. University of California, San Francisco. [Google Scholar]
- Cavalli, V., Vilbois F., Corti M., Marcote M.J., Tamura K., Karin M., Arkinstall S., and Gruenberg J.. 2001. The stress-induced MAP kinase p38 regulates endocytic trafficking via the GDI:Rab5 complex. Mol. Cell. 7:421–432. 10.1016/S1097-2765(01)00189-7 [DOI] [PubMed] [Google Scholar]
- Chan, A.S., Clairfeuille T., Landao-Bassonga E., Kinna G., Ng P.Y., Loo L.S., Cheng T.S., Zheng M., Hong W., Teasdale R.D., et al. 2016. Sorting nexin 27 couples PTHR trafficking to retromer for signal regulation in osteoblasts during bone growth. Mol. Biol. Cell. 27:1367–1382. 10.1091/mbc.E15-12-0851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairfeuille, T., Mas C., Chan A.S., Yang Z., Tello-Lafoz M., Chandra M., Widagdo J., Kerr M.C., Paul B., Mérida I., et al. 2016. A molecular code for endosomal recycling of phosphorylated cargos by the SNX27-retromer complex. Nat. Struct. Mol. Biol. 23:921–932. 10.1038/nsmb.3290 [DOI] [PubMed] [Google Scholar]
- Cui, T.Z., Peterson T.A., and Burd C.G.. 2017. A CDC25 family protein phosphatase gates cargo recognition by the Vps26 retromer subunit. eLife. 6:e24126. 10.7554/eLife.24126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., and Steinberg F.. 2018. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19:679–696. 10.1038/s41580-018-0053-7 [DOI] [PubMed] [Google Scholar]
- Dixon, S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., et al. 2012. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 149:1060–1072. 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti, R.A., Chua J., and Deretic V.. 2003. Induction of p38 mitogen-activated protein kinase reduces early endosome autoantigen 1 (EEA1) recruitment to phagosomal membranes. J. Biol. Chem. 278:46961–46967. 10.1074/jbc.M305225200 [DOI] [PubMed] [Google Scholar]
- Gallon, M., Clairfeuille T., Steinberg F., Mas C., Ghai R., Sessions R.B., Teasdale R.D., Collins B.M., and Cullen P.J.. 2014. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl. Acad. Sci. USA. 111:E3604–E3613. 10.1073/pnas.1410552111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, T.S., and Billadeau D.D.. 2009. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell. 17:699–711. 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, J.K., Zampronio C.G., Jones A.M.E., and Hernandez-Fernaud J.R.. 2018. Updates of the In-Gel Digestion Method for Protein Analysis by Mass Spectrometry. Proteomics. 18:e1800236. 10.1002/pmic.201800236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro, A., Rojas A.L., Rojas R., Murthy N., Effantin G., Kajava A.V., Steven A.C., Bonifacino J.S., and Hurley J.H.. 2007. Functional architecture of the retromer cargo-recognition complex. Nature. 449:1063–1067. 10.1038/nature06216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, B.K., and Agard D.A.. 2010. Conserved tertiary couplings stabilize elements in the PDZ fold, leading to characteristic patterns of domain conformational flexibility. Protein Sci. 19:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky, B.F., Davies B.A., Seaman M.N., McLaughlin S.A., Yoon S., and Emr S.D.. 1997. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol. Biol. Cell. 8:1529–1541. 10.1091/mbc.8.8.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil, G.S., and Davis R.J.. 2016. Cell Signaling and Stress Responses. Cold Spring Harb. Perspect. Biol. 8:a006072. 10.1101/cshperspect.a006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Liu Z., Yang F., Zhou H., Yong X., Yang X., Zhou Y., Xue L., Zhang Y., Liu D., et al. 2019. Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH. Proc. Natl. Acad. Sci. USA. 116:22598–22608. 10.1073/pnas.1909316116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38: 27–28. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Hunter, J.D. 2007. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9:90–95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Jia, D., Gomez T.S., Metlagel Z., Umetani J., Otwinowski Z., Rosen M.K., and Billadeau D.D.. 2010. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl. Acad. Sci. USA. 107:10442–10447. 10.1073/pnas.0913293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, D., Gomez T.S., Billadeau D.D., and Rosen M.K.. 2012. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell. 23:2352–2361. 10.1091/mbc.e11-12-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, D., Zhang J.S., Li F., Wang J., Deng Z., White M.A., Osborne D.G., Phillips-Krawczak C., Gomez T.S., Li H., et al. 2016. Structural and mechanistic insights into regulation of the retromer coat by TBC1d5. Nat. Commun. 7:13305. 10.1038/ncomms13305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Orgaz, A., Kvainickas A., Nägele H., Denner J., Eimer S., Dengjel J., and Steinberg F.. 2018. Control of RAB7 activity and localization through the retromer-TBC1D5 complex enables RAB7-dependent mitophagy. EMBO J. 37:235–254. 10.15252/embj.201797128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, G.L., and Lapadat R.. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 298:1911–1912. 10.1126/science.1072682 [DOI] [PubMed] [Google Scholar]
- Kalvari, I., Tsompanis S., Mulakkal N.C., Osgood R., Johansen T., Nezis I.P., and Promponas V.J.. 2014. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy. 10:913–925. 10.4161/auto.28260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, O., Leneva N., Bykov Y.S., Ariotti N., Teasdale R.D., Schaffer M., Engel B.D., Owen D.J., Briggs J.A.G., and Collins B.M.. 2018. Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature. 561:561–564. 10.1038/s41586-018-0526-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvainickas, A., Jimenez-Orgaz A., Nägele H., Hu Z., Dengjel J., and Steinberg F.. 2017. Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport. J. Cell Biol. 216:3677–3693. 10.1083/jcb.201702137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir, M., Ustunel C., Rajesh S., Kaur J., Moreau D., Gruenberg J., and Overduin M.. 2018. Phosphorylation of conserved phosphoinositide binding pocket regulates sorting nexin membrane targeting. Nat. Commun. 9:993. 10.1038/s41467-018-03370-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Yang F., Liu Z., Wang J., Huang W., Meng W., Billadeau D.D., Sun Q., Mo X., and Jia D.. 2020. Structure of TBC1D23 N-terminus reveals a novel role for rhodanese domain. PLoS Biol. 18:e3000746. 10.1371/journal.pbio.3000746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, M., Gershlick D.C., Vidaurrazaga A., Rojas A.L., Bonifacino J.S., and Hierro A.. 2016. Structural Mechanism for Cargo Recognition by the Retromer Complex. Cell. 167:1623–1635.e14. 10.1016/j.cell.2016.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macé, G., Miaczynska M., Zerial M., and Nebreda A.R.. 2005. Phosphorylation of EEA1 by p38 MAP kinase regulates mu opioid receptor endocytosis. EMBO J. 24:3235–3246. 10.1038/sj.emboj.7600799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey, J.C., Xiao K., Bowman S.L., Mamonova T., Zhang Q., Bisello A., Sneddon W.B., Ardura J.A., Jean-Alphonse F., Vilardaga J.P., et al. 2016. Actin-Sorting Nexin 27 (SNX27)-Retromer Complex Mediates Rapid Parathyroid Hormone Receptor Recycling. J. Biol. Chem. 291:10986–11002. 10.1074/jbc.M115.697045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, K.J., Korswagen H.C., and Cullen P.J.. 2017. The emerging role of retromer in neuroprotection. Curr. Opin. Cell Biol. 47:72–82. 10.1016/j.ceb.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K.E., Faulkner R., Steinberg F., Gallon M., Ghai R., Pim D., Langton P., Pearson N., Danson C.M., Nägele H., et al. 2017. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol. 19:1214–1225. 10.1038/ncb3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, B.R. III, McGee T.D. Jr., Swails J.M., Homeyer N., Gohlke H., and Roitberg A.E.. 2012. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 8:3314–3321. 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- Münz, M., Hein J., and Biggin P.C.. 2012. The role of flexibility and conformational selection in the binding promiscuity of PDZ domains. PLOS Comput. Biol. 8:e1002749. 10.1371/journal.pcbi.1002749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, K., and Han J.. 2000. The p38 signal transduction pathway: activation and function. Cell. Signal. 12:1–13. 10.1016/S0898-6568(99)00071-6 [DOI] [PubMed] [Google Scholar]
- Pelkmans, L., Fava E., Grabner H., Hannus M., Habermann B., Krausz E., and Zerial M.. 2005. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 436:78–86. 10.1038/nature03571 [DOI] [PubMed] [Google Scholar]
- Piotrowski, J.T., Gomez T.S., Schoon R.A., Mangalam A.K., and Billadeau D.D.. 2013. WASH knockout T cells demonstrate defective receptor trafficking, proliferation, and effector function. Mol. Cell. Biol. 33:958–973. 10.1128/MCB.01288-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic, D., Akutsu M., Novak I., Harper J.W., Behrends C., and Dikic I.. 2012. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32:1733–1744. 10.1128/MCB.06717-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, J., Liu Q., Liu Z., Pan Y.Z., Sifuentes-Dominguez L., Stepien K.P., Wang Y., Tu Y., Tan S., Wang Y., et al. 2020. Structural and mechanistic insights into secretagogin-mediated exocytosis. Proc. Natl. Acad. Sci. USA. 117:6559–6570. 10.1073/pnas.1919698117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón, E., Sáez de Guinoa J., Gharbi S.I., Sorzano C.O., Carrasco Y.R., and Mérida I.. 2011. Translocation dynamics of sorting nexin 27 in activated T cells. J. Cell Sci. 124:776–788. 10.1242/jcs.072447 [DOI] [PubMed] [Google Scholar]
- Roy, S., Leidal A.M., Ye J., Ronen S.M., and Debnath J.. 2017. Autophagy-Dependent Shuttling of TBC1D5 Controls Plasma Membrane Translocation of GLUT1 and Glucose Uptake. Mol. Cell. 67:84–95.e5. 10.1016/j.molcel.2017.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N. 2004. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165:111–122. 10.1083/jcb.200312034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N., McCaffery J.M., and Emr S.D.. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665–681. 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N., Harbour M.E., Tattersall D., Read E., and Bright N.. 2009. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122:2371–2382. 10.1242/jcs.048686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N.J., Mukadam A.S., and Breusegem S.Y.. 2018. Inhibition of TBC1D5 activates Rab7a and can enhance the function of the retromer cargo-selective complex. J. Cell Sci. 131:jcs217398. 10.1242/jcs.217398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti, B., Danson C.M., Heesom K.J., and Cullen P.J.. 2017. Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J. Cell Biol. 216:3695–3712. 10.1083/jcb.201703015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti, B., Paul B., Chaudhari K., Weeratunga S., Steinberg F., Gorla M., Heesom K.J., Bashaw G.J., Collins B.M., and Cullen P.J.. 2019. Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins. Nat. Cell Biol. 21:1219–1233. 10.1038/s41556-019-0393-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, V., Yang J., Yin J., Cole R., Tse M., Berman D.E., Small S.A., Petsko G., and Donowitz M.. 2018. Cholera toxin inhibits SNX27-retromer-mediated delivery of cargo proteins to the plasma membrane. J. Cell Sci. 131:jcs218610. 10.1242/jcs.218610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla, A., Fedoseienko A., Giridharan S.S.P., Overlee B.L., Lopez A., Jia D., Song J., Huff-Hardy K., Weisman L., Burstein E., and Billadeau D.D.. 2019. Endosomal PI(3)P regulation by the COMMD/CCDC22/CCDC93 (CCC) complex controls membrane protein recycling. Nat. Commun. 10:4271. 10.1038/s41467-019-12221-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C., Ye M., Liu Z., Cheng H., Jiang X., Han G., Songyang Z., Tan Y., Wang H., Ren J., et al. 2012. Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol. Cell. Proteomics. 11:1070–1083. 10.1074/mcp.M111.012625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen, V., Zhen Y., Zakrzewska M., Haugsten E.M., Wälchli S., Nilsen T., Olsnes S., and Wiedlocha A.. 2008. Phosphorylation of fibroblast growth factor (FGF) receptor 1 at Ser777 by p38 mitogen-activated protein kinase regulates translocation of exogenous FGF1 to the cytosol and nucleus. Mol. Cell. Biol. 28:4129–4141. 10.1128/MCB.02117-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanoska, K., Bertz J., Volkerling A.M., van der Hoven J., Ittner L.M., and Ittner A.. 2018. Neuronal MAP kinase p38α inhibits c-Jun N-terminal kinase to modulate anxiety-related behaviour. Sci. Rep. 8:14296. 10.1038/s41598-018-32592-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, F., Gallon M., Winfield M., Thomas E.C., Bell A.J., Heesom K.J., Tavaré J.M., and Cullen P.J.. 2013. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15:461–471. 10.1038/ncb2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin, P., Lauffer B., Jäger S., Cimermancic P., Krogan N.J., and von Zastrow M.. 2011. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13:715–721. 10.1038/ncb2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourian, L. Jr., Zhao H., and Srikant C.B.. 2004. p38alpha, but not p38beta, inhibits the phosphorylation and presence of c-FLIPS in DISC to potentiate Fas-mediated caspase-8 activation and type I apoptotic signaling. J. Cell Sci. 117:6459–6471. 10.1242/jcs.01573 [DOI] [PubMed] [Google Scholar]
- Trempe, J.F., Sauvé V., Grenier K., Seirafi M., Tang M.Y., Ménade M., Al-Abdul-Wahid S., Krett J., Wong K., Kozlov G., et al. 2013. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 340:1451–1455. 10.1126/science.1237908 [DOI] [PubMed] [Google Scholar]
- Tu, Y., Zhao L., Billadeau D.D., and Jia D.. 2020. Endosome-to-TGN Trafficking: Organelle-Vesicle and Organelle-Organelle Interactions. Front. Cell Dev. Biol. 8:163. 10.3389/fcell.2020.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälchli, S., Skånland S.S., Gregers T.F., Lauvrak S.U., Torgersen M.L., Ying M., Kuroda S., Maturana A., and Sandvig K.. 2008. The Mitogen-activated protein kinase p38 links Shiga Toxin-dependent signaling and trafficking. Mol. Biol. Cell. 19:95–104. 10.1091/mbc.e07-06-0565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., and Kaufman R.J.. 2016. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 529:326–335. 10.1038/nature17041 [DOI] [PubMed] [Google Scholar]
- Wang, J., Fedoseienko A., Chen B., Burstein E., Jia D., and Billadeau D.D.. 2018. Endosomal receptor trafficking: Retromer and beyond. Traffic. 19:578–590. 10.1111/tra.12574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P., and Derynck R.. 2010. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol. Cell. 37:551–566. 10.1016/j.molcel.2010.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J., Yang F., Sun X., Wang S., Gan N., Liu Q., Liu D., Zhang X., Niu D., Wei Y., et al. 2018. Mechanism of inhibition of retromer transport by the bacterial effector RidL. Proc. Natl. Acad. Sci. USA. 115:E1446–E1454. 10.1073/pnas.1717383115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong, X., Hu W., Zhou X., Wang J., Burstein E., and Jia D.. 2018. Expression and purification of the SNX1/SNX6 complex. Protein Expr. Purif. 151:93–98. 10.1016/j.pep.2018.06.010 [DOI] [PubMed] [Google Scholar]
- Yong, X., Zhao L., Deng W., Sun H., Zhou X., Mao L., Hu W., Shen X., Sun Q., Billadeau D.D., et al. 2020. Mechanism of cargo recognition by retromer-linked SNX-BAR proteins. PLoS Biol. 18:e3000631. 10.1371/journal.pbio.3000631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin, T., and Han J.. 2005. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 15:11–18. 10.1038/sj.cr.7290257 [DOI] [PubMed] [Google Scholar]
- Zwang, Y., and Yarden Y.. 2006. p38 MAP kinase mediates stress-induced internalization of EGFR: implications for cancer chemotherapy. EMBO J. 25:4195–4206. 10.1038/sj.emboj.7601297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists antibodies used in this study,
lists reagents used in this study.
lists DNA constructs used in this study.