Abstract
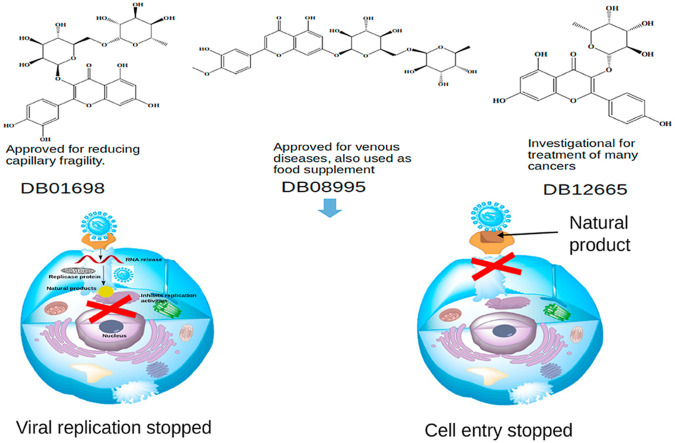
The recent outbreak of SARS-CoV-2 is responsible for high morbidity and mortality rate across the globe. This requires an urgent identification of drugs and other interventions to overcome this pandemic. Computational drug repurposing represents an alternative approach to provide a more effective approach in search for COVID-19 drugs. Selected natural product known to have antiviral activities were screened, and based on their hits; a similarity search with FDA approved drugs was performed using computational methods. Obtained drugs from similarity search were assessed for their stability and inhibition against SARS-CoV-2 targets. Diosmin (DB08995) was found to be a promising drug that works with two distinct mechanisms, preventing viral replication and viral fusion into the host cell. Isoquercetin (DB12665) and rutin (DB01698) work by inhibiting viral replication and preventing cell entry, respectively. Our analysis based on molecular dynamics simulation and MM-PBSA binding free energy calculation suggests that diosmin, isoquercetin, rutin and other similar flavone glycosides could serve as SARS-CoV-2 inhibitor, hence an alternative solution to treat COVID-19 upon further clinical validation.
Keywords: SARS-CoV-2, COVID-19, Natural products, Docking screening, MD simulation, Free energy calculation
Graphical abstract
1. Introduction
The outbreak of the novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) pandemic around the globe have called for an urgent research of drugs to treat the disease. Owing to the pressing health response, drug repurposing presents huge potential towards combating diseases. Drug repurposing is a strategy involved in identifying old approved drugs to treat new diseases [1,2]. This process is effective, efficient, and reliable in drug development compared to de novo approaches [[1], [2], [3]]. Such identified drugs have known pharmacokinetics profile and can be tested in clinical trials which fasten the processes of obtaining new drugs [4]. In an efforts to find new drugs to treat COVID-19 several approved antiviral drugs, [2,4,5] antimalarial drugs such as chloroquine and its derivative hydroxychloroquine, [3]; and [6] antiparasitic drugs [7] and antimicrobial agents such as azithromycin [8] just to mention a few, have been repurposed. Although current research on drug therapeutics and vaccines are yet to produce approved drugs or vaccine against SARS-CoV-2, some promising therapeutics such as remdesivir and galidesivir are in clinical trials [9].
Earlier works on drug repurposing against COVID-19 have targeted the main protease [1,10]. Polypharmacology, a process in drug design where on average a drug has 3 targets is highly recommended [1]. Identification of potential approved drugs with less side effects serve as a strong basis for drug repurposing [1]. Thus, analysis of SARS-CoV-2 molecular therapeutic targets forms an important process of identifying drugs to treat this disease [11]. In an attempt to identify new therapeutics against SARS-CoV-2, we resorted to use previously reported antiviral compounds from our research groups [12,13]. Such small molecules could be important hits towards finding new drugs. The selected hits molecules were used for similarity search for further molecular target evaluation. Molecular targets such as 3C-like protease (3CLpro), responsible for viral replication; RNA-dependent RNA polymerase (RdRp), responsible for viral transcription/replication; papain like protease (PLpro), responsible for cleavage of N-terminus of the replicase poly-protein; human angiotensin converting enzyme 2 (ACE2), responsible for regulating cardiovascular physiology in host, and spike receptor binding domain (RBD)-ACE2, responsible for viral attachment before entering host cells were considered important therapeutic targets in treating COVID-19. It is important to highlight that the SARS-CoV-2 main protease (Mpro) is a cysteine protease with an active site catalytic dyad HIS41 and CYS145 located between domain I and II (Fig. S1) [14,15]. The imidazole group of HIS polarizes and activates the SH group of CYS to form a highly nucleophilic CYSS−/HISH+ ion pair that reacts with the substrate [14].
Virtual screening via docking is an effective method and also a widely used approach in screening large libraries of drugs to determine potential drug candidates. To provide computation efficiency and accuracy, several free energy methods with different scoring functions have been developed and used in both hit identification and optimizations processes [1]. Free energy calculation methods such as thermodynamics integration (TI), [16] molecular mechanics Poisson Boltzmann/Generalized Born surface area (MM-PB(GB)SA), [17] free energy perturbation (FEP) [18] and Linear interaction energy (LIE) [19] are efficient methods in structure based drug design. In this study, we employed computational methods to identify new molecules against SARS-CoV-2. First, we screened antiviral natural products previously reported from Tanzanian medicinal plants against infectious bursal disease virus and newcastle disease virus [12,13]. The obtained hits prompted similarity search for FDA approved drugs that we further subjected to computational inhibitory assays against SARS-CoV-2 proteins. The potential molecules for drug repurposing as reported in this work provides an basis for scientists to develop agents against coronaviruses. Further in vitro and in vivo clinical validation of these molecules is necessary to provide useful information on COVID-19 treatment.
2. Materials and methods
2.1. Virtual screening and molecular docking
2.1.1. Ligand preparations
We started by building a small in-house library comprising of 19 natural products with antiviral properties previously obtained from Tanzanian medicinal plants [12,13]. The 2D and 3D structures of the selected natural products were built, then Gasteiger charges [20] were added and all structures energy minimized using universal force field (UFF) [21] employing steepest descents minimization in Avogadro [22]. The optimized geometric structures were converted to pdbqt file format using Open Babel [23] ready for screening by docking with AutoDock Vina [24].
2.1.2. Protein preparations and docking benchmarking
The study took advantage of the timely crystallized SARS-CoV-2 structural targets for drug design. The protein structures were obtained from the RCSB protein data bank (PDB) [25] with the following PDB IDs; 3C-like protease (3CLpro) (6LU7) [26], RdRp (6M71) [27], PLpro (6W9C) [28], nucleocapsid N-terminal RNA binding protein (6M3M) [29], Nsp9 RNA binding protein (6W4B) [30], native human ACE2 (1R42) [31] and spike RBD-ACE2 (6LZG) [1]. Missing hydrogen at pH 7.4 and Gasteiger charges [20] were added to all protein structures and were converted to pdbqt format as required by AutoDock Vina [24] for docking calculations.
Docking calculations were validated using a set of data with binding free energy as previously reported [32]. A set of experimental binding free energy reported in Refs. [33,34] were used to validate the docking scoring function implemented in our work. Docking calculation was performed to each complex and then the lowest binding free energy (more negative value) of each complex was compared with the experimental data and found to be in a reasonable agreement with a correlation value of r 2 = 0.83 (see Table S1). Besides using experimental data, a co-crystalized ligands of 3CLpro (PDB ID: 6LU7) and (PDB ID: 6W63) were extracted from the crystal binding pose and redocked again to the putative binding pocket to assess whether our method could reproduce experimental binding modes. Redocking gave acceptable RMSD values of ≤ 3 Å for all redocked ligands (Fig. S1). RMSD of ≤ 3 Å was found to be more reliable and can reproduce the experimental binding mode [35]. After validating the docking protocol, an in-house library of 19 natural products (see SI) were blindly screened against SARS-CoV-2 targets, the promising hit compounds with lower binding free energy of −7.5 kcal/mol were selected for further experiments and similarity search. Determination of the best binding energy of small molecules was based on the binding free energy of a co-crystallized ligand that was docked and found to be < −7.5 kcal/mol. In this work, we set the criterion that any ligands with binding free energy ≤ −7.5 kcal/mol similar to inhibition constant of Ki = 3.03 μM to be a potential candidate. Similarity search was performed to repurpose approved drugs for SARS-CoV-2 target proteins. All virtual screening docking calculations were done using AutoDock Vina tool [24] employing a python shell script customized for virtual screening.
2.2. Similarity search
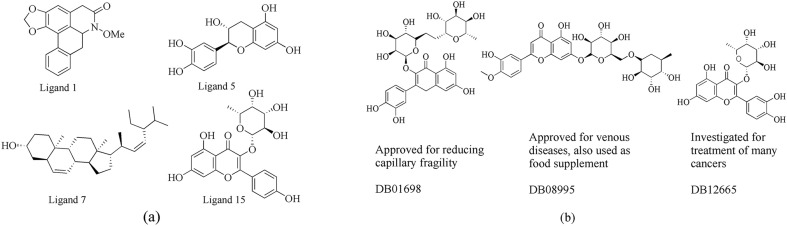
Compounds obtained from docking experiment (Fig. 1 a) were subjected to similarity search. A web tool, swisssimilarity (http://www.swisssimilarity.ch) which comprises of database with approved drugs (such as DrugBank [36], PubChem [37] etc) was used to perform similarity search [38]. An FP2 fingerprint with a Tanimoto score of 0–1 was used, with 0 depicting no similarity and 1 depicting complete identity. Compounds with higher Tanimoto score of ≥ 0.9 were selected for this study.
Fig. 1.
(a) 2D structures of the hit natural product compounds obtained from virtual screening. (b) 2D structures of the FDA approved natural compounds obtained using similarity search, the drugs were similar to ligand 15.
2.3. Quantum mechanical calculations
Compounds obtained from similarity search were subjected to quantum mechanical calculations. The selected geometric structures were optimized using the density functional theory (DFT) method with the B3LYP (Becke three-parameter Lee Yang Parr) in conjunction with the 6-311+G(d,p) basis set [39,40]. The calculated vibrational frequencies ascertained that the structures were minima by the absence of imaginary frequencies. All the calculations were performed using Gaussian16 suite [41] and then the compounds were subjected to molecular docking against all targets to investigate their inhibition effects. Best hits from docking calculations were then selected for molecular dynamics (MD) simulation to evaluate their binding stability in the pockets. Molecules obtained from similarity search are shown in Fig. 1b.
2.4. Molecular dynamics (MD) simulation
All drug molecules with lower binding free energies obtained from docking screening were subjected to MD simulation to assess their time-lapse stability. MD simulation for all complexes was performed using GROMACS v2018 employing all-atom Optimized Potential for Liquid Simulation (OPLS-AA) force field [42,43]. Protein topologies were generated using GROMACS while ligand topologies were generated using LigParGen tool [44]. All systems were placed at the center of cubic box solvated with TIP4P water model [45]. The initial charges were; −5 for Mpro-DB01698 complex, −5 for Mpro-DB08995 complex, −5 for Mpro-DB12665 complex, −26 for RBD-ACE2-DB08995 complex and −5 for ACE2-DB08995 complex. Depending on the charge of each system, sodium ions were added to neutralize the system. All systems underwent two energy minimization steps to remove bad and unfavourable atom contacts from molecular docking experiments. Initially, the systems were energy minimized by using steepest descent algorithm followed by conjugate gradient algorithm and the systems converged at Fmax < 1000 kJ/mol in 50,000 energy minimization steps. Two equilibration phases with position restraints were carried out to relax the system. First, the systems were equilibrated in an NVT ensemble using the V-rescale thermostat [46] for 100 ps. The systems were further equilibrated in the second phase at an NTP ensemble using Parrinello-Rahman barostat at an isotropic pressure of 1 bar for 500 ps [47]. MD production was carried out without position restraints at an NPT ensemble for up to 100 ns depending on the system. During the MD production both pressure and temperature were maintained using Parrinello-Rahman barostat and V-rescale thermostat at 1 bar and 300 K, respectively. In all systems, periodic boundary conditions (PBC) were applied in all directions, Particle Mesh Ewald (PME) method was used to treat long-range electrostatic interactions with a cutoff distance on 11 Å for both electrostatic and van der Waals interactions [48]. Bond length were constrained using LINCS algorithm [49] and an integration time step of 2 fs was used for all system. An extended analysis was then performed using GROMACS built-in and in-house tools. The distances between protein and ligands was measured from specific groups which formed hydrogen bonds, and the free energy was computed as F = −k B Tln(P) [50], where k B is the Boltzmann constant, T is the temperature and P is the probability distribution of the distances. Visualization was done using PyMOL, [51] Chimera [52] and VMD [53].
2.5. Relaxed complex scheme (RCS)
To get more insight into the effects of protein conformation changes on ligand binding affinity, RCS or ensemble based docking was performed as previously described [4]. After performing an MD simulation of the apo and holo protein, a total of 50 snapshots were evenly selected from the trajectory. Docking calculations were performed on each structure and the binding energies were averaged and presented as mean ± SD. All docking calculations were performed using AutoDock Vina tool [24]. In each ensemble structure, the binding site was specified before docking.
2.6. MM-PBSA binding free energy calculations
Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) was calculated using g_mmpbsa tool [17]. The binding free energy (ΔGbind) for protein-ligand interaction is expressed in Equation (1)
| (1) |
| (2) |
| (3) |
| (4) |
where, is the average molecular mechanics energy terms in vacuum which includes bonded and non-bonded interactions, is the free energy of solvation which includes Gpolar and Gnon-polar. γ is a coefficient related to surface tension, SASA is the solvent accessible surface area, b is the fitting parameter and TΔS is the entropic contributions to free energy. In g_mmpbsa an entropic contribution is not considered [17]. The binding free energy was calculated using a single trajectory, where a total of 200 snapshots were evenly extracted at a predetermined intervals, from the first 40 ns and last 40 ns of the trajectory. The solvent dielectric constant was 80, solute dielectric constant was 2, γ was 0.0226778 kJ/Mol/Å2 and b was 3.84928 kJ/Mol, the PB equation was solved by using the linear PBsolver.
3. Results and discussion
Before inferring to any docking calculation, it is very important to understand the limitations of the protocol and tools used. One of the limitations of many docking scoring functions is the ability to reproduce experimental results [54]. Thus, docking methods need to be validated before they can be used in any experiment. One of the most effective ways of validating docking scoring function is to assess its ability to reproduce the binding mode of the small molecule to its receptor determined by root mean square deviation (RMSD). An RMSD value of ≤ 3 Å is an acceptable value in assessing whether the binding mode is reproduced or not [35,55]. In addition to RMSD, comparison to experimental data is another approach used to validate docking methods. In this work, we have used our protocol [4] to validate the docking methods. Redocking of the co-crystalized ligand gave an RMSD value of 2.5 Å which is an acceptable value, this permitted us to continue with the experiments.
After validating the docking methods and identifying the binding sites of each target, ligand virtual screening was carried out against seven SARS-CoV-2 target proteins viz: 3CLpro, PLpro, Nsp9 RNA, Nucleocapside RNA, ACE2, RdRp and spike RBD-ACE2. The selected target proteins represent two broad categories of therapies, those acting to disrupt the virus represented by the first six targets and those blocking the virus from entering the human cell, represented by the last target. Ligand screening was carried out to identify a drug that can disrupt the virus and those that can prevent the virus from entering the host cell. Our in-house library of natural products were screened against each target protein and molecules with binding energies less than −7.5 kcal/mol were considered to have a higher binding affinity with the target protein and were selected for similarity search.
3.1. Targeting inhibition of 3CLpro
3CLpro represents an important therapeutic target in drug development to fight COVID-19. This protein is responsible for mediation of non-structural proteins (Nsps) required for the virus life cycle. The protein is cleaved from poly-protein to release Nsps4-Nsp16 [11]. Inhibition of this protein is essential in preventing viral replication and is a promising target for SARS-CoV-2. 3CLpro is made up of three domains: domain I is formed by amino acid residues 8–101, domain II formed by amino acid residues 102–184 and domain III formed by amino acid residues 201–303 and a loop which connects domain II and III formed by amino acid residues 185–200 [11,56]. The binding pocket of the 3CLpro is situated between domain I and II and is composed of the catalytic dyad CYS145 and HIST41 [11,56]. In order to gain more insight into protein plasticity and conformation changes at the active site, MD simulation of the free protein was carried out. RMSD and root mean square fluctuation (RMSF) analysis showed that, although the active site is composed of long loops it has few fluctuations (Fig. S2).
Screening of our in-house library of natural products against this target showed that four ligands: 1, 5, 15 and 17 possessed low binding free energy of ≤ −7.5 kcal/mol (Fig. 2 a). However, ligand 1 showed a higher affinity with the binding free energy of −8.2 kcal/mol corresponding to 0.93 μM. The four molecules were advanced to the next filter which employed the RCS. A RCS is an important filter as it represents protein flexibility which is a limitation of many docking algorithms, which does not accommodate protein flexibility. In this filter, ligands were re-ranked, some ligands which ranked high when docked to the crystal structure in the ensemble structure ranked least (Fig. 2a). Based on the ensemble holo structure, three ligands: 1, 5 and 15 were selected for similarity search to identify similar molecules. Using a Tanimoto index of 0.9, three glycosylated flavonoids (Fig. 1b) similar to ligand 15 were established and subjected to RCS docking to evaluate their binding affinity to the 3CLpro (Fig. 2a). All three drugs showed lower binding free energy compared to the first screened natural products. Binding mode analysis (Fig. S3) showed the three drugs bound to 3CLpro in the same fashion by forming at least two hydrogen bonds.
Fig. 2.
(a) Binding free energy of the natural product compounds to 3CLpro (b) Probability distribution of RMSD (p(rmsd)) for both ligand and receptor during the simulation time of 100 ns. (c) RMSF differences of apo and bound 3CLpro. (d–e) Distances between specific atoms in drugs and protein amino acid residues that formed hydrogen bonds with DB12665, (f) Distances between specific atoms in drugs and protein amino acid residues that formed hydrogen bonds with DB01698.
Docking calculation of binding free energies is useful in predicting good and bad binders as well as screening large chemical libraries. However, they cannot provide details on the time-lapse stability and are mostly poor in estimating the binding affinity of the complex. To overcome these limitations, MD simulation was carried out to establish the time-lapse stability of the complex and binding affinity of the ligands which was evaluated using the MM-PBSA method. RMSD, RMSF, distances between drugs and protein, and the number of hydrogen bonds were used to analyse the stability of the complexes. RMSD analysis presented in Figure S4, clearly shows that, the system made by DB12665 exhibited some fluctuation during the first 40 ns and remained stable throughout the simulation time. The system made by DB01698 exhibited fluctuations throughout the simulation time, suggesting unstable interactions. However, the RMSD of DB08995 remained stable though it induced conformational fluctuation to the protein. The RMSD probability distribution for DB01265 exhibited single maximum for both ligand and protein (Fig. 2b) suggesting the presence of stable conformation. However, multiple maxima were observed for the protein and ligand in the system of DB01698 and DB08995, which suggests to have unstable interaction. On a closer look, the RMSD for DB08995 calculated after least square fit showed single peak. This analysis suggests that, although DB08995 induced conformation changes to the protein, it remained stable in its pocket. Atomic residues motion and fluctuations are highly related to protein functions. In order to provide information on how the binding of the drugs brought about dynamic changes and motion which influences their function, RMSF difference between apo and holo structures were analysed and presented in Fig. 2c. The binding of the three drugs clearly shows that they induced different motions to the protein. Interestingly, the binding of the DB08995 induced conformation changes at the regions of residues 100–250, whereas such induced fluctuations are not well pronounced in the complexes of DB01698 and DB12665 except at the region around 160 for DB01698 and 125 and 160 for DB12665. Another residue fluctuations for the later two complexes are observed at the region of 50. Such fluctuations were not observed in the former complex. This clearly shows that the binding of DB08995 induced protein motion and dynamics in a different manner when compared to other complexes which suggest that the drug retained its binding pose and changed protein functions. This analysis further suggests that DB12665 and DB08995 could therefore interfere with coronavirus viral RNA synthesis and replication.
In order to explain the dynamic nature of the ligands inside the active site, hydrogen bonds and their distances were measured over the simulation time. The number of hydrogen bonds (Fig. S6b-d) shows that, DB12665 and DB08995 formed a maximum of 8 and 7 hydrogen bond, throughout the simulation time, respectively. The number of hydrogen bonds for DB01698 decreased to 0 during the 40 ns, which suggests that the drug moved out of the pocket. From 50 ns a maximum of 3 hydrogen bonds were formed. Distances between groups of DB12665 which formed hydrogen bonds with residues of 3CLpro were analysed (Fig. 2d–e). DB12665 exhibited changes in the binding pose from the initial docking pose, and remained stable in the second pose. The system made by DB01698 showed higher fluctuations for both receptor and ligand RMSD suggesting to be unstable. Additional analysis based on the number of hydrogen bonds, distances between protein and ligand atoms and snapshots obtained from the trajectory (Fig. 2f, Fig. S7) indicated that DB01698 moved out the pocket after 40 ns. Further analysis on the distance and binding mode showed that DB08995 retained its binding pose (Fig. S6a).
In order to ascertain the observed stability, binding free energy based on MM-PBSA was calculated to evaluate the affinity of the drugs in their active site. MM-PBSA was calculated on a single trajectory using frames from the first 40 and last 40 ns. Based on the last frames, MM-PBSA analysis showed that complex of DB12665 and DB08995 had higher binding affinity than DB01698. Both van der Waals and electrostatic energy terms highly contributed to the interaction of the protein and drugs (Table 1, Table 2 ).
Table 1.
Binding free energy (kJ/mol) for three complexes during the first 40 ns.
| Energy | Complexes |
||
|---|---|---|---|
| DB1265 | DB01698 | DB0895 | |
| ΔEvdW | −116.9 ± 24.4 | −128.3 ± 31.8 | −172.4 ± 17.6 |
| ΔEele | −111.1 ± 56.2 | −59.3 ± 28.3 | −111.9 ± 31.4 |
| ΔEpolar | 161.6 ± 42.3 | 116.9 ± 32.9 | 157.4 ± 35.5 |
| ΔEnon-polar | −16.6 ± 2.0 | −16.5 ± 2.7 | −19.3 ± 1.2 |
| ΔEbinding | −82.9 ± 24.1 | −87.2 ± 27.1 | −146.3 ± 19.6 |
Table 2.
Binding free energy (kJ/mol) for three complexes for the last 40 ns.
| Complexes |
|||
|---|---|---|---|
| Energy | DB1265 | DB01698 | DB0895 |
| ΔEvdW | −128.2 ± 17.4 | −13.2 ± 23.9 | −164.9 ± 13.2 |
| ΔEele | −71.6 ± 16.7 | −6.1 ± 14.2 | −109.6 ± 31.2 |
| ΔEpolar | 184.5 ± 14.0 | 14.0 ± 28.9 | 165.5 ± 36.2 |
| ΔEnon-polar | −18.6 ± 1.0 | −1.8 ± 4.1 | −18.9 ± 1.3 |
| ΔEbinding | −33.9 ± 17.9 | −7.1 ± 24.3 | −127.9 ± 19.5 |
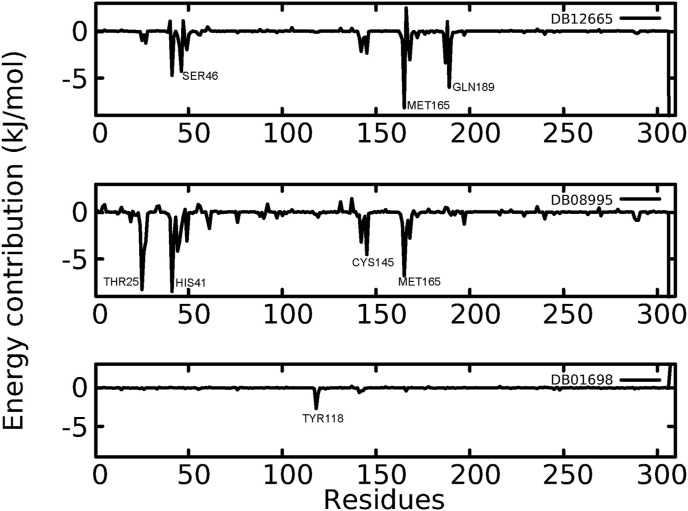
3.1.1. Per-residue energy decomposition analysis
To examine residue contributions to the binding energy of the three ligands, per-residue energy decomposition was performed. The analysis was done using the last 40 ns trajectories and results are presented in Fig. 3 . Interestingly, as observed from the MD simulation and MM-PBSA calculation, the per-residue analysis further showed that during the last 40 ns, residue TYR118 significantly contributed to the interaction with ligand DB01698 having the contribution energy of −3.7 kJ/mol, thus, indicating unstable interaction or weak inhibition of viral replication. On the other hand, a total of 8 residues: THR25 (−8.2 kJ/mol), THR26 (−4.1 kJ/mol), LEU27 (−3.1 kJ/mol), HIS41 (−8.4 kJ/mol), CYS44–4.1 kJ/mol, THR 45 (−3.1 kJ/mol), CYS145 (−4.5 kJ/mol) and MET165 (−6.7 kJ/mol) highly contributed to the interaction with DB08995. It is important to note that DB08995 interacted with the catalytic dyad residues (HIST41 and CYS145) which suggests its strong inhibition. When DB12665 interacted with 3CLpro, a total of 5 residue: SER46 (−4.2 kJ/mol), MET165 (−8.1 kJ/mol), PRO168 (−3.0 kJ/mol), ASP187 (−3.3 kJ/mol) and GLN189 (−5.9 kJ/mol), were found to have highly contributed to the interaction energy. However, interaction between DB08995 and MET165 contributed to lower energy than other residues. Further analysis of per-residue interactions implies that the two drugs could be a potent inhibitor of SARS-CoV-2 targeting viral replication by strongly binding to the catalytic dyad and residues required for substrate binding.
Fig. 3.
Per-residue energy decomposition analysis of the three ligands bound to 3CLpro analysed during the last 40 ns of MD simulation.
3.2. Targeting inhibition of Nsp9 RNA and nucleocapsid RNA
This group of proteins enables the RNA molecules to adapt to their conformational function [57]. Virtual screening was done to identify molecules that can inhibit their functions. Screening inhibitors targeting Nsp9 RNA showed that ligand 7 with binding free energy of −8.3 kcal/mol could potentially inhibit its activity. However, after screening targeting nucleocapsid RNA, all the ligands did not show potential inhibition, except for ligand 7 which showed binding free energy of −7 kcal/mol. It is suggested that ligand 7 could bind selectively to the non-structural proteins RNA as it did not show favourable binding free energy to the main protease (Fig. S5).
3.3. Targeting inhibition of PLpro and RdRp
All the three proteins; 3CLpro, PLpro and RdRp are involved in viral RNA synthesis and replication. PLpro is concerned with cleavages of the N-terminus of the poly-protein replicase to release Nsp1-3 which is required for correcting viral replication [58]. This protein is also responsible for antagonizing hosts innate immune system, [58] making it an important therapeutic target for identification of coronavirus inhibitors. Screening of our in-house natural products and drug obtained from a similarity search showed lower binding energy of −7 kcal/mol (Fig. S8a). The binding free energy suggested that these compounds would not be good inhibitors of PLpro. This motivated us to further investigate other targets responsible for viral RNA synthesis such as RdRp.
RNA-dependent RNA polymerase (RdRp) is an important enzyme of coronavirus responsible for replication and transcription of the virus. The research shows that RdRp is an important target as it is involved in the synthesis of Nsp12-RdRp RNA primer which activates the binding of Nsp12. Thus, targeting RdRp is crucial in the treatment of coronavirus. Virtual screening of natural products from our library and DrugBank showed that drug DB08995 could have potential inhibition with binding free energy −9.1 kcal/mol slightly corresponding to 0.2 μM (Fig. S8a). Natural product (ligand 1) from our library and drug CID442428 also demonstrated inhibition with the binding free energy of −8 and −8.5 kcal/mol, respectively. Binding mode analysis of the crystal structure revealed that DB08995 exhibited multiple binding modes. The binding mode with the lowest binding free energy as shown in Figure S8b did not form any hydrogen bonds with the target protein.
3.3.1. Targeting inhibition of host specific receptor ACE2
Previous studies have shown that enzymatic inhibition of ACE2 has a potential role in treatment of cardiovascular diseases such as hypertension, heart attack, and diabetes, as well as preventing the fusion of corona virus [59,60]. Therefore, we performed screening to target inhibition of ACE2 activity.
Virtual screening targeting inhibition of ACE2 revealed that both natural products from in-house library; ligand 1 and ligand 7, and those from FDA approved drugs; CID442428, DB01698 and DB8995 resulted to protein inhibition with the binding free energy of ≤ −9 kcal/mol (Fig. S8a). When all five compounds were subjected to RCS to evaluate the effects of the protein conformation on the binding energy, it turned out that only DB01698 and DB08995 showed strong inhibition with the binding free energy of −9.3 and −9.4 kcal/mol, respectively (Fig. 4 a). In order to establish the binding stability of DB08995 complex, MD simulation was carried out. RMSD measurement suggested that DB08995 was stable and did not induce conformation to the receptor, although it showed some changes inside the pocked as shown by ligand RMSD (Fig. 4b). Changes in ligand RMSD could be attributed to the large pocket size of the ACE2 receptor. The distance between ligand atoms and receptor residues that formed hydrogen bonds was measured to establish the origin of these changes. As shown in Fig. 4c, the sugar moiety 2 (red colour) exhibited more fluctuation within the pocket and it interacted with residues ASP350, GLU401 and GLU402 between the two helices as shown in Figure S9b. However, the middle ring remained stable with fewer fluctuations within the hydrogen bond distance cut-off. This was further confirmed by the number of hydrogen bonds formed during the simulation time. The number of hydrogen bonds formed is consistent with changes in distances (Fig. S9a).
Fig. 4.
(a) Binding free energies of the six molecules against ACE2. (b) RMSD of ligand and receptor for ACE2 complex. (c) Hydrogen bond distances between specific groups that formed hydrogen bonds in the complex. (d) RCS binding free energy of DB01698 and DB08995 bound to spike RBD-ACE2. (e) RMSD for native and bound spike RBD-ACE2 (e) Hydrogen bonds formed for DB08995-spike RBD-ACE2 complex.
To investigate the stability, binding affinity based on MM-PBSA was calculated and results are presented in Table 3 . Binding free energy suggested that DB08995 strongly inhibited ACE2 activity. Both van der Waals and electrostatics energy terms favourably contributed to the interaction while polar energy terms unfavourably contributed to the interaction. These results suggest that, BD08995 could be potent inhibitor of ACE2 and thus it can prevent coronavirus from entering the host cells.
Table 3.
Binding free energy (kJ/mol).
| Energy | ACE2-DB08995 |
|---|---|
| ΔEvdW | −185.2 ± 24.5 |
| ΔEele | −92.4 ± 42.9 |
| ΔEpolar | 134.8 ± 29.5 |
| ΔEnon-polar | −23.0 ± 1.9 |
| ΔEbinding | −165.9 ± 31.9 |
3.4. Targeting inhibition of spike RBD-ACE2
3.4.1. BD08995 preferentially binds to ACE2 residues at the interface
Many studies have showed that inhibition of the ezymatic activity of ACE2 effectively blocks the interaction of spike SARS-CoV-2 and thus prevents the coronavirus fusion in the human cells [11,[61], [62], [63]]. It has been discovered that the virus spike RBD binds to ACE2 before entering the host [11]. Research also shows that SARS-CoV-2 binds strongly to ACE2 than SARS-CoV [61,64].
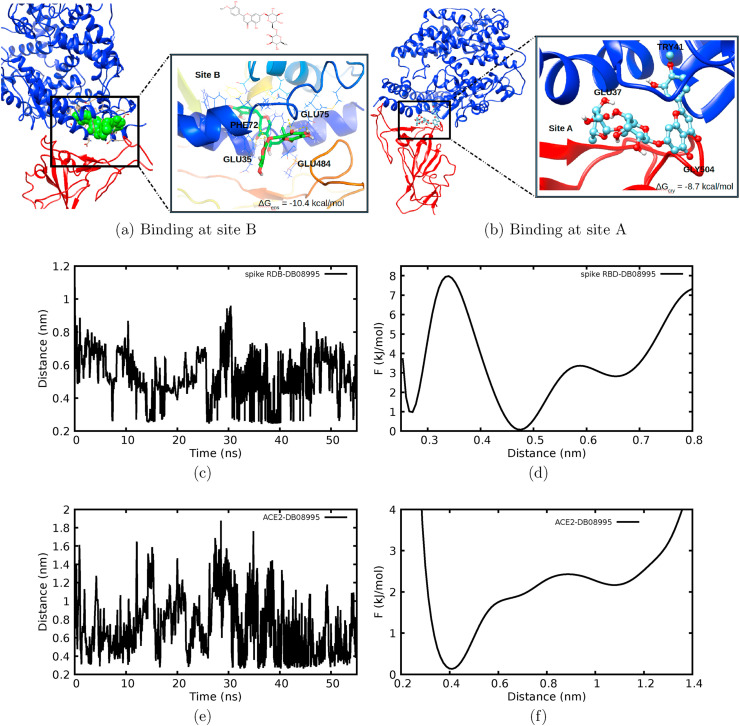
Screening results against the spike RBD-ACE2 showed that two drugs DB01698 and DB08995 could be potent inhibitors. RCS binding free energy showed that the two drugs DB01698 and DB08995 had a mean binding free energy of −9.33 and −9.59 kcal/mol, respectively (Fig. 4d). Interestingly, DB08995 exhibited two binding poses as compared to DB01698 (Fig. 5 a–b) and interacted better with ACE2 residues than DB01698.
Fig. 5.
(a–b) Interaction and binding modes of DB08995 at the spike RBD-ACE2 interface. (c–f) Measured distances between DB08995 with spike RBD-ACE2 with their respective free energies to the right.
3.4.2. Binding of spike RBD to ACE2 does not abrogate the activities of DB08995
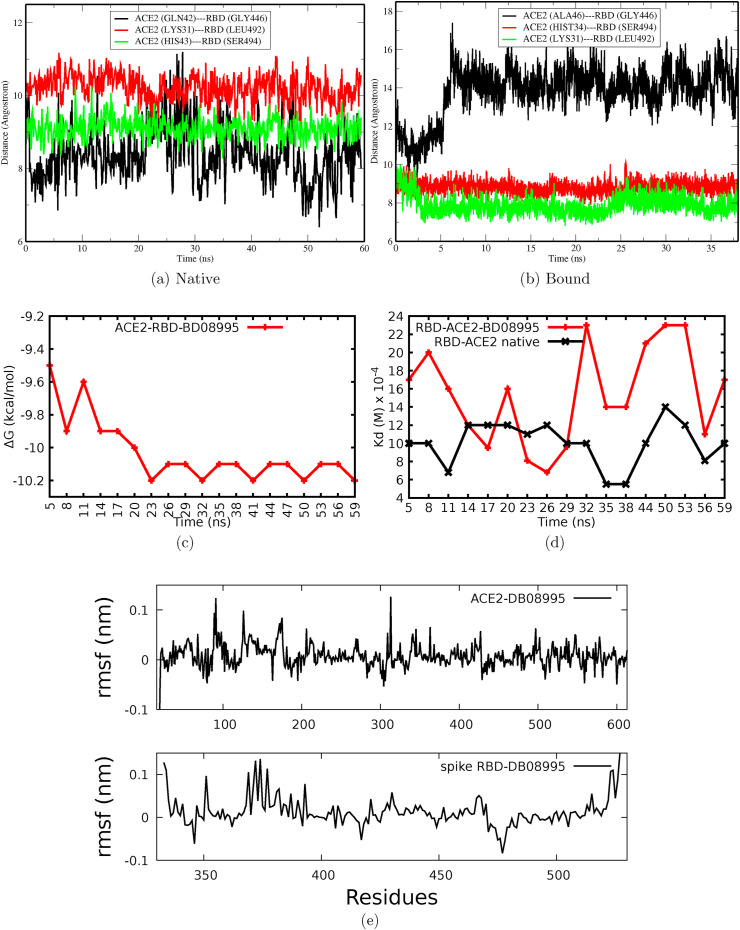
MD simulation was carried out to assess the binding stability of DB08995 and its ability to disrupt the fusion of the coronavirus into the host cells. DB08995 in its binding site B was selected as it showed lower binding free energy. The stability of the complex was assessed by measuring the RMSD, distances between the spike RBD and ACE2, drug and ACE2 hydrogen bonds as well as the binding energy at every 2 ns for the complex.
The measured RMSD for native spike RBD-ACE2 equilibrated to stability within 5 ns with an average RMSD of 0.24 nm. The RMSD further increased up to 0.29 from 25 ns and remained stable until the end of the simulation time. Although the RMSD of the bound spike RBD-ACE2-DB08995 equilibrated quickly, it showed persistent fluctuations until the end of the simulation (Fig. 4e). Comparing RMSD of native and bound spike RDB-ACE2, one can easily observe that DB08995 induced constant fluctuation to the native RBD-ACE2 throughout the simulation time, which suggests that binding of DB08995 could destabilize the spike RBD-ACE2 interaction, and therefore, prevent coronavirus membrane from stable binding and fusion into host cells. Hydrogen bonds formed between RBD-ACE2 and DB08995 are presented in Fig. 4f, which shows that DB08995 exhibited different binding poses. During the first 10 ns the maximum number of hydrogen bonds was 4 and increased up to 9 during 20 ns. The formation of hydrogen bond is important in stabilization of the interaction between the drug and its receptor. The number of hydrogen bonds later decreased to 5 during the end of the simulation time. In order to explain these changes, the distance between the drug and some residue was measured to monitor the stability and changes in binding pose (Fig. 5c–f).
To provide quantitative information, the free energy of the distances was calculated using F = −k B Tln(P(dist)), where F is the free energy, k B is Boltzmann constant, T is temperature and P(dist) is the probability. The free energy showed that DB08995 was bound to spike RBD forming hydrogen bond with residue GLU484, and it exhibited three minima at 0.28, 0.49 and 0.69 nm. Two deep minima at 0.28 and 0.49 nm suggests that, DB08995 bound to GLU484 disrupts its interaction with ACE2. Such inhibition is vital in preventing/altering molecular recognition between spike RBD-ACE. On the other hand, the measured distance between ACE2-DB08995 shows a deep minimum at 0.4 nm which suggests that the drug strongly bound and interacted with ACE2, hence preventing its recognition with the spike RBD.
3.4.3. DB08995 disrupt molecular recognition at the RBD-ACE2 interface
To obtain crucial insight into the disruptive nature of the DB08995 in relation to the spike RBD-ACE2 interface, the distance between RBD and ACE2 was measured for both native and bound protein (Fig. 6 a–b). Binding of DB08995 induced changes in distance between the two proteins. An increase in distance between the measured residues suggests weak interaction induced by the DB08995 at the interface which implies that spike RBD could weakly interact with ACE2 in the presence of DB08995. Time-lapse binding affinity of DB08995 into its cavity and spike RBD-ACE2 dissociation constant were also calculated so as to gain further understanding about the nature of the drug (Fig. 6c–d). The binding affinity of DB08995 in its cavity remained stable. However, the dissociation constant between spike RBD-ACE2 increased upon binding of DB08995 which suggests unstable interaction between spike RBD and ACE2.
Fig. 6.
Interaction between DB08995 with spike RBD-ACE2. (a–b) Distance between RBD and ACE2 for native shown in (a) and bound shown in (b). (c) The binding affinity of DB08995 at the interface at every 3 ns (d) the dissociation constant between RBD and ACE2 for native and bound. (e) RMSF difference between ACE2 and RDB for bound and unbound structures.
The stability of the proteins was further investigated by analysing the atomic residue fluctuations. In addition, the dynamics of the residues were analysed so as to understand how the binding of DB08995 altered the protein fluctuations. In Fig. 6e we present the RMSF of the difference between the native and bound spike RBD-ACE2, which shows that the binding of DB08995 at the interface inhibited residue fluctuations at the region of 404–505 of RBD and induced fluctuations in the region of 370–390. The presence of the spike RBD to the ACE2 further altered atomic fluctuations to the protein. Previously, this region was reported to be involved in spike glycoprotein oligomerization for other coronavirus like murine hepatitis which forms dimeric domain required for receptor binding and entering the cells [65]. These results suggest that DB08995 may change the spike RBD dynamics which influence RBD oligomerization, receptor binding as well as recognition and hence treating the associated disease.
Water is known to play an important role in protein-ligand interaction, protein function and molecular recognition [66]. The role of water in mediating the interaction of drug and protein was also investigated. Figure S9 (left panel) shows that binding of DB08895 into ACE2 and spike RBD-ACE2 (Figure S9 left panel) was further mediated by water. Although water is observed at the interface between spike RBD-ACE2, it is not present in crystal. Such water formed a network of hydrogen bond between DB08995 and the spike RBD. The formed networks were important for DB08995 activity that may facilitate the loss of interaction and molecular recognition between spike RDB and ACE2.
In an effort to identify new drug to fight SARS-CoV-2 which causes COVID-19, a combination of computational methods were applied. Three FDA approved glycosylated flavonoids showed potent inhibition against SARS-CoV-2 targeting different proteins. DB12665 is an investigational drug for treatment of kidney cancer, renal cell carcinoma, advanced renal cell carcinoma, and thromboembolism of vein in pancreatic cancer [36]. This drug has also shown promising antiviral activities against influenza A & B viruses [67] and, therefore, it could be a potential candidate to be repurposed for SARS-CoV-2 targeting viral replication. DB01698 is an approved drug used to decrease capillary fragility [68]. This drug is also reported to have different pharmacological profiles including antiretroviral and antiviral activities [69]. It could also be repurposed against SARS-CoV-2 targeting inhibition of viral fusion into the host cells. DB08995 is an approved drug for venous disease and it also treats various disorders of blood vessels such as hemorrhoids, varicose veins, poor circulation in the legs, and bleeding in the eye or gums. In addition it serves as a food supplement [70]. The present work reveals that DB08995 target SARS-CoV-2 in two different ways; inhibiting viral replication and inhibiting viral fusion into host cells. This drug could further be repurposed for SARS-CoV-2. Recently, DB08995 was reported by other groups to have antiviral properties targeting inhibition on SARS-CoV-2 viral replication in silico [[71], [72], [73]].
3.5. Natural sources of the identified FDA approved drug potential for repurposing
The identified FDA approved drug molecules potential for repurposing are natural products belonging to the group of glycosylated flavonoids. This section provides the sources of these natural products for easy access and remedy against COVID-19. DB08995 is an aglycones flavone derivative [74] mostly found in green tea (Camellia sinensis; Theaceae) and herbal of Rosemary (Rosmarinus officinalis; Lamiaceae) with a concentration < 1–5 mg/serving. DB08995 is also found in juices and orange wines (Citrus sinensis; Rutaceae) in a concentration of 250 mL serving size. It is found in 0.2–0.5 mg/100 g of fresh weight and 1 mg/serving [74]. DB01698 is a citrus glycoside flavonoid found in many plants including green tea, passion flowers, apple and buckwheat [69]. It is an important component of food stuff [69]. DB12665 is highly found in mango (Mangifera indica L.; Anacardiaceae) [75], tea (Camellia sinensis; Theaceae) [76] and in the leaves of custard apple (Annona squamosa L.; Annonaceae) [77]. These computationally identified SARS-CoV-2 inhibitors may serve as cheap alternative remedy to combat COVID-19 once their efficacy is proved since they are plenty and easily available from natural sources.
4. Conclusion
Due to the urgent need to identify drug necessary in fighting COVID-19, we have carried out computational work to quickly identify FDA approved small molecules that could be considered for immediate use. To balance the computational accuracy, a combination of docking screening, molecular dynamics simulation and free energy calculations based on MM-PBSA methods were applied which identified DB12666, DB01698 and DB08995 as potential inhibitors of SARS-CoV-2. The later drug was discovered to work with two distinct mechanisms; targeting the disruption of viral replication/synthesis and inhibition of viral fusion into the host cells. The identified drugs are suggested for quick clinical evaluation to provide an immediate solution against this deadly disease.
Funding
This work was jointly supported by Data-Driven Innovation (DDI) project through CREATES scheme at NM-AIST, Arusha; the University of Dar es Salaam through its Directorate of Research Grant, and the Kenya Education Network (KENET) Mini-Grant 2018/2019.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the Centre for High Performance Computing (CHPC), South Africa, for providing computational resources to this research project. DMS further thanks ICTP for providing access to HPC facility.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2021.107871.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;60(6):3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):1–18. doi: 10.1038/s41421-020-0153-3. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modeling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;105960:2020. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shadrack D.M., Swai H.S., Hassanali A. A computational study on the role of water and conformational fluctuations in Hsp90 in response to inhibitors. J. Mol. Graph. Model. 2020;107510 doi: 10.1016/j.jmgm.2019.107510. 96. [DOI] [PubMed] [Google Scholar]
- 5.Elfiky A.A. Life sciences; 2020. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp): A Molecular Docking Study; p. 117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. Antiviral research; 2020. The FDA-Approved Drug Ivermectin Inhibits the Replication of SARS-CoV-2 in Vitro; p. 104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautret P., Lagier J.-C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Wu D., Koganti R., Lambe U.P., Yadavalli T., Nandi S.S., Shukla D. Vaccines and therapies in development for SARS-CoV-2 infections. J. Clin. Med. 2020;9(6):1885. doi: 10.3390/jcm9061885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosseini F.S., Amanlou M. 2020. Simeprevir, Potential Candidate to Repurpose for Coronavirus Infection: Virtual Screening and Molecular Docking Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyandoro S.S., Nkunya M.H., Cosam J.C., Msoffe P.L. In ovo antiviral potency of the leaf constituents of Tanzanian Toussaintia species against infectious bursal disease virus and newcastle disease virus. Int. J. Biol. Chem. Sci. 2014;8(3):1308–1318. [Google Scholar]
- 13.Nyandoro S.S. In ovo antiviral activity of the constituents of artabotrys monteiroae and artabotrys modestus against infectious bursal disease and newcastle disease viruses. Int. J. Biol. Chem. Sci. 2017;11(6):3075–3085. [Google Scholar]
- 14.Świderek K., Moliner V. Revealing the molecular mechanisms of proteolysis of SARS-CoV-2 M pro by QM/MM computational methods. Chem. Sci. 2020;11(39):10626–10630. doi: 10.1039/d0sc02823a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target, Bioorg. Med. Chem. Lett. 2020;127377:2020. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hummer G. Fast-growth thermodynamic integration: error and efficiency analysis. J. Chem. Phys. 2001;114(17):7330–7337. [Google Scholar]
- 17.Kumari R., Kumar R., Consortium O.S.D.D., Lynn A. g_mmpbsa□ A GROMACS tool for high–throughput MM–PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 18.Shivakumar D., Williams J., Wu Y., Damm W., Shelley J., Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theor. Comput. 2010;6(5):1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- 19.Aqvist J., Marelius J. The linear interaction energy method for predicting ligand binding free energies. Comb. Chem. High Throughput Screen. 2001;4(8):613–626. doi: 10.2174/1386207013330661. [DOI] [PubMed] [Google Scholar]
- 20.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity–a rapid access to atomic charges. Tetrahedron. 1980;36(22):3219–3228. [Google Scholar]
- 21.Rappé A.K., Casewit C.J., Colwell K., Goddard W.A., III, Skiff W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992;114(25):10024–10035. [Google Scholar]
- 22.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminf. 2012;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S., et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X., Zhang B., Jin Z., Yang H., Rao Z. The crystal structure of COVID-19 main protease in complex with an inhibitor N3. Nature. 2020;582(7811):289–293. [Google Scholar]
- 27.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osipiuk J., Jedrzejczak R., Tesar C., Endres M., Stols L., Babnigg G., Kim Y., Michalska K., Joachimiak A. RSCB PDB; 2020. The Crystal Structure of Papain-like Protease of SARS CoV-2. [Google Scholar]
- 29.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10(7):1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan K., Kim Y., Jedrzejczak R., Maltseva N., Endres M., Michalska K., Joachimiak A. RCSB Protein Data Bank; 2020. The Crystal Structure of Nsp9 RNA Binding Protein of SARS CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541) doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baum B., Mohamed M., Zayed M., Gerlach C., Heine A., Hangauer D., Klebe G. More than a simple lipophilic contact: a detailed thermodynamic analysis of nonbasic residues in the S1 pocket of thrombin. J. Mol. Biol. 2009;390(1):56–69. doi: 10.1016/j.jmb.2009.04.051. 2009. [DOI] [PubMed] [Google Scholar]
- 34.Baum B., Muley L., Smolinski M., Heine A., Hangauer D., Klebe G. Non-additivity of functional group contributions in protein–ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry. J. Mol. Biol. 2010;397(4):1042–1054. doi: 10.1016/j.jmb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Brooijmans N. John Wiley and Sons Inc.; New York, NY, USA: 2009. Docking Methods, Ligand Design, and Validating Data Sets in the Structural Genomic Era, Structural Bioinformatics; pp. 635–663. [Google Scholar]
- 36.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zoete V., Daina A., Bovigny C., Michielin O. 2016. SwissSimilarity: a Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening. [DOI] [PubMed] [Google Scholar]
- 39.A. Becke, Density-functional thermochemistry. III. The role of exact exchange (1993) J. Chem. Phys. 98 5648.
- 40.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37(2):785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 41.Frisch A. 25p. gaussian 09W Reference; Wallingford, USA: 2009. [Google Scholar]
- 42.Abraham M., Spoel D., Lindahl E., Hess B., Team T. 2018. GROMACS User Manual Version 2018. [Google Scholar]
- 43.Kaminski G.A., Friesner R.A., Tirado-Rives J., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105(28):6474–6487. [Google Scholar]
- 44.Dodda L.S., Cabeza de Vaca I., Tirado-Rives J., Jorgensen W.L. LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017;45(W1):W331–W336. doi: 10.1093/nar/gkx312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horn H.W., Swope W.C., Pitera J.W., Madura J.D., Dick T.J., Hura G.L., Head-Gordon T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J. Chem. Phys. 2004;120(20):9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 46.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126(1) doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 47.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52(12):7182–7190. [Google Scholar]
- 48.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103(19):8577–8593. [Google Scholar]
- 49.Hess B., Bekker H., Berendsen H.J., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18(12):1463–1472. [Google Scholar]
- 50.Christ C.D., Mark A.E., Van Gunsteren W.F. Basic ingredients of free energy calculations: a review. J. Comput. Chem. 2010;31(8):1569–1582. doi: 10.1002/jcc.21450. [DOI] [PubMed] [Google Scholar]
- 51.DeLano W.L., et al. Pymol: an open-source molecular graphics tool. CCP4 Newslett. Protein Crystallogr. 2002;40(1):82–92. [Google Scholar]
- 52.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W., Dalke A., Schulten K., et al. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 54.David L., Nielsen P.A., Hedstrom M., Norden B. Scope and limitation of ligand docking: methods, scoring functions and protein targets. Curr. Comput. Aided Drug Des. 2005;1(3):275–306. [Google Scholar]
- 55.Hevener K.E., Zhao W., Ball D.M., Babaoglu K., Qi J., White S.W., Lee R.E. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009;49(2):444–460. doi: 10.1021/ci800293n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., et al. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug Carmofur. Nat. Struct. Mol. Biol. 2020:1–4. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 57.Zúñiga S., Sola I., Moreno J.L., Sabella P., Plana-Durán J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357(2):215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan L., Chen Z., Song S., Wang S., Tian C., Xing G., Chen X., Xiao Z.-X., He F., Zhang L. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290(5):3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreutz R., Algharably E.A.E.-H., Azizi M., Dobrowolski P., Guzik T., Januszewicz A., Persu A., Prejbisz A., Riemer T.G., Wang J.-G., et al. Hypertension, the renin–angiotensin system, and the risk of lower respiratory tract infections and lung injury: implications for COVID-19European Society of Hypertension COVID-19 Task Force Review of Evidence. Cardiovasc. Res. 2020;116(10):1688–1699. doi: 10.1093/cvr/cvaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huentelman M.J., Zubcevic J., Hernandez Prada J.A., Xiao X., Dimitrov D.S., Raizada M.K., Ostrov D.A. Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension. 2004;44(6):903–906. doi: 10.1161/01.HYP.0000146120.29648.36. [DOI] [PubMed] [Google Scholar]
- 61.Nami B., Ghanaeian A., Ghanaeian K., Nami N. 2020. The Effect of ACE2 Inhibitor MLN-4760 on the Interaction of SARS-CoV-2 Spike Protein with Human ACE2: a Molecular Dynamics Study. [Google Scholar]
- 62.Zhang G., Pomplun S., Loftis A.R., Loas A., Pentelute B.L. The first-in-class peptide binder to the SARS-CoV-2 spike protein. bioRxiv. 2020 doi: 10.1101/2020.03.19.999318. [DOI] [Google Scholar]
- 63.Huang X., Pearce R., Zhang Y. bioRxiv; 2020. Computational Design of Peptides to Block Binding of the SARS-CoV-2 Spike Protein to Human ACE2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y., Guo Y., Pan Y., Zhao Z.J. 2020. Structure Analysis of the Receptor Binding of 2019-nCoV, Biochemical and Biophysical Research Communications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao X., Feng Y., Chakraborti S., Dimitrov D.S. Oligomerization of the SARS-CoV S glycoprotein: dimerization of the N-terminus and trimerization of the ectodomain. Biochem. Biophys. Res. Commun. 2004;322(1):93–99. doi: 10.1016/j.bbrc.2004.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teng D., Chen J., Li D., Wu Z., Li W., Tang Y., Liu G. Computational insights into molecular activation and positive cooperative mechanisms of FFAR1 modulators. J. Chem. Inf. Model. 2020;60(6):3214–3230. doi: 10.1021/acs.jcim.0c00030. [DOI] [PubMed] [Google Scholar]
- 67.Kim Y., Narayanan S., Chang K.-O. Inhibition of influenza virus replication by plant-derived isoquercetin. Antivir. Res. 2010;88(2):227–235. doi: 10.1016/j.antiviral.2010.08.016. 2010. [DOI] [PubMed] [Google Scholar]
- 68.Shanno R.L. Rutin; a new drug for the treatment of increased capillary fragility. Am. J. Med. Sci. 1946;211:539. [PubMed] [Google Scholar]
- 69.Ganeshpurkar A., Saluja A.K. The pharmacological potential of rutin. Saudi Pharmaceut. J. 2017;25(2):149–164. doi: 10.1016/j.jsps.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Staniewska A. 2016. Safety of Treatment and Dietary Supplementation with dioSmin in Daily doSeS up to 2000 Mg a Day. [Google Scholar]
- 71.Chen Y.W., Yiu C.-P.B., Wong K.-Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research. 2020;9 doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peterson L. 2020. COVID-19 and Flavonoids: in Silico Molecular Dynamics Docking to the Active Catalytic Site of SARS-CoV and SARS-CoV-2 Main Protease. Available at: SSRN 3599426. [Google Scholar]
- 73.Peterson L. 2020. In Silico Molecular Dynamics Docking of Drugs to the Inhibitory Active Site of SARS-CoV-2 Protease and Their Predicted Toxicology and ADME. Available at: SSRN 3580951. [Google Scholar]
- 74.Hostetler G.L., Ralston R.A., Schwartz S.J. Flavones: food sources, bioavailability, metabolism, and bioactivity. Adv. Nutr. 2017;8(3):423–435. doi: 10.3945/an.116.012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh U., Singh D., Singh M., Maurya S., Srivastava J., Singh R., Singh S. Characterization of phenolic compounds in some Indian mango cultivars. Int. J. Food Sci. Nutr. 2004;55(2):163–169. doi: 10.1080/09637480410001666441. [DOI] [PubMed] [Google Scholar]
- 76.Sakakibara H., Honda Y., Nakagawa S., Ashida H., Kanazawa K. Simultaneous determination of all polyphenols in vegetables, fruits, and teas. J. Agric. Food Chem. 2003;51(3):571–581. doi: 10.1021/jf020926l. [DOI] [PubMed] [Google Scholar]
- 77.Panda S., Kar A. Antidiabetic and antioxidative effects of Annona squamosa leaves are possibly mediated through quercetin-3-O-glucoside. Biofactors. 2007;31(3–4):201–210. doi: 10.1002/biof.5520310307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.