Abstract
Passive transfer of convalescent plasma or serum is a time-honored strategy for treating infectious diseases. Human convalescent plasma containing antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently being used to treat patients with coronavirus disease 2019 where clinical efficacy trials are ongoing. Here, we assess therapeutic passive transfer in groups of SARS-CoV-2-infected African green monkeys with convalescent sera containing either high or low anti-SARS-CoV-2 neutralizing antibody titers. Differences in viral load and pathology are minimal between monkeys that receive the lower titer convalescent sera and untreated controls. However, lower levels of SARS-CoV-2 in respiratory compartments, reduced severity of virus-associated lung pathology, and reductions in coagulopathy and inflammatory processes are observed in monkeys that receive high titer sera versus untreated controls. Our data indicate that convalescent plasma therapy in humans may be an effective strategy provided that donor sera contain high anti-SARS-CoV-2 neutralizing titers given in early stages of the disease.
Keywords: SARS-CoV-2, coronavirus, COVID-19, nonhuman primate, viral pneumonia, convalescent serum, antibodies
Graphical abstract
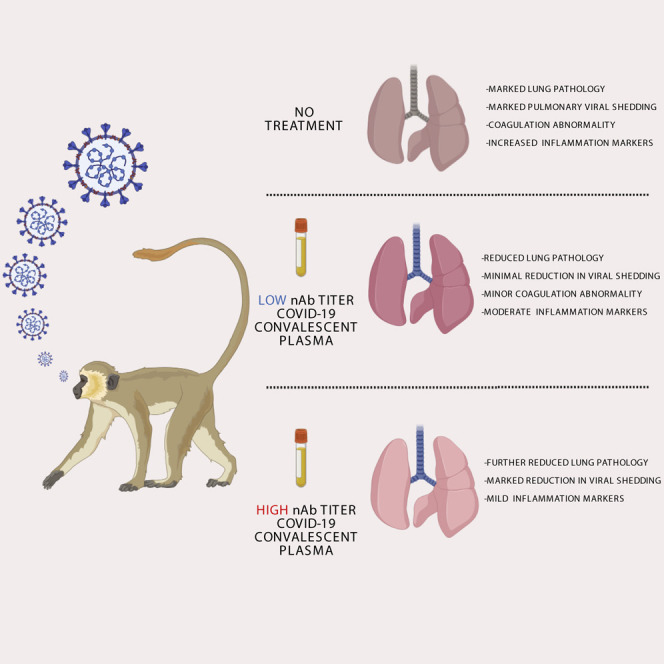
Rapid development of postexposure therapies is necessary for emerging viral threats such as SARS-CoV-2. Cross et al. demonstrate therapeutic reduction of disease severity using convalescent sera with high concentrations of SARS-CoV-2 neutralizing antibodies in the African green monkey model of COVID-19, suggesting this as a possible treatment approach in humans.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can cause a severe, potentially life-threatening viral pneumonia named coronavirus disease 2019 (COVID-19) (World Health Organization, 2020). The COVID-19 pandemic originated in Wuhan, China, and spread across the globe at an explosive rate, leading to over 21 million cases and hundreds of thousands of deaths in just over 6 months’ time. While no currently approved vaccines or therapeutics exist for COVID-19, a battery of medical countermeasures are being developed and assessed in human clinical trials at record speed (Beigel et al., 2020; Jackson et al., 2020; Zhu et al., 2020). Nonetheless, safety and efficacy trials can take considerable time to produce confidence before regulatory approvals are obtained, which creates an immediate need for therapeutic options that may be more accessible—ideally, options with track records of success against related viruses. Transfusion of convalescent blood products has been used in clinical settings for >100 years (Casadevall et al., 2004), including for the treatment of emerging viruses such as Ebola, influenza, and other viruses (Mair-Jenkins et al., 2015).
Immunity to the closely related severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) is understood to be owed, in part, to the development of potent neutralizing antibody responses (Sariol and Perlman, 2020). Indeed, one of the first approaches to treat humans for acute cases that were otherwise unresponsive to standard respiratory virus treatment protocols was the administration of convalescent plasma (CP) from SARS-CoV (Yeh et al., 2005) or MERS-CoV patients (Arabi et al., 2015). Recently, the World Health Organization approved a standardized protocol for the use of CP for the treatment of MERS-CoV (Arabi et al., 2015), yet there is still debate on the feasibility of CP use in the treatment of MERS, as donor antibody titers tend to be too low to produce therapeutic effect (Corti et al., 2016). Nonetheless, with ever-increasing caseloads and no other immediately available options, reports of the use of COVID-19 convalescent plasma (CCP) for the treatment of patients with severe COVID-19 have surfaced, which suggests therapeutic benefit, even when given in some cases of severe disease (Klassen et al., 2020a; Shen et al., 2020). Building on these successes, large-scale clinical trials have been initiated in association with nationwide donation programs (CCPP19, 2020; Malani et al., 2020). In response to the worsening public health crisis, the U.S. Food and Drug Administration issued a federal emergency use authorization (EUA) for the use of CCP on August 23, 2020, to facilitate access to the treatment approach, despite ongoing clinical trials to fully evaluate safety and efficacy (US Food and Drug Administration, 2020).
In parallel with reports of success in humans, hamsters were recently used to experimentally demonstrate the potential of CCP to treat SARS-CoV-2 infection. While informative to demonstrate proof of concept, limited reagents are available to succinctly describe the host responses to infection and treatment in hamsters (Chan et al., 2020; Imai et al., 2020). Non-human primates (NHPs) have long been used to model pathological responses to infection due to their physiological similarity to humans and abundance of cross-reactive reagents, which allow for more detailed analysis than possible in lower vertebrates. A natural extension to this utility is their value in determining predictive efficacy of medical countermeasures such as vaccines or therapies in humans. Several groups have recently described infection with SARS-CoV-2 in a number of NHP species, including rhesus macaques, cynomolgus macaques, baboons, and marmosets (Lu et al., 2020; Rockx et al., 2020; Singh et al., 2020), none of which elicit overt clinical signs of disease reflecting the human condition, making evaluation of therapeutic approaches in these species challenging. Recently, we described a novel African green monkey (AGM) model that recapitulates many of the most salient features of human disease, including severe viral pneumonia, transient coagulopathy, and a prolonged state of recovery (Cross et al., 2020; Woolsey et al., 2021). In this study, we challenged AGMs with SARS-CoV-2 and subsequently treated the animals with different pools of sera derived from AGMs previously infected with the virus. We provide direct evidence for the importance of neutralization potency on the efficacy of convalescent sera to reduce viral burden in the respiratory tract and to reduce systemic and localized evidence of COVID-19 in AGMs.
Results
SARS-CoV-2 infection of AGMs and passive treatment with convalescent serum
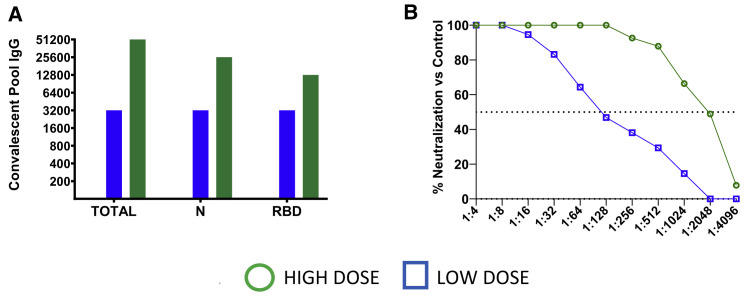
Ten healthy, SARS-CoV-2-naive AGMs were randomized into two treatment cohorts (n = 4 each) and an untreated control cohort (n = 2). All animals were challenged with a target dose of 5.0 × 105 plaque-forming units (PFUs) of SARS-CoV-2 (SARS-CoV-2/INMI1-Isolate/2020/Italy) via combined intranasal (i.n.) and intratracheal (i.t.) inoculation. Ten hours post-challenge, the experimental cohorts were treated intravenously (i.v.) with pooled convalescent sera (6.1 mL/kg) obtained from animals infected with the homologous isolate of SARS-CoV-2 in previous studies (Cross et al., 2020; Woolsey et al., 2021). Animals in one experimental cohort received pooled sera from a group of three AGMs back-challenged 35 days after primary challenge and then euthanized at the scheduled study endpoint 22 days after the back-challenge. To determine the SARS-CoV-2 binding and neutralizing potential of the treatment in vivo, sera from the convalescent sera-treated AGMs were fractionated from whole blood collected 2 and 5 days post-infection (dpi) and assessed for binding by ELISA and neutralizing activity by calculating the dilution of serum that reduced 50% of plaques (PRNT50) (Figures 1A and 1B). The binding titers were 1:51,200 (total virus), 1:25,600 (anti-nucleoprotein), and 1:12,800 (anti-spike receptor binding domain [RBD]), which corresponded to a PRNT50 in pooled sera of ∼1:2,048 for this cohort (designated “high dose”; HD). The second experimental cohort received pooled sera from a group of three AGMs euthanized at the scheduled study endpoint 34 days after challenge with SARS-CoV-2 (Cross et al., 2020); the binding titer was 1:3,200 (total virus, anti-nucleoprotein, and anti-spike RBD), which corresponded to a PRNT50 for pooled sera of ∼1:128 (designated as “low dose”; LD). Animals were longitudinally monitored for clinical signs of illness and euthanized at 5 dpi.
Figure 1.
Characterization of convalescent COVID-19 plasma pools from SARS-CoV-2-infected AGMs
(A) Pooled convalescent plasma was assayed by ELISA for antibodies reactive to SARS-CoV-2 cellular lysates (Total), nucleoprotein (N), or spike protein receptor binding domain (RBD).
(B) PRNT50 assays were performed on pooled convalescent sera from AGMs challenged with the homologous isolate of SARS-CoV-2 in previous studies (Cross et al., 2020; Woolsey et al., 2021).
Clinical disease
In agreement with previous reports (Cross et al., 2020; Hartman et al., 2020; Johnston et al., 2020; Speranza et al., 2020; Woolsey et al., 2021), SARS-CoV-2-infected AGMs in this study showed mild clinical illness (Table 1 ). Shifts in leukocyte populations as compared to pre-challenge baseline counts—specifically, lymphocytopenia, generalized granulocytopenia (neutropenia, eosinopenia, and/or basopenia), and mild to moderate thrombocytopenia—were common to most animals, regardless of cohort, starting approximately 2 dpi (Table 1). Four animals (HD-AGM-3, LD-AGM-3, LD-AGM-4, and C-AGM-2) experienced monocytosis beginning 2–4 dpi; in two of these animals (HD-AGM-3 and LD-AGM-4), this coincided with generalized granulocytosis (neutrophilia, eosinophilia, and/or basophilia). Prothrombin time (PT) was largely unaffected, yet a significantly prolonged coagulation time was noted at 4 dpi for activated partial thromboplastin time (aPTT) in HD versus control animals (p = 0.0459, two-way ANOVA with Tukey’s multiple comparison), decreases in the levels of thrombocytes were more notable in control animals, and increases in circulating fibrinogen were generally more pronounced in the control group compared to either of the experimental groups (Figure S1). These results suggest that while all animals appear to have experienced varying degrees of coagulopathy, treatment with sera with a higher SARS-CoV-2 neutralizing capacity may have partially ameliorated disease in the HD group. However, differences in these parameters were not statistically significant for most time points, because of the small cohort sizes and individual animal variability. Serum markers of renal and/or hepatic function (creatinine [CRE], alanine aminotransferase [ALT], and aspartate aminotransferase [AST]) were mildly elevated in most animals from treatment groups as well as the control group, while C-reactive protein (CRP), a marker of acute systemic inflammation, was mild to moderately (1- to 10-fold over baseline) elevated 4 5 dpi in all but a single animal (LD-AGM-2).
Table 1.
Clinical description and outcome of African green monkeys after SARS-CoV-2 challenge
| Subject no. | Sex | Clinical illness | Clinical pathology |
|---|---|---|---|
| HD-AGM-1 | M | decreased appetite (d4); subject survived to study endpoint (d5) | lymphocytopenia (d2, 3); eosinopenia (d2, 3); basopenia (d2, 3); hypoglycemia (d3, 4); ↑ CRP (d4) |
| HD-AGM-2 | F | none; subject survived to study endpoint (d5) | monocytopenia (d2); eosinopenia (d2, 3); basopenia (d2–4); mild thrombocytopenia (d4); hypoglycemia (d2–5); ↑ CRE (d3, 4); ↑ AST (d3); ↑ CRP (d4) |
| HD-AGM-3 | F | none; subject survived to study endpoint (d5) | monocytosis (d3); neutrophilia (d2, 4, 5); eosinophilia (d2–5); basophilia (d2–5); hypoglycemia (d5); ↑ CRE (d4); ↑ CRP (d4, 5) |
| HD-AGM-4 | M | none; subject survived to study endpoint (d5) | eosinopenia (d2, 3); basopenia (d2, 3); ↑ ALT (d4, 5); ↑ AST (d3–5); ↑ CRP (d4) |
| LD-AGM-1 | M | decreased appetite (4, 5); subject survived to study endpoint (d5) | eosinopenia (d2,3); basopenia (d3); thrombocytopenia (d3, 5); ↑ ALT (d4, 5); ↑ AST (d2–5); ↑ CRP (d4) |
| LD-AGM-2 | F | decreased appetite (d1–5); subject survived to study endpoint (d5) | lymphocytopenia (d5); neutropenia (d2–5); eosinopenia (d2–5); basopenia (d2–5); ↑ ALT (d4, 5); ↑ AST (d3–5) |
| LD-AGM-3 | F | none; subject survived to study endpoint (d5) | lymphocytopenia (d2); neutropenia (d2); eosinopenia (d2–4); basopenia (d2–4); mild thrombocytopenia (d4); monocytosis (d3); ↑ ALT (d3, 4); ↑ AST (d3, 4); hypoamylasemia (d2); ↑↑ CRP (d4); ↑ CRP (d5) |
| LD-AGM-4 | M | tachypnea (d5); subject survived to study endpoint (d5) | monocytosis (d4, 5); neutrophilia (d4); eosinophilia (d4); basophilia (d4); mild thrombocytopenia (d4, 5); ↑ ALT (d4); ↑ AST (d3, 4); ↑↑ CRP (d4) |
| C-AGM-1 | M | none; subject survived to study endpoint (d5) | neutrophilia (d4); eosinophilia (d4); basophilia (d4); mild thrombocytopenia (d5); ↑ ALT (d3); ↑ AST (d3, 5); ↑ CRP (d4, 5) |
| C-AGM-2 | M | none; subject survived to study endpoint (d5) | monocytosis (d2, 4); neutropenia (d4); eosinopenia (d2, 4); basopenia (d2–5); thrombocytopenia (d3); mild thrombocytopenia (d4); ↑ CRE (d4); ↑ AST (d3–5); ↑ CRP (d4, 5) |
Days after the SARS-CoV-2 challenge are in parentheses. All reported findings are in comparison to baseline (d0) values. Decreased appetite is defined as some food but not all food consumed from the previous day. Anorexia is defined as no food consumed from the previous day. Lymphocytopenia, monocytopenia, erythrocytopenia, thrombocytopenia, neutropenia, eosinopenia, and basopenia are defined by a ≥35% drop in numbers of lymphocytes, monocytes, erythrocytes, platelets, neutrophils, eosinophils, and basophils, respectively. Thrombocytopenia is defined as mild if the drop in number of platelets is 25%–34.9%. Lymphocytosis, monocytosis, neutrophilia, eosinophilia, and basophilia are defined by a 100% or greater increase in numbers of lymphocytes, monocytes, neutrophils, eosinophils, or basophils, respectively. Hyperglycemia is defined as a 100% or greater increase in levels of glucose. Hypoglycemia is defined by a ≥25% decrease in levels of glucose. Hypoalbuminemia is defined by a ≥25% decrease in levels of albumin. Hypoproteinemia is defined by a ≥25% decrease in levels of total protein. Hypoamylasemia is defined by a ≥25% decrease in levels of serum amylase. Hypocalcemia is defined by a ≥25% decrease in levels of serum calcium. Hypercapnia was defined as having a partial CO2 > 4 mmHg over d0 baseline values. Increases in ALT, AST, ALP, CRE, CRP, Hct, and Hgb were graded on the following scale: ↑, 1- to 5-fold; ↑↑, >5- to 10-fold; ↑↑↑, >10- to 20-fold; ↑↑↑↑, >20-fold. HD, high dose; LD, low dose; AGM, African green monkey; d, day; M, male; F, female; ALT, alanine aminotransferase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; CRE, creatinine; CRP, C-reactive protein; Hct, hematocrit; Hgb, hemoglobin.
Assessment of antibody levels
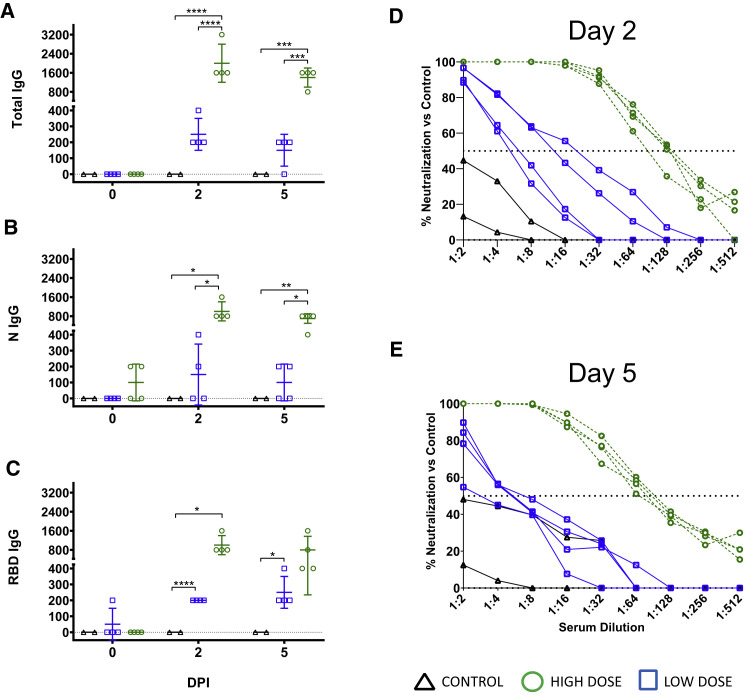
Circulating SARS-CoV-2-specific antibodies to total virus, nucleoprotein, and the RBD of the spike protein were higher in all animals at 2 dpi of the HD group compared to the LD group and untreated subjects, but by day 5, spike RBD titers in 2/4 HD animals began to decline slightly (Figures 2 A–2C). Three of four animals in the HD-treated group had mean PRNT50 titers of ∼1:128 at 2 dpi, while the fourth (HD-AGM-1) had a neutralizing titer between 1:64 and 1:128 (Figure 2D). Neutralizing titers in AGMs that have survived experimental SARS-CoV-2 challenge have previously been demonstrated to be as low as 1:16, suggesting that low neutralizing titers may be negligible when considered as a part of the entire immune response (Cross et al., 2020; Woolsey et al., 2021). Neutralizing antibody titers were markedly lower in LD-treated animals, and a nearly complete lack of neutralizing activity was observed in the untreated control animals at 2 dpi. At 5 dpi, neutralizing antibody titers waned to ∼1:64 in HD-treated animals and between 1:4 and 1:8 in LD-treated animals. A single control animal (C-AGM-1) had similar neutralizing activity at this time point (Figure 2E). The rapid decrease in circulating neutralizing antibody titers by 5 dpi and the lack of robust neutralization in the control and LD-treated animals indicated that much of the neutralizing activity in the HD-treated group was acquired through treatment.
Figure 2.
Serology of SARS-CoV-2-challenged AGMs treated with convalescent sera
(A–C) ELISA binding titers of sera collected on the indicated time points from AGMs enrolled in the present study after challenge with SARS-CoV-2 and passive transfer of convalescent sera for immunoglobulin G (IgG) against SARS-CoV-2 virus cellular lysates. Total (A), nucleoprotein (N) (B), and spike protein receptor binding domain (RBD) (C). Mean ± SD is indicated (bars).
(D and E) PRNT50 assays were performed on pooled convalescent sera from AGMs challenged with the homologous isolate of SARS-CoV-2 in previous studies (Cross et al., 2020; Woolsey et al., 2021) compared with control animals on day 2 post-infection (D) and day 5 post-infection (E). Data shown indicate the percent reduction in SARS-CoV-2 plaque counts (mean of duplicate wells) compared to a control plate (no sera).
Assessment of viral load
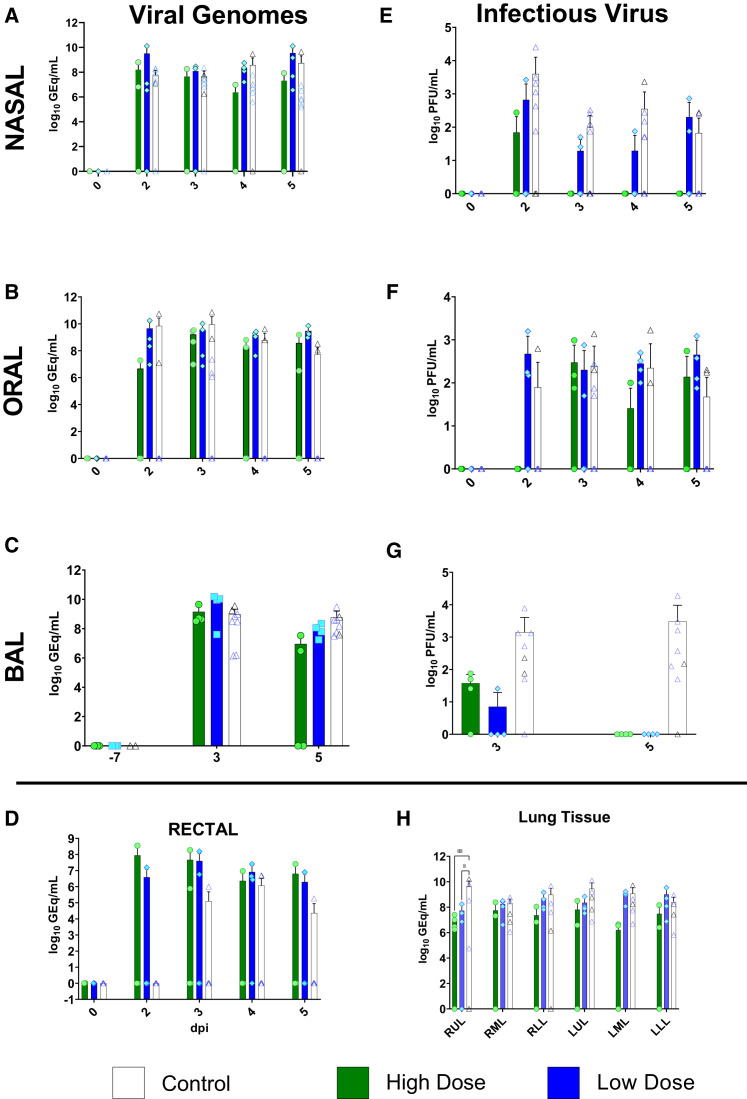
We next assessed viral load in whole blood and mucosal swabs on 0, 2, 3, 4, and 5 dpi; bronchoalveolar lavage (BAL) fluid on −7, 3, and 5 dpi; and lung homogenates at the study endpoint (5 dpi) by both qRT-PCR detection of viral RNA (vRNA) and plaque titration of infectious virus. As previously reported (Cross et al., 2020; Woolsey et al., 2021), there was no circulating SARS-CoV-2 detected in the peripheral blood, as assessed by qRT-PCR of whole blood and plaque titration of the plasma fraction, respectively (data not shown). vRNA was detected in nasal swabs from three animals from the HD-treated group, four animals from the LD-treated group, and all animals from the control group, including 6/6 historical controls (Figure 3 A). Notably, in two animals from the HD-treated group, detection of vRNA was limited to a single day (2 dpi in HD-AGM-2 and 5 dpi in HD-AGM-3). A low amount (∼2.4 log10 PFUs/mL) of infectious SARS-CoV-2 was recovered from nasal mucosa in a single animal from the HD-treated group 2 dpi, while similar amounts of virus were detected on multiple time points for most animals from both the LD-treated and control groups (Figure 3E). Interestingly, while levels of vRNA and infectious virus from the oral mucosa were generally lower and less frequently detected in HD-treated animals, compared to those in LD-treated animals and the two control animals from this study, both vRNA and infectious virus were detected from only 3/6 animals from historical controls, despite animals from both studies being challenged with identical virus stock via identical challenge route (Figures 3B and 3F). Detection of vRNA from the rectal mucosa was similarly variable, present in samples from only 2/4 animals from the HD-treated group, 3/4 from the LD-treated group, and only two control animals (C-AGM-2 and a single HC-AGM), while infectious virus was not recovered from the rectal swabs of any animal (Figure 3D; data not shown). Strikingly, while SARS-CoV-2 vRNA was detected at similar levels in the BAL fluid from all animals at 3 dpi, only 2/4 animals in the HD-treated group had detectable vRNA levels at 5 dpi, compared to all of the animals in both the LD-treated and control groups (Figure 3C), while infectious virus was not recoverable at all from the BAL fluid of both treated groups on day 5. Conversely, infectious virus was detected from both controls from this study and all historical controls (Figure 3G). Genomic vRNA was detected in similar quantities from the lower, middle, and upper sections of the lungs from all animals in all groups (Figure 3H); however, only low levels of infectious virus (approximately 1.5 log PFUs per gram of tissue) were recovered from a single lung lobe from each of the two study control animals and only one of the HD animals (data not shown). We do not have a direct explanation for the lack of detection of infectious virus in the tissue, but given the presence of ongoing inflammatory processes, it is possible that contributions of ongoing antiviral processes (e.g., interferon pathway-related proteins, mucins, responding immune cell activity) present in the tissues at the time of sampling may have played a role. While comparisons of viral load in mucosal compartments failed to reach statistical significance, taken together, these data suggest that treatment with convalescent sera may decrease viral replication and shedding and, thus, mitigate disease and transmission.
Figure 3.
Viral load in mucosal swabs, BAL fluid, and lung tissue from SARS-CoV-2-infected AGMs
SARS-CoV-2 viral load was assessed by plaque titration (B, D, and F) and/or qRT-PCR (A, C, E, G, and H) from mucosal swabs and BAL fluid collected at the indicated time points, and lung tissue harvested at necropsy (5 dpi). For all panels, individual data points represent the mean of two technical replicates from a single assay. Dashed horizontal lines indicate the limit of detection (LOD) for the assay (1,000 genome equivalents [GEq]/mL for qRT-PCR; 25 PFUs/mL for plaque titration). Bars indicate the mean for each cohort at the indicated time point, and values for individual animals within each cohort are indicated as color-coded symbols. Historical control animals (n = 6) from a previous study (Woolsey et al., 2021) utilizing the homologous virus are included for statistical purposes. Error bars indicate the upper bound SD. To fit on a log scale axis, zero values (below LOD) are plotted as “1” (10°); however, statistical comparisons were performed using the original zero values. Statistical significance was assessed by two-way ANOVA with the Geisser-Greenhouse correction without the assumption of sphericity, except for (E), where mixed-effects modeling was used to account for missing historical control values at −7 dpi, and (F), where no correction for sphericity was necessary as only two time points were being compared. Tukey’s post hoc test for multiple comparisons was used to assess differences between cohorts at matched time points. Statistically significant comparisons are indicated by asterisks: (∗0.05 ≤ p ≤ 0.01; ∗∗0.01 ≤ p ≤ 0.001).
Comparative pathology
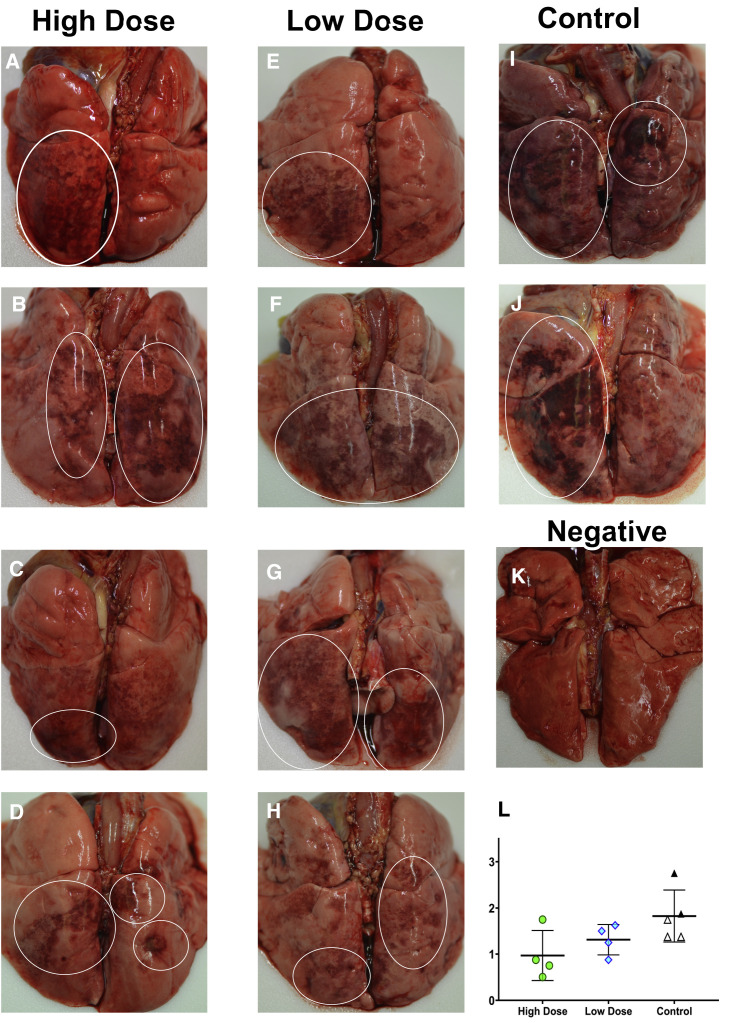
At 5 dpi, all animals were euthanized to gauge viral burden and determine pathological changes in the lungs associated with the treated infection. Multifocal pulmonary consolidation with hyperemia and hemorrhage were noted in all AGMs at 5 dpi. In all AGMs, the most severe lesions were located in the dorsal aspects of the lower lung lobes (Figure 4 ). A board-certified veterinary pathologist approximated gross lesion severity for each lung lobe. Gross lung scores were most severe in untreated control AGMs, followed by the LD-treated animals, and least severe among the HD-treated AGMs (Figure 4L). Mild lymphoid enlargement was noted in one untreated control AGM and two LD-treated AGMs.
Figure 4.
Dorsal view of gross pulmonary lesions in AGMs infected with SARS-CoV-2 with and without treatment
(A–D) Lungs from AGMs treated with high neutralizing antibody titers—HD-AGM-1 (A), HD-AGM-2 (B), HD-AGM-3 (C), and HD-AGM-4 (D)—fail to collapse and present with mild locally extensive pulmonary consolidation with hyperemia and hemorrhage (circled regions).
(E–H) Lungs from AGMs treated with low neutralizing antibody titers—LD-AGM-1 (E), LD-AGM-2 (F), LD-AGM-3 (G), and LD-AGM-4 (H)—fail to collapse and present with moderate locally extensive pulmonary consolidation with hyperemia and hemorrhage (circled regions).
(I and J) Lungs from positive control AGMs C-AGM-1 (I) and C-AGM-2 (J) euthanized 5 dpi after infection with SARS-CoV-2 fail to collapse and present with marked locally extensive pulmonary consolidation with hyperemia and hemorrhage (circled regions).
(K) Dorsal view of control lungs with no significant lesions from SARS-CoV-2-negative AGMs.
(L) Gross examination severity scores by group.
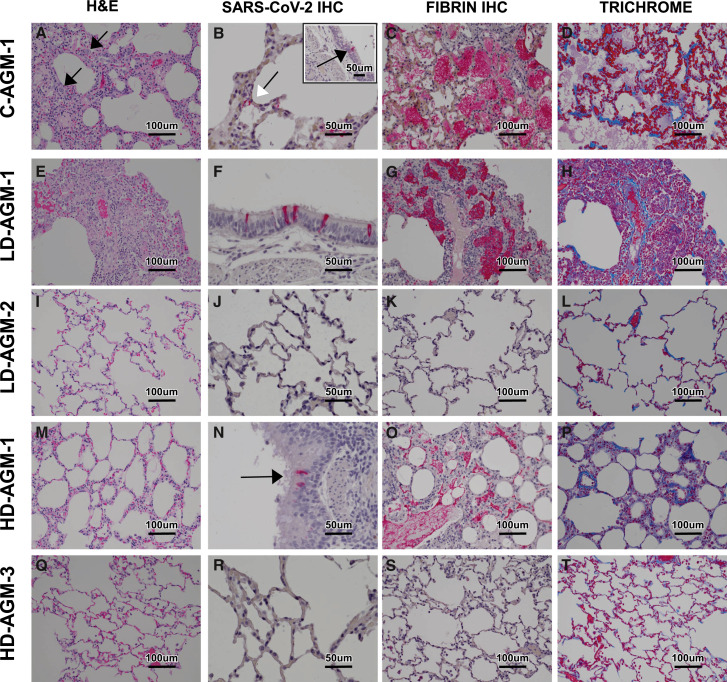
Histologically, the untreated control AGMs euthanized at 5 dpi developed interstitial pneumonia and multifocal alveolar flooding with edema, fibrin, red blood cells, and mixed inflammatory cells, as previously observed (Cross et al., 2020; Woolsey et al., 2021). The interstitial pneumonia was characterized by multifocal moderate expansion of alveolar septae with macrophages, lymphocytes, and fewer numbers of neutrophils (Figure 5 A). Bronchial respiratory epithelium was multifocally ulcerated with associated underlying acute inflammation. Modest expansion of alveolar septae with collagen was noted multifocally (Figure 5D). Colocalization of SARS-CoV-2 antigen with pulmonary lesions was demonstrated with antibodies against SARS-CoV-2 N protein. Positive immunohistochemical (IHC) labeling was noted diffusely within the cytoplasm of respiratory epithelium of the bronchus, type I pneumocytes; type II pneumocytes; and, rarely, alveolar macrophages (Figure 5B).
Figure 5.
Pulmonary histologic changes in AGMs infected with SARS-CoV-2 with and without treatment
(A–D) Lung lobe of positive control C-AGM-2.
(A) Multifocal interstitial pneumonia with multinucleated cells (black arrow).
(B) SARS-CoV-2 IHC-positive (red) pneumoncytes (white arrows) and bronchial respiratory epithelium (inset, black arrow).
(C) Flooded alveolar spaces with fibrin (red).
(D) Modest collagen deposition (blue) in alveolar septum.
(E–H) Lung lobe of low neutralizing antibody titer treatment LD-AGM-1.
(E) Multifocal interstitial pneumonia.
(F) SARS-CoV-2 IHC-positive (red) bronchial respiratory epithelium.
(G) Flooded alveolar spaces with fibrin (red).
(H) Modest collagen deposition (blue) in alveolar septum.
(I–L) Lung lobe of low neutralizing antibody titer treatment LD-AGM-2.
(I) No significant lesions.
(J) No immunolabeling for anti-SARS-CoV-2 antigen.
(K) No immunolabeling for anti-fibrin antigen.
(L) No significant findings.
(M–P) Lung lobe of high neutralizing antibody titer treatment HD-AGM-1.
(M) Mild multifocal interstitial pneumonia.
(N) SARS-CoV-2 IHC-positive (red) bronchial respiratory epithelium.
(O) Partially flooded alveolar spaces with fibrin (red).
(P) Modest collagen deposition (blue) in alveolar septum.
(Q–T) Lung lobe of high neutralizing antibody titer treatment HD-AGM-3.
(Q) No significant lesions.
(R) No immunolabeling for anti-SARS-CoV-2 antigen.
(S) No immunolabeling for anti-fibrin antigen.
(T) No significant findings.
(A, E, I, M, and Q) H&E staining, 20×. (B, B inset, F, J, N, and R) IHC labeling for anti-SARS-CoV2 antigen, 40× (red). (C, G, K, O, and S) IHC labeling for anti-fibrin antigen, 20× (red). (D, H, L, P, and T) Trichrome staining, 20× (blue).
The eight AGMs treated with convalescent sera developed various degrees of multifocal pulmonary lesions. Five treated AGMs, 3/4 from the LD cohort and 2/4 from the HD cohort, developed moderate pulmonary lesions, similar to the untreated control AGMs (Figures 5E, 5G, 5H, 5M, 5O, and 5P). SARS-CoV-2 antigen detection in these AGMs was noted diffusely within the cytoplasm of individual to small clusters of respiratory epithelium of the bronchus (Figures 5F and 5N). The remaining three treated AGMs, one from the LD cohort (LD-AGM-2) and two from the HD cohort (HD-AGM-3 and HD-AGM-2), had minimal multifocal interstitial pneumonia, characterized by minimal expansion of alveolar septae with macrophages; lymphocytes; and, rarely, neutrophils (Figures 5I and 5Q). There was no positive immunolabeling for SARS-CoV-2 in these animals, minimal to no fibrin flooding of alveolar spaces, and no overt expansion of alveolar septae with collagen (Figures 5J–5L and 5R–5T).
Assessment of soluble inflammation markers
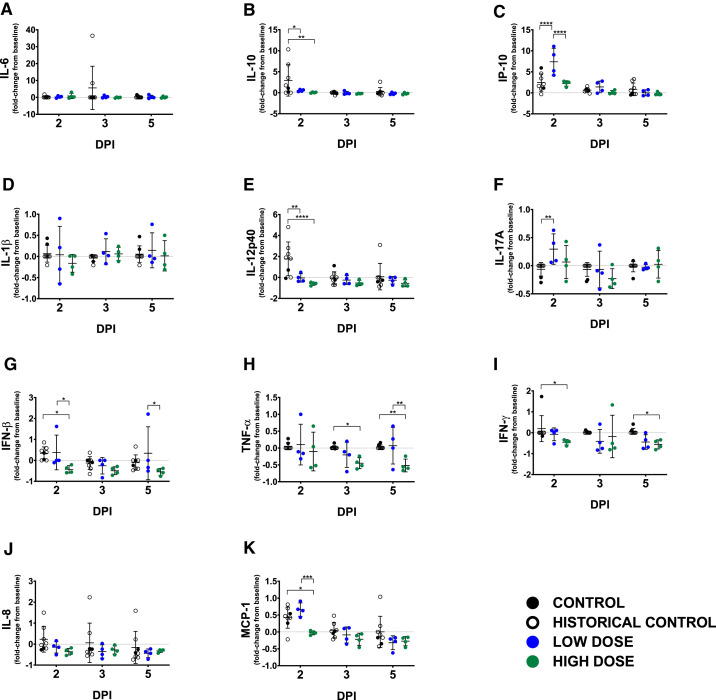
We measured plasma concentrations of a panel of cytokines/chemokines known to be altered in human COVID-19 (Laing et al., 2020). At 2 dpi, interleukin (IL)-10 and IL-12 were significantly elevated in control animals compared to both LD and HD groups: for IL-10, p = 0.0047 (HD group) and p = 0.0210 (LD group); for IL-12p40, p < 0.0001 (HD group) and p < 0.0015 (LD group), two-way ANOVA supported by Tukey’s multiple comparison test (Figures 6B and 6E). Interferon (IFN)-gamma-induced protein 10 (IP-10) was significantly elevated only in the LD cohort (p < 0.0001, compared to control; and p < 0.0001, compared to the HD group; two-way ANOVA supported by Tukey’s multiple comparison test) (Figure 6C). Significant elevations in macrophage chemotactic protein 1 (MCP-1) were observed in all study and historical control animals (p = 0.0109, compared to the HD group; two-way ANOVA supported by Tukey’s multiple comparison test) as well as in the LD group (p = 0.0006, compared to the LD group; two-way ANOVA supported by Tukey’s multiple comparison test) on day 2 (Figure 6K). On day 3, IL-6 was elevated only in two control animals (Figure 6A). Reduced levels of IFN-beta, tumor necrosis factor (TNF) alpha, IFN-gamma, IL-8, and MCP-1 were observed in all of the HD-treated animals as compared to control groups beginning at 2 dpi and trending through the remainder of the study (Figures 6G–6K).
Figure 6.
Circulating levels of inflammatory markers
Fold change from individual subject baselines (0 dpi) were calculated for each subject at the indicated time points. A two-way ANOVA and Tukey’s multiple comparisons test were used to compare each treatment group at the respective time point (∗p < 0.0332; ∗∗p < 0.0021; ∗∗∗p < 0.0002; ∗∗∗∗p < 0.0001). The value shown for individual subjects is the mean from duplicate assays. For each treatment or control group, the mean ± SD is indicated (bars). Historical control animals from a previous study (n = 6) (Woolsey et al., 2021) utilizing the homologous virus were included for statistical purposes. (A) IL-6; (B) IL-10; (C) IP-10; (D) IL-1β; (E) IL-12p40; (F) IL-17A; (G) IFN-β; (H) TNF-α; (I) IFN-γ; (J) IL-8; (K) MCP-1.
Discussion
In response to the dire need for therapeutic options, several human clinical trials have been initiated to evaluate both the safety and the efficacy of passive transfer of CCP to acutely ill COVID-19 patients (Klassen et al., 2020a). Overall, the consensus of these studies is that therapeutic benefit is possible; however, there are a number of caveats associated with doing human studies, including unknown incubation times, effective dose, and/or route(s) of exposure. These shortcomings highlight the need for experimentally controlled studies to help understand potential risks of this approach. Others have demonstrated therapeutic benefit in rodent models of COVID-19, but limited information was gleaned in terms of the pathophysiological parameters affected by treatment with CCP (Chan et al., 2020; Imai et al., 2020). While hyper-immune plasma has been shown to provide therapeutic benefit in related MERS-CoV-infected primates (van Doremalen et al., 2017), to date, there are no reports demonstrating convalescent sera or plasma treatment benefit in NHP models of COVID-19. Here, we provide evidence that convalescent sera with high neutralizing antibody titers can be given as a postexposure treatment using a NHP model that faithfully recapitulates the hallmark features of human COVID-19. Compared to LD-treated or untreated control animals, HD-treated AGMs had lower viral burdens in respiratory compartments, reduced gross and histopathological lesion severity in the lungs, and reductions in several clinical parameters previously shown to be important in human disease, including prolonged coagulation times, elevated fibrinogen, thrombocytopenia, and hypercytokinemia. Differences in clinical parameters of the LD-treated group and untreated control animals from this study or historical control animals were minimal; however, the lack of infectious SARS-CoV-2 in the BAL samples from all of the LD-treated animals and reduced lung pathology suggest that an antiviral effect was present, despite the lower concentration of neutralizing antibodies in the dose of convalescent sera administered. Nonetheless, these data support the need for high-potency neutralizing antibody content in convalescent blood product preparations to achieve maximal therapeutic benefit.
Previous work utilizing passive transfer of potent neutralizing antibody containing plasma derived from a cohort of rhesus macaques vaccinated with a SARS-CoV spike expressing live-attenuated modified vaccinia virus Ankara (ADC-MVA) vaccine resulted in exacerbation of pulmonary disease suggestive of antibody-dependent enhancement (ADE) (Liu et al., 2019), which served to caution the design of therapeutics and vaccines of potential dangers of targeting this antigen for SARS-CoVs. While we characterized the presence of antibodies capable of binding all SARS-CoV-2 proteins, and more specifically, nucleoprotein and the RBD of spike, we did not assess the differential binding potential of all of these targets in the treatments provided. Therefore, we cannot rule out other antiviral mechanisms such as antibody-binding capacity or fragment-crystallizable (FC) region-directed functions associated with antibodies present in these treatments; nor can we account for other soluble factors that may have been present in sera derived from survival of natural infection, which may explain the differences observed in our data versus the passive transfer of vaccine-derived sera. Importantly, as a result of the lack of available therapeutic options and the previous successes with CP in the treatment of SARS-CoV and MERS-CoV, thousands of humans have been treated with CCP through compassionate use or clinical trial access channels where preliminary results suggest that treatment is well tolerated (Joyner et al., 2020). Meta-analysis of the available data suggests a clear benefit as measured by reduced hospitalization times and minimal risk where adverse events were classified as common to CP treatments in general rather than complications related specifically to CCP or ADE (Klassen et al., 2020a; Sun et al., 2020).
Ongoing clinical trials are working to refine the treatment approaches and our understanding of precisely what is needed to improve efficacy in terms of therapeutic dosing of CCP and issues related to safety (Joyner et al., 2020). Several recent studies suggest that timing and dose may be critical to the success of treatment. For example, less therapeutic benefit was observed in patients who began treatment during advanced disease (Bradfute et al., 2020; Gharbharan et al., 2020). Further, a recent large-scale clinical trial in India highlighted the importance of donor CCP potency where the median neutralizing antibody potency of donor CCP was 1:40 (interquartile range = 1:30–1:80), compared to our study wherein both treatment groups had PRNT50 values that were considerably higher (∼1:128 low-dose pool or 1:2,048 high-dose pool). The trial was terminated early, as no clear therapeutic benefit was evident (Agarwal et al., 2020). Conversely, a recent retrospective analysis of 3,082 hospitalized patients saw a reduction in deaths 30 days after treatment in a dose-dependent manner in favor of higher titer CCP administration (Joyner et al., 2021). A smaller study examined 160 patients in a randomized, double-blind, placebo-controlled trial of CCP in older adult patients treated within 72 h of onset of mild COVID-19 symptoms; this study found clear reduction of progression to severe disease (Libster et al., 2021).
Here, we have provided detailed experimental evidence in a relevant animal model of COVID-19 that supports early administration of potent CCP. These findings are supported by recently reported clinical trials that reflect the benefits of this approach. Given the exponential growth of a convalescent COVID-19 population, the availability of CCP as a treatment may serve as a therapeutic bridge until more potent and targeted immunotherapeutics such as monoclonal antibodies or other antiviral drugs become available.
STAR★Methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti Monkey IgG (HRP) antibody | Fitzgerald Industries International | Cat# 43R-IG020HRP, RRID:AB_1287018 Lot# c15032720 |
| SARS Nucleocapsid Protein Antibody | Novus Biologicals | Cat# NB100-56683, RRID:AB_838841 Lot # 111003D-4 |
| Biotinylated Goat Anti-Rabbit IgG antibody | Vector Laboratories | Cat# BA-1000, RRID:AB_2313606 Lot # ZG0122 |
| Anti-fibrin monoclonal mouse primary antibody | Sekisui Diagnostics | Cat# REF 350 Lot# 140714 |
| Goat Anti-Mouse IgG Antibody (H+L), Biotinylated | Vector Laboratories | Cat# BA-9200, RRID:AB_2336171 Lot # ZB0324 |
| Bacterial and virus strains | ||
| SARS-CoV-2 | European Virus Archive | SARS-CoV-2/INMI1-Isolate/2020/Italy |
| Biological samples | ||
| Pooled Convalescent Sera (“High Dose”) | Woolsey et al., 2021 | N/A |
| Pooled Convalescent Sera (“Low Dose”) | Cross et al., 2020 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RT SigmaFast O-phenylenediamine (OPD) substrate | Sigma-Aldrich | Cat# P9187 |
| Alkaline Phosphatase Streptavidin | Vector Laboratories | Cat# SA-5100-1 |
| Bio-Red developer | Bio-Path | Cat# BP-100-FR |
| Critical commercial assays | ||
| ReSARS® CoV-2 (N) IgG ELISA Kit – RUO | Zalgen Labs, LLC | Cat# 10166 |
| ReSARS® CoV-2 (S-RBD) IgG ELISA Kit – RUO | Zalgen Labs, LLC | Cat# 10180-1 |
| QIAamp Viral RNA Mini Kit | QIAGEN | Cat# 52906 |
| QuantiFast Probe RT-PCR Kit | QIAGEN | Cat# 204454 |
| One-Step Trichrome Blue & Red Stain Kit | American Mastertech | Cat# ML7229 |
| Legendplex Multiplex Assay: Human Thrombosis Panel | BioLegend | Cat# 740891 |
| Legendplex Multiplex Assay: NHP Inflammation Panel | BioLegend | Cat# 740389 |
| Piccolo Biochemistry Panel Plus analyzer discs | Abaxis | Cat# 400-7182-1 |
| Vetscan VSPro PT/aPTT Combination test | Abaxis | Cat# 740-7013 |
| Vetscan VSPro Fibrinogen test | Abaxis | Cat# 740-7031 |
| Experimental models: cell lines | ||
| Vero E6 | ATCC | ATCC CRL-1586 |
| Experimental models: organisms/strains | ||
| African green monkey | Worldwide Primates, Inc. | N/A |
| Oligonucleotides | ||
| 2019-nCoV_N2 Forward Primer: TTA CAA ACA TTG GCC GCA AA | U.S. Centers for Disease Control and Prevention | https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html |
| 2019-nCoV_N2 Reverse Primer: GCG CGA CAT TCC GAA GAA | U.S. Centers for Disease Control and Prevention | https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html |
| 2019-nCoV_N2 Probe: FAM-ACA ATT TGC CCC CAG CGC TTC AG-BHQ1 | U.S. Centers for Disease Control and Prevention | https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html |
| Software and algorithms | ||
| Biotek Cytation5 v.3.04 | BioTek (Agilent) | Packaged with Instrument |
| CFX Maestro 1.1 version v4.1.2433.1219 | Bio-Rad | Packaged with Instrument |
| LEGENDplex Data Analysis Software | BioLegend | https://www.biolegend.com/en-us/legendplex |
| BD FACS Diva | BD Biosciences | Packaged with Instrument |
| GraphPad v.9.0.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to Dr. Thomas W. Geisbert (twgeisbe@utmb.edu)
Materials availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Data and software availability
The datasets supporting the current study have not been deposited in a public repository because an appropriate repository for the datasets is not available. Datasets are available from the corresponding author on reasonable request. This study did not generate code.
Experimental model and subject details
Nonhuman primates
Research-naive African green monkeys (Chlorocebus aethiops; n = 10; 6 males, 4 females; St Kitts origin, approximately 3-8 years old) were procured from Worldwide Primates, Inc., and assessed for general health by a board-certified veterinarian upon arrival at UTMB. Prior to the start of the study, all animals underwent quarantine, acclimatization, and socialization periods. All procedures, manipulations, and husbandry involving monkeys were performed in accordance with guidelines and requirements set forth by the UTMB Institutional Animal Care and Use Committee (IACUC) and relevant state and federal authorities. All measurements requiring physical manipulation of the animals were performed under sedation by ketamine.
Virus
The virus (SARS-CoV-2/INMI1-Isolate/2020/Italy) was isolated on January 30, 2020 from the sputum of the first clinical case in Italy, a tourist visiting from the Hubei province of China that developed respiratory illness while traveling. The virus was initially passaged twice (P2) on Vero E6 cells; the supernatant and cell lysate were collected and clarified following a freeze/thaw cycle. This isolate is certified mycoplasma and Foot-and-Mouth Disease virus free. The complete sequence was submitted to GenBank (GenBank: MT066156) and is available on the GISAID website (BetaCoV/Italy/INMI1-isl/2020: EPI_ISL_410545) upon registration. For in vivo challenge, the P2 virus was propagated on Vero E6 cells and the supernatant was collected and clarified by centrifugation making the virus used in this study a P3 stock.
Cell lines
Vero E6 cells (ATCC CRL-1586) were procured from ATCC and maintained with complete EMEM media (10% FBS, 1% penicillin/streptomycin) at 37°C/5% CO2. The sex of Vero E6 cells is female. Cells are routinely screened for mycoplasma contamination. No further validation of the cell line was performed beyond that provided by ATCC.
Method details
Animal challenge
Prior to enrollment in this study, AGMs were tested for sero-reactivity to SARS-CoV-2. All animals were seronegative. Animals were anesthetized with ketamine and inoculated with a target dose of 5.0 × 105 PFU of SARS-CoV-2 (SARS-CoV-2/INMI1-Isolate/2020/Italy) through combined intranasal (i.n.) and intratracheal (i.t.) inoculation, with the dose being divided evenly between both routes. Ten hours post-challenge, the experimental cohorts were treated intravenously (i.v.) with pooled convalescent sera (6.1 ml/kg) obtained from animals infected with the homologous isolate of SARS-CoV-2 in previous studies (Cross et al., 2020; Woolsey et al., 2021). All animals were longitudinally monitored for clinical signs of illness, respiration quality, and clinical pathology. Mucosal swabs were obtained using sterile swabs inserted into the mucosal cavity, gently rotated to maximize contact with the mucosal surface, and deposited into 2.0 mL screw-top tubes containing sterile MEM media supplemented to 10% with FBS. Animals were maintained on a diet of commercial monkey chow, provided daily enrichment, and housed individually with 12 hr:12 hr light:dark cycles with ambient temperature and humidity in a secure BSL-4 facility. The monkeys were monitored daily and scored for disease progression with an internal SARS-CoV-2 humane endpoint scoring sheet approved by the UTMB IACUC. For ethical and logistical reasons, the NHP experiment was only performed once. Animal group size was determined by the expected lethality (low to none, based the limited number of other COVID-19 studies in AGMs and other species of non-human primates), and for ethical reasons, the minimum number of animals by which statistical inferences could be made (n = 4 for low-dose treated cohort, n = 4 for high dose treated cohort, n = 2 for untreated controls). Additionally, we included data from historical untreated controls (n = 6) challenged via the same route, and with the same virus stock and dose from a previous study.
Bronchoalveolar lavage procedure
Animals were anesthetized with ketamine, and the pharynx was sprayed with an anesthetic spray (14% benzocaine/2% butamen/2% tetracaine hydrochloride) for 1 s to ensure proper coating. A 12 French gauge (FR) feeding tube on a wire guide was inserted into the airway, and sterile phosphate buffered saline (PBS) was administered with the animal sitting upright, after which the animal was moved into a supine position. Gentle suction was applied with effort made to retrieve as much of the lavage fluid as possible. This process was repeated until a total of 40 mL of PBS had been applied. Approximately 10-16 mL of lavage fluid was retrieved from each animal in this way. 0.005-0.010 mg/kg of glycopyrrolate was then administered, and the animal was placed on supplementary oxygen and allowed to recover under close observation. BAL fluid was placed on ice and immediately processed for downstream applications.
Hematology and serum biochemistry
Total white blood cell counts, white blood cell differentials, red blood cell counts, platelet counts, hematocrit values, total hemoglobin concentrations, mean cell volumes, mean corpuscular volumes, and mean corpuscular hemoglobin concentrations were analyzed from blood collected in tubes containing EDTA using a Vetscan HM5 hematologic analyzer (Abaxis). Serum samples were tested for concentrations of albumin, amylase, alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), blood urea nitrogen (BUN), calcium, creatinine (CRE), C-reactive protein (CRP), gamma-glutamyltransferase (GGT), glucose, total protein, and uric acid by using a Piccolo point-of-care analyzer and Biochemistry Panel Plus analyzer discs (Abaxis). Partial pressures of CO2 and O2 were obtained using an iSTAT Alinity hematological analyzer (Abbott). For coagulation assays, blood was collected in tubes containing sodium citrate, and assayed with the Vetscan VSPro Fibrinogen or PT/aPTT Combination test cartridges on a Vetscan VSPro hematologic analyzer (Abaxis).
RNA isolation from SARS-CoV-2-infected AGMs
On specified procedure days (days 0, 2, 3, 4, 5), 100 μL of blood was added to 600 μL of AVL viral lysis buffer (QIAGEN) for virus inactivation and RNA extraction. Following removal from the high containment laboratory, RNA was isolated from blood and swabs using the QIAamp viral RNA kit (QIAGEN).
RT-qPCR detection of SARS-CoV-2 vRNA
RNA was isolated from blood and mucosal swabs and assessed using the CDC SARS-CoV-2 N2 assay primers/probe for reverse transcriptase quantitative PCR (RT-qPCR) (Centers for Disease Control and Prevention, 2020). SARS-CoV-2 RNA was detected using One-step probe RT-qPCR kits (QIAGEN) run on the CFX96 detection system (Bio-Rad), with the following cycle conditions: 50°C for 10 minutes, 95°C for 10 s, and 45 cycles of 95°C for 10 s and 55°C for 30 s. Threshold cycle (C T) values representing SARS-CoV-2 genomes were analyzed with CFX Manager Software, and data are presented as GEq. To generate the GEq standard curve, RNA was extracted from supernatant derived from Vero E6 cells infected with SARS-CoV-2/INMI1-Isolate/2020/Italy was extracted and the number of genomes was calculated using Avogadro’s number and the molecular weight of the SARS-CoV-2 genome.
Plaque titration of infectious SARS-CoV-2
Infectious virus was quantitated by plaque assay on Vero E6 cells (ATCC CRL-1586) from all blood plasma and mucosal swabs, and bronchoalveolar lavage (BAL) samples. Briefly, increasing 10-fold dilutions of the samples were adsorbed to Vero E6 cell monolayers in duplicate wells (200 μl). Cells were overlaid with EMEM medium plus 1.25% Avicel, incubated for 2 days, and plaques were counted after staining with 1% crystal violet in formalin. The limit of detection for this assay is 25 PFU/ml.
Serum neutralization assay
Neutralization titers were calculated by determining the dilution of serum that reduced 50% of plaques (PRNT50). A standard 100 PFU amount of SARS-CoV-2 was incubated with two-fold serial dilutions of serum samples for one hour. The virus-serum mixture was then used to inoculate Vero E6 cells for 60 minutes. Cells were overlaid with EMEM medium plus 1.25% Avicel, incubated for 2 days, and plaques were counted after staining with 1% crystal violet in formalin.
ELISA
SARS-CoV-2-specific IgG antibodies were measured in sera by ELISA at the indicated time points. Immunosorbent 96-well plates were coated overnight with each antigen. For total virus-specific IgG, plates were coated with a 1:1000 dilution of irradiated SARS-CoV-2 infected or normal Vero E6 lysate in PBS (pH 7.4) kindly provided by Dr. Thomas W. Ksiazek (UTMB). Nucleoprotein and Spike Receptor Binding Domain (RBD) ELISA kits were kindly provided by Zalgen Labs, LLC. Sera were initially diluted 1:100 and then two-fold through 1:51,200 in 4% BSA in 1 × PBS or in Zalgen-provided reagents. After a one-hour incubation, plates were washed six times with wash buffer (1 x PBS with 0.2% Tween-20) and incubated for an hour with a 1:5000 dilution of horseradish peroxidase (HRP)-conjugated anti-primate IgG antibody (Fitzgerald Industries International). RT SigmaFast O-phenylenediamine (OPD) substrate (Sigma-Aldrich) was added to the wells after six additional washes to develop the colorimetric reaction. The reaction was stopped with 3M sulfuric acid 5-10 minutes after OPD addition and absorbance values were measured at a wavelength of 492nm on a spectrophotometer (Biotek Cytation5 system). Absorbance values were normalized by subtracting uncoated wells from antigen-coated wells at the corresponding serum dilution. End-point titers were defined as the reciprocal of the last adjusted serum dilution with a value ≥ 0.20.
Histopathology and immunohistochemistry
Necropsy was performed on all subjects euthanized at 5 dpi. Tissue samples of all major organs were collected for histopathologic and immunohistochemical (IHC) examination and were immersion-fixed in 10% neutral buffered formalin for > 7 days. Specimens were processed and embedded in paraffin and sectioned at 5 μm thickness. For IHC, specific anti-SARS immunoreactivity was detected using an anti-SARS nucleocapsid protein rabbit primary antibody at a 1:800 dilution for 60 minutes (Novusbio). The tissue sections were processed for IHC using the ThermoFisher Scientific Lab Vision Autostainer 360 (ThermoFisher Scientific). Secondary antibody used was biotinylated goat anti-rabbit IgG (Vector Laboratories) at 1:200 for 30 minutes followed by Vector Streptavidin Alkaline Phosphatase at a dilution of 1:200 for 20 min (Vector Laboratories). Slides were developed with Bio-Red (Biopath) for 7 minutes and counterstained with hematoxylin for one minute. For IHC, specific anti-fibrin was detected using an anti-fibrin monoclonal mouse primary antibody at a 1:3200 dilution for 60 minutes (Sekisui Diagnostics). The tissue sections were processed for IHC using the ThermoFisher Scientific Lab Vision Autostainer 360 (ThermoFisher Scientific). Secondary antibody used was biotinylated goat anti-mouse IgG (Vector Laboratories) at 1:200 for 30 minutes followed by Vector Streptavidin Alkaline Phosphatase at a dilution of 1:200 for 20 min (Vector Laboratories). Slides were developed with Bio-Red (Biopath Laboratories) for 7 minutes and counterstained with hematoxylin for one minute. Tissues were stained following package instructions for collagen with the Trichrome One-Step Blue & Red Stain Kit (American MasterTech Scientific Laboratory Supplies).
Bead-based cytokine immunoassays
Concentrations of immune mediators were determined by flow cytometry using LegendPlex multiplex technology (BioLegend). Serum levels of cytokines/chemokines were quantified using Nonhuman Primate Inflammation 13-plex and Human Thrombosis Panel (1:4 dilution) kits. Samples were processed in duplicate following the kit instructions and recommendations. Following bead staining and washing, 4000 bead events were collected on a FACS Canto II cytometer (BD Biosciences) using BD FACS Diva software. The raw .fcs files were analyzed with BioLegend’s cloud-based LEGENDplex Data Analysis Software.
Quantification and statistical analysis
The data was analyzed and graphed in GraphPad Prism 9.0.0. Lesion severity scores and tissue PCR and plaque assay titers were analyzed using either a one-way ANOVA or a two-way ANOVA supported by Tukey’s multiple comparisons test. ELISAs and LegendPlex assays were also analyzed using a two-way ANOVA and Tukey’s multiple comparisons test. No data points were excluded for our analyses. Specifics of statistical comparisons are detailed in individual figure legends. Significance cut-off values are outlined per data in their respective results sections only where significant differences in groups compared existed.
Acknowledgments
The authors would like to thank the UTMB Animal Resource Center for husbandry support of laboratory animals, Dr. Kevin Melody for assistance with animal studies, Dr. Liana Medina for assistance with processing BAL samples, and Kira Zapalac for assistance with clinical pathology and PCR assays. We also thank Drs. Luis Branco and Matt Boisen for generously providing the SARS-CoV-2 anti-nucleoprotein and spike RBD ELISA assays as well as Dr. Thomas Ksiazek for the irradiated SARS-CoV-2 whole-cell lysate used in ELISAs. The virus used in this publication was kindly provided by the European Virus Archive goes Global (EVAg) project, which has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 653316. The graphical abstract was created using Biorender.com. This study was supported by funds from the Department of Microbiology and Immunology, University of Texas Medical Branch at Galveston, Galveston, TX to T.W.G. Operations support of the Galveston National Laboratory was supported by NIAID grant UC7AI094660.
Author contributions
R.W.C. and T.W.G. conceived and designed the study. D.J.D., J.B.G., and T.W.G. performed the SARS-CoV-2 challenge experiments. R.W.C., D.J.D., J.B.G., and T.W.G. performed animal procedures and clinical observations. K.N.A. and V.B. performed the clinical pathology assays. C.W. processed BAL samples. V.B. performed the SARS-CoV-2 infectivity assays. K.N.A. performed the PCR. N.S.D. performed the immunohistochemistry and in situ hybridization. C.W. performed ELISAs and multiplex assays. K.A.F. performed the necropsies and analysis of the gross pathology, histopathology, immunohistochemistry, and in situ hybridization. All authors analyzed the clinical pathology, virology, and immunology data. R.W.C., A.N.P., K.A.F., and T.W.G. wrote the paper. All authors had access to all of the data and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108837.
Supplemental information
References
- Agarwal A., Mukherjee A., Kumar G., Chatterjee P., Bhatnagar T., Malhotra P., Latha B., Bundas S., Kumar V., Dosi R. Convalescent plasma in the management of moderate COVID-19 in India: An open-label parallel-arm phase II multicentre randomized controlled trial (PLACID Trial) medRxiv. 2020 doi: 10.1101/2020.09.03.20187252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y., Balkhy H., Hajeer A.H., Bouchama A., Hayden F.G., Al-Omari A., Al-Hameed F.M., Taha Y., Shindo N., Whitehead J. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: a study protocol. Springerplus. 2015;4:709. doi: 10.1186/s40064-015-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S. Remdesivir for the Treatment of Covid-19 — Preliminary Report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradfute S.B., Hurwitz I., Yingling A.V., Ye C., Cheng Q., Noonan T.P., Raval J.S., Sosa N.R., Mertz G.J., Perkins D.J. Severe Acute Respiratory Syndrome Coronavirus 2 Neutralizing Antibody Titers in Convalescent Plasma and Recipients in New Mexico: An Open Treatment Study in Patients With Coronavirus Disease 2019. J. Infect. Dis. 2020;222:1620–1628. doi: 10.1093/infdis/jiaa505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Dadachova E., Pirofski L.A. Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2004;2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- CCPP19 . 2020. National COVID-19 Convalescent Plasma Project-Component 3: Clinical Trials.https://ccpp19.org/healthcare_providers/component_3/index.html [Google Scholar]
- Centers for Disease Control and Prevention . 2020. Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes.https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html [Google Scholar]
- Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–2476. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Passini N., Lanzavecchia A., Zambon M. Rapid generation of a human monoclonal antibody to combat Middle East respiratory syndrome. J. Infect. Public Health. 2016;9:231–235. doi: 10.1016/j.jiph.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R.W., Agans K.N., Prasad A.N., Borisevich V., Woolsey C., Deer D.J., Dobias N.S., Geisbert J.B., Fenton K.A., Geisbert T.W. Intranasal exposure of African green monkeys to SARS-CoV-2 results in acute phase pneumonia with shedding and lung injury still present in the early convalescence phase. Virol. J. 2020;17:125. doi: 10.1186/s12985-020-01396-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbharan A., Jordans C.C.E., Geurts van Kessel C., den Hollander J.G., Karim F, Mollema F.P.N., Stalenhoef J.E., Dofferhoff A., Ludwig I., Koster A. Convalescent Plasma for COVID-19. A randomized clinical trial. medRxiv. 2020 doi: 10.1101/2020.07.01.20139857. [DOI] [Google Scholar]
- Hartman A.L., Nambulli S., McMillen C.M., White A.G., Tilston-Lunel N.L., Albe J.R., Cottle E., Dunn M.D., Frye L.J., Gilliland T.H. SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and shedding of infectious virus from both respiratory and gastrointestinal tracts. PLoS Pathog. 2020;16:e1008903. doi: 10.1371/journal.ppat.1008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J. An mRNA Vaccine against SARS-CoV-2 — Preliminary Report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. Published online July 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.C., Jay A., Raymond J.L., Rossi F., Zeng X., Scruggs J., Dyer D., Frick O., Moore J., Berrier K. Development of a Coronavirus Disease 2019 Nonhuman Primate Model Using Airborne Exposure. bioRxiv. 2020 doi: 10.1101/2020.06.26.174128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner M.J., Wright R.S., Fairweather D., Senefeld J.W., Bruno K.A., Klassen S.A., Carter R.E., Klompas A.M., Wiggins C.C., Shepherd J.R.A. Early safety indicators of COVID-19 convalescent plasma in 5000 patients. J. Clin. Invest. 2020;130:4791–4797. doi: 10.1172/JCI140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner M.J., Carter R.E., Senefeld J.W., Klassen S.A., Mills J.R., Johnson P.W., Theel E.S., Wiggins C.C., Bruno K.A., Klompas A.M. Convalescent Plasma Antibody Levels and the Risk of Death from Covid-19. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2031893. Published online January 21, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen S.A., Senefeld J., Johnson P.W., Carter R.E., Wiggins C.C., Shoham S., Grossman B.J., Henderson J.P., Musser J.M., Salazar E. Evidence favouring the efficacy of convalescent plasma for COVID-19 therapy. medRxiv. 2020 doi: 10.1101/2020.07.29.20162917. [DOI] [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Libster R., Pérez Marc G., Wappner D., Coviello S., Bianchi A., Braem V., Esteban I., Caballero M.T., Wood C., Berrueta M., Fundación INFANT–COVID-19 Group Early High-Titer Plasma Therapy to Prevent Severe Covid-19 in Older Adults. N. Engl. J. Med. 2021;384:610–618. doi: 10.1056/NEJMoa2033700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct. Target. Ther. 2020;5:157. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.M., Lim W.S., Makki S., Rooney K.D., Nguyen-Van-Tam J.S., Beck C.R., Convalescent Plasma Study Group The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malani A.N., Sherbeck J.P., Malani P.N. Convalescent Plasma and COVID-19. JAMA. 2020;324:524. doi: 10.1001/jama.2020.10699. [DOI] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol A., Perlman S. Lessons for COVID-19 Immunity from Other Coronavirus Infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.K., Ganatra S.R., Singh B., Cole J., Alfson K.J., Clemmons E., Gazi M., Gonzalez O., Escobedo R., Lee T.-H. SARS-CoV-2 infection leads to acute infection with dynamic cellular and inflammatory flux in the lung that varies across nonhuman primate species. bioRxiv. 2020 doi: 10.1101/2020.06.05.136481. [DOI] [Google Scholar]
- Speranza E., Williamson B.N., Feldmann F., Sturdevant G.L., Pérez-Pérez L., Mead-White K., Smith B.J., Lovaglio J., Martens C., Munster V.J. SARS-CoV-2 infection dynamics in lungs of African green monkeys. bioRxiv. 2020 doi: 10.1101/2020.08.20.258087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Xu Y., He H., Zhang L., Wang X., Qiu Q., Sun C., Guo Y., Qiu S., Ma K. A potentially effective treatment for COVID-19: A systematic review and meta-analysis of convalescent plasma therapy in treating severe infectious disease. Int. J. Infect. Dis. 2020;98:334–346. doi: 10.1016/j.ijid.2020.06.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Food and Drug Administration . 2020. FDA Issues Emergency Use Authorization for Convalescent Plasma as Potential Promising COVID–19 Treatment, Another Achievement in Administration’s Fight Against Pandemic.https://www.fda.gov/news-events/press-announcements/fda-issues-emergency-use-authorization-convalescent-plasma-potential-promising-covid-19-treatment [Google Scholar]
- van Doremalen N., Falzarano D., Ying T., de Wit E., Bushmaker T., Feldmann F., Okumura A., Wang Y., Scott D.P., Hanley P.W. Efficacy of antibody-based therapies against Middle East respiratory syndrome coronavirus (MERS-CoV) in common marmosets. Antiviral Res. 2017;143:30–37. doi: 10.1016/j.antiviral.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey C., Borisevich V., Prasad A.N., Agans K.N., Deer D.J., Dobias N.S., Heymann J.C., Foster S.L., Levine C.B., Medina L. Establishment of an African green monkey model for COVID-19 and protection against re-infection. Nat. Immunol. 2021;22:86–98. doi: 10.1038/s41590-020-00835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus Disease (COVID-2019) Pandemic.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- Yeh K.-M., Chiueh T.-S., Siu L.K., Lin J.-C., Chan P.K.S., Peng M.-Y., Wan H.-L., Chen J.-H., Hu B.-S., Perng C.-L. Experience of using convalescent plasma for severe acute respiratory syndrome among healthcare workers in a Taiwan hospital. J. Antimicrob. Chemother. 2005;56:919–922. doi: 10.1093/jac/dki346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., Li J.-X., Yang B.-F., Wang L., Wang W.-J. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study have not been deposited in a public repository because an appropriate repository for the datasets is not available. Datasets are available from the corresponding author on reasonable request. This study did not generate code.