Abstract
The biosafety level 3 (BSL-3) requirement to culture severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a bottleneck for research. Here, we report a trans-complementation system that produces single-round infectious SARS-CoV-2 that recapitulates authentic viral replication. We demonstrate that the single-round infectious SARS-CoV-2 can be used at BSL-2 laboratories for high-throughput neutralization and antiviral testing. The trans-complementation system consists of two components: a genomic viral RNA containing ORF3 and envelope gene deletions, as well as mutated transcriptional regulator sequences, and a producer cell line expressing the two deleted genes. Trans-complementation of the two components generates virions that can infect naive cells for only one round but does not produce wild-type SARS-CoV-2. Hamsters and K18-hACE2 transgenic mice inoculated with the complementation-derived virions exhibited no detectable disease, even after intracranial inoculation with the highest possible dose. Thus, the trans-complementation platform can be safely used at BSL-2 laboratories for research and countermeasure development.
Keywords: SARS-CoV-2, COVID-19, coronavirus, diagnosis, vaccine, antiviral
Graphical abstract
Zhang et al. develop a trans-complementation system to produce single-round infectious SARS-CoV-2 that recapitulates authentic viral infection and replication. They then show the system can be safely used at biosafety level 2 for high-throughput antiviral testing.
Introduction
Three zoonotic betacoronaviruses have emerged to cause global epidemics or pandemics in less than 20 years: severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, and SARS-CoV-2 in 2019 (Hu et al., 2020). The coronavirus disease 2019 (COVID-19) pandemic has caused unprecedented social and economic disruption. As of January 16, 2021, SARS-CoV-2 had infected over 94 million people, leading to over 2 million deaths (https://www.worldometers.info/coronavirus/). In response to the pandemic, the scientific community has rapidly developed experimental platforms to study COVID-19 and to develop countermeasures. Several groups have established infectious cDNA clones and reporter SARS-CoV-2 to facilitate the development and analysis of first-generation vaccines and therapeutics (Hou et al., 2020; Mulligan et al., 2020; Thi Nhu Thao et al., 2020; Xie et al., 2020a, 2020b). However, since SARS-CoV-2 is a biosafety level 3 (BSL-3) pathogen, the requirement of high containment represents a bottleneck for antiviral and vaccine evaluation. Thus, a BSL-2 cell culture system that recapitulates authentic viral replication is urgently needed.
The genome of SARS-CoV-2 is a positive-sense, single-stranded RNA of approximately 30 kb in length. The SARS-CoV-2 virion consists of an internal nucleocapsid (formed by the genomic RNA coated with nucleocapsid [N] proteins) and an external envelope (formed by a cell-derived bilipid membrane embedded with spike [S], membrane [M], and envelope [E] proteins) (Yao et al., 2020). The genomic RNA encodes open reading frames (ORFs) for replicase (ORF1a/ORF1b), S, E, M, and N proteins, as well as seven additional ORFs for accessory proteins (Hu et al., 2020). Deletion of viral E or other proteins, as well as trans-complementation of the deleted proteins, have been reported for live-attenuated vaccine development for coronaviruses (Almazán et al., 2013). Mutating the transcriptional regulator sequence (TRS), a conserved sequence near the 5′ end of the viral genome and the 5′ end of each downstream ORFs, has also been used for live-attenuated vaccine development (Yount et al., 2006); such a rewired TRS network eliminates the chance of reversion to wild-type (WT) virus through recombination (Graham et al., 2018).
Stable cell lines containing replicons (self-replicating viral RNA genomes with one or more gene deletions) have been developed for many viruses, including coronaviruses (Ge et al., 2007; Hertzig et al., 2004; Khromykh and Westaway, 1997; Lo et al., 2003; Lohmann et al., 1999). Because replicons lack structural genes, they are not infectious and can safely be manipulated in BSL-2 laboratories. For SARS-CoV-2, although a transient replicon system has been established (Kotaki et al., 2021; Xia et al., 2020), no stable replicon cell line has been reported. To overcome this gap, we have developed a single-round infectious SARS-CoV-2 through trans-complementation. The single-round SARS-CoV-2 is engineered with a reporter gene that facilitates high-throughput antiviral screening and neutralizing antibody measurement. We validated the safety of the system in cell cultures, hamsters, and highly susceptible human angiotensin-converting enzyme 2 (hACE2) transgenic mice. Our results suggest that the trans-complementation system can be used safely at BSL-2 laboratories.
Results
A single-round infectious SARS-CoV-2 system
Figure 1A depicts the trans-complementation system to produce single-round infectious SARS-CoV-2. The system contains two components: (1) a viral RNA containing a mNeonGreen (mNG) reporter gene and a deletion of ORF3 and E genes (ΔORF3-E; Figure 1B) and (2) a Vero E6 cell line expressing the ORF3 and E proteins under a doxycycline inducible promoter (Vero-ORF3-E; Figures 1C and 1D). Upon electroporation of ΔORF3-E RNA into Vero-ORF3-E cells and addition of doxycycline, trans-complementation enables production of virions that can continuously infect and amplify on Vero-ORF3-E cells; however, these virions can only infect normal cells for a single round due to the lack of ORF3 and E genes in the packaged RNA genome (Figure 1A).
Figure 1.
Generation of single-round infectious ΔORF3-E mNG virion
(A) A trans-complementation system for SARS-CoV-2. Vero-ORF3-E cells are electroporated with ΔORF3-E mNG RNA. Trans-complementation produces ΔORF3-E mNG virion (left panel), which can infect naive Vero E6 cells for only single round (right panel).
(B) ΔORF3-E mNG virion genome. Both the full-length mNG SARS-CoV-2 genome (top panel) and the ΔORF3-E mNG virion genome (bottom panel) are shown. The genomic fragment 8 (gF8) of reverse transcription PCR (RT-PCR) analysis is indicated above both genomes. The ORF3-E deletion junction is indicated. The WT (green box) and mutant (red box) transcription regulatory sequences (TRSs) are also depicted. The mutated TRS (red) is also engineered at the 5′ end of each of the downstream ORFs.
(C) ORF3-E RNA expression in Vero-ORF3-E cells. Doxycycline (Dox) was used to induce the expression of ORF3-E RNA. RT-PCR analyses were performed on Vero-ORF3-E cells with or without Dox induction as well as on naive Vero E6 cells.
(D) Induction of mCherry expression in Vero-ORF3-E cells. Passage 1 (P1) and 20 (P20) of Vero-ORF3-E cells were induced by Dox to express mCherry fluorescence. Scale bar, 100 μm.
(E) Production of ΔORF3-E mNG virion after electroporation. After electroporating ΔORF3-E mNG RNA into Vero-ORF3-E cells (with Dox), infectious titers of ΔORF3-E mNG virion were measured by infecting Vero-ORF3-E cells. Three sets of repeated experiments are presented with bars representing standard deviations.
(F) Analysis of ΔORF3-E mNG virion infection. Vero E6 or Vero-ORF3-E cells were incubated with WT mNG SARS-CoV-2 or ΔORF3-E mNG virion for 2 h. The cells were washed three times with PBS to remove residual input virus. At 48 h post-infection, the supernatants of the infected cells were transferred to fresh Vero E6 or Vero-ORF3-E cells for a second round of infection. The mNG signals from both rounds of infected cells are presented. Scale bar, 100 μm.
(G) RT-PCR analysis.
Extracellular RNA from the second-round infection from (F) was harvested at 48 h post-infection. Fragment 8 of the viral genome, depicted in (B), was amplified by RT-PCR to confirm the ORF3-E deletion and mNG retention.
Our trans-complementation system is engineered with several safeguards to eliminate WT SARS-CoV-2 production. Besides the ORF3-E deletion, the ΔORF3-E viral RNA contained two additional modifications. (1) The TRS of ΔORF3-E RNA was mutated from the WT ACGAAC to CCGGAT (mutant nucleotides underlined; Figure 1B). Recombination between the TRS-mutated ΔORF3-E RNA with inadvertently contaminating viral RNA would therefore not produce replicative virus (Graham et al., 2018; Yount et al., 2006). (2) An mNG gene was engineered at ORF7 of ΔORF3-E RNA to facilitate the detection of viral replication (Figure 1B). The trans-complementing Vero-ORF3-E cell lines were produced by transducing Vero E6 cells with a lentivirus encoding the following elements (Figure S1 A): a TRE3GS promoter that allows doxycycline to induce ORF3 and E protein expression (Figures 1C, 1D, and S1B), an mCherry gene that facilitates selection of cell lines with high levels of protein expression (Figure S1C), a foot-and-mouth disease virus 2A (FMDV 2A) autocleavage site that enables translation of individual mCherry and viral E protein, and an encephalomyocarditis virus internal ribosomal entry site (EMCV IRES) that bicistronically translates the ORF3 protein. The above design eliminated overlapping sequences between the ORF3-E mRNA and ΔORF3-E viral RNA, thus minimizing homologous recombination during trans-complementation. The Vero-ORF3-E cell line stably expressed the engineered proteins after 20 rounds of passaging, as indicated by the mCherry reporter (Figure 1D).
Figure S1.
Construction of Vero-ORF3-E cell lines, related to Figure 1
(A) Construction of a lentiviral transfer plasmid encoding mCherry, ORF3, and E protein. The sequence of FMDV 2A and its translational break position is indicated by an arrow.
(B) Merged mCherry (red) and nuclei (blue) images of 3 selected clones of Vero-ORF3-E cell lines. Nuclei were stained with Hoechst 33342. Doxycycline induction is indicated. Scale bar, 100 μm.
(C) mCherry expression in doxycycline-induced cells. mCherry-positive cells were quantified using a plate reader. The percentages of mCherry positive cells are presented. The results are presented as means ± standard deviations from six replicates, and more than 105 cells were counted for each clone. Clone 1 was used in the rest of this study.
Electroporation of ΔORF3-E mNG RNA into doxycycline-induced Vero-ORF3-E cells produced virions of ~104 median tissue culture infectious dose (TCID50)/mL titer (Figure 1E). The ΔORF3-E mNG virion exhibited a diameter of ~91 nm under negative staining electron microscopy (Figure S2 A). The ΔORF3-E mNG virion produced in the supernatant could infect Vero-ORF3-E cells for multiple rounds but for only one round on naive Vero E6 (Figures 1F and 1G), Calu-3, or hACE2-expressing A549 cells (A549-hACE2; Figures S2B and S2C). The ΔORF3-E mNG virions did not decrease their infectivity after stored at −80°C for 1 year (data not shown). As controls, WT mNG SARS-CoV-2 could infect cells for multiple rounds (Figures S2B and S2C); transfection of ΔORF3-E mNG RNA into parental Vero E6 cells did not produce any infectious virions (data not shown). These results indicate that the trans-complementation system produces virions that can only infect WT cells for single round.
Figure S2.
Single-round infection of ΔORF3-E mNG virion, related to Figure 1
(A) Negative-staining electron microscopic image of ΔORF3-E mNG virion. Scale bar, 50 nm.
(B) Calu-3 and A549-hACE2 cells (MOI of 1 and 10 for mNG SARS-CoV-2 and ΔORF3-E mNG virion, respectively; viral titers determined on Vero-ORF3-E cells) were infected with mNG SARS-CoV-2 or ΔORF3-E mNG virion for 2 h, after which the cells were washed and cultured in fresh medium. At day 2 post-infection, supernatants of the infected cells were transferred to infect naive Calu-3 and A549-hACE2 for the second round. Fluorescence and phase contrast images for the infected cells are presented. Scale bar, 100 μm.
(C) RT-PCR analysis of viral RNA. Extracellular RNAs from the second round of infection from (B) were harvested at day 2 post-infection and subjected to RT-PCR analysis of viral RNA.
(D) WT mNG SARS-CoV-2 and P10 ΔORF3-E mNG virion (derived from 10 rounds of passaging of ΔORF3-E mNG virion on Vero-ORF3-E cells) were used to infect Vero E6 cells for two rounds as described in Figure 1F. Fluorescence and phase contrast images of infected cells are presented for both the first and second rounds of infections. Scale bar, 100 μm.
(E) RT-PCR analysis of viral RNA extracted from the culture fluids from the second-round infected cells in (D).
Adaptive mutations to improve virion production
To improve the efficiency of the trans-complementation platform, we serially propagated ΔORF3-E mNG virions on Vero-ORF3-E cells for 10 passages (3–4 days per passage) to select for adaptive mutations. The P10 virion replicated to higher titers than the P1 virion on Vero-ORF3-E cells (Figure 2 A), retained the mNG reporter (Figures 2B and 2C), and still infected normal Vero cells for only single round (Figures S2D and S2E). Whole-genome sequencing of the P10 virion revealed three mutations in the nsp1, nsp4, and S genes (Figure 2D). Engineering of these mutations into ΔORF3-E mNG RNA showed that all three were required to enhance the trans-complementation efficiency, producing 106 TCID50/mL of virions (Figures 2A and 2B). These results indicate that (1) adaptive mutations can be selected to improve the yield of single-round virions and (2) WT virus is not produced from the trans-complementation system.
Figure 2.
Adaptive mutations to improve the yield of ΔORF3-E mNG virion production
(A) Viral replication kinetics on Vero-ORF3-E cells. Adaptive mutations (D) were selected by continuously passaging the ΔORF3-E virion on Vero-ORF3-E cells for 10 rounds. For comparing the replication kinetics of the passaged viruses, Vero-ORF3-E cells were infected with the P1 or P10 ΔORF3-E virion, ΔORF3-E virion containing an S mutation in (D) (ΔORF3-E virion mut-S), or ΔORF3-E virion containing all adaptive mutations in nsp1, nsp4, and S in (D) (ΔORF3-E virion mut-All) at an MOI of 0.15. WT mNG SARS-CoV-2 was included as a control. Viral titers in culture supernatants are presented. ANOVA with multiple comparison correction test were performed with ∗p < 0.05; ∗∗p < 0.01. Data are represented as mean ± standard deviation.
(B) mNG-positive cells at 24 and 48 h post-infection from (A). Scale bar, 100 μm.
(C) RT-PCR analysis for single-round infection. For confirming the P10 ΔORF3-E virion remains infectious for only a single round on Vero cells, Vero E6 or Vero-ORF3-E cells were infected with WT mNG SARS-CoV-2 or P10 ΔORF3-E mNG virion for two rounds as described in Figure 1G. Viral RNAs were extracted from the second-round culture fluids and analyzed by RT-PCR. The RT-PCR product, gF8, is indicated in Figure 1B.
(D) Adaptive mutations. Three mutations were identified from whole-genome sequencing of P10 ΔORF3-E mNG virion. No mutation was found in the P1 ΔORF3-E mNG virion.
Exclusion of WT SARS-CoV-2 production
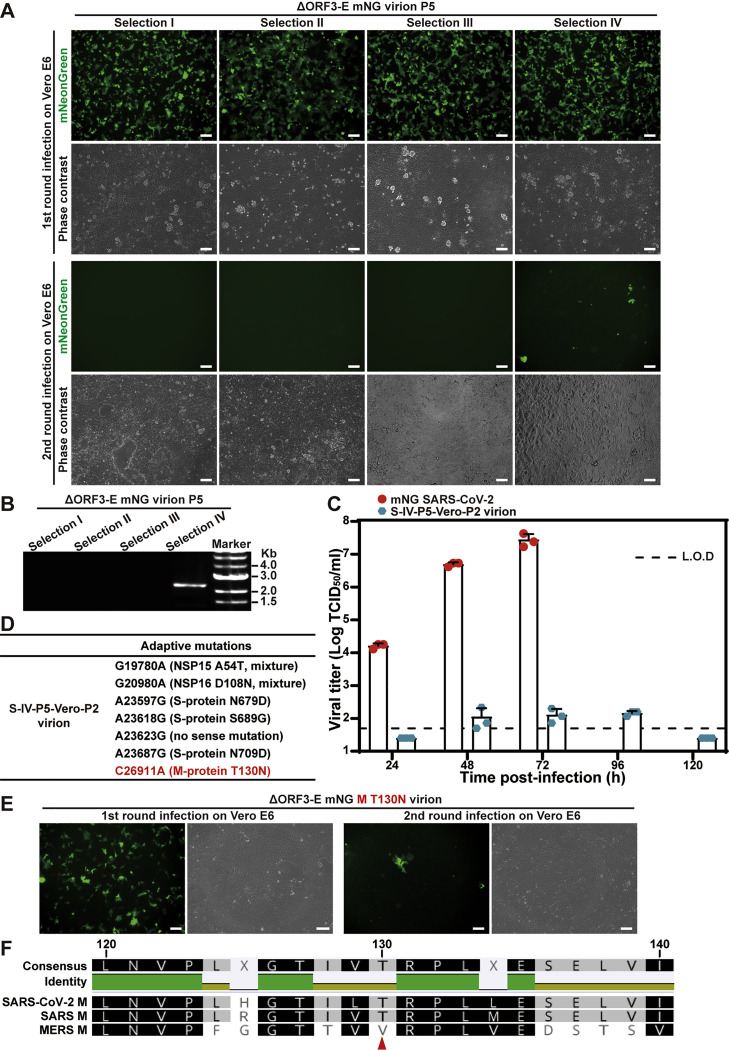
To confirm that no WT SARS-CoV-2 is inadvertently produced during trans-complementation, we performed four additional selections by passaging ΔORF3-E mNG virions on Vero-ORF3-E cells for five rounds. The P5 virions from selections I–III could only infect Vero cells for single round (Figures S3 A and S3B). Unexpectedly, selection IV produced P5 (S-IV-P5) virions that could infect parental Vero E6 cells for more than one round, though at a barely detectable level of ~102 TCID50/mL, which was >100,000-fold lower than the WT mNG SARS-CoV-2 titers (Figure S3C). To remove the single-round virion from the multi-round virion in the S-IV-P5 stock, we passaged the S-IV-P5 virion stock on Vero E6 cells for two rounds, resulting in S-IV-P5-Vero-P2 virion capable of multi-round infection. Full-genome sequencing revealed that the S-IV-P5-Vero-P2 virion retained the ORF3-E deletion but accumulated mutations in nsp15, nsp16, S, and M genes (Figure S3D). Engineering the accumulated mutations into ΔORF3-E mNG RNA showed that the M mutation T130N conferred multi-round infection on Vero cells (Figure S3E). Residue T130 is predicted to be on the intra-virion side of the M protein (Mahtarin et al., 2020; Thomas, 2020) and is conserved in SARS-CoV and SARS-CoV-2 (Figure S3F). The results indicate that, despite an absence of WT SARS-CoV-2 production and a lack of ORF3 and E genes, the trans-complementation system could produce mutant virions capable of infecting parental Vero cells for multiple rounds at a barely detectable level.
Figure S3.
Selection of ΔORF3-E mNG virion capable of inefficiently infecting Vero E6 cells for more than one round, related to Figure 2
Four independently selected P5 ΔORF3-E mNG virions (generated from five rounds of passaging ΔORF3-E mNG virion on Vero-ORF3-E cells) were used to infect naive Vero E6 cells for two rounds as described in Figure 1F.
(A) The P5 ΔORF3-E mNG virion-infected Vero E6 cells were analyzed for mNG signals. Scale bar, 100 μm.
(B) The extracellular RNA from the second-round infected cells were examined for viral RNA by RT-PCR.
(C) Selection IV P5 ΔORF3-E (S-IV-P5) mNG virion could infect Vero cells for multiple rounds. To remove the single-round virion from the multi-round virion in the S-IV-P5 stock, the S-IV-P5 stock was passaged on Vero E6 cells for two rounds, resulting in S-IV-P5-Vero-P2 virion capable of multi-round infection. The replication kinetics of WT mNG SARS-CoV-2 and S-IV-P5-Vero-P2 mNG virion were compared on Vero E6 cells. The cells were inoculated at an MOI of 0.001. Limit of detection, L.O.D. Data are represented as mean ± SD.
(D) Adaptive mutations were identified from the S-IV-P5-Vero-P2 mNG virion.
(E) The T130N mutation from the M protein was engineered into ΔORF3-E mNG virion. The resulting ΔORF3-E mNG M T130N virion was used to infect Vero E6 cells for two rounds. Fluorescence and phase contrast images of the infected cells are shown. Scale bar, 100 μm.
(F) Sequence alignment shows that the M proteins from SARS-CoV and SARS-CoV-2 share the same T130 residue. Red arrow indicates the T130 residue of SARS-CoV-2.
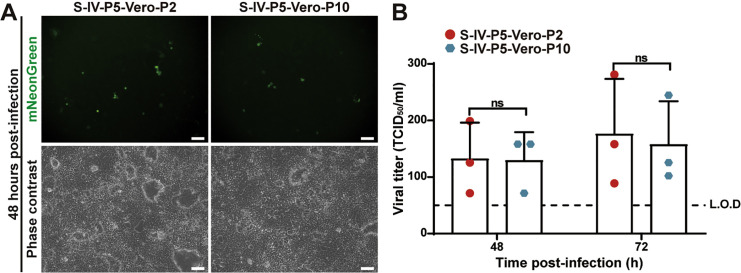
Next, we continuously cultured the S-IV-P5 variant on parental Vero E6 cells for 10 rounds (3–4 days per round) to select for potential virions with improved replication efficiency. However, passage did not improve viral replication on Vero cells (Figure S4 ). The result suggests that, due to the lack of ORF3 and E gene, the S-IV-P5 virion is unlikely to gain efficient multiple-round amplification on normal cells through adaptation.
Figure S4.
No improvement of viral replication of selection IV ΔORF3-E (S-IV-P5) mNG virion after 10 rounds of culturing on Vero E6 cells, related to Figure 2
S-IV-P5 mNG virion was continuously passaged on Vero E6 cells for 10 rounds. The resulting P2 and P10 S-IV-P5 mNG virions (i.e., S-IV-P5-Vero-P2 and S-IV-P5-Vero-P10, respectively) were used to infect Vero E6 cells at an MOI of 0.001. The mNG-positive cells (A) and the growth kinetics of the S-IV-P5-Vero-P2 and S-IV-P5-Vero-P10 virions (B) were compared. Scale bar, 100 μm. We did not use the S-IV-P5-Vero-P1 virion in this experiment because the P1 stock retained some carryover virions derived from the Vero-ORF3-E trans-complementation culture. Viral titers were analyzed by unpaired t test. ns, p > 0.05. Data are represented as mean ± SD.
Safety evaluation of ΔORF3-E virions in vivo
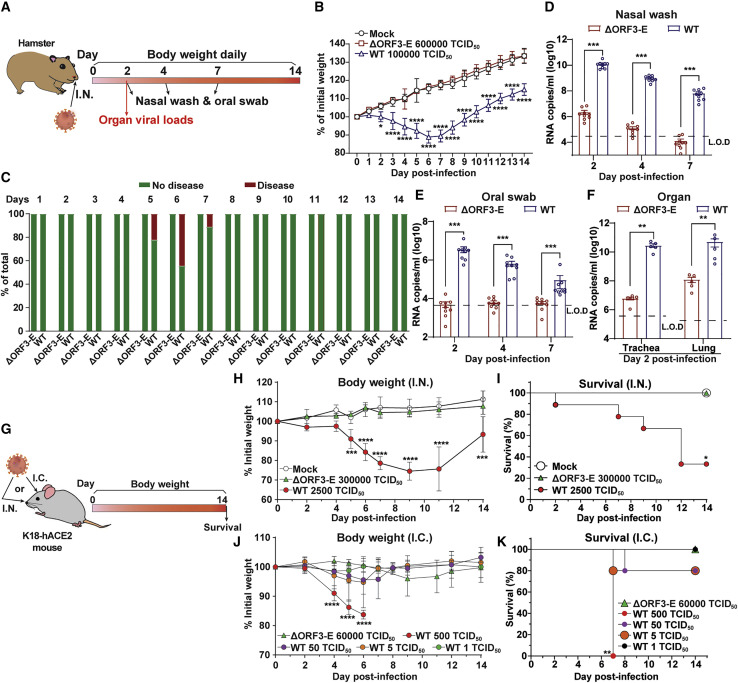
We examined the virulence of ΔORF3-E mNG virion in hamsters and K18-hACE2 transgenic mice (Chan et al., 2020; McCray et al., 2007; Winkler et al., 2020). After intranasal inoculation with 6 × 105 TCID50 of ΔORF3-E mNG virion (the highest possible infecting dose; Figure 3 A), hamsters did not lose weight (Figure 3B) or develop detectable disease (Figure 3C). In contrast, 105 TCID50 of WT SARS-CoV-2-infected hamsters developed weight loss and mild disease (e.g., ruffled fur). The ΔORF3-E mNG virion-infected hamsters contained low levels of viral RNA in nasal washes (Figure 3D) and oral swabs (Figure 3E). Viral RNA levels in the trachea and lungs from the ΔORF3-E virion-infected animals were 5,000- and 400-fold lower than those from the WT virus-infected hamsters, respectively (Figure 3F). Next, we examined the S-IV-P5-Vero-P2 virion, capable of infecting Vero cells for multiple rounds, in hamsters. To maximize the infection dose of S-IV-P5-Vero-P2 virion, we amplified S-IV-P5-Vero-P2 on Vero-ORF3-E cells, producing a virion stock of 5 × 104 TCID50/mL. After intranasal inoculation with 5 × 103 TCID50 of S-IV-P5-Vero-P2 virion (the highest possible dose), hamsters did not lose weight or develop detectable disease (Figures S5 A and S5B). Collectively, the results indicate that both ΔORF3-E mNG virion and S-IV-P5-Vero-P2 virion are highly attenuated and do not disseminate or cause disease in hamsters.
Figure 3.
Safety characterization of ΔORF3-E mNG virion in animal models
(A) Hamster experimental schedule. Four- to five-week-old male Syrian golden hamsters were intranasally (I.N.) inoculated with 105 TCID50 of WT SARS-CoV-2, 6 × 105 TCID50 of ΔORF3-E mNG virion, or PBS mock control. Hamsters were monitored for weight loss, disease, and viral RNA level.
(B) Hamster weight change (n = 9).
(C) Hamster disease (n = 9).
(D) Hamster nasal wash viral RNA level (n = 9).
(E) Hamster oral swab viral RNA level (n = 9).
(F) Viral RNA loads in hamster trachea and lung at day 2 post-infection (n = 5). Limit of detection (L.O.D.) was defined as the RNA copies detected from mock-infected hamster samples. The weight loss data are shown as mean ± standard deviation and statistically analyzed using two-way ANOVA Turkey’s multiple comparison. The genomic RNA levels are presented as mean ± standard error of the mean and analyzed by Mann-Whitney test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
(G–K) Mouse experimental schedule. Seven- to nine-week-old K18-hACE2 mice were inoculated with WT SARS-CoV-2 or ΔORF3-E mNG virion via the I.N. or intracranial (I.C.) route. Mouse weight loss after I.N. (H) or I.C. (J) infection. Body weights were normalized to the initial weight. The means for each group (I.N.: WT SARS-CoV-2 [n = 9], ΔORF3-E mNG virion [n = 4], and mock [n = 4]; I.C.: WT SARS-CoV-2 500 TCID50 [n = 4], 50 TCID50 [n = 5], 5 TCID50 [n = 5], and 1 TCID50 [n = 5] and 6 × 104 TCID50 ΔORF3-E mNG virus [n = 4]) are indicated, with error bars indicating the standard deviation. A mixed-model ANOVA using Dunnett’s test for multiple comparisons was used to evaluate the statistical significance among groups: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Mouse survival after I.N. (I) and I.C. (K) inoculation was analyzed using the Gehan-Breslow-Wilcoxon test. Using groups with 100% survival as a comparator, a Bonferroni correction was applied manually to adjust the threshold for significance (indicated by ∗).
Figure S5.
Safety characterization of S-IV-P5-Vero-P2 virion in hamsters and K18-hACE transgenic mice, related to Figure 3
(A and B) The weight change (A) and disease (B) of hamsters (n = 5) that were intranasally infected with 5,000 TCID50 of S-IV-P5-Vero-P2 virion. A high-titer stock of S-IV-P5-Vero-P2 virion used for this experiment was prepared by amplifying the virion on Vero-ORF3-E cells.
(C) Mouse weight loss after I.N. infection. Mice were infected with 2,500 TCID50 of S-IV-P5-Vero-P2 virion (n = 4) or PBS mock (n = 4) via the I.N. route. The mean ± standard deviations are indicated.
(D) Mouse survival after I.N. infection.
(E) Mouse weight loss after I.C. infection. Mouse were inoculated with 500 TCID50 of S-IV-P5-Vero-P2 virion (n = 4) via the I.C. route. The mean ± standard deviations are indicated.
(F) Mouse survival after I.C. infection. A high titer stock of S-IV-P5-Vero-P2 virion used for this experiment was prepared by amplifying the virion on Vero-ORF3-E cells.
To corroborate the hamster results, we tested ΔORF3-E mNG virion in more susceptible K18-hACE2 mice (Figure 3G). After intranasal inoculation with 3 × 105 TCID50 of ΔORF3-E mNG virion (the highest possible dose), K18-hACE2 mice did not lose weight (Figure 3H) or die (Figure 3I); in contrast, infection with 2.5 × 103 TCID50 of WT SARS-CoV-2 resulted in 25% weight loss and 67% lethality. To increase the stringency of the test, we inoculated K18-hACE2 mice by intracranial injection with 6 × 104 TCID50 of ΔORF3-E mNG virion (the highest possible dose); no morbidity (Figure 3J) or mortality (Figure 3K) was observed. In contrast, mice inoculated by the intracranial route with 500, 50, 5, and 1 TCID50 of WT SARS-CoV-2 developed 100%, 25%, 25%, and 0% mortality, respectively (Figure 3K). Similar to the ΔORF3-E mNG virion, no morbidity or mortality was observed after mice were inoculated by the intranasal or intracranial route with 2.5 × 103 or 5 × 102 TCID50 of S-IV-P5-Vero-P2 virion, respectively (Figures S5C–S5F). Together, the results demonstrate that both single-round ΔORF3-E mNG virion and multiple-round S-IV-P5-Vero-P2 virion lack virulence in K18-hACE2 mice.
High-throughput neutralization and antiviral testing
We adapted ΔORF3-E mNG virion for a high-throughput neutralization and antiviral assay. Figure 4 A outlines the assay scheme in a 96-well plate format. Neutralization titers of 20 convalescent sera from COVID-19 patients were measured by two assays for comparison: the ΔORF3-E mNG virion assay and the gold standard plaque-reduction neutralization test (PRNT). The two assays produced comparable 50% neutralization titers (NT50) for all specimens (Figures 4B and 4C; Table 1 ). In addition, the ΔORF3-E mNG virion assay could also be used to measure the 50% effective neutralizing concentration (EC50) for a monoclonal antibody against SARS-CoV-2 receptor-binding domain (RBD; Figure 4D). Finally, using Remdesivir as a viral polymerase inhibitor, we evaluated the ΔORF3-E mNG virion assay for antiviral testing. Remdesivir exhibited a more potent EC50 on hACE2-A549 cells (0.27 μM; Figure 4E) than that on Vero cells (5.1 μM; Figure 4F). The EC50 discrepancy between the two cell types is likely due to different efficiencies in converting Remdesivir to its triphosphate form, as previously reported (Pruijssers et al., 2020; Xie et al., 2020b). Collectively, the results demonstrate that the ΔORF3-E virion assay can be used for high-throughput neutralization testing and antiviral drug discovery.
Figure 4.
ΔORF3-E mNG virion-based high-throughput neutralization and antiviral testing
(A) Assay scheme in a 96-well format.
(B) Correlation analysis of NT50 values between the ΔORF3-E mNG virion assay and plaque-reduction neutralization test (PRNT). The Pearson correlation coefficiency R2 and p values (two-tailed) are indicated.
(C) Neutralization curves. Representative curves are presented for one negative and three positive sera. The means and standard deviations from two independent experiments are shown.
(D) EC50 of human mAb14 against ΔORF3-E mNG virion infecting Vero CCL81 cells. The mean ± standard deviations from four independent experiments are indicated.
(E) EC50 of Remdesivir against ΔORF3-E mNG virion infecting A549-hACE2 cells.
(F) EC50 of Remdesivir against ΔORF3-E mNG virion on Vero CCL81 cells. For (E) and (F), the mean ± standard deviations from three independent experiments are indicated. The four-parameter dose-response curve was fitted using the nonlinear regression method.
Table 1.
Comparison of neutralization titers between ΔORF3-E mNG virion and PRNT assays
| Serum ID | ΔORF3-E virion-NT50 | PRNT50 |
|---|---|---|
| 1 | <20 | <20 |
| 2 | <20 | <20 |
| 3 | 59 | 80 |
| 4 | 81 | 80 |
| 5 | 169 | 160 |
| 6 | 225 | 200 |
| 7 | 274 | 320 |
| 8 | 353 | 320 |
| 9 | 370 | 320 |
| 10 | 392 | 320 |
| 11 | 394 | 400 |
| 12 | 568 | 320 |
| 13 | 585 | 800 |
| 14 | 666 | 400 |
| 15 | 677 | 640 |
| 16 | 744 | 320 |
| 17 | 909 | 800 |
| 18 | 925 | 640 |
| 19 | 1196 | 800 |
| 20 | 1789 | 1600 |
Discussion
We generated and characterized a trans-complementation system for SARS-CoV-2. The system produced a high yield of single-round infectious ΔORF3-E virion that could be used for neutralization and antiviral testing. An mNG reporter was introduced into the ΔORF3-E virion to indicate viral replication. Depending on research needs, other reporter genes, such as luciferase or GFP, could be engineered into the system. A reliable high-throughput neutralization assay is important for COVID-19 vaccine evaluation and for studying the kinetics of neutralizing antibody levels in post-vaccinated and naturally infected people (Mulligan et al., 2020; Walsh et al., 2020; Widge et al., 2020). Three types of cell-based high-throughput neutralization assays currently are available: (1) pseudovirus assay, which expresses SARS-CoV-2 S protein alone on heterologous viruses, can be performed at BSL-2 laboratories (Case et al., 2020; Zeng et al., 2020); (2) a reporter SARS-CoV-2 assay, which must be performed at BSL-3 laboratories, represents authentic viral infection (Hou et al., 2020; Muruato et al., 2020; Thi Nhu Thao et al., 2020; Xie et al., 2020b); and (3) bona fide fully infectious SARS-CoV-2 by focus reduction neutralization test or PRNT assay (Case et al., 2020; Xie et al., 2021b). The ΔORF3-E mNG virion combines the advantages of each assay type by recapitulating the authentic viral infection for a single round, thus supporting its use at BSL2 laboratories. The ΔORF3-E mNG virion can be readily adapted to investigate vaccine-elicited neutralization against newly emerged SARS-CoV-2 isolates, such as the rapidly spreading United Kingdom and South African strains (Kupferschmidt, 2021; Xie et al., 2021a), by swapping or mutating the S gene.
The trans-complementation system can also be used for high-throughput antiviral screening of large compound libraries. Infection of normal cells with ΔORF3-E mNG virion allows for screening of inhibitors of virus entry, genome translation, and RNA replication, but not virion assembly/release. In contrast, infection of Vero-ORF3-E cells with ΔORF3-E mNG virion can be used to identify inhibitors of all steps of SARS-CoV-2 infection cycle, including virion assembly and release; this system also allows selection for resistance to inhibitors for mode-of-action studies. In addition, the single-round ΔORF3-E virion could be developed as a safe vaccine platform, as previously reported for other coronaviruses (Almazán et al., 2013; Ortego et al., 2002).
Our results support that the trans-complementation system can be performed safely in BSL-2 laboratories. (1) The system produced single-round infectious ΔORF3-E mNG virion that does not infect normal cells for multiple rounds and thus cannot spread in vitro or in vivo. (2) The system did not produce WT virus, even after multiple independent selections. (3) Although an adaptive mutation in M protein was selected to confer multi-round infection on normal cells, the replication level of such virion (i.e., S-IV-P5-Vero-P2) was barely detectable, with infectious titers >100,000-fold lower than the WT SARS-CoV-2. The molecular mechanism of how S-IV-P5-Vero-P2 could infect cells for multiple rounds without the ORF3 and E proteins remains to be defined. (4) Continuous culturing of the S-IV-P5-Vero-P2 virion on naive Vero cells did not improve viral replication. (5) When hamsters and K18-hACE2 mice were infected with the highest possible doses, neither ΔORF3-E mNG virion nor S-IV-P5-Vero-P2 virion caused morbidity or mortality. Even after intracranial infection with the highest possible dose, neither virions caused detectable disease or death in the highly susceptible K18-hACE2 mice. If further safety improvement is needed, more accessory ORFs could be deleted from the ΔORF3-E mNG RNA as accessory proteins are not essential for viral replication (Hu et al., 2020). For our trans-complementation system, it remains to be determined whether trans-supply of ORF3 is essential for a robust production of single-round virion. We trans supplied the ORF3 protein in our SARS-CoV-2 system because (1) ORF3a was previously reported in the virion of SARS-CoV (Shen et al., 2005) and (2) deletion of both ORF3 and E genes was lethal for SARS-CoV (Castaño-Rodriguez et al., 2018).
In summary, we have developed a trans-complementation system for SARS-CoV-2 that likely can be performed at BSL-2 laboratories for COVID-19 research and countermeasure development. Based on the results presented in this study, the Institutional Biosafety Committee of the University of Texas Medical Branch at Galveston has approved the trans-complementation system for BSL-2. Thus, the experimental system could be used by researchers in industry, academia, and government laboratories who lack access to a BSL-3 facility.
Limitations of the study
This study has a few limitations. First, the concept of trans-complementation has been previously established for coronaviruses (Almazán et al., 2013; Ortego et al., 2002), even though the current single-round infectious ΔORF3-E mNG virion will facilitate research by enabling to perform SARS-CoV-2 infection and replication at BSL-2. Second, our study used Vero E6 cells for constructing the Vero-ORF3-E cell line. When propagated on Vero E6 cells, SARS-CoV-2 could accumulate deletions at the furin cleavage site in the S protein (Klimstra et al., 2020; Lau et al., 2020). This cleavage deletion affects the neutralization susceptibility of SARS-CoV-2 and possibly the route of virus entry into cells (Johnson et al., 2020). Although we did not observe furin cleavage deletions when our ΔORF3-E mNG virion was passaged on Vero-ORF3-E cells, this liability could be minimized or eliminated by using other cell lines, such as A549-hACE2 or Vero-TMPRSS2-hACE2 cells. Finally, the current system has the R812R mutation in the S protein that is required to increase the trans-complementation and viral titers of ΔORF3-E mNG virion. This S mutation could affect certain epitope conformation when the single-round ΔORF3-E mNG virion assay is used to examine monoclonal antibody neutralization.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal antibody (mAb14) | Ku et al., 2021 | N/A |
| Bacterial and virus strains | ||
| TransforMax EPI300 Chemically Competent E. coli | Lucigen Corporation | Cat#C300C105 |
| NEB Stable Competent E. coli | New England Biolabs | Cat#C3040I |
| icSARS-CoV-2-mNG reporter virus | Xie et al., 2020a | N/A |
| icSARS-CoV-2 virus | Xie et al., 2020a | N/A |
| Biological samples | ||
| COVID-19 patient serum samples | UTMB Hospital | IRB#20-0070 |
| Chemicals, peptides, and recombinant proteins | ||
| Puromycin | Thermo Fisher Scientific | Cat#A1113802 |
| Doxycycline | Sigma-Aldrich | Cat#D3447 |
| Polybrene | Sigma-Aldrich | Cat#TR-1003 |
| Remdesivir | Gilead Sciences | N/A |
| TRIzol LS Reagent | Thermo Fisher Scientific | Cat#10296028 |
| Critical commercial assays | ||
| Lenti-X Packaging Single Shots (VSV-G) | Takara | Cat#631275 |
| iScript SYBR Green One-Step kit | Bio-Rad | Cat#170-8893 |
| Ingenio Electroporation solution | Mirus Bio LLC | Cat#MIR 50117 |
| T7 mMessage mMachine kit | Thermo Fisher Scientific | Cat#AM1344 |
| QIAquick Gel Extraction Kit | QIAGEN | Cat#28706 |
| SuperScript™ IV One-Step RT-PCR kit | Thermo Fisher Scientific | Cat#12594025 |
| NEBuilder® HiFi DNA Assembly kit | New England Biolabs | Cat#E5520S |
| Deposited data | ||
| Raw image data for electron microscopy | This paper; and Mendeley Data | https://data.mendeley.com/api/datasets/s2dm94z42t/draft/files/5ec65493-deeb-4669-b292-efae0e6539be |
| Experimental models: cell lines | ||
| HEK293T cells | ATCC | Cat#CRL-3216, RRID:CVCL_0063 |
| Vero CCL-81 cells | ATCC | Cat#CCL-81, RRID:CVCL_0059 |
| Vero E6 cells | ATCC | Cat#CRL-1586, RRID: CVCL_0574 |
| Calu-3 cells | ATCC | Cat#HTB-55, RRID:CVCL_0609 |
| A549-hACE2 cells | Mossel et al., 2005 | N/A |
| Vero-ORF3-E cells | This paper | N/A |
| Experimental models: organisms/strains | ||
| Hamster: HsdHan:AURA | Envigo | Cat#8902M |
| Mouse: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J | Jackson Labs | Cat#034860 |
| Oligonucleotides | ||
| Primer pcov-F56-F1 (TATACGAAGTTATA TTCGATGCGGCCGCGTCTCAGAGTGCT TTGGTTTATGATAATAAG) |
Sigma-Aldrich | N/A |
| Primer pncov-R5 (TCGCACTAGAATA AACTCTGAACTC) |
Sigma-Aldrich | N/A |
| Primer pncov-F6 (AGTTCAGAGT TTATTCTAGTGCGAATAATTG CACTTTTGAATATG) |
Sigma-Aldrich | N/A |
| Primer pncov-R6 (ATGGCTAGTGTAACT AGCAAGAATACCAC) |
Sigma-Aldrich | N/A |
| Primer pncov-F7 (GTATTCTTGCTAGTT ACACTAGCCATCCTTACTGCGCTTCG) |
Sigma-Aldrich | N/A |
| Primer pncov-R8 (AGGTCGACTCTAGAGGATCC) | Sigma-Aldrich | N/A |
| Primer cov-21115-F (CATTTGTGGGTTTATACAACAAAAG) | Sigma-Aldrich | N/A |
| Primer TRS2-S-R (GAAAAACA AACATTATCCGGTTAGTTGTT AACAAG) |
Sigma-Aldrich | N/A |
| Primer TRS2-S-F (CTTGTTAACAACTAA CCGGATAATGTTTGTTTTTC) |
Sigma-Aldrich | N/A |
| Primer S-TRS2-M-R (GAAAAACTAATA TAATATTTAATCCGGTTATGTGTAA TGTAATTTGACTCCTTTGAGC) |
Sigma-Aldrich | N/A |
| Primer TRS2-M-F (CCGGATTAAATATT ATATTAGTTTTTCTG) |
Sigma-Aldrich | N/A |
| Primer M-TRS2-R (GTAATAAGAAAG CGTCCGGGATGTAGCAACAGTG) |
Sigma-Aldrich | N/A |
| Primer M-TRS2-F (CACTGTTGCTAC ATCCCGGACGCTTTCTTATTAC) |
Sigma-Aldrich | N/A |
| Primer ORF6-TRS2-mNG-R (CTTTGCTCA CCATATCCGGTTAATCAATCTCC) |
Sigma-Aldrich | N/A |
| Primer ORF6-TRS2-mNG-F (GGAGATTGA TTAACCGGATATGGTGAGCAAAG) |
Sigma-Aldrich | N/A |
| Primer ORF7-TRS2-ORF8-R (CAAGAAA TTTCATATCCGGTTAGGCGTGACAAG) |
Sigma-Aldrich | N/A |
| Primer ORF7-TRS2-ORF8-F (CTTGTCAC GCCTAACCGGATATGAAATTTCTTG) |
Sigma-Aldrich | N/A |
| Primer ORF8-TRS2-N-R (CATTATCAG ACATTTTAGTTTATCCGGTTAGAT GAAATCTAAAACAACACGAACGTC) |
Sigma-Aldrich | N/A |
| Primer TRS2-N-F (CCGGATAAAC TAAAATGTCTGATAATGG) |
Sigma-Aldrich | N/A |
| Primer cov-28501-R (GGTGTTAATT GGAACGCCTTGTCC) |
Sigma-Aldrich | N/A |
| Primer M-T130N-F (CCATGGCACTA TTCTGAACAGACCGCTTCTAGAAAG) |
Sigma-Aldrich | N/A |
| Primer M-T130N-R (CTTTCTAGAAGCG GTCTGTTCAGAATAGTGCCATGG) |
Sigma-Aldrich | N/A |
| Primer 5ʹUTR-TRS2-F (GATCTGTTCTCTAA CCGGATTTTAAAATCTGTGTG) |
Sigma-Aldrich | N/A |
| Primer 5ʹUTR-TRS2-R (CACACAGATTTTA AAATCCGGTTAGAGAACAGATC) |
Sigma-Aldrich | N/A |
| Primer EcoR1-mCherry-F (CACTTCCTAC CCTCGTAAAGAATTCGCCACCATGG TGAGCAAGGGCGAGGAG) |
Sigma-Aldrich | N/A |
| Primer F2A-optE-R (GACACAAAAGA ATACATTGGCCCAGGGTTGGACTCGAC) |
Sigma-Aldrich | N/A |
| Primer F2A-optE-F (CCCTGGGCCAA TGTATTCTTTTGTGTCTGAAG) |
Sigma-Aldrich | N/A |
| Primer EcoR1-Cov-optE-R (GGGGAGGGA GAGGGGCGGGAATTCCTACACCAGCAG GTCGGGGACC) |
Sigma-Aldrich | N/A |
| Primer EcoR1-IRES-F (TAGGAATTCCC GCCCCTCTCCCTCCCCCC) |
Sigma-Aldrich | N/A |
| Primer EMCV-IRES-R (ATTATCATCG TGTTTTTCAAAGGAAAACCACG) |
Sigma-Aldrich | N/A |
| Primer IRES-optORF3-F (GTTTTCCTTT GAAAAACACGATGATAATATGGA CCTGTTCATGAGAATC) |
Sigma-Aldrich | N/A |
| Primer BamH1-Cov-optORF3-R (CTCGCAGGGGAGGTGGTCTGGATCC CTCACAGAGGAACAGATGTGGTGG) |
Sigma-Aldrich | N/A |
| Primer CoV-T7-N-F (ACTGTAATACGA CTCACTATAGGATGTCTGATAATGGA CCCCAAAATC) |
Sigma-Aldrich | N/A |
| Primer polyT-N-R ([T]37AGGCCTGAGTTGAGTCAGCAC) | Sigma-Aldrich | N/A |
| Primer CoV19-N2-F (TTACAAACATTGGCCGCAAA) | Sigma-Aldrich | N/A |
| Primer CoV19-N2-R (GCGCGACATTCCGAAGAA) | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| pUC57-CoV2-F1 | Xie et al., 2020a | N/A |
| pCC1-CoV2-F2 | Xie et al., 2020a | N/A |
| pCC1-CoV2-F3 | Xie et al., 2020a | N/A |
| pUC57-CoV2-F4 | Xie et al., 2020a | N/A |
| pUC57-CoV2-F5 | Xie et al., 2020a | N/A |
| pUC57-CoV2-F6 | Xie et al., 2020a | N/A |
| pCC1-CoV2-F7-mNG | Xie et al., 2020a | N/A |
| pCC1-CoV2-F567-mNG | This paper | N/A |
| pCC1-CoV2-F567-mNG-ΔORF3-E | This paper | N/A |
| pLVX-TetOne-Puro | Takara | Cat#631849 |
| pLVX-ORF3-E | This paper | N/A |
| Synthesized SARS-CoV-2 ORF3 gene (sequence-optimized) | This paper | N/A |
| Synthesized SARS-CoV-2 E gene (sequence-optimized) | This paper | N/A |
| Software and algorithms | ||
| Geneious | Biomatters | N/A |
| ImageJ | NIH | N/A |
| Prism 9.0 software | GraphPad | N/A |
| Illustrator CC | Adobe | N/A |
| Image Lab | Bio-rad | N/A |
| BioRender | BioRender | N/A |
| QuantStudio Software v1.3 | ThermoFisher Scientific | N/A |
| Other | ||
| T4 DNA Ligase | New England Biolabs | Cat#M0202S |
| BsaI restriction endonuclease | New England Biolabs | Cat#R3733S |
| Esp3I restriction endonuclease | New England Biolabs | Cat#R0734S |
| EcoRI restriction endonuclease | New England Biolabs | Cat#R3101S |
| BamHI restriction endonuclease | New England Biolabs | Cat#R3136S |
| Platinum SuperFi II DNA Polymerase | Thermo Fisher Scientific | Cat#12361010 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact, Dr. Pei-Yong Shi (peshi@utmb.edu)
Materials availability
The mNG reporter SARS-CoV-2 has been deposited to the World Reference Center for Emerging Viruses and Arboviruses (https://www.utmb.edu/wrceva) at UTMB for distribution. All reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The uncropped EM images of ΔORF3-E mNG virion have been deposited to Mendeley Data: https://data.mendeley.com/api/datasets/s2dm94z42t/draft/files/5ec65493-deeb-4669-b292-efae0e6539be.
Experimental model and subject details
Cell lines
Vero E6, Vero CCL-81, Calu-3, and HEK293T cells were purchased from the American Type Culture Collection (ATCC) and cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM L-glutamine, 100 U/mL Penicillium-Streptomycin (P/S), and 10% fetal bovine serum (FBS; HyClone Laboratories, South Logan, UT). Vero-ORF3-E cells were maintained in DMEM medium supplemented with 2mM L-glutamine, 100 U/mL P/S, 10% FBS, 0.075% sodium bicarbonate, and 10 μg/mL puromycin. The A549-hACE2 cells were generously provided by Shinji Makino (Mossel et al., 2005) and grown in the culture medium supplemented with 10 μg/mL blasticidin and 10 mM HEPES at 37°C with 5% CO2. Medium and other supplements were purchased from Thermo Fisher Scientific (Waltham, MA).
Selection of Vero-ORF3-E cell line
For packaging the lentivirus, the pLVX-ORF3-E plasmid was transfected into HEK293T cells using the Lenti-X Packaging Single Shots kit (Takara). Lentiviral supernatants were harvested at 72 h post-transfection and filtered through a 0.45 μM membrane (Millipore, Burlington, MA). One day before transduction, Vero E6 cells were seeded in a 6-well plate (4 × 105 per well) with DMEM containing 10% FBS. After 12-18 h, cells were transduced with 2 mL lentivirus for 24 h in the presence of 12 μg/mL of polybrene (Sigma-Aldrich, St. Louis, MO). At 24 h post-transduction, cells from a single well were split into four 10 cm dishes and cultured in medium supplemented with 25 μg/mL of puromycin. The culture medium containing puromycin was refreshed every 2 days. After 2-3 weeks of selection, visible puromycin-resistant cell colonies were formed. Several colonies were transferred into 24-well plates. When confluent, cells were treated with trypsin and seeded in 6-well plates for further expansion. The resulting cells were defined as Vero-ORF3-E P0 cells. For cell line verification, total cellular mRNA was isolated and subject to RT-PCR with primers EcoR1-mCherry-F and BamH1-Cov-optORF3-R, followed by cDNA sequencing of the ORF3-E genes.
Hamsters
Hamster studies were performed in accordance with the guidance for the Care and Use of Laboratory Animals of the University of Texas Medical Branch (UTMB). The protocol (2009087) was approved by the Institutional Animal Care and Use Committee (IACUC) at UTMB. All the hamster operations were performed under anesthesia by isoflurane to minimize animal suffering. Syrian golden hamsters (HsdHan:AURA strain) were purchased from Envigo. Animals were housed in groups and fed standard chow diets. Hamster experiments were performed as described previously (Plante et al., 2020). Briefly, 105 TCID50 of WT SARS-CoV-2, 6 × 105 TCID50 of ΔORF3-E mNG virion, or 5 × 103 TCID50 of S-IV-P5-Vero-P2 virion in 100 μL volume were inoculated into four- to five-week-old male Syrian golden hamsters via the intranasal route. The S-IV-P5-Vero-P2 virion stock was prepared by two rounds of culturing of S-IV-P5 virion (Figure S4A) on Vero E6 cells (to remove single-round infectious virion), followed by propagation on Vero-ORF3-E cells. Fourteen hamsters were used in SARS-CoV-2- and ΔORF3-E mNG virion-infected groups and 5 hamsters were used in S-IV-P5-Vero-P2 virion-infected group. From day 1 to 14 post-infection, hamsters were observed daily for weight change and signs of illness. Five hamsters in WT SARS-CoV-2-, ΔORF3-E mNG virions-, or mock-infected group were sacrificed on day 2 post-infection for lung and trachea collections. Nasal washes and oral swabs of the rest 9 hamsters per group were collected on days 2, 4, and 7 post-infection.
Mice
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Heterozygous K18-hACE c57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from the Jackson Laboratory. Animals were randomized upon arrival at Washington University and housed in groups of < 5 per cage in rooms maintained between 68-74°F with 30%–60% humidity and day/night cycles of 12 h intervals (on 6AM-6PM). Mice were fed standard chow diets. Mice 7-9 weeks of age and of both sexes were used for this study. Intranasal virus inoculations (50 uL/mouse) were performed under sedation with ketamine hydrochloride and xylazine while intracranial virus inoculations (10 μL/mouse) were performed under sedation with isoflurane; all efforts were made to minimize animal suffering.
Human serum specimens
The use of human sera for this study was reviewed and approved by the University of Texas Medical Branch (UTMB) Institutional Review Board. The approved Institutional Review Board protocol number is 20-0070. All human sera were obtained at the UTMB. All specimens were de-identified from patient information. A total of twenty de-identified convalescent sera from COVID-19 patients, previously confirmed by positive viral RT-PCR diagnosis, were tested in this study. Since all specimens were de-identified, information about gender, sex, or age of donors was not available.
Biosafety
All aspects of this study were approved by the Institutional Biosafety Committee of the University of Texas Medical Branch at Galveston before the initiation of this study. Experiments with SARS-CoV-2, trans-complementation, and ΔORF3-E mNG virion were performed in a BSL-3 laboratory by personnel equipped with powered air-purifying respirators.
Method details
Plasmid construction
Seven previously reported subclone plasmids for the assembly of the entire genome of SARS-CoV-2 were used in this study, including pUC57-CoV2-F1, pCC1-CoV2-F2, pCC1-CoV2-F3, pUC57-CoV2-F4, pUC57-CoV2-F5, pUC57-CoV2-F6, and pCC1-CoV2-F7-mNG (Muruato et al., 2020; Xie et al., 2020a). For the convenience of deleting ORF3-E gene, we constructed F5, F6, and F7-mNG fragments into one plasmid. F5, F6, and F7-mNG fragments were amplified from corresponding subclones via PCR with primer pairs pcov-F56-F1/pncov-R5, pncov-F6/pncov-R6, and pncov-F7/pncov-R8, respectively. All PCR products were cloned together into a pCC1 vector through NotI and ClaI restriction sites using the standard restriction digestion-ligation cloning, resulting in subclone pCC1-CoV2-F567-mNG.
To introduce ORF3-E deletion and mutant Transcription Regulatory Sequence (TRS) into pCC1-CoV2-F567-mNG, seven fragments were amplified with primer pairs cov-21115-F/TRS2-S-R, TRS2-S-F/S-TRS2-M-R, TRS2-M-F/M-TRS2-R, M-TRS2-F/ORF6-TRS2-mNG-R, ORF6-TRS2-mNG-F/ORF7-TRS2-ORF8-R, ORF7-TRS2-ORF8-F/ORF8-TRS2-N-R, and TRS2-N-F/cov-28501-R. The seven PCR products were assembled into the pCC1-CoV2-F567-mNG plasmid that were pre-linearized with NheI and XhoI by using the NEBuilder® HiFi DNA Assembly kit (NEB) according to the manufacturer’s instruction, resulting in subclone pCC1-CoV2-F567-mNG-ΔORF3-E. Mutation T130N in M protein was engineered into pCC1-CoV2-F567-mNG-ΔORF3-E with primers M-T130N-F/M-T130N-R via overlap PCR. Mutant TRS was engineered into pCC1-F1 with primers 5ʹUTR-TRS2-F and 5ʹUTR-TRS2-R via overlap PCR.
For making the Vero-ORF3-E cell lines, codon-optimized SARS-CoV-2 ORF3 and E genes were synthesized by GenScript Biotech (Piscataway, NJ). An mCherry reporter Zika virus cDNA plasmid (Shan et al., 2017) was used as a template to amplify the mCherry-F2A gene. For constructing a lentiviral plasmid expressing ORF3 and E protein of SARS-CoV-2, DNA fragments encoding mCherry-F2A, SARS-CoV-2 E, EMCV IRES, and SARS-CoV-2 ORF3 were amplified with primers EcoR1-mCherry-F/F2A-optE-R, F2A-optE-F/EcoR1-Cov-optE-R, EcoR1-IRES-F/EMCV-IRES-R, and IRES-optORF3-F/BamH1-Cov-optORF3-R, respectively. The PCR products then were inserted into a inducible lentiviral vector pLVX-TetOne-Puro (Takara, Mountain View, CA) through EcoRI and BamHI restriction sites, resulting in plasmid pLVX-ORF3-E.
ΔORF3-E mNG cDNA assembly and in vitro RNA transcription
Full-length genome assembly and RNA transcription were performed as described previously with minor modifications (Xie et al., 2020a). Briefly, individual subclones containing fragments of the ΔORF3-E mNG viral genome were digested with appropriated restriction endonucleases and resolved in a 0.8% agarose gel. Specifically, the plasmids containing F1, F2, F3, or F4 fragments were digested with BsaI enzyme, and the plasmid containing F567-mNG-ΔORF3-E fragment was digested with Esp3I enzyme. All fragments were recovered using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany), and total of 5 μg of the five fragments was ligated in an equal molar ratio by T4 DNA ligase (New England Biolabs, Ipswich, MA) at 4°C overnight. Afterward, the assembled full-length genomic cDNA was purified by phenol-chloroform extraction and isopropanol precipitation. ΔORF3-E mNG RNA transcripts were generated using the T7 mMessage mMachine kit (Ambion, Austin, TX). To synthesize the N gene RNA transcript of SARS-CoV-2, the N gene was PCR-amplified by primers CoV-T7-N-F and polyT-N-R from a plasmid containing the F7 fragment (Xie et al., 2020a); the PCR product was then used for in vitro transcription using the T7 mMessage mMachine kit (Ambion).
ΔORF3-E mNG virion production and quantification
Vero-ORF3-E cells were seeded in a T175 flask and grown in DMEM medium with 100 ng/mL of doxycycline. On the next day, 40 μg of ΔORF3-E mNG RNA and 20 μg of N-gene RNA were electroporated into 8 × 106 Vero-ORF3-E cells using the Gene Pulser XCell electroporation system (Bio-Rad, Hercules, CA) at a setting of 270V and 950 μF with a single pulse. The electroporated cells were then seeded in a T75 flask and cultured in the medium supplemented with doxycycline (Sigma-Aldrich) at 37°C for 3-4 days. Virion infectivity was quantified by measuring the TCID50 using an end-point dilution assay as previously reported (Lindenbach, 2009). Briefly, Vero-ORF3-E cells were plated on 96-well plates (1.5 × 104 per well) one day prior to infection. The cells were cultured in medium with doxycycline as described above. ΔORF3-E mNG virions were serially diluted in DMEM medium supplemented with 2% FBS, with 6 replicates per concentration. Cells were infected with 100 μL of diluted virions and incubated at 37°C for 2-3 days. The mNG signals were counted under a fluorescence microscope (Nikon, Tokyo, Japan). TCID50 was calculated using the Reed & Muench method (Reed and Muench, 1938).
To assess viral RNA levels, a quantitative RT-PCR assay was conducted using an iTaq Universal SYBR Green one-step kit (Bio-Rad) on a QuantStudio 7 Flex Real-Time PCR Systems (Thermo fisher) by following the manufacturers’ protocols. Primers CoV19-N2-F and CoV19-N2-R targeting the N gene were used. Absolute RNA copies were determined by standard curve method using in vitro transcribed RNA containing genomic nucleotide positions 26,044 to 29,883 of the SARS-CoV-2 genome.
RNA extraction, RT-PCR, and cDNA sequencing
Supernatants of infected cells were collected and centrifuged at 1,000 g for 10 min to remove cell debris. Clarified culture fluids (250 μl) were mixed thoroughly with 1 mL of TRIzol LS reagent (Thermo Fisher Scientific). Extracellular RNA was extracted per manufacture’s instruction and resuspended in 20 μL of nuclease-free water. RT-PCR was performed using the SuperScript® IV One-Step RT-PCR kit (Thermo Fisher Scientific). Nine cDNA fragments (gF1 to gF9) covering the whole viral genome were generated with specific primers according to the protocol described previously (Xie et al., 2020a). Afterward, cDNA fragments were separated in a 0.8% agarose gel, purified using QIAquick Gel Extraction Kit (QIAGEN), and subjected to Sanger sequencing.
ΔORF3-E mNG virion neutralization assay
For neutralization testing, Vero CCL-81 cells (1.2 × 104) in 50 μL of DMEM containing 2% FBS and 100 U/mL P/S were seeded in each well of black μCLEAR flat-bottom 96-well plate (Greiner Bio-one, Kremsmünster, Austria). At 16 h post-seeding, 30 μL of 2-fold serial diluted human sera were mixed with 30 μL of ΔORF3-E mNG virion (MOI of 5) and incubated at 37°C for 1 h. Afterward, 50 μL of virus–sera complexes were transferred to each well of the 96-well plate. After incubating the infected cells at 37°C for 20 h, 25 μL of Hoechst 33342 Solution (400-fold diluted in Hank’s Balanced Salt Solution; Thermo Fisher Scientific) were added to each well to stain the cell nucleus. The plate was sealed with Breath-Easy sealing membrane (Diversified Biotech, Dedham, MA), incubated at 37°C for 20 min, and quantified for mNG-positive cells using the CellInsight CX5 High-Content Screening Platform (Thermo Fisher Scientific). Infection rates were determined by dividing the mNG-positive cell number to the total cell number. Relative infection rates were obtained by normalizing the infection rates of serum-treated groups to those of non-serum-treated controls. The curves of the relative infection rates versus the serum dilutions (log10 values) were plotted using Prism 9 (GraphPad, San Diego, CA). A nonlinear regression method was used to determine the dilution fold that neutralized 50% of mNG fluorescence (NT50). Each serum was tested in duplicates.
ΔORF3-E mNG virion for mAb and antiviral testing
Vero CCL-81 cells (1.2 × 104) or A549-hACE2 cells in 50 μL of culture medium containing 2% FBS were seeded in each well of black μCLEAR flat-bottom 96-well plate. At 16 h post-seeding, 2- or 3-fold serial diluted human mAb14 (Ku et al., 2021) or Remdesivir were mixed with ΔORF3-E mNG virion (MOI of 1). Fifty microliters of mixtures were transferred to each well of the 96-well plate. After incubating the infected cells at 37°C for 20 h, 25 μL of Hoechst 33342 Solution (400-fold diluted in Hank’s Balanced Salt Solution) were added to each well to stain the cell nucleus. The plate was sealed with Breath-Easy sealing membrane, incubated at 37°C for 20 min. mNG-positive cells were quantified and infection rates were calculated as described above. Relative infection rates were obtained by normalizing the infection rates of treated groups to those of non-treated controls. For Remdesivir, 0.1% of DMSO-treated groups were used as controls. A nonlinear regression method was used to determine the concentration that inhibited 50% of mNG fluorescence (EC50). Experiments were performed in triplicates or quadruplicates.
Transmission Electron Microscopy
Supernatants of infected cells were centrifuged for 10 min at 3,000 g to remove cellular debris. Nickel grids were incubated with clarified supernatants for 10 min followed by glutaraldehyde fixation and 2% uranyl acetate staining. Micrographs were taken using a JEM 1400 (JEOL USA Inc.). Multiple randomly selected fields were imaged.
Bioinformatics analysis
Fluorescence images were processed using ImageJ (Schneider et al., 2012). Virus sequences were download from the NCBI database and aligned using Geneious software. DNA gel images were analyzed using Image Lab software. Statistical graphs or charts were created using the GraphPad Prism 9 software. Figures were created and assembled using BioRender and Adobe illustration (San Jose, CA).
Quantification and statistical analysis
A linear regression model in the software Prism 9 (GraphPad) was used to calculate the NT50 and EC50 values from the ΔORF3-E virion assay. Pearson correlation coefficient and two-tailed p value are calculated using the default settings in the software Prism 9. An unpaired t test (for two-groups comparison) and ANOVA test (for multi-group comparison) were used in statistical analysis (∗, p < 0.05, significant; ∗∗, p < 0.01, very significant; ∗∗∗, p < 0.001, highly significant; ∗∗∗∗, p < 0.0001, extremely significant; ns, p > 0.05, not significant).
Acknowledgments
We thank John Bilello from Gilead for providing Remdesivir and Zhiqiang An from the University of Texas Health Science at Houston for providing mAb14. We thank Q2 Solutions for the sponsored research agreement and scientific input toward this work. P.-Y.S. was supported by NIH grants AI134907 and UL1TR001439; awards from the Sealy & Smith Foundation, Kleberg Foundation, John S. Dunn Foundation, Amon G. Carter Foundation, Gilson Longenbaugh Foundation, and Summerfield Robert Foundation. M.S.D. was supported by R01 AI157155. V.D.M. was supported by NIH grants U19AI100625, R00AG049092, R24AI120942, and a STARs Award from the University of Texas System. S.C.W. was supported by NIH grant R24 AI120942. J.L. is supported by the postdoctoral fellowship from the McLaughlin Fellowship Endowment at UTMB. P.R. and X.X. were partially supported by the Sealy & Smith Foundation.
Author contributions
X.Z., V.D.M., X.X., and P.-Y.S. conceived the study. X.Z., Y.L., J.L., A.L.B., K.S.P., J.A.P., J.Z., H.X., N.E.B., P.R., and X.X. performed the experiments. X.Z., Y.L., A.L.B., P.V.A., P.R., V.D.M., M.S.D., S.C.W., X.X., and P.-Y.S. analyzed the results. P.R. prepared the serum specimens. X.Z., Y.L., A.L.B., V.D.M., M.S.D., S.C.W., X.X., and P.-Y.S. wrote the manuscript.
Declaration of interests
X.Z., X.X., and P.-Y.S. have filed a patent on the trans-complementation system of SARS-CoV-2. M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions.
Published: February 23, 2021
References
- Almazán F., DeDiego M.L., Sola I., Zuñiga S., Nieto-Torres J.L., Marquez-Jurado S., Andrés G., Enjuanes L. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. MBio. 2013;4:e00650-e13. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., et al. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. MBio. 2018;9:e02325-17. doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L., et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge F., Luo Y., Liew P.X., Hung E. Derivation of a novel SARS-coronavirus replicon cell line and its application for anti-SARS drug screening. Virology. 2007;360:150–158. doi: 10.1016/j.virol.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Deming D.J., Deming M.E., Yount B.L., Baric R.S. Evaluation of a recombination-resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Commun. Biol. 2018;1:179. doi: 10.1038/s42003-018-0175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzig T., Scandella E., Schelle B., Ziebuhr J., Siddell S.G., Ludewig B., Thiel V. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J. Gen. Virol. 2004;85:1717–1725. doi: 10.1099/vir.0.80044-0. [DOI] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Xie X., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., Zhang L., et al. Furin Cleavage Site Is Key to SARS-CoV-2 Pathogenesis. bioRxiv. 2020 doi: 10.1101/2020.08.26.268854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Westaway E.G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra W.B., Tilston-Lunel N.L., Nambulli S., Boslett J., McMillen C.M., Gilliland T., Dunn M.D., Sun C., Wheeler S.E., Wells A., et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 2020;101:1156–1169. doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaki T., Xie X., Shi P.Y., Kameoka M. A PCR amplicon-based SARS-CoV-2 replicon for antiviral evaluation. Sci. Rep. 2021;11:2229. doi: 10.1038/s41598-021-82055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku Z., Xie X., Davidson E., Ye X., Su H., Menachery V.D., Li Y., Yuan Z., Zhang X., Muruato A.E., et al. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat. Commun. 2021;12:469. doi: 10.1038/s41467-020-20789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. Fast-spreading U.K. virus variant raises alarms. Science. 2021;371:9–10. doi: 10.1126/science.371.6524.9. [DOI] [PubMed] [Google Scholar]
- Lau S.Y., Wang P., Mok B.W., Zhang A.J., Chu H., Lee A.C., Deng S., Chen P., Chan K.H., Song W., et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D. Measuring HCV infectivity produced in cell culture and in vivo. Methods Mol. Biol. 2009;510:329–336. doi: 10.1007/978-1-59745-394-3_24. [DOI] [PubMed] [Google Scholar]
- Lo M.K., Tilgner M., Shi P.-Y. Potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 2003;77:12901–12906. doi: 10.1128/JVI.77.23.12901-12906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V., Körner F., Koch J., Herian U., Theilmann L., Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Mahtarin R., Islam S., Islam M.J., Ullah M.O., Ali M.A., Halim M.A. Structure and dynamics of membrane protein in SARS-CoV-2. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1861983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossel E.C., Huang C., Narayanan K., Makino S., Tesh R.B., Peters C.J. Exogenous ACE2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J. Virol. 2005;79:3846–3850. doi: 10.1128/JVI.79.6.3846-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Muruato A.E., Fontes-Garfias C.R., Ren P., Garcia-Blanco M.A., Menachery V.D., Xie X., Shi P.Y. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020;11:4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A.J., George A.S., Schäfer A., Leist S.R., Gralinksi L.E., Dinnon K.H., 3rd, Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020;32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C., Muruato A.E., Nunes B.T.D., Luo H., Xie X., Medeiros D.B.A., Wakamiya M., Tesh R.B., Barrett A.D., Wang T., et al. A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat. Med. 2017;23:763–767. doi: 10.1038/nm.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X., Lim S.G., Hong W., Tan Y.J. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi Nhu Thao T., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582:561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Thomas S. The Structure of the Membrane Protein of SARS-CoV-2 Resembles the Sugar Transporter SemiSWEET. Pathog. Immun. 2020;5:342–363. doi: 10.20411/pai.v5i1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2020;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021 doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021 doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Lindesmith L., Baric R.S. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc. Natl. Acad. Sci. USA. 2006;103:12546–12551. doi: 10.1073/pnas.0605438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Evans J.P., Pearson R., Qu P., Zheng Y.M., Robinson R.T., Hall-Stoodley L., Yount J., Pannu S., Mallampalli R.K., et al. Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight. 2020;5:e143213. doi: 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The uncropped EM images of ΔORF3-E mNG virion have been deposited to Mendeley Data: https://data.mendeley.com/api/datasets/s2dm94z42t/draft/files/5ec65493-deeb-4669-b292-efae0e6539be.