Abstract
Coronavirus disease 2019 (COVID-19) is a rapidly evolving infectious/inflammatory disorder which has turned into a global pandemic. With severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as its etiologic agent, severe COVID-19 cases usually develop uncontrolled inflammatory responses and cytokine storm-like syndromes. Measuring serum levels of pro-inflammatory cytokines (e.g., IL-6 and others) as inflammatory biomarkers may have several potential applications in the management of COVID-19, including risk assessment, monitoring of disease progression, determination of prognosis, selection of therapy and prediction of response to treatment. This is especially true for pediatric patients with COVID-19 associated Kawasaki-like disease and similar syndromes. In this report, we review the current knowledge of COVID-19 associated cytokines, their roles in host immune and inflammatory responses, the clinical significance and utility of cytokine immunoassays in adult and pediatric COVID-19 patients, as well as the challenges and pitfalls in implementation and interpretation of cytokine immunoassays. Given that cytokines are implicated in different immunological disorders and diseases, it is challenging to interpret the multiplex cytokine data for COVID-19 patients. Also, it should be taken into consideration that biological and technical variables may affect the commutability of cytokine immunoassays and enhance complexity of cytokine immunoassay interpretation. It is recommended that the same method, platform and laboratory should be used when monitoring differences in cytokine levels between groups of individuals or for the same individual over time. It may be important to correlate cytokine profiling data with the SARS-CoV-2 nucleic acid amplification testing and imaging observations to make an accurate interpretation of the inflammatory status and disease progression in COVID-19 patients.
Keywords: COVID-19, Cytokine, Inflammatory biomarkers, Multiplex cytokine analysis, Kawasaki disease
1. Introduction
Cytokines are hormone-like polypeptides which are produced by different cell types and play essential roles in a broad array of physiologic and pathological processes, including cellular growth and differentiation, wound healing, immunity, inflammation, cardiology, cancer, hematopoiesis, atherogenesis, autoimmune disorders (e.g., rheumatoid arthritis), and neuroimmunologic diseases (e.g., multiple sclerosis) [1], [2], [3], [4], [5], [6], [7], [8], [9]. As intercellular signaling molecules, cytokines may have multiple functions on a variety of cell types and exhibit considerable antagonistic and synergetic effects with other cytokines. They play essential roles in regulating both local and systemic inflammatory responses. Pro-inflammatory cytokines have been considered important immunological and inflammatory biomarkers [2], [10]. Detection and quantification of cytokine levels has become increasingly important in clinical laboratory medicine for assessment of many immunologic and inflammatory disorders as well as infectious diseases [2], [10], [11], [12], [13].
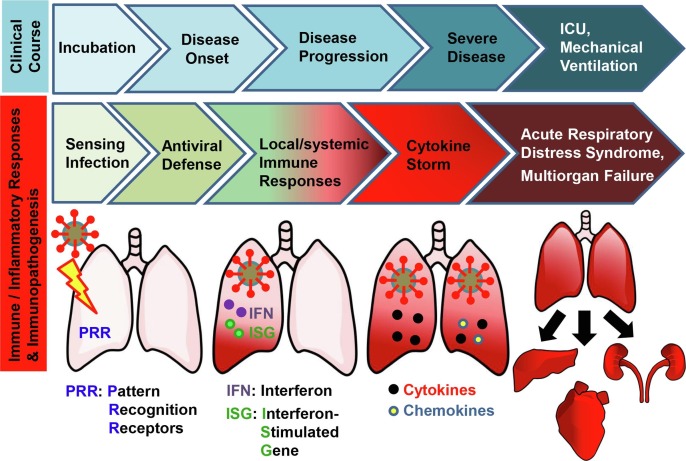
Coronavirus disease 2019 (COVID-19), a novel respiratory infectious disease, was first reported and recognized in December 2019 in the city of Wuhan in Hubei province, China, and evolved rapidly into a pandemic [14], [15], [16], [17]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel human coronavirus (HCoV), has been identified as the causative agent of COVID-19 [14], [15], [16], [17]. The complexity of COVID-19 is notoriously characterized by distinct patterns of disease progression and the unpredictable clinical course of the disease [10], [18]. Individuals contracting COVID-19 may experience the following stages of clinical illness, from an asymptomatic incubation period, disease onset with respiratory symptoms, disease progression, to severe disease phase (Fig. 1 , upper panel) [19]. Severe COVID-19 usually necessitates intensive care unit (ICU) admission and mechanical ventilation [20]. Concomitant with the basic phases of COVID-19 infection, host immune and inflammatory response to SARS-CoV-2 infection may be divided into a local innate immune response phase in the lung(s) (e.g., sensing infection and antiviral defense), later a local/systemic immune response phase, followed by an uncontrolled inflammatory responses and cytokine storm-like syndrome (Fig. 1, middle and lower panel) [19], [21]. Some severe cases may develop acute respiratory distress syndrome (ARDS) and multi-organ failure [3], [20].
Fig. 1.
The clinical course of COVID-19 and host immune and inflammatory responses to SARS-CoV-2 infection. (Top panel) The clinical course of COVID-19 can be categorized into an asymptomatic incubation period, disease onset with respiratory symptoms, disease progression and severe disease phase [19]. Severe COVID-19 usually necessitates intensive care unit (ICU) admission and mechanical ventilation [20]. (Middle panel) Concomitant with the basic phases of COVID-19, host immune and inflammatory responses to SARS-CoV-2 infection may be divided into an early local innate immune response (sensing viral infection and antiviral defense) phase in the lungs, a later local/systemic immune response phase, followed by uncontrolled inflammatory responses and cytokine storm syndromes [19], [21]. Patients with severe cases of COVID-19 may develop acute respiratory distress syndrome (ARDS) and multi-organ failure [20]. (Lower panel) Armed with multi-layered mechanisms, the innate immune system represents a front line of host defense against the viral invasion. Of these, pattern recognition receptors (PRRs) sense viral pathogen-associated molecular patterns (PAMPs), triggering the innate immune signaling pathway that culminate in the production of interferons (IFNs), IFN-stimulated gene (ISG) products and pro-inflammatory cytokines/chemokines [20]. Type I IFNs and ISG products play important roles in early-phase antiviral defense while cytokines/chemokines are responsible for initiation of later inflammatory responses and cytokine storm [20]. The pro-inflammatory cytokine release may also lead to a series of destructive effects on human tissue, ARDS and multi-organ failure [3], [20].
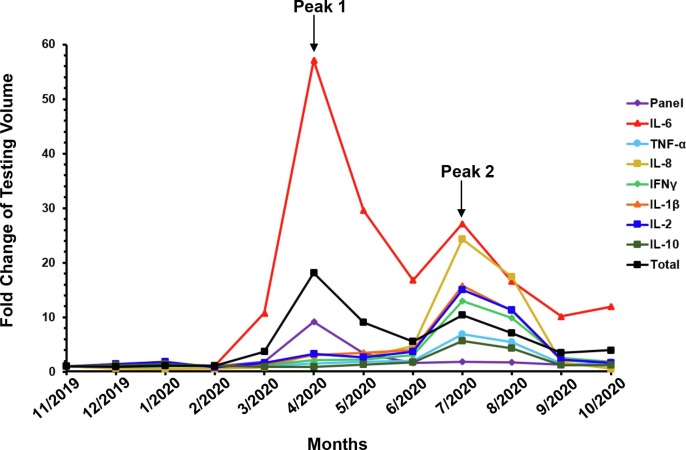
Recent studies have revealed that the hyper-inflammatory response and cytokine storm-like syndrome constitute a major cause of disease severity and death in COVID-19 patients [20], [22], [23], [24]. Early recognition and reduction of cytokine levels may be critical to reduce mortality of severe COVID-19 cases [25], [26]. Measuring serum cytokine levels as inflammatory biomarkers for COVID-19 may complement SARS-CoV-2 nucleic acid amplification testing (NAAT) and help assess the patient inflammatory status, monitor disease progression and stratify patients [26], [27]. Since the onset of the COVID-19 pandemic, our Cellular and Innate Clinical Immunology Laboratory as a part of ARUP Laboratories, the University of Utah owned national reference laboratory, has seen a dynamic change in test volumes for different cytokines (Fig. 2 ). Test order volumes for our panel of 12 cytokines [interleukin (IL)-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12, IL-13, IL-17, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ] and the cytokine receptor (IL-2R) surged in April 2020 (Peak 1), which was concurrent with the outbreak in New York City, NY [28]. The panel test volume then dropped gradually to the pre-COVID-19 levels (before February 2020) in June 2020. Similarly, the testing volumes for IFN-γ, TNF-α, IL-1β, IL-2, IL-8 and IL-10 peaked in July 2020 (Peak 2) and returned to the pre-COVID-19 levels in October 2020. Strikingly, IL-6 and total testing volume participated in both Peaks 1 and 2 and have continued to maintain a significantly higher level than in the pre-COVID-19 time period, e.g., 11.9-fold higher IL-6 tests in October 2020 than November 2019. In this report, we review the current knowledge of COVID-19 associated cytokines, their roles in host immune and inflammatory responses, the clinical significance and utility of cytokine immunoassays in adult and pediatric COVID-19 patients, as well as the challenges and pitfalls in implementation and interpretation of cytokine immunoassays.
Fig. 2.
Dynamic changes in cytokine test volumes at a national reference laboratory (ARUP Laboratories) in response to COVID-19 pandemic. Fold change of test volumes are shown for a cytokine panel (“Panel”; containing 12 cytokines IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12, IL-13, IL-17, TNF-α, IFN-γ and the cytokine receptor IL-2R), the indicated individual cytokines, and the total volume of tests (“Total”) performed at ARUP Laboratories between November 2019 and October 2020. In response to the COVID-19 pandemic, the Panel testing volume surged in April 2020 (Peak 1), which was concurrent with the outbreak in New York City, NY [28]. It then dropped gradually to the pre-COVID-19 levels in June 2020. Similarly, the test volumes for IFN-γ, TNF-α, IL-1β, IL-2, IL-8 and IL-10 peaked in July 2020 (Peak 2) and returned to pre-COVID-19 levels in October 2020. Strikingly, IL-6 and total testing volumes participated in both Peaks 1 and 2 and has continued to maintain a significantly higher level than pre-COVID-19 levels, e.g., 11.9-fold higher IL-6 tests in October 2020 than November 2019.
2. Molecular mechanisms underlying innate immune and inflammatory responses to SARS-CoV-2 infection
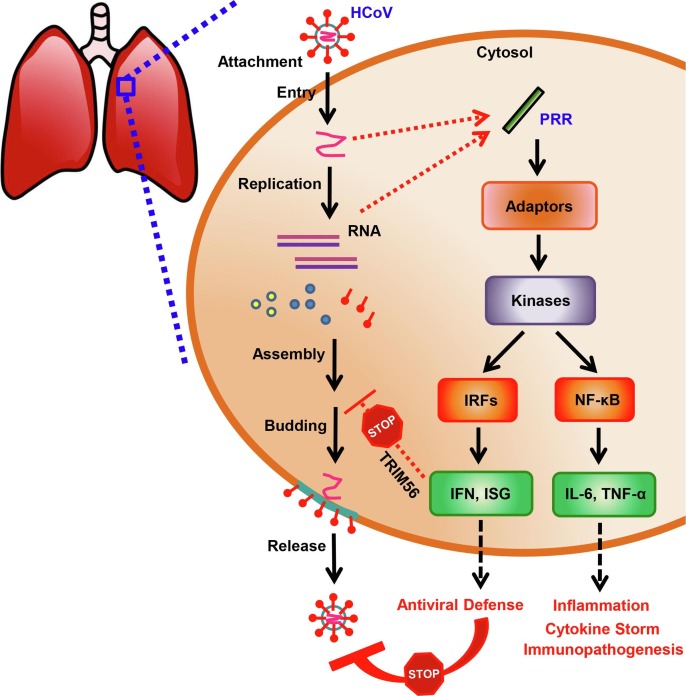
Virus-host interactions play an important role in the clinical course and outcomes of COVID-19 [19], [20], [21], [22], [23]. Host innate immune and inflammatory responses to SARS-CoV-2 infection are critical contributing factors to clinical manifestations and disease pathogenesis [19], [20], [21], [22], [23], [29], [30]. The innate immune system functions as the front line of host defense against HCoV infection, including SARS-CoV-2 [20], [31]. Viral RNA fragments or replication intermediates as viral pathogen-associated molecular patterns (PAMPs) can be sensed by pattern recognition receptors (PRRs) (Fig. 1, lower panel; Fig. 3 ) [20], [31]. PAMP-PRR interactions trigger downstream intracellular signaling pathways that culminate in activation of two types of critical transcription factors - interferon regulatory factors (IRFs) and nuclear factor kappa B (NF-κB) (Fig. 3) [20], [32], [33], [34], [35]. IRF3-mediated IFN and IFN stimulated gene (ISG) products are believed to play pivotal roles in early-phase antiviral defense (Fig. 1, lower panel; Fig. 3) [20], [32], [33], [34], [35]. For example, Tripartite Motif Containing 56, an antiviral ISG, provides direct antiviral activity against HCoV-OC43 propagation by blocking a later step of its life cycle after RNA replication [31]. Type I IFN secreted by the infected cells is responsible for establishing an “antiviral” state in neighboring cells. Whereas, NF-κB-mediated cytokine/chemokine production is indispensable for later inflammatory responses, cytokine storm and immunopathogenesis in different cells and tissues (Fig. 1, lower panel; Fig. 3) [20], [35].
Fig. 3.
Life cycle of human coronavirus (HCoV) and innate immune signaling pathways in response to HCoV infection. (Left and middle) HCoV invades and infects a lung cell after cell attachment and viral entry. Then viral replication occurs, followed by viral assembly, budding and release from the infected cell. (Right) Viral RNA fragments or replication intermediates as viral pathogen-associated molecular patterns (PAMPs) are sensed by pattern recognition receptors (PRRs), thereby triggering innate immune signaling pathways. PAMP-PRR interactions activate adaptor proteins and downstream kinases. The activated innate immune signaling pathways culminate in activation of two types of critical transcription factors - interferon regulatory factors (IRFs) and NF-κB. IRF3-mediated interferon (IFN) and IFN stimulated gene (ISG) products play important roles in antiviral defense. For example, Tripartite Motif Containing 56 (TRIM56), an antiviral ISG, inhibits HCoV-OC43 propagation by targeting a later step of the viral life cycle after RNA replication [31]. Type I IFN secreted by the infected cells is responsible for establishing an “antiviral” state in neighboring cells. Whereas, NF-κB-mediated cytokines/chemokines (e.g., IL-6 and TNF-α) are indispensable for later inflammatory responses, cytokine storm and immunopathogenesis in different cell types and tissues.
3. Clinical significance of measuring cytokine levels for adult patients with COVID-19
Diminished, early innate immune responses have been found in an upper airway gene expression analysis of 93 adult patients with SARS-CoV-2 infection [27]. In another study of 50 COVID-19 patients with various disease severities, impaired type I IFN activity was noted, suggesting type I IFN deficiency in the blood may be a hallmark in severe COVID-19 patients [18]. These two studies indicate that SARS-CoV-2 may be not a potent IFN inducer and that the virus may have evolved an unknown IFN antagonism mechanism to counteract or dampen the early innate immune responses [30].
In contrast, cytokine storm-like syndromes have been reported among adult patients with severe COVID-19 due to innate immune activation [36], [37], [38], [39]. Increased local production of pro-inflammatory cytokines may result in sudden release into the blood, followed by influx of macrophages, neutrophils, and T cells into the site of infection [20], [40]. Cytokine storm-like syndromes may lead to a series of abnormalities and tissue damage, including, but not limited to, lymphocyte dysfunction, granulocyte/monocyte abnormalities, damage of the vascular barrier, capillary damage, diffuse alveolar damage, multi-organ failure, and even death [3], [20], [29]. IL-6 is commonly considered the most important pro-inflammatory cytokine associated with the immunopathogenesis of severe COVID-19. High levels of IL-1 and TNF-α may also be seen in some COVID-19 patients [10], [20]. All these three cytokines are produced after activation of innate immune signaling pathways. It is proposed that modulating the levels of pro-inflammatory cytokines, e.g., IL-6, may alter the course of disease (www.covid19treatmentguidelines.nih.gov). In agreement with this hypothesis, baricitinib (a JAK 1/2 inhibitor and potent anti-inflammatory agent) and tocilizumab (an IL-6 receptor antagonist and inhibitor of cytokine storm) have shown promise in treatment of severe COVID-19 cases [22], [25]. Current NIH COVID-19 Treatment Guidelines, however, do not recommend that IL-6 or IL-1 inhibitors be used for the routine treatment of COVID-19 due to lack of sufficient clinical data describing the efficacy of these drugs (www.covid19treatmentguidelines.nih.gov).
Measuring serum levels of pro-inflammatory cytokines may have several potential applications in the management of COVID-19, including risk assessment, monitoring of disease progression, determination of prognosis, selection of therapy and prediction of response to treatment. IL-6 has been identified as an inflammatory biomarker consistently associated with COVID-19 disease progression [10]. Mandel et al. [41] used IL-6 level to predict 30-day mortality in hospitalized COVID-19 patients and yielded a high sensitivity of 91.7% at the cut-off value of 163.4 pg/ml. Ruan et al. [42] demonstrated that elevated IL-6 levels were significantly associated with adverse clinical outcomes among 150 severe COVID-19 cases. Further, an elevated IL-6 level was shown to be directly correlated with detectable serum SARS-CoV-2 viral load in critically ill COVID-19 patients, suggesting a potential to utilize IL-6 as a therapeutic target in severe patients suffering from an overwhelming inflammatory response [43]. These findings indicate that measuring serum IL-6 level may be helpful to assess the effectiveness of treatment.
Besides analysis of single cytokine IL-6, inflammatory cytokine biosignature depicted by multiplex cytokine profiling was studied to predict COVID-19 severity and survival of COVID-19 patients. Mandel et al. [41] reported that significantly high levels of IL-6 and TNF-α were detected in patients who did not survive severe COVID-19. Del Valle et al. [26] studied the clinical utility of a rapid multiplex cytokine assay measuring serum IL-6, IL-8, TNF-α and IL-1β levels in 1484 hospitalized adult patients with COVID-19. It was found that high-level IL-6, IL-8 and TNF-α in serum upon admission were potent and independent predictors of patient survival. Among them, IL-6 and TNF-α serum levels were validated as independent and significant predictors of disease severity and death in a second cohort of 231 patients. Del Valle et al. [26] proposed that serum IL-6 and TNF-α levels may be helpful in the management and treatment of COVID-19 patients to aid in stratifying patients, guiding resource allocation and selecting therapeutic options.
It has been proposed that alterations in the balance between pro-inflammatory and anti-inflammatory cytokines may also be useful for prediction of COVID-19 clinical progression [44]. McElvaney et al. [44] created an easily-calculated, 5-point linear prognostic score predictor (the Dublin-Boston score) based on IL-6:IL-10 ratio, and found that the Dublin-Boston score and the change in IL-6:IL-10 ratio from day 0 to day 4 yielded a better prediction of clinical outcome in 80 hospitalized COVID-19 patients at day 7 than IL-6 alone. These findings suggest potential applications of an IL-6:IL-10 ratio in assessment of the risk of impending poor outcome, determination of the proper time to escalate care, assistance with decision making on usage of mechanical ventilation, or even considerations for anti-cytokine and other therapies [44].
Interestingly, high levels of some pro-inflammatory cytokines were found specifically associated with men and older COVID-19 patients. In a study by Del Valle et al. [26], among 1298 SARS-CoV-2 positive patients, significantly (P < 0.0001) higher levels of IL-6 were found in men than women, but IL-8, TNF-α or IL-1β did not show any sex preferences. When dividing 1307 patients into three age groups (<50, 50–70, and >70 years old), it was found that the levels of IL-6, IL-8 and TNF-α elevated as the age of patients increased [26].
4. Clinical significance of measuring cytokine levels for pediatric patients with COVID-19
Compared with case reports and studies about adult patients with COVID-19, there are fewer publications on large pediatric cohorts, as children are commonly believed to have a lower incidence and milder course of COVID-19 [30], [45], [46], [47], [48]. These phenomena may be explained by several different characteristics of the pediatric population, including a lower expression of ACE2 receptor in their lungs, higher numbers of regulatory B and T cells, and immune tolerance to the viral infection with less of an inflammatory immune response [30], [47], [48].
In contrast, Kawasaki disease (KD), pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 (PIMS-TS) and multisystem inflammatory syndrome in children (MIS-C) associated with COVID-19 have been reported among pediatric patients with COVID-19 [45], [49], [50], [51], [52], [53]. It is accepted that the latter two syndromes highly resemble KD, an acute vasculitis of childhood and the leading cause of acquired heart disease in developed countries [45], [53]. PIMS-TS, MIS-C and KD share similar symptoms, such as cytokine storm, macrophage activation, hyperinflammatory features and elevated serum pro-inflammatory cytokines, e.g., IL-1, IL-6 and TNF-α [45], [49], [50], [51], [52], [53]. But it is still an open question whether these syndromes fall into the same spectrum of a novel disease or belong to different diseases post SARS-CoV-2 infection. As for laboratory testing for these diseases, both the PIMS-TS Guidance from the Royal College of Pediatrics and Child Health (RCPCH) (https://www.rcpch.ac.uk/resources), and the MIS-C Interim Guidance from the American Academy of Pediatrics (AAP) (https://services.aap.org/en/pages) recommend that expanded laboratory tests for immunological or inflammatory biomarkers be performed to monitor increasing inflammation. According to AAP guidance (https://services.aap.org/en/pages), a multidisciplinary approach involving cardiology, infectious disease, immunology, hematology, rheumatology, pediatric hospital medicine, and critical care, should be used to guide treatment of MIS-C. Intravenous immunoglobulin (IVIG) therapy or anakinra (an IL-1 receptor antagonist) may be applicable for appropriate indications. Immunomodulatory therapy may be considered on a case-by-case basis, according to RCPCH guidance (https://www.rcpch.ac.uk/resources). In a multicenter cohort study of 16 cases with KD-like symptoms, 62% of the patients received a two-tier treatment protocol consisting of an initial IVIG infusion and subsequent treatment with steroid, anakinra or tocilizumab and achieved inflammatory remission [45].
5. Approaches to measuring cytokine levels from serum samples
There are a variety of approaches to measuring cytokine levels from patient sera [2], [54]. The enzyme-linked immunosorbent assay (ELISA) is a routine method for analysis and measurement of secreted cytokines. This assay is classically designed for the analysis of one analyte at a time and, therefore, has a low throughput [54]. ELISA may require a large volume of sample to ensure an accurate detection [54]. If there are more cytokines to be tested, it may require a larger sample volume and more reagents. Running separate ELISA assays for multiple cytokines would be laborious and time-consuming [2]. Chemiluminescent immunoassays (CIA) on automated chemistry analyzers represent another frequently used method for measuring single cytokines. Of note, there are two commercial IL-6 assays which have received Emergency Use Authorization (EUA) approval from U.S. Food and Drug Administration (FDA): Roche Elecsys test (https://www.roche.com/media/releases/med-cor-2020–06-04.htm) and Beckman Coulter Access assay (https://www.beckmancoulter.com/about-beckman-coulter/newsroom/press-releases/2020/q4/2020-october-01-bec-access-il-6-test).
Coincident with the trend that precise and effective disease diagnosis increasingly relies on detection of disease-specific biomarkers [55], the advent of multiplex cytokine analysis overcomes the shortcomings of single-analyte ELISA assays and has become a mainstay diagnostic method for cytokine profiling [2]. Multiplexing allows for measurement of multiple cytokines simultaneously with a reduced volume of patient sample. This undoubtedly reduces the physical burden for some special populations for which sample collection may be difficult, such as pediatric, elderly, and chronically ill patients. There are different state-of-the-art multiplex detection methods, including bead-based multiplex immunoassays (e.g., Cytometric Bead Arrays, Luminex and Bio-Plex Pro), microtitre plate-based arrays [e.g., Mesoscale Discovery (MSD) and Quansys BioSciences Q-Plex], and slide-based arrays (e.g., FastQuant) [54], [56]. Among them, microtitre plate-based MSD platform utilize similar chemistry as an ELISA assay but can be multiplexed, which allow for the detection of up to 10 analytes per well [56]. In contrast, the Luminex Multi-Analyte Profiling (xMAP) technology is characterized by combinational utilization of advanced fluidics, optics, microsphere technology and digital signal processing (www.luminexcorp.com/xmap-technology). ARUP laboratories and the ARUP Institute for Clinical Pathology was one of the first to develop multianalyte cytokine assay testing using the Luminex xMAP Fluorescent Bead assay and offer it for clinical testing [12], [13].
6. Challenges of measuring cytokine levels from serum samples
Interfering substances may cause false negative or positive results for cytokine immunoassays [2], [57], [58]. Heterophile antibodies which cross phyla in their reactivity constitute a well-recognized cause of interference in immunoassays [57]. These antibodies which are usually produced against poorly defined antigens generally show weak avidity but specificity to multispecies. Heterophile antibodies can bind directly to the capture antibody and block the reactive site of the analytes of interest, thereby leading to false-negative results. Another type of interference comes from human anti-animal antibodies in patient serum, e.g., human anti-mouse antibodies (HAMA) [58]. These antibodies may be generated after exposure to animals or animal derived immunoglobulins and may affect some immunoassays that use immunoglobulins derived from the species of interest [58]. In addition, animal derived proteins used as blocking reagents in immunoassays, e.g., bovine serum albumin and casein, may yield high background readings or false-positive results for the cytokine immunoassays [2]. Recent IVIG administration prior to sampling may also lead to equivocal or false positive results for different serologic tests because of the cross-reactivity with different components in the IVIG or dilution of the analytes present in the patient serum [59], [60].
To minimize the interferences, a series of procedures should be done. The amount of assay interference due to heterophile antibodies should be determined during the development of immunoassays [2]. Assay-specific blockers and absorbents should be designed and used to significantly reduce falsely elevated cytokine values while not impacting the standard and control values. In our clinical cytokine assays, patient serum or plasma specimens are diluted and incubated for a short period of time in a serum diluent containing blockers to HAMA and heterophile antibodies prior to the measurement of cytokine concentrations, to eliminate interference.
There are also challenges associated with performing quality control (QC) for multiplex assays. Ellington et al. [61] retrospectively analyzed measurement data of three levels of recombinant protein QC materials for two panels of multiplex assays and found that a substantial portion of QC measurements exceeded target QC ranges with high coefficients of variation, with many plates rejected due to cross-reactivity and/or the QC imprecision. These findings suggest that the conventional univariate QC algorithms may not be suitable for multiplex assays that the FDA has classified as in vitro diagnostic multivariate index assays [61]. This calls for development of a practical, alternative QC algorithm which can monitor analytic variability and meanwhile avoid a high QC failure rate and expansive retesting.
7. Potential pitfalls in interpreting cytokine measurement results for COVID-19
There are several caveats to consider when interpreting cytokine results for COVID-19. First of all, given pro-inflammatory cytokines are also involved in other infectious diseases or immunological disorders, e.g., ARDS and sepsis [11], [62], the specificity of cytokine profiling data for COVID-19 patients is uncertain. Not all severe COVID-19 patients have elevated cytokines and there is some controversy over whether it is truly the same as the cytokine storm observed during CAR T cell therapy, sepsis, macrophage activation syndrome, or hemophagocytic lymphohistiocytosis [38], [63]. In a small-cohort study, Wilson et al. [64] recently found their severe COVID-19 patients did not show higher levels of inflammatory cytokines (IL-1β, IL-1RA, IL-6, IL-8, IL-18 or TNF-α) than critically ill patients with ARDS or sepsis. Though further validation of these findings in a larger cohort study is warranted, these findings suggest that cytokine profiles in plasma may not be specific biomarkers to differentiate between severe COVID-19 and ARDS or sepsis. It may be important to correlate cytokine profiling data with the SARS-CoV-2 NAAT and imaging observations to make an accurate interpretation of inflammatory status and disease progression in COVID-19 patients.
Next, there may be limitations in using a specific concentration of an isolated cytokine measurement for COVID-19 patients. It is known that cytokines play important roles in maintaining a healthy state and baseline levels of different pro-inflammatory cytokines vary among individuals with different immunometabolic comorbidities (e.g., obesity) and immune status, and even among different sample collection times (a trough in the morning) [11], [44]. Comprehensive analysis of multi-cytokine biosignature (e.g., IL-6, TNF-α or IL-10), or a ratio between different cytokines for a shift in inflammatory balance, may provide a more accurate understanding of the inflammatory status of COVID-19 patients than analyzing a single cytokine [26], [44]. It may be also helpful to analyze cytokine levels longitudinally, e.g., at different time points [44].
Last but not least, it should be taken into consideration that biological and technical variables may affect the commutability of cytokine immunoassays and enhance complexity of cytokine immunoassay interpretation [65], [66], [67]. Different laboratories may use different methods, platforms or cut-offs for cytokine profiling, which may lead to inter-laboratory variations affecting cytokine quantification and interpretation. Some assays analyze the abundance or masses of cytokines whereas others measure the biological activities. Thus, it is recommended that the same method, platform and laboratory should be used when monitoring differences in cytokine levels between groups of individuals or for the same individual over time. Since even in the same laboratory, different lots of reagent may also result in minor fluctuations in inter-assay reproducibility (https://www.mlo-online.com/continuing-education/article/13009452). The idea of using a cytokine ratio rather than individual concentrations may overcome some, if not all of the limitations discussed above, but further studies are required to confirm or identify which cytokines should be used and at which point(s) in time.
8. Conclusions
From our review of the currently available literature, several conclusions can be made about the clinical significance of measuring serum cytokine levels for COVID-19. First, measuring serum levels of pro-inflammatory cytokines as inflammatory biomarkers may have several potential applications in the management of COVID-19 patients, including risk assessment, monitoring of disease progression, determination of prognosis, selection of therapy and prediction of response to treatment. This is especially true for pediatric patients with COVID-19 associated PIMS-TS, MIS-C or KD. Second, multiplex cytokine analysis allows for measurement of multiple cytokines simultaneously with a decreased volume, which reduces the physical burden for some special populations for which sample collection may be challenging, such as pediatric, elderly, and chronically ill patients. Last but not least, it may be important to correlate cytokine profiling data with the SARS-CoV-2 NAAT and imaging observations to make an accurate interpretation of inflammatory status and disease progression of COVID-19 patients.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This study was supported by the ARUP Institute for Clinical and Experimental Pathology and formerly by grants from the National Institutes of Health.
References
- 1.Gale M., Jr. Understanding the Expanding Roles of Interferon and Cytokines in Health and Disease. J. Interferon Cytokine Res. 2018;38(5):195–196. doi: 10.1089/jir.2018.29007-mgj. [DOI] [PubMed] [Google Scholar]
- 2.Martins T.B., Pasi B.M., Litwin C.M., Hill H.R. Heterophile antibody interference in a multiplexed fluorescent microsphere immunoassay for quantitation of cytokines in human serum. Clin. Diagn. Lab. Immunol. 2004;11(2):325–329. doi: 10.1128/CDLI.11.2.325-329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaim S., Chong J.H., Sankaranarayanan V., Harky A. COVID-19 and Multiorgan Response. Curr. Probl. Cardiol. 2020;45(8) doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalvakolanu D.V. The “Yin-Yang” of cytokines in cancer. Cytokine. 2019;118:1–2. doi: 10.1016/j.cyto.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Martins T.B., Anderson J.L., Muhlestein J.B., Horne B.D., Carlquist J.F., Roberts W.L., Hill H.R. Risk factor analysis of plasma cytokines in patients with coronary artery disease by a multiplexed fluorescent immunoassay. Am. J. Clin. Pathol. 2006;125(6):906–913. doi: 10.1309/Q3E6-KF0Q-D3U3-YL6T. [DOI] [PubMed] [Google Scholar]
- 6.Martins T.B., Rose J.W., Jaskowski T.D., Wilson A.R., Husebye D., Seraj H.S., Hill H.R. Analysis of proinflammatory and anti-inflammatory cytokine serum concentrations in patients with multiple sclerosis by using a multiplexed immunoassay. Am. J. Clin. Pathol. 2011;136(5):696–704. doi: 10.1309/AJCP7UBK8IBVMVNR. [DOI] [PubMed] [Google Scholar]
- 7.Barker T., Martins T.B., Hill H.R., Kjeldsberg C.R., Trawick R.H., Leonard S.W., Walker J.A., Traber M.G. Vitamins E and C modulate the association between reciprocally regulated cytokines after an anterior cruciate ligament injury and surgery. Am. J. Phys. Med. Rehabil. 2011;90(8):638–647. doi: 10.1097/PHM.0b013e318214e886. [DOI] [PubMed] [Google Scholar]
- 8.Barker T., Leonard S.W., Trawick R.H., Martins T.B., Kjeldsberg C.R., Hill H.R., Traber M.G. Modulation of inflammation by vitamin E and C supplementation prior to anterior cruciate ligament surgery. Free Radic. Biol. Med. 2009;46(5):599–606. doi: 10.1016/j.freeradbiomed.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Barker T., Martins T.B., Kjeldsberg C.R., Trawick R.H., Hill H.R. Circulating interferon-γ correlates with 1,25(OH)D and the 1,25(OH)D-to-25(OH)D ratio. Cytokine. 2012;60(1):23–26. doi: 10.1016/j.cyto.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 2020;57:389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teijaro J.R. Cytokine storms in infectious diseases. Semin. Immunopathol. 2017;39(5):501–503. doi: 10.1007/s00281-017-0640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill H.R., Martins T.B. The flow cytometric analysis of cytokines using multi-analyte fluorescence microarray technology. Methods. 2006;38(4):312–316. doi: 10.1016/j.ymeth.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Martins T.B., Pasi B.M., Pickering J.W., Jaskowski T.D., Litwin C.M., Hill H.R. Determination of cytokine responses using a multiplexed fluorescent microsphere immunoassay. Am. J. Clin. Pathol. 2002;118(3):346–353. doi: 10.1309/N0T6-C56B-GXB2-NVFB. [DOI] [PubMed] [Google Scholar]
- 14.Fauci A.S., Lane H.C., Redfield R.R. Covid-19 - Navigating the Uncharted. N. Engl. J. Med. 2020;382(13):1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng Z., Xing-Lou Y., Xian-Guang W., Ben H., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pneumonia of unknown cause - China: disease outbreak news. Geneva: World Health Organization, January 5, 2020 https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/.
- 18.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall M.W., Joshi I., Leal L., Ooi E.E. Immune modulation in COVID-19: Strategic considerations for personalized therapeutic intervention. Clin. Infect. Dis. 2020:ciaa904. doi: 10.1093/cid/ciaa904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B., Huang S., Yin L. The cytokine storm and COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. COVID-19 cytokine storm: The anger of inflammation. Cytokine. 2020;133 doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noroozi R., Branicki W., Pyrc K., Łabaj P.P., Pospiech E., Taheri M., Ghafouri-Fard S. Altered cytokine levels and immune responses in patients with SARS-CoV-2 infection and related conditions. Cytokine. 2020;133 doi: 10.1016/j.cyto.2020.155143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Titanji B.K., Farley M.M., Mehta A., Connor-Schuler R., Moanna A., Cribbs S.K., O'Shea J., DeSilva K., Chan B., Edwards A., Gavegnano C., Schinazi R.F., Marconi V.C. Use of Baricitinib in Patients with Moderate and Severe COVID-19. Clin. Infect. Dis. 2020:ciaa879. doi: 10.1093/cid/ciaa879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., Marron T.U., Xie H., Patel M., Tuballes K., Van Oekelen O., Rahman A., Kovatch P., Aberg J.A., Schadt E., Jagannath S., Mazumdar M., Charney A.W., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26(10):1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mick E., Kamm J., Pisco A.O., Ratnasiri K., Babik J.M., Castañeda G., DeRisi J.L., Detweiler A.M., Hao S.L., Kangelaris K.N., Kumar G.R., Li L.M., Mann S.A., Neff N., Prasad P.A., Serpa P.H., Shah S.J., Spottiswoode N., Tan M., Calfee C.S., Christenson S.A., Kistler A., Langelier C. Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses. Nat. Commun. 2020;11(1):5854. doi: 10.1038/s41467-020-19587-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson C.N., Baumgartner J., Pichardo C., Toro B., Li L., Arciuolo R., Chan P.Y., Chen J., Culp G., Davidson A., Devinney K., Dorsinville A., Eddy M., English M., Fireteanu A.M., Graf L., Geevarughese A., Greene S.K., Guerra K., Huynh M., Hwang C., Iqbal M., Jessup J., Knorr J., Latash J., Lee E., Lee K., Li W., Mathes R., McGibbon E., McIntosh N., Montesano M., Moore M.S., Murray K., Ngai S., Paladini M., Paneth-Pollak R., Parton H., Peterson E., Pouchet R., Ramachandran J., Reilly K., Sanderson Slutsker J., Van Wye G., Wahnich A., Winters A., Layton M., Jones L., Reddy V., Fine A. COVID-19 Outbreak - New York City, February 29-June 1, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69(46):1725–1729. doi: 10.15585/mmwr.mm6946a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct. Target Ther. 2020;5(1):128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B.M., Hill H.R. Role of host immune and inflammatory responses in COVID-19 cases with underlying primary immunodeficiency: a review. J. Interferon Cytokine Res. 2020;40(12):549–554. doi: 10.1089/jir.2020.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B., Li N.L., Wang J., Shi P.Y., Wang T., Miller M.A., Li K. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J. Virol. 2014;88(23):13821–13835. doi: 10.1128/JVI.02505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Liu B., Wang N., Lee Y.M., Liu C., Li K. TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection. J. Virol. 2011;85(8):3733–3745. doi: 10.1128/JVI.02546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B., Li N.L., Shen Y., Bao X., Fabrizio T., Elbahesh H., Webby R.J., Li K. The C-terminal tail of TRIM56 dictates antiviral restriction of influenza A and B viruses by impeding viral RNA synthesis. J. Virol. 2016;90(9):4369–4382. doi: 10.1128/JVI.03172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y., Li N.L., Wang J., Liu B., Lester S., Li K. TRIM56 is an essential component of the TLR3 antiviral signaling pathway. J. Biol. Chem. 2012;287(43):36404–36413. doi: 10.1074/jbc.M112.397075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K., Li N.L., Wei D., Pfeffer S.R., Fan M., Pfeffer L.M. Activation of chemokine and inflammatory cytokine response in hepatitis C virus-infected hepatocytes depends on Toll-like receptor 3 sensing of hepatitis C virus double-stranded RNA intermediates. Hepatology. 2012;55(3):666–675. doi: 10.1002/hep.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fung M., Babik J.M. COVID-19 in Immunocompromised Hosts: What We Know So Far. Clin. Infect. Dis. 2020:ciaa863. doi: 10.1093/cid/ciaa863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. HLH Across Speciality Collaboration, UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368(6490):473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 39.Schett G., Sticherling M., Neurath M.F. COVID-19: risk for cytokine targeting in chronic inflammatory diseases? Nat. Rev. Immunol. 2020;20(5):271–272. doi: 10.1038/s41577-020-0312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKechnie J.L., Blish C.A. The Innate Immune System: Fighting on the Front Lines or Fanning the Flames of COVID-19? Cell Host Microbe. 2020;27(6):863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandel M., Harari G., Gurevich M., Achiron A. Cytokine prediction of mortality in COVID19 patients. Cytokine. 2020;134 doi: 10.1016/j.cyto.2020.155190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan. China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., Ding J., Li F. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dis. 2020:ciaa449. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McElvaney O.J., Hobbs B.D., Qiao D., McElvaney O.F., Moll M., McEvoy N.L., Clarke J., O'Connor E., Walsh S., Cho M.H., Curley G.F., McElvaney N.G. A linear prognostic score based on the ratio of interleukin-6 to interleukin-10 predicts outcomes in COVID-19. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pouletty M., Borocco C., Ouldali N., Caseris M., Basmaci R., Lachaume N., Bensaid P., Pichard S., Kouider H., Morelle G., Craiu I., Pondarre C., Deho A., Maroni A., Oualha M., Amoura Z., Haroche J., Chommeloux J., Bajolle F., Beyler C., Bonacorsi S., Carcelain G., Koné-Paut I., Bader-Meunier B., Faye A., Meinzer U., Galeotti C., Melki I. Paediatric multisystem inflammatory syndrome temporally associated with SARS-CoV-2 mimicking Kawasaki disease (Kawa-COVID-19): a multicentre cohort. Ann. Rheum. Dis. 2020;79(8):999–1006. doi: 10.1136/annrheumdis-2020-217960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.International Child Health Group, Royal College of Paediatrics & Child Health, Royal College of Paediatrics & Child Heath Impact of the COVID-19 pandemic on global child health: joint statement of the International Child Health Group and the Royal College of Paediatrics and Child Health. Arch. Dis. Child. 2020 doi: 10.1136/archdischild-2020-320652. archdischild-2020-320652. [DOI] [PubMed] [Google Scholar]
- 47.Brodin P. Why is COVID-19 so mild in children? Acta Paediatr. 2020;1 doi: 10.1111/apa.15271. [DOI] [PubMed] [Google Scholar]
- 48.Minotti C., Tirelli F., Barbieri E., Giaquinto C., Donà D. How is immunosuppressive status affecting children and adults in SARS-CoV-2 infection? A systematic review. J. infect. 2020;81(1):e61–e66. doi: 10.1016/j.jinf.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395(10237):1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., Bonanomi E., D'Antiga L. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395(10239):1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belhadjer Z., Méot M., Bajolle F., Khraiche D., Legendre A., Abakka S., Auriau J., Grimaud M., Oualha M., Beghetti M., Wacker J., Ovaert C., Hascoet S., Selegny M., Malekzadeh-Milani S., Maltret A., Bosser G., Giroux N., Bonnemains L., Bordet J., Di Filippo S., Mauran P., Falcon-Eicher S., Thambo J.B., Lefort B., Moceri P., Houyel L., Renolleau S., Bonnet D. Acute Heart Failure in Multisystem Inflammatory Syndrome in Children in the Context of Global SARS-CoV-2 Pandemic. Circulation. 2020;142(5):429–436. doi: 10.1161/CIRCULATIONAHA.120.048360. [DOI] [PubMed] [Google Scholar]
- 52.Licciardi F., Pruccoli G., Denina M., Parodi E., Taglietto M., Rosati S., Montin D. SARS-CoV-2-Induced Kawasaki-Like Hyperinflammatory Syndrome: A Novel COVID Phenotype in Children. Pediatrics. 2020;146(2) doi: 10.1542/peds.2020-1711. [DOI] [PubMed] [Google Scholar]
- 53.Jones V.G., Mills M., Suarez D., Hogan C.A., Yeh D., Segal J.B., Nguyen E.L., Barsh G.R., Maskatia S., Mathew R. COVID-19 and Kawasaki Disease: Novel Virus and Novel Case. Hosp. Pediatr. 2020;10(6):537–540. doi: 10.1542/hpeds.2020-0123. [DOI] [PubMed] [Google Scholar]
- 54.Yu X., Scott D., Dikici E., Joel S., Deo S., Daunert S. Multiplexing cytokine analysis: towards reducing sample volume needs in clinical diagnostics. Analyst. 2019;144(10):3250–3259. doi: 10.1039/c9an00297a. PMID: 31049499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henry N.L., Hayes D.F. Cancer biomarkers. Mol. Oncol. 2012;6(2):140–146. doi: 10.1016/j.molonc.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tighe P., Negm O., Todd I., Fairclough L. Utility, reliability and reproducibility of immunoassay multiplex kits. Methods. 2013;61(1):23–29. doi: 10.1016/j.ymeth.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Boscato L.M., Stuart M.C. Heterophilic antibodies: a problem for all immunoassays. Clin. Chem. 1988;34(1):27–33. [PubMed] [Google Scholar]
- 58.Kricka L.J. Human anti-animal antibody interferences in immunological assays. Clin. Chem. 1999;45(7):942–956. [PubMed] [Google Scholar]
- 59.Ramsay I., Gorton R.L., Patel M., Workman S., Symes A., Haque T., et al. Transmission of hepatitis B Core antibody and Galactomannan enzyme immunoassay positivity via immunoglobulin products: a comprehensive analysis. Clin. Infect. Dis. 2016;63(1):57–63. doi: 10.1093/cid/ciw222. [DOI] [PubMed] [Google Scholar]
- 60.Hanson K.E., Gabriel N., Mchardy I., Hoffmann W., Cohen S.H., Couturier M.R., Thompson G.R., 3rd. Impact of IVIG therapy on serologic testing for infectious diseases. Diagn. Microbiol. Infect. Dis. 2020;96(2) doi: 10.1016/j.diagmicrobio.2019.114952. [DOI] [PubMed] [Google Scholar]
- 61.Ellington A.A., Kullo I.J., Bailey K.R., Klee G.G. Measurement and quality control issues in multiplex protein assays: a case study. Clin. Chem. 2009;55(6):1092–1099. doi: 10.1373/clinchem.2008.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chousterman B.G., Swirski F.K., Weber G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017;39(5):517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- 63.Remy K.E., Mazer M., Striker D.A., Ellebedy A.H., Walton A.H., Unsinger J., Blood T.M., Mudd P.A., Yi D.J., Mannion D.A., Osborne D.F., Martin R.S., Anand N.J., Bosanquet J.P., Blood J., Drewry A.M., Caldwell C.C., Turnbull I.R., Brakenridge S.C., Moldwawer L.L., Hotchkiss R.S. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5(17) doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson J.G., Simpson L.J., Ferreira A.M., Rustagi A., Roque J., Asuni A., Ranganath T., Grant P.M., Subramanian A., Rosenberg-Hasson Y., Maecker H.T., Holmes S.P., Levitt J.E., Blish C.A., Rogers A.J. Cytokine profile in plasma of severe COVID-19 does not differ from ARDS and sepsis. JCI Insight. 2020;5(17) doi: 10.1172/jci.insight.140289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fichorova R.N., Richardson-Harman N., Alfano M., Belec L., Carbonneil C., Chen S., Cosentino L., Curtis K., Dezzutti C.S., Donoval B., Doncel G.F., Donaghay M., Grivel J.C., Guzman E., Hayes M., Herold B., Hillier S., Lackman-Smith C., Landay A., Margolis L., Mayer K.H., Pasicznyk J.M., Pallansch-Cokonis M., Poli G., Reichelderfer P., Roberts P., Rodriguez I., Saidi H., Sassi R.R., Shattock R., Cummins J.E., Jr. Biological and technical variables affecting immunoassay recovery of cytokines from human serum and simulated vaginal fluid: a multicenter study. Anal. Chem. 2008;80(12):4741–4751. doi: 10.1021/ac702628q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mouton W., Albert-Vega C., Boccard M., Bartolo F., Oriol G., Lopez J., Pachot A., Textoris J., Mallet F., Brengel-Pesce K., Trouillet-Assant S. Towards standardization of immune functional assays. Clin. Immunol. 2020;210 doi: 10.1016/j.clim.2019.108312. [DOI] [PubMed] [Google Scholar]
- 67.Albert-Vega C., Tawfik D.M., Trouillet-Assant S., Vachot L., Mallet F., Textoris J. Immune Functional Assays, From Custom to Standardized Tests for Precision Medicine. Front. Immunol. 2018;9:2367. doi: 10.3389/fimmu.2018.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]