The gut microbiota and host genetics are implicated in the pathogenesis of IgAN. Recent studies have confirmed that microbial compositions are heritable (microbiome quantitative trait loci [QTL]).
KEYWORDS: IgA nephropathy, genetics, microbiome quantitative trait loci, gut microbiota, Dialister
ABSTRACT
The gut microbiota has been implicated in immunoglobin A nephropathy (IgAN) because the intestinal immune response is assumed to be one of the disease triggers. Since the microbial composition is heritable, we hypothesize that genetic variants controlling gut microbiota composition may be associated with susceptibility to IgAN or clinical phenotypes. A total of 175 gut-microbiome-associated genetic variants were retrieved from the Genome-Wide Association Study (GWAS) Catalog. Genetic associations were examined in 1,511 patients with IgAN and 4,469 controls. Subphenotype associations and microbiome annotations were undertaken for a better understanding of how genes shaped phenotypes. Likely candidate microbes suggested in genetic associations were validated using 16S rRNA gene sequencing in two independent data sets with 119 patients with IgAN and 45 controls in total. Nine genetic variants were associated with susceptibility to IgAN. Risk genotypes of LYZL1 were associated with higher serum levels of galactose-deficient IgA1 (Gd-IgA1). Other significant findings included the associations between the risk genotype of SIPA1L3 and early age at onset, PLTP and worse kidney function, and AL365503.1 and more severe hematuria. Besides, risk genotypes of LYZL1 and SIPA1L3 were associated with decreased abundances of Dialister and Bacilli, respectively. Risk genotypes of PLTP and AL365503.1 were associated with increased abundances of Erysipelotrichaceae and Lachnobacterium, respectively. 16S data validated a decreased tendency for Dialister and an increased tendency for Erysipelotrichaceae in IgAN. In this pilot study, our results provided preliminary evidence that the gut microbiota in IgAN was affected by host genetics and shed new light on candidate bacteria for future pathogenesis studies.
IMPORTANCE The gut microbiota and host genetics are implicated in the pathogenesis of IgAN. Recent studies have confirmed that microbial compositions are heritable (microbiome quantitative trait loci [QTL]). The relationship between host genetics and the microbiota and the role of the microbiota in IgAN are unclear. We retrieved the GWAS Catalog and associated microbiome QTL in IgAN, observing that nine genetic variants were associated with IgAN susceptibility and some clinical phenotypes. In a consistent way, the decreased abundance of Dialister was associated with higher serum levels of Gd-IgA1, and 16S rRNA gene analysis confirmed the decreased abundance of Dialister in IgAN. These data provided preliminary evidence that the gut microbiota in IgAN was affected by host genetics, which is a new strategy for future pathogenesis and intervention studies.
INTRODUCTION
Immunoglobulin A nephropathy (IgAN) is the most common form of primary glomerulonephritis, but its pathogenesis is not well understood. Current knowledge indicates that defects in IgA1 glycosylation that lead to the formation of immune complexes are at the center of its pathogenesis (1, 2). Genome-wide association studies (GWASs) in IgAN support that galactose-deficient IgA1 (Gd-IgA1) is heritable (3, 4), and they also suggest that some GWAS loci, such as CARD9, TNFSF13, and PSMB8, are shared among IgAN, inflammatory bowel disease (IBD), and bacterial infections (5). These findings highlight an important role for the intestinal immune response to mucosal pathogens in IgAN.
More recent studies directly checking the role of the gut microbiota indeed supported the role of the gut-kidney axis in IgAN, evidenced by studies in both human samples and mouse models (6–8). Functional studies suggested that bacterial infections, as well as some of their metabolites, could induce immune hyperresponsiveness, resulting in the overproduction of IgA and proinflammatory cytokines (9, 10). Moreover, the use of immunosuppressants targeting excessive mucosal immune responses and broad-spectrum antibiotics targeting gut microbes had shown effectiveness in treating IgAN (8, 11).
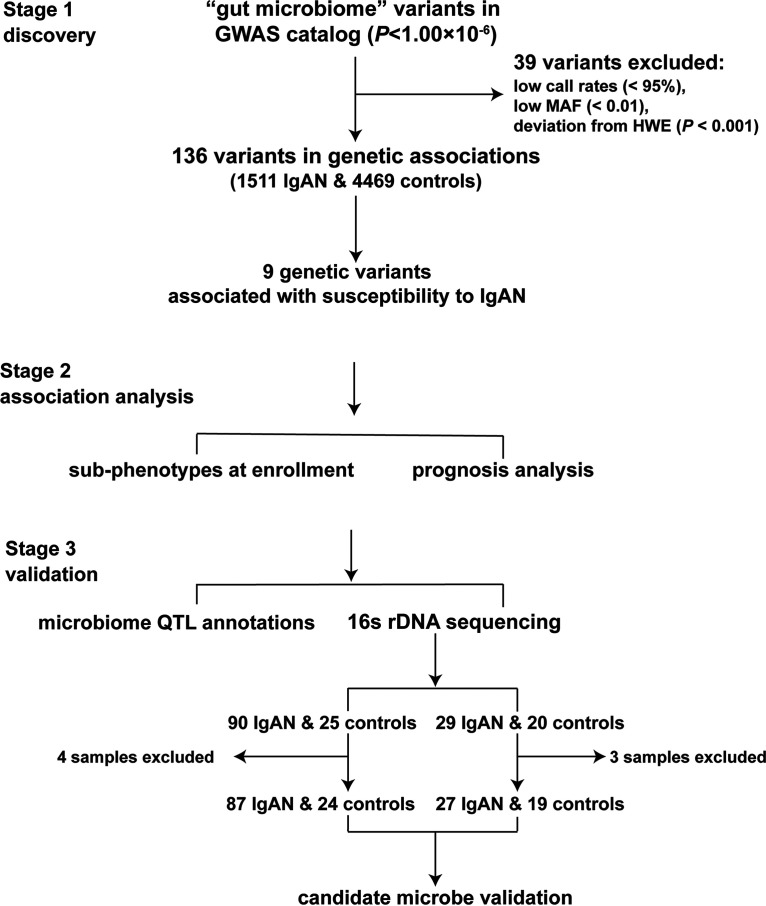
Similar to IBD, while it is known that both host genetics and the microbiome influence the development of the disease, how they precisely interact is less well understood (6, 12, 13). Recent studies have proven that the composition of the gut microbiota is heritable, and host-microbe interactions have a role in shaping the genetic architecture of IBD (14). Thus, in this pilot study, we aim to check the role of the microbiota in the etiology of IgAN in terms of host genetic susceptibility. The flowchart of the present study is shown in Fig. 1.
FIG 1.
Flowchart of the current study. We use a three-step analysis strategy in this study. Abbreviations: HWE, Hardy-Weinberg equilibrium; MAF, minor allele frequency.
RESULTS
Genetic associations between genetic variants affecting the gut microbiome and susceptibility to IgAN.
As described in Materials and Methods, 136 genetic variants were left for genetic associations in this large cohort after quality control. Although we did not observe associations with genome-wide significance (5 × 10−8), nine variants were associated with IgAN with suggestive P values (Table 1). As two single nucleotide polymorphisms (SNPs) (rs1889714 and rs1248290) belonged to the same gene region and were in high linkage disequilibrium (r2 = 1.0), rs1889714, which had a lower P value, was selected for further analysis.
TABLE 1.
The nine SNPs associated with IgAN with P values of <5 × 10−2a
| CHR | SNP | Position (hg19) | Candidate gene(s) | Risk allele | RAF in cases (%) | RAF in controls (%) | P value | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| 6 | rs3010562 | 167765251 | TTLL2 | T | 52.85 | 49.49 | 1.39 × 10−3 | 1.14 (1.05, 1.24) |
| 10 | rs1889714 | 29388639 | LYZL1, LINC01517 | A | 10.86 | 8.98 | 2.79 × 10−3 | 1.24 (1.08, 1.42) |
| 20 | rs6065904 | 44534651 | PLTP | A | 35.02 | 32.07 | 2.88 × 10−3 | 1.14 (1.05, 1.25) |
| 6 | rs9363741 | 68029041 | AL365503.1, AL591004.1 | G | 17.21 | 15.01 | 4.05 × 10−3 | 1.18 (1.05, 1.31) |
| 10 | rs7083345 | 7031144 | AL392086.3 | C | 86.54 | 84.38 | 5.19 × 10−3 | 1.19 (1.05, 1.34) |
| 10 | rs1248290 | 29386906 | LYZL1, LINC01517 | A | 10.67 | 9.26 | 2.41 × 10−2 | 1.17 (1.02, 1.34) |
| 18 | rs11877825 | 10566404 | NAPG, AP001099.1 | G | 79.43 | 77.49 | 2.86 × 10−2 | 1.12 (1.01, 1.24) |
| 8 | rs12541437 | 117918897 | RAD21-AS1, AARD | T | 40.04 | 37.86 | 3.55 × 10−2 | 1.10 (1.01, 1.19) |
| 19 | rs148330122 | 38520324 | SIPA1L3 | C | 92.19 | 90.97 | 4.13 × 10−2 | 1.17 (1.01, 1.36) |
Abbreviations: CHR, chromosome; CI, confidence interval; OR, odds ratio; RAF, risk allele frequency.
Genotype and subphenotype associations at enrollment.
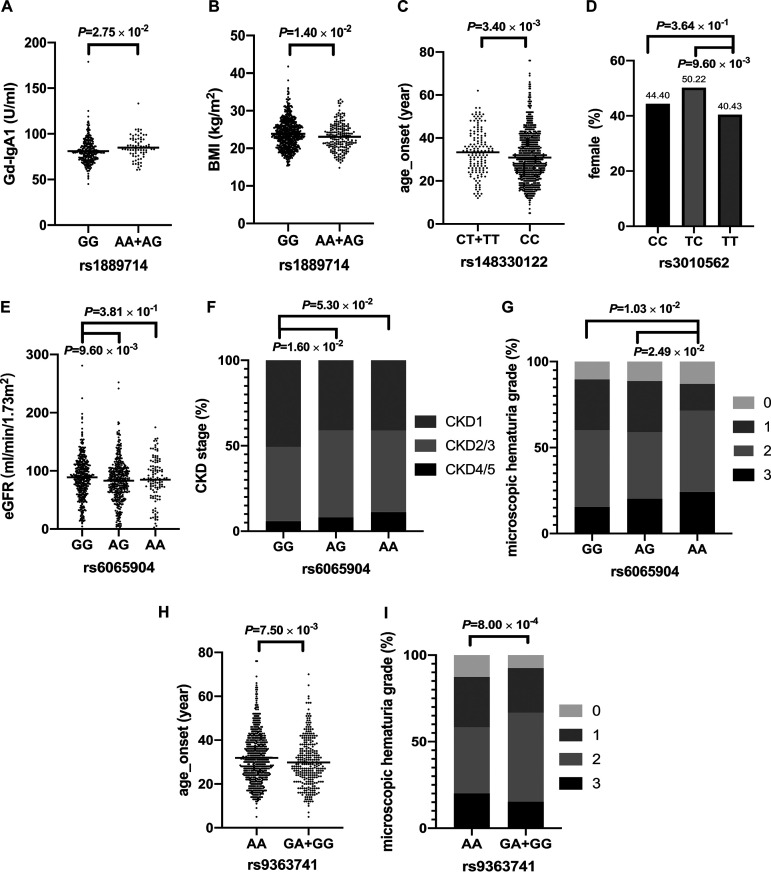
Next, we analyzed the associations between the risk variants and clinical subphenotypes of IgAN (Fig. 2; see also Table S1 in the supplemental material) under additive models unless genotypic counts were <10. The risk genotypes of LYZL1 rs1889714-AA/AG were associated with higher serum levels of Gd-IgA1 and lower body mass index (BMI) (Fig. 2A and B). The risk genotype of SIPA1L3 rs148330122-CC was associated with a younger age at onset (Fig. 2C). The risk genotype of TTLL2 rs3010562-TT was associated with male predominance (Fig. 2D).
FIG 2.
Associations between the genotypes and clinical subphenotypes of IgAN. (A) Patients with rs1889714-AA/AG genotypes (n = 75) had a higher serum level of Gd-IgA1 than did patients with the rs1889714-GG genotype (n = 294) (84.86 ± 12.27 U/ml in rs1889714-AA/AG versus 81.12 ± 13.26 U/ml in rs1889714-GG; the P value was determined by one-way ANOVA with Bartlett’s test for equal variances; the B test statistic is 0.407). (B) Patients with rs1889714-AA/AG genotypes (n = 191) had lower BMI than did patients with the rs1889714-GG genotype (n = 670) (23.08 ± 3.58 kg/m2 in rs1889714-AA/AG versus 23.85 ± 3.89 kg/m2 in rs1889714-GG; the P value was determined by one-way ANOVA with Bartlett’s test for equal variances; the B test statistic is 0.159). (C) Patients with the 148330122-CC genotype (n = 817) had lower age at onset than did patients with rs148330122-CT/TT genotypes (n = 144) (30.00 [interquartile range, 23.00, 38.00] years in rs148330122-CC versus 33.50 [26.00, 41.00] years in rs148330122-CT/TT; the P value was determined by a two-tailed Wilcoxon rank sum test). (D) The proportions of female patients with the rs3010562-TT genotype (n = 282), the rs3010562-TC genotype (n = 454), and the rs3010562-CC (n = 232) genotype were 40.43%, 50.22%, and 44.40%, respectively (the P value was determined by Pearson’s chi-squared test). (E) The eGFR was lower in patients with the rs6065904-AG genotype (n = 425) than in patients with the rs6065904-GG genotype (n = 417) (83.99 [62.95, 102.91] ml/min/1.73 m2 versus 90.36 [67.10, 111.20] ml/min/1.73 m2; the P value was determined by a two-tailed Wilcoxon rank sum test). (F) Patients with the rs6065904-AG genotype (n = 425) had worse kidney function than did patients with the rs6065904-GG genotype (n = 417) (CKD1, 2/3, and 4/5, 40.94%, 51.06%, and 8.00%, respectively, in the rs6065904-AG genotype versus 50.60%, 43.65%, and 5.76%, respectively, in rs6065904-GG; the P value was determined by Pearson’s chi-squared test). (G) Patients with the rs6065904-AA genotype (n = 116) had worse microscopic hematuria than did patients with rs6065904-AG (n = 425) and patients with the rs6065904-GG genotype (n = 415) (the P value was determined by Pearson’s chi-squared test). (H) Patients with rs9363741-GG/GA genotypes (n = 300) had a younger age at onset than did patients with the rs9363741-AA genotype (n = 669) (28.25 [22.00, 36.75] years in rs9363741-GG/GA versus 31.00 [24.00, 39.00] years in rs9363741-AA; the P value was determined by a two-tailed Wilcoxon rank sum test). (I) Patients with the rs9363741-GG/GA genotypes (n = 295) had worse microscopic hematuria than did patients with the rs9363741-AA genotype (n = 666) (MH grades 0, 1, 2, and 3, 7.46%, 25.76%, 51.53%, and 15.25%, respectively, in rs9363741-GG/GA versus 12.61%, 28.98%, 38.29%, and 20.12%, respectively, in rs9363741-AA; the P value was determined by Pearson’s chi-squared test).
Detailed genotype and subphenotype associations among the eight genetic variants and clinical subphenotypes of IgAN. Download Table S1, XLSX file, 0.02 MB (17.1KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Risk genotypes of PLTP rs6065904-AA/AG were associated with worse kidney function and more severe microscopic hematuria (MH) (Fig. 2E to G). The risk genotypes of AL365503.1 rs9363741-GG/GA were associated with a younger age at onset and more severe hematuria (Fig. 2H and I). Of note, the associated clinical subphenotypes, such as age, sex, and BMI, had already been suggested to be strongly associated with the composition of the gut microbiota (15, 16).
Prognosis associations.
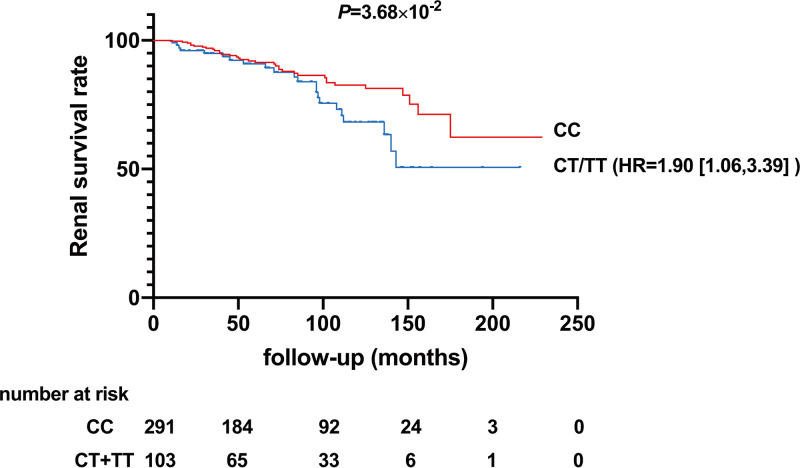
In the current cohort, 409 patients with IgAN were regularly monitored. The median duration of follow-up was 6.5 years. We observed a significant association between genotypes of AL392086.3 and the risk of end-stage renal disease (ESRD), which occurred in 22 out of 103 patients (21.36%) with rs7083345-CT/TT genotypes and 35 out of 291 patients (12.03%) with the CC genotype. Kaplan-Meier analysis showed that the cumulative renal survival rate was significantly lower in patients with rs7083345-CT/TT genotypes (Fig. 3).
FIG 3.
Cumulative renal survival rate in 409 patients with IgAN with regular follow-up information. Kaplan-Meier analysis showed that the cumulative renal survival rate was significantly lower in patients with rs7083345-CT/TT genotypes (the P value was determined by a log rank test). HR, hazard ratio.
Consistent with previous reports (17, 18), the univariate analysis indicated that renal function, Gd-IgA1, and proteinuria were the prognostic factors in IgAN. Moreover, the risk genotype of rs7083345 was observed to be an additional prognostic factor. In the multivariate Cox regression analysis, rs7083345-CT/TT genotypes, lower estimated glomerular filtration rate (eGFR), and higher time-averaged proteinuria were independently associated with poorer renal prognosis (Table 2).
TABLE 2.
Univariate and multivariate analyses of risk factors associated with ESRD in patients with IgANa
| Factor | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| rs7083345-TT/CT | 1.75 (1.03, 2.99) | 3.90 × 10−2 | 1.90 (1.06, 3.39) | 3.00 × 10−2 |
| Age (yrs) | 1.01 (0.98, 1.03) | 5.85 × 10−1 | 0.99 (0.96, 1.03) | 7.03 × 10−1 |
| Sex (female) | 0.67 (0.40, 1.12) | 1.26 × 10−1 | 1.03 (0.57, 1.86) | 9.10 × 10−1 |
| Serum creatinine (μmol/liter) | 1.02 (1.01, 1.02) | <1.00 × 10−3 | ||
| eGFR (ml/min/1.73 m2) | 0.97 (0.96, 0.98) | <1.00 × 10−3 | 0.96 (0.95, 0.97) | <1.00 × 10−3 |
| Gd-IgA1 (U/ml) | 1.02 (1.00, 1.03) | 4.90 × 10−2 | 1.01 (0.99, 1.03) | 2.33 × 10−1 |
| Serum IgA (g/liter) | 0.93 (0.75, 1.15) | 4.99 × 10−1 | ||
| Time-averaged proteinuria (g/g) | 2.03 (1.67, 2.48) | <1.00 × 10−3 | 2.52 (1.89, 3.35) | <1.00 × 10−3 |
| Urine protein excretion (g/24 h) | 1.20 (1.09, 1.31) | <1.00 × 10−3 | ||
| Serum C3 (g/liter) | 0.43 (0.11, 1.70) | 2.29 × 10−1 | ||
| CKD stage | ||||
| 2/3 | 2.95 (1.51, 5.76) | 2.00 × 10−3 | ||
| 4/5 | 24.58 (9.77, 61.80) | <1.00 × 10−3 | ||
P values which are <0.05 are highlighted by boldface type. HR, hazard ratio.
Microbiome QTL annotations.
By data mining of the previous results on microbiome quantitative trait loci (QTL) (Table 3), it was shown that the risk genotypes of rs1889714 (LYZL1) in IgAN were negatively correlated with the abundance of Dialister, which has been reported to be significantly decreased in patients with IgA vasculitis or Crohn’s disease (19, 20). The risk genotypes of rs148330122 (SIPA1L3) and rs7083345 (AL392086.3) in IgAN were reported to be negatively correlated with the abundance of Bacilli, and the risk genotype of rs3010562 (TTLL2) in IgAN was reported to be negatively associated with the abundance of Anaerofilum. These microbiota members were suggested to be beneficial in maintaining intestinal homeostasis (21, 22).
TABLE 3.
Microbiome QTL annotations
| Genetic variant/risk allele in present study | Genetic variant/risk allele in GWAS Catalog | Variant annotation | Associated microbiomea | Beta | P value | GWAS Catalog accession no. |
|---|---|---|---|---|---|---|
| rs1889714-A | rs1889714-A | Intergenic variant | s_Dialister_invisus | 0.41-U decrease | 6.00 × 10−9 | GCST003855 |
| rs1248290-A | rs1248290-A | Intergenic variant | g_Dialister | 0.40-U decrease | 1.00 × 10−8 | GCST003855 |
| rs148330122-C | rs148330122-C | Intron variant | c_Bacilli | 0.48-U decrease | 1.00 × 10−9 | GCST003875 |
| rs7083345-C | rs7083345-T | Intron variant | c_Bacilli | 0.25-U increase | 3.00 × 10−10 | GCST003875 |
| rs3010562-T | rs3010562-G | Intron variant | g_Anaerofilum | 0.67-U increase | 3.00 × 10−7 | GCST003222 |
| rs6065904-A | rs6065904-A | Intron variant | g_Erysipelotrichaceae | 0.19-U increase | 4.00 × 10−6 | GCST003854 |
| rs11877825-G | rs11877825-G | Regulatory region variant | f_Erysipelotrichaceae | 0.27-U decrease | 3.00 × 10−11 | GCST003875 |
| rs9363741-G | rs9363741-G | Intergenic variant | g_Lachnobacterium | 0.86-U increase | 5.00 × 10−7 | GCST003222 |
| rs12541437-T | rs12541437-T | Intergenic variant | g_Lachnobacterium | 0.60-U increase | 2.00 × 10−6 | GCST003223 |
Abbreviations: c_, class; f_, family; g_, genus; s_, species.
On the other hand, the risk genotype of rs6065904 (PLTP) in IgAN was positively associated with the abundance of Erysipelotrichaceae, and risk genotypes of rs9363741 (AL365503.1) and rs12541437 (RAD21-AS1) in IgAN were reported to be positively associated with the abundance of Lachnobacterium (14). Both Erysipelotrichaceae and Lachnobacterium were suggested to be detrimental to gut disease (23, 24).
Gut microbiota in patients with IgAN.
We included two independent data sets to check the diversity and structure of the microbial community using the 16S rRNA technique. After quality control by removing the abnormal samples due to low connectivity (<2.5), we had 87 cases with IgAN (age, 38.78 ± 11.45 years; 47.13% female; BMI, 23.87 ± 3.37 kg/m2) and 24 matched healthy controls (age, 35.79 ± 8.19 years; 50.00% female; BMI, 22.64 ± 2.81 kg/m2) in the first data set. In the second data set, we had 27 cases with IgAN (age, 44.68 ± 12.65 years; 20% female) and 19 healthy controls (age, 31.32 ± 10.59 years; 57.89% female) in 16S rRNA analysis without selection.
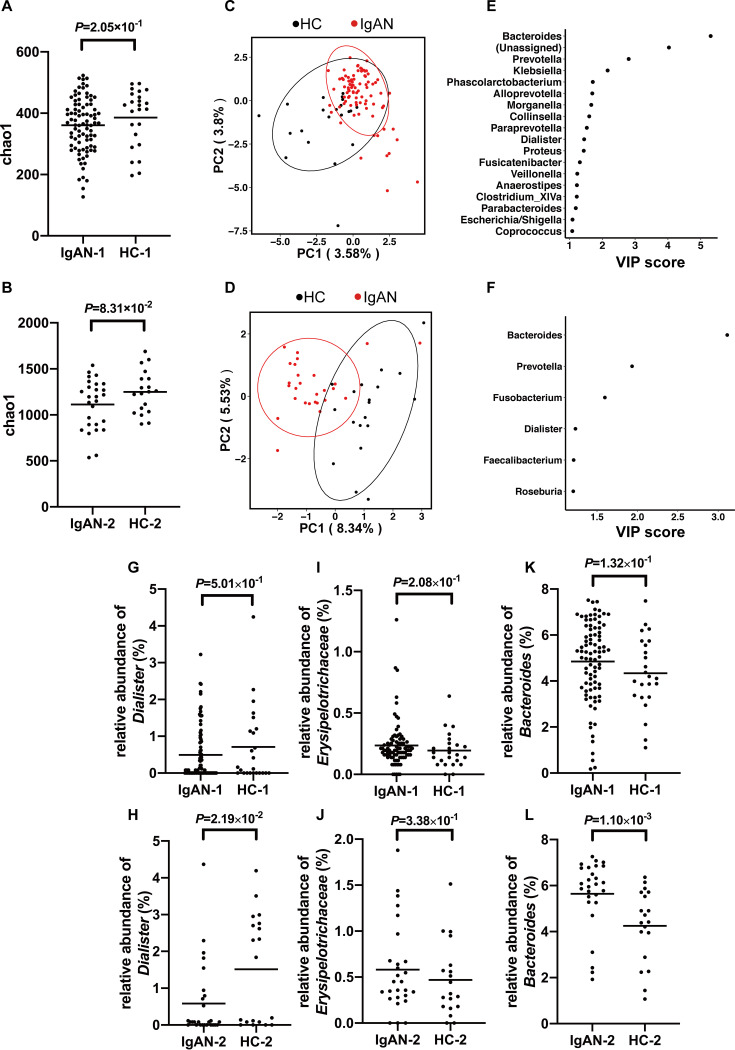
The alpha diversity of bacterial communities was evaluated according to the Chao1 diversity index (Fig. 4A and B). Generally, there was no statistical significance between patients with IgAN and controls in terms of alpha diversity. Partial least-squares discriminant analysis (PLS-DA), a supervised learning method, showed a distinct clustering pattern between samples from cases with IgAN and healthy controls in both data sets (Fig. 4C and D). For both data sets, the top one microbe, which contributed most significantly to differentiate cases from controls, was the same. The variable importance in projection (VIP) scores for Bacteroides were 5.30 and 3.11, respectively (Fig. 4E and F).
FIG 4.
16S rRNA gene analysis in two independent IgAN data sets. (A and B) There was no significant difference in alpha diversity (Chao1) between cases with IgAN and controls in two independent data sets (the P value was determined by a two-tailed Wilcoxon rank sum test). (C and D) PLS-DA score plot of species abundance in samples from individuals with IgAN and healthy controls (HC). It showed a distinct clustering pattern between groups (permutational multivariate analysis of variance with the Bray-Curtis distance metric was used to assess the significance of differences between the two groups [P = 4.5 × 10−2] for data set 1 [C] and for data set 2 [D]). (E and F) VIP scores of PLS-DA. A taxon with a VIP score of ≥1 was considered necessary for the group’s discrimination. The VIP scores of the candidate genus Dialister were 1.45 and 1.23, and the VIP scores of the genus Bacteroides were 5.30 and 3.11, for data set 1 (E) and data set 2 (F), respectively. (G and H) The relative abundance of the genus Dialister tended to decrease in cases compared to healthy controls. (I and J) The relative abundance of the family Erysipelotrichaceae tended to be enriched in patients with IgAN. (K and L) The relative abundance of the genus Bacteroides tended to increase in patients with IgAN. For panels G to L, all the data are expressed as the square root of the relative abundance of the specific bacterium. P values were determined by a two-tailed Wilcoxon rank sum test. PC, principal component.
In confirmation of clues suggested by the genetic associations, Dialister was also observed to play a role in group separation (VIP = 1.45 and 1.23, respectively). To check the difference at the genus level using Wilcoxon rank sum tests, as shown in Fig. 4G and H, the relative abundance of Dialister showed a decreased tendency in IgAN patients, and the relative abundance of Erysipelotrichaceae showed an increased tendency (Fig. 4I and J). For the microbe with the top VIP score, Bacteroides tended to increase in both data sets (Fig. 4K and L). For Bacilli, Anaerofilum, and Lachnobacterium, which were associated with the former genetic associations, no output data were available due to the limited richness of 16S rRNA gene sequencing. The complete results about the relative abundances at the genus level in two independent data sets are shown in Tables S2 and S3 in the supplemental material.
16S rRNA gene data on bacterial taxonomy and relative abundance in the first data set. Download Table S2, XLSX file, 0.1 MB (58.5KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene data on bacterial taxonomy and relative abundance in the second data set. Download Table S3, XLSX file, 0.02 MB (17.1KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
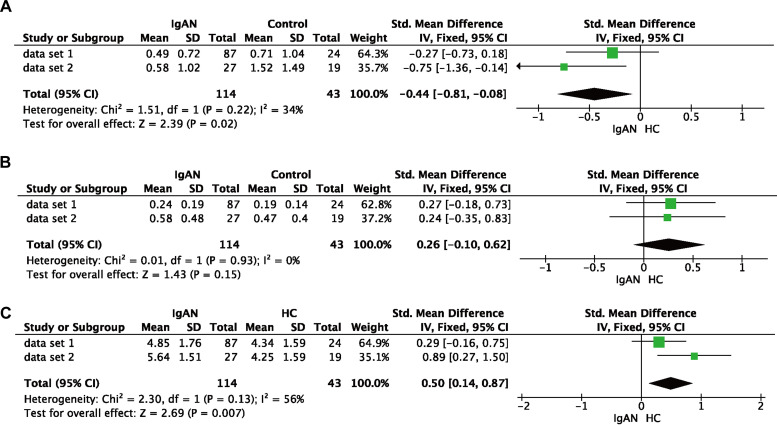
To further confirm the differential abundance, we conducted a meta-analysis for the two data sets (Fig. 5). There was moderate statistical heterogeneity (30% to 60%), but the forest plots showed consistent associations, with a decreased tendency for Dialister and an increased tendency for Bacteroides in IgAN.
FIG 5.
Meta-analysis of the relative abundances of Dialister (A), Erysipelotrichaceae (B), and Bacteroides (C) in two data sets. The relative abundance of Dialister was reduced in patients with IgAN (P = 2.00 × 10−2), while the relative abundance of Bacteroides was increased in patients with IgAN (P = 7.00 × 10−3). However, the relative abundance of Erysipelotrichaceae showed no significant difference (P = 1.50 × 10−1). Abbreviations: CI, confidence interval; IV, inverse variance.
DISCUSSION
The microbial composition was associated with multiple traits (e.g., age, sex, and BMI) and diseases (e.g., cardiovascular disease and IBD) (15, 25, 26). Meanwhile, microbial compositions could be affected by multiple factors such as the environment and host genetics. Genetic variants associated with the microbiome are defined as microbiome QTL. Recent studies have identified microbiome QTL in human diseases, including IBD, cancer, heart disease, and meningitis (27, 28). These microbiome QTL can be regulated by genes involved in microbiome-related pathways, including the immune system, food metabolism, and drug-related systems.
In the current study, we systemically checked the microbiome QTL in reported GWASs. We further investigated associations between reported microbiome QTL and the susceptibility to and severity of IgAN. In a large cohort with 1,511 patients with IgAN and 4,469 controls, we observed that 9 SNPs were associated with susceptibility to IgAN. Genotype-phenotype associations between risk alleles and disease subtypes may provide insight into disease etiology and mechanisms. Intriguingly, these risk variants were observed to be associated with subphenotypes of IgAN, i.e., early age at onset, elevated Gd-IgA1 levels, severe hematuria, and advanced chronic kidney disease (CKD) stage., and in a concordant way, specific risk genotypes in IgAN were associated with decreased abundances of potentially beneficial microbes and increased abundances of potentially harmful microbes, as reported for IBD. Furthermore, one microbiome-associated SNP was also independently associated with renal outcome in IgAN. Along with a gene-centered study, we further checked the validity of the candidate microbiota members by 16S rRNA gene sequencing in two independent data sets to increase statistical power. G*Power 3.1.9.6 software was used to calculate the posterior effect size index using the asymptotic relative efficiency (ARE) method (29). The effect sizes were between 0.25 and 0.30. To estimate the power of the Wilcoxon test for a given F value with the ARE method, we scaled the sample size with the corresponding ARE value. The calculated powers in cohort 1 and cohort 2 were 0.22 and 0.14, respectively. After meta-analysis, the power was 0.34 for the combined samples. According to the results from the two data sets, we confirmed that both microbiota richness/evenness and the abundance of Dialister were decreased in IgAN. However, this candidate microbe was not the most significant in IgAN. In addition, according to the VIP score, we observed that Bacteroides contributed the most in differentiating IgAN from controls. It has been proven that Bacteroides can impact fecal IgA levels (30). However, as Bacteroides was not included in our candidate microbiome QTL, we cannot rule out the role of host genetic impacts. Therefore, by a multistage association strategy in this pilot study, we tentatively confirmed that host genetics had impacts on the gut microbiota, which played a role in both disease susceptibility and severity.
For the function and pathogenesis of the microbiota in IgAN, recent studies suggested that changes of the gut microbiota could alter IgA-mediated immunity, and differences in IgA binding to bacteria have been linked to IBD (31), which shares some similarities in etiology and is also a common comorbidity with IgAN. It is widely recognized that IgA plays a pivotal and special role in IgAN, and our data supported the hypothesis that individuals with IgAN have an altered gut microbiota, which was somewhat genetically determined. Interestingly, the LYZL1 risk genotypes, which were Dialister related, were associated with an elevated level of Gd-IgA1. The relative abundance of the bacterial genus Dialister was reported to be significantly decreased in children with IgA vasculitis (19), in which an increased level of Gd-IgA1 was a common pathogenic mechanism, as in IgAN. Besides, in mouse studies, the c-type lysozyme gene Lyzl1 exhibited antibacterial activity (32), and mouse models targeting LYZL1 and Dialister will be of interest. Studies have highlighted the role of the gut microbiome in IgA or Gd-IgA1 immunity. Specific microbes or metabolites are known to be involved in the class switch of B cells and the production of IgA (7, 30). This study may have significance in future translational medicine. First, we observed that host genetics might affect gut microbiota composition. This genetic information might contribute to risk stratification. Since genetics, instead of diet, were unchanged, we might check whether the tested people were genetically susceptible to abnormal bacterial composition, IgA immunity, and IgAN, especially in risk populations or risk families. Second, a precise link between genetics and the gut microbiota would shed new light on disease pathogenesis. It was speculated that both the mucosal immune response and the gut microbiome participated in the pathogenesis of IgAN. But the precise role of the gut microbiota in mediating the gut-kidney dialog in IgAN has not been clarified. Last but not least, targeting specific bacteria or certain metabolites would have therapeutic significance in IgAN. Thus, future studies integrating host genetics and microbiota may shed some light on precision medicine, possibly by targeting IgA immunity and infections.
Despite a large-scale genetic study on microbiome QTL in IgAN, we should note some limitations. First, there were still no widespread microbiota-related studies in IgAN, not to mention an IgAN disease-specific microbiome QTL study. To guarantee ≥80% power to detect a 1.2-fold-increased risk, we may need at least 2,366 cases and 7,000 controls (see Table S4 in the supplemental material), and we cannot rule out the possibility that other genetic variants had a higher magnitude of risk effects in IgAN, i.e., Bacteroides-associated variants in IgAN. Second, fecal 16S rRNA gene sequencing was limited in precision in microbiota differentiation. Meanwhile, our study did not differentiate between progressor and nonprogressor IgAN patients, whose gut microbiome profiles were different (6). However, consistent with our results, their study also observed decreased alpha diversity and key microbe changes (Bacteroides coprocola, Bacteroides faecis, and Dialister sp.) in IgAN. Thus, a prospective study using metagenomic sequencing to check the relationship between the gut microbiota and disease progression is warranted. Third, we tried our best to minimize the impact of confounding factors, including body weight, BMI, gender, alcohol intake, tobacco use, and diet. Those who were vegetarians or had dietary bias were excluded before enrollment. However, we had to acknowledge that the proportions of some constituents of the diet, such as salt or fatty acids, were not controlled in our cohorts (33–35). Fourth, all the associations may not bear multiple corrections. With 8 independent variants and 13 clinical variables, a conservative Bonferroni threshold to declare a significant association would be 4.81 × 10−4 (0.05/8/13), whereas it may indicate that the functional impact of the gut microbiota was not very high since the occurrence and progression of IgAN were the products of the complex interactions between genetics and the environment. Fifth, both genetic association analysis and 16S rRNA sequencing were conducted in participants with the same genetic background from the same living district (Han Chinese ethnicity). Replication from different ethnicities was the gold standard for evaluating the size and reliability of a genetic finding. As a first attempt to explore potential associations with any available trait, the current data were tempting, but more widespread replications focusing on these eight SNPs would be needed in the future.
Power calculation to estimate the number of patients required with ≥80% power. Download Table S4, XLSX file, 0.01 MB (9.8KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In conclusion, the first pilot microbiome QTL genetic study in IgAN showed that eight independent genetic variants were associated with both the susceptibility to and severity of IgAN. Along with feces microbe confirmation, our data indicated the decreased abundance of potentially beneficial microbes in IgAN, which might shed some light on future interventions.
MATERIALS AND METHODS
SNP selection.
We selected the potentially gut-microbiome-associated genetic variants identified by GWASs by searching the NHGRI GWAS Catalog database as of 5 March 2019 using the keyword “gut microbiome” (36). Finally, we selected 175 genetic variants associated with the gut microbiome (P < 1.00 × 10−6). These genetic variants were associated with alpha/beta diversity, gut microbiota taxa and their relative abundance (37–39). Here, we provide all the related information about these genetic variants in Table S5 in the supplemental material.
Retrieved information for the 136 genetic variants investigated in the current study. Download Table S5, XLSX file, 0.03 MB (35.8KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genotyping and study populations.
For genetic associations, a total of 5,980 participants, comprising 1,511 cases with IgAN and 4,469 healthy controls of Chinese ancestry, were recruited from Peking University First Hospital. The diagnosis of IgAN was confirmed by kidney biopsy with immunofluorescence studies for IgA deposits. Cases with secondary IgAN, such as systematic lupus erythematosus, rheumatic disease, or IgA vasculitis, were excluded. Ethnically and geographically matched healthy controls were voluntarily recruited.
Peripheral blood samples were collected from participants using the anticoagulant EDTA. We obtained the genomic DNA from peripheral blood leukocytes using the salting-out method. Genotyping was performed by using the MassArray system from Sequenom (Compass Biotech, Beijing, China). SNPs with low call rates (<95%), low minor allele frequencies (MAFs) (<0.01), or deviation from Hardy-Weinberg equilibrium (P < 0.001) were excluded.
Clinical variables.
Demographics and clinical data at the time of renal biopsy check included age, gender, BMI, blood pressure, urinary sediment microscopy, 24-h urine protein excretion, serum IgA, complement component 3, and serum creatinine. Microscopic hematuria was defined as ≥3 red blood cells (RBCs) per high-power microscopic field (HPF) in three consecutive urinalyses (40) and was graded as 0 (0 to 3 RBCs/HPF), 1 (3 to 15 RBCs/HPF), 2 (15 to 100 RBCs/HPF), or 3 (>100 RBCs/HPF). Time-averaged proteinuria was defined as the mean of every 6 months of proteinuria measurements. Kidney disease severity was classified into five stages according to the eGFR based on KDIGO guidelines (41) and was grouped as mild (chronic kidney disease stage 1 [CKD1]), moderate (CKD2 and 3), and severe (CKD4 and 5). Gd-IgA1 was detected in 388 cases by a lectin enzyme-linked immunosorbent assay (ELISA) as previously described (42).
Among the cases enrolled, 409 patients were regularly monitored. Clinical outcome of ESRD was defined as an eGFR of <15 ml/min/1.73 m2 or the application of renal replacement therapies, including hemodialysis, peritoneal dialysis, or renal transplantation.
16S rRNA gene sequencing.
16S rRNA gene sequencing was conducted to confirm the likely involved microbes suggested by the genetic associations. In total, we recruited 119 cases with IgAN and 45 healthy controls. Patients with ESRD, secondary IgAN, IBD, and type 2 diabetes mellitus were excluded. For both patients and healthy controls, those who reported the use of antibiotics, microbial agents, or immunosuppressants within 8 weeks before entry were excluded. Besides, those who were vegetarians or had dietary bias were also excluded. Healthy controls had no gastrointestinal diseases.
The propensity score method was used to adjust for known confounding factors, including age, sex, and BMI. Thus, the enrolled participants were divided into two groups. The first data set consisted of 90 cases with IgAN and 25 age/sex/BMI-matched healthy controls, and the second data set consisted of the rest of the 29 cases with IgAN and 20 healthy controls without selection. Fecal samples were deemed biologically representative specimens because of their noninvasion and convenience. The fecal samples from each participant were collected in the hospital and immediately stored at −80°C until 16S rRNA gene sequencing.
Fecal genomic DNA was extracted using the QIAamp Fast DNA stool minikit (catalog number 51604; Qiagen). The targeted V3-V4 hypervariable region of the bacterial 16S rRNA genes was amplified by PCR using the primers 341F (5′-CCTACGGGRSGCAGCAG-3′) and 806R (5′-GGACTACVVGGGTATCTAATC-3′). Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA gel extraction kit (catalog number AP-GX-50; Axygen Biosciences) and quantified using Qubit2.0 (Invitrogen, MA, USA). After the preparation of the library, sequencing was performed on a HiSeq platform to generate paired-end reads of 250 bp (Illumina, CA, USA) for the first data set. The same protocol as the one described above was adopted for the second data set, and the tags were sequenced on the MiSeqDx platform (Illumina, CA, USA).
Data preprocessing.
Consistent methods were used to process all qualified data from the two data sets. The output reads were processed using USEARCH v.10 and in-house scripts (43, 44). The quality of the paired-end Illumina reads was checked by FastQC v.0.11.5 (45) and processed in the following steps by VSEARCH: joining of paired-end reads and relabeling of sequencing names, removal of barcodes and primers and filtering of low-quality reads (Q < 20), and finding of nonredundant reads. Unique reads were denoised into amplicon sequence variants (ASVs)/operational taxonomic units (OTUs). The representative sequences were picked by UPARSEH (46). The OTU table was generated by USEARCH. The taxonomy of the representative sequences was classified with the RDP classifier (47). Analysis of the differential OTU abundance and taxa was performed using the edgeR v3.26.8 package in R v.3.6.1. We subsample (“rarefy”) an OTU table to a fixed number of reads per sample using random subsampling without replacement. Besides, the abnormal observations were filtered if the connectivity was <2.5, using the Weighted Correlation Network Analysis (WGCNA) package (48).
Alpha diversity was evaluated by the Chao1 diversity index. PLS-DA was used to reveal taxonomic changes in different groups, and VIP scores were used to rank the abilities of different taxa to discriminate groups (49). Wilcoxon rank sum tests (two tailed) were conducted to detect differences in relative abundances between the two groups.
Ethics statement.
The study complied with the Declaration of Helsinki and was approved by the Ethics Committee of Peking University First Hospital (Institutional Review Board [IRB] approval numbers 2013[548] and 2019[76]). Written informed consent was obtained from all the patients involved in both genotyping and 16S rRNA gene sequencing.
Statistical analysis.
A two-tailed P value of <0.05 was considered statistically significant. Genetic association analysis was performed using PLINK-1.9 (50). Subphenotype associations were taken under the additive models at the genotypic level. To ensure the stability of the analysis, only if the sample count for the risk homozygous genotype was <10, a dominant model was considered.
Continuous variables in this study were compared using an unpaired t test or analysis of variance (ANOVA) between groups if the variables were normally distributed; otherwise, a Mann-Whitney U test or a Kruskal-Wallis test was performed. Categorical variables were compared using the chi-square test or Fisher’s exact test. Cumulative renal survival rates were calculated according to the Kaplan-Meier method. Univariate and multivariate Cox regression analyses were used to evaluate the risk of ESRD. The statistical analysis was performed with SPSS 26.0 software (SPSS Inc., USA).
As two different sequencing platforms were adopted in this study, we conducted a meta-analysis based on group differences rather than directly combining demultiplexed sequences. Raw data were processed into a normal distribution by calculating the square root of the relative abundance of the specific microbe. Also, further measures of the group differences were processed by Review Manager (version 5.4; Cochrane Library) under the fixed-effect model, and the degree of statistical heterogeneity was quantified by an I2 test.
ACKNOWLEDGMENTS
This work was supported by the Beijing Natural Science Foundation under grant Z190023; the National Science Foundation of China under grants 82022010, 81970613, 81925006, 81670649, and 82070733; the King’s College London-Peking University Health Science Center Joint Institute for Medical Research; the Fok Ying Tung Education Foundation under grant 171030; the Beijing Nova Program Interdisciplinary Cooperation Project under grant Z191100001119004; the Beijing Youth Top-Notch Talent Support Program under grant 2017000021223ZK31; the Clinical Medicine Plus X-Young Scholars Project of Peking University under grant PKU2020LCXQ003; the University of Michigan Health System-Peking University Health Science Center Joint Institute for Translational and Clinical Research under grant BMU2017JI007; and the Chinese Academy of Medical Sciences Research Unit under grant 2019RU023. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We report no conflict of interest.
H.Z. and X.-J.Z. conceived and designed the study; H.Z., J.-C.L., X.-J.Z., and L.-J.L. collaborated in patient recruitment, data acquisition, and organization; S.-F.S., J.-W.H., and Y.-N.W. performed the laboratory analyses; X.-H.X., Y.-F.L., and R.-S.L. performed the 16S rRNA gene sequencing; X.-J.Z., J.-W.H., Y.-N.W., and M.F. analyzed the data; X.-J.Z. and J.-W.H. made the figures; and J.-W.H., X.-J.Z., and H.Z. drafted and revised the manuscript. All authors approved the final version of the manuscript.
REFERENCES
- 1.Berthoux F, Suzuki H, Thibaudin L, Yanagawa H, Maillard N, Mariat C, Tomino Y, Julian BA, Novak J. 2012. Autoantibodies targeting galactose-deficient IgA1 associate with progression of IgA nephropathy. J Am Soc Nephrol 23:1579–1587. doi: 10.1681/ASN.2012010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki H, Moldoveanu Z, Hall S, Brown R, Vu HL, Novak L, Julian BA, Tomana M, Wyatt RJ, Edberg JC, Alarcón GS, Kimberly RP, Tomino Y, Mestecky J, Novak J. 2008. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest 118:629–639. doi: 10.1172/JCI33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomax-Browne HJ, Visconti A, Pusey CD, Cook HT, Spector TD, Pickering MC, Falchi M. 2017. IgA1 glycosylation is heritable in healthy twins. J Am Soc Nephrol 28:64–68. doi: 10.1681/ASN.2016020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gharavi AG, Moldoveanu Z, Wyatt RJ, Barker CV, Woodford SY, Lifton RP, Mestecky J, Novak J, Julian BA. 2008. Aberrant IgA1 glycosylation is inherited in familial and sporadic IgA nephropathy. J Am Soc Nephrol 19:1008–1014. doi: 10.1681/ASN.2007091052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C, Viola BF, Dallera N, Del Vecchio L, Barlassina C, Salvi E, Bertinetto FE, Amoroso A, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boer E, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Di Landro D, Santoro D, Pani A, Polci R, Feriozzi S, Chicca S, Galliani M, et al. 2014. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Angelis M, Montemurno E, Piccolo M, Vannini L, Lauriero G, Maranzano V, Gozzi G, Serrazanetti D, Dalfino G, Gobbetti M, Gesualdo L. 2014. Microbiota and metabolome associated with immunoglobulin A nephropathy (IgAN). PLoS One 9:e99006. doi: 10.1371/journal.pone.0099006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy DD, Kujawa J, Wilson C, Papandile A, Poreci U, Porfilio EA, Ward L, Lawson MA, Macpherson AJ, McCoy KD, Pei Y, Novak L, Lee JY, Julian BA, Novak J, Ranger A, Gommerman JL, Browning JL. 2011. Mice overexpressing BAFF develop a commensal flora-dependent, IgA-associated nephropathy. J Clin Invest 121:3991–4002. doi: 10.1172/JCI45563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chemouny JM, Gleeson PJ, Abbad L, Lauriero G, Boedec E, Le Roux K, Monot C, Bredel M, Bex-Coudrat J, Sannier A, Daugas E, Vrtovsnik F, Gesualdo L, Leclerc M, Berthelot L, Ben Mkaddem S, Lepage P, Monteiro RC. 2019. Modulation of the microbiota by oral antibiotics treats immunoglobulin A nephropathy in humanized mice. Nephrol Dial Transplant 34:1135–1144. doi: 10.1093/ndt/gfy323. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki K, Maruya M, Kawamoto S, Sitnik K, Kitamura H, Agace WW, Fagarasan S. 2010. The sensing of environmental stimuli by follicular dendritic cells promotes immunoglobulin A generation in the gut. Immunity 33:71–83. doi: 10.1016/j.immuni.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y-Y, Chen D-Q, Chen L, Liu J-R, Vaziri ND, Guo Y, Zhao Y-Y. 2019. Microbiome-metabolome reveals the contribution of gut-kidney axis on kidney disease. J Transl Med 17:5. doi: 10.1186/s12967-018-1756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellstrom BC, Barratt J, Cook H, Coppo R, Feehally J, de Fijter JW, Floege J, Hetzel G, Jardine AG, Locatelli F, Maes BD, Mercer A, Ortiz F, Praga M, Sorensen SS, Tesar V, Del Vecchio L, NEFIGAN Trial Investigators. 2017. Targeted-release budesonide versus placebo in patients with IgA nephropathy (NEFIGAN): a double-blind, randomised, placebo-controlled phase 2b trial. Lancet 389:2117–2127. doi: 10.1016/S0140-6736(17)30550-0. [DOI] [PubMed] [Google Scholar]
- 12.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP. 2011. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43:321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ, Sun LD, Sim KS, Li Y, Foo JN, Wang W, Li ZJ, Yin XY, Tang XQ, Fan L, Chen J, Li RS, Wan JX, Liu ZS, Lou TQ, Zhu L, Huang XJ, Zhang XJ, Liu ZH, Liu JJ. 2011. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet 44:178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]
- 14.Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, Ter Steege RWF, Huttenhower C, Dijkstra G, Xavier RJ, Festen EAM, Wijmenga C, Zhernakova A, Weersma RK. 2018. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 67:108–119. doi: 10.1136/gutjnl-2016-312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Cuesta-Zuluaga J, Kelley ST, Chen Y, Escobar JS, Mueller NT, Ley RE, McDonald D, Huang S, Swafford AD, Knight R, Thackray VG. 2019. Age- and sex-dependent patterns of gut microbial diversity in human adults. mSystems 4:e00261-19. doi: 10.1128/mSystems.00261-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X, Zhang M, Xue J, Huang J, Zhuang R, Zhou X, Zhang H, Fu Q, Hao Y. 2018. Body mass index differences in the gut microbiota are gender specific. Front Microbiol 9:1250. doi: 10.3389/fmicb.2018.01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moriyama T, Tanaka K, Iwasaki C, Oshima Y, Ochi A, Kataoka H, Itabashi M, Takei T, Uchida K, Nitta K. 2014. Prognosis in IgA nephropathy: 30-year analysis of 1,012 patients at a single center in Japan. PLoS One 9:e91756. doi: 10.1371/journal.pone.0091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao N, Hou P, Lv J, Moldoveanu Z, Li Y, Kiryluk K, Gharavi AG, Novak J, Zhang H. 2012. The level of galactose-deficient IgA1 in the sera of patients with IgA nephropathy is associated with disease progression. Kidney Int 82:790–796. doi: 10.1038/ki.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Zhang L, Wang Y, Liu X, Zhang H, Liu Y, Shen N, Yang J, Gai Z. 2018. Gut microbiota dysbiosis is associated with Henoch-Schonlein purpura in children. Int Immunopharmacol 58:1–8. doi: 10.1016/j.intimp.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Nagao-Kitamoto H, Kamada N. 2017. Host-microbial cross-talk in inflammatory bowel disease. Immune Netw 17:1–12. doi: 10.4110/in.2017.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo M, Wu F, Hao G, Qi Q, Li R, Li N, Wei L, Chai T. 2017. Bacillus subtilis improves immunity and disease resistance in rabbits. Front Immunol 8:354. doi: 10.3389/fimmu.2017.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. 2015. Role of the normal gut microbiota. World J Gastroenterol 21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, Basic M, Dupont A, Hornef M, von Bergen M, Bleich A, Haller D. 2016. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 65:225–237. doi: 10.1136/gutjnl-2015-309333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yilmaz B, Spalinger MR, Biedermann L, Franc Y, Fournier N, Rossel JB, Juillerat P, Rogler G, Macpherson AJ, Scharl M. 2018. The presence of genetic risk variants within PTPN2 and PTPN22 is associated with intestinal microbiota alterations in Swiss IBD cohort patients. PLoS One 13:e0199664. doi: 10.1371/journal.pone.0199664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown JM, Hazen SL. 2018. Microbial modulation of cardiovascular disease. Nat Rev Microbiol 16:171–181. doi: 10.1038/nrmicro.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schirmer M, Garner A, Vlamakis H, Xavier RJ. 2019. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 17:497–511. doi: 10.1038/s41579-019-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillhouse AE, Myles MH, Taylor JF, Bryda EC, Franklin CL. 2011. Quantitative trait loci in a bacterially induced model of inflammatory bowel disease. Mamm Genome 22:544–555. doi: 10.1007/s00335-011-9343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snijders AM, Langley SA, Kim Y-M, Brislawn CJ, Noecker C, Zink EM, Fansler SJ, Casey CP, Miller DR, Huang Y, Karpen GH, Celniker SE, Brown JB, Borenstein E, Jansson JK, Metz TO, Mao J-H. 2016. Influence of early life exposure, host genetics and diet on the mouse gut microbiome and metabolome. Nat Microbiol 2:16221. doi: 10.1038/nmicrobiol.2016.221. [DOI] [PubMed] [Google Scholar]
- 29.Faul F, Erdfelder E, Lang AG, Buchner A. 2007. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 30.Yang C, Mogno I, Contijoch EJ, Borgerding JN, Aggarwala V, Li Z, Siu S, Grasset EK, Helmus DS, Dubinsky MC, Mehandru S, Cerutti A, Faith JJ. 2020. Fecal IgA levels are determined by strain-level differences in Bacteroides ovatus and are modifiable by gut microbiota manipulation. Cell Host Microbe 27:467–475.e6. doi: 10.1016/j.chom.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA. 2014. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei J, Li S-J, Shi H, Wang H-Y, Rong C-T, Zhu P, Jin S-H, Liu J, Li J-Y. 2013. Characterisation of Lyzls in mice and antibacterial properties of human LYZL6. Asian J Androl 15:824–830. doi: 10.1038/aja.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. 2009. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137:1716–1724.e2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mahler A, Balogh A, Marko L, Vvedenskaya O, Kleiner FH, Tsvetkov D, Klug L, Costea PI, Sunagawa S, Maier L, Rakova N, Schatz V, Neubert P, Fratzer C, Krannich A, Gollasch M, Grohme DA, Corte-Real BF, Gerlach RG, Basic M, Typas A, Wu C, Titze JM, Jantsch J, Boschmann M, Dechend R, Kleinewietfeld M, Kempa S, Bork P, Linker RA, Alm EJ, Muller DN. 2017. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551:585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H. 2019. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, Zhernakova DV, Jankipersadsing SA, Jaeger M, Oosting M, Cenit MC, Masclee AA, Swertz MA, Li Y, Kumar V, Joosten L, Harmsen H, Weersma RK, Franke L, Hofker MH, Xavier RJ, Jonkers D, Netea MG, Wijmenga C, Fu J, Zhernakova A. 2016. The effect of host genetics on the gut microbiome. Nat Genet 48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 38.Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y. 2015. Genome-wide association studies of the human gut microbiota. PLoS One 10:e0140301. doi: 10.1371/journal.pone.0140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, Degenhardt F, Heinsen F-A, Rühlemann MC, Szymczak S, Holm K, Esko T, Sun J, Pricop-Jeckstadt M, Al-Dury S, Bohov P, Bethune J, Sommer F, Ellinghaus D, Berge RK, Hübenthal M, Koch M, Schwarz K, Rimbach G, Hübbe P, Pan W-H, Sheibani-Tezerji R, Häsler R, Rosenstiel P, D’Amato M, Cloppenborg-Schmidt K, Künzel S, Laudes M, Marschall H-U, Lieb W, Nöthlings U, Karlsen TH, Baines JF, Franke A. 2016. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet 48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grossfeld GD, Litwin MS, Wolf JS, Jr, Hricak H, Shuler CL, Agerter DC, Carroll PR. 2001. Evaluation of asymptomatic microscopic hematuria in adults: the American Urological Association best practice policy. Part II: patient evaluation, cytology, voided markers, imaging, cystoscopy, nephrology evaluation, and follow-up. Urology 57:604–610. doi: 10.1016/S0090-4295(01)00920-7. [DOI] [PubMed] [Google Scholar]
- 41.Levey AS, de Jong PE, Coresh J, El Nahas M, Astor BC, Matsushita K, Gansevoort RT, Kasiske BL, Eckardt K-U. 2011. The definition, classification, and prognosis of chronic kidney disease: a KDIGO Controversies Conference report. Kidney Int 80:17–28. doi: 10.1038/ki.2010.483. [DOI] [PubMed] [Google Scholar]
- 42.Zhu L, Zhai YL, Wang FM, Hou P, Lv JC, Xu DM, Shi SF, Liu LJ, Yu F, Zhao MH, Novak J, Gharavi AG, Zhang H. 2015. Variants in complement factor H and complement factor H-related protein genes, CFHR3 and CFHR1, affect complement activation in IgA nephropathy. J Am Soc Nephrol 26:1195–1204. doi: 10.1681/ASN.2014010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Zhang N, Liu YX, Zhang X, Hu B, Qin Y, Xu H, Wang H, Guo X, Qian J, Wang W, Zhang P, Jin T, Chu C, Bai Y. 2018. Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci China Life Sci 61:613–621. doi: 10.1007/s11427-018-9284-4. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Rubio P, Lottaz C, Engelmann JC. 2019. FastqPuri: high-performance preprocessing of RNA-seq data. BMC Bioinformatics 20:226. doi: 10.1186/s12859-019-2799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohart F, Gautier B, Singh A, Le Cao KA. 2017. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol 13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed genotype and subphenotype associations among the eight genetic variants and clinical subphenotypes of IgAN. Download Table S1, XLSX file, 0.02 MB (17.1KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene data on bacterial taxonomy and relative abundance in the first data set. Download Table S2, XLSX file, 0.1 MB (58.5KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene data on bacterial taxonomy and relative abundance in the second data set. Download Table S3, XLSX file, 0.02 MB (17.1KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Power calculation to estimate the number of patients required with ≥80% power. Download Table S4, XLSX file, 0.01 MB (9.8KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Retrieved information for the 136 genetic variants investigated in the current study. Download Table S5, XLSX file, 0.03 MB (35.8KB, xlsx) .
Copyright © 2021 He et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.