Abstract
Background
Circular RNA (circRNA) is involved in the progression of various cancers and has been shown to be an important potential target for cancer therapy. Circ_0020123 has been found to act as oncogene to participate in the malignant progression of non-small cell lung cancer (NSCLC). Therefore, exploring new mechanisms of circ_0020123 regulating NSCLC progression will help us better understand its role in NSCLC.
Methods
Relative expression levels of circ_0020123, microRNA (miR)-142-3p, and zinc-finger protein X-linked (ZFX) in tissues and cells were determined by quantitative real-time PCR (qRT-PCR). Cell proliferation, apoptosis, migration and invasion were assessed using cell counting kit 8 (CCK8) assay, colony formation assay, flow cytometry and transwell assay. Western blot (WB) analysis was used to detect relative protein level. Besides, the interaction between miR-142-3p and circ_0020123 or ZFX was confirmed by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay.
Results
Our results showed that circ_0020123 was upregulated in NSCLC, and its knockdown could suppress NSCLC cell proliferation, migration, invasion, and promote apoptosis. Circ_0020123 was found to interact with miR-142-3p. The inhibition effect of circ_0020123 silencing on NSCLC progression could be reversed by miR-142-3p inhibitor. ZFX could be targeted by miR-142-3p. The silencing of ZFX could hinder the progression of NSCLC and abolish the promotion effect of miR-142-3p inhibitor on NSCLC progression. In addition, circ_0020123 silencing inhibited NSCLC tumorigenesis by the miR-142-3p/ZFX axis.
Conclusion
These findings suggested that circ_0020123 might be a potential therapy target for NSCLC, which could promote NSCLC progression through regulating the miR-142-3p/ZFX axis.
Keywords: non-small cell lung cancer, circ_0020123, miR-142-3p, ZFX
Introduction
Lung cancer (LC) is a malignant tumor with the highest morbidity and mortality worldwide, and its common type is non-small cell lung cancer (NSCLC).1,2 NSCLC has become one of the most threatening malignant tumors to people’s health and life, and it is a disease that seriously affects organ function.3,4 Although the cure rate of early NSCLC is high, the prognosis of patients with advanced NSCLC is still poor.5 At present, NSCLC has entered the era of precision targeted therapy, and individualized molecular targeted therapy has become one of the important methods for the treatment of advanced NSCLC.6,7 Therefore, determining the target of NSCLC treatment is of great significance for improving the cure rate of patients.
Circular RNA (circRNA) is a circular structure transcript, which is mainly produced by the unique splicing mechanism “backsplicing” of the exon or intron sequence of mRNA.8–10 CircRNA has good stability and conservation characteristics, so it can be a biomarker for the treatment of human diseases, including cancer.11,12 CircRNAs have been shown to function as microRNA (miRNA) sponges and can disengage miRNA-mediated gene suppression.10,13,14 At present, numerous studies reveal that circRNA can act as a sponge of miRNA to participate in the regulation of tumor proliferation, metastasis, apoptosis and angiogenesis.15,16 For example, circ_0102231 could sponge miR-145 to accelerate NSCLC proliferation and invasion by promoting RBBP4 expression.17 CircNFIX facilitated NSCLC cell viability, metastasis and glycolysis via regulating the miR-212-3p/ADAM10 axis.18
In the studies related to NSCLC, circ_0020123 (derived from PDZD8 gene) has been proven to play a vital regulatory role in NSCLC malignant progression. Past studies have shown that high circ_0020123 expression is related to the poor prognosis of NSCLC patients, and it can significantly promote the proliferation, metastasis and tumor formation of NSCLC.19–22 Therefore, circ_0020123 is considered to be a biomarker for NSCLC diagnosis and treatment. Exploring more effects and mechanisms of circ_0020123 can provide more evidence for circ_0020123 to become a therapeutic target for NSCLC.
Materials and Methods
Samples Collection
Thirty paired NSCLC tumor tissues and normal non-cancerous tissues from 30 NSCLC patients who had surgery at Huizhou Municipal Central Hospital were collected and rapidly frozen in liquid nitrogen. Written informed consent was obtained from each patient and this study was approved by the Ethics Committee of Huizhou Municipal Central Hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki.
Cell Culture and Transfection
Human NSCLC cells (H1299, H1581 and A549) (ATCC, Manassas, VA, USA) and bronchial epithelial cells (16HBE) (Procell Life Science & Technology, Wuhan, China) were maintained in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Gibco) and 1% Penicillin-Streptomycin (Gibco). All the cells were cultured at 37°C with 5% CO2. After the cell density reached 50–60% in 6-well plates, cell transfection could be carried out. The small interfering RNA (siRNA) of circ_0020123 (si-circ_0020123#1: 5ʹ-ATGACCAGCTTACGTTGAATT-3ʹ; si-circ_0020123#2: 5ʹ-ACCAGCTTACGTTGAATTAAT-3ʹ; si-circ_0020123#3: 5ʹ-GACCAGCTTACGTTGAATTAA-3ʹ) and zinc-finger protein X-linked (ZFX) (si-ZFX#1, F 5ʹ-UUACAUAGCGAAAAUCGGCCC-3ʹ, R 5ʹ-GCCGAUUUUCGCUAUGUAAAU-3ʹ; si-ZFX#2, F 5ʹ-UUUACAUAGCGAAAAUCGGCC-3ʹ, R 5ʹ-CCGAUUUUCGCUAUGUAAAUA-3ʹ; si-ZFX#3, F 5ʹ-AUUUACAUAGCGAAAAUCGGC-3ʹ, R 5ʹ-CGAUUUUCGCUAUGUAAAUAU-3ʹ) or their negative control (si-NC), miR-142-3p mimic and inhibitor (miR-142-3p and in-miR-142-3p) or their negative control (miR-NC and in-miR-NC), as well as the lentivirus short hairpin RNA (shRNA) of circ_0020123 (sh-circ_0020123) and its control (sh-NC) were constructed by GenePharma (Shanghai, China). The oligos (50 nM) were transfected into cells using Lipofectamine 3000 Reagent (Invitrogen, Carlsbad, CA, USA) referring to the manufacturer’s protocol.
Quantitative Real-Time PCR (qRT-PCR)
The RNeasy Mini Kit (Qiagen, Duesseldorf, Germany) was performed to extract the RNA from tissues and cells. The RNA was reverse transcribed into cDNA using M-MLV Reverse Transcriptase Kit (Invitrogen). Basing on the special primer sequences, the cDNA was mixed with SYBR GreenER qPCR SuperMix Universal (Invitrogen) to perform qRT-PCR in PCR system. Relative expression was normalized to GAPDH or U6, and data were analyzed using the 2−ΔΔCT method. The primers were listed as follows: circ_0020123, F 5ʹ-GCCCATCCGAGTCTGTGTAG-3ʹ, R 5ʹ-CAATTGCAGCAGGCGAGTTT-3ʹ; PDZD8, F 5ʹ-GTCCGCCTACTTGTTTGTCAA-3ʹ, R 5ʹ-CACCTCGAAGTCGATCAGCG-3ʹ; miR-142-3p, F 5ʹ-GTCGTATCCAGTGCAGGG-3ʹ, R 5ʹ-CGACGTGTAGTGTTTCCTA-3ʹ; ZFX, F 5ʹ-TTGCTGAAATCGCTGACGAAG-3ʹ, R 5ʹ-GCAATCGGCATGAAGGTTTTGAT-3ʹ; GAPDH, F 5ʹ-TGACTTCAACAGCGACACCCA-3ʹ, R 5ʹ-TGACTTCAACAGCGACACCCA-3ʹ; U6, F 5ʹ-ATACAGAGAAAGTTAGCACGG-3ʹ, R 5ʹ-GGAATGCTTCAAAGAGTTGTG-3ʹ.
Cell Proliferation Analysis
Cell proliferation was assessed by detecting cell viability and cloned cell number using cell counting kit 8 (CCK8) assay and colony formation assay. For CCK8 assay, H1299 and A549 cells (1000 cells/well) were seeded in 96-well plates. After the cells were adherent to the wall, CCK8 solution was added to cells for 4 h at 0, 24, 48 and 72 h, respectively. The optical density (OD) value at 450 nm was measured by a microplate reader. For colony formation assay, H1299 and A549 cells (150 cells/well) were seeded in 6-well plates. After culturing for 14 days, the number of cloned cells (>50 cells) was counted under the microscope after the cells were stained by crystal violet solution (Beyotime, Shanghai, China).
Cell Apoptosis Analysis
Cell apoptosis was analyzed using the Annexin V-FITC Apoptosis Detection Kit (Sigma-Aldrich, St. Louis, MO, USA). Briefly, H1299 and A549 cells were collected and then resuspended with 1 × binding buffer. After the cells were stained with Annexin V-FITC and propidium iodide for 15 min, the apoptotic rate was evaluated by flow cytometry.
Cell Migration and Invasion Analysis
Transwell chambers (Millipore, Billerica, MA, USA) pre-coated with Matrigel was prepared for cell invasion, and non-coated was used for cell migration. H1299 and A549 cells suspended with the serum-free medium were plated in the upper chambers. Complete medium was added into lower chambers. The migrated and invaded cells were fixed and stained with crystal violet solution, and then counted under a microscope (100 ×) after 24 h.
Western Blot (WB) Analysis
Cells were treated with RIPA lysis buffer (Beyotime) to obtain total protein. The protein was separated by 10% SDS-PAGE gel and transferred to PVDF membranes (Millipore). The membrane was blocked by 5% non-fat milk, incubated with primary antibody and secondary antibody (all from Bioss, Beijing, China) in turn. The primary antibodies used in our study including anti-Cyclin D1 (bs-20596R, 1:2,000), anti-Bax (bs-28034R, 1:1,000), anti-MMP-9 (bs-4593R, 1:2,000), anti-ZFX (bs-12306R, 1:1,000) and anti-GAPDH (bs-2188R, 1:1,000). Goat Anti-Rabbit IgG (bs-0295G, 1:20,000) was the secondary antibody. Protein signals were detected by the BeyoECL Star ECL Chemiluminescence Kit (Beyotime) and protein expression was analyzed by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Dual-Luciferase Reporter Assay
The fragments of circ_0020123 and ZFX 3ʹUTR containing the targeted binding sites (wild-type, WT) or mutated sites (mutate-type, MUT) for miR-142-3p were cloned into the pmirGLO vector (Promega, Madison, WI, USA). H1299 and A549 cells were transfected with vectors and miR-142-3p mimic or miR-NC for 48 h. The luciferase activity in H1299 and A549 cells was examined by Dual-Luciferase Reporter Assay System (Promega).
RNA Immunoprecipitation (RIP) Assay
Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) was used for RIP assay. Briefly, H1299 and A549 cells were lysed with RIP buffer, and then the cell lysates were incubated with Magnetic Beads Protein A/G bounded with anti-Ago2 or anti-IgG. The immunoprecipitated RNA was extracted for performing qRT-PCR to determine the enrichment of circ_0020123 and miR-142-3p.
Mice Xenograft Models
Ten male BALB/c nude mice (Vital River, Beijing, China) were divided into the sh-circ_0020123 group (n = 5) and sh-NC group (n = 5). The mice were injected with H1299 cells (2 × 106) transfected with sh-circ_0020123 or sh-NC. Tumor length and width were detected by a vernier caliper to calculate tumor volume every week. After 4 weeks, all mice were sacrificed and the xenograft tumor was removed for other experiments. All animal experimental procedures were approved by the Animal Ethics Committee of Huizhou Municipal Central Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
Statistical analysis was carried out by Graphpad Prism software (La Jolla, CA, USA). All data were presented as means ± standard deviation and all experiments were performed at least 3 times. Comparisons between two groups or multiple groups were performed using Student’s t-test or one-way analyses of variance followed by Tukey post-hoc test. Pearson correlation analysis was used to analyze the correlations among circ_0020123, miR-142-3p and ZFX. Differences were identified to be statistically significant as P < 0.05.
Results
Highly Expressed Circ_0020123 Played an Active Role in NSCLC Progression
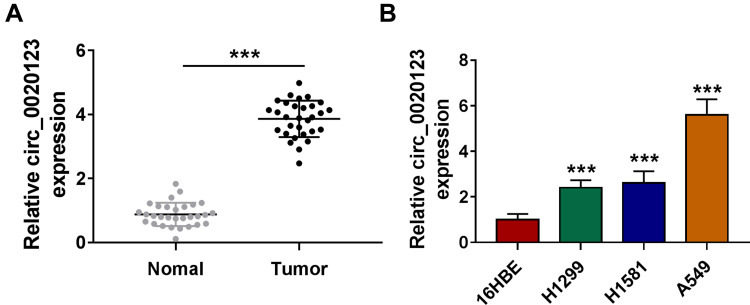
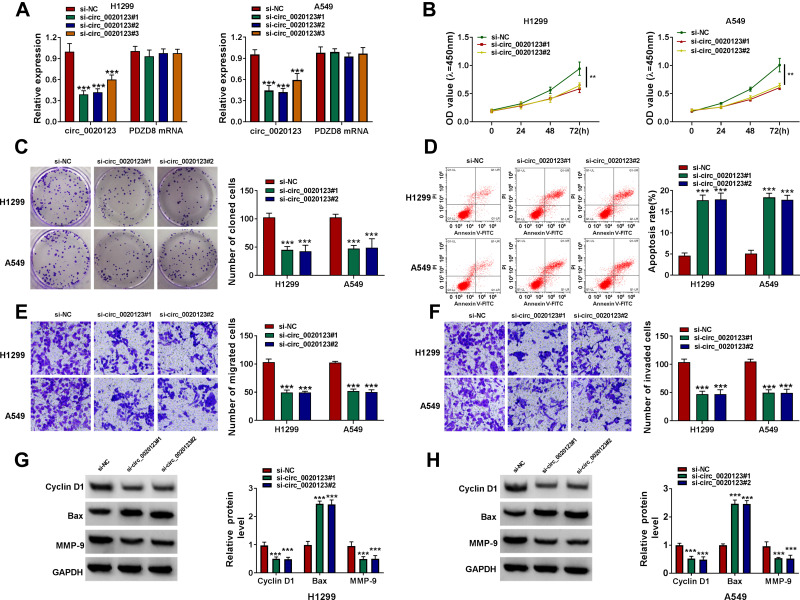
We detected circ_0020123 expression in 30 paired NSCLC tumor tissues and normal non-cancerous tissues and found that circ_0020123 was significantly upregulated in NSCLC tumor tissues (Figure 1A). Also, circ_0020123 expression was markedly enhanced in NSCLC cells (H1299, H1581 and A549) compared with that in 16HBE cells (Figure 1B). To confirm the role of circ_0020123 in NSCLC, we constructed 3 siRNAs for circ_0020123. Through detecting circ_0020123 expression and its linear RNA PDZD8 expression in H1299 and A549 cells transfected siRNAs, we determined that all 3 siRNAs could inhibit circ_0020123 expression without affecting the expression of linear PDZD8 (Figure 2A). Among them, si-circ_0020123#1 and si-circ_0020123#2 had the best effect. Therefore, we examined the biological function of H1299 and A549 cells transfected with si-circ_0020123#1 and si-circ_0020123#2. Our data suggested that circ_0020123 knockdown could inhibit the viability of H1299 and A549 cells (Figure 2B), and suppress the number of cloned H1299 and A549 cells (Figure 2C). Moreover, the apoptosis rate of H1299 and A549 cells was obviously increased after circ_0020123 silencing (Figure 2D). The knockdown of circ_0020123 also repressed the number of migrated and invaded H1299 and A549 cells, as demonstrated by transwell assay (Figure 2E and F). In addition, we discovered that the proliferation marker Cyclin D1 protein level and metastasis marker MMP-9 protein level were obviously decreased, while apoptosis marker Bax protein level was significantly enhanced in H1299 and A549 cells after silencing circ_0020123 (Figure 2G and H). These results showed that circ_0020123 knockdown hindered NSCLC progression, indicating that it might play an oncogenic role in NSCLC.
Figure 1.
Circ_0020123 was upregulated in NSCLC tissues and cells. (A) The expression of circ_0020123 in NSCLC tumor tissues (Tumor, n = 30) and normal non-cancerous tissues (Normal, n = 30) was detected by qRT-PCR. (B) QRT-PCR was used to determine the expression of circ_0020123 in NSCLC cells (H1299, H1581 and A549) and 16HBE cells. ***P < 0.001.
Figure 2.
Interference of circ_0020123 suppressed NSCLC progression. (A) The transfection efficiencies of si-circ_0020123#1, si-circ_0020123#2 and si-circ_0020123#3 were confirmed by detecting circ_0020123 expression using qRT-PCR in H1299 and A549 cells. (B–H) H1299 and A549 cells were transfected with si-NC, si-circ_0020123#1 or si-circ_0020123#2. CCK8 assay (B), colony formation assay (C), flow cytometry (D) and transwell assay (E and F) were used to measure the viability, the number of cloned cells, apoptosis rate, the numbers of migrated and invaded cells. (G and H) WB analysis was used to detect the protein levels of Cyclin D1, Bax and MMP-9. **P < 0.01, ***P < 0.001.
MiR-142-3p Could Be Sponged by Circ_0020123
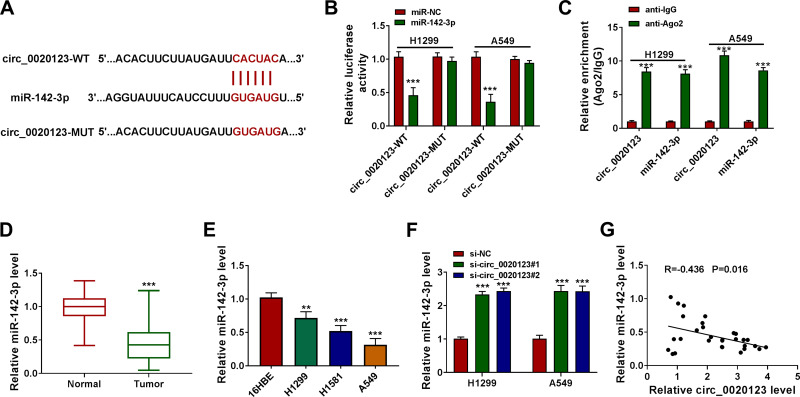
For exploring the new mechanism of circ_0020123 regulated NSCLC progression, the Circinteractome software was used to predict the targeted miRNAs of circ_0020123. After screening, we selected miR-142-3p. The predicted binding sites and designed mutate sites between miR-142-3p and circ_0020123 are shown in Figure 3A. Through dual-luciferase reporter assay, we found that the luciferase activity of circ_0020123-WT vector could be reduced by miR-142-3p mimic, while that of the circ_0020123-MUT vector was not affected by either miR-142-3p mimic or miR-NC (Figure 3B). The results of RIP assay showed that compared to anti-IgG, both circ_0010123 and miR-142-3p could be enriched in anti-Ago2 (Figure 3C). In NSCLC tumor tissues and cells, we found that miR-142-3p was downregulated in NSCLC tumor tissues and cells compared with the corresponding controls (Figure 3D and E). Besides, we also discovered that circ_0020123 silencing could markedly promote miR-142-3p expression in H1299 and A549 cells (Figure 3F), and there had a negatively correlation between circ_0020123 expression and miR-142-3p expression in NSCLC tumor tissues (Figure 3G). These data suggested that circ_0020123 could sponge miR-142-3p.
Figure 3.
MiR-142-3p could be sponged by circ_0020123. (A) The binding sites and the constructed mutate sites of circ_0020123 in miR-142-3p were shown. Dual-luciferase reporter assay (B) and RIP assay (C) were used to confirm the interaction between circ_0020123 and miR-142-3p. (D) QRT-PCR was used to measure miR-142-3p expression in NSCLC tumor tissues (Tumor, n = 30) and normal non-cancerous tissues (Normal, n = 30). (E) The expression of miR-142-3p in NSCLC cells (H1299, H1581 and A549) and 16HBE cells was detected using qRT-PCR. (F) MiR-142-3p expression was examined by qRT-PCR in H1299 and A549 cells transfected with si-NC, si-circ_0020123#1 or si-circ_0020123#2. (G) Pearson correlation analysis was performed to measure the correlation between circ_0020123 and miR-142-3p in NSCLC tumor tissues. **P < 0.01, ***P < 0.001.
MiR-142-3p Inhibitor Reversed the Regulation of Circ_0020123 Silencing on NSCLC Progression
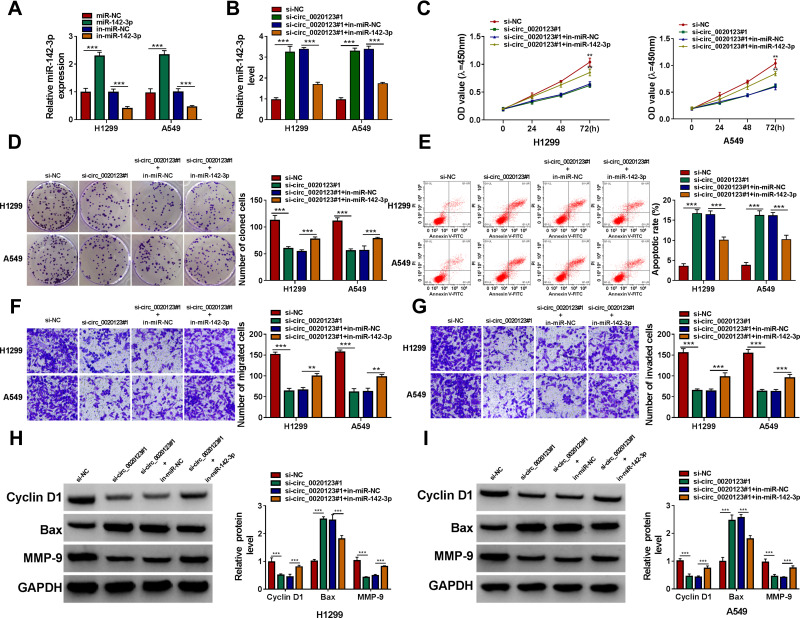
By detecting miR-142-3p expression in H1299 and A549 cells transfected with miR-142-3p mimic and inhibitor, we confirmed the transfection efficiencies of miR-142-3p mimic and inhibitor (Figure 4A). Then, we co-transfected with si-circ_0020123#1 and in-miR-142-3p into H1299 and A549 cells. As presented in Figure 4B, the promotion effect of circ_0020123 silencing on miR-142-3p expression could be reversed by in-miR-142-3p. The detection results of cell viability and colony number showed that the inhibition effect of circ_0020123 knockdown on the viability and the number of cloned cells could be overturned by miR-142-3p inhibitor in H1299 and A549 cells (Figure 4C and D). Flow cytometry results suggested that the promotion effect of circ_0020123 silencing on the apoptosis rate of H1299 and A549 cells could be eliminated by the addition of in-miR-142-3p (Figure 4E). Furthermore, miR-142-3p inhibitor also could abolish the suppressive effect of downregulated circ_0020123 on the migration and invasion of H1299 and A549 cells (Figure 4F and G). Additionally, the increased Cyclin D1 and MMP-9 protein levels and the decreased Bax protein level in the si-circ_0020123#1 + in-miR-142-3p group also revealed that the regulation of circ_0020123 silencing on Cyclin D1, MMP-9 and Bax protein levels could be reversed by miR-142-3p inhibitor (Figure 4H and I). All data showed that circ_0020123 regulated NSCLC progression by sponging miR-142-3p.
Figure 4.
MiR-142-3p inhibitor reversed the regulation of circ_0020123 silencing on NSCLC progression. (A) QRT-PCR was used to examine the expression of miR-142-3p to assess the transfection efficiencies of miR-142-3p mimic and inhibitor in H1299 and A549 cells. (B–H) H1299 and A549 cells were transfected with si-NC, si-circ_0020123#1, si-circ_0020123#1 + in-miR-NC, or si-circ_0020123#1 + in-miR-142-3p. (B) The expression of miR-142-3p was detected by qRT-PCR. The viability, the number of cloned cells, apoptosis rate, the numbers of migrated and invaded cells were determined using CCK8 assay (C), colony formation assay (D), flow cytometry (E) and transwell assay (F and G). (H and I) The protein levels of Cyclin D1, Bax and MMP-9 were measured using WB analysis. **P < 0.01, ***P < 0.001.
ZFX Was a Target of miR-142-3p
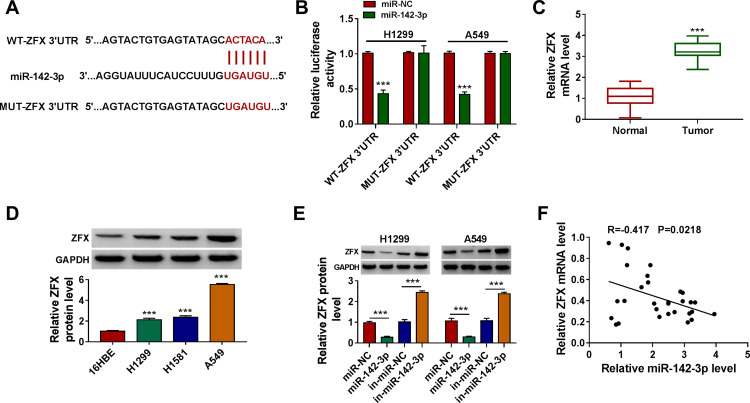
To search for the targets of miR-142-3p, the miRcode tool was used and ZFX 3ʹUTR was found to have complementary binding sites with miR-142-3p (Figure 5A). Dual-luciferase reporter assay results showed that miR-142-3p mimic could inhibit the luciferase activity of WT-ZFX 3ʹUTR vector without affecting that of the MUT-ZFX 3ʹUTR vector (Figure 5B). The highly expressed ZFX was discovered in NSCLC tumor tissues compared to normal non-cancerous tissues (Figure 5C). And the protein level of ZFX was also notably increased in NSCLC cells compared with that in 16HBE cells (Figure 5D). By determining the protein level of ZFX in H1299 and A549 cells transfected with miR-142-3p mimic or inhibitor, we uncovered that the expression of ZFX was inhibited by miR-142-3p overexpression, while promoted by miR-142-3p inhibitor (Figure 5E). In addition, a significant negative correlation was found between ZFX and miR-142-3p expression in NSCLC tumor tissues (Figure 5F). These results indicated that miR-142-3p targeted ZFX in NSCLC.
Figure 5.
ZFX was a target of miR-142-3p. (A) The sequences of WT-ZFX 3ʹUTR and MUT-ZFX 3ʹUTR were presented. (B) The interaction between ZFX and miR-142-3p was verified using dual-luciferase reporter assay. (C) The ZFX mRNA level in NSCLC tumor tissues (Tumor, n = 30) and normal non-cancerous tissues (Normal, n = 30) was detected by qRT-PCR. (D) WB analysis was used to measure the protein level of ZFX in NSCLC cells (H1299, H1581 and A549) and 16HBE cells. (E) After transfected with miR-NC, miR-142-3p, in-miR-NC or in-miR-142-3p into H1299 and A549 cells, the ZFX protein level was determined by WB analysis. (F) The correlation between miR-142-3p and ZFX in NSCLC tumor tissues was evaluated using Pearson correlation analysis. ***P < 0.001.
ZFX Silencing Inhibited NSCLC Cell Proliferation, Metastasis, and Increased Apoptosis
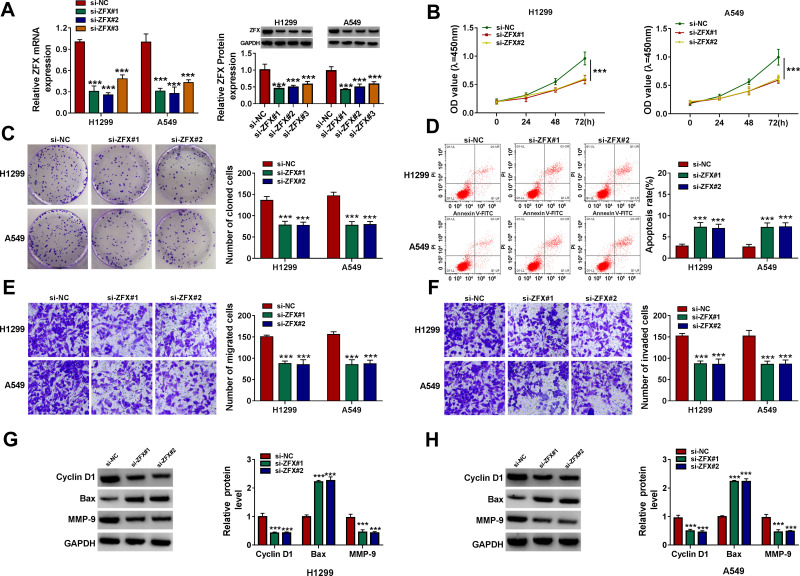
To determine the function of ZFX in NSCLC, we also constructed the siRNAs of ZFX. After transfected si-ZFX#1/#2/#3 into H1299 and A549 cells, the mRNA and protein expression levels of ZFX were markedly decreased (Figure 6A), confirming that the transfection efficiency of ZFX siRNAs was good. Then, we assessed the proliferation, apoptosis, and metastasis of NSCLC cells transfected with si-ZFX#1 and si-ZFX#2. CCK8 assay and colony formation assay results showed that ZFX knockdown could repress the viability and the number of cloned cells in H1299 and A549 cells (Figure 6B and C). In addition, ZFX silencing also promoted the apoptosis rate (Figure 6D), while suppressed the migration and invasion of H1299 and A549 cells (Figure 6E and F). Furthermore, downregulated ZFX could inhibit the protein levels of Cyclin D1 and MMP-9, and enhance the protein level of Bax in H1299 and A549 cells (Figure 6G and H). Our data suggested that ZFX might promote NSCLC progression.
Figure 6.
ZFX silencing inhibited NSCLC proliferation and metastasis. (A) The transfection efficiencies of si-ZFX#1, si-ZFX#2 and si-ZFX#3 were confirmed by detecting ZFX mRNA and protein expression in H1299 and A549 cells using qRT-PCR and WB analysis. (B–H) H1299 and A549 cells were transfected with si-NC, si-ZFX#1 or si-ZFX#2. CCK8 assay (B), colony formation assay (C), flow cytometry (D) and transwell assay (E and F) were employed to examine the viability, the number of cloned cells, apoptosis rate, the numbers of migrated and invaded cells. (G and H) The protein levels of Cyclin D1, Bax and MMP-9 were assessed by WB analysis. ***P < 0.001.
The Promotion Effect of miR-142-3p Inhibitor on NSCLC Progression Could Be Reversed by ZFX Knockdown
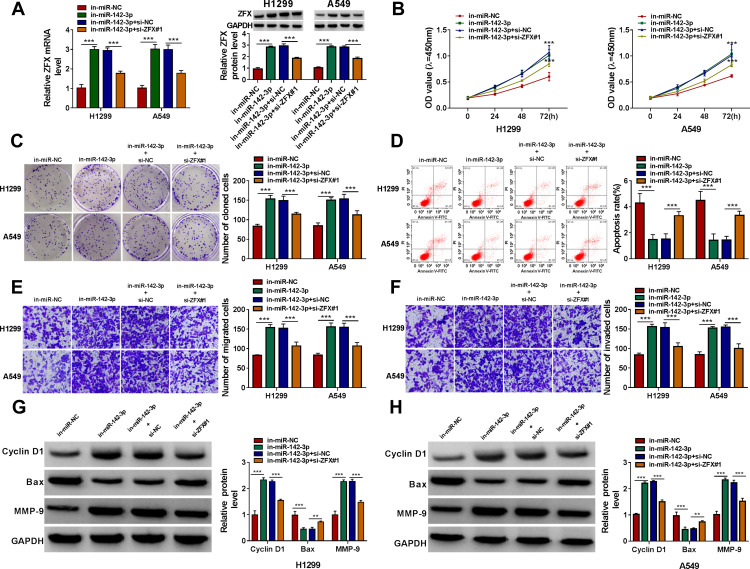
Subsequently, in-miR-142-3p and si-ZFX#1 were co-transfected into H1299 and A549 cells to verify whether miR-142-3p regulated NSCLC progression by targeting ZFX. The detection results of ZFX mRNA and protein expression showed that the addition of si-ZFX#1 could reverse the increasing effect of in-miR-142-3p on ZFX expression (Figure 7A). Functional experiments revealed that silenced ZFX could reverse the enhancing effect of miR-142-3p inhibitor on the viability and the number of cloned cells (Figure 7B and C), and the suppression effect on the apoptosis of H1299 and A549 cells (Figure 7D). Meanwhile, the promotion effect of miR-142-3p inhibitor on the migration and invasion of H1299 and A549 cells also could be abolished by ZFX knockdown (Figure 7E and F). The increasing effect of miR-142-3p inhibitor on Cyclin D1 and MMP-9 protein expression and the inhibition effect of Bax protein expression were also reversed by the knockdown of ZFX in H1299 and A549 cells (Figure 7G and H). These data revealed that miR-142-3p regulated NSCLC progression by targeting ZFX.
Figure 7.
The promotion effect of miR-142-3p inhibitor on NSCLC progression could be reversed by ZFX knockdown. H1299 and A549 cells were transfected with in-miR-NC, in-miR-142-3p, in-miR-142-3p + si-NC or in-miR-142-3p + si-ZFX#1. (A) The mRNA and protein expression of ZFX was measured by qRT-PCR and WB analysis. The viability, the number of cloned cells, apoptosis rate, the numbers of migrated and invaded cells were evaluated by CCK8 assay (B), colony formation assay (C), flow cytometry (D) and transwell assay (E and F). (G and H) WB analysis was employed to detect the protein levels of Cyclin D1, Bax and MMP-9. **P < 0.01, ***P < 0.001.
Circ_0020123 Regulated ZFX by Sponging miR-142-3p
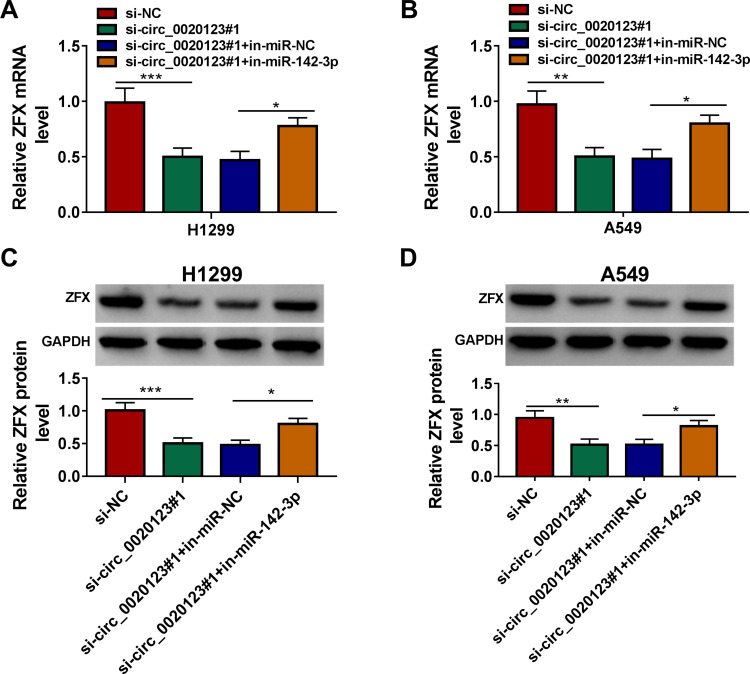
For confirming that circ_0020123 could sponge miR-142-3p to target ZFX, we measured the mRNA and protein expression of ZFX in H1299 and A549 cells co-transfected with si-circ_0020123#1 and in-miR-142-3p. The results indicated that ZFX mRNA and protein expression could be inhibited by circ_0020123 knockdown in H1299 and A549 cells, while these effects could be reversed by miR-142-3p inhibitor (Figure 8A–D). The data suggested that circ_0020123 positively regulated ZFX expression via sponging miR-142-3p.
Figure 8.
Circ_0020123 regulated ZFX by sponging miR-142-3p. H1299 and A549 cells were transfected with si-NC, si-circ_0020123#1, si-circ_0020123#1 + in-miR-NC, or si-circ_0020123#1 + in-miR-142-3p. QRT-PCR (A and B) and WB analysis (C and D) were used to determine the mRNA and protein expression levels of ZFX in H1299 cells and A549 cells. *P < 0.05, **P < 0.01, ***P < 0.001.
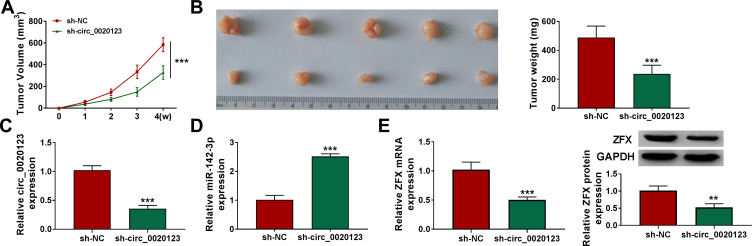
Knockdown of Circ_0020123 Could Hinder NSCLC Tumorigenesis in vivo
Additionally, the subcutaneous xenograft tumors were constructed to explore the role of circ_0020123 in NSCLC tumorigenesis. In the sh-circ_0020123 group, the tumor volume of mice was significantly decreased compared to the sh-NC group (Figure 9A). After removing the tumors for photographing and weighting, we discovered that the tumor size and weight of the mice in the sh-circ_0020123 group were markedly reduced (Figure 9B). Furthermore, we detected circ_0020123, miR-142-3p and ZFX expression in the tumor tissues of mice. The results showed that circ_0020123 expression was inhibited and miR-142-3p expression was promoted in the tumor tissues of the sh-circ_0020123 group (Figure 9C and D). At the same time, the decreased ZFX mRNA and protein expression levels also were observed in the tumor tissues of the sh-circ_0020123 group (Figure 9E). Our results showed that circ_0020123 might promote NSCLC tumorigenesis by regulating the miR-142-3p/ZFX axis.
Figure 9.
Knockdown of circ_0020123 could hinder NSCLC tumorigenesis in vivo. H1299 cells transfected with sh-NC or sh-circ_0020123 were injected into nude mice. (A) Tumor volume was measured every week. (B) After 4 weeks, the tumor was collected, photographed and weighted. (C and D) The expression of circ_0020123 and miR-142-3p was detected by qRT-PCR. (E) The mRNA and protein expression of ZFX in the tumors of mice was assessed using qRT-PCR and WB analysis. **P < 0.01, ***P < 0.001.
Discussion
At present, the important functions of many circRNAs in cancer have been elucidated. However, the continuous presentation of new functions and new mechanisms provides more references for circRNA to be an effective target for cancer. Our study explored a circRNA that has been reported to have a pro-oncogenic effect in NSCLC. Our results revealed that circ_0020123 was remarkably upregulated in NSCLC and its silencing had an inhibition on NSCLC cell proliferation, migration, and invasion, as well as had a promotion on apoptosis. Furthermore, animal experiments revealed that circ_0020123 knockdown markedly inhibited the tumorigenesis of NSCLC in vivo. These evidences suggested that circ_0020123 acted as a tumor promoter in NSCLC, which was consistent with the results previously reported.19–22
Circ_0020123 was found to serve as a ceRNA for miR-495,19 miR-590-5p,20 miR-488-3p21 and miR-144.22 Here, we discovered that miR-142-3p also could be sponged by circ_0020123. MiR-142-3p is believed to be a tumor suppressor involved in cancer progression regulation in many cancer-related studies. Chen et al suggested that circ_0084927 enhanced cervical cancer proliferation and metastasis by sponging miR-142-3p.23 And Shang et al proposed that miR-142-3p also participated in the positive regulation of exosomal circPACRGL on colorectal cancer progression.24 In addition, miR-142-3p had been found to have regulatory effects on chemotherapy sensitivity and cell autophagy in breast cancer.25 In NSCLC, the previous studies revealed that miR-142-3p could hinder cancer progression and promote cancer chemosensitivity.26–28 Consistent with these studies, our data confirmed that miR-142-3p was lowly expressed in NSCLC. The reversal effect of miR-142-3p inhibitor on the function of circ_0020123 knockdown in NSCLC progression revealed that circ_0020123 promoted NSCLC progression by inhibiting miR-142-3p, which once again confirmed the tumor suppressor role of miR-142-3p in NSCLC.
ZFX is a zinc finger protein transcription factor encoded by the X chromosome, which plays a vital role in the self-renewal of peripheral T cell, embryonic stem cells and hematopoietic stem cells.29,30 Not only that,previous studies also revealed that ZFX could regulate cell proliferation, differentiation and apoptosis, and could participate in tumorigenesis.31,32 High expressed ZFX could facilitate the tumor growth of colorectal cancer and was associated with poor prognosis in patients.33 Studies had reported that ZFX was overexpressed in NSCLC, which could promote cancer progression and was associated with lymph node metastasis of NSCLC patients.34–36 Here, we discovered that ZFX was a target of miR-142-3p. Knockdown of ZFX could inhibit NSCLC progression and overturn the enhancing effect of miR-142-3p inhibitor on NSCLC progression, indicating that miR-142-3p targeted ZFX, an oncogene, to restrain NSCLC progression. Besides, we also uncovered that ZFX expression was positively regulated by circ_0020123 in vitro and in vivo. This completed the hypothesis that circ_0020123 mediated NSCLC progression through regulating miR-142-3p/ZFX network.
In summary, our research suggested that circ_0020123 could act as a sponge of miR-142-3p to promote NSCLC progression by upregulating ZFX. Our findings proposed a new mechanism for circ_0020123 mediated NSCLC progression and provided a theoretical basis for the study of circ_0020123 in NSCLC.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Mao Y, Yang D, He J, Krasna MJ. Epidemiology of lung cancer. Surg Oncol Clin N Am. 2016;25(3):439–445. doi: 10.1016/j.soc.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 2.Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103(3):463–473. doi: 10.1016/j.mcna.2018.12.006 [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi: 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 4.Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94(8):1623–1640. doi: 10.1016/j.mayocp.2019.01.013 [DOI] [PubMed] [Google Scholar]
- 5.Rusch VW. Stage III non-small cell lung cancer. Semin Respir Crit Care Med. 2016;37(5):727–735. doi: 10.1055/s-0036-1592112 [DOI] [PubMed] [Google Scholar]
- 6.Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med. 2019;27(148):167–170. [PubMed] [Google Scholar]
- 7.Nagano T, Tachihara M, Nishimura Y. Molecular mechanisms and targeted therapies including immunotherapy for non-small cell lung cancer. Curr Cancer Drug Targets. 2019;19(8):595–630. [DOI] [PubMed] [Google Scholar]
- 8.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 9.Hsiao KY, Sun HS, Tsai SJ. Circular RNA – New member of noncoding RNA with novel functions. Exp Biol Med. 2017;242(11):1136–1141. doi: 10.1177/1535370217708978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336. doi: 10.1016/j.trecan.2020.01.012 [DOI] [PubMed] [Google Scholar]
- 11.Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220. doi: 10.1186/s12967-018-1593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dori M, Bicciato S. Integration of bioinformatic predictions and experimental data to identify circRNA-miRNA associations. Genes. 2019;10(9):642. doi: 10.3390/genes10090642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027 [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Li Z, Zhang M, et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2019;38(1):466. doi: 10.1186/s13046-019-1436-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao X, Li F, Shao J, et al. Circular RNA hsa_circ_0102231 sponges miR-145 to promote non-small cell lung cancer cell proliferation by up-regulating the expression of RBBP4. J Biochem. 2020. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, Zhu Y, Qin Y, Chen Y. CircNFIX acts as a miR-212-3p sponge to enhance the malignant progression of non-small cell lung cancer by up-regulating ADAM10. Cancer Manag Res. 2020;12:9577–9587. doi: 10.2147/CMAR.S272309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bi R, Wei W, Lu Y, et al. High hsa_circ_0020123 expression indicates poor progression to non-small cell lung cancer by regulating the miR-495/HOXC9 axis. Aging. 2020;12(17):17343–17352. doi: 10.18632/aging.103722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Zhao L, Wang Y. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by sponging miR-590-5p to regulate THBS2. Cancer Cell Int. 2020;20:387. doi: 10.1186/s12935-020-01444-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan J, Hao L, Zheng X, Li Z. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by acting as a ceRNA for miR-488-3p to regulate ADAM9 expression. Biochem Biophys Res Commun. 2019;515(2):303–309. doi: 10.1016/j.bbrc.2019.05.158 [DOI] [PubMed] [Google Scholar]
- 22.Qu D, Yan B, Xin R, Ma T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8(8):1387–1402. [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Zhang X, Wang S, Lin X, Xu L. Circ_0084927 facilitates cervical cancer development via sponging miR-142-3p and upregulating ARL2. Cancer Manag Res. 2020;12:9271–9283. doi: 10.2147/CMAR.S263596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang A, Gu C, Wang W, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta1 axis. Mol Cancer. 2020;19(1):117. doi: 10.1186/s12943-020-01235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang L, Fu J, Wang S, et al. MiR-142-3p enhances chemosensitivity of breast cancer cells and inhibits autophagy by targeting HMGB1. Acta Pharm Sin B. 2020;10(6):1036–1046. doi: 10.1016/j.apsb.2019.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Zhou X, Qiao J, Bao A. MiR-142-3p overexpression increases chemo-sensitivity of NSCLC by inhibiting HMGB1-mediated autophagy. Cell Physiol Biochem. 2017;41(4):1370–1382. doi: 10.1159/000467896 [DOI] [PubMed] [Google Scholar]
- 27.Xiao P, Liu WL. MiR-142-3p functions as a potential tumor suppressor directly targeting HMGB1 in non-small-cell lung carcinoma. Int J Clin Exp Pathol. 2015;8(9):10800–10807. [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Tian W, Zhang W, et al. MicroRNA-142-3p/MALAT1 inhibits lung cancer progression through repressing beta-catenin expression. Biomed Pharmacother. 2019;114:108847. doi: 10.1016/j.biopha.2019.108847 [DOI] [PubMed] [Google Scholar]
- 29.Smith-Raska MR, Arenzana TL, D’Cruz LM, et al. The transcription factor Zfx regulates peripheral T cell self-renewal and proliferation. Front Immunol. 2018;9:1482. doi: 10.3389/fimmu.2018.01482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galan-Caridad JM, Harel S, Arenzana TL, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129(2):345–357. doi: 10.1016/j.cell.2007.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Su Z, Huang Y, et al. The Zfx gene is expressed in human gliomas and is important in the proliferation and apoptosis of the human malignant glioma cell line U251. J Exp Clin Cancer Res. 2011;30:114. doi: 10.1186/1756-9966-30-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisberg SP, Smith-Raska MR, Esquilin JM, et al. ZFX controls propagation and prevents differentiation of acute T-lymphoblastic and myeloid leukemia. Cell Rep. 2014;6(3):528–540. doi: 10.1016/j.celrep.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan X, Shan Z, Yan L, et al. High expression of Zinc-finger protein X-linked promotes tumor growth and predicts a poor outcome for stage II/III colorectal cancer patients. Oncotarget. 2016;7(15):19680–19692. doi: 10.18632/oncotarget.7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li K, Zhu ZC, Liu YJ, et al. ZFX knockdown inhibits growth and migration of non-small cell lung carcinoma cell line H1299. Int J Clin Exp Pathol. 2013;6(11):2460–2467. [PMC free article] [PubMed] [Google Scholar]
- 35.Zha W, Cao L, Shen Y, Huang M. Roles of Mir-144-ZFX pathway in growth regulation of non-small-cell lung cancer. PLoS One. 2013;8(9):e74175. doi: 10.1371/journal.pone.0074175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang M, Xu S, Yue W, et al. The role of ZFX in non-small cell lung cancer development. Oncol Res. 2012;20(4):171–178. doi: 10.3727/096504012X13548165987493 [DOI] [PubMed] [Google Scholar]