Abstract
Background
The initial symptoms of patients with COVID-19 are very much like those of patients with community-acquired pneumonia (CAP); it is difficult to distinguish COVID-19 from CAP with clinical symptoms and imaging examination.
Objective
The objective of our study was to construct an effective model for the early identification of COVID-19 that would also distinguish it from CAP.
Methods
The clinical laboratory indicators (CLIs) of 61 COVID-19 patients and 60 CAP patients were analyzed retrospectively. Random combinations of various CLIs (ie, CLI combinations) were utilized to establish COVID-19 versus CAP classifiers with machine learning algorithms, including random forest classifier (RFC), logistic regression classifier, and gradient boosting classifier (GBC). The performance of the classifiers was assessed by calculating the area under the receiver operating characteristic curve (AUROC) and recall rate in COVID-19 prediction using the test data set.
Results
The classifiers that were constructed with three algorithms from 43 CLI combinations showed high performance (recall rate >0.9 and AUROC >0.85) in COVID-19 prediction for the test data set. Among the high-performance classifiers, several CLIs showed a high usage rate; these included procalcitonin (PCT), mean corpuscular hemoglobin concentration (MCHC), uric acid, albumin, albumin to globulin ratio (AGR), neutrophil count, red blood cell (RBC) count, monocyte count, basophil count, and white blood cell (WBC) count. They also had high feature importance except for basophil count. The feature combination (FC) of PCT, AGR, uric acid, WBC count, neutrophil count, basophil count, RBC count, and MCHC was the representative one among the nine FCs used to construct the classifiers with an AUROC equal to 1.0 when using the RFC or GBC algorithms. Replacing any CLI in these FCs would lead to a significant reduction in the performance of the classifiers that were built with them.
Conclusions
The classifiers constructed with only a few specific CLIs could efficiently distinguish COVID-19 from CAP, which could help clinicians perform early isolation and centralized management of COVID-19 patients.
Keywords: COVID-19, clinical laboratory indicators, community-acquired pneumonia, classifier, classification algorithm
Introduction
COVID-19 caused by SARS-CoV-2 infection, which was discovered in early December 2019, has become a global pandemic. As of August 3, 2020, COVID-19 has become widespread in 215 countries, areas, or territories worldwide; it has caused infection in more than 17.9 million people and has resulted in the deaths of more than 686,000 people [1]. The World Health Organization has stated that the spread of COVID-19 may be impeded by early detection, isolation, and the implementation of a robust health care system [2,3]. Nevertheless, the published data indicate that the initial symptoms of COVID-19 in patients are very similar to those in patients with the common cold or influenza. COVID-19 patients exhibit different clinical symptoms, and some of them do not have any symptoms [4-7]. sars-cov-2 infection has a long incubation period, with a median incubation period of 5 to 7 days, which is the chief risk factor for community infection [6,8]. Community-acquired pneumonia (CAP) and COVID-19 have similar clinical and imaging features, but their treatment and infectivity are very dissimilar. Distinguishing COVID-19 from CAP is very important to prevent the spread of COVID-19 and to provide specific treatment.
Some characteristic spectra demonstrated by clinical laboratory indicators (CLIs) of COVID-19 patients have been utilized as auxiliary clues for diagnosis [9]. Previous studies have demonstrated that increased procalcitonin (PCT), lymphocytopenia, and thrombin activation can all be utilized as auxiliary diagnostic indicators of COVID-19 and poor prognostic factors [9-11]. However, they are also correlated with CAP [12-15]. Thus, in accordance with the changes in these indicators, it is impossible to differentiate COVID-19 from CAP. The changes in the neutrophil to lymphocyte ratio, the peak platelet to lymphocyte ratio, lactate dehydrogenase (LDH), C-reactive protein (CRP), and interleukin-6 (IL-6) are considered to be associated with the progression and prognosis of COVID-19 [9], but using the information from the CLIs to give clinicians correct guidance is still a great challenge.
Classifiers established by machine learning (ML) algorithms based on various clinical features, biomarkers, and CLIs are increasingly widely utilized in disease diagnosis and risk prediction [16]. During the COVID-19 pandemic, ML was also widely used to predict, classify, assess, track, and control the spread of SARS-CoV-2 [17,18]. ML can improve diagnostic performance compared with hand-selected biomarkers by selecting relevant biomarkers and more consistently capturing both their relative importance to prediction and their interactions among one another [19]. In this study, we used CLIs to build classifiers with different ML algorithms to distinguish COVID-19 patients from CAP patients; we found that only the feature combinations (FCs) with many specific CLIs rather than the FCs with the most significantly differential CLIs between the two groups could build high-performance classifiers (HPCs).
Methods
Collection of Patients’ Electronic Medical Record Data
The electronic medical records of patients who were admitted to Gong An County People’s Hospital, China, and diagnosed with COVID-19 or CAP from December 2019 to March 2020 were retrieved. The information regarding each patient’s age, sex, clinical symptoms upon admission, medical history, epidemiological history, computed tomography (CT) imaging features, and CLIs were sorted out for retrospective analysis. Only the laboratory test results during admission were included. It was specified that all patients’ data were to be kept confidential, and this data were only to be utilized for comprehensive analysis. No personal information about any patient was mentioned in the paper. This study was approved by the ethics committees from the Guangdong Provincial Hospital of Chinese Medicine (approval No. ZE2020-049-01) with a waiver of informed consent due to the retrospective nature of the study.
Data Description
Diagnosis and clinical classification of COVID-19 were performed according to the Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment (7th edition) [20]. A total of 61 patients with COVID-19 and 60 patients with CAP were enrolled according to the discharge diagnosis on their electronic medical records. There were 3 mild, 47 common, 6 severe, and 5 critical types, which were categorized into two groups for further analysis as follows: COVID19-COM (3 mild and 47 common types) and COVID19-SV (6 severe and 5 critical types). They were matched by age and sex and did not significantly differ in terms of medical history. The main clinical symptoms between CAP and COVID-19 groups were not significantly different.
Primary Analysis
The descriptive analysis of all CLIs was performed between groups or subgroups. Between-group or between-subgroup differences were tested using the statsmodels module from Python (Python Software Foundation) [21]. The Student t test was performed when the distribution of the variables conformed to the normal distribution; otherwise, the Mann-Whitney U test was used. The chi-square test was used to detect differences in baseline data between two groups or subgroups. A value of P<.05 was considered to be significant.
Feature Selection and Data Preprocessing
The CLIs with a missing value ratio greater than 20% were excluded. Only the CLIs with a significant difference between the two groups were selected and used to generate 1,807,780 nonrepetitive random FCs, consisting of one to eight CLIs, by using the combinations iterator in the itertools module from Python [22]. Next, an FC was selected from the FC list one by one to form a new data sheet with the dependent variable (ie, disease type), and 1,807,780 new data sheets were eventually formed. For each new data sheet, the rows with missing values were removed. The remaining rows were then divided into training_dataset and test_dataset using scikit-learn, version 0.23.1 (train_test_split function with test_size = 0.25, random_state = 0). The training data set was used to build the classifier, and the test data set was used to assess the performance. The feature values were standardized using the StandardScaler function in the scikit-learn module before constructing the logistic regression (LR) classifier.
Construction of Classifiers With ML Algorithms in the Scikit-Learn Module
Scikit-learn is a Python module integrating a wide range of state-of-the-art ML algorithms for medium-scale supervised and unsupervised problems [23]. The LR classifier, the random forest classifier (RFC), and the gradient boosting classifier (GBC) have been typically used to construct classifiers in prediction of disease risk, progression, prognosis, and so on [24]. The LR classifier in the sklearn.linear_model is also known as logit regression, maximum-entropy classification, or the log-linear classifier. In this model, the probabilities describing the possible outcomes of a single trial are modeled using a logistic function [24]. The RFC in the sklearn.ensemble module is one of the averaging algorithms in ensemble methods and is a perturb-and-combine technique specifically designed for trees. In the random forest algorithm, each tree in the ensemble is built from a sample drawn with replacement from the training data set. Furthermore, when splitting each node during the construction of a tree, the best split is found either from all input features or from a random subset of size setting with the parameter max_features. In practice, the variance reduction due to the introduction of randomness in the classifier construction is often significant, hence, yielding an overall better model [25,26]. The GBC algorithm, using the sklearn.ensemble function, is a boosting method, in which base estimators are built sequentially. To reduce the bias of the combined estimator, one has to combine several weak models to produce a powerful ensemble. The GBC algorithm builds an additive model in a forward stage-wise fashion, and it allows for the optimization of arbitrary differentiable loss functions [27,28].
In this study, the classifiers were respectively constructed using the LR classifier, RFC, and GBC in the scikit-learn module with the training data set. The model parameter settings were kept as default, except that random_state was modified to “0” for all models and class_weight was modified to “balanced” for the LR classifier and RFC models. The performance of the classifiers was evaluated with the test data set by calculating the recall rate (ie, sensitivity), specificity, accuracy, and area under the receiver operating characteristic curve (AUROC), using the sklearn_metrics.recall_score, sklearn_metrics.precision_score, sklearn_metrics.accuracy_score, and sklearn_metrics.auc functions, respectively. Gini importance was computed using the feature_importance function to measure the importance of each feature in the RFC and the GBC. The higher the Gini importance value, the more important the feature [29]. All the above analyses were performed in Python, version 3.7 (Python Software Foundation).
Results
Basic Characteristics of CAP Group and COVID-19 Group
No significant differences in age and sex were found between CAP and COVID-19 groups (see Table 1); however, the proportions of males in the CAP and COVID-19 groups were 55% (33/60) and 66% (40/61), respectively, and were higher than those of females in both groups. No significant difference in the medical history between the two groups (see Table 1) was observed. Also, no significant difference was found in the proportions of the main clinical symptoms between the two groups, such as fever, cough, fatigue, muscle soreness, and loss of appetite (see Table 1). The average hospitalization days for CAP patients were remarkably lower than those for COVID-19 patients (P<.001). In the CAP group, some patients with pulmonary CT also had imaging features that included patchy hyperdense shadow (11/60, 18%), ground-glass shadow (4/60, 7%), and fibrotic lesion (6/60, 10%). Nonetheless, the chief imaging features of pulmonary CT in the COVID-19 group were patchy hyperdense shadow (25/61, 41%) and ground-glass shadow (9/61, 15%), and many patients (7/61, 11%) had both patchy hyperdense shadow and ground-glass shadow (see Table 1). Among the 61 patients suffering from COVID-19, 3 (5%) had mild symptoms, 47 (77%) had common symptoms, 6 (10%) had severe symptoms, and 5 (8%) had critical symptoms. Fever and cough were the principal symptoms in the early stage of COVID-19, and these accounted for 70% (43/61) and 64% (39/61) of the cases, respectively (see Table 1). Among the CAP patients included in the analysis, no cases of death were found during hospitalization; however, 3 of the 5 (60%) severely ill patients in the COVID-19 group, who were aged 36, 49, and 74 years, died during hospitalization. The 36-year-old patient who died underwent interventricular septal repair in childhood.
Table 1.
Comparison of baseline information between COVID-19 patients and community-acquired pneumonia (CAP) patients.
| Baseline characteristic | CAP patients (n=60) | COVID-19 patients (n=61) | P value | |
| Sex (male), n (%) | 33 (55) | 40 (66) | .27 | |
| Age (years), mean (SD) | 55.72 (18.10) | 50.23 (16.95) | .09 | |
| Hospitalization days, median (IQR) | 9 (7-12) | 21 (13-26) | <.001 | |
| Medical history, n (%) |
|
|
|
|
|
|
Hypertension | 14 (23) | 16 (26) | .83 |
|
|
Diabetes | 2 (3) | 6 (10) | .27 |
|
|
Liver disease | 2 (3) | 3 (5) | .99 |
|
|
Heart disease | 3 (5) | 5 (8) | .72 |
|
|
Exposure history | Unclear | 54 (89) | N/Aa |
|
|
Familial aggregation infectionb | Unclear | 22 (36) | N/A |
| Initial symptoms, n (%) |
|
|
|
|
|
|
Fever | 36 (60) | 43 (70 ) | .26 |
|
|
Cough | 44 (73) | 39 (64) | .33 |
|
|
Myalgia | 4 (7) | 7 (11) | .53 |
|
|
Poor appetite | 5 (8) | 11 (18) | .18 |
|
|
Fatigue | 33 (55) | 24 (39) | .10 |
| Days from onset of symptoms to admission, median (IQR) | Unrecorded | 3 (1-7) | N/A | |
| Imaging features, n (%) |
|
|
|
|
|
|
Patchy high-density opacity | 11 (18) | 25 (41) | .009 |
|
|
Ground-glass opacity | 4 (7) | 9 (15) | .24 |
|
|
Fibrotic lesion | 6 (10) | 3 (5) | .32 |
|
|
Patchy high-density opacity and ground-glass opacity | 0 (0) | 7 (11) | .01 |
| Death cases, n (%) | 0 (0) | 3 (5) | N/A | |
aN/A: not applicable; groups could not be compared because there were no values for the CAP group.
bThere were more than 2 cases of infection after aggregation with family members or relatives.
Characteristic Profile of the CLIs in COVID-19 and CAP
Even though most CLIs had a similar variation trend in both CAP and COVID-19, the extent of change was different. Among more than 60 evaluated CLIs, there were significant differences in 25 CLIs between the two groups (see Table 2). A decrease of lymphocyte, red blood cell (RBC) count, hematocrit or packed-cell volume (PCV), hemoglobin concentration, and mean corpuscular hemoglobin concentration (MCHC) and an increase of neutrophil ratio, prothrombin time (PT), micro-CRP (mCRP), and PCT were observed in both COVID-19 and CAP patients. Furthermore, the neutrophil ratio and levels of PT, mCRP, and PCT in CAP were remarkably higher than those in COVID-19. Levels of lymphocyte, RBC count, PCV, hemoglobin concentration, and MCHC in CAP were significantly lower than those in COVID-19 (see Figure 1). Various erythrocyte-related CLIs—RBC count, PCV, hemoglobin concentration, and MCHC—significantly decreased in both CAP and COVID-19, but there was a greater reduction in CAP patients (see Figure 1). The RBC distribution width–standard deviation (RDW-SD) and RBC mean corpuscular volume (MCV) also indicated prominent differences between CAP and COVID-19 (see Figure 1).
Table 2.
Differences in clinical laboratory indicators (CLIs) between patients with community-acquired pneumonia (CAP) and COVID-19.
| CLI | CAP patients (n=60) | COVID-19 patients (n=61) | P value | ||
|
|
n (%) | Mean (SD) | n (%) | Mean (SD) |
|
| Procalcitonin (ng/mL) | 43 (72) | 0.629 (0.838) | 55 (90) | 0.134 (0.184) | <.001 |
| Monoamine oxidase B (U/L) | 35 (58) | 4.569 (1.748) | 53 (87) | 3.538 (1.592) | .001 |
| Myoglobin (ng/mL) | 14 (23) | 39.179 (29.421) | 23 (38) | 65.794 (87.039) | .04 |
| Micro–C-reactive protein (mg/L) | 41 (68) | 63.943 (64.530) | 13 (21) | 22.568 (29.577) | .004 |
| Prothrombin time (seconds) | 30 (50) | 12.780 (0.873) | 53 (87) | 12.460 (1.107) | .04 |
| Thrombin time (seconds) | 30 (50) | 15.123 (1.565) | 53 (87) | 14.655 (1.422) | .049 |
| Albumin (g/L) | 53 (88) | 35.508 (5.929) | 54 (89) | 37.831 (6.169) | .04 |
| Albumin to globulin ratio | 53 (88) | 1.211 (0.295) | 54 (89) | 1.378 (0.482) | .047 |
| α-L-fucosidase (U/L) | 35 (58) | 17.709 (5.167) | 50 (82) | 22.106 (5.698) | <.001 |
| Uric acid (μmol/L) | 44 (73) | 284.193 (118.608) | 54 (89) | 325.261 (92.914) | .007 |
| Potassium (mmol/L) | 54 (90) | 3.900 (0.462) | 55 (90) | 4.021 (0.392) | .03 |
| White blood count cell (×109/L) | 58 (97) | 8.858 (5.576) | 56 (92) | 5.293 (2.047) | <.001 |
| Neutrophils (%) | 57 (95) | 72.958 (15.544) | 56 (92) | 66.661 (14.013) | .007 |
| Lymphocytes (%) | 56 (93) | 18.646 (13.416) | 56 (92) | 24.014 (11.175) | .002 |
| Neutrophil count (×109/L) | 56 (93) | 6.797 (5.525) | 56 (92) | 3.649 (1.949) | <.001 |
| Monocyte count (×109/L) | 55 (92) | 0.565 (0.337) | 56 (92) | 0.404 (0.194) | .009 |
| Eosinophil count (×109/L) | 55 (92) | 0.111 (0.213) | 56 (92) | 0.053 (0.072) | .03 |
| Basophil count (×109/L) | 55 (92) | 0.021 (0.013) | 56 (92) | 0.015 (0.013) | .002 |
| Red blood cell count (×1012/L) | 56 (93) | 4.028 (0.647) | 56 (92) | 4.284 (0.570) | .008 |
| Hemoglobin concentration (g/L) | 55 (92) | 120.800 (17.326) | 56 (92) | 130.143 (16.888) | .005 |
| Packed-cell volume (hematocrit) (L/L) | 55 (92) | 0.371 (0.052) | 56 (92) | 0.389 (0.049) | .04 |
| Mean red blood cell volume (fL) | 55 (92) | 93.255 (6.662) | 56 (92) | 91.241 (6.501) | .01 |
| Mean corpuscular hemoglobin concentration (g/L) | 55 (92) | 325.473 (8.360) | 56 (92) | 334.482 (13.559) | <.001 |
| Red blood cell distribution width–standard deviation (fL) | 55 (92) | 41.476 (2.573) | 56 (92) | 41.141 (4.082) | .01 |
Figure 1.
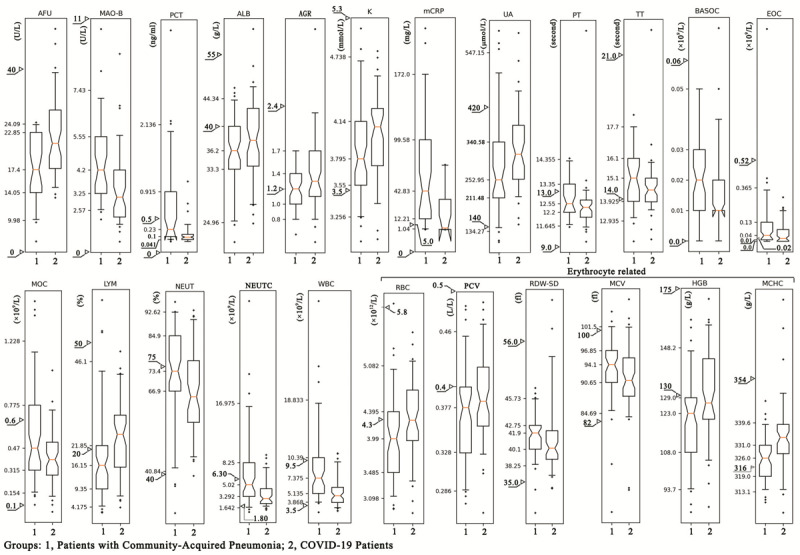
The statistical distribution of the plasma level of the clinical laboratory indicators (CLIs) with a remarkable difference between COVID-19 and community-acquired pneumonia (CAP). The statistical distribution was presented with a box and whisker plot. The horizontal lines within the boxes indicate the median value. The vertical lines extending below and above the boxes represent 5%-95% percentile values. The scale on the y-axis represents the values of the 5th, 25th, 50th, 75th, and 95th percentiles of the CLI in the CAP group. The triangles represent the upper and lower limits of the normal reference range of the laboratory index. AFU: α-L-fucosidase; AGR: albumin to globulin ratio; ALB: albumin; BASOC: basophil count; EOC: eosinophil count; HGB: hemoglobin concentration; K: potassium; LYM: lymphocyte; MAO-B: monoaminoxidase B; MCHC: mean corpuscular hemoglobin concentration; mCRP: micro–C-reactive protein; MCV: mean (red blood cell) corpuscular volume; MOC: monocyte count; NEUT: neutrophil ratio; NEUTC: neutrophil count; PCT: procalcitonin; PCV: packed-cell volume (hematocrit); PT: prothrombin time; RBC: red blood cell count; RDW-SD: red blood cell distribution width–standard deviation; TT: thrombin time; UA: uric acid; WBC: white blood cell count.
Comparing the COVID19-COM and COVID19-SV subgroups, 26 CLIs demonstrated a remarkable difference (see Table 3). In comparison with the COVID19-COM subgroup, LDH, aspartate aminotransferase, fibrinogen content, mCRP, and erythrocyte sedimentation rate increased acutely in the COVID19-SV subgroup, whereas prealbumin, carbon dioxide binding capacity, lymphocytes, and lymphocyte count decreased in the COVID19-SV subgroup (see Multimedia Appendix 1).
Table 3.
Difference in clinical laboratory indicators (CLIs) between patients with common and severe types of COVID-19.
| CLIs | Patients with a common type of COVID-19 (n=50) | Patients with a severe type of COVID-19 (n=11) | P value | ||
|
|
n (%) | Mean (SD) | n (%) | Mean (SD) |
|
| Procalcitonin (ng/mL) | 44 (88) | 0.112 (0.170) | 11 (100) | 0.224 (0.217) | .01 |
| N-terminal pro-B-type natriuretic peptide (pg/mL) | 29 (58) | 366.053 (549.429) | 11 (100) | 534.782 (398.067) | .03 |
| Hypersensitive C-reactive protein (mg/L) | 41 (82) | 23.332 (34.483) | 11 (100) | 72.458 (60.805) | .002 |
| Lactate dehydrogenase (U/L) | 26 (52) | 214.896 (73.319) | 8 (73) | 314.750 (118.755) | .02 |
| D-dimer (mg/L) | 42 (84) | 0.834 (1.115) | 11 (100) | 5.133 (10.399) | .005 |
| Myoglobin (ng/mL) | 16 (32) | 49.221 (60.505) | 7 (64) | 103.674 (127.354) | .02 |
| Cardiac troponin (ng/mL) | 16 (32) | 0.011 (0.003) | 7 (64) | 0.033 (0.041) | .02 |
| Creatine kinase (U/L) | 27 (54) | 81.296 (47.153) | 8 (73) | 202.125 (195.052) | .02 |
| Fibrinogen content (mg/dL) | 42 (84) | 411.905 (104.363) | 11 (100) | 467.455 (76.500) | .03 |
| Aspartate aminotransferase (U/L) | 46 (92) | 29.413 (15.756) | 10 (91) | 45.600 (18.969) | .004 |
| γ-glutamyl transpeptidase (U/L) | 44 (88) | 46.046 (41.609) | 10 (91) | 80.000 (44.229) | .007 |
| Albumin (g/L) | 44 (88) | 38.602 (6.267) | 10 (91) | 34.440 (4.558) | .02 |
| Albumin to globulin ratio | 44 (88) | 1.436 (0.507) | 10 (91) | 1.120 (0.230) | .02 |
| Indirect bilirubin (μmol/L) | 44 (88) | 9.482 (3.841) | 10 (91) | 7.960 (4.336) | .048 |
| Prealbumin (mg/L) | 41 (82) | 180.171 (83.374) | 9 (82) | 125.556 (68.182) | .03 |
| β2-microglobulin (mg/L) | 41 (82) | 1.978 (0.430) | 9 (82) | 2.528 (1.015) | .01 |
| Carbon dioxide binding capacity (mmol/L) | 41 (82) | 25.420 (2.537) | 9 (82) | 22.733 (2.018) | .002 |
| Potassium (mmol/L) | 44 (88) | 4.057 (0.414) | 11 (100) | 3.876 (0.251) | .04 |
| Erythrocyte sedimentation rate (mm/h) | 30 (60) | 55.433 (41.639) | 7 (64) | 87.000 (35.081) | .02 |
| Neutrophils (%) | 45 (90) | 64.496 (13.286) | 11 (100) | 75.519 (14.001) | .02 |
| Lymphocytes (%) | 45 (90) | 25.711 (10.932) | 11 (100) | 17.073 (9.750) | .01 |
| Eosinophils (%) | 45 (90) | 1.236 (1.388) | 11 (100) | 0.391 (1.038) | .009 |
| Eosinophil count (×109/L) | 45 (90) | 0.062 (0.076) | 11 (100) | 0.014 (0.039) | .003 |
| Lymphocyte count (×109/L) | 45 (90) | 1.255 (0.558) | 11 (100) | 0.835 (0.383) | .008 |
| Packed-cell volume (hematocrit) (L/L) | 45 (90) | 0.395 (0.050) | 11 (100) | 0.368 (0.036) | .03 |
| Red blood cell distribution width–coefficient of variation (%) | 45 (90) | 12.658 (1.171) | 11 (100) | 12.873 (0.781) | .03 |
An orderly increase of α-L-fucosidase (AFU), myoglobin, uric acid, and MCHC and an orderly decrease of thrombin time, monocyte count, eosinophil count, RBC MCV, and RDW-SD were observed in CAP, COVID19-COM, and COVID19-SV patients, indicating that these CLIs may be used to distinguish CAP from COVID-19 and may suggest the probability of severe COVID-19 progression (see Multimedia Appendix 2).
Classifiers Constructed From the FCs With Seven to Eight CLIs Could Accurately Distinguish COVID-19 From CAP
The performance of the classifiers gradually improved as the number of CLIs in the FCs increased from one to eight. However, when the number of CLIs in the FCs reached eight, the performance of the classifiers constructed by these FCs no longer significantly improved. The performance of the LR classifier algorithm constructed with the FCs with eight CLIs (ie, 8-CLI combination) was even slightly lower than those constructed by the FCs with seven CLIs (ie, 7-CLI combination). A total of 43 FCs, including five 7-CLI combinations and 38 8-CLI combinations, were determined according to the recall rate. The AUROCs of the classifiers constructed with the LR classifier, RFC, and GBC algorithms were greater than 0.85 (see Multimedia Appendix 3, Table S1). The AUROC and precision-recall curves of the classifiers constructed with the RFC, LR classifier, and GBC algorithms from the representative 7-CLI combination (ie, PCT, albumin to globulin ratio [AGR], uric acid, neutrophil count, basophil count, RBC MCV, and MCHC) showed very high performance and precision in COVID-19 prediction; their AUROCs were 1.0, 0.97, and 0.96, respectively (see Figure 2, A), and their average precision values were 1.0, 0.97, and 0.98, respectively (Figure 2, B). The AUROCs of the classifiers constructed with the RFC, LR classifier, and GBC algorithms from the representative 8-CLI combination (ie, PCT, albumin, uric acid, WBC [white blood cell] count, monocyte count, basophil count, RBC count, and MCHC) were 1.0, 0.90, and 1.0, respectively (see Figure 2, C). The AUROCs of the classifiers constructed with the three algorithms from the 7-CLI combination (ie, agr, afu, lymphocytes, neutrophil counts, eosinophil count, RBC mcv, and mchc) were 0.98, 0.91, and 0.97, respectively (see Figure 2, D). Feature importance results showed that basophil count was the least important in the above two representative CLI combinations, and AFU was the most important in the CLI combinations (see Figure 3). However, when basophil count was substituted with AFU in the two above-mentioned CLI combinations, the performance of the classifiers constructed with the new CLI combinations decreased (see Figure 2, E and F). PCT and AFU were not observed to be in the same CLI combination from which an HPC could be constructed. The evidence above and the fact that only 43 FCs with seven or eight CLIs could be used to build HPCs suggested that only the FCs with specific CLIs can establish HPCs to distinguish COVID-19 from CAP.
Figure 2.
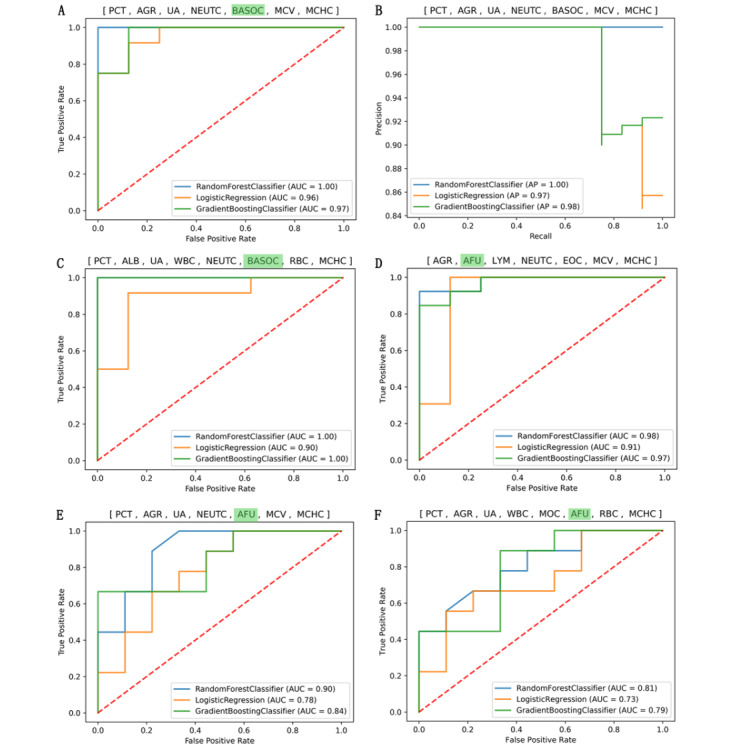
Area under the receiver operating characteristic curve (AUROC) and precision-recall curve plotted for the COVID-19 vs community-acquired pneumonia (CAP) classifiers built with various feature combinations (FCs) of different clinical laboratory indicators (CLIs). At the top of each image is the CLI combination for constructing classifiers using three different classification algorithms. AFU: α-L-fucosidase; AGR: albumin to globulin ratio; ALB: albumin; BASOC: basophil count; EOC: eosinophil count; LYM: lymphocyte; MCHC: mean corpuscular hemoglobin concentration; MCV: mean (red blood cell) corpuscular volume; MOC: monocyte count; NEUTC: neutrophil count; PCT: procalcitonin; RBC: red blood cell count; UA: uric acid; WBC: white blood cell count.
Figure 3.
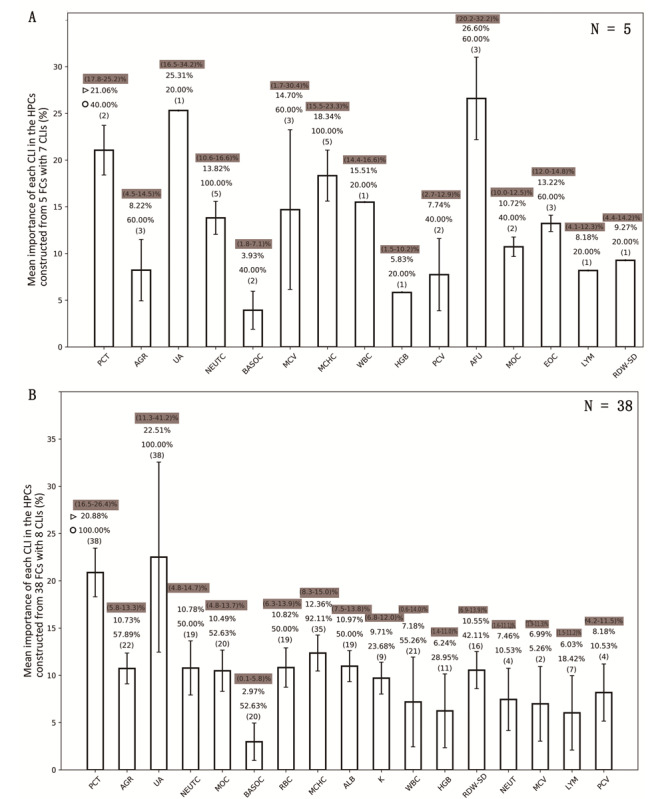
Usage rate and the feature importance of each clinical laboratory indicator (CLI) in the high-performance COVID-19 vs community-acquired pneumonia (CAP) classifiers. (A) The mean feature importance of each CLI in the high-performance classifiers (HPCs) constructed with the 7-CLI combinations. (B) The mean feature importance of each CLI in the HPCs constructed with the 8-CLI combinations. The histogram is represented by mean (SD). The numbers with the shadow backgrounds represent the minimum and maximum values of the feature importance of the CLI. The number indicated with the triangle symbol represents the mean feature importance of CLI in all classifiers. The number indicated with the circle represents the usage rate of the CLI in the HPC. The number in the parentheses indicates how many CLI combinations are capable of constructing the HPCs containing the CLI. AFU: α-L-fucosidase; AGR: albumin to globulin ratio; ALB: albumin; BASOC: basophil count; EOC: eosinophil count; FC: feature combination; HGB: hemoglobin concentration; K: potassium; LYM: lymphocyte; MCHC: mean corpuscular hemoglobin concentration; MCV: mean (red blood cell) corpuscular volume; MOC: monocyte count; NEUT: neutrophil ratio; NEUTC: neutrophil count; PCT: procalcitonin; PCV: packed-cell volume (hematocrit); RBC: red blood cell count; RDW-SD: red blood cell distribution width–standard deviation; UA: uric acid; WBC: white blood cell count.
The importance of different CLIs in classifiers varied greatly, and the importance of the same CLI varied greatly among classifiers constructed by different FCs (see Figure 3). In the HPCs constructed with the 7-CLI combinations, the average feature importance of AFU (26.60%) was the highest, followed by uric acid (25.31%) and PCT (21.06%) (see Figure 3, A). However, in the HPCs constructed with the 8-CLI combinations, the average feature importance of uric acid (22.51%) was the highest, followed by PCT (20.88%) and MCHC (12.36%) (see Figure 3, B). PCT and MCHC were very important to each classifier because they were included, respectively, in 100% (38/38) and 92% (35/38) of the 8-CLI combinations (see Figure 3, B) and in 40% (2/5) and 100% (5/5) of the 7-CLI combinations (see Figure 3, A). Uric acid was also included in all 8-CLI combinations, but its feature importance varied from 11.3% to 41.2% in different classifiers (see Figure 3, B).
Discussion
Principal Findings
The main highlight of this study is that only a few of the common CLIs were required to establish the classifier models to accurately distinguish COVID-19 from CAP. The HPCs could only be constructed by combining several specific CLIs. Among the nearly 2 million FCs with one to eight CLIs, only 43 FCs could be used to construct HPCs with a recall rate greater than 0.9 and an AUROC greater than 0.85 to distinguish COVID-19 from CAP.
Comparison With Prior Work
We have established many COVID-19 versus CAP HPCs with FCs consisting of only CLIs, and almost no similar research results on distinguishing COVID-19 from CAP have been reported. However, many studies have used CLIs to build ML models to help with COVID-19 diagnosis. The prediction performance of these models varied: the accuracy of these models in predicting COVID-19 was between 0.8 and 0.96 [30-32]. In addition, most of the reported ML models for the diagnosis or prediction of COVID-19 have involved more types of variables, such as CT results, clinical symptoms, and CLIs [17,32,33]. Although most of these COVID-19-related ML models were built with more than two ML algorithms, not all models constructed with each algorithm showed high performance. The methods of feature selection that were used in these studies included the recursive feature elimination algorithm [31], causal explanation models [17], and the least absolute shrinkage and selection operator regression [32]. These methods can extract the features that are closely related to the target phenotype, but whether the classifier constructed by the combination of these features has the best performance needs to be determined. The optimized FCs in this study were selected by evaluating the recall rate and AUROC for each FC with one to eight randomly selected CLIs from the differential CLIs between COVID-19 and CAP groups and by constructing classifiers using each FC with the LR classifier algorithm. The FCs that were preliminarily screened were used to build classifiers with RFC and GBC algorithms; finally, only the FCs capable of building the HPC simultaneously with the LR classifier, RFC, and GBC algorithms were selected for the final model construction.
Limitations
As reported earlier, many inflammatory factors, including IL-6 and interleukin-10 (IL-10), are closely related to COVID-19 and have diagnostic value, but neither IL-6 nor IL-10 were detected in the patients of this study. Menni et al [18] reported that loss of smell and taste is a strong predictor for COVID-19. Deviations and omissions may exist in the patients’ self-reported clinical symptoms. Thus, we did not take into account the clinical symptoms when building the classifiers. The possibility that other indicators are more important in constructing COVID-19 versus CAP classifiers was not ruled out. In addition, the sample size included in this study was relatively small, and the classifiers need to be optimized with larger samples before it can be used to distinguish COVID-19 from CAP in practice.
The Rationality of the Research Results
Out of the 43 FCs, 40 contained PCT and MCHC. The feature importance of PCT in each classifier is very high, suggesting that PCT may be a good blood marker to efficiently distinguish COVID-19 from CAP. PCT is one of the markers of lower respiratory tract bacteria and other infections. The US Food and Drug Administration approved the monitoring of the beginning and the entire duration of antibiotic treatment for suspected lower respiratory tract infections based on serum PCT levels [12]. However, the elevation of serum PCT in COVID-19 patients was also reported in many studies [34]. The increase of PCT is a remarkable characteristic of patients with COVID-19 [34]. Increased serum PCT levels in both COVID-19 and CAP patients indicated that the distinction of COVID-19 from CAP could not be made simply on the basis of the increase in PCT. Compared with the normal reference values of the CLIs, the serum levels of most of the CLIs increased or decreased simultaneously in both COVID-19 and CAP patients. Thus, providing references for the diagnosis of COVID-19 or CAP directly in regard to the rise or decrease of the CLIs is difficult. However, we found that the ML classifiers constructed with the FCs with many certain CLIs could distinguish COVID-19 from CAP effectively, suggesting an advantage of ML algorithms in disease classification or diagnosis.
The COVID-19 versus CAP classifiers with the highest performance also involved PCT, MCHC, uric acid, albumin, neutrophil count, monocyte count, basophil count, RBC count, and WBC count, proposing the importance of these CLIs in differentiating COVID-19 from CAP. Few studies have reported the changing trend of MCHC in patients with COVID-19 or CAP, but the results of this study showed that MCHC decreased in both groups and was significantly lower in the CAP group than in the COVID-19 group. The reason for the decrease of MCHC may be closely related to the reduction of iron due to inflammation [35]. The IQRs of uric acid in both COVID-19 and CAP groups were within the normal reference range, but the IQR was significantly higher in the COVID-19 group than in the CAP group. Elevated uric acid is an independent risk factor of renal injury or renal dysfunction; the underlying mechanisms of uric acid elevation are very complicated [36]. The significant difference in uric acid between COVID-19 and CAP may be interpreted as follows: individuals with higher uric acid may be more susceptible to COVID-19 than those with lower uric acid levels. Uric acid exists in all 8-CLI combinations that are capable of constructing high-performance CLIs and has a high feature importance in the classifiers, suggesting that uric acid is another important marker that can distinguish COVID-19 from CAP. Zhou et al reported that albumin significantly decreased in severe and critical COVID-19 patients [37]. Serum albumin level is a good prognostic marker in CAP. A decreased albumin level is closely associated with a higher risk of mortality in patients with CAP [38]. Although albumin decreased remarkably in both COVID-19 and CAP groups, there was still a significant difference between the two groups; the decrease in the CAP group was more obvious than that in the COVID-19 group, which could contribute to the differentiation of COVID-19 from CAP. AFU contributed high feature importance in the HPCs constructed from 7-CLI combinations due to the significant difference in AFU between COVID-19 and CAP. An increase of serum AFU has a certain diagnostic value for primary liver cancer [39]. Thus, the higher AFU in the COVID-19 group than in the CAP group may be explained by the fact that liver injury is more common in COVID-19 than in CAP or that the diversity in AFU levels determines the difference in susceptibility to COVID-19.
Recommendations
Both PCT and AFU contributed high feature importance in the HPCs constructed from the FCs containing PCT or AFU, but the performance of the classifiers constructed from the FCs containing both PCT and AFU decreased remarkably. This result indicated that intrinsic dependence exists among some CLIs that undergo synergistic changes in individuals and can be used to construct HPCs. The internal relationship between CLIs is very complex and difficult to deconstruct. Therefore, the following method may be effective: random selection of different CLIs to construct classifiers with different classification algorithms, followed by the evaluation of the performance of each classifier, and, finally, the discovery of the FCs with certain CLIs that can be used to accurately distinguish COVID-19 from CAP.
Conclusions
The patients suffering from COVID-19 and CAP have their own characteristic profiles of CLIs, and some FCs consisting of seven or eight specific CLIs could build COVID-19 versus CAP HPCs. The usage rate and the feature importance of the CLIs in the HPCs indicated that PCT, MCHC, uric acid, albumin, AGR, neutrophil count, RBC count, monocyte count, and WBC count are the most important indicators that can distinguish COVID-19 from CAP.
Acknowledgments
We are very grateful to Associate Professor Benrong Liu of the Second Affiliated Hospital of Guangzhou Medical University for his guidance in data analysis. We are also very grateful to the doctors and nurses of Gong An County People’s Hospital for their hard work to prevent as well as control COVID-19.
Abbreviations
- AFU
α-L-fucosidase
- AGR
albumin to globulin ratio
- AUROC
area under the receiver operating characteristic curve
- CAP
community-acquired pneumonia
- CLI
clinical laboratory indicator
- COVID19-COM
COVID-19 patient subgroup with mild and common types
- COVID19-SV
COVID-19 patient subgroup with severe and critical types
- CRP
C-reactive protein
- CT
computed tomography
- FC
feature combination
- GBC
gradient boosting classifier
- HPC
high-performance classifier
- IL-6
interleukin-6
- IL-10
interleukin-10
- LDH
lactate dehydrogenase
- LR
logistic regression
- MCHC
mean corpuscular hemoglobin concentration
- mCRP
micro–C-reactive protein
- MCV
mean corpuscular volume
- ML
machine learning
- PCT
procalcitonin
- PCV
packed-cell volume
- PT
prothrombin time
- RBC
red blood cell
- RDW-SD
red blood cell distribution width–standard deviation
- RFC
random forest classifier
- WBC
white blood cell
Appendix
The statistical distribution of the plasma level of the clinical laboratory indicators (CLIs) with a significant difference between COVID19-COM (COVID-19 patient subgroup with mild and common types) and COVID19-SV (COVID-19 patient subgroup with severe and critical types). The statistical distribution was presented with a box and whisker plot. The horizontal lines within the boxes indicate the median value. The vertical lines extending below and above the boxes represent 5%-95% percentile values. The scale on the y-axis represents the 5th, 25th, 50th, 75th, and 95th percentile values of the CLI in the COVID19-COM subgroup. The triangles represent the upper and lower limits of the normal reference range of the laboratory index. The median of the CLI in the COVID19-SV subgroup is also represented in the y-axis. AST: aspartate aminotransferase; CO2CP: carbon dioxide binding capacity; ESR: erythrocyte sedimentation rate; γ-GGT: transglutaminase transpeptidase gamma; FIB: fibrinogen content; LDH: lactate dehydrogenase; LYM: lymphocyte; LYMPH: lymphocyte count; mCRP: micro–C-reactive protein; MYO: myoglobin; NEUT: neutrophil ratio; PA: prealbumin.
The statistical distribution of the plasma level of the clinical laboratory indicators (CLIs) among community-acquired pneumonia (CAP), COVID19-COM (COVID-19 patient subgroup with mild and common types), and COVID19-SV (COVID-19 patient subgroup with severe and critical types). The statistical distribution was presented with a box and whisker plot. The horizontal lines within the boxes indicate the median value. The vertical lines extending below and above the boxes represent 5%-95% percentile values. The scale on the y-axis represents the 5th, 25th, 50th, 75th, and 95th percentile values of the CLI in the CAP group. The triangles represent the upper and lower limits of the normal reference range of the laboratory index. AFU: α-L-fucosidase; EOC: eosinophil count; MCHC: mean corpuscular hemoglobin concentration; MCV: mean (red blood cell) corpuscular volume; MOC: monocyte count; MYO: myoglobin; RDW-SD: red blood cell distribution width–standard deviation; TT: thrombin time; UA: uric acid.
Clinical laboratory indicator (CLI) combinations and the hyper-parameters of the classifiers constructed by different machine learning algorithms from these CLI combinations.
Footnotes
Authors' Contributions: XZH (huangxz020@gzucm.edu.cn) and YX (yujuanxiong@gzucm.edu.cn) share corresponding author duties for this article. XZH and YX made substantial contributions to the study concept and design. YX and PFK were in charge of drafting the manuscript. WD and WH were responsible for obtaining ethical approval, collecting data, and confirming the accuracy of the data. ZZL, QZZ, and YW performed the data analysis and interpretation. All authors agreed on the final version for submission to the journal.
Conflicts of Interest: None declared.
References
- 1.Coronavirus disease (COVID-19) pandemic. World Health Organization. 2020. [2020-05-06]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020 Apr;5(4):536–544. doi: 10.1038/s41564-020-0695-z. http://europepmc.org/abstract/MED/32123347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye F, Xu S, Rong Z, Xu R, Liu X, Deng P, Liu H, Xu X. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. Int J Infect Dis. 2020 May;94:133–138. doi: 10.1016/j.ijid.2020.03.042. https://linkinghub.elsevier.com/retrieve/pii/S1201-9712(20)30174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 Apr 07;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Tian S, Lou J, Chen Y. Familial cluster of COVID-19 infection from an asymptomatic. Crit Care. 2020 Mar 27;24(1):119. doi: 10.1186/s13054-020-2817-7. https://ccforum.biomedcentral.com/articles/10.1186/s13054-020-2817-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen K, Chen R, Tang C, Wang T, Chen P, Xiang J, Li S, Wang J, Liang Z, Peng Y, Wei L, Liu Y, Hu Y, Peng P, Wang J, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zhong N, China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 Apr 30;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020 Mar 26;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. http://europepmc.org/abstract/MED/31995857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, Jung S, Yuan B, Kinoshita R, Nishiura H. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: A statistical analysis of publicly available case data. J Clin Med. 2020 Feb 17;9(2):538. doi: 10.3390/jcm9020538. https://www.mdpi.com/2077-0383/9/2/538/htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terpos E, Ntanasis-Stathopoulos I, Elalamy I, Kastritis E, Sergentanis TN, Politou M, Psaltopoulou T, Gerotziafas G, Dimopoulos MA. Hematological findings and complications of COVID-19. Am J Hematol. 2020 Jul;95(7):834–847. doi: 10.1002/ajh.25829. http://europepmc.org/abstract/MED/32282949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji D, Zhang D, Xu J, Chen Z, Yang T, Zhao P, Chen G, Cheng G, Wang Y, Bi J, Tan L, Lau G, Qin E. Prediction for progression risk in patients with COVID-19 pneumonia: The CALL score. Clin Infect Dis. 2020 Sep 12;71(6):1393–1399. doi: 10.1093/cid/ciaa414. http://europepmc.org/abstract/MED/32271369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Zuo P, Liu Y, Zhang M, Zhao X, Xie S, Zhang H, Chen X, Liu C. Clinical and laboratory predictors of in-hospital mortality in patients with coronavirus disease-2019: A cohort study in Wuhan, China. Clin Infect Dis. 2020 Nov 19;71(16):2079–2088. doi: 10.1093/cid/ciaa538. http://europepmc.org/abstract/MED/32361723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamat IS, Ramachandran V, Eswaran H, Guffey D, Musher DM. Procalcitonin to distinguish viral from bacterial pneumonia: A systematic review and meta-analysis. Clin Infect Dis. 2020 Jan 16;70(3):538–542. doi: 10.1093/cid/ciz545. [DOI] [PubMed] [Google Scholar]
- 13.Robriquet L, Guery B, Fourrier F. Pulmonary coagulopathy as a new target in therapeutic studies of acute lung injury or pneumonia. Crit Care Med. 2006 Sep;34(9):2510; author reply 2510–2511. doi: 10.1097/01.CCM.0000235673.40874.BD. [DOI] [PubMed] [Google Scholar]
- 14.Schultz MJ, Haitsma JJ, Zhang H, Slutsky AS. Pulmonary coagulopathy as a new target in therapeutic studies of acute lung injury or pneumonia--A review. Crit Care Med. 2006 Mar;34(3):871–877. [PubMed] [Google Scholar]
- 15.Ceccato A, Panagiotarakou M, Ranzani OT, Martin-Fernandez M, Almansa-Mora R, Gabarrus A, Bueno L, Cilloniz C, Liapikou A, Ferrer M, Bermejo-Martin JF, Torres A. Lymphocytopenia as a predictor of mortality in patients with ICU-acquired pneumonia. J Clin Med. 2019 Jun 13;8(6):843. doi: 10.3390/jcm8060843. https://www.mdpi.com/2077-0383/8/6/843/htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: Machine learning and the future of medicine. J Intern Med. 2018 Dec;284(6):603–619. doi: 10.1111/joim.12822. doi: 10.1111/joim.12822. [DOI] [PubMed] [Google Scholar]
- 17.Schwab P, DuMont Schütte A, Dietz B, Bauer S. Clinical predictive models for COVID-19: Systematic study. J Med Internet Res. 2020 Oct 06;22(10):e21439. doi: 10.2196/21439. https://www.jmir.org/2020/10/e21439/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menni C, Valdes AM, Freidin MB, Sudre CH, Nguyen LH, Drew DA, Ganesh S, Varsavsky T, Cardoso MJ, El-Sayed Moustafa JS, Visconti A, Hysi P, Bowyer RCE, Mangino M, Falchi M, Wolf J, Ourselin S, Chan AT, Steves CJ, Spector TD. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020 Jul;26(7):1037–1040. doi: 10.1038/s41591-020-0916-2. http://europepmc.org/abstract/MED/32393804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko J, Baldassano SN, Loh P, Kording K, Litt B, Issadore D. Machine learning to detect signatures of disease in liquid biopsies - A user's guide. Lab Chip. 2018 Jan 30;18(3):395–405. doi: 10.1039/c7lc00955k. http://europepmc.org/abstract/MED/29192299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment (7th edition) Beijing, China: China National Health Commission; 2020. Mar 04, [2020-05-11]. http://kjfy.meetingchina.org/msite/news/show/cn/3337.html. [Google Scholar]
- 21.Seabold S, Perktold J. Statsmodels: Econometric and statistical modeling with Python. Proceedings of the 9th Python in Science Conference (SciPy 2010); The 9th Python in Science Conference (SciPy 2010); June 28-July 3, 2010; Austin, TX. 2010. Jul 03, pp. 92–96. https://conference.scipy.org/proceedings/scipy2010/pdfs/seabold.pdf. [DOI] [Google Scholar]
- 22.itertools — Functions creating iterators for efficient looping. The University of Auckland School of Computer Science. [2020-03-08]. https://www.cs.auckland.ac.nz/references/python/3.2.3-docs/library/itertools.html.
- 23.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. https://www.jmlr.org/papers/volume12/pedregosa11a/pedregosa11a.pdf. [Google Scholar]
- 24.Logistic regression, scikit-learn 0.24.1. scikit-learn. [2020-06-12]. https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression.
- 25.Forests of randomized trees, scikit-learn 0.24.1. scikit-learn. [2020-06-12]. https://scikit-learn.org/stable/modules/ensemble.html#forests-of-randomized-trees.
- 26.Breiman L. Random forests. Mach Learn. 2001 Oct;45:5–32. doi: 10.1023/A:1010933404324. https://link.springer.com/content/pdf/10.1023/A:1010933404324.pdf. [DOI] [Google Scholar]
- 27.Gradient tree boosting, scikit-learn 0.24.1. scikit-learn. [2020-06-12]. https://scikit-learn.org/stable/modules/ensemble.html#gradient-boosting.
- 28.Friedman JH. 1999 Reitz Lecture. Greedy function approximation: A gradient boosting machine. Ann Stat. 2001;29(5):1189–1232. https://projecteuclid.org/download/pdf_1/euclid.aos/1013203451. [Google Scholar]
- 29.feature_importances_, scikit-learn 0.24.1. scikit-learn. [2021-02-12]. https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html#sklearn.ensemble.RandomForestClassifier.feature_importances_.
- 30.McDonald SA, Medford RJ, Basit MA, Diercks DB, Courtney DM. Derivation with internal validation of a multivariable predictive model to predict COVID-19 test results in emergency department patients. Acad Emerg Med. 2020 Nov 28;:1–9. doi: 10.1111/acem.14182. https://onlinelibrary.wiley.com/doi/epdf/10.1111/acem.14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao H, Zhang N, Zhang R, Duan M, Xie T, Pan J, Peng E, Huang J, Zhang Y, Xu X, Xu H, Zhou F, Wang G. Severity detection for the coronavirus disease 2019 (COVID-19) patients using a machine learning model based on the blood and urine tests. Front Cell Dev Biol. 2020;8:683. doi: 10.3389/fcell.2020.00683. doi: 10.3389/fcell.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong J, Ou J, Qiu X, Jie Y, Chen Y, Yuan L, Cao J, Tan M, Xu W, Zheng F, Shi Y, Hu B. A tool for early prediction of severe coronavirus disease 2019 (COVID-19): A multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin Infect Dis. 2020 Jul 28;71(15):833–840. doi: 10.1093/cid/ciaa443. http://europepmc.org/abstract/MED/32296824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwab P, DuMont Schütte A, Dietz B, Bauer S. Clinical predictive models for COVID-19: Systematic study. J Med Internet Res. 2020 Oct 06;22(10):e21439. doi: 10.2196/21439. https://www.jmir.org/2020/10/e21439/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z, Cai T, Fan L, Lou K, Hua X, Huang Z, Gao G. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int J Infect Dis. 2020 Jun;95:332–339. doi: 10.1016/j.ijid.2020.04.041. https://linkinghub.elsevier.com/retrieve/pii/S1201-9712(20)30257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y, Hu Z. Lower mean corpuscular hemoglobin concentration is associated with poorer outcomes in intensive care unit admitted patients with acute myocardial infarction. Ann Transl Med. 2016 May;4(10):190. doi: 10.21037/atm.2016.03.42. doi: 10.21037/atm.2016.03.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao X, Wu L, Chen Z. The association between elevated serum uric acid level and an increased risk of renal function decline in a health checkup cohort in China. Int Urol Nephrol. 2018 Mar;50(3):517–525. doi: 10.1007/s11255-017-1732-6. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y, Guo S, He Y, Zuo Q, Liu D, Xiao M, Fan J, Li X. COVID-19 is distinct from SARS-CoV-2-negative community-acquired pneumonia. Front Cell Infect Microbiol. 2020;10:322. doi: 10.3389/fcimb.2020.00322. doi: 10.3389/fcimb.2020.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viasus D, Garcia-Vidal C, Simonetti A, Manresa F, Dorca J, Gudiol F, Carratalà J. Prognostic value of serum albumin levels in hospitalized adults with community-acquired pneumonia. J Infect. 2013 May;66(5):415–423. doi: 10.1016/j.jinf.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Deugnier Y, David V, Brissot P, Mabo P, Delamaire D, Messner M, Bourel M, Legall JY. Serum alpha-L-fucosidase: A new marker for the diagnosis of primary hepatic carcinoma? Hepatology. 1984;4(5):889–892. doi: 10.1002/hep.1840040516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The statistical distribution of the plasma level of the clinical laboratory indicators (CLIs) with a significant difference between COVID19-COM (COVID-19 patient subgroup with mild and common types) and COVID19-SV (COVID-19 patient subgroup with severe and critical types). The statistical distribution was presented with a box and whisker plot. The horizontal lines within the boxes indicate the median value. The vertical lines extending below and above the boxes represent 5%-95% percentile values. The scale on the y-axis represents the 5th, 25th, 50th, 75th, and 95th percentile values of the CLI in the COVID19-COM subgroup. The triangles represent the upper and lower limits of the normal reference range of the laboratory index. The median of the CLI in the COVID19-SV subgroup is also represented in the y-axis. AST: aspartate aminotransferase; CO2CP: carbon dioxide binding capacity; ESR: erythrocyte sedimentation rate; γ-GGT: transglutaminase transpeptidase gamma; FIB: fibrinogen content; LDH: lactate dehydrogenase; LYM: lymphocyte; LYMPH: lymphocyte count; mCRP: micro–C-reactive protein; MYO: myoglobin; NEUT: neutrophil ratio; PA: prealbumin.
The statistical distribution of the plasma level of the clinical laboratory indicators (CLIs) among community-acquired pneumonia (CAP), COVID19-COM (COVID-19 patient subgroup with mild and common types), and COVID19-SV (COVID-19 patient subgroup with severe and critical types). The statistical distribution was presented with a box and whisker plot. The horizontal lines within the boxes indicate the median value. The vertical lines extending below and above the boxes represent 5%-95% percentile values. The scale on the y-axis represents the 5th, 25th, 50th, 75th, and 95th percentile values of the CLI in the CAP group. The triangles represent the upper and lower limits of the normal reference range of the laboratory index. AFU: α-L-fucosidase; EOC: eosinophil count; MCHC: mean corpuscular hemoglobin concentration; MCV: mean (red blood cell) corpuscular volume; MOC: monocyte count; MYO: myoglobin; RDW-SD: red blood cell distribution width–standard deviation; TT: thrombin time; UA: uric acid.
Clinical laboratory indicator (CLI) combinations and the hyper-parameters of the classifiers constructed by different machine learning algorithms from these CLI combinations.


