Abstract
Objective:
Patients with pancreatic cancer (PDAC) who undergo surgical resection and receive effective chemotherapy have the best chance of long-term survival. Unfortunately, we lack predictive biomarkers to guide optimal systemic treatment. Ex-vivo generation of patient-derived organoids (PDO) for pharmacotyping may serve as predictive biomarkers in PDAC. The goal of the current study was to demonstrate the clinical feasibility of a PDO-guided precision medicine framework of care.
Methods:
PDO cultures were established from surgical specimens and endoscopic biopsies, expanded in Matrigel, and used for high-throughput drug testing (pharmacotyping). Efficacy of standard-of-care chemotherapeutics was assessed by measuring cell viability after drug exposure.
Results:
A framework for rapid pharmacotyping of PDOs was established across a multi-institutional consortium of academic medical centers. Specimens obtained remotely and shipped to a central biorepository maintain viability and allowed generation of PDOs with 77% success. Early cultures maintain the clonal heterogeneity seen in PDAC with similar phenotypes (cystic–solid). Late cultures exhibit a dominant clone with a pharmacotyping profile similar to early passages. The biomass required for accurate pharmacotyping can be minimized by leveraging a high-throughput technology. Twenty-nine cultures were pharmacotyped to derive a population distribution of chemotherapeutic sensitivity at our center. Pharmacotyping rapidly-expanded PDOs was completed in a median of 48 (range 18–102) days.
Conclusions:
Rapid development of PDOs from patients undergoing surgery for PDAC is eminently feasible within the perioperative recovery period, enabling the potential for pharmacotyping to guide post-operative adjuvant chemotherapeutic selection. Studies validating PDOs as a promising predictive biomarker are ongoing.
Keywords: Pancreatic cancer, precision medicine, organoids, chemosensitivity, pharmacotyping
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a lethal disease with a limited 5-year overall survival that is driven by nearly universal systemic spread1. Even when amenable to complete surgical resection, systemic disease recurrence is common and the response to chemotherapy is a key driver of overall prognosis2,3. Furthermore, a favorable response to systemic chemotherapy in patients with locally advanced disease (i.e. ‘induction chemotherapy’) may enable surgical utilization along a potentially curative paradigm in selected settings4–6. Two combination chemotherapy regimens are currently viewed as standards for systemic therapy in PDAC, each with an approximately 30% response rate: gemcitabine plus nab-paclitaxel and FOLFIRINOX (5-fluorouracil, folinic acid, irinotecan and oxaliplatin)7–9. Despite these two options as standard of care in PDAC, there are no predictive biomarkers of treatment response available to distinguish patients that may benefit from one or the other. Tractable precision medicine strategies are needed to select an optimal systemic therapy for each patient in order to increase the durability of post-surgical disease-free survival and enable more aggressive surgical utilization in the borderline resectable and locally advanced patient cohort.
There are a number of putative biomarkers for clinical response that have been proposed for PDAC10–12. Most are based upon a gene mutation, gene expression or protein analysis, and are limited by challenges in describing the broader phenotypic behavior resulting from the holistic interplay inherent in cellular processes that drive cancer biology. A system that recapitulates each patient’s tumor as a model organism may enable this broad phenotypic assessment and facilitate rapid ex-vivo personalized drug testing13,14. Options for tumor modeling include two-dimensional cell lines, murine xenografts and three-dimensional patient-derived cultures, commonly termed organoids. Patient-derived organoids (PDOs) are self-organizing, three-dimensional structures that can be cultured from tumor specimens with a high rate of success15. Our previous work demonstrated that PDOs are an efficient model for molecular characterization in PDAC and may aid in the selection of an optimal chemotherapy regimen for each patient16,17.
Several barriers have limited tumor-model based precision medicine approaches. First, the traditional patient-specific models of disease (i.e. 2D cell lines or xenografts) are limited by relatively low rates of success. Second, the time required to generate patient-derived 2D lines and xenografts can often exceed 9–18 months. This is too long for patients to derive benefit in PDAC. In this work, we aim to show the feasibility of PDO-based precision medicine. In order to do this we needed to prove that: (1) PDOs can be established, expanded, and characterized from tissue acquisition protocols that exist within current frameworks of care; (2) the interval between tissue acquisition and ex-vivo pharmacotyping can be expedited to fit within a clinically meaningful time frame; and (3) a population distribution of ex-vivo PDO chemotherapeutic response can be generated to provide data suggesting unique chemotherapeutic sensitivities for each patient. Herein we demonstrate the ability to: (1) generate PDOs from endoscopic biopsies and surgical specimens; (2) complete pharmacotyping prior to start of adjuvant therapies; and (3) derive a population distribution of chemotherapeutic response suggesting unique therapeutic sensitivities for each patient, thus establishing PDO as a potentially important tool in patients undergoing multimodality treatment of localized PDAC.
Methods
Patients and tumor specimen collection
Patients with a presumed or confirmed diagnosis of PDAC were eligible for enrollment onto IRB-approved tissue acquisition protocols at Johns Hopkins Hospital and Massachusetts General Hospital (MGH). For those undergoing surgery, portions of the tumor were harvested by a research pathologist following resection. To evaluate the capacity to establish PDOs from core needle specimens, tissue was obtained during diagnostic endoscopic ultrasound-directed biopsy as part of a multi-institutional clinical trial (NCT03563248). Tissues obtained off-site were shipped on ice overnight via commercial mail in transfer media.
Organoid establishment and maintenance culture
Biopsied tissue was dissociated to single cells with mechanical digestion in human wash media. Surgical tumor pieces were minced with a scalpel and digestion was carried out using a combination of mechanical dissociation and enzymatic digestion. Single cells were suspended in liquid Matrigel, plated in domes and covered with human complete feeding media. Cultures were incubated at 37°C with 21% of O2 and 5% of CO2 and passaged for cell line expansion or characterization biweekly. Complete protocol details are in keeping with prior work performed by our group16.
Clinically relevant characterization: Genomics and gene expression analysis
KRAS mutational status of PDOs were determined using next-generation sequencing (NGS) in methods similar to prior work18. In short, PDO DNA was isolated using the Tissue Preparation System (Siemens Healthineers, Erlangen, Germany) with Versant Tissue Preparation reagents (Siemens Healthineers). NGS was performed on the Ion S5-XL System (Life Technologies, Carlsbad, CA, USA). KRAS mutation calling was performed with the Ion Torrent Variant Caller Plugin (v5.2.1.39) for Torrent Suite software (v5.2). The limit of detection was set at 2%. Fishplot of genomic evolution was visualized using R studio (version 1.2.5033).
RNA extraction was carried out with the AllPrep DNA/RNA/Protein Mini kit (80004, Qiagen, Hilden, Germany). Amplified cDNA was generated using the SMART-Seq® HT Kit (634437, Takara Bio USA, Mountain View, CA, US). PCR product was purified with a 1.8X Agencourt AMPure XP bead cleanup (Beckman Coulter), validated and quantified using the KAPA Library Quantification Kit for Illumina® Platforms (KK4824, Roche, Wilmington, MA) and pooled after tagging at equal concentrations as determined using the KAPA Library Quantification Kit for Illumina® Platforms. Sequencing was performed on a NextSeq 500 system (Illumina) using a high output flow cell and 2 × 75 paired end reads. Reads were aligned to the hg38 genome using the STAR version 2.4.0h aligner (Alex Dobin, https://github.com/alexdobin/STAR) and counted using htseq-count in the intersection-strict mode against the Homo_sapiens.GRCh38.79.gtf annotation table from Ensembl (Hinton, Cambridge, United Kingdom, www.ensembl.org). After normalization, Collisson19 and Moffitt20 classifier gene expression was characterized and illustrated by heat maps.
Clinically relevant characterization: Pharmacotyping
Organoids were dissociated into single cells and plated on a 384-well assay plate in 10% Matrigel. After a 48-hour recovery period, chemotherapeutics were administered using a semi-automated D300e dispenser (Tecan, Männedorf, Switzerland), normalized to 0.5% DMSO. Chemotherapeutics were tested across a logarithmically designed curve: gemcitabine, paclitaxel and irinotecan (range: 8.0 × 10−12 mol/L to 2.0 × 10−6 mol/L), and 5-FU and oxaliplatin (range: 1.0 × 10−8 to 1.0 × 10−4 mol/L). Negative controls included wells with DMSO normalization alone. Cell viability was assessed at 5 days using CellTiter Glo (Promega Corp, Madison, WI, USA)21. Viability curves were fitted using nonlinear logistic regression with Prism v8 (GraphPad Software, La Jolla, CA, US). Normalized area under curve (AUC) was obtained by dividing the AUC value by the maximum area for each concentration range. A population distribution was illustrated by collective analysis of each chemotherapeutic using a violin plot (R studio version 1.2.5033). Pharmacotyping was performed in four biologic replicates, and at least two technical replicates with most performed at early and late passage.
Results
PDOs can be established, expanded and characterized from tissue acquisition protocols that exist within current frameworks of care
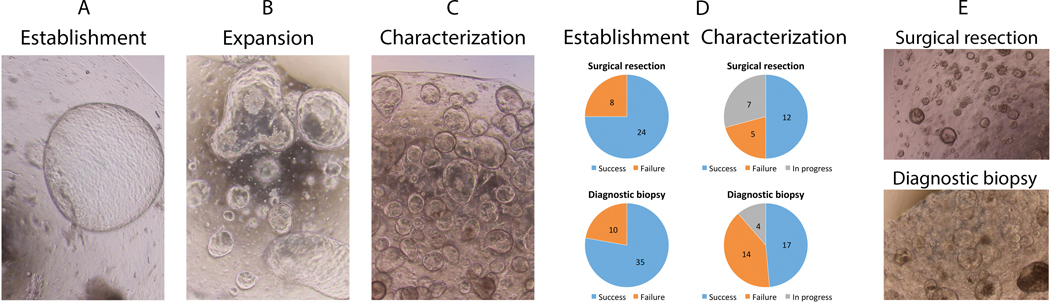
We propose a classification schema for PDO culture maturation in three clinically relevant phases: establishment, expansion, and characterization (Figure 1A–C). The establishment phase describes the emergence of viable ductal organoid structures in the setting of ductal cell selection. Throughout the establishment phase, the culture may contain biomass derived from either normal ducts, PDAC, or both. Expansion is characterized by increasing biomass accumulation, or ‘growth’, in a relatively pure ductal cell culture. This phase varies in duration as each PDO culture has a unique growth rate based on poorly understood growth regulatory mechanisms. The characterization phase is a descriptor used to infer that a critical biomass has been reached allowing for molecular characterization or phenotypic analysis without threatening the continued passage and expansion of the PDO line.
Figure 1.
Phases of organoid culture include (A) establishment of the culture, (B) expansion of the biomass and (C) reaching a critical biomass needed for molecular characterization and stabilization. (D) Patient-derived pancreatic cancer organoids can be established from surgical resection specimens and endoscopic ultrasound-guided diagnostic biopsy cores with high success rate. (E) The late cultures present a characteristic growth pattern that remains stable. The appearance of organoid culture does differ between the sources of original tumor tissue.
In total, patient-derived tumor tissues were received for 76 patients (77 specimens; one patient had both biopsy and surgery). Thirty-two surgical specimens were received, three were received fresh from endoscopic biopsies performed within our institution, and 42 were obtained from our multi-institutional collaboration. Successful establishment occurred in 77% (59/77, Figure 1D) as determined by emergence of organoid structures growing within 1–3 passages of initial plating. Establishment was similar for tissues derived from biopsy (35/45, 78%) and for samples from resection (24/32, 75%; p=n.s.).
The expansion phase appeared to be the most variable amongst our PDO set. Both the phenotype and speed at which lines expand are likely a reflection of the underlying biology of the line and goals for PDO characterization. For example, if aliquots of early passages are committed to downstream molecular analysis, the biomass available for expansion is reduced. Overall, cultures fail to transition from expansion phase to characterization one third of the time (19/59, 32%; Figure 1D). The reasons for failure to expand included 1) “contamination” of the culture by growth of biomass derived from the normal ductal epithelium, 2) suspected microbial contamination and removal from our sterile living biobank, and 3) unclear loss of biomass likely due to disease biology and dependence (or lack thereof) upon growth factors in our PDAC-specific media. At the time of this analysis, 19% remain viable in the expansion phase with plans for characterization. Molecular characterization was initiated for half of the cultures (29/59; 49%) that were successfully established. The phenotypes vary from cystic to solid in nature with the capacity to preserve heterogeneity in phenotype in most cultures, and do not differ between biopsy- or surgery-derived organoids (Figure 1E).
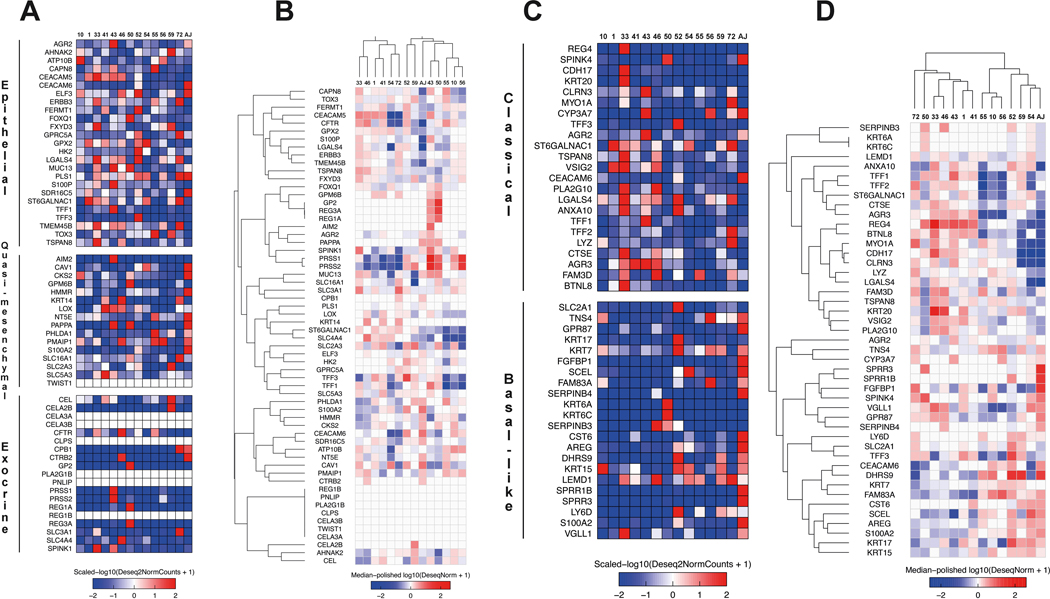
Organoids enable molecular classification through gene expression analysis
As a proof of principle, fourteen PDOs underwent RNA sequencing and supervised gene expression clustering for subtype classification (Figure 2A–D). Similar to work done by others, clustering our ex-vivo lines based upon the strategy devised by Collison et al. 19 revealed the epithelial and quasi-mesenchymal (QM) subtypes (Figure 2A). Though isolated expression was found in some exocrine-like genes, no lines clustered predominantly into this subtype. These data support the notion that PDAC subtyping methods resulting in the original definition of an exocrine-like subtype may have been influenced by normal tissue contamination in samples with low cellularity22.
Figure 2.
PDOs can be utilized to assess PDAC subtype using established methods based upon RNA sequencing. (A) Heat map describing RNA gene expressions used to derive Collison et al. PDAC classification schema19,31. PDO listed above with gene and Collison subtype detailed along the left. (B) Supervised gene expression clustering schema from Collison et al. (C) Heat map detailing gene expression used to derive PDAC subtype according to the Moffitt classification20. (D) Supervised gene expression clustering according to the Moffitt classification.
Given the similarities in supervised clustering done by Collison et al and Moffitt et al., most epithelial lines resembled the classical PDAC subtype using the Moffitt approach (Figure 2C)20. The PDO clustering most strongly with QM, as expected, demonstrated a gene expression profile consistent with the basal-like subtype by Moffitt.
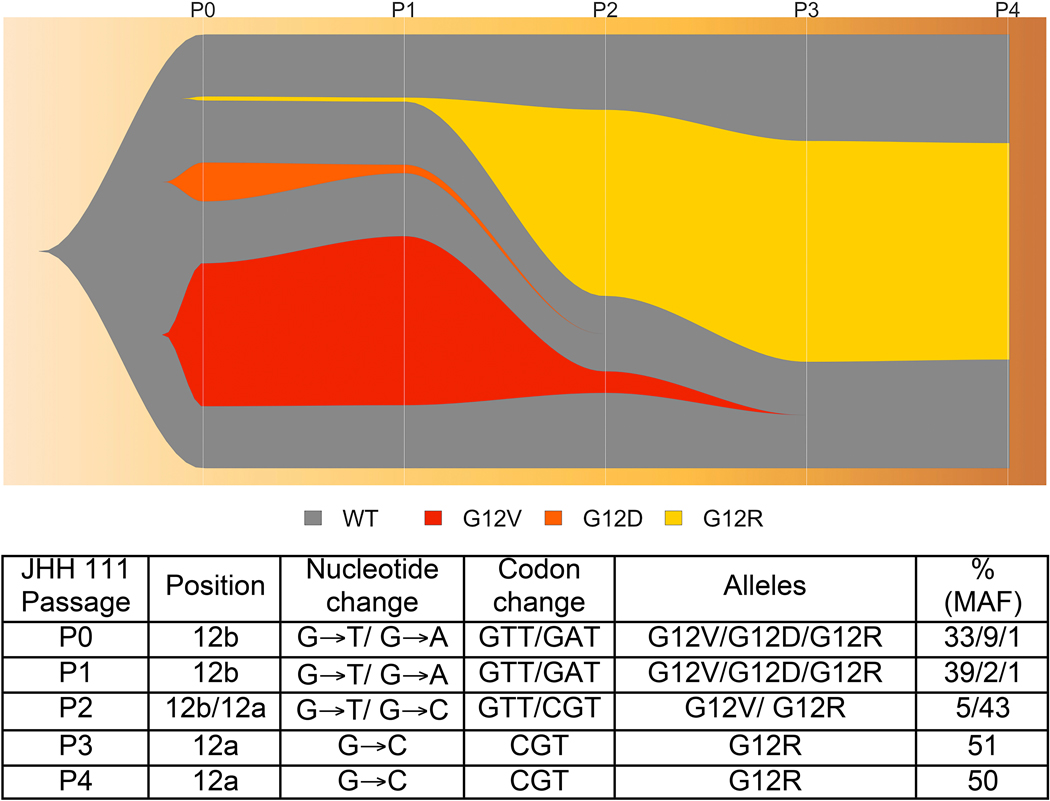
Defining tumor-associated mutation burden and clonality
Targeted sequencing was performed in a laboratory certified for clinical use according to the Clinical Laboratory Improvement Amendments (CLIA) by the Centers for Medicare and Medicaid Services. Confirmation of malignancy and assessment of clonality was undertaken via longitudinal analysis of mutational profile during early organoid culture. The nearly ubiquitous KRAS mutation confirms the organoid culture’s biomass derived from a pancreatic malignancy (Figure 3). While the initial processing of a tumor’s tissue may result in ‘normal’ (i.e. non-cancerous) cell contamination, the biomass present in a successful culture is derived principally from malignant tissue. Mutation profiling and mutant allele frequency (MAF) analysis can be used to assess the capacity to preserve source-tissue heterogeneity. In organoid JHH 111, for example, longitudinal assessment of MAF based upon KRAS from passages 1 to 5 suggested clonal heterogeneity of the original tumor can be maintained in early culture. In the first passage of the PDO culture, variants G12V and G12D and G12R were present with MAF of 33%, 9% and 1%, respectively, in addition to wild type allele. By passage three, variant G12R appeared with 43% allele frequency, and became the dominant clone of the culture by passage 4 (Figure 3). With unbiased collective culture of this line, a dominant clone matures (G12R) in the culture over time. This clone was the only one identified by CLIA-certified somatic testing of bulk primary tumor.
Figure 3.
Patient-derived organoid JHH111 clonal heterogeneity illustrated in fishplot. Notably, the KRAS variant G12R becomes a dominant clone in the culture and is the only clone noted on CLIA testing of the primary tumor.
The interval between tissue acquisition and ex-vivo pharmacotyping can be expedited to fit within a clinically meaningful time-frame
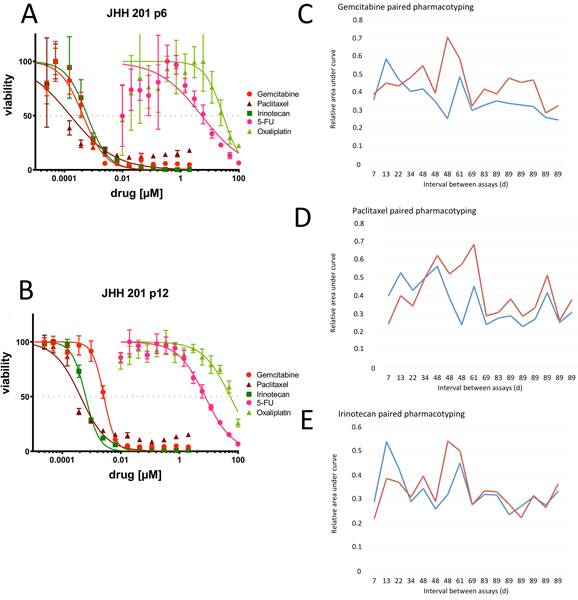
Following our initial efforts to confirm the tumor-derived nature of these organoids (KRAS testing) and the capacity to derive gene-expression based subtype (RNA-seq), we next sought to assess the capacity to perform direct drug-testing (i.e., pharmacotyping) in a clinically relevant time frame. We have previously demonstrated the capacity to pharmacotype organoid lines which are well established and late in passage.16 In this work, we first assessed the stability of pharmacotyping profiles, or drug sensitivity, in early passage PDOs (Figure 4A) as compared to late passage organoids derived from the same tumor (Figure 4B). With clonality established within the first set of passages, we found the dominant clone of a PDO line’s early passage to have a pharmacotyping profile that is similar to the later, more established PDO. This pattern was validated in over a dozen samples with 7 to 89 days between passages (Figures 4C–E). In some, AUC increased marginally at later passage, most noticeably when exposed to gemcitabine. The mean AUC difference between technical replicates at early and late passage was 0.083 (standard deviation ±0.12, Figure 4) in gemcitabine and 0.054 ±0.12 in paclitaxel testing. Irinotecan demonstrated the least variance between early and late testing with a mean difference of 0.009 ±0.076. Work is continuing to investigate the influence of potential batch effects and clonality on these findings.
Figure 4.
Pharmacotyping data from early passage PDOs present similar profiles as from late passage PDOs. (A) PDO 201 pharmacotyping data at passage 6 (p6) with drug response curves generated for the five standard of care agents used in the clinical treatment of PDAC. (B) PDO 201 pharmacotyping data at passage 12 (p12) against the same agents demonstrates similar drug response curves. PDO sensitivity to exposure to (C) gemcitabine, (D) paclitaxel and (E) irinotecan in paired PDO cultures with interval between early (blue) and late (red) pharmacotyping dates, measured in days, marked along the x-axis. Chemotherapeutic sensitivity is represented as an area under the curve (% or 0–1).
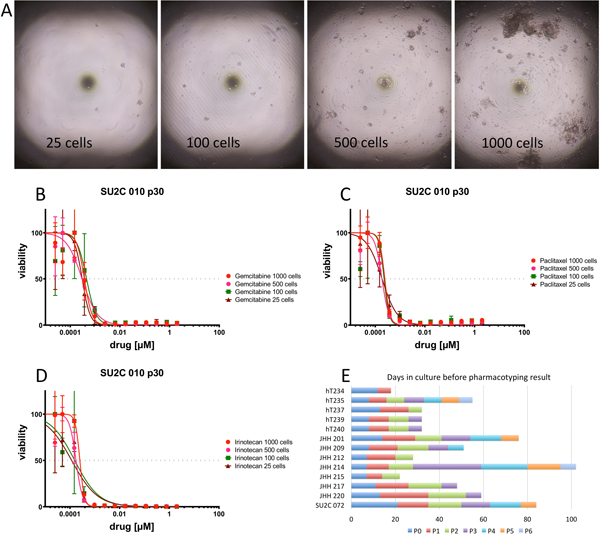
We then assessed the minimum amount of biomass required to carry out pharmacotyping in a high-throughput fashion. With an eye towards liquid handling robotics and drug printer technology we selected a 384-well format and varied biomass from 25 to 1000 cells per well (Figure 5A). Pharmacotyping assessment demonstrated similar drug-sensitivity across all conditions (Figure 5B–D).
Figure 5.
Minimizing biomass needed for pharmacotyping may enable rapid drug testing in a clinical setting. (A) Photomicrograph of high-throughput culture plates containing 25, 100, 500 and 1000 cells per well. Cell survival upon exposure to (B) gemcitabine, (C) paclitaxel and (D) irinotecan with 25, 100, 500 and 1000 cells per well. (E) The interval between clinical specimen acquisition and pharmacotyping can be minimized by leveraging high-throughput technology and minimizing the biomass required for pharmacotyping. Time to pharmacotyping measured in days along x-axis for each PDO. Passage duration noted along bar graph by colors.
Next, we tested the capacity to establish a pharmacotyping profile within a clinically relevant time frame. In a surgery-first approach to disease management, a common clinical goal is to begin adjuvant therapy within 8–12 weeks of the operative date. Thus, we defined a clinically relevant time frame as within 90 days of tumor harvest. We proceeded with a series of patient-derived samples that were processed and expanded with the primary goal of rapid pharmacotyping. Biomass expansion during passage was found to be highly variable between specimens. We suspect this is due to underlying molecular determinants yet to be elucidated. When focused on rapid pharmacotyping, drug testing can usually be performed within the first 5 passages. In our hands, pharmacotyping was possible to perform in as few as 18 days (Figure 5E). The mean time to pharmacotyping was 49 days (range 18–102 d, median 48d). A survey of our pancreatectomy database revealed the median time between surgery and initiation of chemotherapy was 62 days during the same period.
A population distribution of ex-vivo PDO chemotherapeutic response can be generated to provide data suggesting unique chemotherapeutic sensitivities for each patient.
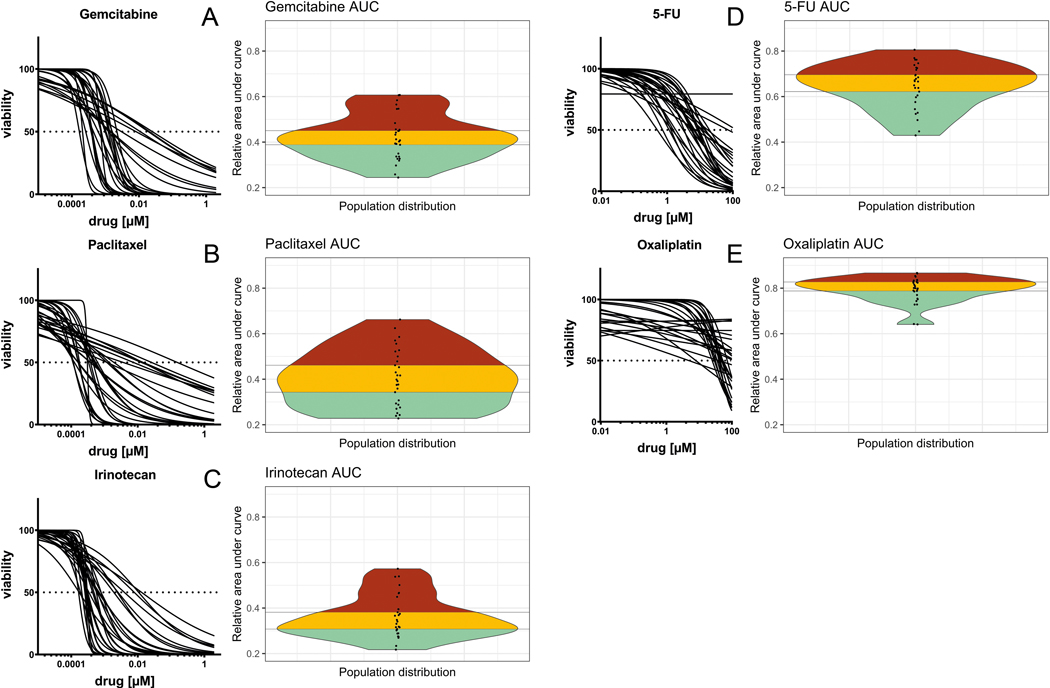
Dose-viability curves for gemcitabine, paclitaxel, irinotecan, 5-FU and oxaliplatin showed substantial inter-patient heterogeneity in chemosensitivity. Normalized AUC describing the relative sensitivity of the patient-derived organoid cultures showed a wide population distribution for all single agents tested (Figure 6A–E). Integrating data from previously conducted clinical trials suggesting chemotherapeutic sensitivity in approximately one-third of patients (and overt resistance in one-third), we clustered the population distribution into thirds to evaluate the capacity to discriminate sensitivity from resistance. Putative gemcitabine sensitivity was reflected by an AUC value of 0.32 ±0.042 (mean ±SD). Putative gemcitabine resistance, in contrast, was defined by an AUC value of 0.54 ±0.060. Similar values can be derived for each chemotherapy: paclitaxel (sensitivity 0.27 ±0.035, resistance 0.56 ±0.059), irinotecan (sensitivity 0.28 ±0.030; resistance 0.48 ±0.063), 5-FU (sensitivity 0.53 ±0.065 ; resistance 0.75 ±0.028), and oxaliplatin (sensitivity 0.73 ±0.052; resistance 0.85 ±0.014). The therapies with the greatest discriminatory capacity using an ex-vivo PDO-guided approach appeared to be gemcitabine, paclitaxel, irinotecan and 5-FU. The oxaliplatin population distribution, in contrast, appeared to have only two outliers amongst a set of otherwise homogenous responses.
Figure 6.
(A-E) Inter-patient variation of chemotherapy response can be captured in the population distribution of PDO cultures. The commonly used chemotherapeutic agents (gemcitabine, paclitaxel, irinotecan, 5-fluorouracil and oxaliplatin) show a spectrum of high (red), intermediate (yellow) and low (cyan) resistance.
A correlation of a gene expression–based subtype to chemotherapeutic sensitivity was seen in our limited subset. As an example, the most striking example of a basal-like PDO was among the most sensitive to gemcitabine, paclitaxel, irinotecan and 5-FU. Enhanced chemotherapeutic sensitivity in basal subtype tumors, particularly to gemcitabine-based regimens, was hypothesized in prior work focused on gene-expression based therapeutic decision making.
Discussion
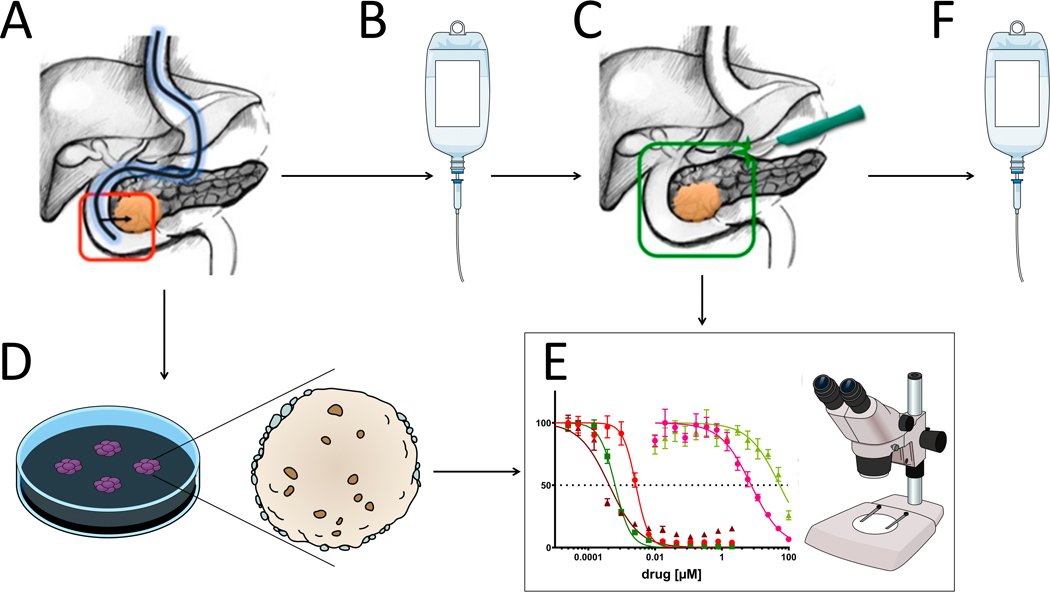
PDO pharmacotyping has potential for future use as a means of personalized therapeutic selection for PDAC patients. In this work, we show the feasibility of PDO-based precision medicine by demonstrating that: (1) PDOs can be established, expanded, and characterized from existing tissue acquisition protocols within current frameworks of care; (2) the interval between tissue acquisition and ex-vivo pharmacotyping can be expedited to fit within a clinically meaningful time frame; and (3) a population distribution of unique ex-vivo PDO chemotherapeutic response discriminate subsets of sensitivity and resistance. Patient-derived tissues, from resection or biopsy, can be utilized to rapidly establish an ex-vivo personalized model of PDAC. Prospective validation of this approach is underway as an integrated translational research component of a multi-center randomized clinical trial (NCT03563248, Figure 7).
Figure 7.
Rationale for precision medicine using patient-derived organoid expansion to characterize tumor.
(A) Pre-treatment core needle biopsies are acquired during a diagnostic endoscopic ultrasound. (B) Patient receives neoadjuvant or induction chemotherapy before (C) surgical resection of the primary tumor. (D) Biopsy-derived tumor cells can be grown into an organoid with a high success rate. A new PDO can be similarly established from the surgical specimen to recapitulate possible changed signature in resistance. (E) After neoadjuvant treatment, clinical response to therapeutic regimen can be compared to chemosensitivity profile of the PDO, and to (F) inform adjuvant chemotherapy selection based on the results of pharmacotyping of the patient-specific culture.
In a subset of PDOs selected for rapid pharmacotyping, the median time required to produce drug response data was shorter than the corresponding time to initiation of chemotherapy in the clinic. The time to pharmacotyping depends upon the biomass diverted from culture to acquire early passage mutation or gene expression data. The time interval can also be shortened if the desire to maintain a line as an immortalized research resource is not a primary aim. When these data are evaluated in aggregate, rapid pharmacotyping may be a clinically tractable strategy for precision medicine in PDAC.
Pharmacotyping our living PDO biobank results in the generation of a population distribution of chemotherapeutic efficacy that appears similar to that seen in our patients and suggests that this ex-vivo distribution may recapitulate the clinical response. In prior work involving 66 patients, 9 were identified in retrospective clinical follow-up16. Five of 9 patients were treated with chemotherapeutics to which their PDO showed unique sensitivity. These patients had better than average progression-free survival. Two of three patients that progressed rapidly had a PDO demonstrating resistance to the therapies selected in their clinical care. These retrospective data are in keeping with other work in gastrointestinal cancers demonstrating correlation between PDO pharmacotype and clinical response25–27.
Though we and others have previously demonstrated that PDOs are able to recapitulate the somatic mutation profile of the primary tumor, the capacity to preserve intratumoral heterogeneity in early passage has been debated27,28. In the current study, we demonstrate that intratumoral heterogeneity is represented in early passage cultures. A dominant clone does emerge in culture. The capacity to propagate with heterogeneity, and the speed at which a dominant clone emerges, is likely dependent upon the starting tissues and the underlying tumor biology. Similar to prior work, we did not encounter clonal drift in longer term cultures (passages 3–5, months 2–5)29,30. These data also suggest an intriguing clinical use for PDOs. As this technique enriches the ductal component, a somatic mutational analysis can be used to detect lower-frequency and targetable mutations that cannot be detected in the diagnostic gene panels from bulk primary tumor due to low cellularity and excess stromal ‘contamination’. On the other hand, in developing an immortalized culture derived from the ductal epithelium alone, the TME and immune infiltrate fail to propagate into later passages. Therefore, when gene-expression subtyping in later passages is performed, the data would not be expected to capture the proposed PDAC subtypes whose methods are dependent upon bulk RNA analysis of whole tumor. This includes the ability to capture the immunogenic subtype originally proposed by Bailey et al.23, and the ability to determine stromal classification24.
There are several limitations to this work. First, this experience highlights several opportunities for improvement in methodology for future consideration. Pitfalls were common during establishment and resulted in the identification of several opportunities for improvement in both logistic and technical domains. Logistic challenges for multi-site collaboration included eliminating variation in collection methodology, streamlining processes to ensure efficient transfer of specimens to commercial shipping services, maintaining viability and sterility through the shipping process, and timely specimen receipt at our centralized laboratory. Technical opportunities for improvement resulted in a reduced reliance upon enzymatic digestion for core needle biopsy specimens. We are also cautious at this juncture to propose current clinical utilization of PDO pharmacotyping. We endorse the position that ethical use requires prospective validation in PDAC. Additionally, using PDOs in clinical decision making will be subject to regulatory requirements and clinical laboratory certification in most jurisdictions. There are several agents in the healthcare environment who have begun to work in this direction in anticipation of prospective validation of PDOs as predictive biomarkers of clinical response. Finally, the pace of organoid growth and pharmacotyping could be further accelerated for routine use in the neoadjuvant or metastatic setting.
A clinical framework, in which PDOs are efficiently generated from surgical specimens or endoscopic biopsies and characterized by pharmacotyping, can be established at academic medical centers with integrated research facilities. Our work demonstrates that this approach is a tractable strategy to facilitate precision medicine in the treatment of pancreatic cancer. Prospective studies to assess correlation between clinical response to chemotherapeutics and PDO pharmacotyping are ongoing.
Acknowledgements
Research is supported by a Stand Up To Cancer – Lustgarten Foundation Pancreatic Cancer Interception Translational Cancer Research Grant (RAB; Grant Number: SU2C-AACR-DT26–17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. The work was also supported by a fellowship grant from Sigrid Juselius Foundation, Helsinki, Finland (TS). We would like to acknowledge our gratitude for technical support in performing these experiments for Reecha Suri and Elisabetta Sereni.
TTS was supported by fellowship grants from Sigrid Juselius Foundation and Instrumentarium Science Foundation. JWZ is supported by 5T32-CA009071. RAB is supported by the National Institutes of Health/National Cancer Institute (K08CA248710) and a Stand Up To Cancer–Lustgarten Foundation Pancreatic Cancer Interception Translational Cancer Research Grant (grant number SU2C-AACR-DT26–17). Stand Up To Cancer (SU2C) is a division of the Entertainment Industry Foundation and funding is administered by the American Association for Cancer Research, the scientific partner of SU2C.
References
- 1.Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2016. National Cancer Institute; Bethesda, MD: Available from: https://seer.cancer.gov/csr/1975_2016/. 2019. [Google Scholar]
- 2.Groot VP, Gemenetzis G, Blair AB, et al. Implications of the Pattern of Disease Recurrence on Survival Following Pancreatectomy for Pancreatic Ductal Adenocarcinoma. Oncol. 2018;25:2475–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groot VP, Blair AB, Gemenetzis G, et al. Recurrence after neoadjuvant therapy and resection of borderline resectable and locally advanced pancreatic cancer. Eur J Surg Oncol. Epub ahead of print 2019. DOI: 10.1016/j.ejso.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 4.He J, Blair AB, Groot VP, et al. Is a Pathological Complete Response Following Neoadjuvant Chemoradiation Associated with Prolonged Survival in Patients with Pancreatic Cancer? Ann Surg. 2018;268:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gemenetzis G, Groot VP, Blair AB, et al. Survival in Locally Advanced Pancreatic Cancer after Neoadjuvant Therapy and Surgical Resection. Ann Surg. 2019;270:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy JE, Wo JY, Ryan DP, et al. Total Neoadjuvant Therapy with FOLFIRINOX in Combination with Losartan Followed by Chemoradiotherapy for Locally Advanced Pancreatic Cancer: A Phase 2 Clinical Trial. JAMA Oncol. 2019;5:1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neoptolemos JP, Palmer DH, Ghaneh P, et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet. 2017;389:1011–1024. [DOI] [PubMed] [Google Scholar]
- 9.Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–25. [DOI] [PubMed] [Google Scholar]
- 10.Aung KL, Fischer SE, Denroche RE, et al. Personalized Medicine and Imaging Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin Cancer Res;24 . Epub ahead of print 2018. DOI: 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter JM, Yeo CJ, Brody JR. Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J Surg Oncol. 2013;107:15–22. [DOI] [PubMed] [Google Scholar]
- 12.Richards NG, Rittenhouse DW, Freydin B, et al. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Ann Surg. 2010;252:499–505. [DOI] [PubMed] [Google Scholar]
- 13.Knudsen ES, Balaji U, Mannakee B, et al. Pancreatic cancer cell lines as patient-derived avatars: Genetic characterisation and functional utility. Gut. 2018;67:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SY, Lee JY, Kim DH, et al. Patient-Derived Cells to Guide Targeted Therapy for Advanced Lung Adenocarcinoma. Sci Rep. 2019;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. [DOI] [PubMed] [Google Scholar]
- 16.Tiriac H, Belleau P, Engle DD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponz-Sarvise M, Corbo V, Tiriac H, et al. Identification of resistance pathways specific to malignancy using organoid models of pancreatic cancer. Clin Cancer Res. 2019;25:6742–6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groot VP, Mosier S, Javed AA, et al. Circulating tumor DNA as a clinical test in resected pancreatic cancer. Clin Cancer Res. 2019;25:4973–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. . Epub ahead of print 2011. DOI: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burkhart RA, Baker LA, Tiriac H. Testing susceptibility of patient-derived organoid cultures to therapies: Pharmacotyping In: Methods in Molecular Biology. Humana Press Inc.; 2018:253–261. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. Electronic address: andrew_aguirre@dfci.harvard.edu , Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 24.Puleo F, Nicolle R, Blum Y, et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology. 2018;155:1999–2013.e3. [DOI] [PubMed] [Google Scholar]
- 25.Ganesh K, Wu C, O’Rourke KP, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med. 2019;25:1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vlachogiannis G, Hedayat S, Vatsiou A, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science (80- ). 2018;359:920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Driehuis E, Van Hoeck A, Moore K, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A. 2019;116:26580–26590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weeber F, Van De Wetering M, Hoogstraat M, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112:13308–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii M, Shimokawa M, Date S, et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell. 2016;18:827–838. [DOI] [PubMed] [Google Scholar]
- 30.Kopper O, de Witte CJ, Lõhmussaar K, et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019;25:838–849. [DOI] [PubMed] [Google Scholar]
- 31.Collisson EA, Bailey P, Chang DK, et al. Molecular subtypes of pancreatic cancer. Nature Reviews Gastroenterology and Hepatology. 2019;16:207–220. [DOI] [PubMed] [Google Scholar]