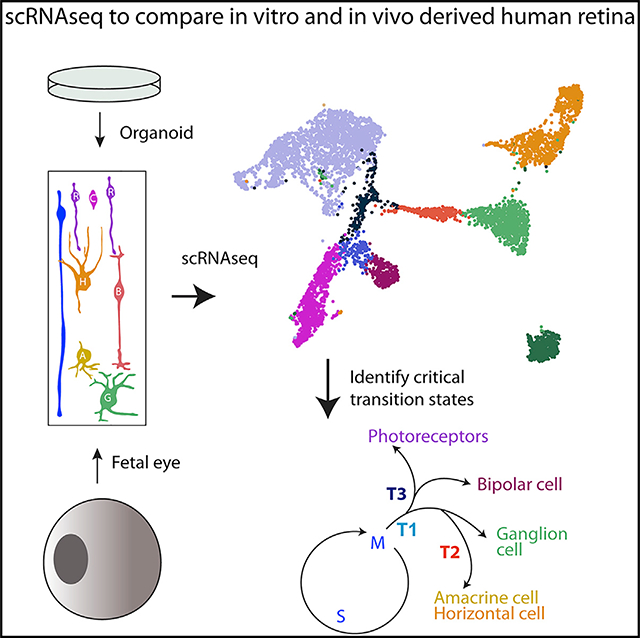
SUMMARY
To study the development of the human retina, we use single-cell RNA sequencing (RNA-seq) at key fetal stages and follow the development of the major cell types as well as populations of transitional cells. We also analyze stem cell (hPSC)-derived retinal organoids; although organoids have a very similar cellular composition at equivalent ages as the fetal retina, there are some differences in gene expression of particular cell types. Moreover, the inner retinal lamination is disrupted at more advanced stages of organoids compared with fetal retina. To determine whether the disorganization in the inner retina is due to the culture conditions, we analyze retinal development in fetal retina maintained under similar conditions. These retinospheres develop for at least 6 months, displaying better inner retinal lamination than retinal organoids. Our single-cell RNA sequencing (scRNA-seq) comparisons of fetal retina, retinal organoids, and retinospheres provide a resource for developing better in vitro models for retinal disease.
In Brief
To determine how closely iPSC-derived retinal organoids model development, Sridhar et al. use scRNA-seq to compare organoids with fetal retina. Despite some defects in inner retinal lamination, organoids closely mimic fetal development in timing and cell composition. Our data provide a resource for improving organoid models for retinal disease.
Graphical Abstract
INTRODUCTION
The vertebrate retina is one of the most well-characterized regions of the central nervous system (Field and Chichilnisky, 2007; Hoon et al., 2014). Similarly, many of the basic mechanisms underlying the generation and differentiation of neurons and glia in the retina of model organisms are known (Kolb et al., 2001; Livesey and Cepko, 2001; Brzezinski and Reh, 2015); however, much less is known about the human fetal retina and its development (for a review, see Hendrickson et al, 2012). Over the last several years, we (and others) have provided a comprehensive molecular atlas of the human fetal retina using RNA sequencing (RNA-seq) (Hoshino et al., 2017; Mellough et al., 2019; Whitmore et al., 2014; Hu et al., 2019; Quinn et al., 2019; Voigt et al., 2019; Kaya et al., 2019). This analysis demonstrated unique characteristics of human retina across major epochs of fetal development; however, assignment of the changes in gene expression to specific cell types was not possible without immunolabeling or in situ hybridization. With the advent of single-cell RNA sequencing (scRNA-seq), genes can now be assigned to specific cells directly (Hu et al., 2019; Quinn et al., 2019; Voigt et al., 2019; Collin et al., 2019; Kim et al., 2019).
In this report, we used scRNA-seq to assess the development of the human retina. At the sampled ages, we identified clusters of progenitor cells and differentiated neurons. Using recently developed computational tools, we focused on three postmitotic transitional cell populations. These transitional cell populations express previously described, developmentally important cell fate determination genes (e.g., ATOH7 and PTF1A) (Fujitani et al., 2006; Jusuf and Harris, 2009; Mu et al., 2005; Yang et al., 2003; Brown et al., 1998); the scRNA-seq data allowed us to find additional co-expressed genes in these clusters that can serve as markers that may be important for these transitional states.
We also assessed how well human pluripotent stem cell (hPSC)-derived retinal organoids compare with fetal retina. hPSC-derived retinal organoids have emerged as potential models for retinal degeneration (Parfitt et al., 2016; Capowski et al., 2014; Phillips et al., 2014; Deng et al., 2018; Gonzalez-Cordero et al., 2017; Saengwimol et al., 2018; Sharma et al., 2017). Organoids are particularly successful in generating photoreceptors; however, inner retinal development is less well characterized (Völkner et al., 2016; Phillips et al., 2012; Zhong et al., 2014; Meyer et al., 2011; Nakano et al., 2012; Wahlin et al., 2017; Fligor et al., 2018; Reichman et al., 2017; Capowski et al., 2019). Our analysis shows that organoids faithfully recapitulate the timing of retinogenesis of the fetal retina, with cellular composition and gene expression comparable across a wide range of developmental stages, despite the relative disorganization of the inner retina.
To determine whether the defects in inner retinal lamination in organoids are due to the tissue culture conditions, we developed a method for fetal retinal culture that mimics aspects of current organoid protocols. Small pieces of fetal retina cultured similarly as retinal organoids form spheres and display more appropriate inner retinal lamination and synaptogenesis than retinal organoids. We postulate that further comparisons of these two related tissue systems will lead to improvements in organoid technology.
RESULTS
scRNA-Seq of Fetal Human Retina Highlights Postmitotic Transitional Cell Populations
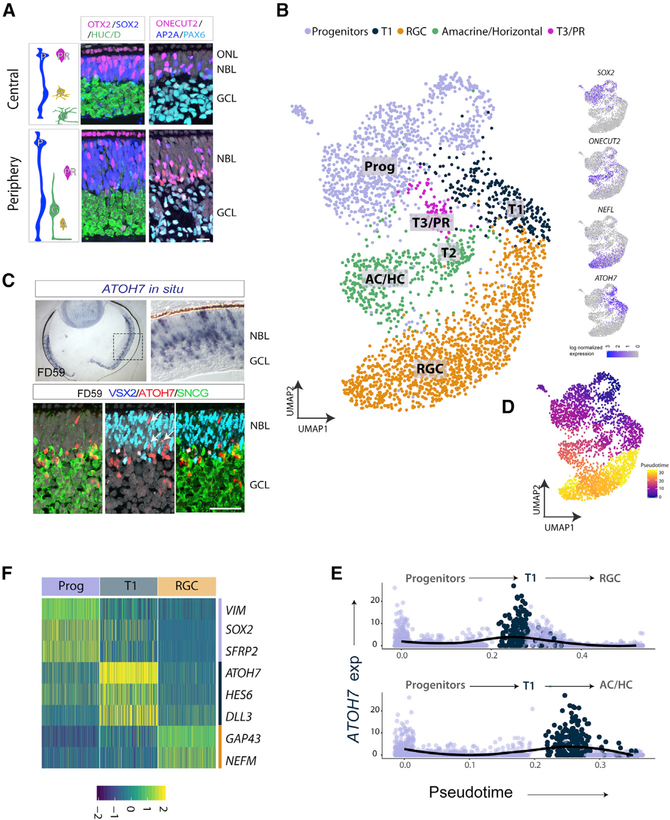
The first age we analyzed in human fetal development was fetal day 59 (FD59). At this age, the presumptive foveal region can already be identified in central temporal retina (Hoshino et al., 2017). Figure 1A shows sections of both central (presumptive foveal region) and peripheral retina at this age. The two predominant cell types in the retina at this stage are retinal progenitors in the neuroblast layer (NBL; SOX2+/PAX6+) and retinal ganglion cells (RGCs;HUC/D+).Smallernumbers of amacrine cells and horizontal cells (TFAP2A+ and ONECUT2+, respectively) and some photoreceptors (OTX2+) are also present at this stage.
Figure 1. scRNAseq of FD59 Retina.
(A) IF of FD59 retina comparing temporal central retina versus periphery, showing RGCs (ganglion cell layer [GCL], HUC/D+ and PAX6+) and progenitors (neuroblast layer [NBL], SOX2+) and some ACs, HCs (AP2A+/ONECUT2+), and PRs (OTX2+) in the ONL. Progenitor cells predominate in peripheral retina.
(B) UMAP plot of FD59 retina, recolored and showing major cell types: progenitors (Progs), transition zone 1 (T1), RGCs, amacrine/horizontal cells (ACs/HCs) and cones (PRs). Right panels: feature plots show expression of some of the genes used to define clusters.
(C) In situ hybridization for ATOH7 at low (left) and high (right) magnification (top panel) and IF for ATOH7 (red), VSX2 (blue), and SCNG (green). Arrows indicate ATOH7+ and VSX2/ SNCG cells.
(D) Pseudotime trajectory with Progs at the root(dark blue loop) and differentiated cells (yellow).
(E) Trajectory analysis from Slingshot, with ATOH7 expression plotted along the individual lineages using UMAP coordinates. T1 cluster cells are plotted in blue, and ATOH7 expression is marked as a solid black line.
(E) (F) Heatmap highlighting genes present in the T1 cluster.
Scale bars, 50 mm. ONL, outer nuclear layer.
Dissociation, scRNA-seq, and Uniform Manifold Approximation and Projection (UMAP) clustering in Seurat (Becht et al., 2018; Butler et al., 2018) allowed identification of the major cell classes with relative numbers closely reflecting the immunofluorescence (IF) analysis (Figure 1B; Tables S1 and S2). The progenitors (blue, SOX2 expression) make up approximately one-third of the total cells. Cell cycle genes are not uniformly distributed in this cluster, but, rather, the G2/M-phase cells fall into a specific region of the progenitor population (as indicated by MKI67 expression and other M-phase markers; Figures S1A and S1B). The differentiated neuron populations show similar ratios as the IF results; ganglion cells are the most abundant, making up approximately one-third of total cells, whereas amacrine cells and horizontal cells are the next most abundant, and OTX2+ photoreceptors were the least abundant of the differentiated neurons (Figure 1B; Figure S1A). Along with progenitor cells and differentiated neurons, the UMAP clustering also revealed a cluster of cells located between the progenitor cells and the differentiated retina cell types. The cells in this cluster do not express high levels of progenitor markers (e.g., SOX2) but are also not yet expressing definitive markers of specific neuronal types (e.g., ONECUT2, NEFL; see also Figure S1). We named the cells in this cluster transition cell population 1 (T1).
The T1 population is characterized by expression of a high level of ATOH7, so we used the expression of this gene to better define this population of cells. Previous studies in mice have shown that this gene is expressed in cells primarily after the final mitotic cell division but has lower expression in the differentiated ganglion cell layer (Brzezinski et al., 2012). Figure 1C (top) shows in situ hybridization for ATOH7 in human retina at FD59, with a similar distribution of labeled cells as reported in mice. ATOH7-expressing cells are located in the NBL but not the ganglion cell layer (Figure 1C, top right). IF analysis shows that most of the ATOH7+ cells (red) are not positive for VSX2 (blue, white arrows), a progenitor marker, but occasionally overlap with the ganglion cell marker SNCG (green).
The T1 population appears to represent an intermediate developmental state. To further explore this possibility, we used Monocle 2.99 software to reorder the cells based on their stage-specific gene expression patterns and reconstructed their developmental timelines (Figure 1D; Cao et al., 2019; Trapnell et al., 2014). This algorithm arranges cells based on pseudotime, where the youngest cells are at pseudotime zero or the beginning/root state. The progenitor cluster, specifically cells expressing markers of the mitotic cell cycle, was designated as starting cells for these analyses. The remaining differentiated clusters were ranked in a similar temporal manner, and developmental lineages were estimated. Our analysis showed two main lineages that linked the FD59 UMAP clusters; a progenitorto-RGC fate or a progenitor-to-amacrine/horizontal fate. Interestingly, a trajectory analysis of individual lineages positioned the T1 cells between progenitor cells and the differentiated neuronal populations (Figure 1D). This is most clear for RGCs and is consistent with lineage studies in mice that have shown that Math5/Atoh7-cre expressing cells can differentiate as any type of retinal neuron (Brzezinski et al., 2012; Le et al., 2006). We also tracked the developmental trajectories in FD59 scRNA-seq data using Slingshot software, which employs a different algorithm, to characterize developmental lineages (Street et al., 2018). ATOH7 expression was analyzed across lineages, from progenitor cells to either RGCs or amacrine/horizontal cells, and Slingshot analysis similarly indicates that ATOH7+ T1 cells contribute to both lineages (Figure 1E). Examination of genes expressed in these populations allowed us to further refine our characterization of the transition states. In addition to high ATOH7 expression, cells in the T1 population have high levels of the Notch pathway components HES6 and DLL3 (Figure 1F; Table S2).
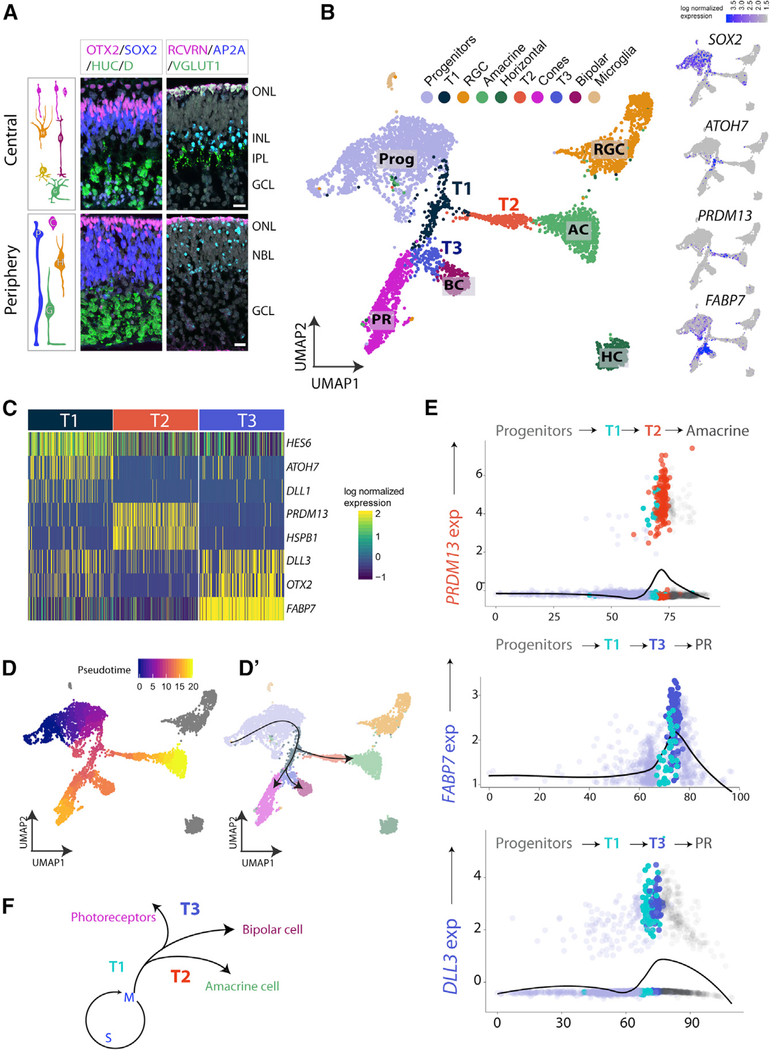
In the next phase of development of the human retina (Figure 2, FD82), the progenitor cells in the central retina have started to generate later-born retinal cell types, including cells that express markers of bipolar cells (OTX2/RCVRN in the inner nuclear layer [INL]), photoreceptors (OTX2/RCVRN in the outer nuclear layer [ONL]), and Müller glia (SOX2 in the INL) (Hoshino et al., 2017); however, the peripheral retina is still dominated by progenitors (SOX2) and RGC, amacrine, or horizontal cells (AP2A/HUC/D) (Figure 2A). We carried out scRNA-seq at FD82 for the central temporal (FD82C) retina (a region that contains the presumptive fovea); the cells form several distinct clusters (Figure 2B). The cell clusters can be identified by their characteristic pattern of gene expression, and the relative numbers of cells of each type approximately match the expected proportions from the IF results (Figure 2B, feature plots). Progenitors and RGCs still make up the largest clusters, but now amacrine cells (ACs) and horizontal cells (HCs) form distinct clusters (TFAP2A, ONECUT2; feature plots in Figure S2A). There is a large increase in the percentage of photoreceptors (PRs), and a distinct population of bipolar cells (BCs) can now be identified. We compared the central retina with peripheral retina at the same stage, and FD82 peripheral (FD82P) retina is more similar to the FD59 retina in cellular composition (Figures S2B and S2B′).
Figure 2. scRNAseq Analysis of FD82 Retina Highlights Three Transition Zones.
(A) IF of central and peripheral FD78 fetal retina(top panels), showing PRs/BCs (OTX2+ cells in the ONL and INL and RCVRN+ cells in the ONL) RGCs (HUC/D+ cells in the GCL), ACs (TFAP2A+), and HCs; the IPL is labeled with VGLUT1. Peripheral retina shows a rudimentary PR layer (OTX2+/ RCVRN+) but no BCs and no defined INL or plexiform layers. Scale bars, 50 μm.
(B) UMAP plot of FD82 central retina. Right panels: feature plots showing the expression of genes characteristic of Progs and transition populations in blue.
(C) Heatmap showing markers of the three transition states.
(D and D’) Monocle pseudotime plot with Progs at the root state and lineages drawn across clusters. Cells shown in gray (RGCs and HCs) were excluded from the trajectory by Monocle.
(E) Slingshot analysis plotting T2-specific PRDM13 expression along the amacrine lineage and T3-specific FABP7 and DLL3 along the PR lineage.
(F) Summary of the transition populations and trajectories seen in central retina at FD82.
In addition to the clusters of differentiated neurons and progenitors, UMAP clustering of the FD82 cells also highlighted transitional cell populations (Figures 2B and S2A). The T1 state, characterized by ATOH7 expression, is similar to the T1 cell cluster found at FD59. At this stage, T1 cells seem to form a root for two additional transition states, T2 and T3, as shown by UMAP clustering. The T2 cells lead to ACs; the cells in this cluster express PRDM13, a gene that promotes AC identity (Bessodes et al., 2017; Goodson et al., 2018; Watanabe et al., 2015). In contrast, T3 cells lie between T1 cells and PR cells or BCs; T3 cells express high levels of FABP7 (Su et al., 2016). Further analysis of these 3 transition states allows for the identification of additional genes co-expressed by transition cells (Figure 2C; Table S3). For example, in addition to ATOH7, T1 cells express HES6, whereas PRDM13+ cells express PTF1A and HSPB1, and T3 cells express high levels of FABP7, OTX2, and DLL3 (Figure S2A).
We further characterized the developmental trajectories of the various cells in these clusters with Monocle and Slingshot, as we did for FD59. Figures 2D and 2D’ show that the transition cell clusters are located between the progenitors and the differentiated neurons in their pseudotime values, with T1 cells having slightly earlier pseudotime values than T2 and T3 cells. Interestingly, the UMAP plot also shows that the RGCs and HCs (gray, Figure 2D) are separate from the other cell transition populations, presumably because they are no longer being generated in the central retina. The results of the Slingshot analysis are shown in Figure 2E; the transition states are again ordered between the progenitor cells and the differentiated neurons, with T1 cells less mature than T2 or T3 cells. For example, the plots in Figure 2E show cells positive for specific genes, like PRDM13, FABP7, and DLL3, as a function of pseudotime for the major cell types being generated at this stage. ACs appear to go through both a T1 and T2 transition state; however, PR cells appear to go through both the T1 and T3 states. Although these putative lineages would need to be validated by more traditional means of lineage tracing, IF analysis of PRDM13 and FABP7 (Figures S3E–S3H) show cells that are neither positive for progenitor markers (e.g., SOX2) or markers of differentiating neurons (AP2A, OTX2), consistent with the pseudotime analysis. Our overall results at this age are summarized in Figure 2F.
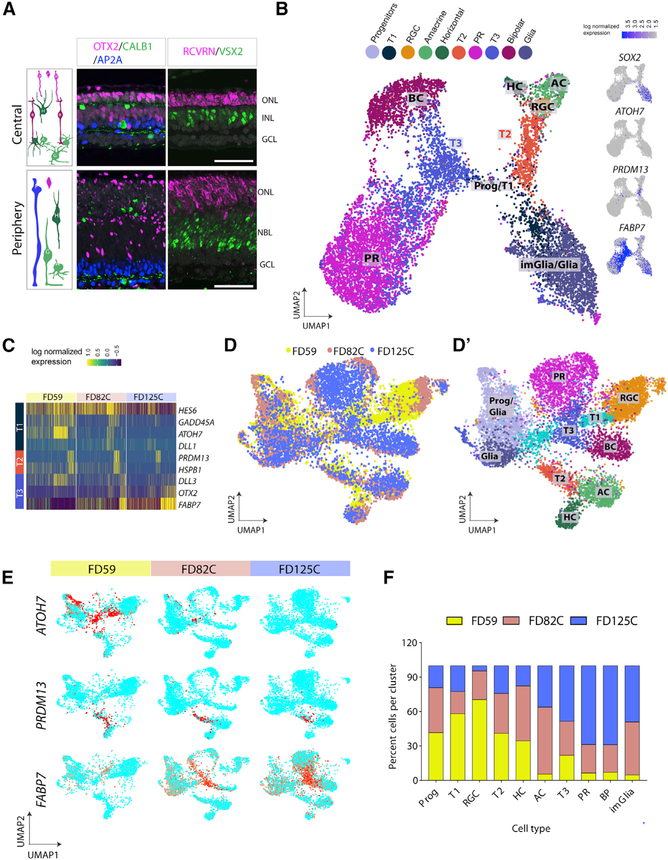
We next carried out scRNA-seq on a third age of fetal human retina, FD125. At this age, the central-most regions of the retina have only a low level of neurogenesis, and all major retinal cell types are present: PRs (OTX2/RCVRN) in the ONL, BCs (OTX2/VSX2/RCVRN) in the INL, and HCs and ACs (AP2A/CALB1; see also Hoshino et al., 2017). The UMAP clustering at FD125 for the central temporal (FD125C) retina shows that PR cells predominate at this age, along with BCs, whereas ACs, HCs, and ganglion cells are much less abundant as a percentage of total cells (Figures 3A and 3B; Table S4). Another striking feature is that there are very few ATOH7+ T1 transition cells present in the central retina at this age (Figure 3B, feature plot of ATOH7). In contrast, there are still substantial numbers of ATOH7 T1 cells in the periphery (Figure S3B), although these are gone by FD163, except at the far peripheral retina (Figures S3C–S3E). The T2 and T3 cell clusters are still apparent in both FD125C and FD125 peripheral (FD125P) retina (Figure 3B; Figures S3A and S3B). T3 cells express FABP7 and are positioned between the progenitors, the BCs, and the PRs, whereas T2 cells still express PRDM13 and are positioned between the progenitor cells and the ACs. Other genes characteristic of these two transition cell populations that were present at FD82 are still among the top genes that discriminate these cell clusters; for example, DLL3 and FABP7 are more highly expressed in T3 cell clusters, whereas PRDM13 is more highly expressed in T2 cells (Figure 3C).
Figure 3. scRNAseq of FD125 Retina.
(A) IF of FD125 near the fovea shows that neurogenesis is complete: a single layer of PRs (ONL, OTX2+/RCVRN+), BCs (INL, OTX2+/VSX2+), and HCs and ACs (CALBINDIN+/TFAP2A+). The periphery at this stage still has a NBL (VSX2+). Scale bars, 50 mm.
(B) UMAP clusters of FD125 central retina (FD125C). Right panels: feature plots showing the lack of ATOH7 (T1 cells); PRDM13 marks T2 cells, and FABP7 marks the T3 population.
(C) Heatmap showing genes characteristic of transition clusters in FD59, FD82, and FD125 retinas; T2 and T3 genes are still expressed at laterages.
(D) CCA plot of fetal ages combined; clusters are plotted by age (left) or retinal cell type (right). RGCs are over-represented in the FD59 sample (yellow), whereas BCs are over-represented in the FD125 sample (blue).
(E) Feature plots showing changes in transition markers across the ages.
(F) The ages that contribute to specific retinal cell types were plotted as percent cells per cluster, colored by age.
The transitional populations we identified in the fetal human retina are also present in the developing mouse retina. We analyzed mouse scRNA-seq data for the presence of these populations using a similar UMAP analysis (Clark et al., 2019; Figure S4). We focused our analysis on embryonic day 16 (E16) (Figures S4A and S4B) and E18 (Figures S4C and S4D) mouse retinal cells because their cellular composition resembles the human FD50 and FD82 central retina, respectively. Although Atoh7 is only detected in very few cells in the mouse (presumably because the sequencing depth is not sufficient), we found a T1-like cluster with cells that express SOX11, GAL, and ELAVL3, genes that are also highly expressed in the human fetal T1 cluster, primarily at FD59 (Figure S4E). Similar to the mouse data, expression of RGS16 and GADD45G increases with time in the human retina, along with NFI genes (Figure S4F).
The change in relative cell composition of the retina, along with changes in the transition cell populations, can be more easily visualized when the three ages are combined using canonical correlation analysis (CCA) (Figures 3D and 3D’). RGCs are more highly represented at FD59; the many yellow dots in Figure 3D show that FD59 cells overlap well with RGCs (orange dots in Figure 3D’). In contrast, PRs predominate at FD125C, as shown by the magenta PR cluster (Figure 3D’) overlapping largely with FD125C cells (Figure 3D, blue). The transition populations change over this period of development (Figures 3E and 3F); the T1 population predominates at FD59, with only a relatively small number of T3 cells. However, by FD125, ATOH7+ cells are not detected in the central retina (Figure 3E; Figure S3D), whereas FABP7-expressing T3 cells now account for the majority of the transition cells in the retina. Interestingly, T2 cells make up approximately the same percentage across the three ages; this suggests that about the same number of inhibitory interneurons (i.e., HCs or ACs) are generated throughout this period of fetal human development. Calculation of percent cells per cluster provided an overall synopsis of the major events of human retinogenesis across these three ages (Figure 3F); progenitors and early-born retinal cells dominate at FD59–FD82C, whereas later-born retinal cells are more highly represented at FD82C–FD125C.
How Well Do Stem Cell-Derived Retinal Organoids Resemble Equivalent Staged Fetal Retina?
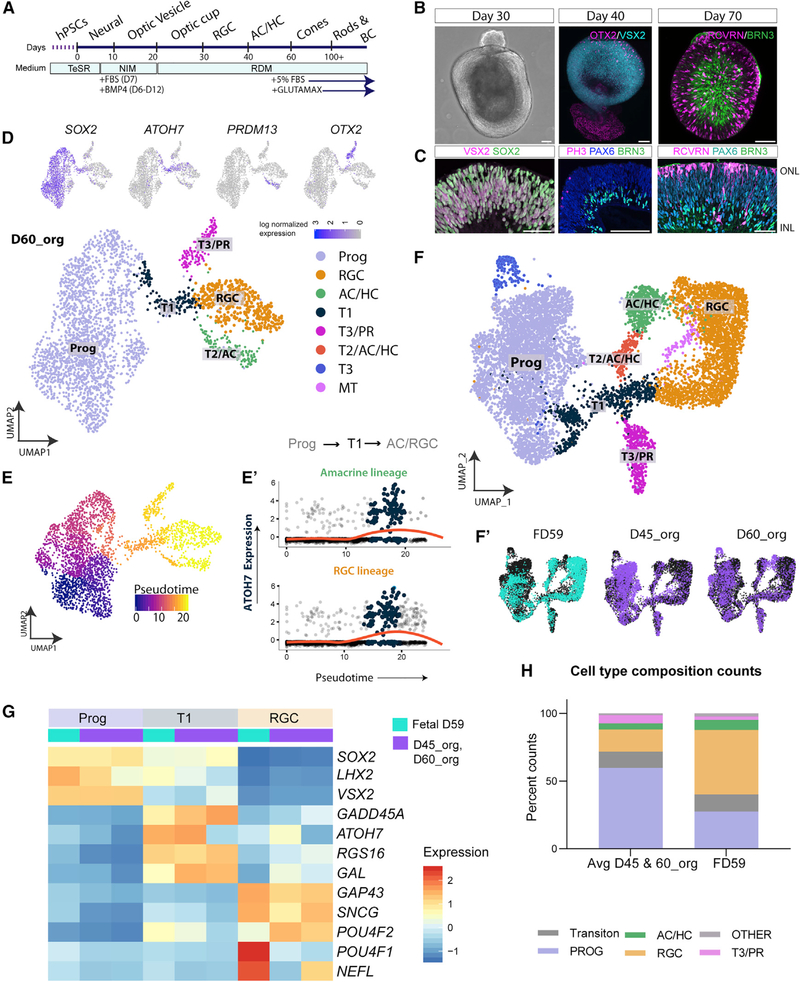
To evaluate how human stem cell-derived retinal organoids compare with the fetal retina, we carried out scRNA-seq of corresponding ages of retinal organoids. We generated retinal organoids with a well-established protocol (Figure 4A) up to approximately 200 days of culture (Fligor et al., 2018; Meyer et al., 2009; Ohlemacher et al., 2015; Zhong et al., 2014; Phillips et al., 2018). In this protocol, the timing of major retinal events approximates that of fetal retina (Figures 4B and 4C); for example, on differentiation day 40 (D40), the first OTX2+ PR cells arise on one side of the organoid (Figure 4B, middle panel), whereas on D70, it is possible to visualize the threelayered structure with BRN3+ RGCs, PAX6+ progenitors, and RCVRN+ PRs (Figures 4B and 4C).
Figure 4. scRNAeq of Early Retinal Organoids.
(A) Differentiation protocol highlighting the order and timing of retinal organoid development.
(B and C) Retinal organoids at 30 days primarily express Prog markers (VSX2/SOX2; C). By 40 days, whole-mount staining shows OTX2+ cells migrating in the neuroepithelial layer, and a small patch of RPE also expresses OTX2 (B, middle panel). RGCs appear (BRN3/PAX6) at 30–40 days (C, middle panel). By 70 days, RCVRN+ PRs are seen in the apical layers, and BRN3+ RGCs occupy the basal layers, as seen in the whole mount (B, right panel) and sections (C, right panel). (D) UMAP plot showing 5 major clusters at day 60: Progs, T1, retinal ganglion cells (RGCs), transition cells 2/ACs/HCs (T2ACs/HCs), and transition cells 3 (T3) and early cone PRs. Inset: the feature plots used to identify clusters.
(E) Pseudotime analysis by Monocle2.99, showing cell ordering with Prog cells at the root.
(E’) Slingshot analysis showing similar ordering of cells. Trajectory analysis shows that ATOH7 expression (y axis) is highest in T1 cells (bump in red line) as cells go from Progs to differentiated cell types.
(F) Integration of organoid (D45, D60) scRNA datasets with the FD59 dataset (Seurat3); cells were reclustered and replotted.
(F’) UMAP plot of the identity of the clusters in (F) (cyan, fetal; purple, organoid).
(G) Heatmap showing cluster averages for key genes, highlighting the expression of cells in the Prog, T1, and RGC clusters, plotted for fetal retina (FD59, cyan) and organoids (D45 and D60, purple).
(H) Each column corresponds to a single dataset. The stacked bar graph shows the cell type composition of the fetal retina and organoids (average of both ages) from (F).
Scale bars in first panels of (B) and (C) represent 100 mm; other scale bars represent 50 mm.
To determine how well the organoids develop, we first carried out scRNA-seq analysis of early stages of retinal organo ids (Figure 4D). We found that the UMAP clustering on organoid D60 gave clusters similar to the FD59 fetal retina (see above). The major cell types at this age are progenitor cells, RGCs, PRs, and ACs/HCs (Figure 4D). In addition, ATOH7+ T1 population cells were also present in the organoids (Figure 4D, feature plot) and expressed high levels of HES6, similar to the FD59 retina (Figures S5A and S5B). We further characterized the developmental trajectories of the various cells in organoids as for fetal retinal cells. In the organoids, T1 cells are positioned in pseudotime between progenitors and differentiated neurons (e.g., RGCs and amacrine/horizontal clusters) similar to what we find in the FD59 retina (Figure 4E, Monocle; Figure 4E’, Slingshot; Figures S5G– S5I’); however, there are some differences between the FD59 and organoid T1 populations. DLL3, a gene that is highly expressed in the FD59 T1 cell cluster, is not enriched in organoid T1 cells (Figures S5A and S5B).
To more closely compare the organoids with FD59 retinal cells, we combined the data from organoids and fetal retina (Figure 4F). We used two samples of organoids, D45 and D60 because it is not clear which organoid age provides the most appropriate comparison with the FD59 sample, given the large central-to-peripheral gradient in development in fetal retina. The datasets largely overlap, and the contributions from each age of organoid and the FD59 fetal sample are shown in Figure 4F’. Although all cell types present in the fetal sample at this age are also present in the organoids, there are some differences in gene expression. Figure 4G shows a heatmap directly comparing the average gene expression among the samples; progenitor genes (e.g., SOX2 and LHX2) are expressed at nearly identical levels in organoids and fetal retina, but several RGC genes show differences (e.g., POU4F1 and N\EFL; Figures S5C and S5D). In addition to these differences in gene expression in the identified populations of retinal cells, we also noted that the ratios of retinal cells are not precisely the same in organoids and fetal retina. For example, there are relatively more progenitor cells and fewer RGCs in organoids than in fetal retina at the same stage (Figure 4H).
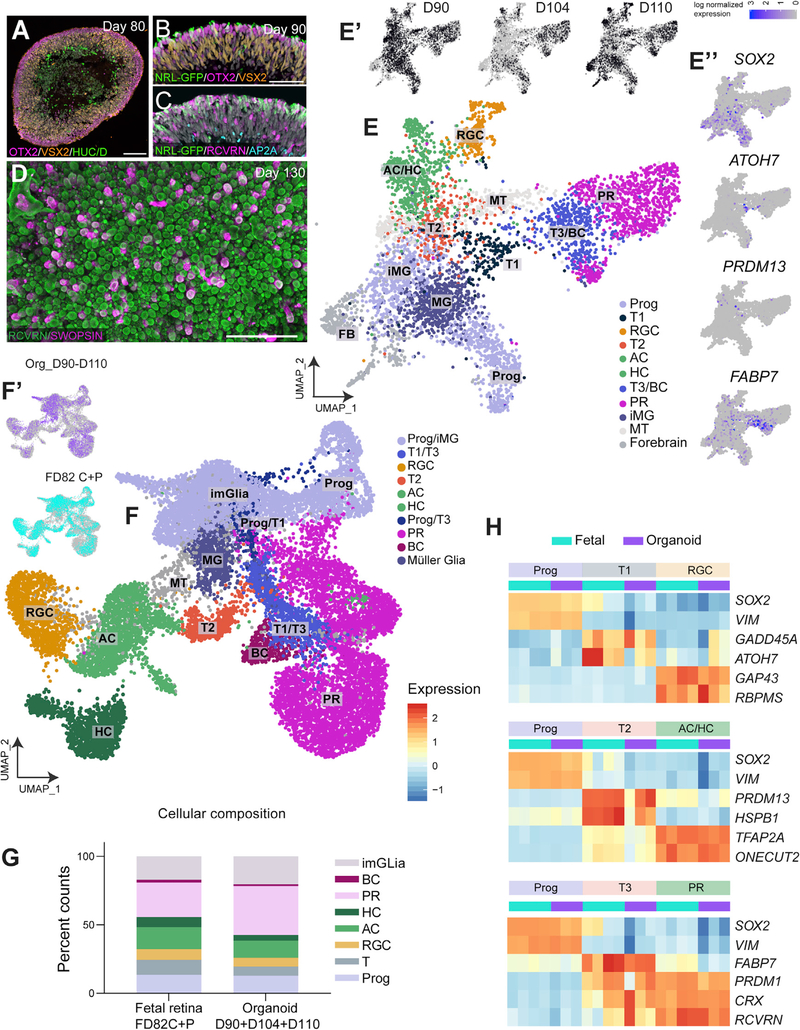
We next compared later stages of retinal development between organoids and fetal retina. At later stages of development (Figures 5A and 5B, D80 and D90), PR layers continue to expand, and the first NRL-GFP+ rod PRs (Figures 5B and 5C) are seen around D90 (Phillips et al., 2018). We observed that the inner layers of organoids become somewhat disorganized at approximately D70–D90. HUC/D+ ACs do not form a continuous and intact layer like the PRs (Figure 5A). Nevertheless, PR development continues quite normally, and SW-OPSIN+ cone PRs emerge in a mosaic-like arrangement in the developing ONL (Figure 5D).
Figure 5. scRNAseq of D100 Retinal Organoids.
(A) D80 organoids contain a PR layer (OTX2, magenta), an NBL (VSX2, orange), and HUC/D+ ganglion cells and ACs (green).
(B and C) Early rods (NRL-GFP) at 90 days of differentiation, along with progenitors (VSX2+) (B) and ACs (AP2A+) (C).
(D) Whole-mount staining at day 130 shows RCVRN+ cone PRs (green), some of which express SWS-OPN (magenta).
(E) UMAP clusters highlighting the cellular composition at this age.
(E’) scRNA-seq analysis of D90, D104, and D110 organoids plotted by origin.
(E”) Feature plots showing Prog and transition cell populations.
(F and F’) Combined scRNA-seq analysis of fetal retina and organoids; FD82 central and periphery (FD82C+P) with organoids (D90, D104, and D110). (F0) shows the composition by origin (organoid, purple; fetal retina, cyan), and (F) shows the UMAP clusters seen at this age.
(G) Stacked plots showing the percent counts from (F), reflecting the composition of the fetal retina and organoids from (F). Counts were calculated for eachdataset and then averaged by fetal or organoid.
(H) Heatmaps plotting the cluster averages of key genes, highlighting the 3 lineages seen at this age. Each column corresponds to a single dataset (cyan, FD82C;P [purple], day 90, day 104, and day 110 organoids).
Scale bars,50μm.. FB, forebrain; MT, mitochondrial cluster.
scRNA-seq analysis of these older organoids shows that they are progressing in their development much like fetal retina, with all main retinal cells present in the organoids (Figure 5E). There are fewer RGCs (gold) and progenitors (cornflower blue, SOX2+) than in the earlier samples (Figure 4) and relatively more PRs (Figure 5E, magenta). At these later ages in organoids, there are fewer ATOH7+ T1 cells but abundant FABP7+ T3 (Figure 5E”) cells.
To directly compare organoid cells with the fetal retina at a similar stage of development, we combined the data from D90–D110 organoid samples with that of FD82 central and peripheral retina (Figures 5F and 5F’). Like the earlier stages of organoids, the cellular composition at this stage is very similar to the corresponding fetal retina stage (Figure 5G); all major cell types can be identified (Figure 5F), including the transition cell populations. We also compared the average gene expression levels of organoid and fetal retinal cells for genes that define specific cell populations, and these are shown as heatmaps in Figure 5H. As for the early stages of organoids, progenitor gene expression is very similar across samples; however, there are some differences between fetal retina and organoids. For example, ATOH7 is more highly expressed in fetal retinal cells than in organoids, whereas GADD45A is more highly expressed in organoids (Figure S5B).
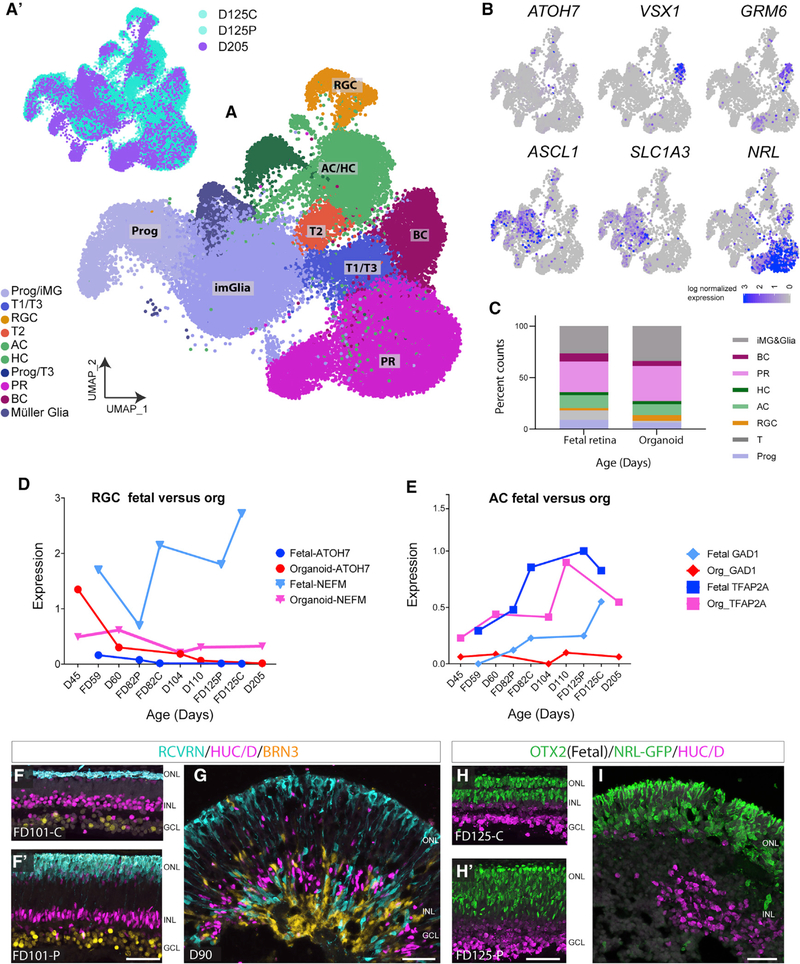
The above analyses indicate that organoids closely model fetal retinal development through FD110. This trend continues even in more mature organoids; we compared scRNA-seq results from FD125 retina with those from D205 organoids (Figure 6), and the cells largely overlap (Figure 6A’). More mature organoids have a higher representation of later-born retinal subtypes; e.g., BCs, rods, and Mùller glia ( Figures 6B and S5J– S5M). In addition, the T2 population is still present (Figure 6A, red), although it is more difficult to locate a cluster of cells with the T1 signature, and only a few ATOH7+ cells remain (Figure 6B). Although there are still some cells with a progenitor profile of gene expression in late-staged organoids (Figure 6B, ASCL1), there is now a large number of cells we identified as Müller glia (Figure 6A, dark blue), based on their expression of RLBP1, AQP4, and SLC1A3 (Figure 6B; Figure S5D). Also at this age, D205 organoids have substantial numbers of BCs, as evidenced by their expression of VSX1, CABP5, and GRM6 (Figure 6B; Figure S5C), and transmission electron microscopy (TEM) of older organoids shows the presence of ribbon synapses in the developing outer plexiform layer (OPL) (Figures S5E and S5F). Quantification of these cell types from FD125 fetal and D205 organoid samples shows that the organoids have a cell composition similar to fetal retina (Figure 6C).
Figure 6. scRNAseq of Older Organoids Compared to Fetal Retina.
(A and A’) Combined scRNA-seq of FD125 central and peripheral datasets with D205 organoids.
(A) UMAP clusters showing the cellular composition across the combined dataset.
(A’) Plots showing the relative contribution of fetal retina (cyan) and organoid (purple) to the clusters in (A).
(B) Feature plots showing the T1 marker ATOH7, bipolar markers (VSX1 and GRM6), Mùller glia markers (RLBP1 and SLC1A3), and rods (NRL).
(C) Percent counts calculated from (A), showing the relative proportions of cells in fetal retina (average of D125C+P) and day 205 organoid.
(D and E) Average expression of key genes for fetal and organoid RGC and AC subsets across all ages.
(E–H) IF comparing lamination across comparative fetal and organoid ages.
(F and F’) FD 101 central and peripheral retina: RCVRN+ PRs (cyan), HUC/D+ ACs (magenta), and BRN3+ ganglion cells (yellow) form distinct layers.
(G) D90 organoid is similar to the FD101 retina. HUC/D and BRN3-td tomato cells (pseudocolored in yellow; cell line from Sluch et al., 2017) show basic lamination, although there is more mixing of cells from other layers.
(H and H’) FD125 central and peripheral retina. OTX2+ PRs and BCs (ONL and INL, green) and HUC/D+ AC and ganglion cells (magenta) show characteristic lamination.
(I) The day 180 organoid has a well-laminated ONL (rod PRs are NRL-GFP+) but less organized INL; HUC/D+ cells (magenta) appear to be clumped together. 50μm.
Although the proportions of the various cell types are similar in organoids and fetal retina of equivalent stages of development, the level of cell-type-specific gene expression shows some differences. Although many genes are expressed at similar levels (Figures S5C and S5D), some genes are differentially expressed in organoids compared with fetal retina (Figures S7A–S7D). For example, ATOH7 has comparable expression in the FD59 fetal retina and the D60 organoid; however, NEFM is expressed at a significantly higher level in the fetal retina at all ages (Figure 6D; Figures S7C and S5). Similar differences are observed in AC-specific genes; GAD1 and TFAP2A are more highly expressed at all ages in fetal retina than in organoids (Figure 6E; Figure S7D).
Although the cellular composition and overall gene expression of organoids largely mirrors that of fetal retina, organoids lose inner retinal organization over time (Figures 6F–6I). Retinal lamination at D90 is comparable with that of the fetal retina (Figures 6F– 6G); however, by D180, the inner retinal cells of organoids are no longer organized as clear lamina despite the fact that the ONL forms a compact layer (Figures 6H–6I).
Retinospheres: Organoid-like Cultures Derived from Fetal Human Retina
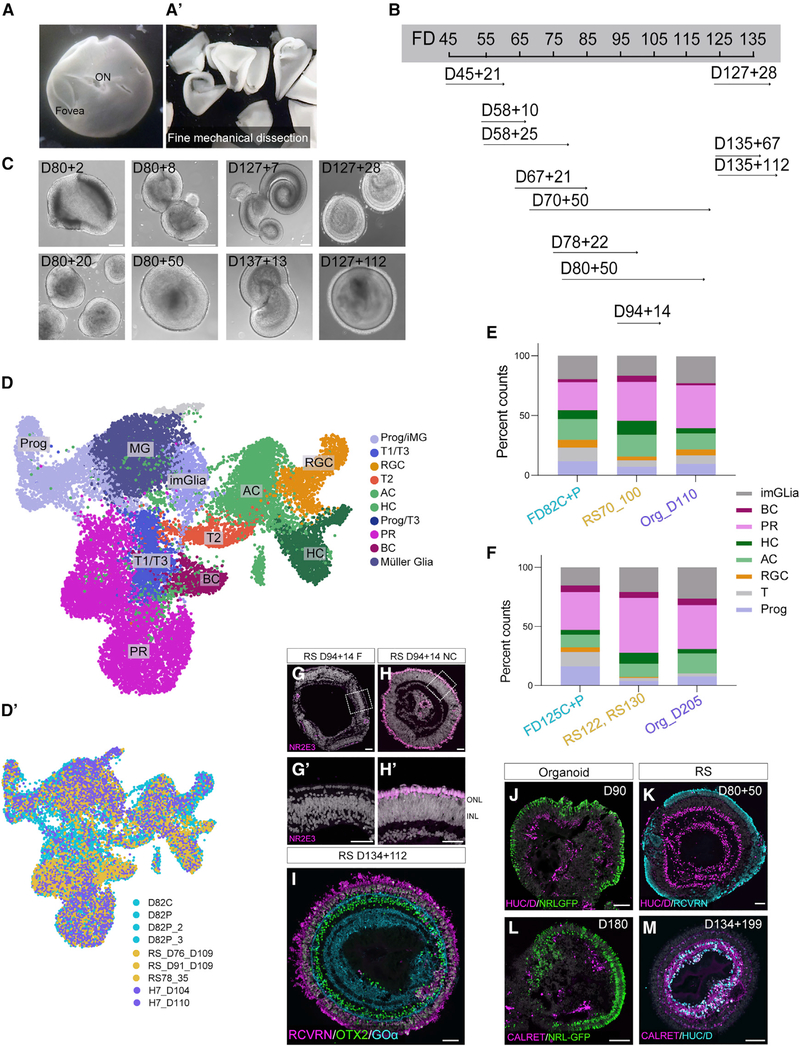
scRNA-seq comparisons of fetal retina with organoids show that the relative cell numbers of the various types of retinal cells generated in organoids are close to those in the fetal retina, even in organoids as old as 200 days. However, several investigators have noted previously that the inner retina of organoids is not well laminated after 100 days (Capowski et al., 2019), although outer retinal lamination is maintained. To determine whether the in vitro culture conditions do not support inner retinal lamination, we cultured fetal retina in a manner very similar to the culture conditions of organoids. The fetal retina was finely dissected into small pieces (within hours post-mortem), approximately the same size as retinal organoids of equivalent age (Figures 7A–B), and these were cultured in the same medium as the organoids (retinal differentiation medium [RDM]; STAR Methods; Meyer et al., 2009). Within the first few days in vitro, the pieces of retina began to round up and form spheres (Figure 7C), which we call retinospheres (to distinguish them from retinal organoids).
Figure 7. Comparison of Fetal Retinal Cultures (Retinospheres) with All Datasets.
(A) FD130 retina with fovea and optic nerve head (ON).
(A’) Pieces of the retina prior to culture.
(B) Age at culture of retinospheres used in this study. FD45+21 refers to a FD45 retina cultured for 21 days.
(C) Retinospheres made from older retinas organized into spheres (D127+7, D137+13) and could be cultured for a month (D127+28) or up to 4 months withprominent outer segments (D127+112).
(D and D’) Combined UMAP plot of retinoshperess, fetal retina, and organoids. (D’) shows UMAP plotted by tissue of origin: retinospheres (RSD76 [RSD, retinosphere age in days] cultured until day 109, RSD91 cultured until day 109, RSD78 cultured until day113), organoids (day 104 and day 110), and fetal retina (FD82 central and periphery).
(E and F) Percent counts from (D) for (E) FD82 retina with equivalent age retinospheres (i.e., retinosphere cultured from D70 to D100) and D110 organoids highlightsimilar composition across all samples. (F) This is also true for older ages of fetal retina (FD125C+P) when compared to older cultures of retinospheres (RSD122 cultured for 14 days and RSD130 cultured for 30 days) and D205 organoids.
(G–H’) Fovea-derived retinospheres lack rods (NR2E3, magenta, G and G’), unlike nasal central (NC)-derived retinospheres (H and H’).
(I) Retinospheres maintained for 100+ days retain lamination, as seen by RCVRN+ PRs (magenta, ONL) and BCs (RCVRN, OTX2, and Goa).
(J and K) RSD80 (K) resemble equivalent age organoids (D90, J) but demonstrate distinct and compact rings of the AC/RGC marker HUC/D.
(L) At later ages, organoids express NRL-GFP, and the ONL is well preserved, but calretinin staining does not form a continuous layer.
(M) After 134 days in vitro, retinospheres continue to maintain inner retinal layers (CALRET/HUC/D).
Scale bars, 50μm.
We successfully carried out this procedure on ~40 fetal retinas from FD45–FD135 (Figures 7B and 7C). Fetal retina of all ages could be maintained as retinospheres, some of which were cultured up to 5 months. Retinospheres could be generated from all regions of the retina, including the presumptive foveal region (Figure 7C). Because retinospheres are cultured under the same conditions as organoids, we postulated that they may show defects in inner retinal lamination. Surprisingly, retinospherescultured for nearly 4 months show excellent lamination and inner retinal preservation, as seen by IF and confocal microscopy (Figures 7I and 7M). TEM of D134 retinospheres cultured to D213 show ribbon synapses in the inner plexiform layer (Figures S6E–S6G). retinospheres made from different regions of the fetal retina retained their original regional identity (Figures 7G–7H’). For example, retinospheres made from the presumptive fovea and the nasal retina from a similar eccentricity developed distinct features of their regional identity (Figures S6A–S6D).
To assess the level of differentiation that occurs in retinospheres, we compared organoids and retinospheres by scRNA-seq. We combined cells from retinospheres with the FD82 retinal sample that included both the central and peripheral regions, along with the D104 organoid (Figures 7D–7E). The UMAP plot of the combined samples shows that the cells fall into similar groups with good overlap among the main cell classes (Figure 7D’); moreover, the relative percentages of cells in retinospheres are very similar to those of the fetal retina and organoids (Figures 7E and 7F). This was true for the younger samples (FD82, RS70_100, D110 organoid), as well as the older samples (FD125, RS130_160, RS122_136 and D205 organoid).
DISCUSSION
In this report, we carried out scRNA-seq of (1) human fetal retina at key developmental stages, (2) retinal organoids derived from hPSCs, and (3) RSs (three-dimensional organoid-like structures derived from fetal retina). We characterized the major cell populations at each stage of retinal development and identified transition cell clusters that fall between the proliferating retinal progenitor cells and the differentiated cell types. Our analysis also shows that retinal organoids have a cellular composition very similar to fetal retina at equivalent stages of development, although it is hard to compare this precisely because of the large central-to-peripheral difference in development in the fetal retina (Hoshino et al., 2017). Despite the similarity in cellular composition between organoids, there are some differences in the levels of some cell-type-specific genes, and organoids typically do not retain their normal inner retinal lamination as they mature. To better understand the differences between hPSC-derived retina and fetal retina, we developed a way to culture the fetal retina under the same conditions as the organoids. This method allows long-term culture of the fetal retina (up to 5 months). In contrast with organoids, we find that RSs maintain inner retinal lamination. Overall, our data represent a comprehensive analysis of the development of cell types in the early fetal human retina and highlight similarities between in vitro- and in vivo-derived retinas.
The molecular makeup of the human fetal retina has been well characterized in recent years. Hoshino et al. (2017) used bulk RNA-seq to provide a transcriptome analysis at critical stages of human retinal development, allowing a better understanding of the dynamics of gene expression and the timing of cell-type-specific genes. However, bulk RNA-seq analysis is limited by the lack of ability to assign specific genes to particular cell types. The single-cell analysis reported here fills this gap. Other reports have recently used scRNA-seq to better characterize the primate retina. For example, Voigt et al. (2019) and Peng et al., 2019 have published single-cell data focused on the adult human and primate retina, respectively, and reported conservation of foveal cones and glial cells between these species (Voigt et al., 2019; Peng etal., 2019). Our data build on these reports by extending the analysis to fetal stages and reveal several important aspects of retinal development not easily observed in bulk RNA-seq analyses.
One of the important features of this report is the description of transition cell populations. These cells appear to be at critical postmitotic junctures “between” the retinal progenitor cells and the differentiated cell types, as shown by UMAP clustering, Monocle (using the Louvain clustering algorithm), and Slingshot. The T1 cell population is characterized by expression of ATOH7; this gene is known in the mouse to be primarily expressed in newly generated postmitotic cells that can differentiate into diverse retinal cell types (Brzezinski et al., 2012). In our data, ATOH7+ T1 cells form a root for all retinal lineages at the early ages of retinal development but are nearly absent from central retina in the older samples. Although transient expression of ATOH7 in recently postmitotic cells from early retina has been well described, the emergence of the cells that express this gene as a distinct cluster has allowed us to discover other genes (e.g., HES6) that are co-expressed in these cells and should provide additional insight into the mechanisms that control cell fate decisions in the early retina. The T2 cell cluster represents cells that lie midway between progenitor cells and differentiated ACs or HCs. These cells are characterized by expression of PRDM13 and PTF1A, a previously described transcription factor known to be critical for development of these cell types (Bessodes et al., 2017; Goodson et al., 2018; Hanotel et al., 2014; Watanabe et al., 2015; Hori et al., 2008; Xiao et al., 2018). Our data indicate that T2 cells may serve as a bi-potential precursor for the main classes of inhibitory interneurons in the retina, and again, the fact that these transition cells form a unique cluster allows us to identify additional gene candidates that might be involved in these cell fates. Last, the analysis identified a cluster of transition cells that appear to serve as precursors for PR cells and BCs, characterized by high levels of FABP7, OTX2, and DLL3. The presence of a bi-potential precursor for these cell types has been inferred from mouse data; for example, deletion of Prdm1/Blimp in mice leads to a transition from PRs to BCs (Brzezinski et al., 2010; Katoh et al., 2010).
An interesting feature of these transition populations is that they all express genes that are associated with cell-autonomous inhibition of the Notch pathway. Several previous reports have shown the importance of the Notch pathway in retinal progenitors (Bae et al., 2000; Chapman et al., 2011; Jhas et al., 2006; Nelson et al., 2009; Nelson and Reh, 2008). Notch1 is active in retinal progenitors, primarily via Dll1, and active Notch induces Hes1/5 expression at sufficiently high levels to maintain the progenitor pool. Exit from the progenitor state to a postmitotic neuron requires Notch inhibition, and inhibition of Notch for even a short time irreversibly commits cells to a neural fate (Nelson et al., 2007). It is therefore quite interesting that one of the most characteristic genes of the T1 cell population is HES6, which inhibits the function of HES1/5 (Bae et al., 2000; Jhas et al., 2006). T1 and T3 cells also express DLL3, which inhibits Notch in a cell-autonomous manner by targeting it for lysosomal degradation in the late endocytic compartment (Chapman et al., 2011). In addition, T2 cells express PTF1A, which is critical for the specification of inhibitory neurons in many areas of the CNS, including the retina, and functions in part by competing with Notch for RBPJ (Hori et al., 2008) a necessary co-factor for Notch signaling. The use of these three different “exit strategies” for inhibiting the Notch pathway in these three lineages of retinal cells may explain some of the cell-type-specific observations made after experimental manipulation in the Notch pathway (Jadhav et al., 2006; Georgi and Reh, 2011) and may underlie mechanisms of cell type diversity. Analysis of the single-cell data indicates that some of these transition genes are expressed in the last cell cycle of the progenitors. There is some overlap between MKI67 (M phase) cells and ATOH7 at FD59. Additionally, there is not much overlap between ATOH7 and OTX2 or ATOH7 and NEUROD1, but we see a lot more overlap with OTX2 and NEUROD1. This potentially indicates that transition cells might be triggering cell cycle exit and activating the differentiation program in the cells.
The fetal human single-cell data are consistent with what has recently been described in a similar study in mice, especially at embryonic stages (Clark et al., 2019). Early neurogenic genes in the mouse, Gal, Sox11, and Elavl3, were also expressed at FD59 in human retina, and the mouse “late progenitor” markers Rgs16 and Fabp7 were also expressed in later-stage progenitor cells in the human fetal retina. The T1 cell population is similar to what Clark et al. (2019) call “neurogenic” progenitors in the mouse; we re-analyzed their data from E16 and E18 using UMAP and were able to find that the T1 clusters at E16 in the mouse and FD59 in humans express many of the same genes. T2 and T3 clusters are also present in the mouse data but are not as well defined as in the human data, possibly because retinogenesis in mice occurs faster than in humans. Clark et al. (2019) also reported that there is an increase in Notch signaling and expression of the members of the Nfi gene family in retinal progenitors near the end of neurogenesis in mice, confirming similar observations made by Nelson et al. (2011) on fluorescence-activated cell sorting (FACS)-isolated progenitors and immature Müller glia. This is also conserved in humans; Notch signaling and NFI family genes increase in late progenitor cells as they transition to immature Müller glia (Figures S7E and S7F).
One of our primary goals in undertaking this study was to evaluate how well organoids compare with the fetal retina. Several studies have compared hPSC-derived retinal organoids with fetal retinal development using IF, bulk RNA-seq (Hoshino et al., 2017, 2019; Welby et al., 2017), and, more recently, scRNA-seq (Kim et al., 2019). In addition, recent RNA-seq comparisons have focused on isolated organoid-derived PRs (Welby et al., 2017; Kim et al., 2019; Kaewkhaw et al., 2015; Phillips et al., 2018) or ganglion cells (Langer et al., 2018). By focusing on particular cell types, these previous studies demonstrated a great deal of similarity in hPSC-derived retinal organoids and fetal retinal development.
By analyzing the overall cellular composition of organoids at various stages, we show more broadly how organoids relate to the fetal retina. Early D60 organoids are remarkably like the FD59 retina; however, although the major cell types are present, they differ from fetal retina in their cell proportions, with increased progenitors and reduced RGCs. At more mature stages of development, organoids and fetal retina grow even more similar in the percentages of the major cell types, although differences were observed in some cell-type-specific genes. We were able to see a clear T1 cell cluster in D60 organoids and cells that clustered with T2 and T3 cells at later stages. Recently, Collin et al. (2019) reported similar results of scRNA-seq using three ages of retinal organoids. Although they did not provide a detailed comparison with human fetal development, the relative cellular composition analyses were very similar to those in our study despite differences in the protocol used to generate the organoids (Figure S5).
Although the organoids are similar to fetal retina, some differences in cell composition and gene expression may be due to limitations of the in vitro system. For example, the fetal retina has a large gradient of development, and at some stages, the central retina can be as much as 6 weeks ahead of the periphery in developmental time (Hendrickson et al., 2012; O’Brien et al., 2003; Hoshino et al., 2017). It is difficult to determine whether organoids represent central or peripheral retina in their timing, which could account for differences in maturation of cell types and their relative numbers. In addition, other sources of variability include the specific cell line used and the details of the protocol. For example, Kaya et al. (2019) used bulk RNA-seq to show that organoids follow normal fetal developmental gene expression, similar to what we report, but they also reported considerable variability across cell lines. Differences in gene expression in organoids may also relate to their lack of endothelial cells and microglia. These non-neural cells may have important roles in developmental processes, including cell proliferation, cell survival, and synapse formation (Silverman and Wong, 2018). Moreover, the culture conditions that are used to maintain organoids in vitro for extended periods of time rely on adequate nutrient and gas exchange, and as the tissue reaches the stage where vasculature perfusion becomes critical, these conditions may be limiting for normal development.
To begin to address the contributions of some of these factors to the appropriate development of the fetal retina, we developed a culture system using human fetal retina that mimics the culture conditions of hPSC-derived retinal organoids. Small pieces of fetal retina self-organize into organoid-like structures in suspension cultures. These RSs develop and maintain inner retinal neurons, including ACs, HCs, and BCs, showing extensive development of the inner plexiform layer and OPL. This method varies from the reaggregate spheres developed by Layer et al. (2002) for embryonic chick retina in that the cells in those cultures were previously dissociated and then allowed to form spheres.
The results with retinospheres provide several insights into the source of the differences between fetal retina and organoids. (1) RGC death alone may not be responsible for inner layer disorganization in organoids. Inner retinal organization and maturation in RSs proceeds very well without RGCs. (2) Extraretinal tissues are not needed to maintain excellent lamination in older retinospheres, although the vasculature or microglia may be required for proper inner plexiform layer (IPL) development because these cell types are present in retinospheres but not organoids. (3) Regional differences in the retina are preserved in RSs; RSs made from fovea retain their unique characteristics for months in vitro.
In summary, the single-cell analysis described in this report provides cell-type-specific information for many of the genes reported in the bulk sequencing publications. The changes in cellular composition that are revealed by scRNA-seq helped us identify three transition clusters as possible postmitotic funnels between progenitors and differentiated neurons. These transition cell populations are conserved between humans and mice and may represent critical steps in the differentiation program. Our results also provide an excellent basis for using scRNA-seq for “staging” retinal organoids and evaluating their similarity to fetal retina. Different organoid protocols can produce somewhat different results, and the scRNA-seq atlas we provide may help to further refine these protocols to more accurately reflect fetal development. Finally, to better understand the differences between development in organoids and fetal retina, we developed an organoid-like in vitro system for fetal retinal culture. Analysis of these RSs revealed that culture conditions alone do not account for inner-layer deficiencies in organoids. Overall, the data presented in this study showcase the ability of scRNA-seq to evaluate multiple datasets of the human retina and retinal organoids in a comprehensive manner and will help to define pathways/genes that could lead to improvements in stem-cell-based organoid models.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Thomas A. Reh (tomreh@uw.edu).
This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Organoid cell culture
Cell culture was done using approved BSL2 practices in a biosafety hood, and all cells were maintained in incubators with 5% CO2. Human pluripotent stem cell lines (H7 BRN3-td Tomato, H9 NRL-GFP, H1) were maintained and differentiated as previously described (Meyer et al., 2009; Zhong et al., 2014; Phillips et al., 2018; Hoshino et al., 2019). Undifferentiated stem cells were maintained as colonies on matrigel coated 6 well plates and supplemented with mTeSR1 (Stem cell technologies) or StemFlex (Thermo Fisher Scientific) media. Upon reaching confluency, cells were passaged for maintenance using dispase (2mg/ml) or ReLeSR (StemCell technologies) using manufacturer’s instructions and split in the ratio of 1:6 every 4–5 days.
For initiating differentiation, confluent colonies of stem cells were passaged with dispase, and allowed to self-aggregate into embryoid bodies (EBs) overnight in T25 flasks with media containing 1:1 of mTeSR1/StemFlex and Neural induction medium (NIM, DMEM/F12, 1% N2 supplement, 1% MEM non-essential amino acids, and 1% Pen strep). By 3 days, EBs were transitioned into complete NIM and maintained in this media up to 20 days of differentiation. To increase the efficiency of retinal induction, BMP4 (0.5μg/μL, R&D systems #314-BP-050) was added at day 6 and was diluted out by day 12 (Phillips et al., 2018). EBs were plated on 6 well plates with 10% FBS on day 7–8 and half medium changes of NIM were done until day 12. Colonies were then dislodged from the plates by day 18–20 either scraping with the back of P1000 tip or by squirting media forcefully with a P1000 tip. The resulting aggregates then self-assembled into retinal organoids withing a couple days and were henceforth maintained in RDM (Retinal differentiation medium, DMEM/DMEM/F12, 2% B27, 1% Pen Strep and 5% FBS). Retinal organoids exhibited a distinct light outer ring and were manually isolated based on their morphology using a P200 pipette, and maintained for long-term cultures in low adhesion 10cm or 6 well plates (Corning). Non-retinal forebrain cells did not exhibit the characteristic retinal morphology, and most of these were manually removed from the cultures by day 40–50. Small pieces of forebrain tissue were occasionally retained in long term cultures, and these were excluded from IF analysis and manually or removed from single cell datasets.
Fetal retina tissue
Human fetal retinal tissue was obtained the Birth Defects Research Laboratory at University of Washington using an approved protocol (UW5R24HD000836). Tissues had no identifiers, and ultrasounds along with physical characteristics such as fetal foot length and crown-rump were used to estimate the age (FitzSimmons et al., 1994).
Retinospheres (in vitro fetal retinal cultures)
Retinospheres were made by dissecting the fetal retina into 300–400 micron pieces with a 30G syringe or mechanically with a pipette or dissecting scissors. These were then maintained in RDM media, as described above for organoids, in low-adhesion 6 well plates/ 10cm dishes. Withing a few days, pieces of the retina rounded up and self-assembled into retinospheres (Figures 7A–7C) For regionspecific retinospheres, parts of the fetal retina were mechanically dissected (similar to the areas used for scRNaseq for the fetal retina) and then made into retinospheres. Retinospheres were cultured using approved BSL2 methods and cells were maintained in 5% CO2 conditions, similar to stem cells.
METHOD DETAILS
1. Tissue dissociation for 10x
Fetal retinas (FD59, FD82 and FD125) were dissected and dissociated using Trypsin for 15–20 minutes. Cell pellets were resuspended in PBS containing 0.04% bovine serum albumin and filtered through a 35μm cell strainer (Fisher, 08–771-23) to remove cell clumps. Organoids or retinospheres were processed similarly.;10–20 organoids or retinospheres were randomly chosen (to account for any variability within the organoids or spheres) and digested using the papain dissociation kit (Worthington, #LK003150).Cells were strained through the cell strainer and counted with a hemocytometer. Typically, 2000–4000 cells of the sample were then input into the 10X protocol. GEM generation, reverse transcription, cDNA amplification, and library construction steps were performed according to manufacturer’s instructions (Chromium Single Cell 3’ v1/v2/v3 platform (10X Genomics, Pleasanton, CA). Sequencing libraries were generated using Illumina 75 cycle high sequencing kit (#20024906) and samples were run on an Illumina NextSeq 500.
2. .scRNaseq analysis
2.1. Data processing
Seurat analysis was performed in R using Seurat (v2.4 Figures 1, 2, and 3) (v3.1 Figures 4, 5, 6, and 7), UMAP (Becht et al., 2018), ggplot2, dplyr. The data was normalized by scaling gene content by cell and log normalizing, and variable features were identified. Datasets combined in Seurat 2 were aligned by performing CCA on the combined datasets, preforming dynamic time warping to align the datasets, and reduced into a lower dimensional space using dimensional reduction (Butler et al., 2018). Datasets combined in Seurat 3 were combined by using mutual nearest neighbors to create anchors, constructing “correction” vectors along the anchors, and transforming data from the second dataset along these vectors as described by Haghverdi et al. (2018); the data was then scaled, PCs were computed, nearest neighbors were determined, and UMAP was performed (Haghverdi et al., 2018). Clustering resolution for all datasets were set so clusters matched well with known cell type markers (Table S5). If a cluster expressed markers from multiple groups the cluster was identified as a hybrid of multiple groups (ex. T1/T3). Cell types were assigned to clusters in the integrated objects. Clusters that expressed forebrain cell markers (such as VSX2-, SOX1/FOXG1+ etc.) were removed before further analysis. Sample pre-processing and integration parameters are recorded in Table S1. Percent cells were calculated by dividing the umber of cells that express the gene of interest in each cluster by the total number of cells in that cluster. Datasets gathered from GEO were processed using the same approach.
2.2. Pseudotime analyses
Monocle 2.99 pseudotime was calculated by setting progenitors at the root state and expression of a gene across select clusters was shown using ggplot2. Slingshot analysis utilized PCA coordinates, with progenitors as the start cluster. For all ages, lineages were redrawn using the cumulative trajectory from Monocle 2.99 and Slingshot. For CCA, the smallest sample size was used as a cutoff to down-sample any large cell datasets and ensure that the same number of cells are sampled across CCA analysis.
3. Sectioning and immunofluorescence
Fetal retinas at younger ages (D50-Day70) were fixed without lens removal with 4% paraformaldehyde (2×20 minutes). Older retinas were first fixed with a small opening in the lens for 20 minutes at room temperature, followed by lens removal and overnight fixation in 4% paraformaldehyde at 4C. Organoids and retinospheres were typically fixed in 4% paraformaldehyde at room temperature for 30 minutes followed by 3 washes in 1X PBS. All samples were then transitioned into 10%, 20% and 30% solutions of sucrose, followed by an additional 30%sucrose +OCT step for the human retina. These were then embedded in OCT and cryosectioned at 14–16 μm.
Immunostaining was performed as previously described (Hoshino et al., 2017). Briefly slides were blocked in 0.5%Triton/10% horse serum for an hour, followed by primary antibody incubation at 4C overnight. The next day, secondary antibody was added after 3 washes of 1X PBS, followed by secondary antibody incubation for an hour at room temperature. Slides were mounted using Fluromount and imaged using the Zeiss 990. For whole mount staining, primary antibody incubation was done for 3–4 days, and secondary was done for 2 hours at room temperature. Samples were then mounted in 2.5% low melting agarose with VectorShield (Vector laboratories, H-1000) using spacers. Image processing was done in ImageJ and Amira (version 6), and Adobe Photoshop.
4. TEM
Samples were fixed in 4% Glutaraldehyde in 0.1M sodium cacodylate buffer and stored at 4C overnight. The tissue was then washed for 5 times 5 minutes each and post fixed in buffered 2% osmium tetroxide for 1 hour on ice. This was followed with washes in ddH20, and en bloc staining in 1% Uranyl Actetate, (aqueous) overnight at 4C. The tissue was washed again next day, and dehydrated in cold 30%, 50%, 70%, and 95% ETOH and then allowed to equilibrate to room temperature. Two changes of 100% ETOH were then done, followed by two changes of propylene oxide. The tissue was then then infiltrated in a 1:1 mixture of propylene oxide: Epon Araldite resin, for 2 hours followed by two changes of fresh Epon Araldite, 2 hours each change. It was then placed in flat embedding molds and polymerized at 60°C overnight.
QUANTIFICATION AND STATISTICAL ANALYSIS
For stacked plots in Figures 4, 5, 6, and 7, percent composition was calculated after integration. To calculate percent composition, each sample was subset from the integrated object, the number of cells in each type within the sample were counted, and the counts were divided by the total number of cells in that sample.
For Figures 6D, 6E, and S7 the fetal and organoid samples were combined separately, and cell type specific clusters were subset from each object. A list of known developmental markers was curated and their expression was represented through feature plots, graphs of cells expressing the gene over time, and average expression. Additionally, t tests were performed on the expression levels of the genes between samples from comparable times for the organoids and fetal retina.
Feature plots had a maximum cutoff of 3. The percent of cells expressing a gene was calculated by taking the number of cells expressing the gene of interest and then dividing by the total number of cells. Average expression was calculated by taking the mean gene expression for each group. When performing t tests of gene expression between integrated objects the objects were merged on the normalized raw counts and scaled together before the test was performed. Bonferroni correction was applied to an uncorrected alpha of 0.05 when testing for significance.
For Figure S7 cell type subsets were taken from the Fetal and Organoid objects, merged together on normalized raw counts, scaled, markers were identified between the fetal and organoid samples, and average expression was calculated for all samples, rows were ordered for readability, lastly gene expression was z-scored by row and plotted in a heatmap
DATA AND CODE AVAILABILITY
The datasets generated during this study are available on GEO: GSE142526. Additional datasets used for comparison are found at GEO: Collin et al. (2019) GSE119893, Kim et al. (2019) GSE119343. Code used in this study are available on Github at https://github.com/Reh-lab/Sridhar-Hoshino-Reh-2019.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ARRESTIN | Novus | Cat# NBP!-37003 RRID: AB_2060085 |
| ATOH7 | Novus | Cat# NBP1-88639; RRID: AB_11034390 |
| BRN3 | Santa Cruz | Cat# sc-6026; RRID: AB_673441 |
| CALB1 | Millipore | Cat# ab1778; RRID: AB_2068336 |
| CALRET | S.W.Ant | Cat# 7699/4; RRID: AB_2313763 |
| ChAT | Millipore | Cat# AB144P; RRID: AB_2079751 |
| FABP7 | Millipore | Cat# ABS184; RRID: AB_10917079 |
| GFP | Abcam | Cat# ab13970; RRID: AB_300798 |
| Go alpha | Millipore | Cat# MAB3073; RRID: AB_94671 |
| HUC/D | Invitrogen | Cat# A-21271; RRID: AB_221448 |
| ML Opsin | Millipore/Chemicon | Cat# AB5407; RRID: AB_177457 |
| NR2E3 | Perseus Proteomics | Cat# PP-H7223-00; RRID: AB_1964331 |
| ONECUT2 | R&D Systems | Cat# AF6294; RRID: AB_10640365 |
| OTX2 | R&D Systems | Cat# BAF1979; RRID: AB_2157171 |
| OTX2 | abcam | Cat# AB21990; RRID: AB_776930 |
| PAX6 | DSHB (Developmental Systems Hybridoma Bank) | Cat# pax6; RRID:AB_528427 |
| PRDM13 | DSHB (Developmental Systems Hybridoma Bank) | Cat# PCRP-PRDM13-2C12; RRID:AB_2618988 |
| PRDM13 | Rabbit | Gift from Dr. Jane Johnson |
| RCVRN | Chemicon | Cat# AB5585; RRID: AB_2253622 |
| RLBP1/CRALBP | Abcam | Cat# Ab15051; RRID: AB_2269474 |
| SNCG | Abnova | Cat# H00006623M01A; RRID: AB_1581209 |
| SW Opsin | Santa Cruz | Cat# Sc-14363; RRID: AB_2158332 |
| SOX2 | Santa Cruz | Cat# SC-17320; RRID: AB_2286684 |
| SOX2 | Abcam | Cat# Ab15830; RRID: Ab15830 |
| TFAP2 alpha | DSHB (Developmental Systems Hybridoma Bank) | Cat# 5E4; RRID: AB_2056333 |
| VGLUT1 | Synaptic Systems | Cat# 135 304; RRID: AB_887878 |
| VSX2 | Santa Cruz | Cat# Sc-21690; RRID: AB_2216006 |
| Donkey anti-mouse 488 | Jackson Immuno | Cat# 715-546-151; RRID: AB_23440850 |
| Donkey anti-mouse 568 | Life Technologies | Cat# A10037;RRID: AB_2534013 |
| Donkey anti-mouse 647 | Jackson Immuno | Cat# 715-605-150; RRID: AB_2340862 |
| Donkey anti-goat 488 | Life Tecnologies | Cat# A11055; RRID: AB_142672 |
| Donkey anti-goat 568 | Life Technologies | Cat# A11057; RRID: AB_142581 |
| Donkey anti-goat 647 | Jackson Immuno | Cat# 705-605-147; RRID: AB_2340437 |
| Donkey anti-rabbit 488 | Life Technologies | Cat# A21206; RRID: AB_141708 |
| Donkey anti-rabbit 568 | Life Technologies | Cat# A100042; RRID: AB_2534017 |
| Donkey anti-rabbit 647 | Thermo Fisher Scientific | Cat# A-31573; RRID: AB_2536183 |
| Donkey anti-sheep 568 | Life Technologies | Cat# A21099; RRID: AB_141474 |
| Donkey anti-sheep 488 | Invitrogen | Cat# A11015; RRID: AB_141362 |
| Donkey anti-sheep 647 | Invitrogen | Cat# A21448; RRID: AB_10374882 |
| Donkey anti-guniea pig 488 | Jackson Immuno | Cat# 706-546-148; RRID: AB_2340473 |
| Donkey anti-guniea pig 594 | Jackson Immuno | Cat# 706-586-148; RRID: AB_2340475 |
| Donkey anti-guniea pig 647 | Jackson Immuno | Cat# 706-606-148; RRID: AB_2340477 |
| DAPI | Sigma | D9542 |
| Biological Samples | ||
| Fetal eyes | Birth Defects Research Laboratory at the University of Washington | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BMP4 | R&D systems | Cat# 314-BP-050 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# RT-15710 |
| Trizol | Invitrogen | Cat# 15596026 |
| Critical Commercial Assays | ||
| Chromium Single Cell 3′ v1/v2/v3 Platform | 10X Genomics | N/A |
| Illumina 75 Cycle High Sequencing Kit | Illumina | #20024906 |
| Papain Dissociation Kit | Worthington | #LK003150 |
| Deposited Data | ||
| Single cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures | This paper | GEO: GSE142526 |
| Experimental Models: Cell Lines | ||
| H1-hESC | WiCell | RRID: CVCL_9771 |
| H7-BRN3-tdTomato, hESC | WiCell (Sluch et al., 2017) | RRID:CVCL_9772 |
| H9-NRLGFP, hESC | WiCell (Phillips et al., 2018) | RRID: CVCL_9773 |
| Software and Algorithms | ||
| Adobe Creative Suite 4 | Adobe | N/A |
| ImageJ | NIH | N/A |
| Microsoft Office 2013 | Microsoft | N/A |
| Monocle 2.99 R package | Trapnell et al., 2014 | https://github.com/cole-trapnell-lab/monocle-release |
| Slingshot 1.2.0 R package | Street et al., 2018 | https://bioconductor.org/packages/release/bioc/html/slingshot.html |
| Seurat 3.1.0 R package | Stuart et al., 2019 | https://github.com/satijalab/seurat |
| Seurat 2.3.4 R package | Butler et al., 2018 | https://github.com/satijalab/seurat |
| umap-learn 0.3.7 Python library | Becht et al., 2018 | https://github.com/lmcinnes/umap |
| Other | ||
| Deconstructing Retinal Organoids: single cell RNA-Seq reveals the cellular components of human pluripotent stem cell-derived retina | Collin et al., 2019 | GEO: GSE119893 |
| Generation, transcriptome profiling, and functional validation of single cells from cone-enriched human retinal organoids | Kim et al., 2019 | GEO: GSE119343 |
Highlights.
scRNA-seq of human retina highlights key developmental transition states
hPSC-derived retinal organoids mimic fetal retinal cellular composition
Inner layers in organoids exhibit differences in gene expression and organization
Fetal retinal cultures resemble organoids but maintain better inner lamination
ACKNOWLEDGMENTS
We would like to thank the Reh and Bermingham-McDonogh lab members and Dr. Joseph A. Brzezinski IV for technical advice and critical feedback. We would also like to thank Edward Parker and Sharm Knetch for assistance with TEM processing and image acquisition. We thank Dr. Donald Zack and Dr. David Gamm for sharing the H7 BRN3-tdTomato and NRL-GFP cell lines, respectively. This project was funded by an ISCRM training grant (to A.S.) and a distinguished investigator award (to T.A.R.) from the Paul G. Allen Family Foundation. Fetal retina tissue samples with no identifiers were made available through the Birth Defects Research Laboratory at University of Washington (UW5R24HD000836).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.01.007.
REFERENCES
- Bae S, Bessho Y, Hojo M, and Kageyama R (2000). The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 127, 2933–2943. [DOI] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44. [DOI] [PubMed] [Google Scholar]
- Bessodes N, Parain K, Bronchain O, Bellefroid EJ, and Perron M (2017). Prdm13 forms a feedback loop with Ptf1a and is required for glycinergic amacrine cell genesis in the Xenopus Retina. Neural Dev. 12, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, and Glaser T (1998). Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development 125, 4821–4833. [DOI] [PubMed] [Google Scholar]
- Brzezinski JA, and Reh TA (2015). Photoreceptor cell fate specification in vertebrates. Development 142, 3263–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JAT 4th, Lamba DA, and Reh TA (2010). Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JAT 4th, Prasov L, and Glaser T (2012). Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol 365, 395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski EE, Simonett JM, Clark EM, Wright LS, Howden SE, Wallace KA, Petelinsek AM, Pinilla I, Phillips MJ, Meyer JS, et al. (2014). Loss of MITF expression during human embryonic stem cell differentiation disrupts retinal pigment epithelium development and optic vesicle cell proliferation. Hum. Mol. Genet 23, 6332–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski EE, Samimi K, Mayerl SJ, Phillips MJ, Pinilla I, Howden SE, Saha J, Jansen AD, Edwards KL, Jager LD, et al. (2019). Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 146, dev171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman G, Sparrow DB, Kremmer E, and Dunwoodie SL (2011). Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum. Mol. Genet, 20, 905–916. [DOI] [PubMed] [Google Scholar]
- Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE, et al. (2019). Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 102, 1111–1126.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin J, Queen R, Zerti D, Dorgau B, Hussain R, Coxhead J, Cockell S, and Lako M (2019). Deconstructing Retinal Organoids: Single Cell RNASeq Reveals the Cellular Components of Human Pluripotent Stem Cell-Derived Retina. Stem Cells 37, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WL, Gao ML, Lei XL, Lv JN, Zhao H, He KW, Xia XX, Li LY, Chen YC, Li YP, et al. (2018). Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients. Stem Cell Reports 10, 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field GD, and Chichilnisky EJ (2007). Information processing in the primate retina: circuitry and coding. Annu. Rev. Neurosci 30, 1–30. [DOI] [PubMed] [Google Scholar]
- FitzSimmons J, Fantel A, and Shepard TH (1994). Growth parameters in mid-trimester fetal Turner syndrome. Early Hum. Dev 38, 121–129. [DOI] [PubMed] [Google Scholar]
- Fligor CM, Langer KB, Sridhar A, Ren Y, Shields PK, Edler MC, Ohlemacher SK, Sluch VM, Zack DJ, Zhang C, et al. (2018). Three-Dimensional Retinal Organoids Facilitate the Investigation of Retinal Ganglion Cell Development, Organization and Neurite Outgrowth from Human Pluripotent Stem Cells. Sci. Rep 8, 14520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, et al. (2006). Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development 133, 4439–4450. [DOI] [PubMed] [Google Scholar]
- Georgi SA, and Reh TA (2011). Dicer is required for the maintenance of notch signaling and gliogenic competence during mouse retinal development. Dev. Neurobiol 71, 1153–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Cordero A, Kruczek K, Naeem A, Fernando M, Kloc M, Ribeiro J, Goh D, Duran Y, Blackford SJI, Abelleira-Hervas L, et al. (2017). Recapitulation of Human Retinal Development from Human Pluripotent Stem Cells Generates Transplantable Populations of Cone Photoreceptors. Stem Cell Reports 9, 820–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson NB, Nahreini J, Randazzo G, Uruena A, Johnson JE, and Brzezinski JAT 4th. (2018). Prdm13 is required for Ebf3+ amacrine cell formation in the retina. Dev. Biol 434, 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanotel J, Bessodes N, Thé lie A, Hedderich M, Parain K, Van Driessche B, Brandaõ Kde.O., Kricha S, Jorgensen MC, Grapin-Botton A, et al. (2014). The Prdm13 histone methyltransferase encoding gene is a Ptf1aRbpj downstream target that suppresses glutamatergic and promotes GABAergic neuronal fate in the dorsal neural tube. Dev. Biol 386, 340–357. [DOI] [PubMed] [Google Scholar]
- Hendrickson A, Possin D, Vajzovic L, and Toth CA (2012). Histologic development of the human fovea from midgestation to maturity. Am. J. Ophthalmol 154, 767–778.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon M, Okawa H, Della Santina L, and Wong RO (2014). Functional architecture of the retina: development and disease. Prog. Retin. Eye Res 42, 44–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Cholewa-Waclaw J, Nakada Y, Glasgow SM, Masui T, Henke RM, Wildner H, Martarelli B, Beres TM, Epstein JA, et al. (2008). A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 22, 166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Ratnapriya R, Brooks MJ, Chaitankar V, Wilken MS, Zhang C, Starostik MR, Gieser L, La Torre A, Nishio M, et al. (2017). Molecular Anatomy of the Developing Human Retina. Dev. Cell 43, 763–779.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Horvath S, Sridhar A, Chitsazan A, and Reh TA (2019). Synchrony and asynchrony between an epigenetic clock and developmental timing. Sci. Rep 9, 3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wang X, Hu B, Mao Y, Chen Y, Yan L, Yong J, Dong J, Wei Y, Wang W, et al. (2019). Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 17, e3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav AP, Mason HA, and Cepko CL (2006). Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133, 913–923. [DOI] [PubMed] [Google Scholar]
- Jhas S, Ciura S, Belanger-Jasmin S, Dong Z, Llamosas E, Theriault FM, Joachim K, Tang Y, Liu L, Liu J, and Stifani S (2006). Hes6 inhibits astrocyte differentiation and promotes neurogenesis through different mechanisms. J. Neurosci 26, 11061–11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf PR, and Harris WA (2009). Ptf1a is expressed transiently in all types of amacrine cells in the embryonic zebrafish retina. Neural Dev. 4, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkhaw R, Kaya KD, Brooks M, Homma K, Zou J, Chaitankar V, Rao M, and Swaroop A (2015). Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks. Stem Cells 33, 3504–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Omori Y, Onishi A, Sato S, Kondo M, and Furukawa T (2010). Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J. Neurosci 30, 6515–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya KD, Chen HY, Brooks MJ, Kelley RA, Shimada H, Nagashima K, de Val N, Drinnan CT, Gieser L, Kruczek K, et al. (2019). Transcriptome-based molecular staging of human stem cell-derived retinal organoids uncovers accelerated photoreceptor differentiation by 9-cis retinal. Mol. Vis 25, 663–678. [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lowe A, Dharmat R, Lee S, Owen LA, Wang J, Shakoor A, Li Y, Morgan DJ, Hejazi AA, et al. (2019). Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc. Natl. Acad. Sci. USA 116, 10824–10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H, Nelson R, Ahnelt P, and Cuenca N (2001). Cellular organization of the vertebrate retina. Prog. Brain Res 131, 3–26. [DOI] [PubMed] [Google Scholar]
- Langer KB, Ohlemacher SK, Phillips MJ, Fligor CM, Jiang P, Gamm DM, and Meyer JS (2018). Retinal Ganglion Cell Diversity and Subtype Specification from Human Pluripotent Stem Cells. Stem Cell Reports 10, 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer PG, Robitzki A, Rothermel A, and Willbold E (2002). Of layers and spheres: the reaggregate approach in tissue engineering. Trends Neurosci. 25, 131–134. [DOI] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, and Brown NL (2006). Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev. Biol 295, 764–778. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, and Cepko CL (2001). Vertebrate neural cell-fate determination: lessons from the retina. Nat. Rev. Neurosci 2, 109–118. [DOI] [PubMed] [Google Scholar]
- Mellough CB, Bauer R, Collin J, Dorgau B, Zerti D, Dolan DWP, Jones CM, Izuogu OG, Yu M, Hallam D, et al. (2019). An integrated transcriptional analysis of the developing human retina. Development 146, dev169474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JS, Shearer RL, Capowski EE, Wright LS, Wallace KA, McMillan EL, Zhang SC, and Gamm DM (2009). Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 106, 16698–16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JS, Howden SE, Wallace KA, Verhoeven AD, Wright LS, Capowski EE, Pinilla I, Martin JM, Tian S, Stewart R, et al. (2011). Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells 29, 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, and Klein WH (2005). A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev. Biol 280, 467–481. [DOI] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, and Sasai Y (2012). Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785. [DOI] [PubMed] [Google Scholar]
- Nelson BR, and Reh TA (2008). Relationship between Delta-like and proneural bHLH genes during chick retinal development. Dev. Dyn 237, 1565–1580. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Georgi SA, Lan MS, and Reh TA (2007). Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev. Biol 304, 479–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Ray CA, Hayashi T, Bermingham-McDonogh O, and Reh TA (2009). Acheate-scute like 1 (Ascl1) is required for normal delta-like (Dll) gene expression and notch signaling during retinal development. Dev. Dyn 238, 2163–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Ueki Y, Reardon S, Karl MO, Georgi S, Hartman BH, Lamba DA, and Reh TA (2011). Genome-wide analysis of Mu€ller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS ONE 6, e22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KM, Schulte D, and Hendrickson AE (2003). Expression of photoreceptor-associated molecules during human fetal eye development. Mol. Vis 9, 401–409. [PubMed] [Google Scholar]
- Ohlemacher SK, Iglesias CL, Sridhar A, Gamm DM, and Meyer JS (2015). Generation of highly enriched populations of optic vesicle-like retinal cells from human pluripotent stem cells. Curr. Protoc. Stem Cell Biol 32, 1H.8.1–1H.8.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt DA, Lane A, Ramsden CM, Carr AJ, Munro PM, Jovanovic K, Schwarz N, Kanuga N, Muthiah MN, Hull S, et al. (2016). Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell 18, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Wallace KA, Dickerson SJ, Miller MJ, Verhoeven AD, Martin JM, Wright LS, Shen W, Capowski EE, Percin EF, et al. (2012). Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest. Ophthalmol. Vis. Sci 53, 2007–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Perez ET, Martin JM, Reshel ST, Wallace KA, Capowski EE, Singh R, Wright LS, Clark EM, Barney PM, et al. (2014). Modeling human retinal development with patient-specific induced pluripotent stem cells reveals multiple roles for visual system homeobox 2. Stem Cells 32, 1480–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Capowski EE, Petersen A, Jansen AD, Barlow K, Edwards KL, and Gamm DM (2018). Generation of a rod-specific NRL reporter line in human pluripotent stem cells. Sci. Rep 8, 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn PM, Buck TM, Mulder AA, Ohonin C, Alves CH, Vos RM, Bialecka M, van Herwaarden T, van Dijk EHC, Talib M, et al. (2019). Human iPSC-Derived Retinas Recapitulate the Fetal CRB1 CRB2 Complex Formation and Demonstrate that Photoreceptors and Mu€ller Glia Are Targets of AAV5. Stem Cell Reports 12, 906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman S, Slembrouck A, Gagliardi G, Chaffiol A, Terray A, Nanteau C, Potey A, Belle M, Rabesandratana O, Duebel J, et al. (2017). Generation of Storable Retinal Organoids and Retinal Pigmented Epithelium from Adherent Human iPS Cells in Xeno-Free and Feeder-Free Conditions. Stem Cells 35, 1176–1188. [DOI] [PubMed] [Google Scholar]
- Saengwimol D, Rojanaporn D, Chaitankar V, Chittavanich P, Aroonroch R, Boontawon T, Thammachote W, Jinawath N, Hongeng S, and Kaewkhaw R (2018). A three-dimensional organoid model recapitulates tumorigenic aspects and drug responses of advanced human retinoblastoma. Sci. Rep 8, 15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma TP, Wiley LA, Whitmore SS, Anfinson KR, Cranston CM, Oppedal DJ, Daggett HT, Mullins RF, Tucker BA, and Stone EM (2017). Patient-specific induced pluripotent stem cells to evaluate the pathophysiology of TRNT1-associated Retinitis pigmentosa. Stem Cell Res.(Amst.) 21, 58–70. [DOI] [PubMed] [Google Scholar]
- Silverman SM, and Wong WT (2018). Microglia in the Retina: Roles in Development, Maturity, and Disease. Annu. Rev. Vis. Sci 4, 45–77. [DOI] [PubMed] [Google Scholar]
- Sluch VM, Chamling X, Liu MM, Berlinicke CA, Cheng J, Mitchell KL, Welsbie DS, and Zack DJ (2017). Enhanced Stem Cell Differentiation and Immunopurification of Genome Engineered Human Retinal Ganglion Cells. Stem Cells Transl. Med 6, 1972–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, and Dudoit S (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3RD, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Tan QS, Parikh BH, Tan A, Mehta MN, Sia Wey Y, Tun SB, Li LJ, Han XY, Wong TY, et al. (2016). Characterization of Fatty Acid Binding Protein 7 (FABP7) in the Murine Retina. Invest. Ophthalmol. Vis. Sci 57, 3397–3408. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt AP, Whitmore SS, Flamme-Wiese MJ, Riker MJ, Wiley LA, Tucker BA, Stone EM, Mullins RF, and Scheetz TE (2019). Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp. Eye Res 184, 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vö lkner M, Zschä tzsch M, Rostovskaya M, Overall RW, Busskamp V, Anastassiadis K, and Karl MO (2016). Retinal Organoids from Pluripotent Stem Cells Efficiently Recapitulate Retinogenesis. Stem Cell Reports 6, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlin KJ, Maruotti JA, Sripathi SR, Ball J, Angueyra JM, Kim C, Grebe R, Li W, Jones BW, and Zack DJ (2017). Photoreceptor Outer Segment-like Structures in Long-Term 3D Retinas from Human Pluripotent Stem Cells. Sci. Rep 7, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Sanuki R, Sugita Y, Imai W, Yamazaki R, Kozuka T, Ohsuga M, and Furukawa T (2015). Prdm13 regulates subtype specification of retinal amacrine interneurons and modulates visual sensitivity. J. Neurosci 35, 8004–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welby E, Lakowski J, Di Foggia V, Budinger D, Gonzalez-Cordero A, Lun ATL, Epstein M, Patel A, Cuevas E, Kruczek K, et al. (2017). Isolation and Comparative Transcriptome Analysis of Human Fetal and iPSC-Derived Cone Photoreceptor Cells. Stem Cell Reports 9, 1898–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore SS, Wagner AH, DeLuca AP, Drack AV, Stone EM, Tucker BA, Zeng S, Braun TA, Mullins RF, and Scheetz TE (2014). Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp. Eye Res 129, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D, Liu X, Zhang M, Zou M, Deng Q, Sun D, Bian X, Cai Y, Guo Y, Liu S, et al. (2018). Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat. Commun 9, 2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, and Gan L (2003). Math5 determines the competence state of retinal ganglion cell progenitors. Dev. Biol 264, 240–254. [DOI] [PubMed] [Google Scholar]
- Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH, Peters A, Park TS, Zambidis ET, Meyer JS, et al. (2014). Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun 5, 4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available on GEO: GSE142526. Additional datasets used for comparison are found at GEO: Collin et al. (2019) GSE119893, Kim et al. (2019) GSE119343. Code used in this study are available on Github at https://github.com/Reh-lab/Sridhar-Hoshino-Reh-2019.