Abstract
In vivo RNA structure analysis has become a powerful tool in molecular biology, largely due to the coupling of an increasingly diverse set of chemical approaches with high-throughput sequencing. This has resulted in a transition from single target to transcriptome-wide approaches. However, these methods require sequencing depths that preclude studying low abundance targets, which are not sufficiently captured in transcriptome-wide approaches. Here we demonstrate that enrichment of low abundance targets before reverse transcription broadens the range of molecules analyzed and results in improved analysis for low abundance transcripts. In addition, this method is compatible with any choice of chemical adduct or read-out approach. We combine this method with inducible expression of an RBP of interest to study an autoregulated event in the pre-mRNA of the splicing factor, muscleblind-like splicing regulator 1 (MBNL1) in a cellular context.
Keywords: RNA structure, SHAPE-MaP, ultraconserved elements, muscleblind
INTRODUCTION
RNA molecules have been identified to play critical roles in nearly every aspect of basic and disease biology. Crucial to RNA's role is its ability to fold into complex two and three-dimensional structures. Further, as an RNA is made and traverses throughout the cell in its lifetime, it is constantly interacting with RNA binding proteins (RBPs), which can alter structure and function. As such, characterizing RNA structure, its interaction with proteins, and its changes to structure when bound to a protein is extremely important, but very challenging, especially in the cellular setting.
The majority of work characterizing RNA structure has been limited to work in vitro. However, extensions of structure probing methods are beginning to be used in the complex cellular environment. The development of new chemistries that expand the repertoire of interactions that can be probed (for reviews, see Strobel et al. 2018; Mitchell et al. 2019a) and its combination with high-throughput sequencing approaches are expanding the limitations of RNA structure probing methods. For example, it is now possible to probe all four ribonucleobases at the Watson–Crick interface by combining dimethyl sulfate (DMS) (Peattie and Gilbert 1980; Schroder et al. 1998) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) (Mitchell et al. 2019b; Wang et al. 2019a). Another powerful approach is to interrogate the ribose sugar backbone, independently of nucleobase identity as in selective 2′hydroxyl acylation and primer extension (SHAPE) and several SHAPE reagents have been developed for in vivo use including 1-methyl-7-nitroisatoic anhydride (1M7) (Smola et al. 2015a) and 2-methylnicotinic acid imidazolide (NAI) (Spitale et al. 2013). The addition of these chemical adducts results in either a block to reverse transcription (RT-stop) or the introduction of a mutation at the site, depending on the methodology used. The location and abundance of either the stop or mutation is identified by high-throughput sequencing. Various combinations of chemical probes and read-out strategies allow for multiple approaches for RNA structure analysis reviewed in (Strobel et al. 2018).
Mutational readthrough approaches such as SHAPE-MaP (Smola et al. 2015b) and DMSMaPseq (Zubradt et al. 2017), which permit the interrogation of more than one adduct per molecule, allows structural changes within the same molecule to be assayed and have emerged as effective technologies for abundant, short, or highly structured targets. They have been successfully leveraged to examine several purified or capsid-associated viral genome structures (Siegfried et al. 2014; Larman et al. 2017; Dethoff et al. 2018; Kutchko et al. 2018; Guo et al. 2020; Tomezsko et al. 2020; Zhou and Routh 2020). In a cellular context, abundant mRNAs such as beta actin (Woods et al. 2017), IFNL3 (Lu et al. 2015), and chloroplast mRNAs (Gawronski et al. 2020), or well-expressed lncRNAs such as XIST (Smola et al. 2016), PAN (Sztuba-Solinska et al. 2017), MEG3 (Sherpa et al. 2018), and GAS5 (Frank et al. 2020) have been studied in detail. These approaches have also been leveraged for transcriptome-wide studies in vivo (Zubradt et al. 2017; Mustoe et al. 2018; Wang et al. 2019b). However, given that broad target coverage necessitates a sacrifice in read depth, low abundance RNA species are not captured on this scale. Gaps in the coverage include both lowly expressed fully processed RNAs and many intronic sequences within pre-mRNAs. Introns are information-rich regions and in addition to the requisite signals for recruiting spliceosomal machinery, they may contain regulatory elements including intronic splicing enhancers (ISEs) and intronic splicing silencers (ISSs) and additional regulatory RNAs such as lncRNAs, snoRNAs, and miRNA can also be resident within introns. The information contained in this missing segment of the transcriptome presents a gap in our knowledge of the role of structure in RNA metabolism.
While many introns are constitutively spliced, alternative splicing occurs in 90%–95% of human genes (Pan et al. 2008; Wang et al. 2008). The balance between alternative isoforms is driven largely by interactions with RBPs. An individual RBP may have many potential binding sites on an RNA, intermixed with binding sites for numerous other RBPs, resulting in combinatorial systems of binding sites and interacting factors that allow exquisite control of cellular pathways reviewed in Ule and Blencowe (2019). Regulation of alternative splicing is a powerful driver of cellular biology as isoform changes can alter the function, localization or stability of the encoded protein and/or the RNA itself. For discussion of many excellent studies please see reviews by (Kelemen et al. 2013; Zheng and Black 2013; Mockenhaupt and Makeyev 2015). RNA mis-splicing is now understood to drive pathology in many human diseases as reviewed in (Scotti and Swanson 2016; Montes et al. 2019). Coordinated networks of alternative splicing have been demonstrated to play powerful roles in cellular metabolism. One elegant example is the regulation of the SR protein family of splicing regulators. In addition to their protein coding isoforms, each family member can also produce an alternatively spliced isoform that is sensitive to degradation by nonsense mediated decay (AS-NMD) (Lareau et al. 2007; Ni et al. 2007). The regulated inclusion of exons containing premature termination codons (PTCs) in the SR RNA transcript is itself controlled by interactions with SR proteins, often in an autoregulatory manner. These PTC-containing exons are often embedded in ultraconserved elements (UCEs) in the genome, which were originally defined as >200 nt of 100% conservation between the human, mouse and rat genomes (Bejerano et al. 2004). Exon-spanning UCEs are enriched in genes encoding RNA-binding proteins and splicing factors (Bejerano et al. 2004) and are often associated with a regulatory alternative splicing event. The extreme conservation of these regulatory elements as well as the interconnectedness of the network underscores the importance of controlling splicing factor expression in the cell.
Alternative splicing is also a potent regulator of development, reviewed in (Baralle and Giudice 2017; Gallego-Paez et al. 2017), with widespread changes between fetal and adult isoforms, often in specific temporal or spatial patterns. For example, widespread intron retention is a naturally occurring phenomenon in various developmental contexts, reviewed in (Jacob and Smith 2017). An intriguing study has recently demonstrated that coordinated alternative splicing of a pair of enzymes with opposing functions in the GlcNAc pathway can control a large set of detained intron splicing events in response to the cellular status of O-GlcNAc (Tan et al. 2020). O-GlcNAcase (OGA) and O-linked N-acetylglucosamine transferase (OGT) alternate between a productive protein coding form and an AS-NMD form in response to alternative splicing events in a reciprocal manner. The OGT splicing event is also found embedded in a UCE and this conserved, highly regulated pair of events permits the cells to transduce changes in metabolic state into widespread alterations in splicing and gene expression. In all of these examples, little is known about structural contributions to regulatory decisions.
Given the importance of intron regulation to cellular functions, the inability to widely assess structures in low abundance RNAs is a barrier in the field. While individual binding motifs have been derived for many RBPs (Ray et al. 2013; Dominguez et al. 2018), there are often many more instances of a particular consensus sequence than appear to be utilized. As binding sites are typically predicted based on linear RNA sequence, an understanding of the local topology of the RNA around the binding site within the cell would aid in discriminating between biologically active and inactive motifs and would enhance our understanding of regulation of alternative splicing events. RNA structures or RBP binding within the intronic regions can positively or negatively regulate access to these sites, influencing which isoforms of mRNA and its encoded protein are produced, ultimately having a profound effect on cellular functions, reviewed in Bartys et al. (2019). One example is cardiac troponin T (cTNT) where alternative splicing of exon 5 is controlled via competition between muscleblind-like splicing regulator 1 (MBNL1) and U2AF65 (Warf et al. 2009). The 3′ end of the cTNT upstream intron 4 may be single-stranded or it can form a stem–loop structure. Interaction with U2AF65, which promotes inclusion of exon 5, requires a single-stranded region, whereas MBNL1 interacts with the stem–loop structure to promote exclusion of exon 5. This event clearly defines the importance of the interplay between RNA structure and RBP binding.
A major challenge in interrogating low abundance RNA transcripts during or before splicing is understanding the contribution of RBPs to RNA structure and also changes in RNA structure due to protein recognition. Further, designing systematic and controlled experimental systems for the analysis of presplicing transcripts and their interactions with proteins, through the controlled expression of a given RBP, have yet to be demonstrated in the literature.
Here, we report the development of an enrichment protocol for very low abundance targets that is compatible with all sequencing-based RNA structure methodologies. As a proof of concept, we chose SHAPE-MaP to examine an autoregulated alternative splicing event within the splicing factor MBNL1. This region contains a 212 nucleotide (nt) UCE; however, there is no premature termination codon introduced upon alternative splicing in the Mbnl1 transcript. Instead of affecting the RNA levels, the inclusion of exon 5 alters the subcellular localization of MBNL1 protein, thereby influencing its ability to act as a splicing regulator. MBNL1 is important for the transition from fetal to adult splicing patterns and its loss of function is a driver of pathology in myotonic dystrophy type 1 and 2 (for review, see Scotti and Swanson 2016).
For the first time, we demonstrate the utility of controlled protein expression merged with RNA structure probing in living cells. We utilize a mouse embryonic fibroblast (MEF) cell line with doxycycline-inducible MBNL1 expression to examine changes in local structure from upstream of the branchpoint in intron 4 to the end of a UCE in intron 5, including a previously defined 90 nt intronic control sequence with multiple MBNL1 binding sites (Gates et al. 2011; Wagner et al. 2016).
Our results demonstrate that Mbnl1 exon 5 is highly structured in cells but also revealed an unanticipated alteration in the RNA structure of the branchpoint region, which indicates MBNL binding reduces, or blocks, branchpoint region accessibility to regulate alternative splicing of exon 5. Overall, these results reveal novel aspects of RNA structure-controlled protein recognition in splicing and also present an experimental framework for interrogating low abundance RNAs or RNA–protein interactions in living cells through structure probing.
RESULTS
MBNL1 exon 5 autoregulation
A prominent example of RBP-mediated regulation is provided by the MBNL protein family (MBNL1, MBNL2, and MBNL3), an evolutionarily conserved set of factors that function in the developmental regulation of RNA alternative splicing (Ho et al. 2004; Pascual et al. 2006), alternative polyadenylation (Batra et al. 2014), and localization (Wang et al. 2012). Loss of MBNL1 activity results in reversion to fetal splicing patterns and disease-associated pathology in the neuromuscular disorder myotonic dystrophy types 1 and 2 (DM1 and DM2), where expression of a dense array of MBNL1 YGCY binding sites results in protein sequestration and spliceopathy (for review, see Sznajder and Swanson 2019).
Among its many RNA targets, MBNL1 autoregulates the inclusion of a 54 nt alternative exon in its own pre-mRNA (exon 5) (Gates et al. 2011), with high levels of MBNL1 protein resulting in skipping of exon 5 (Fig. 1A). Inclusion of exon 5 adds an additional 18 amino acids that encode an essential segment of a multipartite nuclear localization sequence (NLS) resulting in a change in the protein's subcellular localization to predominantly nuclear. The consensus splicing signals that govern this event are atypical with a far-distal branchpoint positioned at −141, and an AAG 3′ splice site (Fig. 1B). Interestingly, this region of the genome also harbors a 212 nt UCE that spans exon 5 and includes an intronic sequence on either side. The preservation of this sequence reinforces the functional importance of this autoregulatory alternative splicing event within MBNL1.
FIGURE 1.

Experimental design to study autoregulation of MBNL1 exon 5 in Mbnl1−/−: Mbnl2−/− MEF cell line. (A) Schematic of the MBNL1 autoregulation of exon 5. Constitutive flanking exons are depicted in blue while alternative exon 5 is shown in purple. The lengths of the flanking introns are shown in nucleotides. Binding of MBNL1 protein upstream of exon 5 inhibits its inclusion into mRNA. (B) Atypical splicing signals controlling splicing of intron 4. This event contains a far-distal branchpoint 141 nt from the weak AAG 3′splice site. The position of the ultraconserved element (UCE) is shown by a rectangle. The green vertical marks denote positions of YGCY binding motifs for MBNL1, and the purple box shows the location of exon 5. The relative locations of the forward (FP) and reverse primers (RP) are shown with triangles. (C) Sequence of the MBNL1 region analyzed in this study, highlighting the motifs involved in autoregulation of exon 5. The green highlights depict the YGCY motifs, the purple highlight is exon 5, and the branchpoint and splice sites flanking exon 5 are shown in orange. Sequences within the boxes represent primer binding sites. (D) Experimental design for in cell RNA structure probing. Mbnl1−/−;Mbnl2−/− double knockout MEF cells were reconstituted with a doxycycline-inducible MBNL1 which allows precise control over MBNL1 protein expression and then treated with DMSO or NAI as indicated. (E–J) Induced MBNL1 regulates alternative splicing. RT-PCR analysis of MEFs shows MBNL1 protein is functioning as expected and can inhibit (Mbnl1, Add3) (E–H) or enhance (Plod2) (I,J) inclusion of an alternative exon. Panels E, G, and I are duplicate RT-PCR reactions analyzed by capillary electrophoresis (C,E), and the corresponding percent spliced in quantifications (ψ) are shown on the right (F,H, and J).
Our previous work found that MBNL1 protein negatively regulates the inclusion of exon 5 by interacting with a 90 nt region between the branchpoint and the 3′ splice site of intron 4 (Gates et al. 2011; Wagner et al. 2016). This region contains multiple MBNL1 binding YGCY motifs (Fig. 1B,C) and deletion of this 90 nt region in a minigene splicing reporter resulted in a shift to near complete inclusion of exon 5 into the mRNA, and resistance to regulation by MBNL1 protein expression. Smaller deletions scanning this region demonstrated that an 18 nt deletion in the middle of this sequence that harbors a single MBNL1 binding site (deletion 3) also resulted in near complete inclusion of exon 5. The key nature of this single binding site was not predicted by linear sequence alone as the clustered motifs contained in this sequence were anticipated to have broader effects, therefore we were interested in probing this region to assess the contribution of RNA structure to the regulation of this splicing event.
As cotranscriptional RBP assembly and the stoichiometry of the interacting partners is central to splicing regulation, we chose to study endogenous Mbnl1 pre-mRNA transcripts. This strategy avoids variations across cell populations due to uneven uptake of transfected plasmids and potential carry over of plasmids into downstream sequencing pipelines. Our laboratory has developed an inducible Mbnl1−/−; Mbnl2−/− double knockout mouse embryonic fibroblast (MEF) cell line where MBNL1 protein expression is controlled by an integrated doxycycline-inducible MBNL1 construct, which allows precise control of MBNL1 protein expression (Fig. 1D). The Mbnl1 knockout removes the translational start sites for the protein, but the downstream transcript is intact thereby allowing analysis of endogenous Mbnl1 transcripts in this system and the response of exon 5 inclusion to MBNL1 protein levels remains intact in these cells with and without doxycycline (Fig. 1E,F).
For our initial experiment, we treated a set of MEFs with doxycycline for 18 h to induce MBNL1 expression, while maintaining an untreated set for comparison. These cells were then subjected to SHAPE treatment with 2′-methylnicotinic acid imidazolide (NAI) or DMSO only as described in Materials and Methods. Before performing the full experimental pipeline, we confirmed that MBNL1 expression was regulating alternative splicing as expected under our conditions. DMSO treated RNA samples were used for RT-PCR analysis of the MBNL1 sensitive splicing targets Add3 (Fig. 1G) and Plod2 (Fig. 1I), an exclusion and an inclusion event, respectively. The quantitation of the percent spliced in (PSI) for the alternative cassette exon is shown for each target (Fig. 1H,J). These results demonstrate that both inclusion and exclusion activities are occurring as anticipated upon induction of MBNL1 expression with doxycycline.
Our initial experiment utilized a two-pronged approach of performing a transcriptome-wide experiment using rRNA depleted samples, as well as a targeted amplicon approach for Mbnl1 from the sample RNA samples, as previously described (Smola et al. 2015b). The region of Mbnl1 probed starts upstream of the branchpoint in intron 4, spans exon 5 and ends after the UCE in intron 5 (mm10, chr3: 60,614,506–560,614,916) (Fig. 1C). Unsurprisingly, the transcriptome-wide data was dominated by snRNAs, lncRNAs such as Rn7SK and Rpph1, the RNA component of RNase P and abundant mRNAs such as Actb, and Col1a1. We analyzed Rn7SK and Rpph1 to validate our SHAPE-MaP pipeline and the data generated was high quality with low variation and high reproducibility (Fig. 2A,B). Broadly, SHAPE-Map reactivities are determined by the frequency of adduct-induced mutations in a particular position compared to untreated controls, and reactivities are categorized as high (blue), medium (orange) or low (black). For in depth mathematical details please refer to (Siegfried et al. 2014). Positions with high reactivity are more frequently modified by the NAI adduct and represent accessible nucleotides within the structure across the transcript population average. We mapped our reactivity data onto consensus structure models of Rn7SK (Fig. 2D; Marz et al. 2009) and Rpph1 (Fig. 2E; Esakova and Krasilnikov 2010) and found good concordance between the structures and our SHAPE-MaP data.
FIGURE 2.
SHAPE-MaP reactivity profiles from MEF cells without doxycycline treatment. (A,B) 100 nt regions of SHAPE-MaP profiles from transcriptome-wide profiling data from a region of Rn7SK (A) or Rrph1(B). High reactivity positions are shown in blue, medium in orange, and low in black. (C) SHAPE-MaP profile of MBNL1 amplicon from same RNA samples as above. (D,E) MEF transcriptome-wide SHAPE-MaP reactivity data mapped onto consensus structural models of Rn7SK (D) or Rpph1 (E).
Given that Mbnl1 is not sufficiently detected in the transcriptome-wide data set, we analyzed the Mbnl1 data from the amplicon-based library preparations. In contrast, the SHAPE-MaP reactivity profile was noisy, exhibiting high variation in the structure probing data, despite having a similar (Rn7SK) or greater (Rpph1) read depth to the RNA test cases from the transcriptome-wide library (Fig. 2). One major difference among these RNAs is their endogenous abundance within the cell. The power of mutational read-through strategies such as SHAPE-MaP relies on the ability to achieve significant read depths across a region of interest, where mutations within the reads encode the information about the flexibility of the RNA. The accuracy of this method mandates that the library contains a reasonable sampling of the treated population to enable encoding of all of the structural information into the library. During treatment with a SHAPE reagent, 2′ hydroxyl-acylation occurs in regions where the RNA is flexible and can adopt the proper conformation for the reaction to occur (Merino et al. 2005). This results in covalent adducts that mark the position of the flexible nucleotide and during reverse transcription, in the presence of manganese to reduce the fidelity of the reverse transcriptase, mutations will be introduced into the DNA at the positions of these adducts (Siegfried et al. 2014). As a result, all of the information about the structural complexity of a sample of interest is already encoded at this step. In the case of an intronic target in a lowly expressed transcript, the number of target molecules that would be included in a total RNA sample for a typical reverse transcription reaction does not sufficiently capture the landscape of potential mutations resulting in under-sampling of the data (Fig. 3). One approach would be to increase the amount of total RNA into the reaction; however, the maximum recommended input of total RNA for a typical SuperScript II reaction is 5 µg and these enzymatic reactions do not scale up efficiently. In many cases, it is not feasible to increase the input sufficiently to meaningfully increase the presence of a low abundance target in the reaction. Therefore, we decided to develop a method to enrich for the presence of low abundance RNA in our SHAPE-MaP pipeline before the information is encoded during reverse transcription.
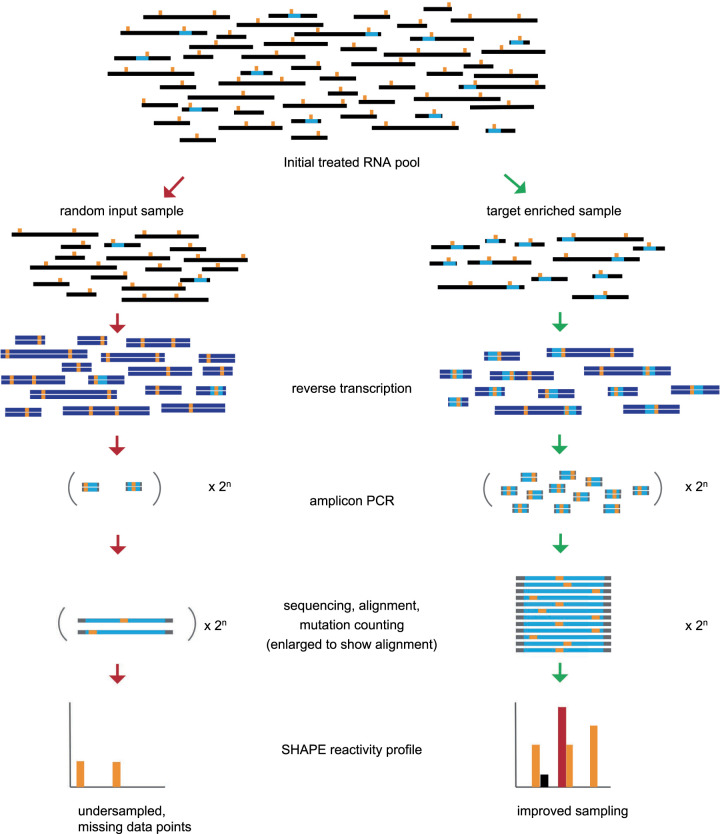
FIGURE 3.
Model of signal improvement for low abundance sequences due to enrichment of target before the reverse transcription step. Random length RNAs are depicted as black lines while the sequence of interest is shown in light blue. The location of the chemical adducts are shown as orange vertical lines. Double-stranded DNA molecules are shown in dark blue with the target sequence in light blue and location of mutations shown in orange. Gray ends of amplicons represent primer sequences that do not contain structural information.
Development of an enrichment strategy for low abundance targets compatible with sequencing-based structure probing
We chose to use a hybridization-capture approach to enrich for the MBNL1 regulatory region. We designed a set of anti-sense oligos that tile across the sequence plus an extra 50 bp on each side to account for decreased sequencing depths at the ends of the amplicon due to Nextera-based library construction. We used the online oligo probe designer at singlemoleculefish.com to design an array of 20 nt anti-sense oligos, to which we then appended a 3′ Biotin-TEG. This resulted in a set of 17 antisense oligos for our target that were combined to make a pool for hybridization (Fig. 4A). Given that MBNL2 is a close paralog of MBNL1 and also contains the UCE, we would expect that Mbnl2 transcripts will also be captured in the hybridization process. This is not problematic as the goal of hybridization is not purification, but rather to enrich for the presence of the target. The presence of other transcripts at this point is expected both due to direct interaction with the support matrix and low-level cross talk of the oligos. The priority for this step is to capture as much of the target population as possible because, given the stochastic nature of the NAI modification, each individual MBNL1 transcript can have a different mutation profile. Increasing the number of target molecules in the reaction promotes the sampling of multiple positions across the region and increases the structural information content. The specificity for the target is introduced at the amplicon step of library preparation, which uses intronic primers unique to the MBNL1 paralog that selects for its pre-mRNA form (arrows, Fig. 4A).
FIGURE 4.

Target enrichment by hybridization capture and validation. (A) Design for enrichment of the MBNL1 target sequence. A set of 17 anti-sense biotinylated DNA oligos were selected to tile across part of intron 4, all of exon 5, and the start of intron 5. The oligos are positioned to capture a slightly larger region than the desired amplicon size. The block arrows indicate the position of the intronic primers used to create the amplicon in the downstream library preparation steps. (B) Hybridization capture protocol. First, to facilitate sample processing when the target sequence is found in a long RNA, samples may be fragmented using a Zn2+ based approach to sizes larger than the region of interest. The samples are then cleaned and concentrated before hybridization to a set of biotinylated oligos. The RNA:oligo hybrids are incubated with magnetic streptavidin beads before being captured on a magnet. The supernatant is removed, and the remaining beads are solubilized in TRIzol, chloroform extracted, and the enriched RNA is recovered on an RNA clean up column. The numbers denote the samples that correspond with the lane numbers in C. (C) RT-PCR detection of the MBNL1 amplicon after enrichment. The reactions were programmed with 1 µg of input RNA (lane 1), 1 µg of RNA from postbinding supernatant (lane 2), or half of eluted RNA from target enrichment protocol (lane 3). The expected amplicon size is 459 bp.
Given that the MBNL1 pre-mRNA is ∼100 kb in length, we initially wanted to fragment our RNA to facilitate hybridization capture and downstream sample handling. Typical fragmentation in sequencing protocols tend to be magnesium based and is not ideal for SHAPE-MaP, which is based on replacing magnesium with manganese for reverse transcription. Therefore, we used a zinc-based approach and initially tested conditions on untreated total RNA samples to define conditions that would result in fragment sizes larger than our desired amplicon (>500 nt) as the manufacturer's instructions produce much smaller fragments.
The fragmented RNA was then hybridized to the oligo pool in a formamide-based buffer for 4 h, as described in Materials and Methods (Fig. 4B). Magnetic streptavidin beads washed in the hybridization buffer were added to the samples for 30 min before capture on a magnetic stand. The postbinding RNA sample was retained to assess the efficiency of the capture, while the streptavidin beads were immediately resuspended in TRIzol to permit isolation of the enriched RNA population. The enriched sample and the depleted postbinding sample were both recovered using NucleoSpin RNA XS Clean Up kits and the target-enriched RNA was eluted in a small volume to permit the entire sample to be introduced into the reverse transcription reaction. In Figure 4C, the results of RT-PCR for the Mbnl1 amplicon are shown where the reverse transcription reactions were programmed with 1 µg of RNA (lanes 1,2) or half of the eluted RNA obtained from the enrichment procedure (lane 3). Comparing the signal from the input RNA sample (lane 1) to the post binding RNA sample (lane 2) shows the successful capture of the MBNL1 transcripts and their transfer to the enriched pool (lane 3).
MBNL1 protein expression influences RNA structure near the branchpoint
After optimizing the enrichment procedure on the untreated RNA, we combined our approach with SHAPE-Map to analyze the MBNL1 intron4/exon5 regulatory region. MEFs were plated subconfluently and the following day half of the plates were dosed with doxycycline for 18 h to induce MBNL1 protein expression and subsequently SHAPE treated with NAI or DMSO. The MEFs were grown in sufficient quantities to permit 150 µg of total RNA to be utilized for each condition during the hybridization capture procedure. This results in a 30× increase in representation of our target transcripts into the reverse transcription step and strongly impacts the mutational landscape sampling. This amount can be scaled up or down according to the relative abundance of a target of interest. The enriched RNA populations were then used to make SHAPE-MaP amplicon libraries as described in Materials and Methods. Libraries were sequenced on a NextSeq 500 and the sequencing analysis was performed using the ShapeMapper 2 pipeline.
Comparing the identical sub regions of the reactivity profiles with or without target enrichment (Fig. 5A) shows that the data from the enriched RNA samples are greatly improved, with low variation in the structure probing data and tight error bars across the top of each position. The target enriched SHAPE-MaP reactivity profiles generated in the absence or presence of MBNL1 protein expression are depicted in Figure 5B. The location of the MBNL1 binding sites from our previous study are shown by the green boxes, while exon 5 is marked with a purple box and the ultraconserved element is marked in gray. Initial scanning of the profiles broadly shows extensive similarity between the presence and absence of MBNL1; however, in both cases the UCE looks generally less reactive than the surrounding sequence. Comparison of SHAPE reactivities between conditions can be achieved using ΔSHAPE analysis (Smola et al. 2015a). We applied it to our SHAPE-MaP data in the presence and absence of MBNL1 expression, averaging over a 3 nt sliding window to account for local signal fluctuations. We compared three different MBNL1 expressing data sets that were each treated with a different lot of NAI. As shown in Figure 4C, the three sets show a consistent decrease in reactivity near the exon proximal YGCY motifs, which is not unexpected if the MBNL1 protein is binding in this region. The surprising finding was the striking MBNL1-dependent change in reactivity at nucleotide 63 of the amplicon, which corresponds to the major far-distal branchpoint within intron 4. Closer inspection of this region shows that while some nucleotides retain identical reactivity in the presence or absence of MBNL1, there are 6 nt showing significant changes with three becoming more reactive, and three becoming less reactive including the branchpoint adenosine (Fig. 6A, arrows). There are no MBNL1 binding sites immediately adjacent to this branchpoint, so the decrease in reactivity upon MBNL1 expression is likely due to remodeling effects in response to protein binding.
FIGURE 5.
Shape reactivity profiles after enrichment protocol. (A) Comparison of a subsection of the reactivity profiles with or without target enrichment. Nucleotide numbering refers to the position within the amplicon, data is derived from samples in the absence of MBNL1 overexpression. High reactivity positions are shown in blue, medium in orange, and low in black. Error bars are shown in black. (B) SHAPE-MaP reactivity profiles of the MBNL1 amplicon in the presence or absence of MBNL1 protein. The ultraconserved region is overlined in gray, the YGCY motifs are marked in green, and exon 5 is shown in purple. Error bars are shown in black at the top of each position. (C) Comparison of SHAPE-MaP reactivities between conditions using ΔSHAPE analysis. Three different MBNL1 expressing data sets were compared, each treated with a different batch of NAI. Regions with positive ΔSHAPE values shown in light blue indicate greater flexibility upon MBNL1 induction, while the negative ΔSHAPE values shaded in dark blue are more constrained. The green boxes indicate the position of the YGCY binding motifs, and the purple box represents exon 5.
FIGURE 6.
Structural alterations in regions of particular interest. (A) Changes in structure near the far-distal branchpoint in response to MBNL1 protein. The SHAPE-MaP profile of the branchpoint is shown in the presence and absence of MBNL1 protein. The light gray shading indicates the location of the major branchpoint adenosine. Nucleotides whose reactivity is changing are marked with arrows. The direction of the arrow depicts the direction of change (more or less reactive) in the presence of MBNL1 protein. (B) Changes in accessibility of the YGCY motifs upon addition of MBNL1. The locations of the YGCY motifs are depicted in green. Nucleotides that change in these regions are marked with blue dots in both panels and the direction of change is indicated with black arrows in the lower panel. (C) Comparison of exon 5 structure in cells and in vitro. Superfold was used to model the structure of exon 5 from the MEFs and is compared to our previous in vitro model on the left. Filled gray nucleotides correspond to the flanking 3′ and 5′ splice sites. The SHAPE-MaP reactivity of exon 5 in the absence of MBNL1 protein is modeled on the predicted structure from MEFs. The arrowheads show changes in reactivity upon addition of MBNL1 protein with the colors representing three different experiments. Filled arrowheads show positions that are more reactive in the presence of MBNL1 protein, while open arrowheads mark positions that are less reactive.
We also examined the other consensus splicing signals. Downstream from the branchpoint, the U-rich sequences of the polypyrimidine tract are largely nonreactive, likely due to protein contacts. The adenine of the AG dinucleotide at the 3′ ss increases its reactivity in the presence of MBNL1, but the adjacent sequences remain similar in both conditions, therefore stearic hinderance does not appear to be responsible for skipping of exon 5 in the presence of MBNL1. Interestingly, upon MBNL1 expression there is increased reactivity just downstream from the 5′ splice site of intron 5 in positions where the U1 snRNA would be expected to contact the pre-mRNA.
Given our previous knowledge about the binding of MBNL1 across the sites in intron 4, we were particularly interested in examining changes in reactivity due to the presence of MBNL1. Across the 91 nt regulatory region, 26 nt change reactivity in response to MBNL1 with 11 increasing and 15 decreasing in reactivity (Fig. 6B). Local impacts at the sites of MBNL1 binding are observed in most of the YGCY motifs, except for the first cluster which is not accessible for SHAPE modification even in the absence of MBNL1 suggesting either a protein protection or an RNA structure that precludes MBNL1 accessing that site in vivo. We cannot exclude the possibility that MBNL1 takes the place of a different protein at this position, but deletion of this region in a minigene context did not impact inclusion rates of exon 5. Of those nucleotides that decrease their reactivity, 80% (12/15) are located at a YGCY motif ±2 nt. A region of flexibility encompassing nucleotides 135–158 is the longest consecutive flexible sequence within the UCE. In the in vitro enzymatic probing assay, this is the region that was the most sensitive to the addition of recombinant MBNL1 protein, resulting in reduced cleavage across the region (Gates et al. 2011). In contrast, our in vivo data shows no impact on nucleotides 136–148, where there are no YGCY binding motifs, but does show an MBNL1 expression dependent decrease in reactivity of nucleotides 149–153, which is the position of the key MBNL1 YGCY motif. This may reflect the difference in MBNL1 expression levels in the two systems as well as the presence of the authentic cellular environment for the in vivo data. In the absence of other interactors in vitro, recombinant MBNL1 may bind the authentic site then interact with additional MBNL1 molecules to sterically hinder the access of the enzymes to these nearby sites. Our previous in vitro deletion 3 mutant removes nucleotides 140–158 which our new data suggests would strongly impact the flexibility of this autoregulatory region. This flexibility may allow the pre-mRNA to adopt different conformations that may promote exclusion of exon 5, including topologies that allow easier access for MBNL1 to the dominant binding site within the deletion 3 region.
In addition to information about the structural content of the intronic regulatory region, we were interested in examining exon 5. Our previous in vitro studies suggested that exon 5 is highly structured (Gates et al. 2011), and we were interested whether this structure was preserved in the endogenous RNA within the cell. For direct comparison to the in vitro data, we used Superfold (Smola et al. 2015b) to model the SHAPE-MaP data in the absence of doxycycline. This shows that with the exception of the first 5 nt, exon 5 is highly structured and is folded into two stem–loop structures in cells (Fig. 6C), which bring the two splice sites into close proximity. Stem–loop I (SLI) harbors two guanine bulges in a 9 nt stem and this structure was completely concordant between in vitro and in vivo experiments, suggesting that it folds independently of protein chaperones and is stable in the presence of cellular proteins and metabolites. The terminal loop of SLI contains a protected 3 nt sequence suggesting either RNA:protein interaction or distal RNA:RNA interactions. Interestingly, the four reactive nucleotides (Fig. 6C, nt 223–224, 226–227) on the transition from the loop to the stem changed their reactivity upon MBNL1 addition. The modeling of stem–loop II (SLII) differs between the in vitro and in cell experiments. The folding is driven by the base-pairing assignments for CC and GG dinucleotide pairs at positions 235/236, 241/242, 246/247, 253/254, and 256/257. The high SHAPE reactivity of C247 in the MEF data precludes the formation of the C247–G256 base pair assigned in the in vitro derived structure model. Superfold remodeled this region to reflect our in vivo SHAPE-MaP data, with reactivity assigned to an internal bulge as well as the terminal loop. The in-cell experiments are population based, so it is also possible that a dynamic exchange between these structures is occurring.
Interestingly, upon induction of MBNL1 protein expression, we see similar changes across three experimental replicates (Fig 6C), with decreased reactivity of the G211 bulged nucleotide, increased reactivity of bases in the transition into the loop of SLI (C217, A219) and decreased reactivity of bases on the exit from the loop (A223, C224, G226). We also see a flipped reactivity of the bases in the loop of SLII (C247, U248). These changes are observed repeatedly and likely represent local remodeling in response to adjacent binding of MBNL1 to the exon proximal YGCY motifs that consistently show reduced activity in the ΔSHAPE analysis. The highly structured nature of exon 5 is intriguing both because of potential contributions to splicing regulation but also because this highly structured region occurs within a UCE, which were previously thought to be less prone to forming structure (Sathirapongsasuti et al. 2011).
DISCUSSION
Low abundance RNA sequences such as introns have high information content with respect to regulation of RNA metabolism and the outcomes of splice site selection can have profound effects on cellular metabolism. Thus, it is important to develop the molecular tools required to analyze these regions. Previous strategies have been developed to enrich the RNA populations in structure probing methods. icSHAPE uses azide-modified NAI (Spitale et al. 2015; Flynn et al. 2016) while SHAPE Selection (SHAPES) uses a N-propanone isatoic anhydride (NPIA) (Poulsen et al. 2015), and both reagents allow covalent coupling to biotin and an updated variant of Structure-seq2 incorporates a biotinylated dNTP during reverse transcription (Ritchey et al. 2017). After cDNA synthesis, the reactions are enriched on streptavidin beads, thereby reducing the background from the bulk unmodified RNA. However, highly expressed transcripts would still dominate this population and as selection occurs after the RT step this has the potential to miss information from target molecules when transcripts are limiting. Our hybridization capture enrichment prior to the encoding of structural information at the reverse transcription step broadly samples the mutational landscape and results in a strong improvement in signal to noise ratio. Our method is also simpler than DMS/SHAPE-LMPCR (Kwok et al. 2013) to study low abundance in vivo RNA targets in that is does not require ligation steps or optimization of gene-specific reverse transcription primers for each target and does not require gel analysis. Our approach also supports multiplexing many targets and is compatible with all sequencing-based RNA structure probing methods independent of selected probing chemistries, making it broadly applicable to studying low abundance RNA targets. We also herein reported the first use of controlled expression of an RNA binding protein in cells to interrogate RNA structure and RNA–protein interactions.
This technique allowed us to successfully capture structural information about the autoregulatory region controlling inclusion of MBNL1 exon 5 in cells. First, we found that exon 5 itself, which is embedded in the UCE, is highly structured with two stem–loops. The first stem consists of an 8 bp stem with two G-bulges followed by an AU-rich loop and is identical in cells and in vitro suggesting that its formation does not require protein chaperones and is very stable in the presence of cellular factors and metabolites. This finding is in contrast to a previous study using in silico predictions to suggest that UCEs are resistant to folding. Bulges in stem–loops can modulate the stability of stem–loop structures and often serve as recognition motifs for interacting factors. We also found local, reproducible shifts in the exon structure in response to MBNL1 expression. This structural information can be used to inform future experiments in the minigene contexts to understand what role, if any, the structure of exon 5 plays in its regulation.
An unanticipated finding was that increased MBNL1 protein expression had the largest impact on the RNA structure near the far-distal (−141) branchpoint region of the intron. Typically, branchpoints are found much closer to the 3′ ss, with 80% of detected U2-type branchpoints falling within a window of −49 to −20 (Pineda and Bradley 2018). Only 4.6% of constitutive introns contain far-distal branchpoints (>100 nt upstream); however, nearly 14% of introns upstream of regulated cassette exons have far-distal branchpoints, implying that the position of the branchpoint is involved in regulation of some cassette exons (Pineda and Bradley 2018). An MBNL1-regulated alteration in structure near the branchpoint is not unprecedented as our study on cTNT intron 4 showed that MBNL1 and U2AF65 compete for binding to a stem–loop structure proximal to the branchpoint capturing the RNA in alternate conformations (Warf et al. 2009). The presence of MBNL1 binding prevents U2AF65 association resulting in skipping of the downstream exon. However, in the cTNT intron there are MBNL1 YGCY motifs flanking the predicted polypyrimidine tract. In the case of MBNL1 autoregulation of exon 5, there are no binding motifs adjacent to the branchpoint or flanking the putative polypyrimidine tract, so impacts on this region would not be predicted and were not apparent in our original in vitro probing data. Thus, our ability to structure probe this low abundance intronic region in cells has uncovered a new line of inquiry for MBNL1 autoregulation.
While these local regions within the RNA were structurally impacted by MBNL1 expression, much of the region was not strongly influenced, as visualized in the ΔSHAPE data (Fig. 4C). While there are examples of global RNA rearrangements in response to ligand binding, it is not unusual for effects to be relatively local. Riboswitches can be broadly categorized into two different groups based on whether their rearrangements upon ligand binding are relatively local (Type I) or also result in global changes in the structure (Type II) reviewed in (Montange and Batey 2008). Binding of the RBP Pumilio-1 to the 3′-UTR of tumor suppressor p27 causes a local change in RNA structure that permits the association of miR-221 and mIR-222, resulting in the transition of the cells from a quiescent to a proliferative state (Kedde et al. 2010). The sites of local structural influence uncovered in this study will guide our future research into the mechanism of MBNL1 mediated autoregulation of exon 5.
Countless RNA regulatory steps are mediated through sequences that are not amenable to structural investigation by existing techniques due to low expression levels such as introns and many lncRNAs. Other low abundance RNAs remain a functional enigma, such as stable intronic sequence RNAs (Gardner et al. 2012; Talhouarne and Gall 2018), and additional structural information may provide clues to their biogenesis or function. Our enrichment strategy can be easily streamlined into existing RNA structure protocols and provides the opportunity to begin to address important biological questions related to low abundance RNA targets.
MATERIALS AND METHODS
Cell culture
The double-dosing MEF cell line (ddMEF) was generated from a parental Mbnl1−/−: Mbnl2−/− MEF cell line. A doxycycline inducible GFP-tagged MBNL1 (NCBI accession number NP_066368) was stably integrated using a PiggyBac transposon system. Briefly, 24 h after transfection using TransIT-LT1(Mirus), cells were selected for puromycin resistance (4 µg/mL) and allowed to recover for several days. The pool of cells was treated with 1 µg/µL doxycycline (Sigma Aldrich) to induced expression of GFP-MBNL1 and cells were sorted for high GFP expression using FACSAria II cell sorter (BD Biosciences). Individual clones were isolated, and the populations expanded in the presence of puromycin (2 µg/mL). A single clonal population was then transfected as above to achieve integration of ponasterone-A inducible mOrange-RBFOX1 (NCBI accession NP_061193). Cells were treated with 10 µM ponasterone-A, sorted for mOrange expression, and individual clones isolated and expanded in the presence of puromycin (2 µg/mL). Individual clones were selected for experimental use based on GFP-MBNL1 and mOrange-RBFOX1 expression across a range of doxycycline and ponasterone A concentrations, respectively. Cells were cultured in DMEM (Gibco) with 10% fetal bovine serum (Sigma) and 1× Pen/Strep at 37°C and 5% CO2.
In vivo SHAPE modification
MEFs were plated at 1.8 × 106 into 150 mm cell culture dishes, with eight plates per condition. The following day, MBNL1 expression was induced with 1 µg/mL of doxycycline for 18–20 h (doxycycline hyclate, Sigma-Aldrich) in half of the plates. Cells were rinsed with 1× PBS and gently harvested from the plates using TrypLE (Gibco). Aliquots of 15 × 106 cells were prepared and centrifuged at 400g for 3 min. The cell pellets were resuspended to a volume of 465 µL in 1× PBS and 10 µL of SUPERase-In RNase Inhibitor was added to each reaction to deter degradation in the samples during treatment. SHAPE samples were treated with 25 µL of 2M NAI (Millipore Sigma, #03-310) for a final concentration of 100 mM, and control cells were treated with 25 µL of anhydrous dimethyl sulfoxide. Reactions were incubated at 37°C with rotation for 12 min. Reactions were stopped by centrifuging the cells to pellets, removing the supernatant and resuspending the samples in 1 mL of TRIzol (Life). RNA was extracted using the DirectZol RNA Miniprep Plus kit (Zymo) according to manufacturer's instructions, including an on-column DNase treatment. Samples were eluted into 50 µL of nuclease-free water. To ensure no genomic DNA carry-over, samples were then further treated with TURBO DNase Enzyme for 30 min at 37°C (TURBO DNA-free, Invitrogen). Reactions were stopped using a DNase inactivation reagent in slurry form as per manufacturer's instructions, allowing removal of the enzyme and divalent cations such as magnesium and calcium by centrifugation. RNA was quantitated and stored at −80°C until use.
Low abundance target enrichment
A total of 150 µg of total RNA for each condition (no doxycycline with DMSO, no doxycycline with NAI, doxycycline with DMSO, and doxycycline with NAI) was partitioned into 2 PCR tubes containing 75 µg in a volume of 87 µL nuclease-free water. A total of 10 µL of 10× RNA fragmentation reagent containing a buffered zinc solution (Thermo Fisher, AM8740) was added to the samples, which were then incubated at 70°C in a preheated thermocycler with a heated lid for 2.5 min. A total of 10 µL of Stop solution (200 mM EDTA) was added to stop the fragmentation and samples were placed on ice. Fragmented samples were cleaned and concentrated using NucleoSpin RNA XS Clean Up kits (Machary Nagel) according to manufacturer's protocol. Each column was eluted with 20 µL of RNase-free water and replicates were combined into one sample for each condition. Volumes were adjusted with RNase-free water to a final volume of 40 µL when necessary, and 80 µL of fresh hybridization buffer (50 mM Tris pH 7.0, 750 mM NaCl, 1 mM EDTA, 1% SDS, 15% formamide) was added to each sample along with 2 µL of SUPERase-IN and 200 pmol (1 µL) of the pool of antisense oligos. Reactions were incubated at 37°C with rotation for 4 h. During this time, 50 µL of streptavidin beads per sample were washed in hybridization buffer diluted to 2/3 strength to mimic the binding conditions (Dynabeads MyOne Streptavidin C1, Invitrogen). Reactions were brought to 350 µL with diluted hybridization buffer and 50 µL of washed beads were added to each sample, along with 3 µL of SUPERase-IN. Samples were incubated for 30 min at 37°C with rotation, after which the samples were placed in a magnetic stand to capture the beads. The supernatant was removed and the samples were resuspended in 500 µL of TRIzol to extract bound RNAs. A total of 100 µL of chloroform was added to each sample, vortexed to mix and centrifuged for 15 min at 21,000g at 4°C. The aqueous phase was transferred to a new tube and the RNA was captured using the NucleoSpin XS RNA Clean Up Kit. Samples were eluted with 11 µL of nuclease-free water. In initial experiments the postcapture supernatant was retained to check for depletion status of our target and those RNA samples were also captured with the NucleoSpin XS RNA Clean Up kit following the manufacturer's instruction.
Hybridization oligos
Oligos were designed using the online designer at https://www.biosearchtech.com/stellarisdesigner. Oligos were ordered from Integrated DNA Technologies. Upon receipt, each oligo was resuspended to 200 pmol/µL and then a pool of all 17 oligos was created for the hybridization capture.
MBNL1-as1: 5′-GGCACTATTGAGTTGTTATT-3′BioTEG
MBNL1-as2: 5′-ACAAGACTAAGCATGCACAA-3′BioTEG
MBNL1-as3: 5′-TCATCGGAATGCCATATACA-3′BioTEG
MBNL1-as4: 5′-GCATTTTGGGTAGGTGAGAA-3′BioTEG
MBNL1-as5: 5′-AAACAGCAAGCAGAGGTGCA-3′BioTEG
MBNL1-as6: 5′-TGGGGTTCAAGCGCATTAAC-3′BioTEG
MBNL1-as7: 5′-CGAGCACATGATGGCAATGG-3′BioTEG
MBNL1-as8: 5′-CTGAGTCTTAATTAGCAGGC-3′BioTEG
MBNL1-as9: 5′-TCGCTTCAGTGATTTGACAG-3′BioTEG
MBNL1-as10: 5′-CATAGTACCAGGTCAAAGGT-3′BioTEG
MBNL1-as11: 5′-ATGCCAAGCTAAAAGGTGAA-3′BioTEG
MBNL1-as12: 5′-CCTGTATCTACAATAAAGCT-3′BioTEG
MBNL1-as13: 5′-ACCAAAACCAAACCAAACCA-3′BioTEG
MBNL1-as14: 5′-CCACACTCAGATTTTCATTG-3′BioTEG
MBNL1-as15: 5′-TTCTGCTCATTTTTTCAGGA-3′BioTEG
MBNL1-as16: 5′-TCTATGTATGCATTTTAGGT-3′BioTEG
MBNL1-as17: 5′-CAGTCTTTCATTGTACCTTA-3′BioTEG
SHAPE-MaP library preparation and data processing
A 5× reverse transcription buffer (250 mM Tris-HCl pH = 8.3, 375 mM KCl, 15 mM MnCl) was freshly prepared. A total of 10 µL of the enriched RNA sample was combined with 1 µL of 10 mM dNTPs and 1 µL random nonamers at 200 ng/µL (cat # and double check concentration) and incubated at 65°C for 5 min in a thermocycler. Tubes were quick cooled on ice and then 4 µL of 5× reverse transcription buffer, 2 µL of 0.1 M DTT, and 1 µL of SUPERase-IN were added before incubation at 25°C for 2 min. A total of 1 µL of SuperScript II reverse transcriptase (Invitrogen) was added and the samples were incubated for 25°C for 10 min, 42°C for 3 h, 70°C for 15 min followed by holding at 12°C. We then used primers on either side of our region of interest to generate amplicons for the sequencing library. The amplicon sequence spans genomic coordinates chr3:60,614,481–460,614,939 (mm10), with the structure probing data collected from chr3:60,614,506–560,614,916.
MBNL1 amplicon primers:
Forward: 5′-CTCAATAGTGCCTTTATTGTGCATG-3′
Reverse: 5′-CAGGAAACCACACTCAGATTTTC-3′
We made a master mix consisting of 10 µL Q5 Hot Start Buffer (NEB), 1 µL 10 mM dNTPs, 2.5 µL of 10 µM forward primer, 2.5 µL of 10 µM reverse primer, 0.5 µL Q5 Hot Start DNA polymerase, and 28.5 µL of water. Each sample received 45 µL from the master mix and 5 µL of cDNA from the Mn2+ reverse transcription reaction. The reactions were denatured at 98°C for 30 sec, then subjected to 25 cycles of 98°C for 10 sec, 65°C for 30 sec, 72°C for 20 sec, followed by a final extension at 72°C for 2 min. The PCR reactions were cleaned and concentrated with the MinElute PCR purification kit (Qiagen) and eluted in 10 µL of water. The concentrations were determined with the high sensitivity DNA Qubit kit (Qiagen) and 1 ng of amplicon was diluted in 5 µL of water for library preparation with the Nextera XT kit. A total of 10 µL of Tagment DNA buffer and 5 µL of Amplicon Tagment mix were added to the amplicon samples and incubated at 55°C for 10 min, and then cooled to 10°C. A total of 5 µL of Neutralize Tagment Buffer was added and samples were incubated for 5 min at room temperature. Unique barcodes were then added to each sample by PCR using the 25 µL tagmented DNA sample, 15 µL of Nextera PCR mix, 5 µL of Index Primer 1 and 5 µL of Index Primer 2 from the Nextera XT primer kit. The PCR program was as follows: 72°C for 3 min, 95°C for 30 sec, 12 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec, followed by a final extension of 72°C for 5 min and holding at 10°C. Reactions were purified using a 0.75× AMPure XP bead clean up (Beckman Coulter). Beads were vortexed to ensure uniformity before use and 37.5 µL were added to each PCR reaction. Samples were pipetted up and down thoroughly to mix and incubated for 5 min at room temperature. The samples were placed on a magnetic stand and the reactions were allowed to clear (∼2 min) before removing the supernatant and washing twice with 200 µL of freshly prepared 80% ethanol. All traces of ethanol were removed with a fine tip vacuum. Samples were removed from the magnet and resuspended in 17 µL of nuclease-free water to elute the libraries from the beads. After incubating at room temperature for 2 min the samples were placed on the magnetic stand and allowed to clear. Fifteen microliters of the eluate were transferred to a new tube and represents the finished libraries. The concentration of the libraries was determined using a high sensitivity Qubit kit and the library quality and average fragment size was assessed using a fragment analyzer (Agilent). Libraries were sequenced on a NextSeq 500 using the 300 cycle Mid-output kit (Illumina).
All libraries were processed with the ShapeMapper2 (v2.1.3) pipeline using the Mus musculus mm10 genome coordinates. For the MBNL1 amplicon, the reference fasta sequence provided marked the primers in lowercase to exclude them from the SHAPE-MaP analysis. The MBNL1 amplicon spans 459 nt at mm10 chr3:60,614,481–60,614,939. The sequences for the 7SK and Rpph1 controls were extracted using the command fetch in python module pysam (https://github.com/pysam-developers/pysam) using the mm10 genomic coordinates (Rn7SK chr9:78175303–78175633, Rpph1 chr14:50807449–50807767) and converted to fastq files using the bamtofastq module within BEDTools (Quinlan and Hall 2010). All comparisons were performed between NAI-treated cells and DMSO only controls. The ΔSHAPE analysis was performed using the python script deltaSHAPE.py (weekslab.com/software) with the following arguments to trim primer sequences and color the significant differences (–mask5 25 –mask3 23 –pdf –colorfill -o #OUTFILENAME #INPUTFILENAME.map). Structure prediction of exon 5 was performed with SuperFoldv1.0 (weekslab.com/software). The structures in Figures 2D,E and 6C were rendered using StructureEditorv6.2 (rna.urmc.rochester.edu/GUI/html/StructureEditor.html). Data associated with this study is available under GEO accession number GSE159719.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health, grant number R01GM121862.
Author contributions: J.B. conceptualized the method and conducted all experiments except for the RT-PCR validation. M.H. performed the RT-PCR. O.M., E.W., and M.H. created the ddMEF inducible cell line. J.B. wrote the manuscript with editing contributions from R.C.S., M.S.S., and J.A.B.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.077263.120.
REFERENCES
- Baralle FE, Giudice J. 2017. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol 18: 437–451. 10.1038/nrm.2017.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartys N, Kierzek R, Lisowiec-Wachnicka J. 2019. The regulation properties of RNA secondary structure in alternative splicing. Biochim Biophys Acta Gene Regul Mech 1862: 194401 10.1016/j.bbagrm.2019.07.002 [DOI] [PubMed] [Google Scholar]
- Batra R, Charizanis K, Manchanda M, Mohan A, Li M, Finn DJ, Goodwin M, Zhang C, Sobczak K, Thornton CA, et al. 2014. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell 56: 311–322. 10.1016/j.molcel.2014.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325. 10.1126/science.1098119 [DOI] [PubMed] [Google Scholar]
- Dethoff EA, Boerneke MA, Gokhale NS, Muhire BM, Martin DP, Sacco MT, McFadden MJ, Weinstein JB, Messer WB, Horner SM, et al. 2018. Pervasive tertiary structure in the dengue virus RNA genome. Proc Natl Acad Sci 115: 11513–11518. 10.1073/pnas.1716689115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, et al. 2018. Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell 70: 854–867 e859. 10.1016/j.molcel.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Krasilnikov AS. 2010. Of proteins and RNA: the RNase P/MRP family. RNA 16: 1725–1747. 10.1261/rna.2214510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Zhang QC, Spitale RC, Lee B, Mumbach MR, Chang HY. 2016. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat Protoc 11: 273–290. 10.1038/nprot.2016.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Kavousi N, Bountali A, Dammer EB, Mourtada-Maarabouni M, Ortlund EA. 2020. The lncRNA growth arrest specific 5 regulates cell survival via distinct structural modules with independent functions. Cell Rep 32: 107933 10.1016/j.celrep.2020.107933 [DOI] [PubMed] [Google Scholar]
- Gallego-Paez LM, Bordone MC, Leote AC, Saraiva-Agostinho N, Ascensao-Ferreira M, Barbosa-Morais NL. 2017. Alternative splicing: the pledge, the turn, and the prestige: the key role of alternative splicing in human biological systems. Hum Genet 136: 1015–1042. 10.1007/s00439-017-1790-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner EJ, Nizami ZF, Talbot CC Jr, Gall JG. 2012. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev 26: 2550–2559. 10.1101/gad.202184.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates DP, Coonrod LA, Berglund JA. 2011. Autoregulated splicing of muscleblind-like 1 (MBNL1) Pre-mRNA. J Biol Chem 286: 34224–34233. 10.1074/jbc.M111.236547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronski P, Palac A, Scharff LB. 2020. Secondary structure of chloroplast mRNAs in vivo and in vitro. Plants (Basel) 9: 323 10.3390/plants9030323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LT, Adams RL, Wan H, Huston NC, Potapova O, Olson S, Gallardo CM, Graveley BR, Torbett BE, Pyle AM. 2020. Sequencing and structure probing of long RNAs using MarathonRT: a next-generation reverse transcriptase. J Mol Biol 432: 3338–3352. 10.1016/j.jmb.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. 2004. Muscleblind proteins regulate alternative splicing. EMBO J 23: 3103–3112. 10.1038/sj.emboj.7600300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AG, Smith CWJ. 2017. Intron retention as a component of regulated gene expression programs. Hum Genet 136: 1043–1057. 10.1007/s00439-017-1791-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. 2010. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020. 10.1038/ncb2105 [DOI] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. 2013. Function of alternative splicing. Gene 514: 1–30. 10.1016/j.gene.2012.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchko KM, Madden EA, Morrison C, Plante KS, Sanders W, Vincent HA, Cruz Cisneros MC, Long KM, Moorman NJ, Heise MT, et al. 2018. Structural divergence creates new functional features in alphavirus genomes. Nucleic Acids Res 46: 3657–3670. 10.1093/nar/gky012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. 2013. Determination of in vivo RNA structure in low-abundance transcripts. Nat Commun 4: 2971 10.1038/ncomms3971 [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929. 10.1038/nature05676 [DOI] [PubMed] [Google Scholar]
- Larman BC, Dethoff EA, Weeks KM. 2017. Packaged and free satellite tobacco mosaic virus (STMV) RNA genomes adopt distinct conformational states. Biochemistry 56: 2175–2183. 10.1021/acs.biochem.6b01166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YF, Mauger DM, Goldstein DB, Urban TJ, Weeks KM, Bradrick SS. 2015. IFNL3 mRNA structure is remodeled by a functional non-coding polymorphism associated with hepatitis C virus clearance. Sci Rep 5: 16037 10.1038/srep16037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, Bensaude O. 2009. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol 26: 2821–2830. 10.1093/molbev/msp198 [DOI] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231. 10.1021/ja043822v [DOI] [PubMed] [Google Scholar]
- Mitchell D III, Assmann SM, Bevilacqua PC. 2019a. Probing RNA structure in vivo. Curr Opin Struct Biol 59: 151–158. 10.1016/j.sbi.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D III, Renda AJ, Douds CA, Babitzke P, Assmann SM, Bevilacqua PC. 2019b. In vivo RNA structural probing of uracil and guanine base-pairing by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). RNA 25: 147–157. 10.1261/rna.067868.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockenhaupt S, Makeyev EV. 2015. Non-coding functions of alternative pre-mRNA splicing in development. Semin Cell Dev Biol 47–48: 32–39. 10.1016/j.semcdb.2015.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange RK, Batey RT. 2008. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 37: 117–133. 10.1146/annurev.biophys.37.032807.130000 [DOI] [PubMed] [Google Scholar]
- Montes M, Sanford BL, Comiskey DF, Chandler DS. 2019. RNA splicing and disease: animal models to therapies. Trends Genet 35: 68–87. 10.1016/j.tig.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe AM, Busan S, Rice GM, Hajdin CE, Peterson BK, Ruda VM, Kubica N, Nutiu R, Baryza JL, Weeks KM. 2018. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell 173: 181–195 e118. 10.1016/j.cell.2018.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr. 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718. 10.1101/gad.1525507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R. 2006. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation 74: 65–80. 10.1111/j.1432-0436.2006.00060.x [DOI] [PubMed] [Google Scholar]
- Peattie DA, Gilbert W. 1980. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci 77: 4679–4682. 10.1073/pnas.77.8.4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda JMB, Bradley RK. 2018. Most human introns are recognized via multiple and tissue-specific branchpoints. Genes Dev 32: 577–591. 10.1101/gad.312058.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LD, Kielpinski LJ, Salama SR, Krogh A, Vinther J. 2015. SHAPE selection (SHAPES) enrich for RNA structure signal in SHAPE sequencing-based probing data. RNA 21: 1042–1052. 10.1261/rna.047068.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey LE, Su Z, Tang Y, Tack DC, Assmann SM, Bevilacqua PC. 2017. Structure-seq2: sensitive and accurate genome-wide profiling of RNA structure in vivo. Nucleic Acids Res 45: e135 10.1093/nar/gkx533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathirapongsasuti JF, Sathira N, Suzuki Y, Huttenhower C, Sugano S. 2011. Ultraconserved cDNA segments in the human transcriptome exhibit resistance to folding and implicate function in translation and alternative splicing. Nucleic Acids Res 39: 1967–1979. 10.1093/nar/gkq949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder AR, Baumstark T, Riesner D. 1998. Chemical mapping of co-existing RNA structures. Nucleic Acids Res 26: 3449–3450. 10.1093/nar/26.14.3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. 2016. RNA mis-splicing in disease. Nat Rev Genet 17: 19–32. 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherpa C, Rausch JW, Le Grice SF. 2018. Structural characterization of maternally expressed gene 3 RNA reveals conserved motifs and potential sites of interaction with polycomb repressive complex 2. Nucleic Acids Res 46: 10432–10447. 10.1093/nar/gky722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11: 959–965. 10.1038/nmeth.3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Calabrese JM, Weeks KM. 2015a. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry 54: 6867–6875. 10.1021/acs.biochem.5b00977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. 2015b. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10: 1643–1669. 10.1038/nprot.2015.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci 113: 10322–10327. 10.1073/pnas.1600008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. 2013. RNA SHAPE analysis in living cells. Nat Chem Biol 9: 18–20. 10.1038/nchembio.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. 2015. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519: 486–490. 10.1038/nature14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel EJ, Yu AM, Lucks JB. 2018. High-throughput determination of RNA structures. Nat Rev Genet 19: 615–634. 10.1038/s41576-018-0034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznajder LJ, Swanson MS. 2019. Short tandem repeat expansions and RNA-mediated pathogenesis in myotonic dystrophy. Int J Mol Sci 20: 3365 10.3390/ijms20133365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztuba-Solinska J, Rausch JW, Smith R, Miller JT, Whitby D, Le Grice SFJ. 2017. Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res 45: 6805–6821. 10.1093/nar/gkx241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talhouarne GJS, Gall JG. 2018. Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc Natl Acad Sci 115: E7970–E7977. 10.1073/pnas.1808816115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ZW, Fei G, Paulo JA, Bellaousov S, Martin SES, Duveau DY, Thomas CJ, Gygi SP, Boutz PL, Walker S. 2020. O-GlcNAc regulates gene expression by controlling detained intron splicing. Nucleic Acids Res 48: 5656–5669. 10.1093/nar/gkaa263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomezsko PJ, Corbin VDA, Gupta P, Swaminathan H, Glasgow M, Persad S, Edwards MD, McIntosh L, Papenfuss AT, Emery A, et al. 2020. Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 582: 438–442. 10.1038/s41586-020-2253-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Blencowe BJ. 2019. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell 76: 329–345. 10.1016/j.molcel.2019.09.017 [DOI] [PubMed] [Google Scholar]
- Wagner SD, Struck AJ, Gupta R, Farnsworth DR, Mahady AE, Eichinger K, Thornton CA, Wang ET, Berglund JA. 2016. Dose-dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet 12: e1006316 10.1371/journal.pgen.1006316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. 2012. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150: 710–724. 10.1016/j.cell.2012.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PY, Sexton AN, Culligan WJ, Simon MD. 2019a. Carbodiimide reagents for the chemical probing of RNA structure in cells. RNA 25: 135–146. 10.1261/rna.067561.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang M, Wang T, Zhang Y, Zhang X. 2019b. Genome-wide probing RNA structure with the modified DMS-MaPseq in Arabidopsis. Methods 155: 30–40. 10.1016/j.ymeth.2018.11.018 [DOI] [PubMed] [Google Scholar]
- Warf MB, Diegel JV, von Hippel PH, Berglund JA. 2009. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci 106: 9203–9208. 10.1073/pnas.0900342106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CT, Lackey L, Williams B, Dokholyan NV, Gotz D, Laederach A. 2017. Comparative visualization of the RNA suboptimal conformational ensemble in vivo. Biophys J 113: 290–301. 10.1016/j.bpj.2017.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Black DL. 2013. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet 29: 442–448. 10.1016/j.tig.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Routh A. 2020. Mapping RNA-capsid interactions and RNA secondary structure within virus particles using next-generation sequencing. Nucleic Acids Res 48: e12 10.1093/nar/gkz1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. 2017. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods 14: 75–82. 10.1038/nmeth.4057 [DOI] [PMC free article] [PubMed] [Google Scholar]