Abstract
The toxins that some bacteria secrete to kill off rival species can also generate mutations that help toxin-resistant populations adapt to new environments.
Research organism: E. coli, Other
Related research article de Moraes MH, Hsu F, Huang D, Bosch DE, Zeng J, Radey MC, Simon N, Ledvina HE, Frick JP, Wiggins PA, Peterson SB, Mougous JD. 2021. An interbacterial DNA deaminase toxin directly mutagenizes surviving target populations. eLife 10:e62967. doi: 10.7554/eLife.62967
Bacterial communities are often comprised of numerous different species which either co-exist in harmony or compete with each other for resources. To gain an upper hand on the competition, some bacteria have developed a form of needle-like machinery called a type VI secretion system (or T6SS for short) that injects toxic proteins directly into their rivals (Basler et al., 2012; Hood et al., 2010; Klein et al., 2020; Russell et al., 2011). It is generally thought that the toxins secreted by T6SS decapacitate the target organism by impairing important processes such as cell wall synthesis, ATP production and DNA replication (Ahmad et al., 2019; Jurėnas and Journet, 2020; Whitney et al., 2015). While these toxins are clearly involved in anti-bacterial warfare, it is unclear whether T6SS can also facilitate symbiotic relationships within bacterial communities.
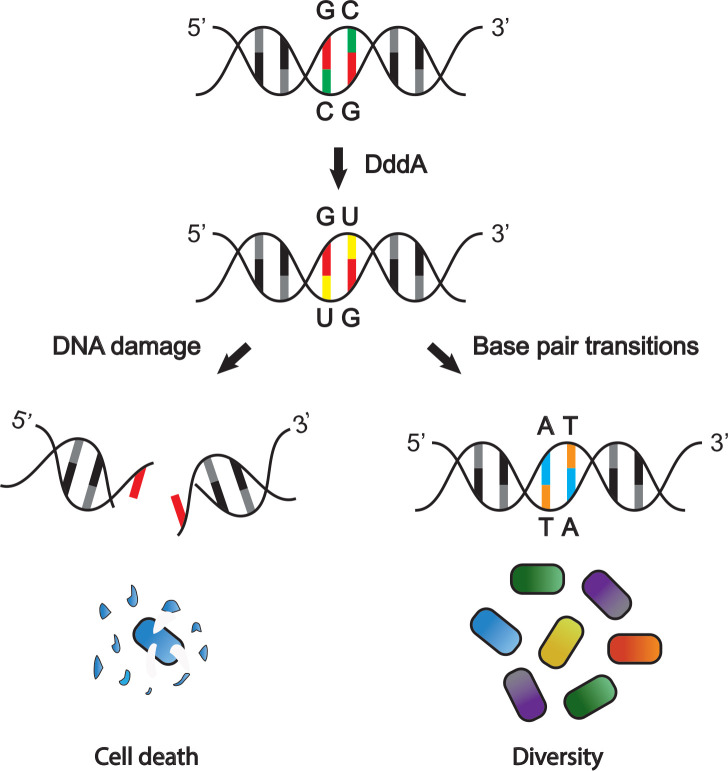
Recently, a collaboration between groups led by Joseph Mougous (University of Washington) and David Liu (Harvard University and the Broad Institute) discovered that the T6SS of Burkholderia cenocepacia secretes a toxin called DddA that catalyzes the removal of an amino group from cytosine, converting it to uracil (Mok et al., 2020). In most cells, these enzymes – known as cytosine deaminases – are important for maintaining the levels of nucleotide precursors in cells (Neuhard, 1968). While most cytosine deaminases catalyze this reaction in single-stranded DNA, DddA is the first enzyme found to convert cytosine to uracil in double-stranded DNA. However, uracil is normally only found in RNA, where it pairs with another nucleotide base called adenosine: in DNA, adenosine pairs with thymine, whereas cytosine pairs with guanine in both DNA and RNA (Figure 1).
Figure 1. The DddA toxin from B. cenocepacia affects other bacterial species in different ways.
DddA is a toxin that removes an amino group from cytosine (C; green), converting it into uracil (U; yellow) in chromosomal DNA (top). In some bacterial species uracil is then removed by the DNA repair machinery (left), which can lead to double-stranded DNA breaks and ultimately cell death. In bacteria resistant to the toxic effects of DddA, DNA breaks do not occur (right): instead, uracil is converted into thymine (T; orange), which causes guanine (G; red) to convert to adenosine (A; blue). This results in genetic variation within the targeted population, further diversifying the community of bacteria.
Now, in eLife, Mougous and colleagues – including Marcos de Moraes as first author – report that as well as destroying bacteria, DddA also provides a selective advantage for some bacteria in the community (de Moraes et al., 2021). The team (who are based at the University of Washington) found that when the bacterium B. cenocepacia injects DddA, uracil accumulates in the DNA of the targeted bacteria. Since uracil is normally only found in RNA, its presence triggers a repair mechanism that attempts to remove it from the DNA. However, this repair mechanism can lead to a break in one strand of the DNA, and if there are too many breaks in close proximity, they can lead to double-stranded breaks which stop DNA replication and result in bacterial cell death (Figure 1; D'souza and Harrison, 2003; Wallace, 2014). Indeed, de Moraes et al. found that the DddA delivered by B. cenocepacia suppresses the viability of several other bacterial species, including Pseudomona aeruginosa and Burkholderia thailandensis.
Although DddA causes double-stranded DNA breaks and cell death in most bacterial species, some disease-causing bacteria – including Eschericia coli and Salmonella enterica – are able to resist its detrimental effects. To better understand this observation, de Moraes et al. examined the long-term effects of DddA on these bacteria. They found that rather than excising the uracil that had replaced cytosine, the DNA replication machinery in the infected bacteria converts the uracil into a different nucleotide, thymine. As thymine pairs with adenosine in DNA, C-G pairs throughout the genome get replaced with T-A pairs (Figure 1).
An unanticipated consequence of this exchange was that the bacteria not killed by DddA acquired resistance to the antibiotic rifampicin. Further experiments revealed that other deaminase toxins similar to DddA were also able to introduce mutations in single-stranded DNA, suggesting this may be a widespread mechanism within bacterial populations (de Moraes et al., 2021).
It is generally thought that DNA mutations that arise during natural selection are caused by errors during chromosome replication or by exogenous factors such as chemicals and ionizing stress (Schroeder et al., 2018). Some bacteria rapidly adapt by importing fragments of foreign DNA from the environment and integrating them into their genome. This mechanism has been proposed to increase the genetic variation of populations, which provides a selective advantage to bacteria living in challenging environments or competing with other species (Dubnau and Blokesch, 2019; Mell and Redfield, 2014). The findings of de Moreas et al. suggest a similar mechanism in which the mutations produced by deaminase toxins help to diversify the population, creating new variants that can rapidly adapt to environmental changes.
There are several exciting avenues of research resulting from these findings. First, it is unclear why some bacteria are susceptible to DddA intoxication, whereas others are resistant. Defining these resistance mechanisms could help reveal the biological scenarios in which T6SS helps communities of bacteria adapt to their environment. Second, it will be interesting to determine how DddA toxins alter the composition of bacterial communities: DddA intoxication could eliminate competing species, while the genetic variations induced in resistant species may help enhance the long-term fitness of the community. Lastly, new insights into these topics may increase our understanding of human diseases where pathogenic bacteria come into contact with communities of microbes in the body, such as inflammatory bowel disease and cystic fibrosis, which could ultimately lead to better treatments (Buffie and Pamer, 2013; Spiewak et al., 2019).
Biographies
Maarten De Jong is in the Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, United States
Neal M Alto Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, United States
Competing interests
No competing interests declared.
References
- Ahmad S, Wang B, Walker MD, Tran HR, Stogios PJ, Savchenko A, Grant RA, McArthur AG, Laub MT, Whitney JC. An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature. 2019;575:674–678. doi: 10.1038/s41586-019-1735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nature Reviews Immunology. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'souza DI, Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Research. 2003;31:4573–4581. doi: 10.1093/nar/gkg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moraes MH, Hsu F, Huang D, Bosch DE, Zeng J, Radey MC, Simon N, Ledvina HE, Frick JP, Wiggins PA, Peterson SB, Mougous JD. An interbacterial DNA deaminase toxin directly mutagenizes surviving target populations. eLife. 2021;10:e62967. doi: 10.7554/eLife.62967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Blokesch M. Mechanisms of DNA uptake by naturally competent bacteria. Annual Review of Genetics. 2019;53:217–237. doi: 10.1146/annurev-genet-112618-043641. [DOI] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host & Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurėnas D, Journet L. Activity, delivery, and diversity of type VI secretion effectors. Molecular Microbiology. 2020;575:14648. doi: 10.1111/mmi.14648. [DOI] [PubMed] [Google Scholar]
- Klein TA, Ahmad S, Whitney JC. Contact-dependent interbacterial antagonism mediated by protein secretion machines. Trends in Microbiology. 2020;28:387–400. doi: 10.1016/j.tim.2020.01.003. [DOI] [PubMed] [Google Scholar]
- Mell JC, Redfield RJ. Natural competence and the evolution of DNA uptake specificity. Journal of Bacteriology. 2014;196:1471–1483. doi: 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok BY, de Moraes MH, Zeng J, Bosch DE, Kotrys AV, Raguram A, Hsu F, Radey MC, Peterson SB, Mootha VK, Mougous JD, Liu DR. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhard J. Pyrimidine nucleotide metabolism and pathways of thymidine triphosphate biosynthesis in Salmonella typhimurium. Journal of Bacteriology. 1968;96:1519–1527. doi: 10.1128/JB.96.5.1519-1527.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JW, Yeesin P, Simmons LA, Wang JD. Sources of spontaneous mutagenesis in bacteria. Critical Reviews in Biochemistry and Molecular Biology. 2018;53:29–48. doi: 10.1080/10409238.2017.1394262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiewak HL, Shastri S, Zhang L, Schwager S, Eberl L, Vergunst AC, Thomas MS. Burkholderia cenocepacia utilizes a type VI secretion system for bacterial competition. MicrobiologyOpen. 2019;8:e774. doi: 10.1002/mbo3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SS. Base excision repair: a critical player in many games. DNA Repair. 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Quentin D, Sawai S, LeRoux M, Harding BN, Ledvina HE, Tran BQ, Robinson H, Goo YA, Goodlett DR, Raunser S, Mougous JD. An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor tu for delivery to target cells. Cell. 2015;163:607–619. doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]