Abstract
Hematopoietic stem cell transplantation (HSCT) is a curative therapy for patients with malignant hematologic diseases. Killer immunoglobin-like receptor (KIR) expressed by NK cells is closely associated with the transplant outcomes, and it has been widely explored and debated for a few decades. Recently published studies have revealed that inhibitory KIRs (iKIRs) are educated by their cognate human lymphocyte antigen (HLA) ligands, and that decreased iKIR-HLA pairs post-transplantation may indicate a reduced NK cell function and impaired control of the primary disease. However, this theory still needs to be validated by additional clinical studies. Here we conducted a retrospective analysis of 246 patients who received haploidentical (haplo)-HSCT at our treatment center between January 2015 and June 2018. Our data suggests that decreased iKIR-HLA C pair post-HSCT correlated with a significantly higher risk of relapse [hazard risk (HR) = 2.95, p = 0.019] and reduced overall survival (OS) (HR = 3.74, p = 0.001) and disease-free survival (DFS) (HR = 4.05, p = 0.0004) in patients with myeloid disease. In conclusion, decreased iKIR-HLA C pair should be avoided during anti-thymocyte globulin (ATG)-based haplo-HSCT, especially for patients with myeloid disease.
Keywords: KIR, hematopoietic stem cell transplantation, iKIR-HLA model, relapse, survival
Introduction
Natural killer (NK) cells act as the first line of defense in the immune system. They can rapidly recognize autologous cells and eliminate non-self-components without prior antigen presentation (1, 2). Multiple receptors expressed on NK cells have been implicated in the regulation of their function, with a particular focus on the activities of killer immunoglobin-like receptors (KIRs).
It is well accepted that KIR genes and receptors can be divided into inhibitory and activating functions based on their diverse activities (3). Inhibitory KIRs (iKIRs) bind human lymphocyte antigen (HLA) class I molecules in a specific manner, KIR2DL1 recognizes HLA-C2 group allies, KIR2DL2 and KIR2DL3 recognize HLA-C1 group allies, KIR3DL1 recognizes HLA-Bw4 group allies, and KIR3DL2 recognizes HLA-A3/A11 allies. Activating KIRs (aKIRs) such as KIR2DS1, KIR2DS2, and KIR2DS4 recognize HLA C2, HLA C1, and HLA A11, respectively, but the ligands of the remaining KIRs remain largely unknown. Based on their chromosomal locations, KIR genes can be further identified as centromeric (cen) or telomeric (tel) genes. In addition, KIR genotype AA is made up of only one aKIR gene: KIR2DS4, while KIR genotype B/x is made up of a number of more variable aKIR genes (4).
Normally, autoimmune activation is inhibited because autologous cells express at least one inhibitory HLA ligand; however, tumor transformed cells downregulate HLA expression and/or upregulate activating signals that may trigger NK cell activation (5, 6). Following allogeneic hematopoietic stem cell transplantation (allo-HSCT), donor-derived NK cells may be activated as the recipients may not express the same inhibitory HLA ligands as the donor, preventing their association with the donor iKIRs. This has led to widespread speculation that NK cell alloreactivity in graft versus host (GVH) direction may provide additional benefits to tumor-killing strategies.
The Perugia group first established the KIR ligand-ligand model (also known as the KIR ligand model) based on HLA phenotype differences between donors and recipients. In this model, they assumed that donor-derived NK cells might kill recipient cells because the HLA ligands presented by the donor might be absent in the recipient. When they evaluated T cell-depleted (TCD) transplants without post-transplant immunosuppression, they were able to show that KIR ligand mismatch between donor-recipient pairs provided some protective effect against relapse, especially in patients with acute myeloid leukemia (AML) (7). Further development of KIR-typing technology allowed researchers to develop the receptor-ligand model (also known as the missing ligand model), which was used to evaluate the compatibilities between donor iKIRs and recipient HLA ligands. Results using this model suggested that the receptor-ligand model was a more accurate predictor for relapse risk than the KIR ligand model in leukemia patients (8). Additionally, Cooley et al. reported that KIR B/x donors significantly improved the relapse-free survival (RFS) rates for recipients with AML when compared to donors with a KIR AA genotype, suggesting that aKIRs may play a critical role in reducing relapse (9). Following these observations, numerous clinical studies have explored the impact of KIR on transplant outcomes. However, a large variability was found in these results and several factors may be responsible for these discrepancies, including disease type, transplant regimen, donor-recipient relationship, graft source and graft composition, etc (10–12).
In the last few decades, our understanding of NK cell reconstitution and KIR education has evolved a great deal. Pioneer studies in this field have found that reconstituted NK cells are highly immature and exhibit compromised cytotoxicity against leukemia cells in the early phases following transplantation. Afterward, these NK cells gradually acquire receptors and KIR reconstitution can take between several months and even years (13, 14). Importantly, a variety of data has suggested that NK cells acquire specific functionality only after engagement between the iKIRs and their cognate ligands. However, NK cells expressing iKIRs without cognate ligands (non-self KIR) are hyporesponsive and referred to “uneducated cells” (15, 16). Further, the education process mediated by cognate ligands is not restricted to autologous NK cells, but has also been demonstrated in donor-derived reconstituted NK cells after HSCT (17–19).
Recently, the Nowak team proposed the iKIR-HLA model to explore the optimal donor. Since the HLA environment may be altered after transplantation (from donor to recipient), the variations in iKIR-HLA pairs could be divided into three groups (decreased group: cognate iKIR-HLA pairs present in donor but absent in recipient; unchanged group: cognate iKIR-HLA pairs present both in donor and recipient; increased group: cognate iKIR-HLA pairs present in recipient but absent in donor). Consistent results from their studies showed that decreased iKIR-HLA pairs post transplantation correlated with a higher risk of relapse and inferior overall survival (OS), indicating that poor NK cell education resulted in weaker graft versus leukemia (GVL) effects (20–22). To further investigate the effects of these KIR interactions on transplant outcomes, we designed a retrospective study to evaluate a cohort of 246 patients, and evaluated our clinical outcomes using the iKIR-HLA model, the receptor-ligand model and KIR gene content.
Methods
Patients
This retrospective study was comprised of 246 patients with hematological malignancies. All transplants were performed between January 2015 and June 2018 and all methodologies applied during this study were consistent with the Declaration of Helsinki. The protocol was approved by the Ethics Review Committee of the First Affiliated Hospital of Zhejiang University and informed consent was obtained from each patient before transplantation. The authors had full access to the data and assume responsibility for its authenticity.
KIR and HLA Typing
Peripheral blood mononuclear cells were collected from recipients and their donors prior to transplantation and used for HLA and KIR testing. Alleles in the HLA-A, -B, and -C loci were determined using high-resolution HLA typing and KIR gene analysis was performed using the PCR-SSO method (KIR SSO Genotyping Test; OneLamda, Canoga Park, CA, USA).
Transplant Protocol
Most patients were subjected to a myeloablative conditioning (MAC) regimen that included administration of cytarabine (4 g/m2/d IV on days −10 to −9), busulfan (Bu) (3.2 mg/kg/d IV on days −8 to −6), cyclophosphamide (Cy) (1.8 g/m2/d IV on days −5 to −4), methyl-N-(2-chloroethyl)-N-cyclohexyl-N-nitrosourea (Me-CCNU) (250 mg/m2 orally on day −3), and antithymocyte globulin Fresenius [ATG-F; 2.5 mg/(kg d) IV on days −5 to −2] or ATG Genzyme [ATG-G; 1.5 mg/(kg d) IV on days −5 to −2]. The other patients were subjected to reduced-intensity conditioning (RIC) that consisted of exposure to fludarabine 30 mg/m2/d IV between days −10 and −5, Bu 3.2 mg/kg/d IV between days −6 and −5, and ATG-F 5 mg/(kg d) IV between days −4 and −1 or ATG-G 2.5 mg/(kg d) IV between days −4 and −1. All patients received G-CSF mobilized peripheral blood stem cells and no graft was subjected to ex vivo T-cell depletion. Graft versus host disease (GVHD) prophylaxis consisted of cyclosporin A (CsA) or Tacrolimus (Tac), with methotrexate (MTX) and low-dose mycophenolate mofetil (MMF) (23, 24).
Definitions
Relapse was defined as disease reoccurrence in bone marrow and/or extramedullary sites. Non-relapse mortality (NRM) was defined as death from any cause apart from relapse. Overall survival (OS) was defined as the time from transplant until death or last follow up, and disease-free survival (DFS) was defined as survival without relapse. Patients were classified as low/intermediate risk or high/very high risk based on the refinement of the disease risk index (DRI) (25). Diagnosis of acute and chronic GVHD (aGVHD and cGVHD) was made using established criteria (26, 27). The viral loads for Epstein-Barr virus (EBV) and cytomegalovirus (CMV) were monitored weekly for the first 3 months after transplantation, biweekly from the fourth to the sixth month post-transplant, and monthly from the seventh to the twelfth month post-transplant. Viremia was defined as a viral load in excess of 5 × 102 copies/ml.
Statistical Analysis
All clinical data were analyzed using SPSS 22.0 (IBM, Armonk, NY, USA) and R project 3.6.1 software (http://www.r-project.org). The clinical features for the samples were presented as median or percentage values. OS and DFS were calculated using the Kaplan–Meier method and compared using the log-rank test. The cumulative incidences of EBV viremia, CMV viremia, aGVHD, cGVHD, relapse, and NRM were estimated via the competing-risks model and compared using the Gray test. All variables with a p-value of <0.10 in the univariate analysis were then included in the multivariate analysis. Results were considered statistically significant when p < 0.05.
Results
Characteristics of Patients and Donors
The clinical features of these 246 donor-patient pairs are summarized in Table 1 . In this retrospective study, 142 (57.7%) patients with myeloid disease and 104 (42.3%) patients with lymphoid disease received haplo-HSCT at our center. Disease types included acute myeloid leukemia (AML, n = 115), myelodysplastic syndrome (MDS, n = 22), myeloproliferative neoplasm (MPN, n = 5), acute lymphoblastic leukemia (ALL, n = 93), and lymphoma (n = 11). The median age of the patients and donors in these groups were 30 years (range, 9–50 years) and 35 years (range, 11–59 years), respectively. The median mononuclear (MNC) cell and CD34+ cell counts in the grafts were 15.34 × 108/kg (range, 2.97–59.80 × 108/kg) and 6.30 × 106/kg (range, 0.27–34.37 × 106/kg), respectively. A total of 233 (94.7%) patients received the MAC regimen and 13 (5.3%) received the RIC regimen. ATG-F was used in 205 (83.3%) patients while the other 41 (16.7%) received ATG-G as part of their conditioning regimen. One hundred eighty-one (73.5%) patients received haplo-HSCT during their first remission (CR1); 73 (29.7%) patients were defined as high or very high risk based on the refinement of DRI (49 in the myeloid cohort and 24 in the lymphoid cohort, 34.5 vs 23.1%, p = 0.053). Most patients expressed HLA C1C1 or HLA C1C2 and only 4.5% presented with a HLA C2C2 ligand.
Table 1.
Clinical features of patients, donors, and transplants.
| Variables | All patients (246) | Myeloid cohort (142) | Lymphoid cohort (104) |
|---|---|---|---|
| Median patient age (years) | 30 (9–60) | 33 (9–60) | 24 (13–56) |
| Median donor age (years) | 35 (11–59) | 32 (11–55) | 38 (13–59) |
| Median MNC cells (×10 E8/kg) | 15.34 (2.97–59.80) | 14.36 (2.97–59.80) | 15.61 (5.80–46.14) |
| Median CD34+ cells (×10 E6/kg) | 6.30 (0.27–34.37) | 6.06 (0.27–34.37) | 7.03 (1.77–22.87) |
| Median follow up time (years) | 3.0 (0.1–5.5) | 3.0 (0.2–5.5) | 2.9 (0.1–5.5) |
| Patient sex | |||
| Male Female |
136 (55.3) 110 (44.7) |
77 (54.2) 65 (45.8) |
59 (56.7) 45 (43.3) |
| Donor/Patient sex combination | |||
| Female/Male Other combinations |
44 (17.9) 202 (82.1) |
27 (19.0) 115 (81.0) |
17 (16.3) 87 (83.7) |
| ABO blood mismatch | |||
| Identical Mismatch |
131 (53.3) 115 (46.7) |
73 (51.4) 69 (48.6) |
58 (55.8) 46 (44.2) |
| Diagnosis | / | / | |
| AML MDS MPN ALL Lymphoma |
115 (46.7) 22 (8.9) 5 (2.0) 93 (37.8) 11 (4.5) |
||
| Disease status at HSCT | |||
| CR1 >CR1 |
181 (73.5) 65 (26.4) |
99 (69.7) 43 (30.3) |
82 (78.9) 22 (21.2) |
| Disease risk index | |||
| Low/Int risk High/Very high risk |
173 (70.3) 73 (29.7) |
93 (65.5) 49 (34.5) |
80 (76.9) 24 (23.1) |
| Conditioning regimen | |||
| MAC RIC |
233 (94.7) 13 (5.3) |
133 (93.7) 9 (6.3) |
100 (96.2) 4 (3.8) |
| ATG | |||
| ATG-F ATG-G |
205 (83.3) 41 (16.7) |
117 (82.4) 25 (17.6) |
88 (84.6) 16 (15.4) |
| HLA ligands of patients | |||
| A3/A11+
Bw4+ C1/C1 C1/C2 C2/C2 |
115 (46.7) 148 (60.2) 167 (67.9) 68 (27.6) 11 (4.5) |
66 (46.5) 90 (63.4) 95 (66.9) 42 (29.6) 5 (3.5) |
49 (47.1) 58 (55.8) 72 (69.2) 24 (23.1) 6 (5.8) |
| Receptor-ligand (R-L) model | |||
| R-L A3/A11 mismatch R-L Bw4 mismatch R-L C mismatch |
131 (53.3) 96 (39.0) 176 (71.5) |
76 (53.5) 49 (34.5) 98 (69.0) |
55 (52.9) 47 (45.2) 78 (75.0) |
| Donor KIR genotype | |||
| AA B/x BA BB |
143 (58.1) 103 (41.9) 76 (30.9) 27 (11.0) |
81 (57.0) 61 (43.0) 45 (31.7) 16 (11.3) |
62 (59.6) 42 (40.4) 31 (29.8) 11 (10.6) |
| Donor activating KIR gene | |||
| KIR2DS1+
KIR2DS2+ KIR2DS3+ KIR2DS4+ KIR2DS5+ KIR3DS1+ |
83 (33.7) 46 (18.7) 41 (16.7) 238 (96.7) 57 (23.2) 85 (34.6) |
49 (34.5) 27 (19.0) 24 (16.9) 135 (95.1) 36 (25.4) 51 (35.9) |
34 (32.7) 19 (18.3) 17 (16.3) 103 (99.0) 21 (20.2) 34 (32.7) |
| iKIR-HLA pairs variation | |||
| A3/A11 | |||
| Decreased (D) Unchanged (U) Increased (I) |
40 (16.2) 166 (67.5) 40 (16.2) |
22 (15.5) 98 (69.0) 23 (16.2) |
18 (17.3) 68 (65.4) 17 (16.3) |
| Bw4 | |||
| Decreased (D) Unchanged (U) Increased (I) |
26 (10.6) 189 (76.8) 31 (12.6) |
14 (9.9) 109 (76.8) 19 (13.4) |
12 (11.5) 80 (76.9) 12 (11.5) |
| C | |||
| Decreased (D) Unchanged (U) Increased (I) |
43 (17.5) 163 (66.3) 40 (16.3) |
20 (14.1) 98 (69.0) 24 (16.9) |
23 (22.1) 65 (58.7) 16 (15.4) |
| EBV viremia | 90 (36.6) | 41 (28.9) | 49 (47.1) |
| CMV viremia | 160 (65.0) | 93 (65.5) | 67 (64.4) |
| aGVHD | |||
| Grade 0 Grade I Grade II Grade III Grade IV |
92 (37.4) 71 (28.9) 55 (22.4) 12 (4.9) 16 (6.5) |
55 (38.7) 44 (31.0) 30 (21.1) 7 (4.9) 6 (4.2) |
37 (35.6) 27 (26.0) 25 (24.0) 5 (4.8) 10 (9.6) |
| cGVHD | |||
| Not included No Mild Moderate Severe |
7 (2.8) 139 (56.5) 58 (23.6) 26 (10.6) 16 (6.5) |
2 (1.4) 76 (53.5) 39 (27.5) 14 (9.9) 11 (7.7) |
5 (4.8) 63 (60.6) 19 (18.3) 12 (11.5) 5 (4.8) |
| relapse | 55 (22.4) | 28 (16.9) | 27 (26.0) |
| NRM | 14 (5.7) | 4 (2.8) | 10 (9.6) |
| OS | 185(75.2) | 115 (82.4) | 70 (67.3) |
| DFS | 177 (72.0) | 110 (80.3) | 67 (64.4) |
AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; ALL, acute lymphoblastic leukemia; MNC, mononuclear; CR1, first complete remission; MAC, myeloablative conditioning; RIC, reduced-intensity conditioning; ATG, Antithymocyte Globulin; EBV, Epstein-Barr virus; CMV, cytomegalovirus; aGVHD, acute graft versus host disease; cGVHD, chronic graft versus host disease; NRM, non-relapse mortality; OS, overall survival; DFS, disease-free survival.
Of the 246 donors, 143 (58.1%) were KIR genotype AA, 76 (30.9%) were KIR BA, and 27 (11.0%) were KIR BB. Receptor-ligand (R-L) mismatches at the HLA-A3/A11 locus, HLA-Bw4 locus, and HLA-C locus were identified in 53.3, 39.0, and 71.5% of the donor-recipient pairs, respectively. After transplantation, 40 (16.2%) patients experienced a decrease in their iKIR-HLA A3/A11 pair, 26 (10.6%) exhibited decreased iKIR-HLA Bw4 pair, and 43 (17.5%) had decreased iKIR-HLA C (C1 or C2) pair.
EBV and CMV Viremia
During the first 180 days after HSCT, 90 (36.6%) patients developed EBV viremia. Disease category (myeloid or lymphoid) (p = 0.001), ATG source (p = 0.0003), and patient sex (p = 0.029) were identified as potent factors influencing EBV viremia ( Table 2 ). Multivariate analysis suggested that myeloid disease [hazard risk (HR) = 0.48, p = 0.0005] was a protective factor for EBV viremia, while ATG-G (HR = 2.58, p < 0.0001) and sex (male patients (HR = 1.57, p = 0.042)) were independent risk factors for EBV viremia ( Table 3 ). In lymphoid disease, KIR2DS2+ donors were found to exhibit a higher incidence of EBV viremia when compared with KIR2DS2− donors (63.2 vs 43.5%, p = 0.078), but this was not identified to be an independent effect in the multivariate analysis.
Table 2.
Univariate analysis of factors that influence transplant outcomes.
| Outcome and potent factors, % | All patients | P | Myeloid cohort | P | Lymphoid cohort | P |
|---|---|---|---|---|---|---|
| 1. EBV viremia* | ||||||
| Myeloid vs Lymphoid | 28.9 vs 47.1 | 0.001 | ||||
| ATG-G vs ATG-F | 58.6 vs 32.2 | 0.0003 | 48.0 vs 24.8 | 0.009 | 75.0 vs 42.1 | 0.003 |
| Male vs Female | 42.7 vs 29.1 | 0.029 | 35.1 vs 21.5 | 0.087 | 52.5 vs 40.0 | 0.177 |
| KIR2DS2+ vs KIR2DS2− | 43.5 vs 35.0 | 0.199 | 25.0 vs 29.7 | 0.643 | 63.2 vs 43.5 | 0.078 |
| 2. CMV viremia* | ||||||
| ATG-G vs ATG-F | 78.1 vs 62.4 | 0.003 | 72.0 vs 64.1 | 0.199 | 87.5 vs 60.4 | 0.001 |
| KIR2DS1+ vs KIR2DS1− | 67.5 vs 63.8 | 0.935 | 77.6 vs 59.1 | 0.029 | 52.9 vs 70.0 | 0.030 |
| KIR2DS3+ vs KIR2DS3− | 63.4 vs 65.4 | 0.695 | 79.2 vs 62.7 | 0.191 | 41.2 vs 69.0 | 0.057 |
| KIR3DS1+ vs KIR3DS1− | 67.9 vs 63.6 | 0.997 | 78.0 vs 58.7 | 0.041 | 52.9 vs 70.0 | 0.030 |
| R-L C (mismatch vs match) | 62.5 vs 71.4 | 0.079 | 63.3 vs 70.5 | 0.267 | 61.5 vs 73.1 | 0.151 |
| 3. Grade II-IV aGVHD* | ||||||
| High/Very high risk vs Low/Int risk | 41.1 vs 30.6 | 0.080 | 38.8 vs 25.8 | 0.068 | 45.8 vs 36.3 | 0.454 |
| MAC vs RIC | 35.2 vs 7.7 | 0.041 | 31.6 vs 11.1 | 0.178 | 40.0 vs 0.0 | 0.125 |
| R-L C (mismatch vs match) | 31.25 vs 40.0 | 0.256 | 29.6 vs 31.8 | 0.853 | 33.3 vs 53.9 | 0.095 |
| 4. Moderate to severe cGVHD* | ||||||
| KIR2DS2+ vs KIR2DS2− | 6.5 vs 19.2 | 0.048 | 7.4 vs 20.3 | 0.133 | 5.3 vs 17.7 | 0.197 |
| KIR2DS3+ vs KIR2DS3− | 7.3 vs 18.7 | 0.083 | 12.5 vs 19.0 | 0.487 | 0.0 vs 18.4 | 0.057 |
| 5. 3-yr CIR | ||||||
| Myeloid vs Lymphoid | 17.3 vs 26.2 | 0.087 | ||||
| High/Very high risk vs Low/Int risk | 32.9 vs 16.1 | 0.002 | 26.5 vs 12.4 | 0.031 | 45.8 vs 20.4 | 0.009 |
| CR1 vs >CR1 | 16.5 vs 33.9 | 0.002 | 13.6 vs 25.7 | 0.070 | 23.5 vs 36.4 | 0.202 |
| iKIR-HLA C variation (D vs U+I) | 38.1 vs 17.5 | 0.005 | 40.0 vs 13.5 | 0.004 | 35.8 vs 23.5 | 0.317 |
| 6. 3-yr NRM | ||||||
| Myeloid vs Lymphoid | 2.8 vs 9.9 | 0.024 | ||||
| High/Very high risk vs Low/Int risk | 1.4 vs 7.7 | 0.057 | 2.0 vs 3.2 | 0.683 | 0.0 vs 12.8 | 0.070 |
| iKIR-HLA C variation (D vs U+I) | 4.7 vs 6.0 | 0.745 | 10.0 vs 1.6 | 0.037 | 0.0 vs 12.7 | 0.078 |
| KIR2DS3+ vs KIR2DS3− | 0.0 vs 7.0 | 0.086 | 0.0 vs 3.4 | 0.362 | 0.0 vs 11.8 | 0.145 |
| KIR B/x vs KIR AA | 2.9 vs 7.9 | 0.112 | 0 vs 4.9 | 0.079 | 7.1 vs 11.7 | 0.497 |
| 7. 3-yr OS | ||||||
| Myeloid vs Lymphoid | 81.6 vs 67.7 | 0.016 | ||||
| ATG-G vs ATG-F | 75.5 vs 75.8 | 0.861 | 92.0 vs 79.3 | 0.147 | 49.2 vs 71.0 | 0.041 |
| High/Very high risk vs Low/Int risk | 67.0 vs 79.3 | 0.036 | 71.4 vs 87.1 | 0.029 | 58.3 vs 70.5 | 0.202 |
| CR1 vs >CR1 | 79.7 vs 64.5 | 0.010 | 85.8 vs 72.0 | 0.053 | 72.4 vs 50.0 | 0.025 |
| iKIR-HLA C variation (D vs U+I) | 65.1 vs 77.9 | 0.093 | 55.0 vs 86.0 | 0.0006 | 73.9 vs 65.9 | 0.418 |
| 8. 3-yr DFS | ||||||
| Myeloid vs Lymphoid | 79.9 vs 63.9 | 0.006 | ||||
| ATG-G vs ATG-F | 73.2 vs 73.3 | 0.835 | 78.3 vs 88.0 | 0.293 | 50.0 vs 66.5 | 0.085 |
| High/Very high risk vs Low/Int risk | 65.7 vs 76.2 | 0.080 | 71.4 vs 84.4 | 0.066 | 54.2 vs 66.8 | 0.218 |
| CR1 vs >CR1 | 76.7 vs 63.0 | 0.024 | 83.3 vs 72.0 | 0.107 | 67.7 vs 50.0 | 0.085 |
| iKIR-HLA C variation (D vs U+I) | 57.3 vs 76.5 | 0.016 | 50.0 vs 84.9 | 0.0001 | 64.2 vs 63.9 | 0.813 |
*Estimations of cumulative incidence are given at 100 days post-HSCT for aGVHD; 180 days post-HSCT for EBV and CMV viremia; 3 years post-HSCT for cGVHD.
Potent factors with p < 0.10 were in bold type.
Table 3.
Multivariate analysis of factors that influence transplant outcomes.
| Outcomes and significant factors | All patients | Myeloid cohort | Lymphoid cohort | |||
|---|---|---|---|---|---|---|
| P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | |
| 1. EBV viremia* | ||||||
| ATG-G vs ATG-F | <0.0001 | 2.58 (1.61–4.13) | 0.007 | 2.51 (1.28–4.93) | 0.004 | 2.66 (1.37–5.14) |
| Male vs Female | 0.042 | 1.57 (1.02–2.41) | ||||
| Myeloid vs Lymphoid | 0.0005 | 0.48 (0.31–0.72) | ||||
| 2. CMV viremia* | ||||||
| ATG-G vs ATG-F | 0.008 | 1.70 (1.15–2.51) | 0.005 | 1.76 (1.19–2.59) | ||
| 3. 3-yr CIR | ||||||
| CR1 vs >CR1 | 0.029 | 0.53 (0.30–0.94) | ||||
| iKIR-HLA C variation (D vs U+I) | 0.033 | 1.95 (1.06–3.61) | 0.019 | 2.95 (1.19–7.27) | ||
| 4. 3-yr OS | ||||||
| Myeloid vs Lymphoid | 0.006 | 0.49 (0.29–0.82) | ||||
| CR1 vs >CR1 | 0.004 | 0.46 (0.27–0.78) | 0.029 | 0.45 (0.22–0.92) | ||
| iKIR-HLA C variation (D vs U+I) | 0.001 | 3.74 (1.66–8.39) | ||||
| 5. 3-yr DFS | ||||||
| Myeloid vs lymphoid | 0.003 | 0.47 (0.28–0.77) | ||||
| CR1 vs >CR1 | 0.009 | 0.51 (0.31–0.84) | 0.034 | 0.47 (0.24–0.94) | ||
| iKIR-HLA C variation (D vs U+I) | 0.0004 | 4.05 (1.87–8.80) | ||||
*Estimations of cumulative incidence are given at 100 days post-HSCT for aGVHD; 180 days post-HSCT for EBV and CMV viremia.
Significant factors with P < 0.05 were in bold type.
The CI for CMV viremia within 180 days of transplant was 65.0% (78.1% in patients treated with ATG-G and 62.4% in patients treated with ATG-F, p = 0.003). Donor-patient pairs with R-L mismatch at HLA-C locus tended to experience a lower CI for CMV viremia than did donor-patient R-L C matched pairs (62.5 vs 71.4, p = 0.079). The multivariate analysis revealed that only ATG-G was an independent risk factor for CMV viremia (HR = 1.70, p = 0.008).
aGVHD and cGVHD
Following transplantation, 83 (33.7%) developed grade II–IV aGVHD (aGVHD2–4). As expected, a significant reduction in aGVHD2–4 occurrence was found in patients receiving RIC conditioning compared with patients receiving MAC conditioning (7.7 vs 35.2%, p = 0.041). Patients with low and intermediate risk also experienced a lower CI of aGVHD2–4 (30.6 vs 41.1%, p = 0.080). However, none of these factors remained significant in the multivariate analysis. In lymphoid cohort, there was a trend that donor-patient pairs with R-L mismatch on HLA-C locus experienced a lower aGVHD2–4 (33.3 vs 53.9%, P = 0.095).
Among patients who survived more than 100 days after transplantation, 100 (41.8%) patients developed cGVHD and 42 (17.6%) of them had moderate to severe cGVHD. Univariate analysis identified KIR2DS2 (p = 0.048) and KIR2DS3 (p = 0.083) as two potent protective factors for moderate to severe cGVHD. Nevertheless, no such correlations were found in the multivariate analysis.
Relapse and NRM
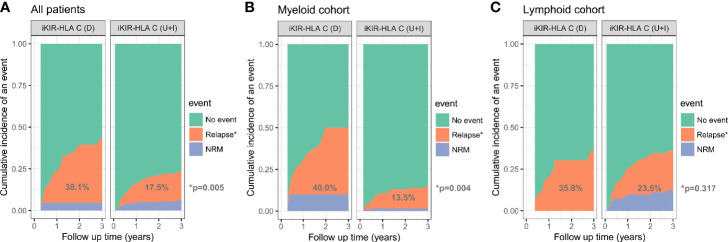
After a median follow up time of 3.0 years (yr) (range, 0.1–5.5 yr), 55 (22.4%) patients experienced relapse. Patients with lymphoid disease experienced a higher 3-yr relapse rate than patients with myeloid disease (26.2 vs 17.3%, p = 0.087). The CI for 3-yr relapse was also higher in patients with high/very high-risk disease (32.9 vs 16.1%, p = 0.002). Patients who received HSCT at CR1 experienced a lower 3-yr relapse rate than the other group (16.5 vs 33.9%, p = 0.002). No significant differences in relapse rate were found using the receptor ligand model and activating KIRs. However, decreased iKIR-HLA C pair were associated with a higher risk for 3-yr relapse (38.1 vs 17.5%, p = 0.005). When analyzed separately, the discrepancy in relapse rates was more evident in the myeloid cohort (40.0 vs 13.5%, p = 0.004) than in the lymphoid cohort (35.8 vs 23.5%, p = 0.317) ( Figure 1 ). Multivariate analysis revealed that CR1 (HR = 0.53, P = 0.029) and decreased iKIR-HLA C pair (HR = 1.95, P = 0.033) were independent factors for relapse for the entire cohort, and the adverse effects of decreased iKIR-HLA C pair on the 3-yr relapse rate was more evident in myeloid disease (HR = 2.95, p = 0.019).
Figure 1.
Cumulative incidence of relapse among all patients (A), the myeloid cohort (B), and the lymphoid cohort (C), based on the variation (D, decreased; U, unchanged; I, increased) in iKIR-HLA C pair post-transplantation.
A total of 14 (5.7%) patients experienced NRM at a median follow-up time of 0.3 yr (range, 0.1–2.8 yr), 6 (2.4%) patients died of severe GVHD (5 aGVHD and 1 cGVHD), 7 (2.8%) patients died from severe infection (6 pulmonary infections and 1 sepsis), and 1 (0.4%) patient with primary poor graft function (28) died from an intracranial hemorrhage. No variables were found to be significant predictors of NRM.
OS and DFS
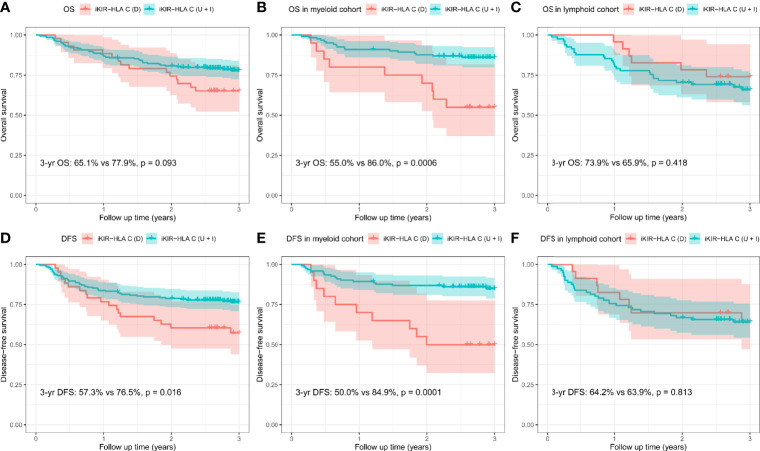
The CI for 3-yr OS was 75.6% for all patients. Disease category (p = 0.016), disease status (p = 0.010), and disease risk index (p = 0.036) were all found to influence 3-yr OS in the univariate analysis. In addition, the 3-yr OS rate in transplants with decreased iKIR-HLA C pair was shown to be 65.1% [95% confidence interval (CI): 52.3–81.0%], which was lower than those with unchanged or increased iKIR-HLA C pair (77.9%, 95% CI: 72.3–83.9%, p = 0.093), and the negative impact of decreased iKIR-HLA C pair was more apparent in the myeloid cohort [55.0% (95% CI: 37.0–81.8%) vs 86.0 (95% CI: 80.0–92.4%), p = 0.0006] than in the lymphoid cohort [73.9% (95% CI: 58.0–94.2%) vs 65.9% (95% CI: 56.1–77.3%), p = 0.418] ( Figures 2A–C ). In the lymphoid cohort, patients who received ATG-G prior to transplantation experienced a lower 3-yr OS (49.2 vs 71.0%, p = 0.041). Multivariate analysis identified myeloid disease (HR = 0.49, p = 0.006) and CR1 (HR = 0.46, p = 0.004) as protective factors for 3-yr OS. CR1 in the lymphoid cohort (HR = 0.45, p = 0.029) remained significant in multivariate analysis, and decreased iKIR-HLA C pair conferred a poorer 3-yr OS in the myeloid cohort (HR = 3.74, p = 0.001).
Figure 2.
Overall survival (OS) and disease-free survival (DFS) rates for all patients (A, D), the myeloid cohort (B, E), and the lymphoid cohort (C, F), based on the variation (D, decreased; U, unchanged; I, increased) in their iKIR-HLA C pair post-transplantation.
In addition, dramatically reduced 3-yr DFS was observed when iKIR-HLA C pair was decreased both in the entire cohort [57.3% (95% CI: 44.0–74.6%) vs 76.5% (95% CI: 70.8–82.6%), p = 0.016] and the myeloid cohort [50.0% (95% CI: 32.3–77.5%) vs 84.9% (95% CI: 78.6–91.6%), p = 0.0001]. For patients with lymphoid disease, variation in iKIR-HLA C pair was not associated with DFS [64.2% (95% CI: 47.0–87.8%) vs 63.9% (95% CI: 54.2–75.4%), p = 0.813] ( Figures 2D–F ). In the multivariate analysis, myeloid disease (HR = 0.47, p = 0.003) and CR1 (HR = 0.51, p = 0.009) were shown to be independent factors influencing DFS. A significantly reduced DFS was also observed in myeloid patients with decreased iKIR-HLA C pair (HR = 4.05, p = 0.0004).
Discussion
There has been a longstanding debate about the impact of KIR alloreactivity on clinical outcomes. It was only recently revealed that reconstituted KIR are educated by HLA ligands and that the loss of the cognate ligands dampens NK cell functions (17–19). This means that searching for donors who exhibit the greatest NK cell function in recipients rather than “match or mismatch” would be a more reliable measure for predicting transplant success.
Previously, Nowak et al. proposed the iKIR-HLA model that could be used to predict transplant outcomes (20–22). Among the multiple interactions between the iKIRs and HLA ligands, we identified that only decreased iKIR-HLA C pair post transplantation was a negative indicator for relapse and survival, especially in patients with myeloid disease. Nevertheless, variations in iKIR-HLA A3/A11 pair and iKIR-HLA Bw4 pair did not influence the treatment outcomes.
It is widely accepted that almost all HLA C molecules are recognized by iKIRs. However, only a minority of HLA B and HLA A epitopes act as KIR ligands (29–31). Similarly, all patients in our cohort expressed at least one HLA C ligand, while the HLA Bw4 and A3/A11 ligands were expressed at a frequency of 60.2 and 46.7%, respectively. This suggests that the HLA C ligands play a dominant role in KIR education (32). Given this, reconstituted NK cells with decreased iKIR-HLA C pair may exhibit impaired anti-tumor effects (18, 19). In addition, the expression levels of HLA A and B ligands on normal cells are more than tenfold higher than that of the HLA C (33), this means that when cancerous cells downregulate HLA class I antigens to escape immune surveillance, the stability of the self-tolerance mediated by iKIR-HLA C interactions is more vulnerable to be broken. In other words, HLA-C may play a major role in missing-self recognition and modulate NK cell activation. Moreover, Pende et al. found that lymphoblastic leukemias express a higher surface density of HLA class I molecules than myeloid leukemias (34). Verheyden et al. went on to test the expression of HLA ligands in normal T cells, AML cells, B-ALL cells, and B-chronic lymphoid leukemic (B-CLL) cells. Interestingly, only HLA C were dramatically downregulated on all types of leukemic cells as compared with their healthy control, with this downregulation being the most apparent in AML cells (35). Makanga et al. demonstrated that CD57+ and KIR+ NK cells from healthy individuals exhibited the highest degree of cytotoxicity against AML blasts, while ALL targets were less susceptible to KIR+ NK subsets compared with NKG2A+ NK subsets (36). On the basis of previous studies, we hypothesize that KIR may have a minor impact on the elimination of lymphoblastic leukemias, and patients with myeloid disease are more likely to benefit from well KIR-educated NK cells.
In many European studies, aKIRs, especially KIR2DS1 (37–39) and KIR2DS2 (40, 41), have been shown to be associated with improved survival or reduced relapse. Yet, as reported in several studies from East Asia (42–45), aKIRs were not found to grant any survival advantage or relapse protection to the patients in our cohort. One reason for this may be the genetic differences between these different ethnic groups. Single et al. revealed that almost 46.7% Europeans express the KIR2DS2 gene, and 66.5% present the HLA C2 ligand for KIR2DS1 (46). However, both the KIR2DS2 gene (18.7%) and the HLA C2 ligand (32.1%) were expressed at much lower frequencies in this study. The KIR2DS1 gene frequency in our cohort was also a bit lower than those of the European populations (33.7 vs 37.8%). Thus, we speculate that KIR2DS1 may have a reduced chance of activation resulting from the absence of its cognate ligand, and that the beneficial impact of KIR2DS2 on transplant outcomes may be more apparent in a larger cohort of Chinese patients.
Additionally, we could not find evidence of any significant association between receptor ligand mismatch and clinical outcomes. Since mature donor lymphocytes are mostly eliminated following ATG treatment, the transient expression of alloreactive NK cells in the recipients may not be sufficient to influence GVHD (47–49). After which the reconstituted NK cells expressing non-self KIRs may not exhibit enough cytotoxicity to eliminate the remaining leukemic cells (17–19).
In summary, we conclude that when using ATG-based haplo-HSCT, deceased iKIR-HLA C pair should be avoided during donor selection, especially for patients with myeloid disease. The exact role of the aKIRs in the Chinese population still needs to be explored in future studies.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Review Committee of the First Affiliated Hospital of Zhejiang University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
HH designed the study and supervised the analyses and manuscript preparation. YZ, FG, and YW collected and analyzed the data, YZ and FG wrote the manuscript. All authors discussed and interpreted the results. All authors contributed to the article and approved the submitted version. YZ and FG contributed equally to this work and should be considered as co-first authors.
Funding
This work was supported by the National Natural Science Foundation of China (81670148 and 81730008) and Key Project of Science and Technology Department of Zhejiang Province (2019C03016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Shanghai Tissuebank Diagnostics Co., Ltd. and Zhejiang Blood Center for valuable assistance in HLA and KIR genotyping.
References
- 1. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol (2008) 9(5):503–10. 10.1038/ni1582 [DOI] [PubMed] [Google Scholar]
- 2. Caligiuri MA. Human natural killer cells. Blood (2008) 112(3):461–9. 10.1182/blood-2007-09-077438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pegram HJ, Andrews DM, Smyth MJ, Darcy PK, Kershaw MH. Activating and inhibitory receptors of natural killer cells. Immunol Cell Biol (2011) 89(2):216–24. 10.1038/icb.2010.78 [DOI] [PubMed] [Google Scholar]
- 4. Manser AR, Weinhold S, Uhrberg M. Human KIR repertoires: shaped by genetic diversity and evolution. Immunol Rev (2015) 267(1):178–96. 10.1111/imr.12316 [DOI] [PubMed] [Google Scholar]
- 5. Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol (2001) 1(1):41–9. 10.1038/35095564 [DOI] [PubMed] [Google Scholar]
- 6. Sivori S, Vacca P, Del Zotto G, Munari E, Mingari MC, Moretta L. Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell Mol Immunol (2019) 16(5):430–41. 10.1038/s41423-019-0206-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science (2002) 295(5562):2097–100. 10.1126/science.1068440 [DOI] [PubMed] [Google Scholar]
- 8. Leung W, Iyengar R, Turner V, Lang P, Bader P, Conn P, et al. Determinants of antileukemia effects of allogeneic NK cells. J Immunol (2004) 172(1):644–50. 10.4049/jimmunol.172.1.644 [DOI] [PubMed] [Google Scholar]
- 9. Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood (2009) 113(3):726–32. 10.1182/blood-2008-07-171926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao F, Ye Y, Gao Y, Huang H, Zhao Y. Influence of KIR and NK Cell Reconstitution in the Outcomes of Hematopoietic Stem Cell Transplantation. Front Immunol (2020) 11:2022. 10.3389/fimmu.2020.02022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heidenreich S, Kroger N. Reduction of Relapse after Unrelated Donor Stem Cell Transplantation by KIR-Based Graft Selection. Front Immunol (2017) 8:41. 10.3389/fimmu.2017.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Elssen C, Ciurea SO. NK Cell Alloreactivity in Acute Myeloid Leukemia in the Post-transplant Cyclophosphamide Era. Am J Hematol (2020) 95(12):1590–8. 10.1002/ajh.25983 [DOI] [PubMed] [Google Scholar]
- 13. Vago L, Forno B, Sormani MP, Crocchiolo R, Zino E, Di Terlizzi S, et al. Temporal, quantitative, and functional characteristics of single-KIR-positive alloreactive natural killer cell recovery account for impaired graft-versus-leukemia activity after haploidentical hematopoietic stem cell transplantation. Blood (2008) 112(8):3488–99. 10.1182/blood-2007-07-103325 [DOI] [PubMed] [Google Scholar]
- 14. Russo A, Oliveira G, Berglund S, Greco R, Gambacorta V, Cieri N, et al. NK cell recovery after haploidentical HSCT with posttransplant cyclophosphamide: dynamics and clinical implications. Blood (2018) 131(2):247–62. 10.1182/blood-2017-05-780668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity (2006) 25(2):331–42. 10.1016/j.immuni.2006.06.013 [DOI] [PubMed] [Google Scholar]
- 16. Belanger S, Tu MM, Rahim MM, Mahmoud AB, Patel R, Tai LH, et al. Impaired natural killer cell self-education and “missing-self” responses in Ly49-deficient mice. Blood (2012) 120(3):592–602. 10.1182/blood-2012-02-408732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boudreau JE, Liu XR, Zhao Z, Zhang A, Shultz LD, Greiner DL, et al. Cell-Extrinsic MHC Class I Molecule Engagement Augments Human NK Cell Education Programmed by Cell-Intrinsic MHC Class I. Immunity (2016) 45(2):280–91. 10.1016/j.immuni.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Landtwing V, Raykova A, Pezzino G, Beziat V, Marcenaro E, Graf C, et al. Cognate HLA absence in trans diminishes human NK cell education. J Clin Invest (2016) 126(10):3772–82. 10.1172/JCI86923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao XY, Yu XX, Xu ZL, Cao XH, Huo MR, Zhao XS, et al. Donor and host coexpressing KIR ligands promote NK education after allogeneic hematopoietic stem cell transplantation. Blood Adv (2019) 3(24):4312–25. 10.1182/bloodadvances.2019000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rogatko-Koros M, Mika-Witkowska R, Bogunia-Kubik K, Wysoczanska B, Jaskula E, Koscinska K, et al. Prediction of NK Cell Licensing Level in Selection of Hematopoietic Stem Cell Donor, Initial Results. Arch Immunol Ther Exp (Warsz) (2016) 64(Suppl 1):63–71. 10.1007/s00005-016-0438-2 [DOI] [PubMed] [Google Scholar]
- 21. Graczyk-Pol E, Rogatko-Koros M, Nestorowicz K, Gwozdowicz S, Mika-Witkowska R, Pawliczak D, et al. Role of donor HLA class I mismatch, KIR-ligand mismatch and HLA:KIR pairings in hematological malignancy relapse after unrelated hematopoietic stem cell transplantation. HLA (2018) 92(Suppl 2):42–6. 10.1111/tan.13386 [DOI] [PubMed] [Google Scholar]
- 22. Nowak J, Gwozdowicz S, Graczyk-Pol E, Mika-Witkowska R, Rogatko-Koros M, Nestorowicz K, et al. Epstein-Barr virus infections are strongly dependent on activating and inhibitory KIR-HLA pairs after T-cell replate unrelated hematopoietic stem cell transplantation, the principles, and method of pairing analysis. HLA (2019) 94(Suppl 2):40–8. 10.1111/tan.13770 [DOI] [PubMed] [Google Scholar]
- 23. Luo Y, Xiao H, Lai X, Shi J, Tan Y, He J, et al. T-cell-replete haploidentical HSCT with low-dose anti-T-lymphocyte globulin compared with matched sibling HSCT and unrelated HSCT. Blood (2014) 124(17):2735–43. 10.1182/blood-2014-04-571570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao Y, Luo Y, Shi J, Cai Z, Huang H. Second-generation tyrosine kinase inhibitors combined with stem cell transplantation in patients with imatinib-refractory chronic myeloid leukemia. Am J Med Sci (2014) 347(6):439–45. 10.1097/MAJ.0000000000000186 [DOI] [PubMed] [Google Scholar]
- 25. Armand P, Kim HT, Logan BR, Wang Z, Alyea EP, Kalaycio ME, et al. Validation and refinement of the Disease Risk Index for allogeneic stem cell transplantation. Blood (2014) 123(23):3664–71. 10.1182/blood-2014-01-552984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris AC, Young R, Devine S, Hogan WJ, Ayuk F, Bunworasate U, et al. International, Multicenter Standardization of Acute Graft-versus-Host Disease Clinical Data Collection: A Report from the Mount Sinai Acute GVHD International Consortium. Biol Blood Marrow Transplant (2016) 22(1):4–10. 10.1016/j.bbmt.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jagasia MH, Greinix HT, Arora M, Williams KM, Wolff D, Cowen EW, et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group report. Biol Blood Marrow Transplant (2015) 21(3):389–401.e1. 10.1016/j.bbmt.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao Y, Gao F, Shi J, Luo Y, Tan Y, Lai X, et al. Incidence, Risk Factors, and Outcomes of Primary Poor Graft Function after Allogeneic Hematopoietic Stem Cell Transplantation. Biol Blood Marrow Transplant (2019) 25(9):1898–907. 10.1016/j.bbmt.2019.05.036 [DOI] [PubMed] [Google Scholar]
- 29. Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol (2013) 13(2):133–44. 10.1038/nri3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A (1993) 90(24):12000–4. 10.1073/pnas.90.24.12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol (2010) 185(7):4238–51. 10.4049/jimmunol.1001494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anderson SK. Molecular evolution of elements controlling HLA-C expression: Adaptation to a role as a killer-cell immunoglobulin-like receptor ligand regulating natural killer cell function. HLA (2018) 92(5):271–8. 10.1111/tan.13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Apps R, Meng Z, Del Prete GQ, Lifson JD, Zhou M, Carrington M. Relative expression levels of the HLA class-I proteins in normal and HIV-infected cells. J Immunol (2015) 194(8):3594–600. 10.4049/jimmunol.1403234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pende D, Spaggiari GM, Marcenaro S, Martini S, Rivera P, Capobianco A, et al. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112). Blood (2005) 105(5):2066–73. 10.1182/blood-2004-09-3548 [DOI] [PubMed] [Google Scholar]
- 35. Verheyden S, Ferrone S, Mulder A, Claas FH, Schots R, De Moerloose B, et al. Role of the inhibitory KIR ligand HLA-Bw4 and HLA-C expression levels in the recognition of leukemic cells by Natural Killer cells. Cancer Immunol Immunother (2009) 58(6):855–65. 10.1007/s00262-008-0601-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makanga DR, Da Rin de Lorenzo F, David G, Willem C, Dubreuil L, Legrand N, et al. Genetic and Molecular Basis of Heterogeneous NK Cell Responses against Acute Leukemia. Cancers (Basel) (2020) 12(7):1927. 10.3390/cancers12071927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schellekens J, Rozemuller EH, Petersen EJ, van den Tweel JG, Verdonck LF, Tilanus MG. Activating KIRs exert a crucial role on relapse and overall survival after HLA-identical sibling transplantation. Mol Immunol (2008) 45(8):2255–61. 10.1016/j.molimm.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 38. Mancusi A, Ruggeri L, Urbani E, Pierini A, Massei MS, Carotti A, et al. Haploidentical hematopoietic transplantation from KIR ligand-mismatched donors with activating KIRs reduces nonrelapse mortality. Blood (2015) 125(20):3173–82. 10.1182/blood-2014-09-599993 [DOI] [PubMed] [Google Scholar]
- 39. Tordai A, Bors A, Kiss KP, Balassa K, Andrikovics H, Batai A, et al. Donor KIR2DS1 reduces the risk of transplant related mortality in HLA-C2 positive young recipients with hematological malignancies treated by myeloablative conditioning. PLoS One (2019) 14(6):e0218945. 10.1371/journal.pone.0218945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Impola U, Turpeinen H, Alakulppi N, Linjama T, Volin L, Niittyvuopio R, et al. Donor Haplotype B of NK KIR Receptor Reduces the Relapse Risk in HLA-Identical Sibling Hematopoietic Stem Cell Transplantation of AML Patients. Front Immunol (2014) 5:405. 10.3389/fimmu.2014.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Babor F, Peters C, Manser AR, Glogova E, Sauer M, Potschger U, et al. Presence of centromeric but absence of telomeric group B KIR haplotypes in stem cell donors improve leukaemia control after HSCT for childhood ALL. Bone Marrow Transplant (2019) 54(11):1847–58. 10.1038/s41409-019-0543-z [DOI] [PubMed] [Google Scholar]
- 42. Zhao XY, Huang XJ, Liu KY, Xu LP, Liu DH. Prognosis after unmanipulated HLA-haploidentical blood and marrow transplantation is correlated to the numbers of KIR ligands in recipients. Eur J Haematol (2007) 78(4):338–46. 10.1111/j.1600-0609.2007.00822.x [DOI] [PubMed] [Google Scholar]
- 43. Yabe T, Matsuo K, Hirayasu K, Kashiwase K, Kawamura-Ishii S, Tanaka H, et al. Donor killer immunoglobulin-like receptor (KIR) genotype-patient cognate KIR ligand combination and antithymocyte globulin preadministration are critical factors in outcome of HLA-C-KIR ligand-mismatched T cell-replete unrelated bone marrow transplantation. Biol Blood Marrow Transplant (2008) 14(1):75–87. 10.1016/j.bbmt.2007.09.012 [DOI] [PubMed] [Google Scholar]
- 44. Park S, Kim K, Jang JH, Kim SJ, Kim WS, Kang ES, et al. KIR alloreactivity based on the receptor-ligand model is associated with improved clinical outcomes of allogeneic hematopoietic stem cell transplantation: Result of single center prospective study. Hum Immunol (2015) 76(9):636–43. 10.1016/j.humimm.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 45. Hosokai R, Masuko M, Shibasaki Y, Saitoh A, Furukawa T, Imai C. Donor Killer Immunoglobulin-Like Receptor Haplotype B/x Induces Severe Acute Graft-versus-Host Disease in the Presence of Human Leukocyte Antigen Mismatch in T Cell-Replete Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant (2017) 23(4):606–11. 10.1016/j.bbmt.2016.12.638 [DOI] [PubMed] [Google Scholar]
- 46. Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet (2007) 39(9):1114–9. 10.1038/ng2077 [DOI] [PubMed] [Google Scholar]
- 47. Leung W, Iyengar R, Leimig T, Holladay MS, Houston J, Handgretinger R. Phenotype and function of human natural killer cells purified by using a clinical-scale immunomagnetic method. Cancer Immunol Immunother (2005) 54(4):389–94. 10.1007/s00262-004-0609-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grullich C, Ziegler C, Finke J. Rabbit anti T-lymphocyte globulin induces apoptosis in peripheral blood mononuclear cell compartments and leukemia cells, while hematopoetic stem cells are apoptosis resistant. Biol Blood Marrow Transplant (2009) 15(2):173–82. 10.1016/j.bbmt.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 49. Bosch M, Dhadda M, Hoegh-Petersen M, Liu Y, Hagel LM, Podgorny P, et al. Immune reconstitution after anti-thymocyte globulin-conditioned hematopoietic cell transplantation. Cytotherapy (2012) 14(10):1258–75. 10.3109/14653249.2012.715243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.