Graphical abstract
Inhibition of ADP-ribose-1 monophosphatase enzyme of COVID-19 by a copper complex can inhibit coronavirus infection.

Keywords: COVID-19, Chloroquine, Hydroxychloroquine, Copper complex, ADP-ribose-1 monophosphatase enzyme
Abstract
The pandemic by COVID-19 is hampering everything on the earth including physical and mental health, daily life and global economy. At the moment, there are no defined drugs, while few vaccines are available in the market to combat SARS-CoV-2. Several organic molecules were designed and tested against the virus but they did not show promising activity. In this work we designed two copper complexes from the ligands analogues with chloroquine and hydroxychloroquine. Both the ligands and complexes were well characterized by using various spectroscopic, thermal and X-ray diffraction techniques. Both the complexes as well as ligands were screened through in silico method with the chloroquine and hydroxychloroquine which essentially proved pivotal for successful understanding towards the target protein and their mechanism of action. The results indicated that the balanced hydrophobic and polar groups in the complexes favor their binding in the active site of the viral ADP-ribose-1 monophosphatase enzyme over the parent organic molecules.
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the cause of COVID-19 pandemic [1]. The transmission stages of the virus are asymptomatic, moderate, extreme and critical. The major symptoms associated with these infections are high fever, pneumonia, encephalopathy, dry cough and dyspnea and in severe cases, it may causes heart failure and kidney injury [2]. Presently, there are no defined drugs, although few vaccines are available in the market with a hope to combat SARS-CoV-2 deadly viral infection. It is well know that the viral proteases have long been shown to be effective targets of antiviral therapies. This is the reason why most of the antiviral drugs have been used for the COVID-19 treatment at an early stage of diagnosis [3].
Navarro et al. reported that the incorporation of metal centers into organic pharmacophores displayed better anti-malarial activity than chloroquine alone, against the chloroquine-susceptible strain which clearly compliment the molecular diversity created by purely organic scaffolds [4]. Quinoline moiety based anti-malarial drug such as chloroquine and hydroxychloroquine have been approved for the Food and Drug Administration (FDA) for the treatment of COVID-19 [5]. These medicines are only use for emergency purposes because of their some dose related toxic effect in critical cases [5]. However, these medicines are safe if taken within clinically instructed dose limits. Therefore, development of new advanced molecules is on demand.
Interestingly, metal-based drugs offer potential benefits over the various non-steroidal anti-inflammatory drugs (NSAIDs), broad-spectrum fluoroquinolone and ornidazole drugs and possess improved pharmacological and toxicological behaviors, exemplifying the role of the metal ions [6], [7], [8]. This is because of the fact that metal can coordinate ligands in a particular geometries with favorable thermodynamic and kinetic properties. Thus allowing: (i) synergistic effects of the ligands and the coordination residue, (ii) the tailoring/tuning of the molecule to recognize and interact with specific biological target, (iii) protection from the enzymatic degradation of the drug and (iv) superior delivery of pharmacophore through cell membranes and hence, pharmacokinetics profile of the drug (absorption, distribution and excretion etc) may get altered upon metal ion complexation [9]. Among the transition metals, copper is a physiologically important endogenous metal ion and cofactor of several enzymes involved in oxidative metabolism, ceruloplasmine, superoxide dismutase, etc that carry out fundamental biological functions, important in cellular growth and development [10].
Some time it has been observed that metal complex of drugs molecules or allied ligands performed more efficiently than that of the compound itself. The Mn2+ complex of curcumin (a well-known anti-bacterial) show strong activity against multiple strain of both gram-positive and gram-negative bacteria [11]. A large number of copper(II) complexes have also been successfully tested in various diseases including cancer [12]. Recently, Cu2+-curcumin [13] and Cu2+-ornidazole [6] complexes have efficiently been designed to target nucleic acid.
Recently, several authors have brought interest in copper well prior to the Covid-19 viral epidemic as an alternate source to make the immune system competent to fight somehow against the SARS‐CoV‐2 [14]. It has been recognized that copper can boost the host's immune system response against pathogens via cell-mediated. immunity such as T helper cells, B cells, neutrophils natural killer (NK) cells, and macrophages [15].
All the above facts are prompted us to design new compounds for COVID-19 with the help of structural knowledge of chloroquine and hydroxychloroquine (HCQ). In this case, the designed organic molecules should be capable of coordination with Cu2+ to increase their activity. In recent time, computer aided drug designing is a reliable and most used methodology [16], [17], [18], [19]. This technology can be utilized to find COVID-19 target protein for the designed Cu-complexes and explore the structure activity relationship of them. Synthesis of these molecules through a simple and cost-effective method are also very challenging task.
2. Experimental section
2.1. Reagents and materials
Reagent-grade 2-(bromomethyl) benzonitrile and 8-hydroxyquinoline were purchased from Sigma Aldrich. K2CO3, Cu(NO3)2·3H2O, potassium iodide and NaOH were obtained from Thermo Fisher scientific, India. Solvents were purified prior to use following standard methods.
2.2. Methods and instrumentation
Microanalyses for the compounds were performed using a CE–440 elemental analyzer (Exeter Analytical Inc.). Infrared spectra were obtained (KBr disk, 400–4000 cm−1) on a Perkin–Elmer Model 1320 spectrometer. ESI mass spectra were recorded on a WATERS Q–TOF Premier mass spectrometer. The NMR (1H and 13C NMR) analysis were run on a Bruker Avance-300 spectrometer in DMSO‑d 6 at room temperature. Thermal gravimetric analysis of complex 1 and 2 was determined using Mettler Toledo thermogravimetric analyzer (TGA) with built in gas controller (TGA2 SF/1100) and fitted with XP1U TGA balance (ultra-micro balance), in the temperature range 25–800 °C under nitrogen atmosphere at a heating rate of 10 °C/min.
2.3. Synthesis
2.3.1. Synthesis of 2-(quinolin-8-yloxy)(methyl)benzonitrile (L1)
The ligand L1 was synthesized according to the earlier reported procedure [20] as; 8-hydroxyquinoline (2 g, 13.7 mmol) and dry K2CO3 (5 g, 36.17 mmol) were mixed in a round-bottom flask under an inert atmosphere, and acetonitrile (60 mL) added to the above mixture. The mixture was allowed to stirring for 60 min at 90 °C. The mixture was treated with 2-(bromomethyl) benzonitrile (2.69 g, 13.7 mmol), and the resulting solution refluxed for 24 h. After completion of the reaction, the solution was allowed to cool to room temperature and the mixture was poured slowly in ice cold water (100 mL) with constant stirring to give a white muddy solid precipitate that was collected by filtration and dried under vacuum. Block-shaped pale yellow crystals of L1 were collected from the slow evaporation of ethanolic solution after 2 weeks. Yield: 3.3 g (70%); Melting Point: 120 °C; Elemental analysis (%): Calcd. For C17H12N2O (260.09): C, 78.44; H, 4.65; N, 10.76; Found: C, 78.54; H, 4.56; N, 10.64; IR (cm−1): 3398(s), 2948(m), 2922(m), 2835(w), 2224(w), 1619(w), 1564(w), 1503(m), 1468(m), 1379(m), 1319(m), 1262(m), 1177(w),1105(m), 1027(s), 983(w), 952(w), 893(w), 856(w), 820(m), 794(m), 753(m), 713(w), 668(w), 547(w), 418(w); 1H NMR (DMSO–d 6, 300 MHƶ, δ; ppm, J; Hƶ): 5.66 (s, 2H, –CH2–), 7.13 (t, J = 2.5, ArH), 7.55–7.44 (m, 4H, ArH), 7.64 (t, J = 7.5, 1H; ArH), 7.73 (d, J = 7.8, 1H, ArH), 7.94 (d, J = 7.8, 1H, ArH); 8.23 (d, J = 8.1, 1H, ArH) and 9.05 (d, J = 2.7, 1H, ArH); 13C NMR (75MHƶ, DMSO–d 6, δ, ppm): 68.2, 110.4, 110.7, 117.1, 120.7, 121.8, 126.9, 128.4, 128.5, 129.7, 132.7, 133.4, 136.8, 139.5, 140.4, 149.2 and 153.4; ESI-MS: m/z (100%) 261.1046 [M+1] (Supplementary materials Figs. S1-S4).
2.3.2. Synthesis of 2-((quinolin-8-yloxy)methyl)benzoic acid (L2)
The ligand L2 was achieved by hydrolyzing L1 (2 g, 7.68 mmol) by 6(N) NaOH solution (100 mL) under refluxing condition for 24 h. The resulting solution was allowed to cool 5 °C and acidified with the 6(N) HCl to obtained light pale yellow solid. This compound was collected by filtration and air dried. Block-shaped pale yellow crystals of L2 were collected from the slow evaporation of ethanolic solution after 2 weeks. Yield: 1.71 g (85%); Melting Point: 201 °C; Elemental analysis (%): Calcd. For C17H13NO3 (279.09): C, 73.11; H, 4.69; N, 5.02 Found: C, 73.98; H, 4.77; N, 5.10; IR(cm−1): 3382(m), 3191(w), 3058(w), 2921(m), 2856(m), 1654(s), 1620(w), 1596(w), 1572(w), 1502(s), 1470(w), 1404(w), 1371(s), 1312(s), 1265(s), 1185(m), 1115(s), 1066(m), 1015(w), 827(s), 801(s), 772(s), 742(s), 724(m), 654(m), 622(w), 576(w), 505(w), 458(w). 1H NMR (DMSO–d 6, 300 MHƶ, δ; ppm, J; Hƶ): 5.50 (s, 2H, –CH2–), 5.70 (br. s, 1H, –OH), 7.34 (d, J = 7.2, 1H, ArH), 7.45–7.41 (m, 2H, ArH), 7.69–7.48 (m, 3H, ArH), 7.72–7.76 (m, 2H, ArH), 8.28 (br. s, 1H, ArH) and 8.97 (s, 1H, ArH); 13C NMR (75MHƶ, DMSO–d 6, δ, ppm): 70.0, 111.4, 120.5, 121.7, 127.4, 128.7, 128.9, 129.6, 130.6, 133.2, 133.3, 136.4, 136.6, 138.8, 148.6, 153.1 and 171.3; ESI-MS: m/z (100%) 279.1136 [M+1] (Supplementary materials Figs. S5-S8).
2.3.3. Synthesis of copper(II) complex {[Cu(L1)(L2)(q)]·NO3·2H2O} (1)
A mixture containing L1 (0.02 g), L2 (0.02 g) and 8-hydroxyquinoline (Hq) (0.02 g) was dissolved in 3 mL methanol in a round bottom flask and added Cu(NO3)2·3H2O (0.08 g) dissolved in 2 mL water solution dropwise, then added 3 drops of NH3 solution and allow to stirrer for 6 h at 80 °C. The resulting solution was cool down and filtered out, then kept to slow evaporation at room temperature. After 2–3 week green block shape crystals were obtained and washed with cold ethanol, which was suitable for data collection of single crystal X-ray. Yield: 75%. Melting Point: 186 °C. Elemental analysis (%): Calcd. For C43H35CuN5O10: C, 61.09; H, 4.17; N, 8.28%; Found: C, 61.32; H, 4.35, N, 8.35%. IR(cm−1): 3376(s), 3270(w), 3184(s), 3061(m), 2929(m), 2859(w), 2223(w), 1986(w), 1899(w), 1818(w), 1650(s), 1614(m), 1568(w), 1497(s), 1465(s), 1374(s), 1323(s), 1261(s), 1180(m), 1112(s), 1047(w), 1006(w), 965(w), 826(s), 802(w), 772(m), 740(s), 649(m), 620(w), 577(w), 547(m). TGA decomposition pattern (three steps weight loss): near 100 °C (loss of two water molecules); 150–200 °C (loss of lattice nitrate and coordinated 8-hydroxyquinoline groups); plateau around 600 °C (decomposition of the framework) (Supplementary materials; Figs. S9 and S10).
2.3.4. Synthesis of copper(II) complex {[Cu(L2)2 (q)]·NO3·2H2O} (2)
Synthesis of complex 2 was similar to the above method of complex 1. Herein, a mixture containing L2 (0.04 g) and 8-hydroxyquinoline (Hq) (0.04 g) was dissolved in 3 mL methanol in a round bottom flask and Cu(NO3)2·3H2O (0.08 g) dissolved in 2 mL water then added into above mixture and stirrer after that added 3 drops of NH3 solution and allowed to stirrer for 6 h at 80 °C. The resultant solution was cool down to room temperature and filtered, then kept to crystal grow by slow evaporation. After 10 days green cubic shape crystal was obtained and washed with cold ethanol, data collection of suitable crystal was determined by single crystal X-ray. Yield: 65% Melting Point: 270 °C. Elemental analysis (%): Calcd. For C43H36CuN4O12: C, 60.25; H, 4.35; N, 6.57%; Found: C, 59.75; H, 4.19, N, 6.48%. IR(cm−1): 3475(s), 3348(w), 3111(w), 3068(w), 2924(m), 2855(m), 1684(s), 1659(s), 1616(m), 1574(s), 1502(s), 1462(s), 1377(s), 1319(m), 1260(s), 1183(w), 1098(w), 966(m), 877(w), 827(s), 771(s), 732(w), 632(m), 580(w), 515(w); TGA decomposition pattern (three steps weight loss): near 100 °C (loss of two water molecules); 150–200 °C (loss of lattice nitrate and coordinated 8-hydroxyquinoline groups); plateau around 600 °C (decomposition of the framework) (Supplementary materials; Figs. S11 and S12).
2.4. Single crystal X-ray studies
The diffraction data was collected with a Bruker SMART APEX CCD diffractometer using monochromatic Mo-Kα radiation (λ = 0.71073 Å) at 100(2) K. The linear absorption coefficients, and the anomalous dispersion corrections were mentioned from the International Tables for X–ray crystallography [21]. The data integration and reduction were worked out with SAINT software [22]. Empirical absorption correction was applied to the collected reflections with SADABS [23]. Using Olex2 [24], the structure was solved with the olex2.solve [25] structure solution program using Charge Flipping and refined with the olex2.refine [25] refinement package using Gauss-Newton minimisation. All hydrogen atoms were located in difference Fourier maps in the structures and refined isotropically. All non–H atoms were refined anisotropically. The crystal and refinement data are collected in Table S1 for ligands L1 and L2 and Table S2 for complexes 1 and 2, respectively. The selected bond distances and angles for ligands are given in Table S3 (for L1) and Table S4 (for L2) and Table S5 (for complex 1) and Table S6 (for complex 2).
2.5. Molecular docking
Energy minimized structure of chloroquinoline, HCQ, L1 and L2 which was obtained from DFT optimization and were utilized for molecular docking with the spike protein (PDB: 6VSB), receptor-binding domain of chimeric (PDB: 6VW1), RNA dependent RNA polymerase (PDB: 6M71), ADP-ribose-1 monophosphatase (PDB: 6VXS), main protease (PDB: 6LU7), non-structural protein 3 (PDB: 6W02) and 9 (PDB: 6W4B) protein structure. For metal complexes, we used the structural information found from single crystal X-ray crystallography. The AutoDock 4.2.0 applications employed for all the docking studies with the help of Autodock tools. Structural preparation of all the compounds and proteins were done by the MGL Tools. Addition of hydrogens to all the atoms of proteins as well as molecules before the partial atomic charges (Gasteiger charges) calculation for them were performed through the MGL Tools. The proteins structures are kept as rigid and all the small molecules are allowed as flexible during all the docking studies. Using a Lamarckian genetic algorithm inside the 126 × 126 × 126 Å3 grid box with 1000 exhaustiveness parameter and 20 000 precise docking step the calculations were performed.
3. Result and discussion
3.1. Designing of complexes 1 and 2
The chloroquine and hydroxychloroquine are differing by an alkyl chain hydroxyl group (Fig. 1 ). For protein binding of ligand, CH-π interaction play a vital role and it is also found in polar carbohydrate protein interactions [26]. Therefore, the 4-aminoquinoline moiety of these molecules is key for the protein binding, so we took 8-hydroxyquinoline molecule for our study. Both the chloroquine and hydroxychloroquine contain nine carbon side chains with hetero atoms. Considering these feature we replaced the side chain with 2-cyanobenzyl and 2-carboxylbenzyl groups (for good CH-π interaction) to get 2-((quinolin-8-yloxy)methyl)benzonitrile (L1) and 2-((quinolin-8-yloxy)methyl)benzoic acid (L2), respectively.
Fig. 1.
Designing of ligands L1 and L2 by mimicking chloroquine and hydroxychloroquine.
Examining the three dimensional structures of protein targets of SARC-CoV-2 and physico-pharmaceutical properties of chloroquine and hydroxychloroquine, we chose the –CN and COOH groups for the ligand. Some of the protein targets of the virus contains nucleophilic and cationic amino acids in their active sites which can involve nucleophilic reaction with –CN group and salt bridge like interaction with –COOH. In this way these groups can facilitate binding of the complexes. The physicochemical, lipophilicity, water solubility, pharmacokinetics and druglikeness properties of these molecules (chloroquine and hydroxychloroquine, L1 and L2) are very much similar. This is why it is very efficient mimicking of chloroquine and hydroxychloroquine. It is very interesting to note that the values of the above mentioned parameters of L1 and L2 are very close resemblance to chloroquine and hydroxychloroquine, respectively. Thus the –CN and –COOH functionality of ligand facilitate the accessibility of complexes in the body tissue. All these parameters of these complexes are summarized in the Table S7.
3.2. Crystal description of ligand L1 and L2
Crystal structures of L1 and L2 were confirmed by the crystallographic data which shows that ligands crystallized in monoclinic, tetragonal crystal system with P21/c and I41/a space group, respectively. The asymmetric unit of L1 and L2 have shown non-planar structure with dihedral angle 79.570(32)° and 14.032(54)°, respectively. 8-hydroxyquinoline moiety has planar geometry and 2-(Bromomethyl)benzonitrile is non-planar and hence, L1 crystallized as non-planar geometry. However, in L2, hydrolysis of nitrile group to convert into carboxylate group is approaching to planar molecule (Fig. 2 a and b).
Fig. 2.
Asymmetric unit of ligands (a) L1 and (b) L2.
Ligand L1 and L2 are comprised of same organic moiety with different functional groups. Ligand L1 is stabilized via hydrogen bonding (C17-N2…H10b = 2.669 Å, C15-H15…N1 = 2.691 Å, C17-N2…H6 = 2.657 Å) and weak interaction such as π-π, C—H…π, which gives a arabesque like pattern along b-axis (Fig. 3 a and b). Similarly, L2 is also stabilized via strong hydrogen bonding and other weak interaction such as O2-H2…O1 = 2.111, C11-H11…O2 = 2.639 Å, C12-H12…O1 = 2.286 Å hydrogen bonding, C—H…π (C17-H17…C13 = 2.717 Å) and C3-H3…H16-C16 = 2.346 Å weak interaction and form a herringbone (45°) like supramolecular framework along c-axis (Fig. 4 a and b).
Fig. 3.
A view of (a) crystal packing structure of ligand L1 which showing arabesque like pattern and (b) formation of supramolecular structure via hydrogen bonding and other weak interactions along with crystallographic b-axis.
Fig. 4.
A view of (a) crystal packing showing herringbone pattern and (b) supramolecular network formation via H-bonding and other weak interactions along with c-axis in ligand L2.
Ligand L1 contain nitrile functional group which act as coordinating agent with beneficial properties and play a promising key role in biological drug development, whereas ligand L2 has carboxylic group that act as chelating agent. Chelating groups are shown more efficient performance against harmful cells via strong interaction with diseases cell and release the toxicity of cells. Functionality of both groups have more significance regarding biological application on the basis of theoretical modeling.
3.3. Crystal description of copper(II) complexes 1 and 2
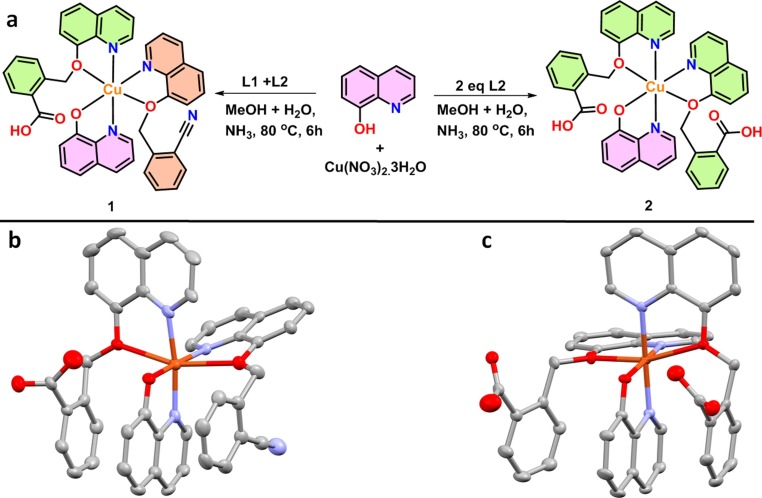
Mixing of Cu(NO3)2·3H2O, 8-hydroxyquinoline, L1 and L2 in 1:1:1:1 ratio under appropriate conditions, it give a pale yellow solid of complex 1 and by omitting L2 with 1:1:2 ratio under suitable reaction conditions the mixture produce the complex 2 (Fig. 5 a). Both the complexes were finally characterized with the help of single crystal X-ray crystallography. Complex 1 crystallizes in triclinic crystal system with P-1 space group. The asymmetric unit consisting of one Cu(II) ion, one L1 ligand, one L2 ligand and one 8-hydroxyquinoline ligand besides one nitrate lattice and two water lattice molecules. The metal ion exhibit distorted octahedral CuN3O3 configuration (Fig. 5b) with ligation from three N atoms via N1 of L2, N2 of L2 and N3 of 8-hydroxyquinoline {(Cu1-N1 = 2.024(4), Cu1-N2 = 1.976(4), Cu1-N3 = 2.024(4) Å} (Fig. 2b), similarly Cu-O coordination from three O atom of O3 of L2, O4 of 8-hydroxyquinoline and O5 of L1 {(Cu1-O3 = 2.254(3), Cu1-O4 = 1.947(3), Cu1-O5 = 2.508(3) Å} respectively (Fig. 5b).
Fig. 5.
(a) Synthesis of complex 1 and 2, ortep representation of asymmetric unit of (b) complex 1 and (c) complex 2.
Complex 2 crystallized in triclinic crystal system with P-1 (2) space group. The asymmetric unit consisting of one Cu(II) metal ion, two unit of L2 ligands and one 8-hydroxyquinoline ligand besides one nitrate and two water lattice molecules moieties. The metal ion exhibit distorted octahedral (CuN3O3) configuration geometry (Fig. 5c) with coordination sphere from three N atom of one 8-hydroxyquinoline and two L2, similarly three O atom of one 8-hydroxyquinoline and two L2 ligand. The coordination of O and N with Copper (II) ion is {(Cu1-N1 = 2.0489(18), Cu1-N2 = 2.0138(18), Cu1-N3 = 2.0041(17), and Cu1-O3 = 2.4725(15), Cu1-O4 = 1.9285(15), Cu1-O5 = 2.3386(16)Å}.
3.4. Drug-likeness properties of the compounds
It is important to know that the drug-likeness properties of all the aforesaid compounds and complexes before any screening. The results are summarized in the Table 1 . Both the complexes were failed to attain only one Lipinski’s rule (Molecular weight < 500, hydrogen acceptor < 10, hydrogen donor < 5, and LogP < 5) which is molecular wait. However, they are apt of beliciting pharmacological response. Due to the presence of the carboxylic acid –COOH group and over all ionic nature both the complexes are water soluble and experimentally determined logS values of both the complexes are given in the Table 1.
Table 1.
Drug-likeness properties of the compounds.
| Sl. No | Compounds | M.W (150–500 g/mol) | H-acceptors (≤10) | H-donors (≤5) | Log P (0.7–5.0) | No. of violations (Rule of 5) | TPSA (20–130 Å2) | Rotatable bonds (<9) | Log S (> − 6) | Fraction Csp3 (>0.25) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Chloroquinoline | 285.43 | 2 | 1 | 4.15 | 0 | 28.16 | 7 | −3.95 | 0.5 |
| 2 | HCQ | 287.40 | 3 | 2 | 3.32 | 0 | 48.39 | 7 | −3.37 | 0.47 |
| 3 | L1 | 260.29 | 3 | 0 | 3.04 | 0 | 45.91 | 4 | −3.88 | 0.06 |
| 4 | L2 | 279.29 | 4 | 1 | 2.87 | 0 | 59.42 | 4 | −3.79 | 0.06 |
| 5 | Complex-1 | 747.28 | 6 | 1 | 4.50 | 1 | 103.57 | 5 | −10.30 | 0.05 |
| 6 | Complex-2 | 766.28 | 7 | 2 | 4.32 | 1 | 117.08 | 6 | −10.22 | 0.05 |
3.5. Finding of protein target of SARS-CoV-2
The RNA of SARS-CoV-2 translate two classes of proteins such as structural proteins and non-structural proteins. The first category consist with nucleocapsid, envelope, membrane and trimeric spike proteins. These proteins help to build and maintain the structure and shape of the virus play most important role in the infection process. Whereas the second category includes sixteen proteins which come in to existence from polyprotein PP1a and PP1b. They are also very essential for viral metabolism processes like translation. Therefore, any one of these proteins can be the target for all these compounds (chloroquine, hydroxychloroquine, L1, L2, complex 1 and 2).
The pathogenesis superintendence protein of SARS-CoV-2 are our potential drug target for the compounds. For this reason, we have chosen spike protein (PDB: 6VSB), receptor-binding domain of chimeric (PDB: 6VW1), RNA dependent RNA polymerase (PDB: 6 M71), ADP-ribose-1 monophosphatase (PDB: 6VXS), main protease (PDB: 6LU7), non-structural protein 3 (PDB: 6 W02) and 9 (PDB: 6W4B) for the screening of these compounds.
3.6. Screening of the complexes against SARS-CoV-2.
We have screened all the six compounds with the above mentioned seven viral proteins and the data associated to the binding energy is summarized in the Table 2 . The chloroquinoline and HCQ preferentially binds with ADP-ribose-1 monophosphatase (PDB: 6VXS). For this protein, the study is blind docking. The compounds bind at the active site of the protein and the change of Gibb’s free energy for the binding are −5.95 and −6.26 kcal/mol. The ligands (L1 and L2) designed from the structural analogy of chloroquinoline and HCQ which were not preformed so well as these parent compounds. It should be stated that, unlike L1 and others, L2 show higher preference towards main protease enzyme.
Table 2.
Binding affinity (kcal/mol) of the compounds with the essential proteins of SARS-CoV-2.
| Sl No | Compounds | 6W4B | 6VXS | 6VW1 | 6VSB | 6M71 | 6LU7 | 6W02 |
|---|---|---|---|---|---|---|---|---|
| 1 | Chloroquinoline | −4.68 | −5.95 | −5.17 | −4.60 | −5.58 | −4.42 | −5.21 |
| 2 | HCQ | −5.51 | −6.26 | −5.36 | −6.24 | −5.89 | −4.95 | −5.46 |
| 3 | L1 | −4.19 | −4.73 | −4.24 | −3.97 | −4.55 | −3.79 | −4.13 |
| 4 | L2 | −3.85 | −4.14 | −3.87 | −4.02 | −4.08 | −4.26 | −3.54 |
| 5 | Complex 1 | −6.39 | −7.62 | −7.05 | −5.98 | −5.41 | −7.13 | −6.83 |
| 6 | Complex 2 | −7.88 | −9.05 | −8.51 | −7.23 | −8.62 | −8.73 | −7.56 |
Like chloroquinoline and HCQ, the complexes 1 and 2 show binding affinity towards ADP-ribose-1 monophosphatase. However, the binding affinity of both the complexes is very high with respect to the reference compounds. The ΔG° values of complex 1 and complex 2 are −7.62 and −9.05 kcal/mol, respectively. Therefore, complex 2 bind more strongly with the ADP-ribose-1 monophosphatase than that of complex 1. The Table 2 also indicates that these complexes have another binding preference and the protein is the main protease. This affinity may be the result of the presence of ligand L2 in both the complexes. The complex 2, contain two unit of L2, show more negative ΔG° value for main protease with respect to complex 1 which have only one L2 unit. Therefore, the study shows that the complex 2 can act as an anti-COVID-19 agent through the inhibiting the ADP-ribose-1 monophosphatase and main protease enzymes.
The ADP-ribose-1 monophosphatase is a α-helix enriched protein containing 165 amino acid residues. It has one hydrophobic pocket (i.e. active site) and few surface binding sites. Both the complexes were bound at the active site like chloroquinoline and HCQ. It should be mentioned here that both the compounds (chloroquinoline and HCQ) prefer hydrophobic interactions. However, in the case of HCQ, the side-chain hydroxyl group involved in the hydrogen bonding (Fig. S13a and S13b).
The ligand with aromatic moiety increases the hydrophobicity after ligation with Cu2+ which may favor to bind at the hydrophobicity loving active site of the protein. It is clear from the large green surface on the molecular electro static potential (MEP) which indicates that both the complexes are sufficiently hydrophobic in nature (Fig. S14a and S14b). In the case of complex 1, the D22, G48, V49, A52, A129 and F156 amino acid residues are in the contact with the aromatic part (Fig. 6 a). For complex 2, the total number interacting amino acids are higher with respect to the complex 1 and these amino acids are I23, A38, V49, L126, A129, A154, V155 and L160 (Fig. 6b). It is very interesting to note that the mode of binding is only CH-π interactions between aromatic parts of the complexes and amino acids (Fig. S15a and S15b).
Fig. 6.
Docking poses of (a) complex 1 and (b) complex 2 in the active site of the ADP-ribose-1 monophosphatase of COVID-19.
It should be mentioned here that the topological polar surface area (TPSA; Table S7), the sum of surfaces of polar atoms in a molecule, is high in complex 2 with respect to complex 1. The complex 2 is slightly more polar with hydrophobic surface than complex 1. Therefore, an amphiphilic molecule 2 is good protein binder and hydrophobicity is not only the factor to it.
4. Conclusions
In summary of this work, it can be stated that we successfully designed and synthesized two copper complexes against COVID-19. In this work, we designed copper complexes with the help of structural information of the chloroquine and HCQ. We compared the drug likeness and pharmaco-physical properties of the complexes with the organic molecules. Among various important structural and non-structural viral proteins, we recognized the ADP-ribose-1 monophosphatase enzyme as a protein host for the investigated molecules. Complex 2 shows strong binding affinity for the protein. Therefore, considering all the results, it is clear that the Complex 2 has enormous potential as an anti-COVID-19 agent.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The Department of Applied Chemistry, AMU and Department of Chemistry, Jadavpur University have gratefully acknowledged for research facility. The authors are grateful to Researchers Supporting Project number (RSP-2020/288), King Saud University, Riyadh, Saudi Arabia and UGC Start-up grant and TEQIP-III, ZHCET, Aligarh Muslim University for funding this work.
Author contributions
N.S. and M.A. designed, evaluated and follow-up all the experiments and calculations. AA performed all the synthesis. N.S. performed the docking calculations. M.A. and A.A. helped in bioavailability calculation and analysis. N.S., M.A and M.S. wrote the manuscript with assistance and feedback from all other. All authors of the manuscript checked the theoretical results and approved of the manuscript as the final version. All authors agree the corresponding authors as the representative person for handling this manuscript.
Footnotes
Supplementary crystallographic data for ligands (L1 and L2)andcomplexes (1 and 2), respectively. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif. Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioorg.2021.104772.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2, Nat. Microbiol. 5 (2020) 536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed]
- 2.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabby M.I.I. Current drugs with potential for treatment of COVID-19: a literature review. J. Pharm. Pharm. Sci. 2020;23:58–64. doi: 10.18433/jpps31002. [DOI] [PubMed] [Google Scholar]
- 4.Navarro M., Castro W., Madamet M., Amalvict R., Benoit N., Pradines B. Metal-chloroquine derivatives as possible anti-malarial drugs: evaluation of anti-malarial activity and mode of action. Malar. J. 2014;13:471. doi: 10.1186/1475-2875-13-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derwand R., Scholz M. Does zinc supplementation enhance the clinical efficacy of chloroquine/hydroxychloroquine to win today’s battle against COVID-19? Med. Hypotheses. 2020;142:109815. doi: 10.1016/j.mehy.2020.109815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nandy P., Das S. In situ reactivity of electrochemically generated nitro radical anion on Ornidazole and its monomeric Cu(II) complex with nucleic acid bases and calf thymus DNA. Inorg. Chim. Acta. 2020;501:119267. doi: 10.1016/j.ica.2019.119267. [DOI] [Google Scholar]
- 7.Manning T.J., Leggett T., Jenkins D., Furtado I., Phillips D., Wylie G., Bythell B.J., Zhang F. Structural and some medicinal characteristics of the copper(II)–hydroxychloroquine complex. Bioorg. Med. Chem. Lett. 2013;23:4453–4458. doi: 10.1016/j.bmcl.2013.05.041. [DOI] [PubMed] [Google Scholar]
- 8.Katsarou M.E., Efthimiadou E.K., Psomas G., Karaliota A., Vourloumis D. Novel copper(II) complex of N-Propyl-norfloxacin and 1,10-Phenanthroline with enhanced antileukemic and DNA nuclease activities. J. Med. Chem. 2008;51:470–478. doi: 10.1021/jm7013259. [DOI] [PubMed] [Google Scholar]
- 9.Tamasi G., Serinelli F., Consumi M., Magnani A., Casolaro M., Cini R. Release studies from smart hydrogels as carriers for piroxicam and copper(II)–oxicam complexes as anti-inflammatory and anti-cancer drugs. X-ray structures of new copper(II)–piroxicam and –isoxicam complex molecules. J. Inorg. Biochem. 2008;102:1862–1873. doi: 10.1016/j.jinorgbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Hordyjewska A., Popiołek Ł., Kocot J. The many “faces” of copper in medicine and treatment. BioMetals. 2014;27:611–621. doi: 10.1007/s10534-014-9736-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha T., Kumar P., Sepay N., Ganguly D., Tiwari K., Mukhopadhyay K., Das S. Multitargeting antibacterial activity of a synthesized Mn2+ complex of curcumin on gram-positive and gram-negative bacterial strains. ACS Omega. 2020;5:16342–16357. doi: 10.1021/acsomega.9b04079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santini C., Pellei M., Gandin V., Porchia M., Tisato F., Marzano C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014;114:815–862. doi: 10.1021/cr400135x. [DOI] [PubMed] [Google Scholar]
- 13.Saha T., Singha S., Kumar S., Das S. Spectroscopy driven DFT computation for a structure of the monomeric Cu2+-curcumin complex and thermodynamics driven evaluation of its binding to DNA: pseudo-binding of curcumin to DNA. J. Mol. Struct. 2020;1221:128732. doi: 10.1016/j.molstruc.2020.128732. [DOI] [Google Scholar]
- 14.Andreou A., Trantza S., Filippou D., Sipsas N., Tsiodras S. COVID-19: the potential role of copper and N-acetylcysteine (NAC) in a combination of candidate antiviral treatments against SARS-CoV-2. Vivo (Brooklyn) 2020;34:1567–1588. doi: 10.21873/invivo.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raha S., Mallick R., Basak S., Duttaroy A.K. Is copper beneficial for COVID-19 patients? Med. Hypotheses. 2020;142:109814. doi: 10.1016/j.mehy.2020.109814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shehroz M., Zaheer T., Hussain T. Computer-aided drug design against spike glycoprotein of SARS-CoV-2 to aid COVID-19 treatment. Heliyon. 2020;6:e05278. doi: 10.1016/j.heliyon.2020.e05278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onawole A.T., Sulaiman K.O., Kolapo T.U., Akinde F.O., Adegoke R.O. COVID-19: CADD to the rescue. Virus Res. 2020;285:198022. doi: 10.1016/j.virusres.2020.198022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macalino S.J.Y., Gosu V., Hong S., Choi S. Role of computer-aided drug design in modern drug discovery. Arch. Pharm. Res. 2015;38:1686–1701. doi: 10.1007/s12272-015-0640-5. [DOI] [PubMed] [Google Scholar]
- 19.Gaudêncio S.P., Pereira F. A computer-aided drug design approach to predict marine drug-like leads for SARS-CoV-2 main protease inhibition. Mar. Drugs. 2020;18:633. doi: 10.3390/md18120633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali A., Mishra S., Kamaal S., Alarifi A., Afzal M., Das Saha K., Ahmad M. Evaluation of catacholase mimicking activity and apoptosis in human colorectal carcinoma cell line by activating mitochondrial pathway of copper(II) complex coupled with 2-(quinolin-8-yloxy)(methyl)benzonitrile and 8-hydroxyquinoline. Bioorg. Chem. 2021;106:104479. doi: 10.1016/j.bioorg.2020.104479. [DOI] [PubMed] [Google Scholar]
- 21.International Tables for X-Ray Crystallography, Vol. III, Kynoch Press, Birmingham, England, 1952, pp. 257–269.
- 22.SAINT, version 6.02; Bruker AXS: Madison, WI, 1999.
- 23.G.M. Sheldrick, SADABS, Empirical Absorption Correction Program, University of Göttingen, Göttingen, Germany, 1997.
- 24.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009;42:339–341. doi: 10.1107/S0021889808042726. [DOI] [Google Scholar]
- 25.Bourhis L.J., Dolomanov O.V., Gildea R.J., Howard J.A.K., Puschmann H. The anatomy of a comprehensive constrained, restrained refinement program for the modern computing environment – Olex2 dissected. Acta Crystallogr. Sect. A Found. Adv. 2015;71:59–75. doi: 10.1107/S2053273314022207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asensio J.L., Ardá A., Cañada F.J., Jiménez-Barbero J. Carbohydrate-aromatic interactions. Acc. Chem. Res. 2013;46:946–954. doi: 10.1021/ar300024d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.