Abstract
Malassezia spp. are common eukaryotic yeasts that colonize mammalian skin. Recently, we and others have observed that Malassezia globosa and Malassezia restricta can be found in the intestines in the context of certain diseases, including Crohn’s disease and pancreatic cancer. In order to better understand the nature of innate inflammatory responses to these yeasts, we have evaluated inflammatory responses induced by M. restricta and M. globosa in mouse bone marrow-derived macrophages (BMDM) and dendritic cells (BMDC). While Malassezia yeasts induce proinflammatory cytokine production from both macrophages and dendritic cells, the levels of production from BMDC were more pronounced. Both M. restricta and M. globosa activated inflammatory cytokine production from BMDC in large part through Dectin2 and CARD9 signaling, although additional receptors appear to be involved in phagocytosis and activation of reactive oxygen production in response to the yeasts. Both M. restricta and M. globosa stimulate production of pro-IL-1β as well as activation of the NLRP3 inflammasome. NLRP3 inflammasome activation by Malassezia fungi requires SYK signaling, potassium efflux and actin rearrangement. Together, the data further our understanding of the coordinated involvement of multiple innate immune receptors in recognizing Malassezia globosa and Malassezia restricta and orchestrating phagocyte inflammatory and antimicrobial responses.
Keywords: phagocytosis, reactive oxygen species, IL-1β, yeast, innate immunity
Summary sentence
Malassezia restricta and Malassezia globosa stimulate production of cytokines and chemokines from phagocytes and activate the NLRP3 inflammasome via multiple receptors and signaling pathways.
INTRODUCTION
The gastrointestinal tract is a symbiotic environment established by the interactions between mammalian intestinal epithelial and immune cells, and the microbiota.1,2 Intestinal microbes are essential for health of the gut and development of the immune system. Also, changes in the gut microbiota have been associated with diverse diseases including inflammatory bowel disease (IBD). While most studies have focused on defining normal and disease-associated bacterial members of the gut microbiota, there is an increasing appreciation that other microbes, including fungi, may also play important roles in health and disease.3–5
We initially reported finding diverse fungal species in fecal material of mice and humans and finding that immunity to intestinal fungi is important in models of intestinal inflammation.6 More recently, we examined intestinal mucosa-associated fungal populations in patients with Crohn’s disease and healthy controls.7 We observed a strong association of Malassezia spp. (M. restricta & M. globosa) with Crohn’s disease. Subsequently, Miller and coworkers observed a remarkable appearance of gut-derived Malassezia in pancreatic tumors8, further supporting the notion that Malassezia is associated with intestinal dysbiosis-driven diseases. In Crohn’s disease, we found that increases in intestinal Malassezia were especially prevalent in patients carrying a previously-characterized single nucleotide polymorphism in CARD9 that confers increased risk of developing Crohn’s disease.9–11 CARD9, a signaling adaptor protein connecting C-type lectin receptor signaling with activation of NF-κB, is essential for host defense against fungi, and the disease-associated change is a nonsynonymous polymorphism in exon 2 of CARD9 leading to production of a CARD9S12N variant of the protein12. Together, these data led us to hypothesize that Malassezia might exacerbate intestinal inflammation in a CARD9-dependent manner, and we observed this to be true in the mouse acute DSS model of colitis.
Malassezia spp. are members of the Basidiomycota phyla of fungi and, while all other members of the Ustilaginomycotina subdivision are plant pathogens, Malassezia are commensal skin microbes found on nearly all warm-blooded animals. In humans, Malassezia is known to cause folliculitis and dandruff.13,14 An important feature of Malassezia genomes is the loss of key enzymes required for lipid metabolism, including fatty acid synthase, Δ9desaturase, and Δ2,3enoyl CoA isomerase.15 Therefore, they cannot produce fatty acids themselves and need lipids from the environment for growth. In the skin, they harvest lipids from sebum in hair follicles through secretion of a host of lipases and phospholipases. These enzymes can release unsaturated free fatty acids from sebum lipids including oleic acid and arachidonic acid that can be inflammatory.16 That Malassezia spp. might be important in intestinal inflammation is unexpected.
Due to the prevalence of Malassezia spp. on skin and its association with skin disorders, most of our limited understanding of how immune cells respond to Malassezia is focused on skin responses and, for technical and historical reasons, mostly on species other than M. globosa and M. restricta.17–20 To extend this work and focus on gut-associated species and systemic immune cells, we have here systematically characterized inflammatory responses induced by M. restricta and M. globosa in bone marrow-derived macrophages (BMDM) and dendritic cells (BMDC). While Malassezia yeasts induce proinflammatory cytokine production from both macrophages and dendritic cells, the levels of production from BMDC were more pronounced. Both M. restricta and M. globosa activated inflammatory cytokine production from BMDC primarily through Dectin2 and CARD9 signaling, however, the data indicate there may be some redundancy in the recognition with other innate immune receptors. Both M. restricta and M. globosa were able to stimulate production of pro-IL-1β as well as activate the NLRP3 inflammasome in BMDM and BMDC. NLRP3 inflammasome activation by Malassezia fungi requires SYK signaling, potassium efflux and actin rearrangement. From these data we are able to better understand the inflammatory properties of these gut-associated Malassezia spp.
MATERIALS AND METHODS
Fungal and Bacterial Growth
All experiments using S. aureus were preformed using mutant S. aureus (ΔoatA-SA) kindly provided by Fiedrich Götz.21,22 S. aureus was grown in THB media overnight at 37°C with agitation. S. aureus was then subcultured in THB at a dilution of 1 to 100 for 2.5 hours at 37°C with agitation until the culture reaches log phase growth. Bacteria were washed and diluted to OD600=0.4, determined to be 6.7×107cfu/ml. M. restricta (ATCC MYA-4611) and M. globosa (ATCC MYA-4612) were grown under static conditions for 2–3 days at 30oC in modified Dixon media containing glycerol monostearate. C. albicans (ATCC 90028) was grown overnight under static conditions at 30oC in Sabouraud dextran broth. Live yeasts were harvested and washed 3 times in media before counting. Fixed yeasts were harvested, washed 3 times with sterile PBS and fixed in 2% paraformaldehyde for 1 hour at room temperature. Cells were then washed 6 times with media and counted on a hemocytometer.
Mice and Cell culture
Dectin1−/−, Dectin2−/−, and CARD9−/− mice were bred and housed under SPF conditions in the Cedar-Sinai Medical Center animal facility and handled in accordance with approved IACUC procedures. C57BL/6 and MyD88−/− mice were purchased from Jackson Laboratories (Bar Harbor, Maine). Bone marrow derived macrophages (BMDM) and dendritic cells (BMDC) were generated from femurs and tibias by culturing for 7–9 days in complete RPMI 1640 (5 mM glucose, 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine; Mediatech) supplemented with 50 ng/ml human recombinant M-CSF or mouse recombinant GM-CSF (Peprotech). BMDM and BMDC were plated the day before experiments 1×105/well on 96-well plate, 4×105/well on 24-well plate, or 2.5×105/well 48-well plate. For inflammasome experiments cells were primed with 100 ng/ml LPS (S. minnesota, Invivogen) for 4 hr. Then stimulated with ATP 5 mM (Sigma), 1 μg/ml pdA:dT (Invivogen) was complexed with Lipofectamine 2000 (ThermoFisher) for 30 min at room temperature and 10ul was added per well (96-well plate). Fungi were washed with PBS and then hand counted to determine the fungi/ml. Infections were done by adding indicated MOIs of bacteria or fungi onto cells and spinning at 450xg for 1 min. The S. aureus infection was allowed to progress for 30 min, then 100 μg/ml gentamycin was added to the media to limit growth of extracellular bacteria, supernatants were harvested at designated times, cytokine levels were measured by IL-1β, TNFα, and IL-6 ELISA (Biolegend) or Mouse Macrophage/Microglia 13-plex LegendPlex (Biolegend) (limit of detection according to the assay instructions. The limit of detection for all ELISAs is 15.625 pg/ml. The limit of detection for the LegendPlex assay is 2.44 pg/ml.
Immunoblotting
At designated timepoints, cells were lysed in 1x Nupage SDS loading buffer (ThermoFisher) or cells were stimulated in Optimem serum-free media and supernatants were harvested. Supernatants were concentrated using 5 μl of Strataclean resin (Agilent Technologies) per 500 μl of supernatant the agitated at low speed for 30 min at room temperature, resin was pelleted at 2500xg for 2 min, then boiled in 20 μl 1x Nupage SDS loading buffer. Samples were run on Nupage precast gels in MOPS running buffer and transferred to PVDF. Membrane was blocked in Odyssey blocking buffer (Li-Cor) 1–2 hr at room temperature. Blots were stained with primary antibodies overnight at 4°C. Antibodies: 1:1000 anti-IL-1β (AF-401-NA, R&D), 1:1000 anti-NLRP3 (Cryo-2, Adipogene), 1:500 anti-caspase-1 p10 (M-20,Santa Cruz), 1:2000 anti-tubulin (TUB2.1, Sigma).
Phagocytosis assay
BMDCs were plated the day before at 4×105/well on 24-well non-tissue culture treated plates (Falcon). Live yeasts (400×106/ml) were labelled with cell proliferation dye eFluor™ 670 (eBioscience) at 5 μM final concentration in PBS for 10 minutes at 37oC followed by washing 4 times in media. Labelled yeasts were added to BMDC at a MOI of 5 and spun down onto cells at 450xg for 1 min and then incubated at 37°C for 5, 15, or 30 minutes. After the incubation time, wells were washed 3 times with PBS and BMDCs were lifted from the wells using PBS with Ca2+ containing proteinase K (1:100) (Sigma P5568). Cells were stained with FITC-labeled anti-CD45 (1:200) (Biolegend) for 30min on ice. Cells were then fixed in 2% paraformaldehyde at RT for 20 minutes washed, resuspended in FACS buffer and analyzed on a BD LSR2 flow cytometer (BD). Flow cytometry analysis was performed using FlowJo software (TreeStar).
Reactive oxygen assay
BMDC were plated in opaque white NUNC 96-well plates at 5×104/well. Cells were stimulated with live fungi at an MOI of 5 in the presence of 100μM luminol (Sigma). Luminescence was measured for 5 seconds every 9 minutes for 20 cycles on a Clariostar plate reader.
Statistics
All statistics are done by Student t-test with triplicated stimulated wells of cells* p≤.05, ** p≤0.01,*** p≤0.001. All experiments were done a minimum of three independent times unless stated in the figure legend.
RESULTS
Gut-associated Malassezia species induce proinflammatory cytokine products from macrophages and dendritic cells
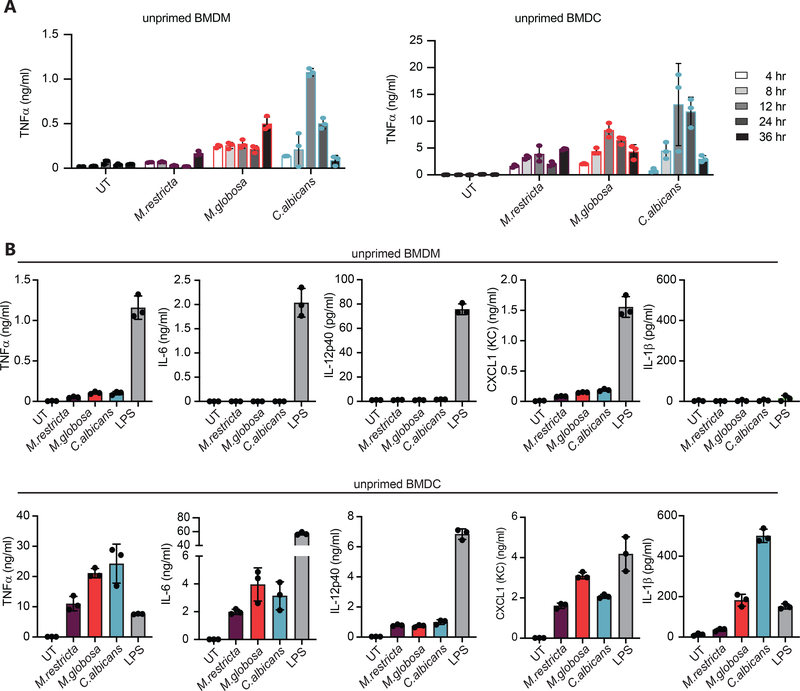
In our previous work we found increased burdens of Malassezia restricta and Malassezia globosa in the intestinal mucosa of patients with Crohn’s disease, and we noted in vitro that fixed M. restricta could induce TNFα and IL-6 from mouse BMDM and BMDC.7 Here we have extended these observations to evaluate responses to live M. restricta and M. globosa yeasts. When we stimulated BMDM or BMDC with M. restricta or M. globosa compared to Candida albicans we find that all the fungal species induced production TNFα in a time dependent manner (Fig. 1A). To more thoroughly characterize the inflammatory responses, we performed LegendPlex multiplex cytokine and chemokine analysis (Biolegend) of supernatants from cells stimulated with live fungi. Malassezia species consistently induced TNFα and CXCL1 (KC) from both macrophages and dendritic cells (Fig. 1B). We only detected measurable increase in production of IL-12p40, IL-6 and IL-1β in BMDC (Fig. 1B). We did not detect production of TGF-β1, IL-18, or IL-12p70. Together the data show that macrophages and dendritic cells encountering live M. restricta and M. globosa mount individual proinflammatory responses.
Figure 1. Malassezia induces a limited panel of cytokines from macrophages and dendritic cells.
(A) BMDM and BMDC treated with live M. restricta, M. globosa, or C. albicans at MOI=5. Supernatants were collected at designated time points to determine cytokine by ELISA. (B) Production of the indicated cytokines was assessed by LegendPlex multiplex analysis in supernatants from cells stimulated as in A, or with 100 ng/ml LPS, for 24 hr. LegendPlex data are representative of two independent experiments.
Inflammatory responses to live Malassezia restricta and globosa is predominantly dependent on Dectin2 and CARD9 signaling
Our initial examination of the inflammatory responses to M. restricta and M. globosa showed that inflammatory cytokine production was more prominent in dendritic cells than in macrophages. Because of this, and together with the fact that dendritic cell subsets have been shown to be vital for sensing and inflammatory signaling in response to fungi in the gut23,24, we focused our subsequent experiments on dendritic cell responses.
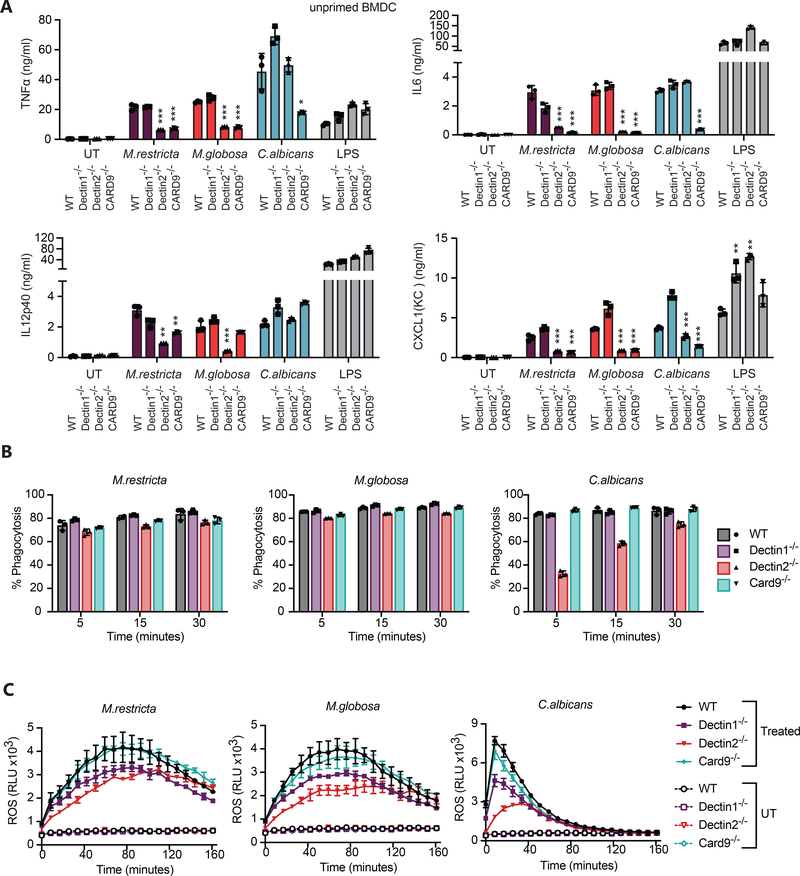
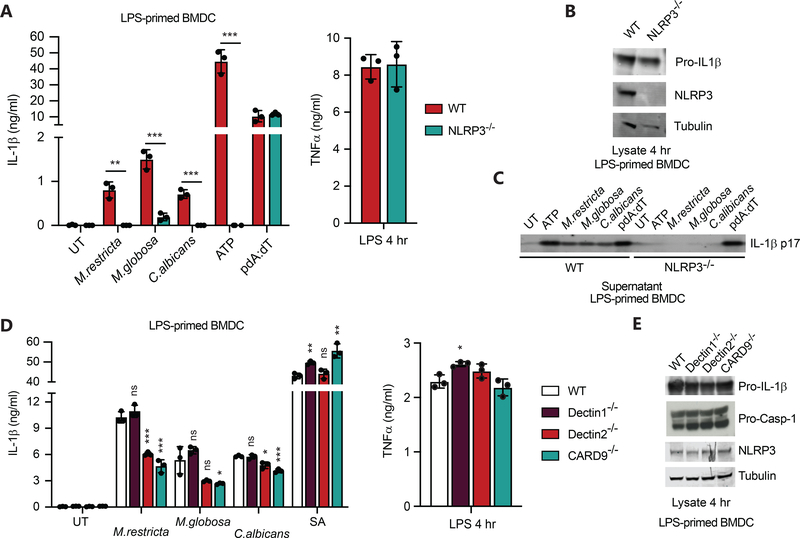
To determine which fungal receptors might contribute to the inflammatory response to Malassezia, we infected BMDC deficient for the C-type lectin receptors Dectin1 or Dectin2 or the signaling molecule CARD9 with live M. restricta, M. globosa, or C. albicans. Following exposure to live M. restricta we found that TNFα, IL-6, IL-12p40 and CXCL1 were only partially dependent on Dectin1 signaling but were strongly regulated by both Dectin2 and CARD9 (Fig. 2A). In addition, we also found that Dectin2 and Card9 were predominantly responsible for G-CSF and IL-10 production (Supplemental material Fig. 1A).
Figure 2. Role of CARD9 signaling in dendritic cell responses to Malassezia.
(A) BMDC from wild type and the indicated knockout mice were treated with live M. restricta, M. globosa, or C. albicans at MOI=5 or with 100 ng/ml LPS. Supernatants were collected after 24 hr, and production of the indicated cytokines was measured by LegendPlex. (B) BMDC from wild type and the indicated knockout mice were incubated for the indicated periods of time with eFluor 670 fluorescently labeled yeasts, and phagocytosis was quantified by flow cytometry. (C) BMDC from wild type and the indicated knockout mice were treated with live M. restricta or M. globosa (MOI=5) or not (UT = untreated), and production of reactive oxygen species (ROS) was measured by luminol-enhanced chemiluminescence (RUL=relative light units). The experiments in this figure are representative of two independent experiments. All data represent measurements done in triplicate ± SD, and statistics were done by student t-test * p≤0.05, ** p≤0.01,*** p≤0.001.
Dectin-1 is known to trigger phagocytosis and activation of reactive oxygen production, so we tested whether these receptors might be involved in these responses to Malassezia. We observed only the mildest effect on efficiency of phagocytosis of M. restricta or M. globosa in Dectin2−/− cells, but no observable impact in the absence of Dectin1 or CARD9 in dendritic cells (Fig. 2B and Supplemental Material Fig. 1B). We also observed that both M. restricta or M. globosa trigger production of reactive oxygen species by BMDC and found CARD9 to be dispensable for this response in dendritic cells (Fig. 2C and Supplemental Material Fig. 1C). However, we did see a consistent decrease in ROS production in Dectin2−/− BMDC. The importance of Dectin1 was less clear as it varied between experiments but looks to play the mildest role in M. restricta and M. globosa ROS production. In contrast, it is clearly important for detection of depleted-zymosan, a predominantly β-glucan particle (Fig. 2C and Supplemental Material Fig. 1C). Together these data indicate that multiple receptors contribute to the varied responses of dendritic cells to M. restricta and M. globosa and that additional dominant phagocytic receptor(s) have yet to be identified.
Live Malassezia fungi induce cytokine production that is not dependent on TLR signaling and induce significantly less cell death than C. albicans
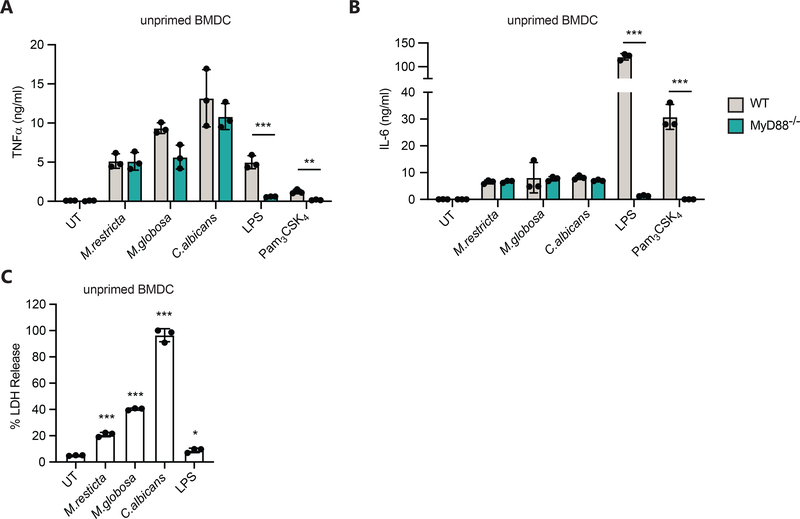
While C-type lectins are the dominant phagocytic receptors for the detection of fungi, inflammatory responses often include signaling through Toll-like receptors (TLR). To further characterize the receptors involved in responses to live Malassezia we infected BMDC from mice deficient for myeloid differentiation primary response 88 (MyD88), an adaptor molecule that is essential for all but intracellular TLR4 and TLR3 signaling25. To our surprise TLR signaling through MyD88 plays little role in the detection of Malassezia, at least for the production of TNFα and IL-6 (Fig. 3A–B). It is possible that MyD88-TLR signaling is important for specific cytokines, and this will need to be addressed in future work. We also assessed if Malassezia is toxic to BMDC. It is well known that C. albicans in cell culture media shifts to a hyphal form of growth that can disrupt cell integrity after phagocytosis. While most Malassezia spp. can form hyphae, this is kind of growth is rare, and we did not see any visual evidence of hyphal formation in our cell culture experiments. However, after infection for 24 hours, cell death, as measured by LDH release, was elevated in Malassezia-treated BMDC compared to untreated (UT) or LPS-treated BMDC (Fig. 3C). The cell death induced by Malassezia was significantly less than that caused by infection with C. albicans, which consistently induced 100% cell death by 24 hours (Fig. 3C). These results help to clarify dynamics of the myeloid cell responses to Malassezia infection.
Figure 3. Cytokine responses to live Malassezia fungi do not require TLR signaling.
(A-C) BMDC from the wild type and MyD88−/− mice were treated with live M. restricta, M. globosa, or C. albicans at MOI=5 or with 100 ng/ml LPS or 1 μg/ml Pam3CSK4. Supernatants were collected after 24 hr and production of TNFα (A) and IL-β (B) was measured by ELISA, or cell death was measured by lactate dehydrogenase release (LDH) (C). All data represent measurements done in triplicate ± SD, and statistics were done by student t-test * p≤0.05, ** p≤0.01, *** p≤0.001.
Live Malassezia fungi induce a more potent IL-6 and IL-1β inflammatory response than killed Malassezia fungi
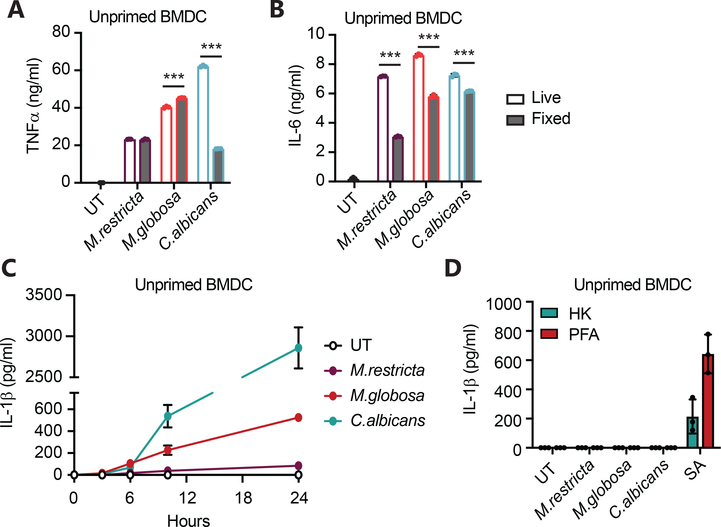
In our previous work observed that formaldehyde fixed Malassezia induced inflammatory cytokines from macrophages and dendritic cells suggesting that yeast do not need to be alive to stimulate responses.7 However, to directly assess the contribution of yeast viability to innate immune sensing, we compared inflammatory cytokine production by BMDC and BMDM stimulated with live or fixed M. restricta or M. globosa. We found that live and dead yeasts stimulated similar production of TNFα from dendritic cells (Fig. 4A). However, IL-6 production is reduced when M. restricta and M. globosa are fixed (Fig. 4B). In contrast, TNFα production in response to C. albicans was reduced by more than 60% when the fungi were fixed but IL-6 was slightly reduced by fixation (Fig. 4A–B). These data suggest that the availability of ligands may be different in live compared to dead fungi as well as between different fungal species, which may contribute to the unique inflammatory responses elicited by different fungi.
Figure 4. Live Malassezia fungi are more potent cytokine producers than dead fungi.
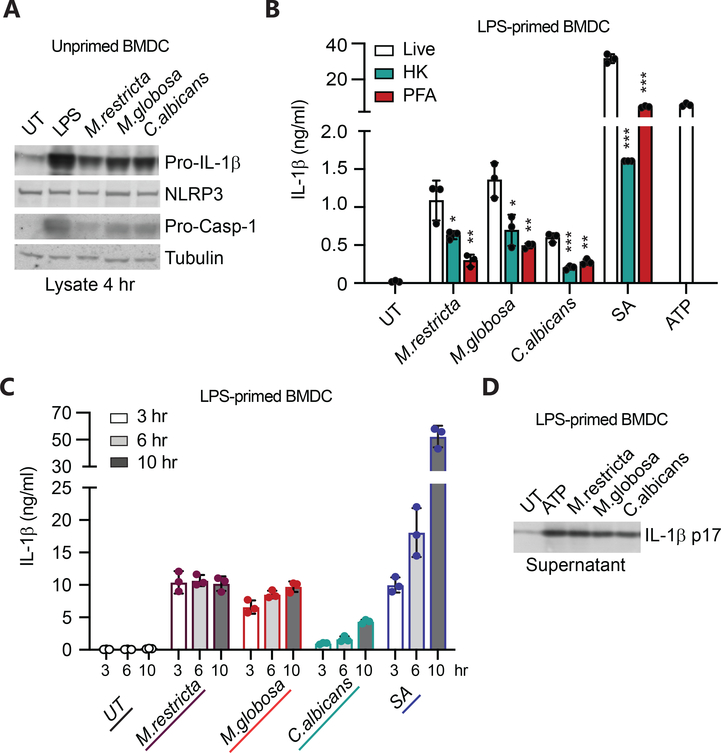
(A-B) BMDC were treated with live or PFA-fixed fungi at MOI=5 for 24 hr (6 hr. live C. albicans) and cytokines were determined by LegendPlex. (C) Unprimed BMDC were treated with the indicated fungi or not (UT = untreated) at MOI=5, supernatants were collected at designated time points, and IL-1β was measured by ELISA. (D) Unprimed BMDC were treated with either heat-killed (HK) or paraformaldehyde fixed (PFA) fungi or with S. aureus (SA) at MOI=5 for 6 hr, and IL-1β was measured in supernatants by ELISA. All data represent measurements done in triplicate ± SD, and statistics were done by student t-test * p≤0.05, ** p≤0.01, *** p≤0.001.
Production of IL-1β, however, was strongly influenced by yeast viability. During our multiplex cytokine experiments we observed small amounts of IL-1β production in the presence of Malassezia (Fig. 1B), so we performed a time course experiment in unprimed BMDC to extend the results. Compared to live C. albicans, which was a potent inducer of IL-1β, live M. globosa stimulated low levels of IL-1β secretion by unprimed BMDC (Fig. 4C), and M. restricta induced little or no IL-1β. In contrast, dead (fixed) M. globosa failed entirely to induce IL-1β production (Fig. 4D). S. aureus, used as a control in this experiment, does not need to be live to induce IL-1β secretion.
IL-1β production is important for induction and maintenance of Th17 T cells, which are important in colitis, and we had previously observed elevation of Th17 T cell responses in the intestines of mice gavaged with Malassezia prior to DSS-induced colitis. 7 We were therefore interested to more fully understand the mechanisms by which Malassezia induce or fail to induce IL-1β.
Malassezia fungi inflammasome priming and activation occur at discordant times
Production of mature functional IL-1β is a two-step process, first requiring activation of transcriptional and translational production of cytosolic immature pro-IL-1β protein that is subsequently processed into mature cytokine and secreted upon activation of any one of a number of NLR proteins leading to the assembly of the caspase-1-activating, multi-protein signaling aggregate, called the inflammasome.26–32 Given the poor induction of IL-1β from BMDC by Malassezia fungi that we observed, we examined whether Malassezia adequately “prime” the cells for expression of pro-IL-1β and the necessary inflammasome components. When we treated BMDC with live Malassezia, C. albicans or LPS for 4 hours we observed expression of pro-IL-1β, NLRP3, and pro-caspase-1 in the cell lysates (Fig. 5A). The induction by the fungal species was not as potent as LPS, however all the necessary inflammasome components were produced.
Figure 5. Malassezia fungi can prime an inflammasome response and induce caspase-1 activation, but in the timing is discordant.
(A) Unprimed BMDM were left untreated (UT) or treated with 100 ng/ml LPS or fungi at MOI=5. Cell lysates were harvested at 4 hr, and protein expression determined by immunoblotting. (B) LPS-primed BMDC (4 hr) were treated with live or killed M. restricta, M. globosa, or C. albicans at MOI=5 for 3 hr, S. aureus (SA) MOI=5 for 6 hr, or 5 mM ATP for 2 hr. IL-1β was measured in the supernatants by ELISA. (C) LPS-primed BMDC (4 hr) were treated with live fungi or S. aureus as in B and supernatants were collected at designated time points. (D) LPS-primed BMDC (4 hr) were treated with 5 mM ATP for 2h or live fungi at MOI=5 for 3 hr. Cleaved IL-β p17 was assessed in the supernatants by immunoblotting. HK=heat-killed, PFA=1% paraformaldehyde fixed, SA= S. aureus. All data represent measurements done in triplicate ± SD and statistics were done by student t-test * p≤0.05, ** p≤0.01, *** p≤0.001.
To specifically examine the capacity of each of the fungi to induce inflammasome assembly and IL-1β cleavage, we “primed” a cohort of BMDM with LPS for 4 hours to induce uniform pro-IL-1β production and expression of inflammasome components. We then treated the cells with live, heat-killed (HK), or PFA-fixed (PFA) fungi, S. aureus, or ATP (positive control). In this context, we found that M. restricta and M. globosa were as good or better than C. albicans at inducing secretion of mature IL-1β (Fig. 5B). As has previously been reported for C. albicans,30 we noted that heat-killed or PFA-fixed Malassezia induced inflammasome activation and secretion of mature IL-1β much less efficiently than live organisms.
When we examined the kinetics of inflammasome activation and IL-1β secretion from LPS-primed BMDC following treatment with live fungi, we found that Malassezia rapidly (within 3 hours) induced maximum IL-1β levels (Fig. 5C). In contrast, C. albicans and S. aureus more slowly triggered inflammasome activation. In both cases, we confirmed that the IL-1β being released into the supernatants was mature cleaved IL-1β p17 by immunoblotting (Fig. 5D).
Together, these results suggest that Malassezia fungi induce a rapid and transient inflammasome activation signal. This could explain the relatively poor induction of IL-1β secretion in unprimed cells given that the early and transient inflammasome activation signal would precede expression of pro-IL-1β protein.
Malassezia fungi activate the NLPR3 inflammasome by a partially Dectin2 and CARD9-dependent mechanism
Previous work with C. albicans fungi found that activation of the NLRP3 inflammasome was largely responsible for IL-1β production.28–32 We similarly observed that secretion of mature IL-1β in LPS-primed BMDC response to live M. restricta and M. globosa was lost in NLRP3−/− cells (Fig. 6A). By contrast, activation of the AIM2 inflammasome by poly(dA:dT) was not affected by loss of NLRP3. NLRP3 deficiency did not affect priming by LPS as determined by LPS-induced secretion of TNFα in culture supernatants (Fig. 6A) and expression of pro-IL-1β in cell lysates (Fig. 6B). We also confirmed that the NLRP3-dependent IL-1β secreted in response to the yeasts was mature IL-1β by measuring IL-1β p17 by immunoblotting culture supernatants (Fig. 6C).
Figure 6. Malassezia induces production of IL-1β through the NLRP3 inflammasome and is partially dependent on Dectin-2 and CARD9.
(A) BMDC from wild type and NLRP3 knockout mice were primed with LPS (100 ng/ml, 4 hr) and stimulated with the indicated yeasts (MOI=5) for 3 hr or S. aureus (MOI=5) for 6 hr, and IL-1β and TNFα secretion in the supernatants was measured by ELISA. (B) Cell lystates from LPS-primed cells prepared as in (A) were immunoblotted for the indicated proteins. (C) Cell culture supernatants from cells stimulated as in (A) were immunoblotted for mature IL-1β p17. (D) BMDC from wild type and the indicated knockout mice were primed with LPS (100 ng/ml, 4 hr) and stimulated as in (A), and IL-1β and TNFα secretion in the supernatants was measured by ELISA. (E) Cell lysates from LPS-primed cells prepared as in A were immunoblotted for the indicated proteins. All data represent measurements done in triplicate ± SD, and statistics were done by student t-test * p≤0.05, ** p≤0.01, *** p≤0.001.
To try to understand which receptors might trigger activation of the inflammasome in response to M. restricta and M. globosa, we measured IL-1β secretion from LPS-primed BMDC from Dectin1, Dectin2, or CARD9 deficient mice (Fig. 6D). IL-1β production was not affected by Dectin-1 deficiency, but it was partially blocked in Dectin-2- and CARD9-deficient cells. Dectin-1, Dectin-2, and CARD9 deficiency did not affect priming of the cells by LPS as determined by LPS-induced secretion of TNFα in culture supernatants (Fig. 6D) and expression of pro-IL-1β and inflammasome components in cell lysates (Fig. 6E).
Together, these data suggest that in addition to signaling for cytokine production via a C-type lectin/CARD9 pathway, M. restricta and M. globosa also trigger some of the observed NLRP3 inflammasome activation via this pathway.
Inflammasome activation by Malassezia is dependent on SYK-signaling, potassium efflux and actin rearrangement
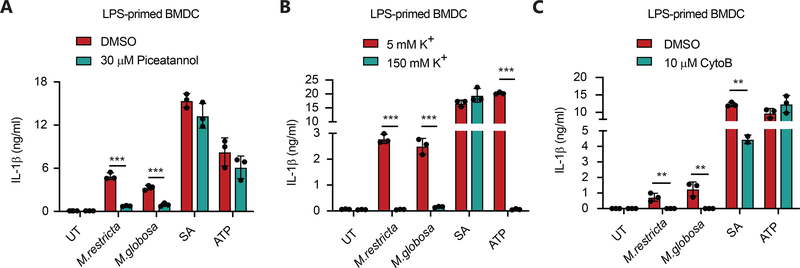
There are conflicting reports describing how fungi can activate the NLRP3 inflammasome and production of IL-1β, so we examined the importance of several mechanisms implicated inflammasome activating pathways. Previous work has suggested that the tyrosine-protein kinase SYK (spleen tyrosine kinase) activated downstream of C-type lectin receptors may play a role in NLRP3 activation.19,24,28 After LPS-priming, we pretreated BMDC with the SYK inhibitor, piceatannol, before exposing them to live fungi and found that SYK was important specifically for responses to both M. restricta and M. globosa-induced IL-1β production but not S. aureus- or ATP-induced IL-1β production (Fig. 7A).
Figure 7. Malassezia-induced inflammasome activation is partially dependent on SYK signaling, potassium efflux and actin rearrangement.
(A) BMDC from wild type mice were primed with LPS (100 ng/ml, 4 hr) and stimulated with the indicated yeasts (MOI=5) for 3 hr or S. aureus (MOI=5) for 6 hr, or ATP (5 mM) for 30 min in the presence or absence of the SYK inhibitor Piceatannol. IL-1β secretion in the supernatants was measured by ELISA. (B) BMDC from wild type mice were primed with LPS (100 ng/ml, 4 hr) and stimulated as in (A) in the presence of the indicated levels of extracellular potassium. IL-1β secretion in the supernatants was measured by ELISA. (C) BMDC from wild type mice were primed with LPS (100 ng/ml, 4 hr) and stimulated as in (A) in the presence or absence of the actin polymerization inhibitor cytochalasin B. IL-1β secretion in the supernatants was measured by ELISA. All data represent measurements done in triplicate ± SD and statistics were done by student t-test * p≤0.05, ** p≤0.01,*** p≤0.001.
One model of activation of NLRP3 inflammasome assembly involves the efflux of potassium from cells.33 We tested this model by incubated LPS-primed BMDC in isotonic Ringer’s buffer containing either normal concentrations of extracellular potassium (5 mM K+) or concentrations of potassium equivalent to the cytosol (150 mM K+) disrupting the gradient of potassium and thus preventing any potassium efflux. When LPS-primed BMDC treated with live M. restricta and M. globosa were both inhibited by high concentrations of extracellular potassium (Fig. 7B). This is the same response seen with the NLRP3 activator ATP, previously shown to be dependent on potassium efflux, but contrasts with S. aureus, which we have previously shown is potassium efflux-independent (Fig. 7B).34
We also examined whether phagocytosis of yeasts was important for NLRP3 activation in response to Malassezia. We treated LPS-primed BMDC with the actin polymerization inhibitor cytochalasin B before infecting with live M. restricta or M. globosa. We observed a significant inhibition of IL-1β production (Fig. 7C) suggesting that phagocytosis, or at the very least actin rearrangement, is necessary for the activation of NLRP3 by Malassezia yeasts.
DISCUSSION
Several recent studies have found that under certain conditions the skin fungus Malassezia becomes abundant in the intestinal tract where it is associated with exacerbated inflammation7 and can transit to the pancreas and exacerbate pancreatic cancer.8 While more is known about immune responses to Malassezia in the skin, little is currently known about how systemic phagocytes recognize and respond to the major gut-associated Malassezia spp., M. restricta and M. globosa. We have examined the inflammatory responses of conventional bone marrow-derived macrophages and dendritic cells to the live M. restricta and M. globosa and found that they induced a broad range of proinflammatory cytokines and chemokines including TNFα, IL-6, IL-1β IL-12p40, IL-10 and CXCL1. While BMDM were able to produce some cytokines, the induction was substantially lower than from BMDC. This may be a consequence of reduced levels of CARD9 expression in macrophages or other regulatory factors known to increase the inflammatory response in GM-CSF-treated cells.35 Cytokine and chemokine production was largely, although not entirely, dependent on signaling by the C-type lectin receptor Dectin-2 via the signaling protein CARD9. We expected TLR signaling might synergize with the C-type lectin receptors as has been shown for responses to some microbes36,37, however, when we examined responses in MyD88−/− BMDC that will be deficient in most TLR signaling, we saw little to no impact on Malassezia-induced TNFα and IL-6 production. Given that we do not see complete abrogation of responses to Malassezia in Dectin2−/− BMDC, we imagine additional pattern recognition receptors are involved, but further work will need to be done to determine the identity of those receptors.
Upon engaging M. restricta and M. globosa BMDC phagocytose the yeasts and activate production of reactive oxygen species. While phagocytosis of Malassezia was not impacted in Dectin1, Dectin2 or Card9 knockout BMDC, we observed reduced reactive oxygen species production in Dectin-2−/− cells. Together these observations illustrate the requirement for coordinated engagement of multiple innate immune receptors to orchestrate the full responses of macrophages and dendritic cells to Malassezia.
Malassezia can induced production of pro-IL-β from dendritic cells, however, in unprimed cells production of active IL-1β is low and requires a minimum of 10 hours of stimulation. Our observations suggest that priming (stimulation of transcription and translation of pro-IL-1β) is a relatively slow process, requiring 10 or more hours. In contrast, triggering inflammasome activation by M. restricta and M. globosa is relatively fast and transient, being complete in 3 hours or less. Together, the data suggest that IL-1β production is poor in unprimed cells exposed to the yeasts because priming and inflammasome activation signals are misaligned in time. However, in dendritic cells previously exposed to a priming stimulus, as for example during repeated exposure to fungi or other microbial stimuli, Malassezia can trigger NLRP3 inflammasome activation and release of mature IL-1β. Also, freshly isolated peritoneal macrophages, intestine tissue and keratinocytes exhibit an already primed state expressing pro-IL1β, raising questions as to how often the timing of a “priming” step is relevant during the course of infection and host defense in vivo.38–40
Our observations raise several questions that will require further study. First, the data suggest that Malassezia might be an excellent model for dissecting how multiple pattern recognition receptors coordinate the different responses of dendritic cells to contact with the microbe. Identifying the family of dominant receptors influencing these processes will be illuminating and may provide deeper insights to mechanisms influencing diseases of the gut. Second, it is unclear if tissue specific myeloid cells will respond differentially to different fungi or if they utilize different subsets of receptors to detect fungi. While there have been comparisons different species within Candida and Malassezia genus13,41, there have not been comparisons in different tissue-specific myeloid cells, which will be important for understanding the different responses seen at different sights of infection. Third, the mechanisms by which SYK signaling, actin polymerization, and, to a lesser degree, CARD9 signaling influence NLRP3 inflammasome activation by M. restricta and M. globosa warrant further study. Previous work on C-type lectin receptor signaling suggests that blocking phagocytosis with cytochalasin results in prolonged SYK activation42, so we might have expected if SYK signaling is important for inflammasome activation that cytochalasin B would have enhanced rather than blocked inflammasome activation and IL-1β production. However, it is also possible that actin rearrangement is necessary for steps downstream of SYK signaling for inflammasome activation. SYK signaling in response to C. albicans was shown to been important for NLRP3 activation28, however, C. albicans induced NLRP3 activation was also dependent on hyphae formation.30 We did not observe hyphae formation during M. restricta and M. globosa infection in vitro during our studies suggesting that these fungi may activate the NLRP3 inflammasome by a different combination of signals. It is also unclear how Malassezia triggers potassium efflux from cells. These topics will need to be addressed in future experiments.
The data presented here further our understanding of the innate pro-inflammatory responses of myeloid phagocytes to M. restricta and M. globosa yeasts and some of the underlying mechanisms. In the context of colitis, the increased presence of Malassezia in the gut could result in increased inflammation when the yeast come in contact with phagocytic cells, contributing to the persistent inflammation that is the hallmark of inflammatory bowel diseases. Our recent work noted a specific enrichment of Malassezia in the colonic mucosa of patients with Crohn’s disease,7 although in other studies and models additional fungi have been implicated in colonic inflammation including C. tropicalis and C. albicans6,43,44. Given the complex nature of the microbiome, it is unclear if specific fungal species drive colitis, or if the disruption of the gut epithelial layer and dysbiosis allow for fungal species to expand where they would not normally. It is also possible that the balance of fungal to bacterial recognition leads to improper levels of inflammation in colitis. It will be important in the future to examine whether tissue specific subsets of macrophages and dendritic cells respond differently to different fungi. Understanding these interactions and the roles of gut fungi in colitis may lead to development of novel strategies for targeted therapeutic interventions in select patients.
Supplementary Material
Supplemental Figure 1. Role of CARD9 signaling in dendritic cell responses to Malassezia. (A) BMDC were treated and cytokines measured as in Figure 2A. (B) Representative flow cytometry plots of BMDC phagocytosis of e670 fluorescently labeled Malassezia at 5 min. post treatment as represented in Figure 2B. CD45+ staining was used identify BDMC from free fungi. (C) BMDC from wild type and the indicated knockout mice were treated with depleted zymosan (Treated) or not (UT = untreated), and production of reactive oxygen species (ROS) was measured by luminol-enhanced chemiluminescence (RUL=relative light units). All data represent measurements done in triplicate ± SD, and statistics were done by student t-test * p≤0.05, ** p≤0.01,*** p≤0.001.
ACKNOWLEDGEMENTS
This study was supported by grants to D.M.U. from the NIH (R01 DK093426 and P01 DK046763) and the Janis and William Wetsman Chair in Inflammatory Bowel Disease.
ABBREVIATIONS
- IBD
inflammatory bowel disease
- BMDM
bone marrow-derived macrophages
- BMDC
bone marrow-derived dendritic cells
- HK
heat-killed
- PFA
paraformaldehyde fixed
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors declare no conflict of interest.
REFERENCES
- 1.Paterson MJ, Oh S, Underhill DM Host-microbe interactions: commensal fungi in the gut. Curr Opin Microbiol. 2017;40:131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rooks MG and Garrett WS Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Limon JJ, Skalski JH, Underhill DM Commensal Fungi in Health and Disease. Cell Host Microbe. 2017;22:156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li XV, Leonardi I, Iliev ID Gut Mycobiota in Immunity and Inflammatory Disease. Immunity. 2019;50:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard ML and Sokol H The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2019;16:331–345. [DOI] [PubMed] [Google Scholar]
- 6.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, Iliev ID, Skalski JH, Brown J, Landers C, Borneman J, Braun J, Targan SR, McGovern DPB, Underhill DM Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe. 2019;25:377–388.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, Guo Y, Saxena A, Vardhan M, Diskin B, Wang W, Leinwand J, Kurz E, Kochen Rossi JA, Hundeyin M, Zambrinis C, Li X, Saxena D, Miller G The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574:264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, Stevens C, Alikashani A, Ladouceur M, Ellinghaus D, Torkvist L, Goel G, Lagace C, Annese V, Bitton A, Begun J, Brant SR, Bresso F, Cho JH, Duerr RH, Halfvarson J, McGovern DP, Radford-Smith G, Schreiber S, Schumm PL, Sharma Y, Silverberg MS, Weersma RK, Quebec IBDGC, Consortium NIG, International IBDGC, D’Amato M, Vermeire S, Franke A, Lettre G, Xavier RJ, Daly MJ, Rioux JD Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, Fennell T, Kirby A, Latiano A, Goyette P, Green T, Halfvarson J, Haritunians T, Korn JM, Kuruvilla F, Lagace C, Neale B, Lo KS, Schumm P, Torkvist L, National Institute of, D., Digestive Kidney Diseases Inflammatory Bowel Disease Genetics, C., United Kingdom Inflammatory Bowel Disease Genetics, C., International Inflammatory Bowel Disease Genetics, C., Dubinsky MC, Brant SR, Silverberg MS, Duerr RH, Altshuler D, Gabriel S, Lettre G, Franke A, D’Amato M, McGovern DP, Cho JH, Rioux JD, Xavier RJ, Daly MJ Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, Bevova M, Nijmeijer RM, van ‘t Slot R, Heijmans R, Boezen HM, van Heel DA, van Bodegraven AA, Stokkers PC, Wijmenga C, Crusius JB, Weersma RK Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartjes L and Ruland J CARD9 Signaling in Intestinal Immune Homeostasis and Oncogenesis. Front Immunol. 2019;10:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol. 2016;55:494–504. [DOI] [PubMed] [Google Scholar]
- 14.Thayikkannu AB, Kindo AJ, Veeraraghavan M Malassezia-Can it be Ignored? Indian J Dermatol. 2015;60:332–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theelen B, Cafarchia C, Gaitanis G, Bassukas ID, Boekhout T, Dawson TL Jr. Malassezia ecology, pathophysiology, and treatment. Med Mycol. 2018;56:S10–S25. [DOI] [PubMed] [Google Scholar]
- 16.Sparber F and LeibundGut-Landmann S Host Responses to Malassezia spp. in the Mammalian Skin. Front Immunol. 2017;8:1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balato A, Paoletti I, De Gregorio V, Cantelli M, Ayala F, Donnarumma G Tacrolimus does not alter the production of several cytokines and antimicrobial peptide in Malassezia furfur-infected-keratinocytes. Mycoses. 2014;57:176–83. [DOI] [PubMed] [Google Scholar]
- 18.Kesavan S, Walters CE, Holland KT, Ingham E The effects of Malassezia on pro-inflammatory cytokine production by human peripheral blood mononuclear cells in vitro. Med Mycol. 1998;36:97–106. [PubMed] [Google Scholar]
- 19.Kistowska M, Fenini G, Jankovic D, Feldmeyer L, Kerl K, Bosshard P, Contassot E, French LE Malassezia yeasts activate the NLRP3 inflammasome in antigen-presenting cells via Syk-kinase signalling. Exp Dermatol. 2014;23:884–9. [DOI] [PubMed] [Google Scholar]
- 20.Walters CE, Ingham E, Eady EA, Cove JH, Kearney JN, Cunliffe WJ In vitro modulation of keratinocyte-derived interleukin-1 alpha (IL-1 alpha) and peripheral blood mononuclear cell-derived IL-1 beta release in response to cutaneous commensal microorganisms. Infect Immun. 1995;63:1223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55:778–87. [DOI] [PubMed] [Google Scholar]
- 22.Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Gotz F, Liu GY, Underhill DM Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe. 2010;7:38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, Pla J, Iliev ID CX3CR1(+) mononuclear phagocytes control immunity to intestinal fungi. Science. 2018;359:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Lopez M, Iborra S, Conde-Garrosa R, Mastrangelo A, Danne C, Mann ER, Reid DM, Gaboriau-Routhiau V, Chaparro M, Lorenzo MP, Minnerup L, Saz-Leal P, Slack E, Kemp B, Gisbert JP, Dzionek A, Robinson MJ, Ruperez FJ, Cerf-Bensussan N, Brown GD, Bernardo D, LeibundGut-Landmann S, Sancho D Microbiota Sensing by Mincle-Syk Axis in Dendritic Cells Regulates Interleukin-17 and −22 Production and Promotes Intestinal Barrier Integrity. Immunity. 2019;50:446–461.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai T and Akira S Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. [DOI] [PubMed] [Google Scholar]
- 26.Lamkanfi M and Dixit VM Mechanisms and Functions of Inflammasomes. Cell. 2014;157:1013–1022. [DOI] [PubMed] [Google Scholar]
- 27.Martinon F, Burns K, Tschopp J The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–26. [DOI] [PubMed] [Google Scholar]
- 28.Gross O, Poeck H, Bscheider M, Dostert C, Hannesschlager N, Endres S, Hartmann G, Tardivel A, Schweighoffer E, Tybulewicz V, Mocsai A, Tschopp J, Ruland J Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–6. [DOI] [PubMed] [Google Scholar]
- 29.Hise AG, Tomalka J, Ganesan S, Patel K, Hall BA, Brown GD, Fitzgerald KA An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe. 2009;5:487–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joly S, Ma N, Sadler JJ, Soll DR, Cassel SL, Sutterwala FS Cutting edge: Candida albicans hyphae formation triggers activation of the Nlrp3 inflammasome. J Immunol. 2009;183:3578–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joly S and Sutterwala FS Fungal pathogen recognition by the NLRP3 inflammasome. Virulence. 2010;1:276–80. [DOI] [PubMed] [Google Scholar]
- 32.van de Veerdonk FL, Joosten LA, Devesa I, Mora-Montes HM, Kanneganti TD, Dinarello CA, van der Meer JW, Gow NA, Kullberg BJ, Netea MG Bypassing pathogen-induced inflammasome activation for the regulation of interleukin-1beta production by the fungal pathogen Candida albicans. J Infect Dis. 2009;199:1087–96. [DOI] [PubMed] [Google Scholar]
- 33.Mathur A, Hayward JA, Man SM Molecular mechanisms of inflammasome signaling. J Leukoc Biol. 2018;103:233–257. [DOI] [PubMed] [Google Scholar]
- 34.Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M, Underhill DM Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan. Cell. 2016;166:624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodridge HS, Shimada T, Wolf AJ, Hsu YM, Becker CA, Lin X, Underhill DM Differential use of CARD9 by dectin-1 in macrophages and dendritic cells. J Immunol. 2009;182:1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue M and Shinohara ML Clustering of pattern recognition receptors for fungal detection. PLoS Pathog. 2014;10:e1003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kagan JC and Barton GM Emerging principles governing signal transduction by pattern-recognition receptors. Cold Spring Harb Perspect Biol. 2014;7:a016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brough D and Rothwell NJ Caspase-1-dependent processing of pro-interleukin-1beta is cytosolic and precedes cell death. Journal of cell science. 2007;120:772–81. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Fu S, Sun S, Li Z, Guo B Inflammasome activation has an important role in the development of spontaneous colitis. Mucosal Immunology. 2014;7:1139–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sand J, Haertel E, Biedermann T, Contassot E, Reichmann E, French LE, Werner S, Beer H-D Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death & Disease. 2018;9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navarro-Arias MJ, Hernández-Chávez MJ, García-Carnero LC, Amezcua-Hernández DG, Lozoya-Pérez NE, Estrada-Mata E, Martínez-Duncker I, Franco B, Mora-Montes HM Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect Drug Resist. 2019;12:783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, Weiss A, Vasilakos JP, Underhill DM Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature. 2011;472:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panpetch W, Hiengrach P, Nilgate S, Tumwasorn S, Somboonna N, Wilantho A, Chatthanathon P, Prueksapanich P, Leelahavanichkul A Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes. 2020;11:465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Role of CARD9 signaling in dendritic cell responses to Malassezia. (A) BMDC were treated and cytokines measured as in Figure 2A. (B) Representative flow cytometry plots of BMDC phagocytosis of e670 fluorescently labeled Malassezia at 5 min. post treatment as represented in Figure 2B. CD45+ staining was used identify BDMC from free fungi. (C) BMDC from wild type and the indicated knockout mice were treated with depleted zymosan (Treated) or not (UT = untreated), and production of reactive oxygen species (ROS) was measured by luminol-enhanced chemiluminescence (RUL=relative light units). All data represent measurements done in triplicate ± SD, and statistics were done by student t-test * p≤0.05, ** p≤0.01,*** p≤0.001.