Abstract
Background and Objective.
Metabolomics deals with the identification and quantification of small molecules (metabolites) in biological samples. As metabolite levels can reflect normal or altered metabolic pathways, their measurement provides information to improve the understanding, diagnosis and management of diseases. Despite its immense potential, metabolomics applications to pain research have been sparse. This paper describes current metabolomics techniques, reviews published human metabolomics pain research, and compares successful metabolomics research in other areas of medicine with the goal of highlighting opportunities offered by metabolomics to advance pain medicine.
Databases and Data Treatment.
Non-systematic review.
Results.
Our search identified 19 studies that adopted a metabolomics approach in: fibromyalgia (7), chronic widespread pain (4), other musculoskeletal pain conditions (5), neuropathic pain (1), complex regional pain syndrome (1) and pelvic pain (1). The studies used either mass spectrometry or nuclear magnetic resonance. Most are characterized by small sample sizes. Some consistency has been found for alterations in glutamate and testosterone metabolism, and metabolic imbalances caused by the gut microbiome.
Conclusions.
Metabolomics research in chronic pain is at its infancy. Most studies are at the pilot stage. Metabolomics research has been successful in other areas of medicine. These achievements should motivate investigators to expand metabolomics research to improve understanding of the basic mechanisms of human pain, as well as provide tools to diagnose, predict and monitor chronic pain conditions. Metabolomics research can lead to the identification of biomarkers to support the development and testing of treatments, thereby facilitating personalized pain medicine.
INTRODUCTION
The growing area of metabolomics, also sometimes referred to as metabolite profiling or metabonomics, deals with the identification and quantification of small molecules, which are commonly known as metabolites. Thousands of metabolites have been identified in a variety of biological samples, including human and animal cells, tissues, fluids, as well as plants and food, and many others. Metabolites are the downstream products of genes, transcripts and enzyme functions. Their levels can therefore represent measures of normal or altered metabolic pathways and therefore reflect the metabolic status of the system.
While the vast progress in the fields of genomics and proteomics has provided a wealth of information for understanding the factors that regulate cellular functions, the study of metabolite concentrations and fluxes offer complementary insight into the functions at the cellular, sub-cellular and systems level (Patti et al., 2012). The ability to measure hundreds or even thousands of metabolites in a single measurement offers an attractive avenue to detect altered metabolite levels due, e.g., to changes in gene expression or protein levels, and often related to a variety of biological stresses such as the effects of drugs, toxins and environmental exposures. Furthermore, the individual biological variations measured through metabolite levels offer the opportunity to monitor the course of numerous diseases. All these factors can contribute to an improved, personalized medicine approach (Holmes et al., 2008).
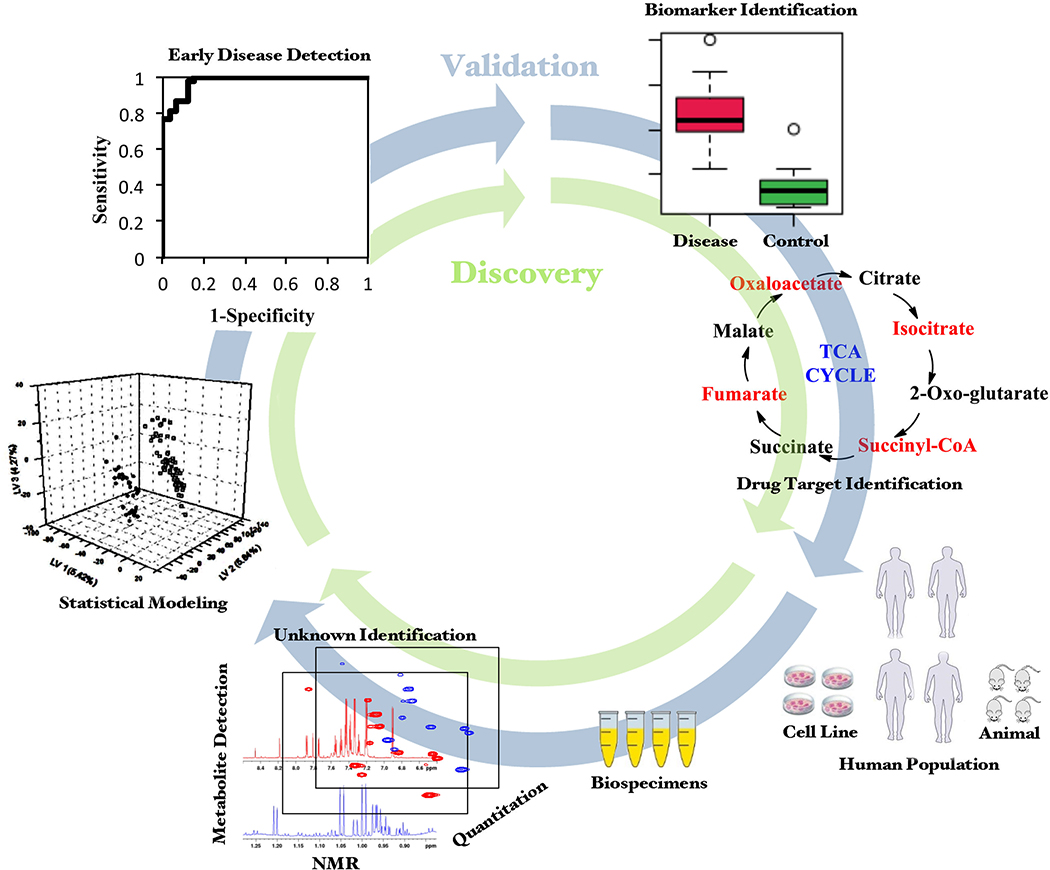
Metabolomics applications have spread to numerous disciplines including the life, food and plant sciences, toxicology, environmental science and drug development (Nagana Gowda and Raftery, 2013; Johnson et al., 2016; Wishart, 2016a; Viant et al., 2019; Gonzalez-Dominguez et al., 2020; Steck and Murphy, 2020). The application to life sciences has been the major focus of metabolomics, driven by the urgent need to understand and manage numerous major human diseases including but not limited to cancer (Nagana Gowda and Raftery, 2013; Armitage and Ciborowski, 2017), diabetes (Guasch-Ferre et al., 2016; Newgard, 2017) and cardiovascular diseases (Ussher et al., 2016; Newgard, 2017; Ruiz-Canela et al., 2017). The applications utilize a variety of bio-specimens from humans as well as animal and cell models of human diseases focused on improving the mechanistic understanding, diagnosis and management of various diseases (Figure 1).
Figure 1.
Translational process from metabolomics analysis to applications in diagnosis, prognosis and treatment.
However, to date, the application of metabolomics approaches to pain research has been sparse. Chronic pain remains a leading cause of suffering, disability and enormous economic burden worldwide (Gaskin and Richard, 2012; GBD 2017 Disease and Injury Incidence and Prevalence Collaborators, 2018). The understanding of mechanisms of human pain, as well as the diagnosis and treatment of pain conditions, are still highly unsatisfactory. Metabolomics research has the potential to improve our understanding of the mechanisms underlying human pain conditions, provide diagnostic and predictive tools to improve personalized pain medicine, and support the development of mechanism-specific pain treatments.
CURRENT STATUS OF METABOLOMICS RESEARCH IN AREAS OTHER THAN PAIN
To date, a large body of work documents the discovery of potential biomarkers and provides greater insights into the pathogeneses of numerous human diseases. Several studies have also focused on translating the findings to early detection, therapy prediction and prognosis, monitoring treatment and recurrence detection. Therapeutic target drug discovery is another area that has seen major translational efforts (Wishart, 2016b). Numerous recent reviews highlight in a great detail the developments made in these areas so far and the remaining challenges (Nagana Gowda and Raftery, 2013; Liggi and Griffin, 2017; Lopez-Lopez et al., 2018). Some brief examples are provided below.
Investigations of diabetes have grown enormously since the early report of altered metabolite levels in biological fluids from diabetic patients (Nicholson et al., 1984). Both Type 1 and Type 2 diabetes have been the focus of numerous investigation with a goal of identifying a variety of biomarkers and their association with perturbed metabolic pathways (Friedrich, 2012). Early studies identified branched chain amino acids as playing an important role in obesity and the development of insulin resistance (Newgard et al., 2009). Studies focused on assessing the risk of developing diabetes have shown that branched chain amino acids can be predictive of Type 2 diabetes as early as 5 years before disease onset (Wang et al., 2011). Further, to better assess the risk of diabetes, there has been significant interest to combine metabolomics with other omics areas (Liggi and Griffin, 2017).
Metabolomics studies of almost all types of cancers have resulted in the identification of numerous potential metabolite biomarkers (Nagana Gowda and Raftery, 2013; Lopez-Lopez et al., 2018). For example, elevated choline metabolites and lower glycerophosphocholine levels are shown to have diagnostic value for breast cancer. Acylcarnitines and metabolites associated with tryptophan metabolism were found to be linked with kidney cancer. Lower levels of citrate in seminal fluid is found to be linked with prostate cancer. Similarly, biomarkers for other cancer types including gliomas, ovarian cancer, liver cancer, lung cancer, pancreatic cancer, endometrial cancer, bladder cancer, esophageal cancer and colon cancer have been reported However, few if any biomarkers have been sufficiently validated for clinical use.
Heart disease has been one of the major subjects of metabolomics investigations and, to date, several metabolites have been shown to have potential diagnostic value. For example, studies have found metabolites such as choline, trimethylamine N-oxide (TMAO) and betaine to be predictive of cardiovascular disease (Wang et al., 2011). Separately, studies have shown that medium-chain acylcarnitines, short-chain dicarboxylacylcarnitines, long-chain dicarboxylacylcarnitines and branched-chain amino acids are associated with mortality due to cardiovascular disease (Shah et al., 2012). Studies have also shown that measurement of blood lipids enable identification of several (15 to 18) lipoprotein classes and subclasses of lipid particles that predict the risk of cardiovascular disease (Otvos et al., 1991), and this approach has been FDA approved for clinical use. The findings have led to new avenues for clinical assessment and management of atherosclerotic cardiovascular disease (Otvos et al., 1991; Jeyarajah et al., 2006).
In the same way, a large number of other diseases including inborn errors of metabolism (Sandlers, 2017), neurological disorders including Alzheimer’s disease and Parkinson’s disease (Kaddurah-Daouk and Krishnan, 2009; Saiki et al., 2017; Wang et al., 2020), and infectious diseases (Pacchiarotta et al., 2012; Zurfluh et al., 2018) have been the subjects of metabolomics investigations, with promising results for improved methods of early disease detection.
Clearly, such achievements would be of enormous relevance in pain medicine. In the area of prognostic biomarkers, identifying patients at risk of developing chronic pain would facilitate the development and testing of preventive strategies. In the area of predictive biomarkers, identifying potential responders and non-responders would improve the efficacy and safety of interventions. These goals have been so far difficult to achieve (Schliessbach et al., 2018; Klyne et al., 2019; Muller et al., 2019). Unlike other areas of medicine, the field of diagnostic biomarkers in pain management is almost unexplored. To date, pain is classified and diagnosed mostly based on phenotypic characteristics (Dworkin et al., 2016; Treede et al., 2019). While pain remains a subjective experience, there is an urgent need to develop mechanism-based diagnosis of pain conditions to support a mechanism-targeted treatment approach. Metabolomics research has the potential to support this goal.
AIMS
This paper reviews human pain studies that adopted a metabolomics approach, i.e., using a comprehensive metabolic profiling or investigating broad sets of metabolites that result from defined cellular processes. In order to provide perspective, we briefly describe current metabolomics techniques and methodologies, and highlight some successful metabolomics research in other areas of medicine (see above). The potential of metabolomics to provide biomarkers that improve personalized pain medicine and facilitate drug discovery, to ultimately reduce the burden of chronic pain, is emphasized. Our paper does not review pain research on single metabolites. Its ultimate goal is to highlight opportunities offered by metabolomics to advance the field of pain medicine.
METABOLOMICS METHODS
Metabolomics studies broadly involve two major steps: (1) analysis of a large number of metabolites in parallel from biological specimens such as biofluids and tissue, and (2) analysis of metabolite data utilizing advanced statistical and/or pathway analysis methods. While the analysis of metabolites provides identity as well as relative or absolute concentrations of metabolites, data analysis enables mechanistic understanding and identification of biomarkers and drug targets.
The two most widely used analytical platforms for analysis of metabolites are mass spectrometry (MS) and nuclear magnetic resonance spectroscopy (NMR). Both MS and NMR enable analysis of a large number of metabolites in a single measurement with high throughput. There are numerous techniques within each platform that enable access to a broad array of metabolites (aqueous, lipophilic, stable, unstable, volatile, non-volatile, etc.), and the characteristics of the platforms are complimentary to one another (Raftery, 2014; Nagana Gowda and Raftery, 2019).
The obtained metabolite data are subjected to analysis using statistical methods. Both univariate analysis such as Student’s t-test and multivariate analysis methods are widely used. While univariate analysis compares one metabolite at a time to distinguish, for example, between patient and control groups, multivariate analysis considers concentrations of many or even all metabolites in the analysis. Broadly, the methods are divided into two major major groups: (1) unsupervised analysis methods and (2) supervised analysis methods. For unsupervised analysis methods, the information of the sample group (example, control or disease) to which the sample belongs is not provided. Such analysis is generally used for identification of outlier samples, in discovery research and for hypothesis generation. On the other hand, for supervised analysis, the information of the ample group is provided as part of the analysis. Supervised analysis methods are useful for developing predictive models but require extensive validation.
Another area that utilizes the obtained metabolites is metabolic pathway analysis. It is a powerful approach for identifying altered pathways due to biological stresses such as disease, drugs, or toxicity. Further, it provides mechanistic insights into altered enzymatic reactions, along with an improved understanding of the molecular basis of disease and other biological effects. Clues to help identify drug targets for therapy development can be gleaned. Pathway analyses map experimentally obtained changes in metabolite profile to specific and statistically significant metabolic pathway alterations that can then provide links between genes, proteins and metabolites for a broader mechanistic understanding. Metabolites-based pathway analysis can also be integrated with other omics data such as microarray and proteomics to further illuminate the altered pathways and unravel disease mechanisms; such analysis incorporates a number of types of interactions such as protein signaling, metabolite signaling, drug action and xenobiotic pathways (Pinu et al., 2019).
For further details on various methods used in metabolomics, the readers are encouraged to refer to the literature (Nagana Gowda et al., 2015; Wishart, 2016a; Trivedi et al., 2017).
METABOLOMICS RESEARCH IN PAIN MEDICINE
To review the published metabolomics research in pain medicine, a search on pubmed.gov was performed, using the terms “pain” and “metabolomics”, with no filter. As mentioned in the Aims section, the focus was on studies that adopted a metabolomics approach, i.e., those that used a comprehensive metabolic profiling or investigated broad sets of metabolites and resulted from defined cellular processes. Papers focusing on single metabolites were not considered. From the list of retrieved investigations, we excluded animal studies, studies on acute pain, in healthy volunteers, focusing on single metabolites, and studies on pain related to angina pectoris. In the selected papers, the list of references was scanned using the same inclusion and exclusion criteria.
The initial search yielded 256 papers on March 25, 2020 with the search terms of metabolomics and pain. After applying the exclusion criteria, 14 papers remained for evaluation. The references of these papers yielded 5 additional studies. Thus, 19 papers were included in the present review. These studies are presented below and summarized in Table 1, according to pain syndrome and sorted by publication year.
Table 1.
Studies retrieved by the search and fulfilling the inclusion criteria, according to pain syndrome and sorted by publication year.
| Aim | Sample type | Sample size | Method | Main findings | Reference |
|---|---|---|---|---|---|
| Fibromyalgia (FM) and Chronic Widespread Pain (CWP) | |||||
| To evaluate serum purine metabolite levels in patients with FM | Serum | 22 FM, 22 healthy controls | LC / UV-vis | Significantly higher levels of inosine, hypoxanthine and xanthine, and significantly lower levels of adenosine in FM. Negative correlation of Fibromyalgia Impact Questionnaire with adenosine levels. | Fais et al, 2013 |
| To analyze the plasma metabolic profile of patients with FM aiming to discriminate FM patients from controls | Plasma | 22 FM, 21 controls | LC-Q-TOF/MS | Lysophosphocholines (lysoPCs), phosphocholines and ceramides discriminated patients from controls. An interaction analysis suggested that lipids oxidative fragmentation generates an overproduction of lysoPCs, which may contribute to pain hypersensitivity. | Caboni et al, 2014 |
| To analyze the urine metabolic profile of patients with FM | Urine | 18 FM, 11 family members, 10 age-related individuals without FM, 20 healthy young subjects | NMR spectroscopy | Increases in metabolites related to the gut microbiome (hippuric, succinic and lactic acids). The combination of succinic acid, taurine and creatine correlated with pain and fatigue symptoms. | Malatji et al, 2017 |
| To analyze the urine metabolic profile of patients with FM | Urine | Same samples as for above paper | GC-MS | FM patients displayed 14 significantly increased metabolites associated with energy metabolism and gut metabolites. | Malatji et al, 2019 |
| To study the ability of metabolic profile to differentiate FM from rheumatoid arthritis (RA) and osteoarthritis (OA) | Dried blood spots | 14 FM, 15 RA, 12 OA | IRMS | Changes in tryptophan catabolism discriminated the fibromyalgia group from the other groups. | Hackshaw et al, 2013 |
| To study the ability of metabolic profile to differentiate FM from rheumatoid arthritis (RA), osteoarthritis (OA) and systemic lupus erythematosus (SLE) | Dried blood spots | 50 FM, 29 RA, 19 OA, 23 SLE | Vibrational spectroscopy, uHPLC-PDA-MS/MS | Protein backbones and pyridine-carboxylic discriminated the fibromyalgia group from the other groups. | Hackshaw et al, 2019 |
| To study microbiome, serum metabolome, circulating cytokines and miRNAs in FM | Faeces, blood samples | 105 FM, 54 healthy controls | UPLC-MS | Elevated serum levels of glutamate in FM and reduction in bacteria from Bifidobacterium and Lactobacillus genera (which transform glutamate into GABA). | Clos Garcia et al, 2019 |
| Nontargeted metabolomics screening in CWP, including 324 metabolites | Plasma | 2444 Twins | LC/MS | Identification of epiandestrone sulfate as potential biomarker. | Livshits et al, 2015 |
| To identify molecular genetic factors underlying the heritability of frailty and its correlation with CWP | Plasma | 3626 Twins | LC-MS | The effect of epiandestrone sulfate on frailty seemed to be mediated through CWP (see above paper by same authors). |
Livshits et al, 2018 |
| To identify biomarkers for fatigue in CWP | Plasma and serum | 2055 females with fatigue (621 with CWP, 1434 without CWP | UPLC / Tandem MS | No association between fatigue and metabolites in twins without CWP; in the CWP group, patients with fatigue had lower levels of eicosapentaenoate (EPA) ω-3 fatty acid. | Freidin et al, 2018 |
| To determine systemic differences in serum metabolome in localized and chronic widespread pain | Serum | 16 CWP, 30 Neck-shoulder pain, 39 healthy controls | GC-TOF-MS | Identified metabolites in neck pain were related to carbohydrate and energy metabolism. Weak systematic difference between subjects with CWP and healthy controls. |
Hadrevi et al, 2015 |
| Osteoarthritis (OA) | |||||
| To evaluate if metabolomics profiling can distinguish synovial fluid samples of knee OA patients from normal human cadaveric knee joints |
Synovial fluid | 55 knee OA patients, 13 normal human cadaveric knee joints | H-NMR GC-MS |
Fructose and citrate were increased in OA samples as compared to controls. O-acetylcarnitine, N-phenylacetylglycine, methionine, ethanol, creatine, malate, ethanolamine, 3-hydroxybutyrate and hexanoylcarnitine were decreased in OA samples. |
Mickiewicz et al, 2015 |
| To identify biomarkers for knee OA | Plasma | 64 OA patients, 45 controls (discovery stage); 72 OA patients, 72 healthy controls (validation stage) | UPLC/MS | Arginine was significantly lower in knee OA patients. | Zhang et al, 2016 |
| Other Musculoskeletal Pain Conditions | |||||
| To compare metabolomics in the muscle interstitial fluid in trapezius myalgia | Micro-dialysate from muscle | 10 Trapezius myalgia, 10 controls | GC-MS | L-leucine and pyroglutamic acid were more abundant in muscles of subjects with trapezius myalgia. | Hadrevi et al, 2013 |
| To study levels of ornithine and closely related amino acids in subjects with persistent musculoskeletal pain | Whole blood or plasma | 76 with persistent pain, 221 with non-persistent pain, 61 no pain | LC/MS | Ornithine levels were increased in the group with persistent pain, as compared with the other two groups. | Mantyselka et al, 2017 |
| To identify changes in plasma N-glycosylation pattern as markers if inflammation in chronic low back pain (CLBP) | Plasma | 1128 CLBP, 126 acute inflammation undergoing abdominal surgery, 760 healthy controls | HILIC-UPLC | Significant changes in different N-glycan structures in CLBP, consistent with N-glycosylation changes typically observed in chronic inflammation. | Trbojević et al, 2018 |
| Neuropathic Pain and Complex Regional Pain Syndrome (CRPS) | |||||
| To determine if urine metabolomics differentiate neuropathic from nociceptive pain | Urine | 25 neuropathic pain, 12 nociceptive pain, 37 pain-free controls | H NMR | Classification of neuropathic and nociceptive pain with high sensitivity and specificity, more so for neuropathic than for nociceptive pain. Upregulation of phosphocholine, alanine and taurine in neuropathic pain, suggestive of damage of nervous system tissues. | Finco et al, 2016 |
| To evaluate plasma levels of amino acids in CRPS | Plasma | 160 CRPS, 60 healthy controls | HPLC | Changes in plasma levels of amino acids involved in glutamate receptor activation and in amino acids associated with immune function in CRPS. | Alexander et al, 2013 |
| Pelvic Pain | |||||
| To identify urine metabolites in interstitial cystitis/bladder pain syndrome (IC/BPS) | Urine | 40 IC/BPS, 40 healthy subjects | HPLC | Etio-S distinguished IC/BPS from controls with sensitivity and specificity > 90%. The levels of this metabolite were correlated with symptom severity. | Parker et al, 2016 |
Abbreviations, alphabetically: CLBP: chronic low back pain; CRPS: Complex Regional Pain Syndrome; CWP: chronic widespread pain; FM: fibromyalgia; GC: gas chromatography; HILIC-UPLC: hydrophilic interaction ultra-performance liquid chromatography; HPLC: high performance liquid chromatography; HRMAS: high resolution magic angle spinning; IC/BPS: interstitial cystitis/bladder pain syndrome; IRMS: mid-infrared microspectroscopy; LC: liquid chromatography; MS: mass spectroscopy; NMR: nuclear magnetic resonance; OA: osteoarthritis; Q-TOF: quadruple time of flight; RA: rheumatoid arthritis; SLE: systemic lupus erythematosus; TOF: time of flight; UPLC: ultra performance liquid chromatography. UV-vis: Ultraviolet-visible spectrophotometry.
Fibromyalgia and Widespread Pain
Most of the metabolomics studies identified by our search were on fibromyalgia and widespread pain. Fais et al. studied the serum levels of purine metabolites in 22 fibromyalgia patients and 22 healthy controls (Fais et al., 2013). The main rationale for this study was the role of purines in pain transmission and the antinociceptive effect of adenosine when released into the extracellular space. Significantly higher levels of inosine, hypoxanthine and xanthine, and significantly lower levels of adenosine were observed in fibromyalgia patients, compared to controls. Analysis of the Fibromyalgia Impact Questionnaire revealed a negative correlation with adenosine levels. The authors postulate an increase activity of adenosine deaminase (ADA), leading to an excessive conversion of adenosine to inosine. Adenosine has known antinociceptive properties (Sawynok, 2016), therefore a low extracellular concentration could cause lowered pain threshold and central sensitization in fibromyalgia.
Caboni et al. analyzed the plasma metabolic profile of 22 patients with fibromyalgia and 21 controls (Caboni et al., 2014). The authors focused on oxidative stress with lipid peroxidation. The study found mainly that lysophosphocholines (lysoPCs), phosphocholines and ceramides discriminated patients from controls. In addition, the authors modeled an interaction of lysophosphocholines with the PAFr (Platelet Activating Factor - Receptor). Activation of the PAF/PAFr system leads to pain hypersensitivity, allodynia and production of pro-inflammatory cytokines. The results suggested that lipid oxidative fragmentation generates an overproduction of lysoPCs. LysoPCs act as PAF-like bio-activators, which in turn may contribute to pain hypersensitivity in fibromyalgia.
Malatji et al. performed untargeted urine metabolomics in 18 fibromyalgia patients and three control groups (family members, age-related individuals with no fibromyalgia, and healthy young individuals). In a first publication based on NMR, they found significant increases in metabolites related to the gut microbiome (hippuric, succinic and lactic acids) (Malatji et al., 2017). The combination of three metabolites (succinic acid, taurine and creatine) correlated with pain and fatigue symptoms. In a second publication on the same samples, based on GC-MS, fibromyalgia patients displayed 14 significantly increased metabolites associated with energy metabolism and gut metabolites (Malatji et al., 2019). The authors inferred that energy utilization and disrupted microbiota metabolites may be involved in fibromyalgia. A disrupted gut-brain axis may lead to altered central pain processing and associated fibromyalgia symptoms, such as fatigue.
Hackshaw et al. used mid-infrared micro-spectroscopy in blood samples to differentiate 15 patients with fibromyalgia from patients with rheumatoid arthritis and osteoarthritis (Hackshaw et al., 2013). Rheumatoid arthritis and osteoarthritis patients were metabolically similar, whereas the biochemical characteristics of fibromyalgia patients were distinctive. The authors stressed the ability of changes in tryptophan catabolism to differentiate patients with fibromyalgia from patients with rheumatoid arthritis and osteoarthritis. The analysis included multiple correlations with patient symptoms, but the low sample size exposed the study to the risk of false positive results. In a later study, the same group used vibrational spectroscopy to compare 50 fibromyalgia patients with patients with rheumatoid arthritis, osteoarthritis and systemic lupus erythematosus (Hackshaw et al., 2019). Protein backbones and pyridine-carboxylic discriminated fibromyalgia patients from the other groups. The meaning of this finding in terms of pathophysiology of fibromyalgia remains unclear.
Clos-Garcia et al. adopted a combined omics approach to study the microbiome, the serum metabolome, circulating cytokines and miRNAs in feces and blood samples of 105 fibromyalgia patients and 54 healthy controls (Clos-Garcia et al., 2019). The study was based on the concept that the gut microbiome affects metabolic activity, providing an insight into the pathogenesis of fibromyalgia. The serum metabolome analysis revealed elevated levels of glutamate in fibromyalgia, an excitatory neurotransmitter involved in pain sensitization. The microbiome analysis revealed a reduction in bacteria from Bifidobacterium and Lactobacillus genera, which transform glutamate into GABA. This is a possible explanation for the elevation of glutamate serum levels in fibromyalgia. The findings of this study have some consistency with the aforementioned studies by Malatji et al (Malatji et al., 2017; Malatji et al., 2019), supporting a role of alterations in the gut microbiome in the pathophysiology of fibromyalgia.
In two large studies using the TwinsUK discovery sample that included monozygotic and dizygotic twins, Livshits et al. examined a panel of 364 plasma metabolites in patients with chronic widespread pain (Livshits et al., 2015; Livshits et al., 2018). After adjusting for covariates and false discovery rate, 6 metabolites showed significant associations with chronic widespread pain. In particular, epiandrosterone sulphate displayed the strongest association, with lower levels indicating higher risk of chronic widespread pain. A genome-wide association study of epiandrosterone sulphate levels revealed multiple highly associated SNPs. The study could not establish whether the altered steroid metabolism lies in the causal pathway or is rather a consequence of the disease but identified epiandrosterone sulphate as potential biomarker to detect individuals at risk of chronic widespread pain.
In another analysis of the same discovery sample set, the authors aimed to identify biomarkers for fatigue (Freidin et al., 2018). No association between fatigue and metabolites was identified in twins without chronic widespread pain, whereas in the patients with chronic widespread pain lower levels of eicosapentaenoate (EPA), a ω−3 fatty acid, were observed in patients with fatigue. Analysis based on the receiver operating characteristics (ROC) curve identified a combination of 15 metabolites with an area under the curve (AUC) for fatigue of 75% (95% CI 69–80%). Because EPA was not associated with chronic widespread pain, the results suggest that decreases in EPA levels are specific for fatigue associated with this pain condition. As EPA has an anti-inflammatory action, the results support a role of downregulation of endogenous anti-inflammatory pathways in promoting fatigue in this patient population.
Hadrevi et al. analyzed serum blood samples from 30 subjects with neck pain, 16 with chronic widespread pain and 39 healthy controls (Hadrevi et al., 2015). Identified metabolites in neck pain were related to carbohydrate and energy metabolism, mainly amino acids, cholesterol, glycerol-3-phosphate, α-tocopherol (vitamin E) and threonic acid (vitamin C metabolite). The increase is carbohydrates leads to more production of saturated and unsaturated fatty acids, another finding of the study, which in turn are precursors for the synthesis of bioactive fatty acid amides related to inflammation. There was only a weak systematic difference between subjects with chronic widespread pain and healthy controls.
Osteoarthritis
Mickiewicz et al. (Mickiewicz et al., 2015) analyzed synovial fluid samples collected from symptomatic chronic knee osteoarthritis patients and normal human cadaveric knee joints. The samples were analyzed using both NMR and GC-MS, and the results of the two procedures were integrated in a statistical model. The study found an increase in fructose and citrate in osteoarthritis synovial fluid samples, whereas O-acetylcarnitine, N-phenylacetylglycine, methionine, ethanol, creatine, malate, ethanolamine, 3-hydroxybutyrate and hexanoylcarnitine were decreased in the same samples. The authors interpreted the increased fructose levels as sign of hypoxia of the knee joint. The increased concentration of citrate as well the decreased levels of malate, 3‐hydroxybutyrate, creatine and ethanol were interpreted as indicators of perturbation and dysregulation in the energy production and demand.
Zhang et al. conducted a two-stage validation study on knee osteoarthritis, with a discovery phase (64 patients and 45 controls) and a validation phase (72 patients and 72 controls) (Zhang et al., 2016). Six plasma metabolites were significantly associated with knee osteoarthritis. Arginine depletion was the most significant finding, with a receiver operating characteristic (ROC) area under the curve (AUC) of 0.984. The analysis of the metabolome provided some explanations and interpretation of this impressive finding. First, an excessive catabolism of arginine to substances that promote cartilage repair, such as ornithine, proline, and polyamines was interpreted by the authors as an attempt to repair the damaged cartilage. In addition, the authors found some evidence that arginine inhibits substances that promote cartilage breakdown. Therefore, one possible effect of arginine depletion is over-activity of such substances, ultimately contributing to osteoarthritis.
Other musculoskeletal pain conditions
Hadrevi et al. performed a metabolomics analysis of trapezius muscle by microdialysis. They found leucine and pyroglutamic acid to be significantly more abundant in muscles of subjects with trapezius myalgia (Hadrevi et al., 2013). Pyroglutamic acid is considered a reservoir for glutamate, which is involved in peripheral nociception (Cairns et al., 2001). The increase in leucine has been interpreted as indicator of increased remodeling of the myalgic muscle.
Mantyselka et al. studied the circulating levels of ornithine and related amino acids in a population of patients with various musculoskeletal pain conditions (Mantyselka et al., 2017). Ornithine levels were found to be increased in the group with persistent musculoskeletal pain, as compared with individuals with no pain or non-persistent pain. The mechanisms underlying the increase in ornithine remained unclear. The authors discussed the role of ornithine in the metabolism of glutamate, an excitatory neurotransmitter with crucial role in pain facilitation (Woolf and Thompson, 1991), and in the repair of tissues through synthesis of proline.
In a large multicenter study on chronic low back pain, Trbojević et al hypothesized the presence of an underlying inflammatory process via plasma N-glycosylation and analyzed the plasma of 1128 patients and 760 healthy controls (Trbojević-Akmačić et al., 2018). A cohort of 126 patients with acute inflammation undergoing abdominal surgery served as a further control group. Significant changes in different N-glycan structures were identified in chronic low back patients, consistent with N-glycosylation changes typically observed in chronic inflammation. Plasma glycomics is therefore a potential biomarker to identify subsets of patients with chronic low back pain characterized by inflammatory processes.
Neuropathic Pain and Complex Regional Pain Syndrome (CRPS)
Finco et al. investigated urine metabolomics to determine differences between patients with neuropathic pain (n=25), nociceptive pain (n=12) and pain-free controls (n=37) (Finco et al., 2016). The global urine metabolic profile allowed the classification of neuropathic and nociceptive pain with high sensitivity and specificity, more so for neuropathic than for nociceptive pain. With regard to specific metabolites, the study found an upregulation of phosphocholine, alanine and taurine in neuropathic pain, suggestive of damage of nervous system tissues. Noticeably, plasma phosphocholine and urinary taurine were found to be elevated also in the above-mentioned studies on fibromyalgia patients (Caboni et al., 2014; Malatji et al., 2017).
In a study on 160 CRPS patients and 60 pain-free controls, Alexander et al. found changes in plasma levels of amino acids involved in glutamate receptor activation and in amino acids associated with immune function (Alexander et al., 2013). In particular, the main findings were increases in plasma glutamate and aspartate, a modest decrease in plasma tryptophan, along with a significant increase in the kynurenine to tryptophan ratio, and a decrease in arginine. The increase in glutamate levels are consistent with above-mentioned metabolomics studies in other pain conditions. Glutamate receptors are present in nerve terminals and their stimulation induces hyperalgesia in neuropathic pain (Jang et al., 2004), suggesting that elevated glutamate levels in patients contributes to the typical feature of hyperalgesia in CRPS.
Pelvic Pain
We found one study on pelvic pain by Parker et al. that applied MS-based global metabolite profiling to urine specimens from a cohort of female subjects with interstitial cystitis or bladder pain syndrome (Parker et al., 2016). The analyses identified multiple metabolites that discriminated patients from controls. Etiocholan-3α-ol-17-one sulfate (Etio-S), a sulfoconjugated 5-β reduced isomer of testosterone, displayed a sensitivity and specificity > 90%. Among patients, the levels of this metabolite were correlated with symptom severity, such as pain and number of painful body sites. The result was unexpected and of difficult interpretation with the current knowledge. However, there is some consistency between this finding and the above-mentioned association of epiandrosterone, a precursor of testosterone, with chronic widespread pain (Livshits et al., 2015).
ACHIEVEMENTS, GAPS AND PERSPECTIVES
The field of metabolomics has now passed 20 years in its existence. During this time, major advances in analytical technologies have greatly expanded the size and quality of the coverage of the metabolome and offer new avenues for investigations of a broad set of metabolic pathways and networks (Raftery, 2014; Nagana Gowda and Raftery, 2019). Advances in software and databases increasingly offer complex data analysis and interpretation.
Applications have spread to numerous areas including early disease diagnosis, toxicology, pharmacology, drug target identification and development that are quite relevant to pain medicine studies. The major focus in human disease studies has been on the discovery of metabolite biomarkers for diagnosing, predicting, preventing and managing human diseases, as well as on developing better metabolic-related mechanistic understanding of these diseases. As a consequence, to date, a large body of literature documents the discovery of potential biomarkers and provides greater insights into pathogeneses of numerous human diseases. The outcomes of many studies indicate at least some potential for clinical translation, although validation work remains a challenging step in the translational process. A somewhat more recent development is the effort to target metabolic irregularities and vulnerabilities in an effort to develop new drugs (Popovici-Muller et al., 2018).
The present review has identified few human pain studies that have specifically adopted a comprehensive metabolomics approach, indicating that the field of metabolomics has been underrepresented in pain research. Most of the studies that were identified are characterized by small sample sizes, and as such, the results have to be taken with caution. The majority of studies have been performed in fibromyalgia and widespread pain.
Limited consistent picture emerges from the results. Pathways leading to elevated glutamate levels either systemically or locally have been shown in different conditions, specifically in fibromyalgia (Clos-Garcia et al., 2019), CRPS (Alexander et al., 2013), trapezius myalgia (Hadrevi et al., 2013), and mixed musculoskeletal pain conditions (Mantyselka et al., 2017). This consistency across pain conditions is of interest, given the well-known role of glutamate signaling in both central and peripheral sensitization (Woolf and Thompson, 1991; Cairns et al., 2001; Jang et al., 2004). There may be a role for glutamate metabolomics as marker of nociception in different pain conditions. Three studies converged to the conclusion that alterations in the gut microbiome may cause metabolic imbalances that are associated with fibromyalgia (Malatji et al., 2017; Clos-Garcia et al., 2019; Malatji et al., 2019). Finally, altered metabolism of testosterone pathways have been independently associated with bladder pain syndrome / interstitial cystitis (Parker et al., 2016) and chronic widespread pain (Livshits et al., 2015). Interestingly, bladder pain syndrome / interstitial cystitis is frequently associated with widespread pain (Thu et al., 2019).
The high number of analyses in small patient populations exposes most studies to the risk of false positive results. This may be one of the explanations for the limited consistency of results across investigations. Clearly, additional validation studies are warranted in many cases.
Studies have mostly analyzed the ability of metabolites to discriminate patients with chronic pain from healthy controls or from patients with different pain conditions. Positive results do not necessarily imply that the identified metabolites are involved in the pathophysiology of the pain condition. A link between a metabolic pathway and the pathophysiology requires further mechanistic investigation, such as modulation of the metabolic pathway. This may also be accomplished with animal models, whereby the translational process goes from human research back to basic science and then further progresses to human clinical validation. We are not aware of metabolomics studies that have adopted this translational approach in pain medicine.
Accordingly, metabolomics findings of case-control studies do not indicate per se that identified metabolites are biomarkers of metabolic pathways relevant to pain. For this, several steps in the biomarker validation process are required, as established by guidelines and reviews (FDA/NIH Evidentiary Criteria Writing Group; Hayes, 2015; Robb et al., 2016; Burke and Grizzle, 2017; Leptak et al., 2017), and depicted in Figure 1.
CONCLUSIONS
Metabolomics research in chronic pain is at its infancy. Most studies that adopted a metabolomics approach can be considered as pilot studies. The last years have witnessed substantial advances in analytical technologies, data analysis, detection sensitivity and ability to investigate a very wide range of metabolic pathways using relatively non-invasive procedures such as blood draws and urine samples. These advances render metabolomics research suitable for large-scale investigations in human pain conditions. Metabolomics research has been successful in other areas of medicine. Those achievements should motivate investigators to expand metabolomics research to improve understanding of the basic mechanisms of human pain, as well as to provide tools to diagnose, predict and monitor chronic pain conditions. This research can lead to the identification of biomarkers to support the development and testing of treatments, thereby facilitating personalized and precision pain medicine.
Significance:
The review identified 19 studies that use a metabolomics approach to investigate human chronic pain. Metabolomics pain research has an enormous potential and can contribute to a better understanding of the basic mechanisms of human pain, as well as provide tools to diagnose, predict and monitor chronic pain conditions. This research can lead to the identification of biomarkers to support the development and testing of treatments, thereby facilitating personalized pain medicine.
ACKNOWLEDGEMENTS
NG and DR thank the NIH (P30 AR074990) for partial funding for this work.
Funding: D. Raftery and G.A. Nagana Gowda thank the NIH (P30 AR074990) for the financial support.
Footnotes
Conflict of interest: No author has any conflict of interest.
REFERENCES
- Alexander GM, Reichenberger E, Peterlin BL, Perreault MJ, Grothusen JR, & Schwartzman RJ (2013). Plasma amino acids changes in complex regional pain syndrome. Pain research and treatment, 2013, 742407–742407. 10.1155/2013/742407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage EG, & Ciborowski M (2017). Applications of metabolomics in cancer studies. Advances in Experimental Medicine and Biology, 965, 209–234. 10.1007/978-3-319-47656-8_9 [DOI] [PubMed] [Google Scholar]
- Burke HB, & Grizzle WE Clinical validation of molecular biomarkers in translational medicine In: Srivastava S, editor. Biomarkers in cancer screening and early detection: John Wiley & Sons; 2017. p. 256–266. [Google Scholar]
- Caboni P, Liori B, Kumar A, Santoru ML, Asthana S, Pieroni E, Fais A, Era B, Cacace E, Ruggiero V, & Atzori L (2014). Metabolomics analysis and modeling suggest a lysophosphocholines-paf receptor interaction in fibromyalgia. Public Library of Science One, 9, e107626 10.1371/journal.pone.0107626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BE, Hu JW, Arendt-Nielsen L, Sessle BJ, & Svensson P (2001). Sex-related differences in human pain and rat afferent discharge evoked by injection of glutamate into the masseter muscle. Journal of Neurophysiology, 86, 782–791. 10.1152/jn.2001.86.2.782 [DOI] [PubMed] [Google Scholar]
- Clos-Garcia M, Andrés-Marin N, Fernández-Eulate G, Abecia L, Lavín JL, van Liempd S, Cabrera D, Royo F, Valero A, Errazquin N, Vega MCG, Govillard L, Tackett MR, Tejada G, Gónzalez E, Anguita J, Bujanda L, Orcasitas AMC, Aransay AM, Maíz O, López de Munain A, & Falcón-Pérez JM (2019). Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine, 46, 499–511. 10.1016/j.ebiom.2019.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin RH, Bruehl S, Fillingim RB, Loeser JD, Terman GW, & Turk DC (2016). Multidimensional diagnostic criteria for chronic pain: Introduction to the acttion-american pain society pain taxonomy (aapt). The Journal of Pain, 17, T1–9. 10.1016/j.jpain.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Fais A, Cacace E, Corda M, Era B, Peri M, Utzeri S, & Ruggiero V (2013). Purine metabolites in fibromyalgia syndrome. Clinical Biochemistry, 46, 37–39. 10.1016/j.clinbiochem.2012.09.009 [DOI] [PubMed] [Google Scholar]
- FDA/NIH Evidentiary Criteria Writing Group. (October 10, 2016). “Framework for defining evidentiary criteria for biomarker qualification.” from https://fnih.org/sites/default/files/final/pdf/Evidentiary%20Criteria%20Framework%20Final%20Version%20Oct%2020%202016.pdf. [Google Scholar]
- Finco G, Locci E, Mura P, Massa R, Noto A, Musu M, Landoni G, d’Aloja E, De-Giorgio F, Scano P, & Evangelista M (2016). Can urine metabolomics be helpful in differentiating neuropathic and nociceptive pain? A proof-of-concept study. Public Library of Science One, 11, e0150476 10.1371/journal.pone.0150476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidin MB, Wells HRR, Potter T, Livshits G, Menni C, & Williams FMK (2018). Metabolomic markers of fatigue: Association between circulating metabolome and fatigue in women with chronic widespread pain. Biochimica et Biophysisica Acta. Molecular Basis of Disease, 1864, 601–606. 10.1016/j.bbadis.2017.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich N (2012). Metabolomics in diabetes research. Journal of Endocrinology, 215, 29–42. 10.1530/JOE-12-0120 [DOI] [PubMed] [Google Scholar]
- Gaskin DJ, & Richard P (2012). The economic costs of pain in the united states. The Journal of Pain, 13, 715–724. 10.1016/j.jpain.2012.03.009 [DOI] [PubMed] [Google Scholar]
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators (2018). Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the global burden of disease study 2017. The Lancet, 392, 1789–1858. 10.1016/S0140-6736(18)32279-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Dominguez R, Jauregui O, Mena P, Hanhineva K, Tinahones FJ, Angelino D, & Andres-Lacueva C (2020). Quantifying the human diet in the crosstalk between nutrition and health by multi-targeted metabolomics of food and microbiota-derived metabolites. International Journal of Obesity (London). 10.1038/s41366-020-0628-1 [DOI] [PubMed] [Google Scholar]
- Guasch-Ferre M, Hruby A, Toledo E, Clish CB, Martinez-Gonzalez MA, Salas-Salvado J, & Hu FB (2016). Metabolomics in prediabetes and diabetes: A systematic review and meta-analysis. Diabetes Care, 39, 833–846. 10.2337/dc15-2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackshaw KV, Aykas DP, Sigurdson GT, Plans M, Madiai F, Yu L, Buffington CAT, Giusti MM, & Rodriguez-Saona L (2019). Metabolic fingerprinting for diagnosis of fibromyalgia and other rheumatologic disorders. Journal of Biological Chemistry, 294, 2555–2568. 10.1074/jbc.RA118.005816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackshaw KV, Rodriguez-Saona L, Plans M, Bell LN, & Buffington CA (2013). A bloodspot-based diagnostic test for fibromyalgia syndrome and related disorders. Analyst, 138, 4453–4462. 10.1039/c3an36615d [DOI] [PubMed] [Google Scholar]
- Hadrevi J, Bjorklund M, Kosek E, Hallgren S, Antti H, Fahlstrom M, & Hellstrom F (2015). Systemic differences in serum metabolome: A cross sectional comparison of women with localised and widespread pain and controls. Scientific Reports, 5, 15925 10.1038/srep15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadrevi J, Ghafouri B, Sjors A, Antti H, Larsson B, Crenshaw AG, Gerdle B, & Hellstrom F (2013). Comparative metabolomics of muscle interstitium fluid in human trapezius myalgia: An in vivo microdialysis study. European Journal of Applied Physiology, 113, 2977–2989. 10.1007/s00421-013-2716-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes DF (2015). Biomarker validation and testing. Molecular Oncology, 9, 960–966. 10.1016/j.molonc.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Wilson ID, & Nicholson JK (2008). Metabolic phenotyping in health and disease. Cell, 134, 714–717. 10.1016/j.cell.2008.08.026 [DOI] [PubMed] [Google Scholar]
- Jang JH, Kim DW, Sang Nam T, Se Paik K, & Leem JW (2004). Peripheral glutamate receptors contribute to mechanical hyperalgesia in a neuropathic pain model of the rat. Neuroscience, 128, 169–176. 10.1016/j.neuroscience.2004.06.040 [DOI] [PubMed] [Google Scholar]
- Jeyarajah EJ, Cromwell WC, & Otvos JD (2006). Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clinics in Laboratory Medicine, 26, 847–870. 10.1016/j.cll.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Johnson CH, Ivanisevic J, & Siuzdak G (2016). Metabolomics: Beyond biomarkers and towards mechanisms. Nature Reviews Molecular Cell Biology, 17, 451–459. 10.1038/nrm.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, & Krishnan KR (2009). Metabolomics: A global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology, 34, 173–186. 10.1038/npp.2008.174 [DOI] [PubMed] [Google Scholar]
- Klyne DM, Moseley GL, Sterling M, Barbe MF, & Hodges PW (2019). Are signs of central sensitization in acute low back pain a precursor to poor outcome? The Journal of Pain, 20, 994–1009. 10.1016/j.jpain.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Leptak C, Menetski JP, Wagner JA, Aubrecht J, Brady L, Brumfield M, Chin WW, Hoffmann S, Kelloff G, Lavezzari G, Ranganathan R, Sauer J-M, Sistare FD, Zabka T, & Wholley D (2017). What evidence do we need for biomarker qualification? Science Translational Medicine, 9 10.1126/scitranslmed.aal4599 [DOI] [PubMed] [Google Scholar]
- Liggi S, & Griffin JL (2017). Metabolomics applied to diabetes-lessons from human population studies. Int J Biochem Cell Biol, 93, 136–147. 10.1016/j.biocel.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Livshits G, Macgregor AJ, Gieger C, Malkin I, Moayyeri A, Grallert H, Emeny RT, Spector T, Kastenmüller G, & Williams FMK (2015). An omics investigation into chronic widespread musculoskeletal pain reveals epiandrosterone sulfate as a potential biomarker. Pain, 156, 1845–1851. 10.1097/j.pain.0000000000000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits G, Malkin I, Bowyer RCE, Verdi S, Bell JT, Menni C, Williams FMK, & Steves CJ (2018). Multi-omics analyses of frailty and chronic widespread musculoskeletal pain suggest involvement of shared neurological pathways. Pain, 159, 2565–2572. 10.1097/j.pain.0000000000001364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lopez A, Lopez-Gonzalvez A, Barker-Tejeda TC, & Barbas C (2018). A review of validated biomarkers obtained through metabolomics. Expert Review of Molecular Diagnostics, 18, 557–575. 10.1080/14737159.2018.1481391 [DOI] [PubMed] [Google Scholar]
- Malatji BG, Mason S, Mienie LJ, Wevers RA, Meyer H, van Reenen M, & Reinecke CJ (2019). The gc-ms metabolomics signature in patients with fibromyalgia syndrome directs to dysbiosis as an aspect contributing factor of fms pathophysiology. Metabolomics, 15, 54 10.1007/s11306-019-1513-6 [DOI] [PubMed] [Google Scholar]
- Malatji BG, Meyer H, Mason S, Engelke UFH, Wevers RA, van Reenen M, & Reinecke CJ (2017). A diagnostic biomarker profile for fibromyalgia syndrome based on an nmr metabolomics study of selected patients and controls. BMC Neurology, 17, 88 10.1186/s12883-017-0863-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantyselka P, Ali-Sisto T, Kautiainen H, Niskanen L, Viinamaki H, Velagapudi V, & Lehto SM (2017). The association between musculoskeletal pain and circulating ornithine: A population-based study. Pain Medicine, 18, 1145–1151. 10.1093/pm/pnw285 [DOI] [PubMed] [Google Scholar]
- Mickiewicz B, Kelly JJ, Ludwig TE, Weljie AM, Wiley JP, Schmidt TA, & Vogel HJ (2015). Metabolic analysis of knee synovial fluid as a potential diagnostic approach for osteoarthritis. Journal of Orthopedic Research, 33, 1631–1638. 10.1002/jor.22949 [DOI] [PubMed] [Google Scholar]
- Muller M, Curatolo M, Limacher A, Neziri AY, Treichel F, Battaglia M, Arendt-Nielsen L, & Juni P (2019). Predicting transition from acute to chronic low back pain with quantitative sensory tests-a prospective cohort study in the primary care setting. European Journal of Pain, 23, 894–907. 10.1002/ejp.1356 [DOI] [PubMed] [Google Scholar]
- Nagana Gowda GA, Gowda YN, & Raftery D (2015). Expanding the limits of human blood metabolite quantitation using nmr spectroscopy. Analytical Chemistry, 87, 706–715. 10.1021/ac503651e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda GA, & Raftery D (2013). Biomarker discovery and translation in metabolomics. Current Metabolomics, 1, 227–240. 10.2174/2213235X113019990005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda GA, & Raftery D Nmr based metabolomics-methods and protocols In: Walker J, editor. Methods in molecular biology. New York: Springer; 2019. p. [Google Scholar]
- Newgard CB (2017). Metabolomics and metabolic diseases: Where do we stand? Cell Metabolism, 25, 43–56. 10.1016/j.cmet.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS Jr., Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, & Svetkey LP (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism, 9, 311–326. 10.1016/j.cmet.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, O’Flynn MP, Sadler PJ, Macleod AF, Juul SM, & Sonksen PH (1984). Proton-nuclear-magnetic-resonance studies of serum, plasma and urine from fasting normal and diabetic subjects. The Biochemical Journal, 217, 365–375. 10.1042/bj2170365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos JD, Jeyarajah EJ, & Bennett DW (1991). Quantification of plasma lipoproteins by proton nuclear magnetic resonance spectroscopy. Clinical Chemistry, 37, 377–386. https://www.ncbi.nlm.nih.gov/pubmed/2004444 [PubMed] [Google Scholar]
- Pacchiarotta T, Deelder AM, & Mayboroda OA (2012). Metabolomic investigations of human infections. Bioanalysis, 4, 919–925. 10.4155/bio.12.61 [DOI] [PubMed] [Google Scholar]
- Parker KS, Crowley JR, Stephens-Shields AJ, van Bokhoven A, Lucia MS, Lai HH, Andriole GL, Hooton TM, Mullins C, & Henderson JP (2016). Urinary metabolomics identifies a molecular correlate of interstitial cystitis/bladder pain syndrome in a multidisciplinary approach to the study of chronic pelvic pain (mapp) research network cohort. EBioMedicine, 7, 167–174. 10.1016/j.ebiom.2016.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, & Siuzdak G (2012). Innovation: Metabolomics: The apogee of the omics trilogy. Nature Reviews Molecular Cell Biology, 13, 263–269. 10.1038/nrm3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinu FR, Beale DJ, Paten AM, Kouremenos K, Swarup S, Schirra HJ, & Wishart D (2019). Systems biology and multi-omics integration: Viewpoints from the metabolomics research community. Metabolites, 9 10.3390/metabo9040076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici-Muller J, Lemieux RM, Artin E, Saunders JO, Salituro FG, Travins J, Cianchetta G, Cai Z, Zhou D, Cui D, Chen P, Straley K, Tobin E, Wang F, David MD, Penard-Lacronique V, Quivoron C, Saada V, de Botton S, Gross S, Dang L, Yang H, Utley L, Chen Y, Kim H, Jin S, Gu Z, Yao G, Luo Z, Lv X, Fang C, Yan L, Olaharski A, Silverman L, Biller S, Su S-SM, & Yen K (2018). Discovery of ag-120 (ivosidenib): A first-in-class mutant idh1 inhibitor for the treatment of idh1 mutant cancers. ACS Medicinal Chemistry Letters, 9, 300–305. 10.1021/acsmedchemlett.7b00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery D Mass spectrometry in metabolomics-methods and protocols In: Walker J, editor. Methods in molecular biology. New York: Springer; 2014. p. [Google Scholar]
- Robb MA, McInnes PM, & Califf RM (2016). Biomarkers and surrogate endpoints: Developing common terminology and definitions. JAMA, 315, 1107–1108. 10.1001/jama.2016.2240 [DOI] [PubMed] [Google Scholar]
- Ruiz-Canela M, Hruby A, Clish CB, Liang L, Martinez-Gonzalez MA, & Hu FB (2017). Comprehensive metabolomic profiling and incident cardiovascular disease: A systematic review. Journal of Americam Heart Association, 6 10.1161/JAHA.117.005705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki S, Hatano T, Fujimaki M, Ishikawa KI, Mori A, Oji Y, Okuzumi A, Fukuhara T, Koinuma T, Imamichi Y, Nagumo M, Furuya N, Nojiri S, Amo T, Yamashiro K, & Hattori N (2017). Decreased long-chain acylcarnitines from insufficient beta-oxidation as potential early diagnostic markers for parkinson’s disease. Scientific Reports, 7, 7328 10.1038/s41598-017-06767-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandlers Y (2017). The future perspective: Metabolomics in laboratory medicine for inborn errors of metabolism. Translational Research, 189, 65–75. 10.1016/j.trsl.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Sawynok J (2016). Adenosine receptor targets for pain. Neuroscience, 338, 1–18. 10.1016/j.neuroscience.2015.10.031 [DOI] [PubMed] [Google Scholar]
- Schliessbach J, Siegenthaler A, Butikofer L, Vuilleumier P, Juni P, Stamer U, Arendt-Nielsen L, & Curatolo M (2018). Predicting drug efficacy in chronic low back pain by quantitative sensory tests. European Journal of Pain, 22, 973–988. 10.1002/ejp.1183 [DOI] [PubMed] [Google Scholar]
- Shah SH, Sun JL, Stevens RD, Bain JR, Muehlbauer MJ, Pieper KS, Haynes C, Hauser ER, Kraus WE, Granger CB, Newgard CB, Califf RM, & Newby LK (2012). Baseline metabolomic profiles predict cardiovascular events in patients at risk for coronary artery disease. American Heart Journal, 163, 844–850 e841. 10.1016/j.ahj.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Steck SE, & Murphy EA (2020). Dietary patterns and cancer risk. Nature Reviews Cancer, 20, 125–138. 10.1038/s41568-019-0227-4 [DOI] [PubMed] [Google Scholar]
- Thu JHL, Vetter J, & Lai HH (2019). The severity and distribution of nonurologic pain and urogenital pain in overactive bladder are intermediate between interstitial cystitis and controls. Urology, 130, 59–64. 10.1016/j.urology.2019.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trbojević-Akmačić I, Vučković F, Vilaj M, Skelin A, Karssen LC, Krištić J, Jurić J, Momčilović A, Šimunović J, Mangino M, De Gregori M, Marchesini M, Dagostino C, Štambuk J, Novokmet M, Rauck R, Aulchenko YS, Primorac D, Kapural L, Buyse K, Mesotten D, Williams FMK, van Zundert J, Allegri M, & Lauc G (2018). Plasma n-glycome composition associates with chronic low back pain. Biochimica et Biophysica Acta (BBA) - General Subjects, 1862, 2124–2133. 10.1016/j.bbagen.2018.07.003 [DOI] [PubMed] [Google Scholar]
- Treede RD, Rief W, Barke A, Aziz Q, Bennett MI, Benoliel R, Cohen M, Evers S, Finnerup NB, First MB, Giamberardino MA, Kaasa S, Korwisi B, Kosek E, Lavand’homme P, Nicholas M, Perrot S, Scholz J, Schug S, Smith BH, Svensson P, Vlaeyen JWS, & Wang SJ (2019). Chronic pain as a symptom or a disease: The iasp classification of chronic pain for the international classification of diseases (icd-11). Pain, 160, 19–27. 10.1097/j.pain.0000000000001384 [DOI] [PubMed] [Google Scholar]
- Trivedi DK, Hollywood KA, & Goodacre R (2017). Metabolomics for the masses: The future of metabolomics in a personalized world. New Horizons in Translational Medicine, 3, 294–305. 10.1016/j.nhtm.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussher JR, Elmariah S, Gerszten RE, & Dyck JR (2016). The emerging role of metabolomics in the diagnosis and prognosis of cardiovascular disease. Journal of the American College of Cardiology, 68, 2850–2870. 10.1016/j.jacc.2016.09.972 [DOI] [PubMed] [Google Scholar]
- Viant MR, Ebbels TMD, Beger RD, Ekman DR, Epps DJT, Kamp H, Leonards PEG, Loizou GD, MacRae JI, van Ravenzwaay B, Rocca-Serra P, Salek RM, Walk T, & Weber RJM (2019). Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nature Communications, 10, 3041 10.1038/s41467-019-10900-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wei R, Xie G, Arnold M, Kueider-Paisley A, Louie G, Mahmoudian Dehkordi S, Blach C, Baillie R, Han X, De Jager PL, Bennett DA, Kaddurah-Daouk R, & Jia W (2020). Peripheral serum metabolomic profiles inform central cognitive impairment. Scientific Reports, 10, 14059 10.1038/s41598-020-70703-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, & Gerszten RE (2011). Metabolite profiles and the risk of developing diabetes. Nature Medicine, 17, 448–453. 10.1038/nm.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS (2016a). Emerging applications of metabolomics in drug discovery and precision medicine. Nature Reviews Drug Discovery, 15, 473 10.1038/nrd.2016.32 [DOI] [PubMed] [Google Scholar]
- Wishart DS (2016b). Emerging applications of metabolomics in drug discovery and precision medicine. Nature Reviews Drug Discovery, 15, 473–484. 10.1038/nrd.2016.32 [DOI] [PubMed] [Google Scholar]
- Woolf CJ, & Thompson SWN (1991). The induction and maintenance of central sensitation is dependent on n-methyl-d-aspartic acid receptor activation: Implications for the treatment of post-injury pain hypersensitivity states. Pain, 44, 293–299. [DOI] [PubMed] [Google Scholar]
- Zhang W, Sun G, Likhodii S, Liu M, Aref-Eshghi E, Harper PE, Martin G, Furey A, Green R, Randell E, Rahman P, & Zhai G (2016). Metabolomic analysis of human plasma reveals that arginine is depleted in knee osteoarthritis patients. Osteoarthritis Cartilage, 24, 827–834. 10.1016/j.joca.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Zurfluh S, Baumgartner T, Meier MA, Ottiger M, Voegeli A, Bernasconi L, Neyer P, Mueller B, & Schuetz P (2018). The role of metabolomic markers for patients with infectious diseases: Implications for risk stratification and therapeutic modulation. Expert Review of Anti-infective Therapy, 16, 133–142. 10.1080/14787210.2018.1426460 [DOI] [PubMed] [Google Scholar]