Abstract
Vanderbilt University Medical Center implemented pharmacogenomics testing with the Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT) initiative in 2010. This tutorial reviews the laboratory considerations, technical infrastructure and programmatic support required to deliver panel-based PGx testing across a large health system with examples and experiences from the first decade of the PREDICT initiative. From the time of inception, automated clinical decision support (CDS) has been a critical capability for delivering pharmacogenomic (PGx) results to the point-of-care. Key features of the CDS include human-readable interpretations and clinical guidance that is anticipatory, actionable, and adaptable to changes in the scientific literature. Implementing CDS requires that structured results from the laboratory be encoded in standards-based messages that are securely ingested by electronic health records (EHRs). Translating results to guidance also requires an informatics infrastructure with multiple components: (1) to manage the interpretation of raw genomic data to “star allele” results to expected phenotype, (2) to define the rules which associate a phenotype with recommended changes to clinical care, and (3) to manage and update the knowledge base. Knowledge base management is key to processing new results with the latest guidelines, and to ensure that historical genomic results can be reinterpreted with revised CDS. We recommend that these components be deployed with institutional authorization, programmatic support, and clinician education to govern the CDS content and policies around delivery.
Keywords: pharmacogenomics, pharmacogenetics, clinical decision support, personalized medicine, precision medicine
INTRODUCTION
One critical goal of pharmacogenomics (PGx) is to use a patient’s genetic information to optimize and tailor drug therapy to improve treatment outcomes.1 PGx implementation has important potential clinical and economic benefits, such as preventing adverse events, improving drug efficacy and reducing unnecessary treatment trials.2,3 Moreover, PGx is an accessible and mature approach to implementing precision medicine and thus can serve as a model for integrating precision medicine into routine clinical practice.4 The Clinical Pharmacogenetics Implementation Consortium (CPIC) has created guidelines to facilitate the use of PGx results to tailor a wide range of therapies, including analgesics, antidepressants and common cardiovascular agents, indicating the applicability of PGx in the general patient population.5,6
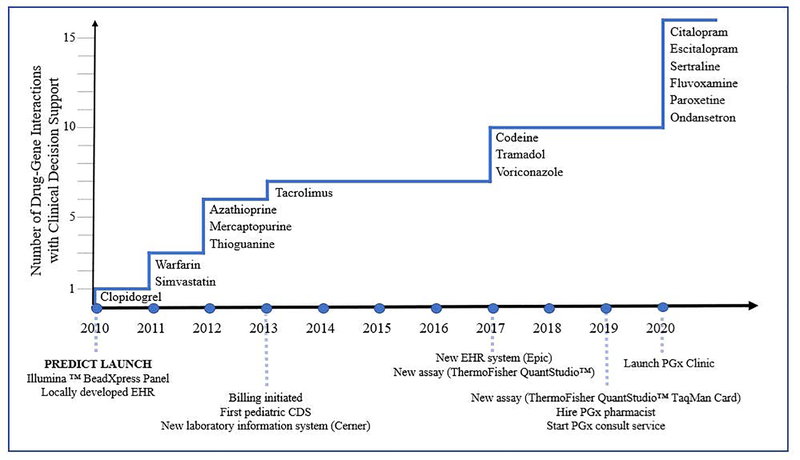
Given the improvements in genomic technology and decreased costs of testing, the main requirement for PGx implementation across health systems is an efficient method to bring PGx test results and recommendations to the point-of-care.7 Several NIH funded initiatives, including the Electronic Medical Records and Genomics (eMERGE) and Implementing GeNomics In practice (IGNITE) networks, have encouraged early adoption of PGx among at least 27 institutions, including Vanderbilt University Medical Center (VUMC), and affiliates in the United States.8 The Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT) program, one of the first preemptive pharmacogenomic initiatives, began in 2010, starting with clinical decision support (CDS) for one drug-gene interaction (DGI) (clopidogrel - CYP2C19) and evolving to coverage for 16 DGIs today (Figure 1).9,10 After an initial period of offering institutionally-supported, preemptive pharmacogenomic testing, the PREDICT program transitioned to a billing model including reimbursement by insurance when available. Since this transition, providers order testing concurrent with a prescription with pharmacogenomic relevance. As a panel-based test, the additional genes are reported and available as preemptive results for future prescriptions. In addition to increasing the number of DGIs, the PREDICT program has incorporated updates in PGx recommendations, implemented a new laboratory platform, and transitioned all results and CDS functionality to a new electronic health record (EHR) system. VUMC is an academic medical center that serves a range of specialty and primary care as well as adults and pediatric patients. We present here our decade-long experience and knowledge gained from programmatic changes in the hopes that the program can inform clinical PGx implementation at other sites.
Figure 1.
Timeline of VUMC PREDICT initiative illustrating growth of coverage for drugs with CDS (top) as well as major transitions and milestones in the program (bottom). EHR – electronic health record; CDS – clinical decision support; PGx – Pharmacogenomics.
Surveys of practicing clinicians and their patients indicate a high level of interest in implementing PGx testing.11–13,14 However, clinicians have also described specific barriers that need to be addressed to support improved workflow and actionability15. One such critical barrier is the lack of provider familiarity with standard PGx result reporting including the star allele nomenclature, phenotype interpretations, along with the associated CPIC or FDA supporting information. Recent FDA actions discourage PGx testing laboratories from providing prescribing recommendations, which may exacerbate prescriber frustrations.16 In addition, with the use of multi-gene panel testing and the longitudinal applicability of PGx results, many test results gathered may not be immediately actionable at the time of testing. Each patient’s drug list changes over time, and so clinical interpretation becomes most useful when disseminated at point-of-care.8 “End-to-end” CDS, defined below, is one way to address these critical implementation barriers.17, 18 Effective use of PGx results, facilitated by end-to-end CDS, has the potential to maximize the clinical and economic benefits from PGx testing.
For the purposes of this manuscript, end-to-end CDS refers to a health information pipeline that translates the results of a raw PGx panel assay into a set of phenotype interpretations and guidance on specific medication or dosage selection. This pipeline helps automate the delivery of PGx results to the point-of-care, at the time providers are actively making prescribing decisions within the EHR. The objective of the pipeline is to manage PGx results and interpretations across the tested population.19 Therefore, decision support oriented toward ordering PGx testing, or CDS that does not incorporate a patient’s PGx test result is out of scope of this tutorial, although these features may be valuable components of clinical PGx implementation. Throughout the tutorial, PGx results (i.e. genotypes) are referred to as results while PGx phenotypes (e.g. metabolizer status) are referred to as interpretations. Guidance suggesting an alternative drug, dose adjustment, or consultation is referred to as recommendations.
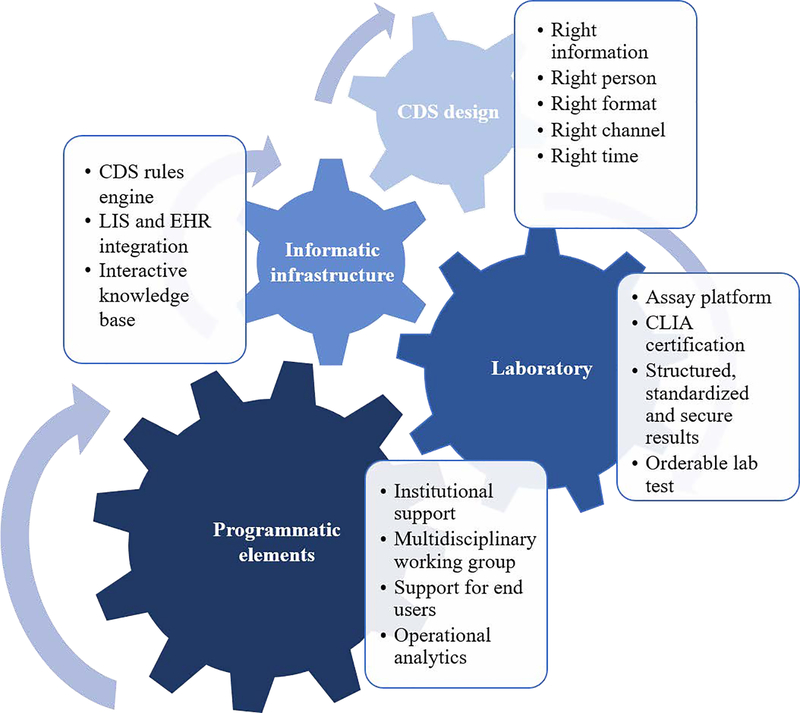
Automated CDS is paramount for routine clinical use of PGx results by transforming static results into interpretations that are anticipatory, actionable, and adaptable, and does so immediately upon the test result becoming available. The CDS implementation is built into workflows to anticipate the information clinicians need at point-of-care. It distills complex PGx results into actionable recommendations for clinicians. It adapts by updating for the evolving body of knowledge and the discovery of new variants. This is consistent with the long term goal of precision medicine as well as the PREDICT initiative to develop the infrastructure for incorporating genomic results into the EHR and to make recommendations available to clinicians at point-of-care. 9,20 The aim of this tutorial is to demonstrate the framework for end-to-end CDS for institutions interested in clinical implementation of PGx in routine care. Examples and experiences from the PREDICT initiative will be used to illustrate approaches for end-to-end CDS. This tutorial will begin with laboratory considerations, followed by components of an informatics infrastructure to support knowledge management, integration of PGx recommendations at point-of-care, and end with programmatic components to support successful implementation (Figure 2).
Figure 2.
Key components for implementing end-to-end pharmacogenomic CDS. CDS – clinical decision support; LIS – laboratory information system; EHR – electronic health record; CLIA – Clinical Laboratory Improvement Amendments.
GENERATION OF LABORATORY RESULTS
PGx testing is most often performed within a high complexity molecular genetics or molecular pathology laboratory. US clinical laboratories that support patient care are regulated by the Centers for Medicare and Medicaid Services through the Clinical Laboratory Improvement Amendments (CLIA).21,22 Laboratories can also pursue additional accreditation and become accredited by professional societies such as The College of American Pathologists (CAP).23 Certification by these groups implies strict guidelines to ensure analytical accuracy, precision, sensitivity and specificity of the result for laboratory developed tests. It also serves to minimize laboratory errors and ensures procedures for correct and standardized interpretation and reporting of results. These guidelines encompass all aspects of laboratory testing such as laboratory safety, leadership and testing personnel, test validation and precision, and reporting of patient results.22
Testing Platform Considerations
Choosing a testing platform is a crucial decision affecting multiple aspects of a PGx program and if CDS can be implemented. Commercially available send out tests typically do not facilitate end-to-end automated CDS due to barriers including turnaround time (TAT), need for storage of discrete, standardized results, and demand for flexibility over time. The TAT is an important consideration to maximize the impact for patient management. In house testing has advantages over send out tests and is performed by VUMC. The laboratory instrumentation or testing platform chosen for PGx analysis will be dependent upon several variables and should be based on the specific needs of the institution and their program. This requires careful forethought with clear goals and objectives for the program to choose the correct instrumentation and understand the required resources needed to launch clinical testing. One of the most important considerations in this process is the anticipated number of patients. For sample throughput greater than 50 per day, the QuantStudio™ 12K Flex Real-Time OpenArray Polymerase Chain Reaction (PCR) platform may be considered.24 This system is robust, accurate and uses TaqMan® chemistry.24 For lower throughput, a variety of other TaqMan® based platforms are available from single tube to medium-throughput assays. The number of variants to be included on the panel also influences choice of platform. Both immediate and long term needs should be considered, recognizing the potential for scalability and flexibility of the assay to grow and evolve as both the demand for testing and or the number of clinically significant genes and variants increases.
Next Generation Sequencing (NGS) platforms utilizing PGx gene panels enable the interrogation of the widest array of clinically useful gene variants with rare and less characterized gene variants also identified. These additional findings offer the potential for clinical and basic research. However, like other platforms there are some disadvantages to this technology. NGS assays have extended TATs, increased labor and reagent expenses, and lower sample throughput which may make it a less viable option for some programs that have limited resources and require quick TAT. In contrast to NGS testing, some platforms offer rapid analysis, (e.g. <4 hours), for a limited number of genes and clinically significant variants (e.g. XT-8 genotype assay for CYP2C19 or CYP2C9 and VKORC1).25 The variants tested for such platforms should be examined to see if appropriate for the institution’s specific population as the exclusion of variants common in a target population (e.g. one of non-European ancestry) may result in misclassification of patients and inappropriate CDS guidance.
Clinical Validity and Quality Control
Considering all the previously mentioned parameters, clinical validity and quality control of the assay are imperative for use in patient care. An example using the VUMC Molecular Diagnostics Laboratory will demonstrate this process. The laboratory began with an assay that included a large number of non-clinically actionable genes and variants which was not incorporated into CDS and transitioned into one where all selected genes and variants can be used clinically. The laboratory performs a Laboratory Developed Test (LDT) utilizing the TaqMan® Array Cards run on the QuantStudio™ 7 Flex Real-Time PCR instrument. To increase efficiency, a microplate mover performs automated loading of prepared cards into the instrument.24 LDTs (sometimes called Laboratory Developed Procedures) require extensive validation prior to clinical testing of patient samples in accordance with CLIA and CAP certification.22,26 The VUMC assay card is configured for the simultaneous detection of 45 unique variants associated with 10 genes per patient sample (Table S1). Of note, a separate Copy Number TaqMan® assay for CYP2D6 is performed for each patient using a 96 well plate format.
In addition to the 45 pharmacovariants, the multi-gene assay includes two internal controls, CYP2C19*2 and CYP3A5*3, run in duplicate but at distant locations within the card to serve as quality control matrices. Further, a gender specific assay is used to discriminate between the presence of X and Y chromosome specific DNA sequences (Amelogenin) to verify the expected sex of each patient.27 TaqMan® Array cards are purchased with PCR primers and associated VIC and FAM dye-labeled probes corresponding to each gene variant pre-spotted at locations 1 through 48 within the array card.24 The configuration of the card is determined by the laboratory and uses predesigned PCR primers and probes available from the manufacturer. The assay is performed using patient DNA extracted from a peripheral blood specimen at a concentration of 15 ng/μL according to instructions provided by the manufacturer.28 Special attention must be given for some patient populations. For allogeneic stem cell transplant (SCT) patients, it is important to ensure that the blood sample was collected prior to transplant, thus ensuring that the sample represents the recipient’s DNA rather than that of the donor. Similarly, if the patient has undergone liver transplant, the PGx test results obtained from a blood sample may not be relevant for hepatic metabolism. For SCT and diagnoses associated with leukopenia (e.g. oncology patients during some treatment phases) a sufficient white blood cell count is required to enable recovery of an adequate amount of DNA for testing. A patient’s diplotype at a particular locus is determined by reviewing the collective fluorescence emission of the VIC and FAM probe values measured by the instrument for all variants interrogated for a specific gene. Although all 45 gene variants are validated to be used in the clinical setting, at this time not all have CDS. The results that do not yet have CDS developed and approved by the institution are masked prior to testing and not viewed by testing personnel or released to providers. Whether to mask results when there is no CDS is also an important consideration to contemplate for PGx programs.
Pharmacogenomic Test Results Generated
Results should aim to be structured, standardized, and secure. Discrete and structured results make CDS and other manipulations of the results feasible. Unstructured results, such as pdf or free text entry of results, have limited ability to be transformed into CDS without ongoing manual effort. At VUMC, the genotype and the CYP2D6 copy number results are entered by laboratory staff as discrete results within the laboratory report in the laboratory information system (LIS), Cerner Millennium Helix® module.28 Standardization of PGx nomenclature is particularly relevant as changing laboratory testing platforms will often cause differences in the format of the results. The next section on informatics infrastructure will provide guidance on the multistep process of transforming laboratory results to CDS.
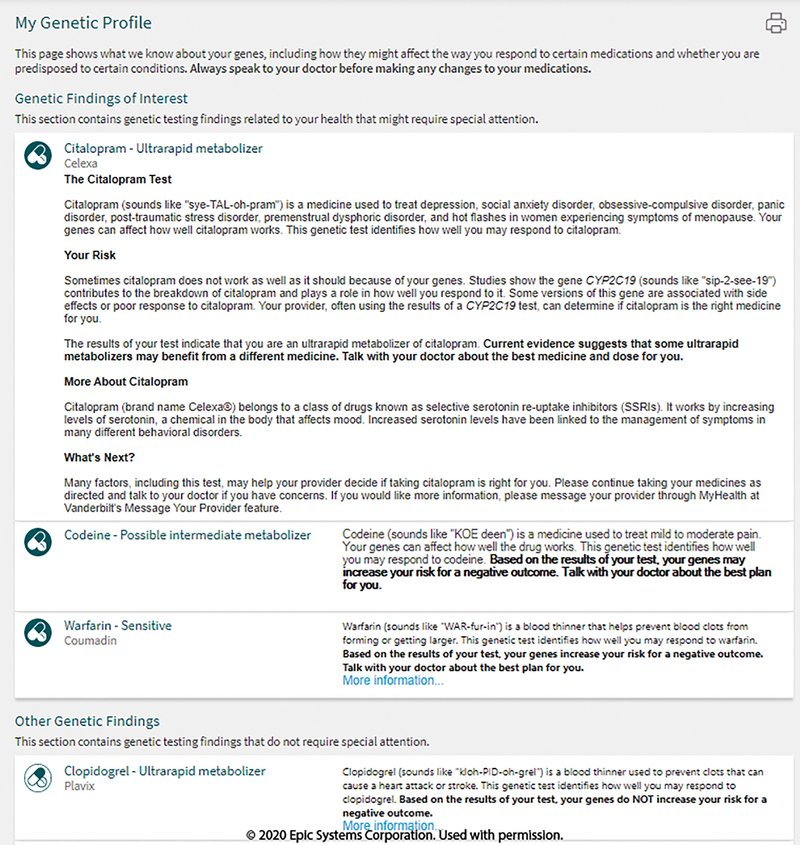
If the PGx program seeks to provide enduring CDS for patients as care is continued in the health system, standard nomenclature such as that proposed by CPIC assist with longitudinal CDS despite technical platform changes.29 Lastly, enduring CDS will require that the patients’ results be stored in a secure database such that patient confidentiality is protected. These results are accessible to clinicians through the patient’s EHR (Figure S1, S2). Care is also taken to ensure that patients themselves also have access to the results. At VUMC patient friendly versions of the PGx results and interpretations are available through the patient-facing portal, MyHealthAtVanderbilt (MHAV) (Figure 3). Collectively, these diverse presentations of the patients’ PGx results illustrate the need for backward-compatible data structures and nomenclature.
Figure 3.
Patient-facing pharmacogenomic results. My Health at Vanderbilt (MHAV) is a patient portal that providers result and interpretation information using patient friendly language. This is automatically populated when pharmacogenomic results are in the electronic health record. Image are copyright of Epic Systems Corporation and used with permission.
INFORMATICS INFRASTRUCTURE AND KNOWLEDGE MANAGEMENT
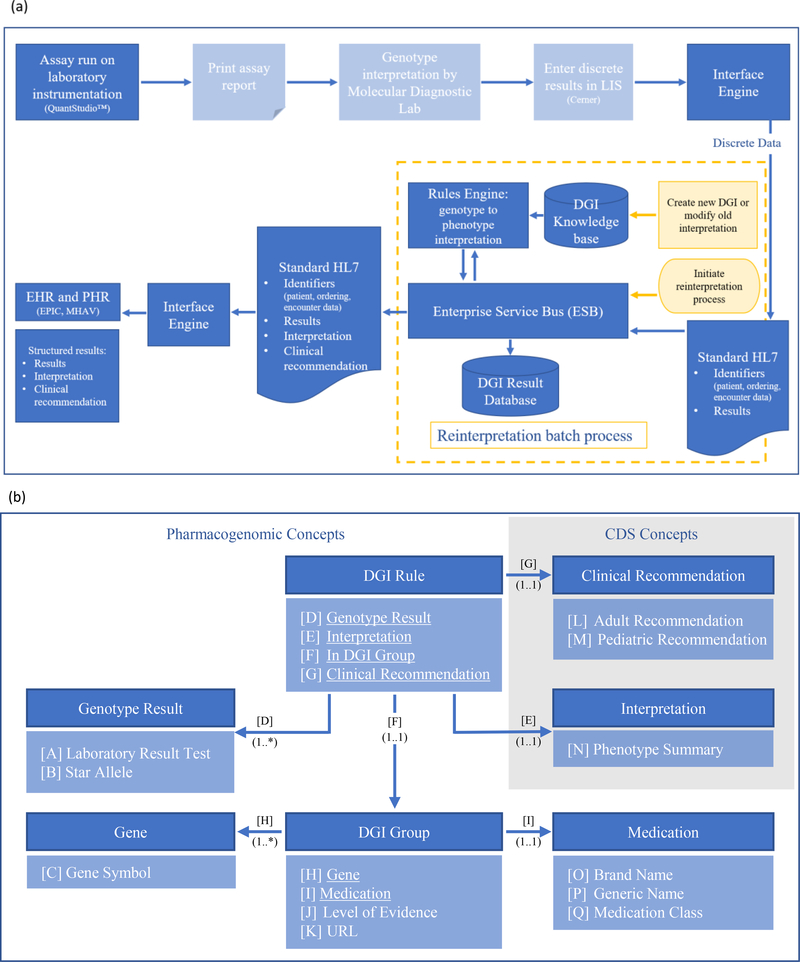
An integrated informatics infrastructure is essential for delivering end-to-end provider and patient-facing PGx based CDS. The PREDICT informatics infrastructure uses two different processes, one is for newly tested patients and another for patients with existing results. Figure 4a depicts the components of the infrastructure. The specific components mentioned here are for illustrative purposes and institution as well as EHR specific considerations should be made.
Figure 4.
Knowledge management for pharmacogenomics data.
(a) An overview of the PREDICT informatics infrastructure. Genotype resulting and phenotype interpretation of new patients is shown in the blue boxes, with light shading indicating manual steps and dark shading indicating automated steps. Yellow boxes indicate steps for phenotype reinterpretation for patients with existing results. The DGI knowledge base stores knowledge content required for interpretation and clinical recommendations. The DGI result database stores longitudinal patient specific results and interpretation which allows for real-time data reprocessing. LIS – laboratory information system; DGI – drug-gene interaction; HL7 – Health Level Seven standards; EHR – electronic health record; PHR – patient health record; MHAV – My Health at Vanderbilt.
(b) Domain Model of the PREDICT knowledge base high-level concepts represented as a unified modeling language class diagram. This class diagram illustrates the concepts and relationships of the PREDICT domain. Concepts of the domain are depicted by dark blue shading with attributes and relationships (underlined) listed below the name of each concept in light blue shading. Associations across the elements are represented by labeled arrows. DGI Group concept represents the drug-gene associations, while DGI Rule links this concept to genotype results, interpretations and associated clinical recommendations. Once stored in a particular patient’s EHR, interpretations fire the CDS upon ordering the medication represented within the DGI Group concept, while associated patient and provider facing clinical recommendations provide the appropriate guidance. DGI – drug-gene interaction; CDS – clinical decision support; URL – uniform resource locator.
For new results, the Molecular Diagnostics Lab processes and verifies a result for each specimen (Figure 4a). Genomic results are subsequently interfaced with the EHR system to store discrete results, allowing for clinician retrieval and consumption by the CDS logic. At VUMC the discrete variant results are stored in Epic as lab result records. The associated interpretations and clinical recommendations are made available to clinicians and patients via genomic indicators, an Epic module (Figure S2). Before storage in the EHR, the discrete results are processed by an integration architecture that is comprised of the DGI knowledge base and the corresponding rules engine referencing the knowledge base on a patient by patient basis. Both components are integrated with rest of the infrastructure via an enterprise service bus (ESB), which facilitates the real-time data exchange. The DGI knowledge base is the “source of truth” for genotype to phenotype interpretations. The knowledge base is built with a drug-centric structure, enabling association of specific genetic results to specific CDS for each drug. At the time of knowledge base conception, enzyme activity scores were not included, but will be incorporated into future versions to support the diversity of potential recommendations. Use of an activity scoring system aids in nuanced translation of genotype results into interpretations (e.g. CYP2D6). 30
The knowledge base is curated and maintained by VUMC’s PGx experts based on CPIC guidelines and other sources such as the dbSNP and Pharmacogene Variation Consortium (Pharmvar.org) database. By referencing the DGI knowledge base, the rules engine provides an interpretation for each patient specific genotype result for each drug. Both the discrete results and corresponding interpretations are subsequently relayed to the EHR system and stored as a laboratory report and interpretation report (Figure S1). Appropriate configuration of the EHR system and storage of these results triggers the creation of the associated genomic indicators serving as a mechanism to identify the type of recommendation based on the phenotype delivered by the CDS.
Although genetic test results should not change over time, the interpretations and/or recommendations associated with those results may change as knowledge evolves. A mechanism must be in place to update old interpretations with new CDS based on current evidence (reprocessing). The reprocessing infrastructure uses the same architectural components to enable the interpretation process to run retrospectively on all patients with stored discrete PGx results in the DGI results database (Figure 4a). Reinterpretation of existing results is based on changes in clinical evidence published by CPIC and will be discussed in more detail in the Updates and Reinterpretation section below. When the knowledge base is updated the rules engine can reference it to run a batch process to update the interpretation results for all patients impacted by the changes. The updated interpretation results will be stored in the EHR system and may change the genomic indicators. This process allows for PGx based CDS to provide recommendations reflecting the latest evidence.
A critical consideration for designing a PGx based CDS system, such as PREDICT, is the design of the knowledge base driving these CDS. When applied correctly, knowledge representation and management principles provide transparency and analyzability of knowledge assets, which ultimately leads to effective and efficient management of the content driving the CDS. One of these methods is domain modeling, during which the appropriate representation of a particular clinical domain, in this case PGx, is explicitly captured. Formal knowledge representation provides means for a precise documentation of the important domain concepts, with appropriate levels of details, allowing for effective knowledge curation by subject matter experts while maintaining content integrity. Figure 4b illustrates the domain model of the PREDICT knowledge base.
UPDATES AND REINTERPRETATION
PGx implementation is a dynamic and evolving process. The beginning years of PREDICT implementation faced limited CPIC guidelines and curation of knowledge represented a major need and time investment. Since the launch of PREDICT, CPIC has published 24 different DGI guidelines. These guidelines are routinely evaluated and 18 different guideline updates have been published within the last decade (Table S2).31 Clinical implementation of PGx requires a robust and flexible infrastructure capable of adjusting to changes in the identification of genetic variants and interpretation of results to make evidence-based CDS recommendations. Modular and coordinated PGx program management components, where each component is optimized to accomplish a key feature in the process (e.g. resulting and reporting, evidence-based knowledge curation, CDS development and integration, etc.) facilitates a successful PGx program. This approach helps to streamline the workflows for programmatic changes and facilitate segmentation of workstreams required for deploying new content. We will briefly review two of our recent or ongoing programmatic updates and will discuss our strategy and experiences with supporting the growth of a clinical PGx program.
Updating with New Content
Adding new CDS to a PGx portfolio is a key strategy for enhancing the clinical impact of a program. New CDS provides support for new clinical areas and patient populations and thus should be calibrated for the institution’s clinical needs and patient demographics. The PREDICT program has expanded its testing portfolio to include new DGIs since inception. The program leadership considers three key factors when selecting the direction of this expansion: 1) institutional patient population; 2) clinical needs of end users; and 3) existing programmatic resources.
Implementing effective PGx requires a partnership among the patient, provider, and the PGx program. Evaluations of putative new CDS consider the overall prevalence of many key variants within the patient population served.32–34 Local clinicians provide the most accurate assessment of local clinical needs. The PREDICT program initiates conversations to identify clinical champions within our institution to determine enthusiasm and desire for PGx support. This activity helps us understand how our clinical partners prioritize their needs, and it establishes lasting relationships with clinical champions who are important partners for educational, implementation, and expansion efforts. Quantitative institutional assessments complement these conversations with clinicians; for example, prescribing data inform the potential number of patients who could be tested and potentially benefit from CDS alerts. Taking into account these factors, existing programmatic infrastructure is considered to ensure an efficient use of resources. For example, PREDICT launched with testing of CYP2C19 and CDS for clopidogrel; recent expansions in the program have leveraged this existing infrastructure by adding new CDS for citalopram, escitalopram, and sertraline (all CYP2C19 substrates). The modular build of the program required updates to the knowledge base, LIS system result template, and the EHR configurations, and reprocessing of patients with existing genotype results to generate new DGI indicators, with the molecular diagnostic assays already in place to support these additions. Therefore, judicious expansions and updates to PGx implementation require revaluation of the patient population, conversations with clinical partners as well as internal evaluations.
Expanding support within an existing patient population can be accomplished by incorporating additional CDS associated with a specific gene of interest. In the CYP2C19 example provided above, patients with legacy CYP2C19 results were reprocessed to enable CDS for three new drugs. Reprocessing required a coordinated reevaluation of the original laboratory results through our modular end-to-end CDS pipeline.
Reinterpretation of Results
Updating existing CDS content is required to maintain compliance with evolving CPIC guidelines and CLIA/CAP standards and maximize benefits to patients. Recent guidelines from CPIC have sought to standardize the method for interpreting CYP2D6 genotypes into phenotypes. These guidelines will impact future PGx CDS by 1) reassigning a new activity value for CYP2D6 *10 allele, and 2) realigning activity score-based phenotype classifications (Figure S3). The realignment of activity score classifications now accommodates an allele value of 0.25 and provides more nuance between CYP2D6 intermediate and normal metabolizers as well as CYP2D6 normal and ultrarapid metabolizers.29 As evidence for new interpretations and clinical recommendations for existing ones evolve, these changes get incorporated into the knowledge base. Updates to the knowledge base subsequently leads to changes in the interpretation report, genomic indicators and finally updates to the CDS.
Reinterpreting historical results will change existing CDS, and we anticipate that as PGx knowledge evolves, additional reinterpretations will become available. The PREDICT program has developed a complementary patient and provider approach to follow up on programmatic updates or reinterpretations. Communicating critical changes that occur after reprocessing are essential, especially if there is action to be taken in either adjusting or changing a patient’s medication. We have collaborated with our institutional Pharmacy and Therapeutics committee to establish templated communiques to be issued when necessary during reprocessing. These communication rubrics are specific for each DGI based on the safety profile and timeliness of the PGx information and will be delivered to providers if their patients are re-categorized from actionable to non-actionable, and vice versa. As updates are performed, our team receives reports of all patients with changes who have the relevant drug on their active medication list. Directed outreach to treating providers will then offer guidance, such as: “This patient was previously tested for genetic variants associated with drug metabolism using the PREDICT test. Based on new interpretations of the genotype, updated actionable recommendations pertaining to X gene and Y drug are available. If you have questions, please consult with the pharmacogenomics pharmacist now on staff. If you (re)order a drug with actionable pharmacogenomic information, you will be given a selection of alternatives via a best practice advisory message.” This active management helps ensure the safety of our patients while providing the treating healthcare teams with appropriate context and information surrounding the revised recommendation to guide their practice.
INTEGRATING PHARMACOGENOMIC RECOMMENDATIONS AT POINT-OF-CARE
Implementation of end-to-end CDS for PGx is a resource-heavy endeavor that requires both initial and ongoing investments in the laboratory and informatics infrastructure. To maximize return on those investments, results should inform clinical decision making longitudinally across diverse encounters with the health system. For example, for a 55-year-old man undergoing percutaneous coronary intervention, PGx testing can inform antiplatelet therapy after his procedure. Storage of his CYP2C19 results in the EHR allows triggering of CDS years later when his primary care provider prescribes citalopram for an episode of major depressive disorder. The primary care provider may not have viewed the PGx results in the patient’s chart from years past, but CDS provides the result, the interpretation, and therapeutic recommendations at point-of-care. Ideally, the patient is also reminded of the result by the provider and goes home knowing his medication therapy was personalized. This example scenario involves only results from one gene, although testing for a panel of genes is now common and showcases how PGx results contribute to recommendations for multiple drugs.
CDS Design
Careful and targeted CDS construction is essential for actively engaging providers. If the CDS does not reach the right person at the right place, the right time, and with the right content, the clinical utility of the test suffers. Practical issues to consider when designing PGx-driven CDS are summarized using the Agency for Healthcare Research and Quality’s “Five Rights” framework.35 In addition, cdskg.org is another source to consider for additional examples of PGx CDS.36
The right person: The CDS design process should start with identification of the audience or end users. End users of the CDS should be individuals affected by the content of the recommendation and who will potentially be prescribing the drug or reviewing drug orders (e.g. physicians, physician assistants, nurse practitioners, and pharmacists). Although patients can also be end users and benefit from CDS, our main focus has been on providers.
The right CDS intervention format: CDS alerts or best practice advisory (BPAs – these are specific to the Epic EHR) are the most common disruptive clinical interventional format. PGx-driven BPAs are triggered by an order or prescription for a target drug in the setting of an actionable phenotype. (Figures 5, 6, and 7) Care should be taken to reduce BPA frequency to fire when necessary to avoid alert fatigue. For example, if an end user has already acknowledged a BPA for a patient, a second BPA could be suppressed if the end user prescribes the drug again for the same patient in the future. BPAs that have recommendations regarding initiation doses of drugs should be minimized to firing only for patients not previously prescribed the drug. Of note, interruptive CDS alerts may not be appropriate for some PGx scenarios. For example, inhaled anesthetic agents are typically administered in the operating room and typically not preceded by an order in the EHR. In our EHR system, when malignant hyperthermia risk due to genetic variants in RYR1 or CACNA1S are documented in the patient’s problem list, a series of flags and warnings in the anesthesia care systems are enabled. In addition, all PGx interpretations should be accessible within the patient’s EHR for end users to use prior to the prescribing process (Figure S1, S2).
-
The right information: The most relevant results, interpretations, and additional clinical data considered for the prescribing decision should be displayed using the easiest representation for all elements. Recommendations should be crafted with input from institutional pharmacists and clinicians to provide recommendations tailored to local clinical practice and drug formulary. Ideally, the end user should not have to exit out of the CDS alert to find relevant clinical information or to enact the recommendation. Thus, alerts can be built to include options to order alternative drugs, remove a non-recommended drug, order complementary laboratory tests (e.g. creatine kinase for simvastatin), link to additional resource (e.g. Mydruggenome.org, CPIC, drug database) and order for additional support (e.g. PGx consult).
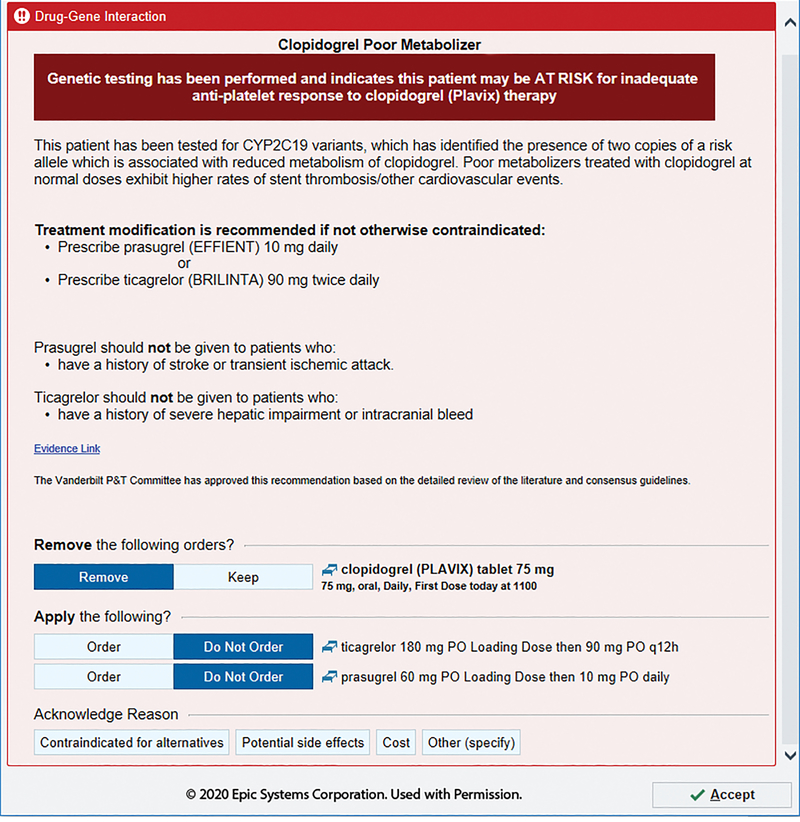
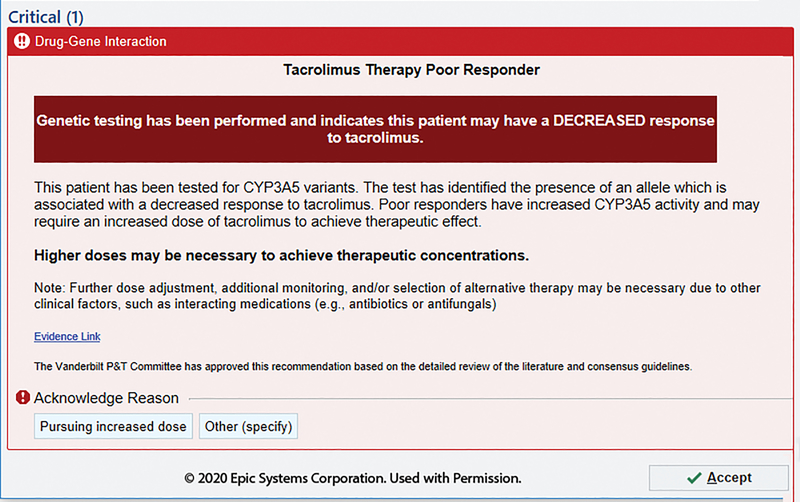
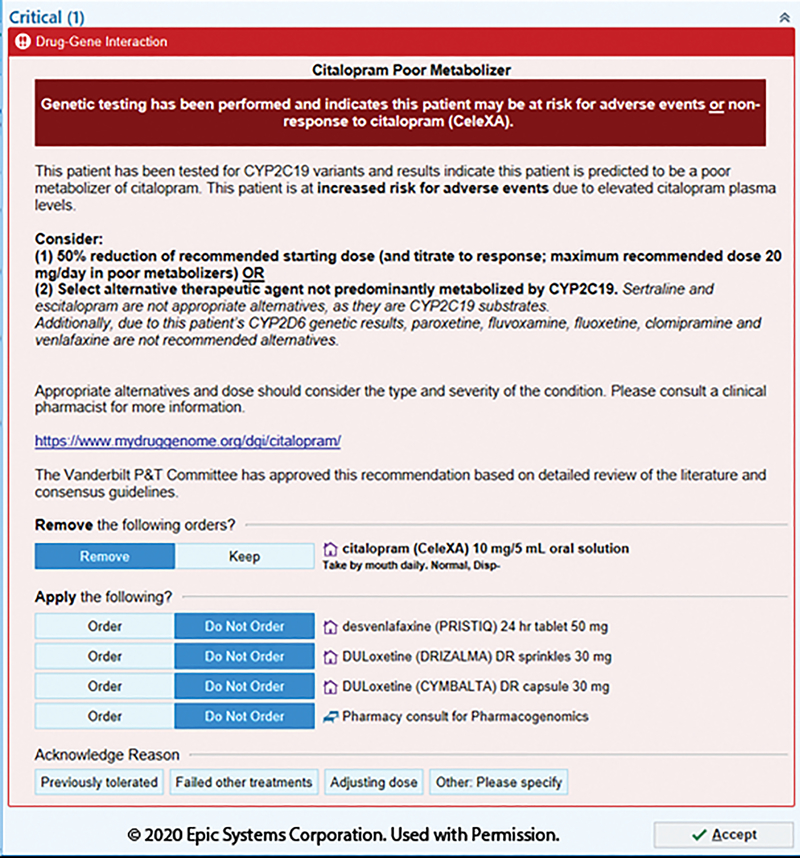
Different DGIs require different content; a CYP2C19-driven clopidogrel BPA may include the contraindications for the alternative drugs (Figure 5), whereas a CYP3A5-driven tacrolimus BPA includes a recommendation on altering dose (Figure 6). Indications of drugs are difficult to incorporate into CDS and can be considered within the clinical recommendations. For example, tacrolimus may be used for different indications and not all of which have evidence to support specific dose changes (Figure 6). Language must be carefully chosen, again in consultation with the end users. For example, stating CYP3A5 “normal metabolizers” require an increase dose of tacrolimus may cause more confusion than using “extensive metabolizer” (old terminology) or even a nonstandard terminology such as “poor responder” (Figure 6). The intermediate metabolizer status is often a non-intuitive term and is an example of why adding in an associated risk helps provide perspective. In addition, when recommending alternative drugs, the relevant genes for alternative drug should be accounted for within the CDS. For example, a concomitant CYP2C19 poor metabolizer and CYP2D6 ultrarapid metabolizer would receive recommendations with a limited list of alternative antidepressants because both genetic results are taken into consideration within the CDS (Figure 7).
The right channel: Having the CDS and BPA firing within the EHR provides an effective method to communicate recommendations, as this is coupled to electronic prescribing. In addition, patient-facing portals can enable patients to access results and share them with health care providers outside of our health care system. Patients can view PREDICT results in the MHAV patient portal where genetic findings of interest are presented in lay language (Figure 3). This not only serves as a channel to educate patients about their results but encourages sharing of the results with providers outside of the Vanderbilt network. In addition, with Epic’s Care Everywhere functionality, genomic indicators can be shared with other Epic organizations. When configured, genomic indicators are included in the data Care Everywhere sends and receives so they not only can be manually accessed by clinicians to make decisions but also used as building blocks to trigger locally developed CDS alerts.
The right time in workflow: PGx interpretations are intended to inform prescribing decisions, having CDS alerts at the time of the start of a prescribing action is most efficient for most scenarios. Upstream alerts (e.g. alerting providers at the time of opening a patient’s chart) would result in excessive alerting and contribute to alert fatigue. On the other hand, firing CDS at the end of the prescribing process, (after the prescriber has chosen the drug, dose, route and frequency) may reduce compliance with the alert compared to alerts provided at the moment the target drug was selected.
Figure 5.
Clopidogrel alert for adult CYP2C19 poor metabolizers. Relative and absolute contraindications of alternative drugs are listed for consideration for end users. Loading and maintenance doses are available for alternative drugs listed. Image is copyright of Epic Systems Corporation and used with permission.
Figure 6.
Tacrolimus alert for adult CYP3A5 normal or extensive metabolizers. Modification of standard metabolizer status terminology to “poor responder” to simplify complexity in interpretation language used. Image is copyright of Epic Systems Corporation and used with permission.
Figure 7.
Citalopram alert for adult CYP2C19 poor metabolizers with actionable CYP2D6 metabolizer status. Modification of alternative drugs to account for additional CYP2D6 metabolizer status. Additional consult option is available due to the complexity of clinical scenarios possible. Image is copyright of Epic Systems Corporation and used with permission.
PROGRAMMATIC COMPONENTS OF SUCCESS
Institutional and programmatic factors can influence the success of an end-to-end PGx CDS system. Five particularly critical elements include: 1) Institutional support, as PGx implementation requires significant investment; 2) Multidisciplinary team, as implementation and maintenance of a PGx program requires ongoing effort; 3) Integration of PGx testing into workflows that support the needs of the end users; 4) Ongoing clinical and educational support for end users to support use and knowledge of updates; and 5) Ongoing feedback with pre-defined metrics of success.
1. Institutional support for implementation
VUMC has a long-standing and enduring commitment to personalized patient care. PGx is one key step toward this goal. The success of a PGx program is critically tied to enterprise leadership, the clinicians, and the institutional committees. The implementation and maintenance of PGx programs requires all levels of institutional support. These ingrained ties then lead to effective and meaningful bidirectional communication on programmatic evolution and strategic direction. The process of gaining institutional support also ensures programmatic alignment to the specific needs of the institution and the end users. Collaboration with clinical groups and researchers provides valuable data regarding the anticipated and observed number of patients influenced by testing and the impact of the clinical recommendation on end users prior to implementation. Additionally, having all CDS recommendations vetted and approved by an established and independent committee such as a Pharmacy and Therapeutics committee gives end users greater confidence in the clinical utility of the results. This could be aided by having a credible executive sponsor introduce the initiative. Equally, executive sponsors and Health IT stakeholders will be essential for navigating ongoing programmatic maintenance, as new evidence require revisions to CDS and new technologies (e.g. new laboratory assays or new EHR updates) will require recalibration of the PGx pipeline. If testing is conducted in house, the laboratory can provide expertise on identifying patient variants relevant in the institute’s population.
2. Multidisciplinary team for ongoing PGx program management
A dedicated and standing working group of multidisciplinary stakeholders can provide diverse perspectives on the most effective method of CDS implementation for PGx and to steer the program in the long term. This working group can consist of leaders of the institution as well as experts that aid in the internal- and customer-facing development of the CDS. Examples of such experts include molecular diagnostics professionals, health informaticists, clinical pharmacologists, clinical pharmacists, product and project managers, biostatisticians, and clinicians practicing in relevant fields. Institutional leaders should be diverse, and could include members of the executive team, Health IT leaders, or independent healthcare committees. This committee of multidisciplinary stakeholders handles knowledge base maintenance to necessitate reprocessing of interpretations, updating CDS to meet changes in guidelines, ongoing clinical engagement or education, or even operational monitoring and quality improvements associated with the CDS. In addition, clinical experts are needed to evaluate and design CDS content for PGx implementation and expansion of the program. When choosing a DGI to implement, committee and clinical experts should consider the likelihood of the patient population carrying an actionable variant, the strength of evidence to support a clinical action, ease of following the recommendation for end users, and the type of patient outcome that is associated with the recommendation. All of these factors help define the clinical utility and impact on the patient population.
3. Integration into workflows
The workflow to implement PGx should be straightforward to the end users from the point of ordering to the point-of-care. PGx-driven CDS alerts can provide interpretations and recommendations at point-of-care to guide prescribing as described above. There are also several strategies to reduce barriers at the point of ordering. CDS may be used to identify patients with imminent medical necessity (reactive testing), and/or those where the PGx test is anticipatory (preemptive testing) of a future exposure. While the latter approach can counteract issues of long TAT, reimbursement for preemptive testing is currently uncertain at best. Another effective strategy is to include the PGx test in existing order sets and clinical protocols so that testing becomes part of routine clinical care. In addition, the orderable laboratory panel should be linked to multiple related search terms (synonyms in the library of orderable) so that it is easy for providers to find. For example, our PGx panel search terms include the individual gene names, as well as “PREDICT,” “Pharmacogenomics,” “Pharmacogenetics,” “PGx,” and “PDx.”
Establishing the process of PGx testing in the institution involves addressing topics such as accessibility to testing, support available at point of ordering, and the financial structure necessary to sustain the program. For the test to be used in routine clinical care you may need to consider resource and reimbursement factors. These factors will ultimately influence whether the test is ordered inpatient or outpatient, what the required TAT is, which type of providers should be ordering the test, and which patients get testing.
4. Ongoing clinical and educational support for end users
Although all the previously mentioned support for end users may help with the ordering process, clinician comfort with ordering PGx testing is driven mostly by confidence in their ability to interpret and use the test results. Carefully crafted CDS can raise confidence, but additional support such as a clinic or consult service will provide further reassurance that providers will not be responsible for interpreting a whole panel of genes or optimizing therapy outside of their scope of practice. A consult service or clinic can also assist with educating and counseling patients, which providers may not have the time or expertise to perform. We also recommend providing targeted continuing education sessions to end users in specific clinical fields, including both providers and pharmacists. Although there may be organizational support and integration in normal workflows, ongoing educational support is a key factor to promote testing and compliance with the CDS recommendations. Without providing educational support to end users, PGx results remains a foreign tool or one not present at all in the clinician’s toolbox.
5. Feedback and metrics of success
It is imperative to establish a method to collect programmatic feedback. Operational analytics are critical for executing data-driven decisions across the PGx program. These decisions will impact forecasting and planning, user engagement and test utilization, as well as quality management. These data can be related to the CDS, the diagnostic laboratory, the end users, and if de-identified – the patient community.
Readily available data include CDS metrics such as frequency of CDS alerts and provider actions (Figure 8a–b). Our PGx CDS alerts require acknowledgement, providing de-identified responses to each alert that can be aggregated across providers (Figure 8a). These data demonstrate that many of the users will adjust their treatment regimen to align with the PGx guidance; however, many providers will also decline the CDS recommendation, acknowledging a pre-specified reason or annotating their own reason such as “previously tolerated” and “short term management.” Attention to common responses can inform revisions to the CDS to increase efficiency (e.g. adding acknowledgement reasons). BPA data are important for understanding the successes and opportunities for programmatic refinement. Testing data can also be used to direct broader programmatic development. Patient demographics, PGx testing data (e.g. date of testing, volume of tests, TAT, ordering provider), and PGx results data (e.g. metabolizer status, variant allele prevalence, number of actionable variants) will help the program identify and meet benchmarks for success or improvement. PREDICT employs a Tableau data visualization tool to ensure programmatic operational support. For example, we have found that ~50% of our tested patient population carries at least 2 PGx risk variants (Figure 8b); these data demonstrating the value of a panel-based PGx testing program. Optimizing the value of a PGx program relative to its cost is crucial. These data facilitate financial analyses such as institutional cost savings relative to decreased send out genetic tests or therapeutic drug monitoring laboratory tests. Additionally, clinical adverse events or hospitalization stays could be compared between tested and untested patients in a pragmatic trial such as the PGx trial being conducted by the Incorporating Genomics in Practice (IGNITE) network. Clinical utility as well as provider and patient satisfaction should be considered when contemplating metrics for success. These types of surveys are ideal quality improvement initiatives and the data will help a program customize itself for its patients and clinical users.
Figure 8.
Programmatic operational data aids CDS developments and program improvement
(a) Provider interaction with the PREDICT clopidogrel BPA from 2019 until 2020. The clopidogrel BPA requires acknowledgement, and the percent distribution of responses are provided. BPA – best practice alert.
(b) The number of actionable pharmacogenetic variants within the tested population from 2010–2020. PGx - pharmacogenomics
DISCUSSION
End-to-end CDS delivers numerous advantages for routine clinical use of PGx results as it transforms static results into interpretations that are anticipatory, actionable, and adaptable. In this tutorial we establish a framework for implementing end-to-end CDS and address key considerations for this process. We acknowledge the multitude of methods in implementing PGx CDS and the unique advantages and disadvantages with each method35. This tutorial serves as a general overview of the one such method, with additional details for specific key components and critical elements for success. In addition, the considerations mentioned throughout may be applicable for implementation of other genomic results in addition to PGx.
There has been tremendous evolution in technology to aid in the use of PGx recommendations although more can be done to further the goal of incorporating PGx into routine clinical care. An example of this is the configuration of the EHR to enable intuitive use of PGx recommendations. The Epic genomic indicator has revised from including summary statements of the literature and FDA label to one that now allows for a simple summary of the recommendation – although hovering over an icon is necessary (Figure S2). Evidence for PGx and the standardization of terminology continue to evolve, as illustrated by updates to CYP2D6 nomenclature. In addition, evidence in the future can dictate substrate specific interactions for medications which will require flexibility in the informatics infrastructure to accommodate this complexity (e.g. in vitro data suggest substrate-dependent activity for CYP2D6*17).37,38 Phenoconversion caused by drug-drug-gene interactions is also an emerging area for automated CDS. Currently our CDS does not take into consideration phenoconversion due to concomitant strong inducers/inhibitors, although pharmacists are aware of the phenomenon and PGx consults can be ordered. Incorporation of drug-drug-gene interactions increase complexity of CDS builds and will require a larger commitment for ongoing maintenance.
In addition, we recognize that next generation sequencing, including whole genome and whole exome sequencing, is evolving and will become more accessible in the future. New technologies such as these have potential to improve use and access to pharmacogenomics results. However, accurate assessment of pharmacovariants from next generation sequencing and incorporation of these data into clinical care currently present significant challenges. New resources are maturing and those developed by CPIC, Pharmacogene Variation Consortium and EU’s Ubiquitous Pharmacogenomics with implementation functionality as a goal will impact the field significantly.
The future of PGx and precision medicine holds promising improvements for patient care with evolution in technology and methods of application aiding in PGx implementation reaching its full potential.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to recognize Dr. Jeff Balser, President and CEO of Vanderbilt University Medical Center, who strongly supports personalized medicine initiatives at VUMC including PREDICT. The authors also express gratitude for executive sponsorship provided by Dr. Gordon Bernard, Dr. Dan Roden, and Director Jill Pulley. The authors would like to thank and acknowledge the Medical Laboratory Scientists within the VUMC Molecular Diagnostics Laboratory for performing patient testing associated with the PREDICT initiative. Finally, this work would not have been possible without the support from HealthIT leadership and expertise provided by HealthIT analysts, knowledge engineers, and developers who play an instrumental role in the design, implementation, and support of the informatics infrastructure.
Funding information: Support for this work is provided by Vanderbilt University Medical Center and funding from the National Institutes of Health, National Human Genome Research Institute (NIH/NHGRI) grants U01HG010232 and U01HG007253 to JFP and SLV. The pharmacogenomic program is supported in part by institutional funding and by the Vanderbilt Clinical and Translational Science Awards (CTSA) grant UL1TR000445 from NCATS/NIH.
Footnotes
Conflicts of interest: As an Associate Editor of Clinical Pharmacology & Therapeutics, Sara Van Driest was not involved in the review or decision process for this paper. All other authors declared no competing interests for this work.
REFERENCES
- 1.Roden DM et al. Pharmacogenomics. Lancet 394, 521–532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plumpton CO, Roberts D, Pirmohamed M & Hughes DA A Systematic Review of Economic Evaluations of Pharmacogenetic Testing for Prevention of Adverse Drug Reactions. Pharmacoeconomics 34, 771–793 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Roden DM et al. Benefit of pre-emptive pharmacogenetic information on clinical outcome. Clin Pharmacol Ther 103, 787–794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shurin SB & Nabel EG Pharmacogenomics--ready for prime time? N. Engl. J. Med 358, 1061–1063 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Scott SA et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther 94, 317–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hicks JK et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther 98, 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Relling MV & Evans WE Pharmacogenomics in the clinic. Nature 526, 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krebs K & Milani L Translating pharmacogenomics into clinical decisions: do not let the perfect be the enemy of the good. Hum. Genomics 13, 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pulley JM et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther 92, 87–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson JF et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet. Med 15, 833–841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abou Diwan E, Zeitoun RI, Abou Haidar L, Cascorbi I & Khoueiry Zgheib N Implementation and obstacles of pharmacogenetics in clinical practice: An international survey. Br J Clin Pharmacol 85, 2076–2088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson JF et al. Attitudes of clinicians following large-scale pharmacogenomics implementation. The Pharmacogenomics Journal 16, 393–398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haga S, Burke W, Ginsburg G, Mills R & Agans R Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet 82, 388–394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel HN, Ursan ID, Zueger PM, Cavallari LH & Pickard AS Stakeholder views on pharmacogenomic testing. Pharmacotherapy 34, 151–165 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Unertl KM, Field JR, Price L & Peterson JF Clinician Perspectives on Using Pharmacogenomics in Clinical Practice. Per Med 12, 339–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Center for Devices and Radiological Health. Warning Letter Inova Genomics Laboratory - 577422 - 04/04/2019. <https://www.fda.gov/inspections-compliance-enforcement-and-criminal-investigations/warning-letters/inova-genomics-laboratory-577422-04042019> (2019). Accessed 1 June 2020.
- 17.Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE & Hoffman JM Integrating pharmacogenomics into electronic health records with clinical decision support. Am J Health Syst Pharm 73, 1967–1976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuldiner A et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Overcoming Challenges of Real-World Implementation. Clin Pharmacol Ther 94, 207–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman JM et al. Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC). J Am Med Inform Assoc 23, 796–801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson JF et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet. Med. 15, 833–841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genzen JR, Mohlman JS, Lynch JL, Squires MW & Weiss RL Laboratory-Developed Tests: A Legislative and Regulatory Review. Clin. Chem. 63, 1575–1584 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Centers for Medicare and Medicaid Services. Clinical Laboratory Improvement Amendments (CLIA). <https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA> (2020). Accessed 1 June 2020.
- 23.Hoeltge GA Accreditation of Individualized Quality Control Plans by the College of American Pathologists. Clin. Lab. Med. 37, 151–162 (2017). [DOI] [PubMed] [Google Scholar]
- 24.ThermoFisher Scientific. <https://www.thermofisher.com/us/en/home.html> (2020). Accessed 1 June 2020.
- 25.GenMark Diagnostics. XT-8 Panels — Comprehensive Multiplex Panels. <https://www.genmarkdx.com/solutions/panels/xt-8-panels/>(2020). Accessed 1 June 2020.
- 26.Ferreira-Gonzalez A et al. Revisiting oversight and regulation of molecular-based laboratory-developed tests: a position statement of the Association for Molecular Pathology. J Mol Diagn 16, 3–6 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Tzvetkov MV, Meineke I, Sehrt D, Vormfelde SV & Brockmöller J Amelogenin-based sex identification as a strategy to control the identity of DNA samples in genetic association studies. Pharmacogenomics 11, 449–457 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Cerner. Laboratory. <https://www.cerner.com/solutions/laboratory> (2020). Accessed 1 June 2020.
- 29.Caudle KE et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci 13, 116–124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaedigk A, Dinh JC, Jeong H, Prasad B & Leeder JS Ten Years’ Experience with the CYP2D6 Activity Score: A Perspective on Future Investigations to Improve Clinical Predictions for Precision Therapeutics. J Pers Med 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical Pharmacogenetics Implementation Consortium. Guidelines. <https://cpicpgx.org/guidelines/> (2020). Accessed 1 July 2020.
- 32.Suarez-Kurtz G & Parra EJ Population Diversity in Pharmacogenetics: A Latin American Perspective. Adv. Pharmacol. 83, 133–154 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, De T, Zhong Y & Perera MA The Advantages and Challenges of Diversity in Pharmacogenomics: Can Minority Populations Bring Us Closer to Implementation? Clin. Pharmacol. Ther. 106, 338–349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popejoy AB Diversity In Precision Medicine And Pharmacogenetics: Methodological And Conceptual Considerations For Broadening Participation. Pharmgenomics Pers Med 12, 257–271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freimuth R et al. Implementing Genomic Clinical Decision Support for Drug‐Based Precision Medicine. CPT Pharmacometrics Syst Pharmacol 6, 153–155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CDS KnowledgeBase. Library. < https://cdskb.org/library/> (2020). Accessed 1 July 2020.
- 37.Bogni A, Monshouwer M, Moscone A, et al. Substrate specific metabolism by polymorphic cytochrome P450 2D6 alleles. Toxicol In Vitro 19, 621–629 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Shen H, He MM, Liu H, et al. Comparative metabolic capabilities and inhibitory profiles of CYP2D6.1, CYP2D6.10, and CYP2D6.17. Drug Metab Dispos 35, 1292–1300 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.