Abstract
Bronchodilator (BD) drugs are commonly prescribed for treatment and management of obstructive lung function present with diseases such as asthma. Administration of BD medication can partially or fully restore lung function as measured by pulmonary function tests. The genetics of baseline lung function measures taken before BD medication have been extensively studied, and the genetics of the BD response itself have received some attention. However, few studies have focused on the genetics of post-BD lung function. To address this gap, we analyzed lung function phenotypes in 1103 subjects from the Study of African Americans, Asthma, Genes, and Environment, a pediatric asthma case–control cohort, using an integrative genomic analysis approach that combined genotype, locus-specific genetic ancestry, and functional annotation information. We integrated genome-wide association study (GWAS) results with an admixture mapping scan of three pulmonary function tests (forced expiratory volume in 1 s [FEV1], forced vital capacity [FVC], and FEV1/FVC) taken before and after albuterol BD administration on the same subjects, yielding six traits. We identified 18 GWAS loci, and five additional loci from admixture mapping, spanning several known and novel lung function candidate genes. Most loci identified via admixture mapping exhibited wide variation in minor allele frequency across genotyped global populations. Functional fine-mapping revealed an enrichment of epigenetic annotations from peripheral blood mononuclear cells, fetal lung tissue, and lung fibroblasts. Our results point to three novel potential genetic drivers of pre- and post-BD lung function: ADAMTS1, RAD54B, and EGLN3.
Keywords: admixture, African American, asthma, GWAS, integrative genomic analysis, lung function
1 |. INTRODUCTION
Asthma is a disease characterized by episodic obstruction of airways that affects nearly 339 million people worldwide (The Global Asthma Network, 2018) and is the most common chronic disease among children. Asthma constitutes a massive global economic burden, representing $81.9 billion in medical costs in the United States alone (Nurmagambetov et al., 2018). As a complex disease, asthma results from both environmental and genetic factors, with genetic heritability estimates ranging from 0.35 to 0.90 (Ober & Yao, 2011). The advent of genome-wide association studies (GWAS; Risch & Merikangas, 1996), combined with progressively larger sample sizes in recent years, has enabled researchers to query the genetic basis of asthma at unprecedented scale, with numerous loci identified in autoimmune and inflammatory pathways (Demenais et al., 2018). However, these loci account for a small portion of asthma liability (Demenais et al., 2018).
Pulmonary function tests are recommended to guide the diagnosis of asthma and monitor patient status (Asthma and Allergy Foundation of America, 2019). During these tests, patients breathe through a spirometer that captures key measures of lung function, including the forced expiratory volume in 1 s (FEV1), which measures initial forced exhalatory capacity; the forced vital capacity (FVC), which measures the maximum total volume of air that a patient can forcibly exhale; and their ratio (FEV1/FVC). Lung function measures can be population-normalized according to expected lung function values that account for age, sex, height, and ethnicity of the patient (Hankinson et al., 1999). Spirometric measurements can be taken both before bronchodilator (BD) treatment (pre-BD) and after (post-BD) to further understand lung function status. Historically, baseline lung function is measured with pre-BD measures, but among people with asthma, post-BD lung function may best reflect lung health (Brehm et al., 2015).
While the genetic contribution to asthma and lung function has been extensively studied via GWAS, most analyses have relied on subjects of European descent (Demenais et al., 2018; Johansson et al., 2019; Pickrell et al., 2016; Z. Zhu et al., 2018). This overrepresentation of ethnically white subjects in biomedical research has impaired the generalizability of genetic studies of complex disease (Burchard, 2014; Bustamante et al., 2011; Popejoy & Fullerton, 2016). Ethnic differences in lung function, particularly between non-Hispanic African Americans and European Americans, have been reported for over 40 years (Binder et al., 1976; Glindmeyer et al., 1995; Hsi et al., 1983; Rossiter & Weill, 1974; Schwartz et al., 1988). Ethnic disparities in lung function were attributed to population differences in sitting height, as increased height leads to increased lung capacity. However, adjustment for sitting height only explains 42%–50% of ethnic differences in lung function between African Americans and European Americans (Harik-Khan et al., 2004), suggesting that a simplistic reduction to ethnic differences in height cannot account for the observed disparity in lung function. Unequal socioeconomic conditions were also thought to contribute to ethnic differences in lung function (Braun, 2015; Quanjer, 2013, 2015), but socioeconomic factors only account for 7%–10% of unexplained variance (Harik-Khan et al., 2004). Self-identified race or ethnicity are commonly used in the clinic to interpret lung function measures, but these are not ideal variables for understanding genetic differences in lung function between populations. Kumar et al. (2010) observed that the proportion of global African genetic ancestry is inversely correlated with lung function. Spear et al. (2019) later observed population differences among African Americans, Mexican Americans, and Puerto Ricans in BD drug response to albuterol, the short-acting β2-adrenergic receptor agonist that is the most commonly prescribed drug for the treatment of acute asthma symptoms. Specifically, Spear et al. performed admixture mapping, a technique designed to identify regions of the genome where locus-specific ancestry drives variation in a disease trait (Shriner, 2013) that has been helpful in studies of complex diseases, including asthma and breast cancer (Féjerman et al., 2012; Pino-Yanes et al., 2015). However, admixture mapping studies comparing baseline and post-BD lung function have not yet been performed in African Americans. In this study, we address this gap in knowledge by evaluating the effect of locus-specific ancestry on both pre- and post-BD lung function measures in a pediatric case–control cohort of non-Hispanic African Americans children and adolescents.
2 |. METHODS
2.1 |. Ethics statement and data availability
Data from the Study of African Americans, Asthma, Genes, and Environments (SAGE) cohort were used for this study. The data that support the findings of this study are available in the NCBI dbGaP repository under ascension number phs000921.v1.p1. Data from SAGE were approved for human subjects research under expedited review by IRB 10–02877 at the University of California, San Francisco with reference #244919. All subjects gave written consent for genotyping, phenotyping, and data usage for general research use.
2.2 |. Study population
SAGE is a case–control cross-sectional cohort study of genetics and gene–environment interactions in non-Hispanic African American children and adolescents in the United States. SAGE includes detailed clinical, social, and environmental data on both asthma and asthma-related conditions. Full details of the SAGE study protocols are described in detail elsewhere (Borrell et al., 2013; Mak et al., 2018; Nishimura et al., 2013; Thakur et al., 2013). Briefly, SAGE was initiated in 2006 and recruited participants with and without asthma through a combination of clinic- and community-based recruitment centers in the San Francisco Bay Area. All participants in SAGE self-identified as African American and self-reported that all four grandparents were non-Hispanic African American. To reduce population heterogeneity resulting from very recent admixture, we only analyzed subjects who affirmatively self-identified as having been born in the United States and whose parents were also born in the United States.
Pulmonary function tests were taken before administration of albuterol BD medication for all individuals, both those with and without asthma. Post-BD spirometry measures were performed only for individuals with asthma. Analyses of pre-BD lung function measures included all 1103 asthma cases and controls with complete covariate information. Post-BD analyses were performed on the 831 asthma cases with post-BD measurements.
2.3 |. Genotyping and quality control
DNA was isolated from whole blood collected from SAGE participants at the time of study enrollment as described previously (Borrell et al., 2013). DNA was extracted using the Wizard® Genomic DNA Purification kits (Promega). Samples were genotyped with the Affymetrix Axiom LAT1 array (World Array 4, Affymetrix).
Genotype quality control was performed in PLINK v1.9 (Chang et al., 2015). Of the 772,703 genotyped variants, 111,901 single nucleotide polymorphisms (SNPs) were excluded from analysis due to genotype missingness more than 5% (n = 28,211), minor allele frequency (MAF) less than 1% (n = 80,420) or deviation from Hardy-Weinberg expectations (HWE) at p < 0.001 (n = 3270). The final set included 660,802 genotyped markers (Table S1).
Genotyped SNPs were submitted to the Michigan Imputation Server (Das et al., 2016), phased using EAGLE v2.3 (Loh et al., 2016), and imputed from the 1000 Genomes Project reference panel (The 1000 Genomes Project Consortium, 2015) using Minimac3 (Das et al., 2016). Imputed SNPs with imputation R2 < .3, with deviation from HWE p < 10−4, or with MAF < 1% were discarded. Of the 47,101,126 imputed SNPs, a total of 31,146,322 were culled due to either low MAF (n = 31,095,418) or deviation from HWE (n = 50,904). All variants in the imputed set showed a genotype missingness of no more than 5%. The final number of SNPs used in association analyses was 15,954,804 (Table S1).
2.4 |. Outcome phenotypes
Pulmonary function testing was performed at the time of recruitment according to the American Thoracic Society/European Respiratory Society standards (Miller et al., 2005; Pellegrino et al., 2005; Wanger et al., 2005) with a KoKo PFT Spirometer (nSpire Health Inc.). Spirometry was performed both before and 15 min after administration of four puffs of albuterol (90 μg per puff) through a 5-cm plastic mouthpiece from a standard metered-dose inhaler. Patients were assessed for the following spirometric measures before and after BD drug usage (pre-BD and post-BD, respectively): (a) FEV1, (b) FVC, and (c) FEV1/FVC. A total of six phenotypes were assessed for genotype association: pre-BD FEV1 (pre-FEV1), pre-BD FVC (pre-FVC), pre-BD FEV1/FVC (pre- FEV1/FVC), post-BD FEV1 (post-FEV1), post-BD FVC (post-FVC), and post-BD FEV1/FVC (post-FEV1/FVC). All phenotype values were normalized based on the expected lung function values calculated from the Hankinson equations (Hankinson et al., 1999), which account for age, sex, height, and self-reported ethnicity. Phenotype distributions were checked for normality and to detect outliers. Outliers were determined using the method of Tukey fences (John Tukey, 1977). For each phenotype, we computed the first quartile value (Q1), the third quartile value (Q3), and the interquartile range (IQR). We declared as outliers all values outside of the range
Individuals with outlier values for a phenotype were removed from association analyses for that phenotype.
2.5 |. Covariates
2.5.1 |. Age, sex, and body mass index (BMI)
Biometric covariates such as age, sex, BMI, and height were measured directly at time of recruitment. BMI was categorized into underweight, normal, overweight, and obese, according to CDC guidelines for defining childhood obesity (Barlow, 2007; Cote et al., 2013; Whitlock et al., 2005). An overweight status was defined as a BMI at or above the 85th percentile for the general population of children of the same sex and in the same age group. An obese status was defined as a BMI at or above the 95th percentile. Underweight individuals (bottom 5th percentile, n = 9) were excluded from analysis.
2.5.2 |. Asthma status
Case status was defined as physician-diagnosed asthma supported by reported asthma medication use and symptoms of coughing, wheezing, or shortness of breath in the 2 years preceding enrollment.
2.5.3 |. Maternal educational attainment
Maternal educational attainment was measured at recruitment and included in analyses to control for socioeconomic status. It was coded as total years of education completed from the first grade: for example, a complete K-6 education was 6 years, a complete high school education was 12 years, and any additional years (college or trade school and beyond) were counted as 1 year each.
2.5.4 |. Genetic ancestry
Previous literature on the genetics of non-Hispanic African Americans has observed global genetic ancestry proportions of 73.2% West African, 24.0% European, and 0.8% Amer-indigenous (Bryc et al., 2015), strongly suggesting that a reference panel of African and European was sufficient for accurate global genetic ancestry estimates in SAGE. Global genetic ancestry was estimated for each individual with the ADMIXTURE software (Alexander et al., 2009) in supervised learning mode assuming one West African and one European ancestral population, with HapMap Phase III YRI and CEU populations as references (The International HapMap 3 Consortium, 2010). Local ancestry estimation was performed with RFMix (Maples et al., 2013; Spear et al., 2019) using the same two-way ancestry reference from HapMap Phase III.
2.5.5 |. Estimation of genetic relatedness and genotype principal components
Genetic relatedness matrices (GRMs) were generated in R using GENESIS (Gogarten et al., 2019), which provides a computational pipeline for handling complex population structure using principal components analysis (PCA). We used PCAir (Conomos et al., 2015) to correct for distant population structure accounting for relatedness, and PC-Relate (Conomos et al., 2016) to adjust for genetic relatedness in recently admixed populations. The resulting principal components provide better correction for population stratification in admixed populations compared to standard PCA on genotypes (Patterson et al., 2006). Additionally, since GRMs produced by GENESIS are corrected for cryptic relatedness, the resulting association test statistics do not suffer inflation resulting from confounding relatedness in our sample.
2.6 |. Genetic association analyses
Genotype association testing was performed with the MLMA-LOCO algorithm from GCTA (Yang et al., 2011, 2014) to correct for population structure using GRMs generated with GENESIS. Association testing of outcome phenotypes with allele dosages at 15,954,804 biallelic SNPs was performed with a “leave one chromosome out” model to avoid double-fitting tested variants. Other variables included in models were age, sex, BMI, maternal educational attainment, and three genotype principal components. Models of pre-BD also included asthma status.
The suggestive and significant association thresholds for each outcome phenotype were determined by the effective number of independent statistical tests (Meff) calculated with CODA (Plummer et al., 2006). CODA computes Meff using the autocorrelation of p values from GWAS. This produces population-specific Bonferroni thresholds that account for correlation between statistical tests without increasing the Type I error rate (Sobota et al., 2015). For our analyses, Meff ranged from 488,819 to 507,975 (Table S2). The Bonferroni corrected genome-wide significance threshold was computed as 0.05/Meff, while the suggestive threshold was computed as (1/Meff), yielding a single pair of thresholds for all six outcome phenotypes considered: p < 1.99 × 10−6 for suggestive association, and p < 9.95 × 10−8 for significant association (Table S2).
Admixture mapping analyses were performed using linear regression models in R and local ancestry calls from RFMix for 454,322 genotyped SNPs. Counts of 0, 1, or 2 alleles of African descent were computed for each person at each SNP. Phenotypes were then regressed onto ancestral allele counts for each SNP while including age, sex, height, BMI, maternal educational attainment (as a proxy for socioeconomic status), and global African genetic ancestry proportion as covariates. Analyses with pre-BD outcome measures also included asthma status as a covariate.
2.7 |. Fine-mapping genetic associations
Functional fine-mapping with PAINTOR (Kichaev et al., 2014) was used to identify putative causal variants in novel loci deemed statistically significant by admixture mapping. PAINTOR applies a Bayesian probabilistic framework to integrate functional annotations, association summary statistics (Z-scores), and linkage disequilibrium information for each locus to prioritize the most likely causal variants in a given region. Functional annotations were selected per locus as recommended by the authors of PAINTOR (Kichaev, 2017). A subset of lung- and blood-related functional annotations from the Roadmap Epigenomics Project (Roadmap Epigenomics Consortium et al., 2015) and the ENCODE Consortium (ENCODE Project Consortium, 2012) were assessed for their individual improvement to the posterior probability of causality; the top five minimally correlated annotations were selected for each locus.
2.8 |. Annotation tools
The NHGRI/EBI GWAS Catalog (Buniello et al., 2019), Ensembl Genome Browser release 98 (Cunningham et al., 2019) and gnomAD browser v3.0 (Karczewski et al., 2020) were used to look up known associations at significant loci according to our analyses. Annotation lookups in the gnomAD browser v3.0 used hg38 coordinates translated from our hg19-aligned genotypes via liftOver (Hinrichs et al., 2006). Data management, statistical analysis, and figure generation made extensive use of GNU parallel (Tange, 2018) and several R packages, including data.table, doParallel, optparse, ggplot2, and the tidyverse bundle (Davis, 2020; Dowle & Srinivasan, 2020; Microsoft Corporation & Weston, 2019; Wickham, 2016; Wickham & Garrett, 2017).
3 |. RESULTS
3.1 |. Cohort characteristics
Characteristics of all SAGE participants included in analyses are shown in Table 1. Distributions of each lung function measure stratified by case–control status and BD administration (pre-BD vs. post-BD) are shown in Figure S1. FVC showed no significant difference between asthma cases and controls (Kruskal-Wallis p = .073), while stratification by case–control status yielded significantly different distributions for FEV1 (Kruskal-Wallis p = 4.8 × 10−7) and FEV1/FVC (Kruskal-Wallis p = 1.5 × 10−7). Among cases, statistically significant differences were observed between distributions of pre-BD and post-BD measures of FEV1 (Kruskal-Wallis p = 1.2 × 10–38), FVC (Kruskal-Wallis p = 5.4 × 10–16), and FEV1/FVC (Kruskal-Wallis p = 4.0 × 10–29), illustrating a measurable effect of BD medication on lung function.
TABLE 1.
Summary statistics of phenotypes and covariates from the SAGE cohort
| Characteristics | Cases | Controls | Total |
|---|---|---|---|
| Subjects (n) | 831 | 272 | 1,103 |
| Age (year) | 14.1 (3.66) | 16.3 (3.77) | 14.7 (3.8) |
| Female (n) | 406 | 166 | 572 |
| Height (cm) | 158 (14.34) | 162.4 (13.26) | 159.1 (14.2) |
| African ancestry (%) | 80.4 (0.1) | 79.6 (0.1) | 80.2 (0.1) |
| Maternal education (yr) | 12.4 (1.47) | 12.2 (1.5) | 12.3 (1.48) |
| Obesity status | |||
| Obese (n) | 276 | 74 | 350 |
| Nonobese (n) | 555 | 198 | 753 |
| Pre-FEV1 | 103 (13.79) | 98.1 (13.02) | 99.3 (13.77) |
| Pre-FVC | 103.4 (12.84) | 105.1 (13.09) | 103.8 (12.92) |
| Pre-FEV1/FVC | 95.1 (9.35) | 98.4 (8.2) | 95.9 (9.19) |
| Post-FEV1 | 107 (13.44) | n/a | n/a |
| Post-FVC | 109 (14.42) | n/a | n/a |
| Post-FEV1/FVC | 99 (7.83) | n/a | n/a |
Note: Displayed numbers are either counts (n) or averages followed by standard errors in parentheses. Units are listed where appropriate. An “n/a” appears where measurements were taken on cases only
Abbreviations: FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity.
The global African genetic ancestry proportion in our sample varied from 30.7% to 100%, with an average proportion of 80.2% (Figure S2), concordant with empirically observed averages (Baharian et al., 2016). Global ancestry contained the same information as the first genotype principal component (R2 = 0.984, Figure S3).
3.2 |. Genetic association testing finds novel and known loci
Figure 1 shows results from GWAS performed on pre-BD phenotypes (pre-FEV1, pre-FVC, and pre-FEV1/FVC) and post-BD phenotypes (post-FEV1, post-FVC, and post-FEV1/FVC) using linear mixed modeling. The association results showed no evidence of genomic inflation, with genetic control λ ranging from 0.98 to 0.99 (Table S2 and Figure S4). Table 2 lists the 18 genome-wide significant associations found, each associated with exactly one of the six lung function measures. An additional 252 variants were suggestively associated with at least one phenotype (Tables S3–S8). Of the 18 variants, 4 variants on chromosome 13 in a region spanned by the gene ATP8A2 were associated with pre-FEV1/FVC (Figure S6). Two variants on chromosome 16 that were associated with pre-FVC flanked the promoter region of IRX3 (Figure S7). A third variant associated with pre-FVC was located on chromosome 20 near THBD (Figure S8), a gene linked to venous thromboembolism in African American and Afro-Caribbean individuals (Hernandez et al., 2016). Two variants associated with Post-FVC were in a gene-rich region on chromosome 19 (Figure S9), with the peak near TMIGD2 and SHD, while eight other variants pointed to a second gene-rich region on chromosome 11 near CXCR5 and HYOU1 (Figure S10). Post-FEV1/FVC was associated with a region on chromosome 15 near the genes AKAP13 and ADAMTS7P4 (Figure S11).
FIGURE 1.
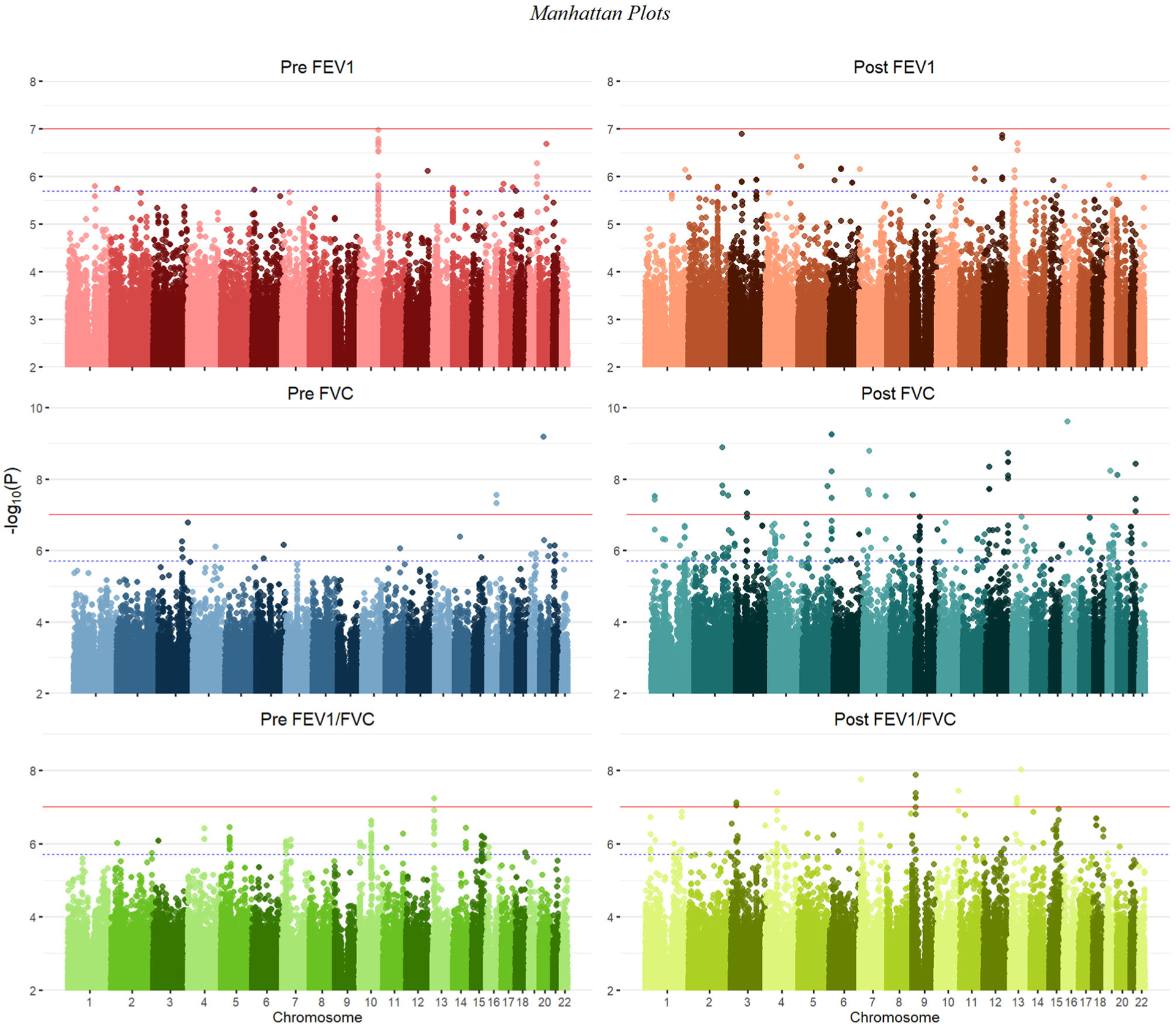
Manhattan plots summarizing GWAS p values for all six lung function phenotypes. The solid red line denotes genome-wide significance (p < 9.95 × 10−8), while the dashed blue line marks the suggestive threshold (p < 1.99 × 10−6), per CODA calculations. Variants with a p value greater than 0.05 were deemed uninformative and therefore not plotted
TABLE 2.
Significant association results from GWAS
| Phenotype | Chr | Position (bp) | A1 | A2 | MAF | β | SE | p value | Genes |
|---|---|---|---|---|---|---|---|---|---|
| Post-FEV1/FVC | 15 | 85798401 | A | G | 0.018 | −6.7846 | 1.27 | 9.05 × 10−8 | ADAMTS74P, AKAP13 |
| Post-FVC | 11 | 118932913 | C | T | 0.010 | 18.9094 | 3.29 | 9.47 × 10−9 | CXCR5, HY0U1 |
| Post-FVC | 11 | 118902275 | C | T | 0.010 | 17.9775 | 3.29 | 4.83 × 10−8 | CXCR5, HYOU1 |
| Post-FVC | 11 | 118905095 | A | G | 0.010 | 17.9775 | 3.29 | 4.83 × 10−8 | CXCR5, HY0U1 |
| Post-FVC | 11 | 118905316 | T | C | 0.010 | 17.9775 | 3.29 | 4.83 × 10−8 | CXCR5, HYOU1 |
| Post-FVC | 11 | 118906065 | A | G | 0.010 | 17.9775 | 3.29 | 4.83 × 10−8 | CXCR5, HY0U1 |
| Post-FVC | 11 | 118906240 | T | C | 0.010 | 17.9775 | 3.29 | 4.83 × 10−8 | CXCR5, HY0U1 |
| Post-FVC | 11 | 118906745 | C | G | 0.010 | 17.9775 | 3.29 | 4.83 × 10−8 | CXCR5, HYOU1 |
| Post-FVC | 11 | 118907923 | G | T | 0.010 | 18.2978 | 3.41 | 7.85 × 10−8 | CXCR5, HYOU1 |
| Post-FVC | 19 | 4289259 | T | C | 0.106 | 5.1672 | 0.967 | 9.21 × 10−8 | TMIGD2, SHD |
| Post-FVC | 19 | 4291817 | T | C | 0.111 | 5.2674 | 0.963 | 4.52 × 10−8 | TMIGD2, SHD |
| Pre-FEV1/FVC | 13 | 26235394 | G | A | 0.010 | −10.5334 | 1.94 | 5.80 × 10−8 | ATP8A2 |
| Pre-FEV1/FVC | 13 | 26247080 | G | A | 0.010 | −10.5334 | 1.94 | 5.80 × 10−8 | ATP8A2 |
| Pre-FEV1/FVC | 13 | 26262378 | T | G | 0.011 | −10.5325 | 1.94 | 5.81 × 10−8 | ATP8A2 |
| Pre-FEV1/FVC | 13 | 26268604 | A | C | 0.011 | −10.5325 | 1.94 | 5.81 × 10−8 | ATP8A2 |
| Pre-FVC | 20 | 22900228 | G | A | 0.010 | 18.0953 | 2.93 | 6.77 × 10−10 | THBD |
| Pre-FVC | 16 | 54327903 | G | A | 0.023 | 9.8543 | 1.78 | 2.83 × 10−8 | IRX3, FTO |
| Pre-FVC | 16 | 54327610 | A | G | 0.034 | 8.3689 | 1.53 | 4.85 × 10−8 | IRX3, FTO |
Note: The p values for all SNPs listed here met the significance threshold of 9.95 × 10−8. SNPs were specified by chromosome (Chr) and physical position in base pairs (bp).
Abbreviations: A1, major allele; A2, minor allele; B, effect size; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; Genes, any genes within proximity of the associated variants; MAF, minor allele frequency; SE, standard error of the estimate of β.
Among the suggestive associations, a variant on chromosome 12 associated with post-FEV1 was near BTBD11 (Figure S12), a gene previously associated with post-FEV1, post-FEV1/FVC, and ΔFEV1, the change in lung function due to BD administration (Hardin et al., 2016; Lutz et al., 2015), as well as BMI (Kichaev et al., 2019). A suggestive association with pre-FEV1 on chromosome 12 fell near SCARB1 (Figure S13), which was previously associated with FEV1 and FVC (Wyss et al., 2018) and HDL cholesterol levels (Wojcik et al., 2019). Another suggestive association with pre-FEV1 on chromosome 20 was near the gene PTPRT (Figure S14), which was previously associated with thromboembolism susceptibility in 5334 African American individuals (Heit et al., 2017).
3.3 |. Admixture mapping identified five novel loci not found by GWAS
Table 3 shows five regions where admixture proportions were statistically significantly associated with one of the six phenotypes. The three pre-BD phenotypes (pre-FEV1, pre-FVC, pre-FEV1/FVC) were each associated with one region, while post-FVC was associated with two distinct regions. Post-FEV1 and post-FEV1/FVC had no significant associations. None of the regions overlapped with those significant in our GWAS, and none showed large deviations from mean genome-wide African genetic ancestry. A small region on chromosome 21 that was significantly associated with pre-FEV1 flanked the genes ADAMTS1 and ADAMTS5 (Figure S15). The region on chromosome 4 associated with pre-FVC pointed to two candidate genes, RCHY1 and THAP6, that had no prior lung disease associations (Figure S17). A region on chromosome 19 associated with pre-FEV1/FVC spanned the genes ZNF557 and INSR (Figure S18). Post-FVC was associated with two regions, one on chromosome 8 spanning the genes ESRP1, INTS8, TP53INP1, and NDUFAF6 (Figure S19), and another on chromosome 14 encompassing EGLN3 and SNX6 (Figure S20).
TABLE 3.
Admixture mapping in SAGE identified five regions with statistically significant association to at least one phenotype
| Locus | Phenotype | Chr | Start (bp) | End (bp) | Length (kb) | Threshold | SNPAdmix (n) | SNPGWAS (n) | AFR (%) | Genes |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Pre-FEV1 | 21 | 28,209,667 | 28,240,392 | 30.72 | 1.03 × 10−4 | 4 | 215 | 78.7 | ADAMTS1 |
| 2 | Pre-FVC | 4 | 75,555,658 | 76,873,740 | 1318.08 | 9.64 × 10−5 | 102 | 7905 | 79.8 | RCHY1, THAP6 |
| 3 | Pre-FEV1/FVC | 19 | 7,068,207 | 7,127,294 | 59.09 | 1.04 × 10−4 | 18 | 376 | 80.4 | INSR, ZNF557 |
| 4 | Post-FVC | 8 | 95,387,941 | 95,820,594 | 432.65 | 9.93 × 10−5 | 45 | 2822 | 80.8 | ESRP1, INTS8, TP53INP1, NDUFAF6 |
| 5 | Post-FVC | 14 | 34,283,561 | 34,595,061 | 311.50 | 9.93 × 10−5 | 95 | 1982 | 80.5 | EGLN3, SNX6 |
Note: The regions are arbitrarily numbered from 1 to 5 and defined by phenotype, chromosome (Chr), physical starting point (in base pairs), and end point (in base pairs). Physical positions are given in hg19 coordinates. The total length of the region is given in kilobasepairs (kb). The threshold for statistical significance is given for each region. The column “Genes” lists genes physically within and near the associated regions.
Abbreviations: AFR, the percentage (%) of local genetic ancestry of African origin; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; SNP, single nucleotide polymorphism; SNPAdmix, counts the number (n) of genotyped SNPs from admixture mapping that met the significance threshold; SNPGWAS, the total number of GWAS SNPs (genotyped and/or imputed) in the admixture mapping region.
3.4 |. Functional fine-mapping found three novel putatively causal loci for lung function phenotypes
Table 4 lists the most probable causal SNP for each of the five admixture mapping loci according to PAINTOR. SNP rs13615 showed the highest probability of causality (0.630) with pre-FEV1 on locus 1 (Figure 2). This variant falls within the 3′-untranslated region (3′-UTR) of ADAMTS1, suggesting that ADAMTS1 drives the admixture mapping association and not its physical neighbor ADAMTS5. The MAF of rs13615 in African and African diaspora populations was lower than every other global population (2.6% AFR vs. 7.0%–54.5% other populations, gnomAD v3; see Figure S21). The SNP rs10857225 emerged as the most likely causal variant (probability 0.361) for the association of pre-FVC with locus 2 on chromosome 4 (Figure 3). This variant is located within an intron of the gene THAP6, suggesting that THAP6 is more likely the causal gene behind the association with pre-FVC. In contrast to locus 1, the MAF of rs10857225 is highest in global African populations and markedly lower in other global populations (59.1% AFR vs. 28.1%–38.4% other populations, gnomAD v3). Locus 3 on chromosome 19 associated with pre-FEV1/FVC, and locus 4 on chromosome 8 associated with post-FVC, showed little information gain from functional fine-mapping. The driving variant for locus 3, SNP rs72986681, was located in the 3′-UTR of ZNG557, but showed a low probability of causality (0.168, Figure S22). The most probable marker for locus 4, the SNP rs2470740, which is located in intron 2 of RAD54B, showed an even lower probability of causality (0.109, Figure S23). Functional fine-mapping of locus 5, a region on chromosome 14 associated with post-FVC, yielded the SNP rs1351618 with a moderate probability of causality (.390, Figure 4). rs1351618 is located in an intron of EGLN3. As with locus 2, rs1351618 had a much higher MAF in populations of African ancestry versus other global populations (12.4% AFR vs. <2.2% other populations, gnomAD v3).
TABLE 4.
Results from PAINTOR highlighting the most probably causal SNPs for each locus as defined by admixture mapping
| Locus | Phenotype | Chr | Position (bp) | SNP | Ref allele | Alt allele | MAF (%) | P-value | Annotation | Pr (causal) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Pre-FEV1 | 21 | 28,209,667 | rs13615 | A | G | 2.55 | 6.95 × 10−3 | ADAMTS1, 3′-UTR | 0.630 |
| 2 | Pre-FVC | 4 | 76,464,584 | rs10857225 | A | C | 59.07 | 3.11 × 10−6 | THAP6, Intron Variant | 0.361 |
| 3 | Pre-FEV1/FVC | 19 | 7,087,789 | rs72986681 | G | A | 1.54 | 7.25 × 10−4 | ZNF557, 3′-UTR | 0.168 |
| 4 | Post-FVC | 8 | 95,399,551 | rs2470740 | A | T | 23.82 | 2.04 × 10−3 | RAD54B, Intron Variant | 0.109 |
| 5 | Post-FVC | 14 | 34,531,633 | rs1351618 | C | T | 12.40 | 1.99 × 10−4 | EGLN3, Intron Variant | 0.390 |
Note: As in Table 3, the loci are arbitrarily numbered from 1 to 5 and defined by phenotype and chromosome. The physical position (in basepairs) of the most likely causal SNP is given in hg19 coordinates. MAFs are taken from global populations from the gnomAD server v3. The displayed p values are from our discovery GWAS. Pr(causal) is computed from PAINTOR.
Abbreviations: 3′-UTR, 3′-untranslated region; Chr, chromosome; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; MAF, minor allele frequency; Pr (causal), probability of causality; SNP, single nucleotide polymorphism.
FIGURE 2.

A CANVIS plot of results from PAINTOR functional fine-mapping for locus 1, an association with pre-FEV1 on chromosome 21. The SNP rs13615, which sits in the 3′-UTR of the gene ADAMTS1, attains a posterior probability of causality of 0.630. The panels show, from top to bottom, the posterior probability of causality; the five most informative functional annotations; GWAS p values; and local linkage disequilibrium expressed as a signed Pearson correlation. 3′-UTR, 3′-untranslated region; FEV1, forced expiratory volume in 1 s; SNP, single nucleotide polymorphism
FIGURE 3.
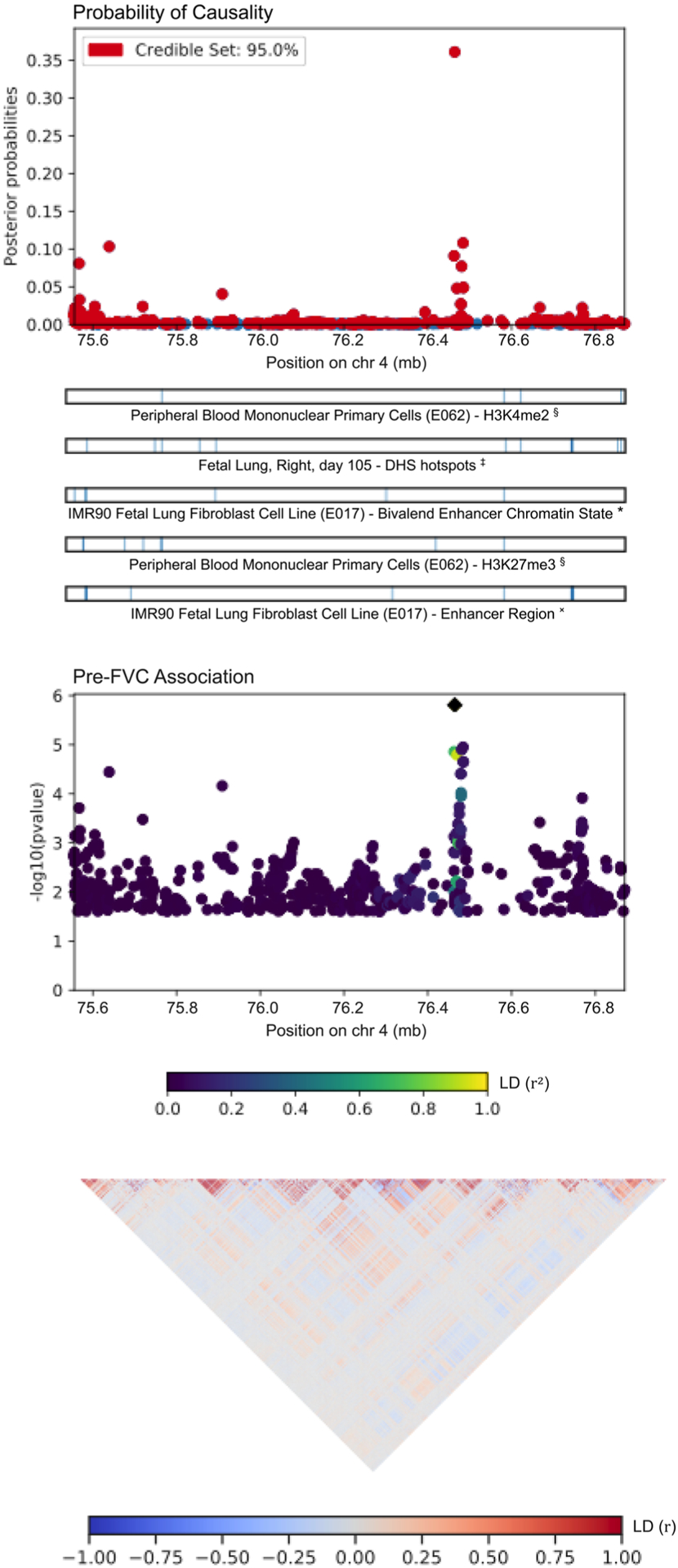
PAINTOR results for locus 2, an association on chromosome 4 with pre-FVC. The sentinel SNP, rs10857225, corresponds with a GWAS peak that does not pass Bonferroni correction for statistical significance. The highlighted peak tags the intron of the gene THAP6. FVC, forced vital capacity; SNP, single nucleotide polymorphism
FIGURE 4.
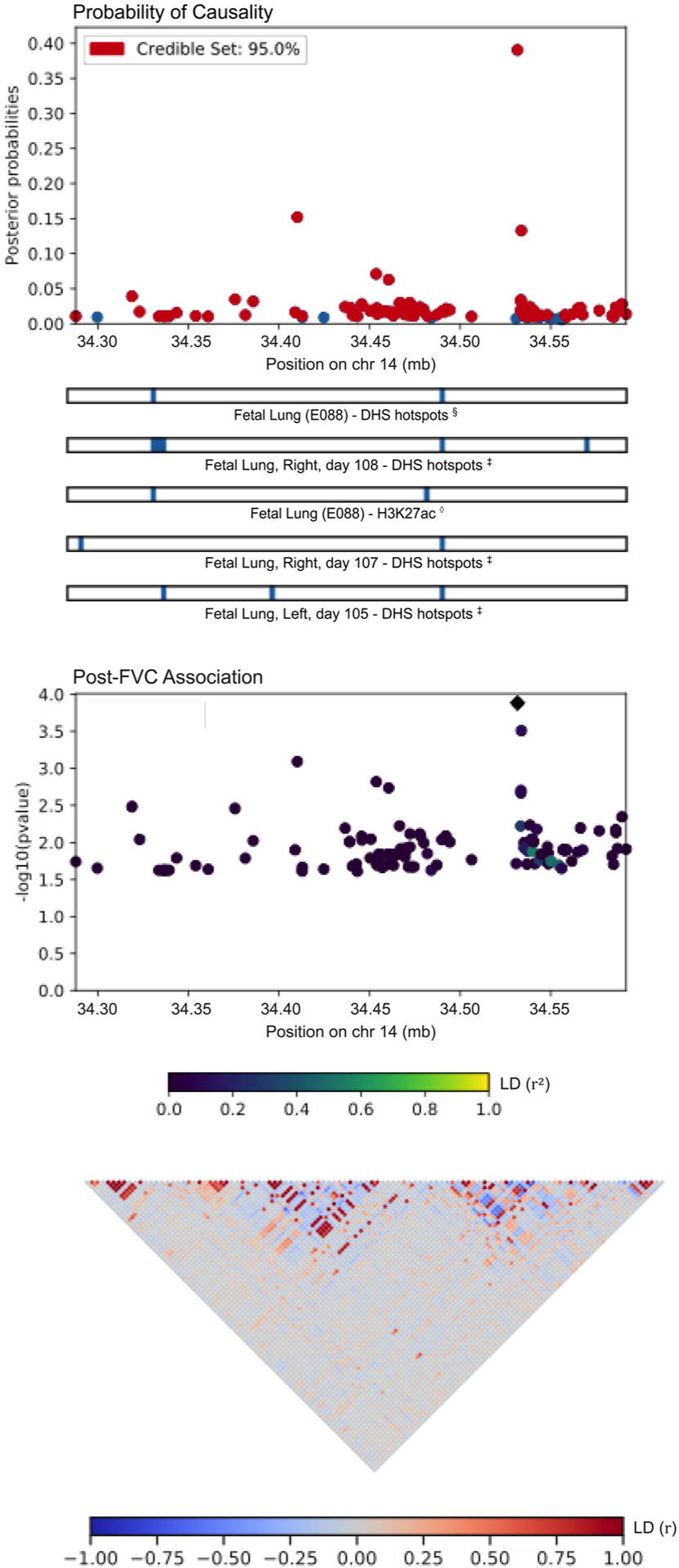
PAINTOR fine-mapping results for locus 5, corresponding to a region on chromosome 14 associated with post-FVC. The most likely causal SNP, rs1351618, tags an intron of the gene EGLN3. FVC, forced vital capacity; SNP, single nucleotide polymorphism
4 |. DISCUSSION
We analyzed the genetic basis of six lung function phenotypes in 1103 non-Hispanic African American children with and without asthma. The phenotypes consisted of three standard spirometric measures—FEV1, FVC, and FEV1/FVC—measured before and after administration of BD medication. Our GWAS identified 18 genome-wide significant loci, while our integrative genetic analysis approach that layered GWAS, admixture mapping, and functional fine-mapping identified another five putatively causal loci that could drive differences between pre- and post-BD lung function.
The four variants on chromosome 13 associated with pre-FEV1/FVC pointed to ATP8A2 as a candidate gene. ATP8A2 encodes an ATPase involved in phospholipid transport and is highly expressed in brain tissue, testes, and the adrenal glands, and to a lesser degree in the lung (Fagerberg et al., 2014). Mutations in ATP8A2 have been linked to several neurological disorders (Martín-Hernández et al., 2016). Two variants on chromosome 16 that were associated with pre-FVC point to IRX3 as a candidate gene. IRX3 encodes a homeobox protein crucial for neural development, and its promoters previously showed a long-range interaction with the FTO gene. Expression levels of the FTO gene are known to influence BMI and are of great interest in type II diabetes and obesity research (Smemo et al., 2014). Post-FVC showed associations on two chromosomes. Notable genes near the association peak on chromosome 19 included MAP2K2 and ZBTB7A, genes associated with variation in the two genes closest to the association peak, TMIGD2 and SHD, have not been previously associated with any traits. TMIGD2 is involved in T-cell costimulation and the immune response through an interaction with BMI, visceral adiposity, and eosinophil counts (Kichaev et al., 2019; Pulit et al., 2019; Rüeger et al., 2018); and CHAF1A, HDGFL2, PLIN4, ANKRD24, MPND, and SH3GL1, previously associated with corpuscular volume and hemoglobin concentration (Astle et al., 2016; Kichaev et al., 2019; van Rooij et al., 2017). Interestingly, HHLA2, suggesting that it could possibly play an immune or allergic response role in lung function (Y. Zhu et al., 2013). Among the genes within or near the chromosome 11 peak, two emerge as potentially key loci. The first is CXCR5, which has been linked to increased risk of childhood onset asthma (Ferreira et al., 2019; Johansson et al., 2019; Pividori et al., 2019) and respiratory disease (Kichaev et al., 2019), as well as related allergic and immunological conditions such as eczema, leukocyte count, rheumatoid arthritis, and Sjögren’s syndrome (Ferreira et al., 2017; Kichaev et al., 2019; Laufer et al., 2019; Lessard et al., 2013). The second is HYOU1, which has been associated with BMI and post-FEV1/FVC (Lutz et al., 2015; Pulit et al., 2019). Post-FEV1/FVC was associated with two genes, AKAP13 and ADAMTS7P4. AKAP13 has been previously associated with numerous conditions, including interstitial lung disease and psoriasis in European individuals (Fingerlin et al., 2013; Tsoi et al., 2015) as well as weight, BMI, and cardiovascular traits such as blood pressure and hemoglobin count in multiple populations (Giri et al., 2019; Kichaev et al., 2019). ADAMTS7P4 was previously associated with red blood cell volume (Kichaev et al., 2019). The statistically suggestive association of post-FEV1 with BTBD11 pointed to previous associations with various lung function measures, including post-FEV1, post-FEV1/FVC, and ΔFEV1 (Hardin et al., 2016; Lutz et al., 2015). These previously detected associations were based on much larger sample sizes than what was available to us: the associations with post-FEV1 and post-FEV1/FVC found by Lutz et al. were discovered in a population of 10,094 European and 3260 African American smokers with chronic obstructive pulmonary disorder (COPD), while the association with ΔFEV1 found by Hardin et al. was based on 5766 Europeans and 811 African Americans with COPD, suggesting that our inability to reach genome-wide significance in our sample was due to insufficient statistical power.
Among significant and suggestive GWAS loci, the association of post-FVC with variants in or near CXCR5 and HYOU1 is the only one that replicates known lung function loci: CXCR5 was previously associated with asthma (Ferreira et al., 2019; Johansson et al., 2019; Pividori et al., 2019), and HYOU1 was previously associated with post-FEV1/FVC (Lutz et al., 2015). The association with post-FEV1/FVC comes from an adult COPD cohort ascertained by smoking status; in contrast, SAGE is a pediatric asthma cohort. The mechanism by which HYOU1 affects lung function in both youth and adults is unclear. Nevertheless, the overlap of post-BD pulmonary function measures at this locus suggests that the region encompassing CXCR5 and HYOU1 plays a role in lung disease among people with obstructive lung function.
Admixture mapping identified five genomic regions where variation in genetic ancestry was significantly associated with phenotypic variation. Locus 1 on chromosome 21 spanned the genes ADAMTS1 and ADAMTS5, which encode extracellular proteases within the same protein family but with different consequences for disease. Although both genes have been linked to blood protein levels (Suhre et al., 2017), ADAMTS1 has been associated with pre-FVC (Kichaev et al., 2019) and is expressed in arterial, adipose, and lung tissue, while ADAMTS5 is not appreciably expressed in the lung (Figure S16). Further fine-mapping with PAINTOR places the most likely causal SNP (rs13615) within the 3′-UTR of ADAMTS1. Although the region is sparsely genotyped, and follow-up with whole genome sequencing data in this region is recommended, these results suggest that ADAMTS1 may be functionally related to lung function. Interestingly, the association of ADAMTS1 with pre-FVC (Kichaev et al., 2019) was discovered in a European sample of substantially larger size than our cohort, highlighting the ability of admixture mapping to detect associations in scenarios with low statistical power. Locus 3, spanning a region on chromosome 19 that was associated with pre-FEV1/FVC, contains the genes ZNF557 and INSR. ZNF557 has not been previously associated with any traits, while INSR is the well-known insulin receptor that has been previously associated with childhood onset asthma in our own cohort (White et al., 2016), as well as blood pressure levels, triglyceride levels, HDL cholesterol levels, and hypothyroidism across multiple populations (Bentley et al., 2019; Ehret et al., 2016; Kichaev et al., 2019; Klarin et al., 2018). Post-FVC showed two distinct admixture mapping signals. The first region on chromosome 8, which we call Locus 4, includes the genes ESRP1, INTS8, TP53INP1 and NDUFAF6 was previously associated with type II diabetes and eosinophil counts (Kichaev et al., 2019; Mahajan et al., 2018). The second region, Locus 5, spans EGLN3 and SNX6, both of which show previous associations with blood phenotypes such as blood pressure and hematocrit levels (Astle et al., 2016; Evangelou et al., 2018).
Overall, evaluation of our GWAS and admixture mapping lung function results suggests that genetics of this trait underlie some pleiotropy observed across pulmonary, hematological, cardiovascular, and obesity-related traits. Such pleiotropy has been observed in UK BioBank participants: as lung function decreases, BMI and type II diabetes incidence increases, as well as levels of eosinophils and neutrophils, both of which are common biomarkers for allergic disease (Figure S25; McInnes et al., 2019). The link between obesity and lung function is particularly interesting since obesity is a known asthma comorbidity, and lung function may play a role in obese asthma (Baffi et al., 2015; Gruchała-Niedoszytko et al., 2015). Our findings suggest that genetically based differences in lung function may provide a link between obesity and asthma.
It is curious that the regions identified by admixture mapping and subjected to functional fine-mapping did not overlap with the statistically significant GWAS loci. We attribute this in part to the different types of information used by each approach: GWAS analyzes how allelic variation affects a trait, while admixture mapping analyzes the phenotypic consequences of variation in genetic ancestry. Our integrative approach deprioritized results solely from GWAS, an approach driven by the fact that a supermajority of individual GWAS results fall in noncoding regions of the genome and are consequently notoriously difficult to interpret. By integrating GWAS summary statistics with loci identified via admixture mapping, we found that three of the admixture mapping-based loci—ADAMTS1, THAP6, and EGLN3—had evidence of causal effects. Each of the sentinel SNPs tagging these genes showed a notable difference in ancestral allele frequency: populations of African descent had either the highest or the lowest MAF among all global populations, likely the result of admixture mapping prioritizing loci that varied by genetic ancestry. None of these loci have been previously associated with lung traits, highlighting the strength of our integrative analysis. The association with EGLN3 is particularly curious since it has been previously associated with a variety of traits, including heart rate response to β-blocker therapy (Shahin et al., 2018). Short-acting β−2 agonists such as albuterol selectively target β−2 receptors in the lungs, while the first-generation β-blockers taken for cardiac conditions bind to both β−1 and β−2 receptors, affecting the heart as well the lungs. Bronchospasm and FEV1 reduction are clinically significant side effects of first-generation β−1 selective and nonselective β-blockers for cardiac conditions. Consequently, these nonselective β-blockers must be initiated with caution and close monitoring in patients with asthma (Christiansen & Zuraw, 2019). β-blockers lower blood pressure by reducing heart rate and cardiac contractility and are less effective in people with high levels of African genetic ancestry (Brewster & Seedat, 2013; Whelton et al., 2018). It has been previously observed that African Americans with asthma demonstrate lower BD drug response than European Americans (Blake et al., 2008), suggesting a possible pharmacological interaction between β−2 receptors and African ancestry. Furthermore, EGLN3 is strongly expressed in cardiac tissue, suggesting that EGLN3 could possibly influence post-FVC through cardiac phenotypes (Figure S24). Further functional studies are required to elucidate the role of EGLN3 on lung function and BD drug response.
This study has some important limitations. First, while our data set includes rich phenotyping of pulmonary traits with socioeconomic and biometric measures, it still constitutes a somewhat small sample by modern measures. The tradeoff between rich phenotyping and increased sample size is not trivial, particularly for studies of populations traditionally underrepresented in genetic research, a challenge that still plagues large studies like NHLBI TOPMed and NIH Million Veterans Program. Our approach of layering multiple types of genomic information serves as a partial workaround. However, by layering GWAS with admixture mapping and functional fine-mapping, we restricted our focus to regions where variants showed differential ancestry, which excluded our strongest GWAS hits. While PAINTOR can provide evidence of putatively causal markers that do not necessarily meet strict genome-wide thresholds of significance, it was not designed with admixed populations in mind. We applied suitably rigorous statistical stringency to both our GWAS and our admixture mapping results by thresholding to the effective number of independent tests as estimated by the CODA software, an approach designed to produce population-specific significance thresholds (Sobota et al., 2015), so we are confident that these regions are indeed correlated to the phenotype. Nevertheless, further research is needed to understand how the hierarchical Bayesian model in the PAINTOR inference engine behaves in regions of heterogeneous ancestry. Finally, if the truly causal variant is not genotyped, then PAINTOR and similar fine-mapping frameworks suffer a performance hit that is difficult to rectify without ultra-fine genotyping in the locus of interest (e.g., from whole genome sequencing). We suspect that our relatively low probabilities of causality can be attributed to a confluence of differential ancestry, small sample size, and insufficiently resolved fine-mapping, which could be improved in future studies by using whole-genome sequencing data instead of imputed genotypes.
Our integrative analysis approach leverages available functional annotations and genetic ancestry estimates in the absence of molecular data to yield some promise for discovery of novel loci. Our study is limited to three tiers—genotypes, genetic ancestry, and functional annotations—and makes use of gene expression results from GTEx v8. However, it does not directly incorporate any transcriptomic, metabolomic, proteomic, or methylomic information. As large multiomic data sets from NHLBI TOPMed, UK Biobank, and the NIH Million Veterans Program become available, the need for integrative genomic approaches to studying complex diseases will increase. Future multiomic models of complex diseases, including obstructive lung function disorders, may deliver on the promise of precision medicine and provide actionable clinical translation of biomedical and pharmacogenomic insights into novel therapies.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported in part by the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, the Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II, the National Heart, Lung, and Blood Institute (NHLBI) grants R01HL117004, R01HL128439, R01HL135156, X01HL134589, R01HL141992, R01HL104 608, R01HL141845, and U01HL138626, the National Human Genome Research Institute (NHGRI) grants U01HG007419 and U01HG009080, the National Institute of Environmental Health Sciences grants R01ES015794, R21ES24844, the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) grant R01HD085993, the National Institute on Minority Health and Health Disparities (NIMHD) grants P60MD006902, R01MD010443, and R56MD013312, and the Tobacco-Related Disease Research Program under Award Numbers 24RT-0025 and 27IR-0030. Research reported in this article was funded by the National Institutes of Health Common Fund and Office of Scientific Workforce Diversity under three linked awards RL5GM118984, TL4GM118986, 1UL1GM118985 administered by the National Institute of General Medical Sciences (NIGMS). The authors wish to acknowledge the following SAGE co-investigators for subject recruitment, sample processing and quality control: Luisa N. Borrell, DDS, PhD, Emerita Brigino-Buenaventura, MD, Adam Davis, MA, MPH, Michael A. LeNoir, MD, Kelley Meade, MD, Fred Lurmann, MS and Harold J. Farber, MD, MSPH. The authors also wish to thank the staff and participants who contributed to the SAGE study. This manuscript uses results and visualization provided by the Stanford Global Biobank Engine. The authors would like to thank the Rivas Lab for making the resource available. P. C. G. was additionally funded by NHGRI training grant T32HG000044 to the Department of Genetics at Stanford University. K. L. K. was additionally supported by NHLBI grant supplement R01HL135156-S1, the UCSF Bakar Computational Health Sciences Institute, the Gordon and Betty Moore Foundation grant GBMF3834, and the Alfred P. Sloan Foundation grant 2013-10-27 to UC Berkeley through the Moore-Sloan Data Sciences Environment initiative at the Berkeley Institute for Data Science (BIDS). The logistical space, technical support, administrative assistance, and indefatigable good humor of the members and staff at BIDS are gratefully acknowledged. E. Y. L., L. S. B., and A. K. L. were supported by a National Research Service Award grant T32GM007546 from the NIGMS. M. G. C. was additionally supported by NIH MARC U-STAR grant T34GM008574 at San Francisco State University. M. J. W. was additionally supported by NHLBI grant supplement R01HL117004-S1, an NIGMS Institutional Research and Academic Career Development Award K12GM081266, and an NHLBI Research Career Development Award K01HL140218.
Funding information
Gordon and Betty Moore Foundation, Grant/Award Number: GBMF3834; National Institute of Environmental Health Sciences, Grant/Award Numbers: R01ES015794, R21ES24844; Tobacco-Related Disease Research Program, Grant/Award Numbers: 24RT-0025, 27IR-0030; National Human Genome Research Institute, Grant/Award Numbers: T32HG000044, U01HG007419, U01HG009080; Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/Award Number: R01HD085993; National Institute on Minority Health and Health Disparities, Grant/Award Numbers: P60MD006902, R01MD010443, R56MD013312; National Heart, Lung, and Blood Institute, Grant/Award Numbers: K01HL140218, R01HL104608, R01HL117004, R01HL117004-S1, R01HL128439, R01HL135156, R01HL135156-S1, R01HL141845, R01HL141992, U01HL138626, X01HL134589; National Institute of General Medical Sciences, Grant/Award Numbers: 1UL1GM118985, K12GM081266, RL5GM118984, T32GM007546, TL4GM118986; Alfred P. Sloan Foundation, Grant/Award Number: 2013-10-27
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in the NCBI dbGaP repository under accession number phs000921.v1.p1.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- Alexander DH, Novembre J, & Lange K (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19(9), 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthma and Allergy Foundation of America. (2019). Asthma diagnosis. https://www.aafa.org/lung-function-tests-diagnose-asthma/
- Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, Lambourne JJ, Sivapalaratnam S, Downes K, Kundu K, Bomba L, Berentsen K, Bradley JR, Daugherty LC, Delaneau O, … Soranzo N (2016). The allelic landscape of human blood cell trait variation and links to common complex disease. Cell, 167(5), 1415–1429e19. 10.1016/j.cell.2016.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffi CW, Winnica DE, & Holguin F (2015). Asthma and obesity: Mechanisms and clinical implications. Asthma Research and Practice, 1, 1 10.1186/s40733-015-0001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharian S, Barakatt M, Gignoux CR, Shringarpure S, Errington J, Blot WJ, Bustamante CD, Kenny EE, Williams SM, Aldrich MC, & Gravel S (2016). The great migration and African-American genomic diversity. PLOS Genetics, 12(5), e1006059 10.1371/journal.pgen.1006059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow SE (2007). Expert committee recommendations regarding the prevention, assessment, and treatment of child and adolescent overweight and obesity: Summary report. Pediatrics, 120(Suppl 4), S164–S192. 10.1542/peds.2007-2329C [DOI] [PubMed] [Google Scholar]
- Bentley AR, Sung YJ, Brown MR, Winkler TW, Kraja AT, Ntalla I, Schwander K, Chasman DI, Lim E, Deng X, Guo X, Liu J, Lu Y, Cheng C-Y, Sim X, Vojinovic D, Huffman JE, Musani SK, Li C, … Cupples LA (2019). Multiancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nature Genetics, 51(4), 636–648. 10.1038/s41588-019-0378-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder RE, Mitchell CA, Schoenberg JB, & Bouhuys A (1976). Lung Function among Black and White children. American Review of Respiratory Disease, 114(5), 955–959. 10.1164/arrd.1976.114.5.955 [DOI] [PubMed] [Google Scholar]
- Blake K, Madabushi R, Derendorf H, & Lima J (2008). Population pharmacodynamic model of bronchodilator response to inhaled albuterol in children and adults with asthma. Chest, 134(5), 981–989. 10.1378/chest.07-2991 [DOI] [PubMed] [Google Scholar]
- Borrell LN, Nguyen EA, Roth LA, Oh SS, Tcheurekdjian H, Sen S, Davis A, Farber HJ, Avila PC, Brigino-Buenaventura E, LeNoir MA, Lurmann F, Meade K, Serebrisky D, Rodriguez-Cintron W, Kumar R, Rodriguez-Santana JR, Thyne SM, & Burchard EG (2013). Childhood obesity and asthma control in the GALA II and SAGE II studies. American Journal of Respiratory and Critical Care Medicine, 187(7), 697–702. 10.1164/rccm.201211-2116OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun L (2015). Race, ethnicity and lung function: A brief history. Canadian Journal of Respiratory Therapy, 51(4), 99–101. [PMC free article] [PubMed] [Google Scholar]
- Brehm JM, Man Tse S, Croteau-Chonka DC, Forno E, Litonjua AA, Raby BA, Chen W, Yan Q, Boutaoui N, Acosta-Pérez E, Avila L, Weiss ST, Soto-Quiros M, Cloutier MM, Hu D, Pino-Yanes M, Wenzel SE, Spear ML, Kolls JK, … Celedón JC (2015). A genome-wide association study of post-bronchodilator lung function in children with asthma. American Journal of Respiratory and Critical Care Medicine, 192(5), 634–637. 10.1164/rccm.201501-0047LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster LM, & Seedat YK (2013). Why do hypertensive patients of African ancestry respond better to calcium blockers and diuretics than to ACE inhibitors and β-adrenergic blockers? A systematic review. BMC Medicine, 11, 141 10.1186/1741-7015-11-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, Durand EY, Macpherson JM, Reich D, & Mountain JL (2015). The genetic ancestry of African Americans, Latinos, and European Americans across the United States. American Journal of Human Genetics, 96(1), 37–53. 10.1016/j.ajhg.2014.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, … Parkinson H (2019). The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Research, 47(D1), D1005–D1012. 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard EG (2014). Medical research: Missing patients. Nature News, 513(7518), 301–302. 10.1038/513301a [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Burchard EG, & De la Vega FM (2011). Genomics for the world. Nature, 475(7355), 163–165. 10.1038/475163a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, & Lee JJ (2015). Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience, 4(1), 7 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen SC, & Zuraw BL (2019). Treatment of hypertension in patients with asthma. New England Journal of Medicine, 381(11), 1046–1057. 10.1056/NEJMra1800345 [DOI] [PubMed] [Google Scholar]
- Conomos MP, Miller MB, & Thornton TA (2015). Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genetic Epidemiology, 39(4), 276–293. 10.1002/gepi.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos MP, Reiner AP, Weir BS, & Thornton TA (2016). Model-free estimation of recent genetic relatedness. The American Journal of Human Genetics, 98(1), 127–148. 10.1016/j.ajhg.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote AT, Harris KC, Panagiotopoulos C, Sandor GGS, & Devlin AM (2013). Childhood obesity and cardiovascular dysfunction. Journal of the American College of Cardiology, 62(15), 1309–1319. 10.1016/j.jacc.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, Cummins C, Davidson C, Dodiya KJ, Gall A, Girón CG, Gil L, Grego T, Haggerty L, Haskell E, … Flicek P (2019). Ensembl 2019. Nucleic Acids Research, 47(D1), D745–D751. 10.1093/nar/gky1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh P-R, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, … Fuchsberger C, (2016). Next-generation genotype imputation service and methods. Nature Genetics, 48, 1284–1287. 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL (2020). optparse: Command Line Option Parser. R package (Version 1.6.6) [Computer software]. https://CRAN.R-project.org/package=optparse
- Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmüller J, Ang W, Barr RG, Beaty TH, Becker AB, Beilby J, Bisgaard H, Bjornsdottir US, Bleecker E, Bønnelykke K, Boomsma DI, Bouzigon E, Brightling CE, Brossard M, Brusselle GG, … Nicolae DL (2018). Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nature Genetics, 50(1), 42–53. 10.1038/s41588-017-0014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowle M, & Srinivasan A (2020). data.table: Extension of data.frame [Computer software]. https://Rdatatable.gitlab.io/data.table
- Ehret GB, Ferreira T, Chasman DI, Jackson AU, Schmidt EM, Johnson T, Thorleifsson G, Luan J, Donnelly LA, Kanoni S, Petersen A-K, Pihur V, Strawbridge RJ, Shungin D, Hughes MF, Meirelles O, Kaakinen M, Bouatia-Naji N, Kristiansson K, … Munroe PB (2016). The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nature Genetics, 48(10), 1171–1184. 10.1038/ng.3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489(7414), 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, Ng FL, Evangelou M, Witkowska K, Tzanis E, Hellwege JN, Giri A, Velez Edwards DR, Sun YV, Cho K, … Million Veteran Program (2018). Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nature Genetics, 50(10), 1412–1425. 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA-K, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, … Uhlén M (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & Cellular Proteomics, 13(2), 397–406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, Helmer Q, Tillander A, Ullemar V, van Dongen J, Lu Y, Rüschendorf F, Esparza-Gordillo J, Medway CW, Mountjoy E, Burrows K, Hummel O, Grosche S, Brumpton BM, … Paternoster L (2017). Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nature Genetics, 49(12), 1752–1757. 10.1038/ng.3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MAR, Mathur R, Vonk JM, Szwajda A, Brumpton B, Granell R, Brew BK, Ullemar V, Lu Y, & Jiang Y, 23 and Me Research Team, eQTLGen Consortium, BIOS Consortium, Magnusson PKE, Karlsson R, Hinds DA, Paternoster L, Koppelman GH, & Almqvist C (2019). Genetic architectures of childhood- and adult-onset asthma are partly distinct. The American Journal of Human Genetics, 104(4), 665–684. 10.1016/j.ajhg.2019.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, Loyd JE, Cosgrove GP, Lynch D, Groshong S, Collard HR, Wolters PJ, Bradford WZ, Kossen K, Seiwert SD, du Bois RM, Garcia CK, Devine MS, Gudmundsson G, … Schwartz DA (2013). Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nature Genetics, 45(6), 613–620. 10.1038/ng.2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Féjerman L, Chen GK, Eng C, Huntsman S, Hu D, Williams A, Pasaniuc B, John EM, Via M, Gignoux C, Ingles S, Monroe KR, Kolonel LN, Torres-Mejía G, Pérez-Stable EJ, Burchard EG, Henderson BE, Haiman CA, & Ziv E (2012). Admixture mapping identifies a locus on 6q25 associated with breast cancer risk in US Latinas. Human Molecular Genetics, 21(8), 1907–1917. 10.1093/hmg/ddr617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, Torstenson ES, Kovesdy CP, Sun YV, Wilson OD, Robinson-Cohen C, Roumie CL, Chung CP, Birdwell KA, Damrauer SM, DuVall SL, Klarin D, Cho K, Wang Y, … Edwards TL (2019). Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nature Genetics, 51(1), 51–62. 10.1038/s41588-018-0303-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glindmeyer HW, Lefante JJ, McColloster C, Jones RN, & Weill H (1995). Blue-collar normative spirometric values for Caucasian and African-American men and women aged 18 to 65. American Journal of Respiratory and Critical Care Medicine, 151(2 Pt 1), 412–422. 10.1164/ajrccm.151.2.7842200 [DOI] [PubMed] [Google Scholar]
- Gogarten SM, Sofer T, Chen H, Yu C, Brody JA, Thornton TA, Rice KM, & Conomos MP (2019). Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics, 10.1093/bioinformatics/btz567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruchała-Niedoszytko M, Niedoszytko M, Sanjabi B, van der Vlies P, Niedoszytko P, Jassem E, & Małgorzewicz S (2015). Analysis of the differences in whole-genome expression related to asthma and obesity. Polskie Archiwum Medycyny Wewnetrznej, 125(10), 722–730. 10.20452/pamw.3109 [DOI] [PubMed] [Google Scholar]
- Hankinson JL, Odencrantz JR, & Fedan KB (1999). Spirometric reference values from a sample of the general U.S. population. American Journal of Respiratory and Critical Care Medicine, 159(1), 179–187. 10.1164/ajrccm.159.1.9712108 [DOI] [PubMed] [Google Scholar]
- Hardin M, Cho MH, McDonald M-L, Wan E, Lomas DA, Coxson HO, MacNee W, Vestbo J, Yates JC, Agusti A, Calverley PMA, Celli B, Crim C, Rennard S, Wouters E, Bakke P, Bhatt SP, Kim V, Ramsdell J, … COPDGene Investigators—clinical centers (2016). A genome-wide analysis of the response to inhaled β2-agonists in chronic obstructive pulmonary disease. The Pharmacogenomics Journal, 16(4), 326–335. 10.1038/tpj.2015.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harik-Khan RI, Muller DC, & Wise RA (2004). Racial difference in lung function in African-American and White children: Effect of anthropometric, socioeconomic, nutritional, and environmental factors. American Journal of Epidemiology, 160(9), 893–900. 10.1093/aje/kwh297 [DOI] [PubMed] [Google Scholar]
- Heit JA, Armasu SM, McCauley BM, Kullo IJ, Sicotte H, Pathak J, Chute CG, Gottesman O, Bottinger EP, Denny JC, Roden DM, Li R, Ritchie MD, & de Andrade M (2017). Identification of unique venous thromboembolism-susceptibility variants in African-Americans. Thrombosis and Haemostasis, 117(4), 758–768. 10.1160/TH16-08-0652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez W, Gamazon ER, Smithberger E, O’Brien TJ, Harralson AF, Tuck M, Barbour A, Kittles RA, Cavallari LH, & Perera MA (2016). Novel genetic predictors of venous thromboembolism risk in African Americans. Blood, 127(15), 1923–1929. 10.1182/blood-2015-09-668525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, Hillman-Jackson J, Kuhn RM, Pedersen JS, Pohl A, Raney BJ, Rosenbloom KR, Siepel A, Smith KE, Sugnet CW, … Kent WJ (2006). The UCSC genome browser database: Update 2006. Nucleic Acids Research, 34(Database issue), D590–D598. 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsi BP, Hsu KHK, & Jenkins DE (1983). Ventilatory functions of normal children and young adults: Mexican-American, white, and black. III. Sitting height as a predictor. The Journal of Pediatrics, 102(6), 860–865. 10.1016/S0022-3476(83)80012-2 [DOI] [PubMed] [Google Scholar]
- Johansson Å, Rask-Andersen M, Karlsson T, & Ek WE (2019). Genome-wide association analysis of 350 000 Caucasians from the UK Biobank identifies novel loci for asthma, hay fever and eczema. Human Molecular Genetics, 28(23), 4022–4041. 10.1093/hmg/ddz175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey John. (1977). Exploratory data analysis. Addison-Wesley. [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, … Genome Aggregation Database Consortium. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581(7809), 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G (2017). PAINTOR v3.0. https://github.com/gkichaev/PAINTOR_V3.0
- Kichaev G, Bhatia G, Loh P-R, Gazal S, Burch K, Freund MK, Schoech A, Pasaniuc B, & Price AL (2019). Leveraging polygenic functional enrichment to improve GWAS power. American Journal of Human Genetics, 104(1), 65–75. 10.1016/j.ajhg.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G, Yang W-Y, Lindstrom S, Hormozdiari F, Eskin E, Price AL, Kraft P, & Pasaniuc B (2014). Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLOS Genetics, 10(10), e1004722 10.1371/journal.pgen.1004722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, Gagnon DR, DuVall SL, Li J, Peloso GM, Chaffin M, Small AM, Huang J, Tang H, Lynch JA, Ho Y-L, Liu DJ, Emdin CA, Li AH, … Assimes TL (2018). Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nature Genetics, 50(11), 1514–1523. 10.1038/s41588-018-0222-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Seibold MA, Aldrich MC, Williams LK, Reiner AP, Colangelo L, Galanter J, Gignoux C, Hu D, Sen S, Choudhry S, Peterson EL, Rodriguez-Santana J, Rodriguez-Cintron W, Nalls MA, Leak TS, O’Meara E, Meibohm B, Kritchevsky SB, … Burchard EG (2010). Genetic ancestry in lung-function predictions. The New England Journal of Medicine, 363(4), 321–330. 10.1056/NEJMoa0907897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer VA, Tiwari HK, Reynolds RJ, Danila MI, Wang J, Edberg JC, Kimberly RP, Kottyan LC, Harley JB, Mikuls TR, Gregersen PK, Absher DM, Langefeld CD, Arnett DK, & Bridges SL (2019). Genetic influences on susceptibility to rheumatoid arthritis in African-Americans. Human Molecular Genetics, 28(5), 858–874. 10.1093/hmg/ddy395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard CJ, Li H, Adrianto I, Ice JA, Rasmussen A, Grundahl KM, Kelly JA, Dozmorov MG, Miceli-Richard C, Bowman S, Lester S, Eriksson P, Eloranta M-L, Brun JG, Gøransson LG, Harboe E, Guthridge JM, Kaufman KM, Kvarnström M, … Sivils KL (2013). Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren’s syndrome. Nature Genetics, 45(11), 1284–1292. 10.1038/ng.2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, & Price AL (2016). Reference-based phasing using the Haplotype Reference Consortium panel. Nature Genetics, 48(11), 1443–1448. 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz SM, Cho MH, Young K, Hersh CP, Castaldi PJ, McDonald M-L, Regan E, Mattheisen M, DeMeo DL, Parker M, Foreman M, Make BJ, Jensen RL, Casaburi R, Lomas DA, Bhatt SP, Bakke P, Gulsvik A, Crapo JD, … COPDGene Investigators (2015). A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genetics, 16, 138 10.1186/s12863-015-0299-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, Cook JP, Schmidt EM, Wuttke M, Sarnowski C, Mägi R, Nano J, Gieger C, Trompet S, Lecoeur C, … McCarthy MI (2018). Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nature Genetics, 50(11), 1505–1513. 10.1038/s41588-018-0241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak ACY, White MJ, Eckalbar WL, Szpiech ZA, Oh SS, Pino-Yanes M, Hu D, Goddard P, Huntsman S, Galanter J, Wu AC, Himes BE, Germer S, Vogel JM, Bunting KL, Eng C, Salazar S, Keys KL, Liberto J, … NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium (2018). Whole-genome sequencing of pharmacogenetic drug response in racially diverse children with asthma. American Journal of Respiratory and Critical Care Medicine, 197(12), 1552–1564. 10.1164/rccm.201712-2529OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples BK, Gravel S, Kenny EE, & Bustamante CD (2013). RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. American Journal of Human Genetics, 93(2), 278–288. 10.1016/j.ajhg.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Hernández E, Rodríguez-García ME, Camacho A, Matilla-Dueñas A, García-Silva MT, Quijada-Fraile P, Corral-Juan M, Tejada-Palacios P, de Las Heras RS, Arenas J, Martín MA, & Martínez-Azorín F (2016). New ATP8A2 gene mutations associated with a novel syndrome: Encephalopathy, intellectual disability, severe hypotonia, chorea and optic atrophy. Neurogenetics, 17(4), 259–263. 10.1007/s10048-016-0496-y [DOI] [PubMed] [Google Scholar]
- McInnes G, Tanigawa Y, DeBoever C, Lavertu A, Olivieri JE, Aguirre M, & Rivas MA (2019). Global Biobank Engine: Enabling genotype-phenotype browsing for biobank summary statistics. Bioinformatics, 35(14), 2495–2497. 10.1093/bioinformatics/bty999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Microsoft Corporation & Weston S (2019). doParallel: Foreach Parallel Adaptor for the ‘parallel’ Package. R package (Version 1.0.15) [Computer Software]. https://CRAN.R-project.org/package=doParallel
- Miller MR, Crapo R, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, Enright P, van der Grinten CPM, Gustafsson P, Jensen R, Johnson DC, MacIntyre N, McKay R, Navajas D, Pedersen OF, Pellegrino R, Viegi G, Wanger J, & ATS/ERS Task Force (2005). General considerations for lung function testing. The European Respiratory Journal, 26(1), 153–161. 10.1183/09031936.05.00034505 [DOI] [PubMed] [Google Scholar]
- Nishimura KK, Galanter JM, Roth LA, Oh SS, Thakur N, Nguyen EA, Thyne S, Farber HJ, Serebrisky D, Kumar R, Brigino-Buenaventura E, Davis A, LeNoir MA, Meade K, Rodriguez-Cintron W, Avila PC, Borrell LN, Bibbins-Domingo K, Rodriguez-Santana JR, … Burchard EG (2013). Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. American Journal of Respiratory and Critical Care Medicine, 188(3), 309–318. 10.1164/rccm.201302-0264OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmagambetov T, Kuwahara R, & Garbe P (2018). The economic burden of asthma in the United States, 2008–2013. Annals of the American Thoracic Society, 15(3), 348–356. 10.1513/AnnalsATS.201703-259OC [DOI] [PubMed] [Google Scholar]
- Ober C, & Yao T-C (2011). The genetics of asthma and allergic disease: A 21st century perspective. Immunological Reviews, 242(1), 10–30. 10.1111/j.1600-065X.2011.01029.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, & Reich D (2006). Population structure and eigenanalysis. PLOS Genetics, 2(12), e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino R, Viegi G, Brusasco V, Crapo RO, Burgos F, Casaburi R, Coates A, van der Grinten CPM, Gustafsson P, Hankinson J, Jensen R, Johnson DC, MacIntyre N, McKay R, Miller MR, Navajas D, Pedersen OF, & Wanger J (2005). Interpretative strategies for lung function tests. The European Respiratory Journal, 26(5), 948–968. 10.1183/09031936.05.00035205 [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, & Hinds DA (2016). Detection and interpretation of shared genetic influences on 42 human traits. Nature Genetics, 48(7), 709–717. 10.1038/ng.3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino-Yanes M, Thakur N, Gignoux CR, Galanter JM, Roth LA, Eng C, Nishimura KK, Oh SS, Vora H, Huntsman S, Nguyen EA, Hu D, Drake KA, Conti DV, Moreno-Estrada A, Sandoval K, Winkler CA, Borrell LN, Lurmann F, … Burchard EG (2015). Genetic ancestry influences asthma susceptibility and lung function among Latinos. The Journal of Allergy and Clinical Immunology, 135(1), 228–235. 10.1016/j.jaci.2014.07.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pividori M, Schoettler N, Nicolae DL, Ober C, & Im HK (2019). Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: Genome-wide and transcriptome-wide studies. The Lancet. Respiratory Medicine, 7(6), 509–522. 10.1016/S2213-2600(19)30055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer M, Best N, Cowles K, & Vines K (2006). CODA: Convergence diagnosis and output analysis for MCMC. R News, 6(1), 7–11. [Google Scholar]
- Popejoy AB, & Fullerton SM (2016). Genomics is failing on diversity. Nature, 538(7624), 161–164. 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, Yengo L, Ferreira T, Marouli E, Ji Y, Yang J, Jones S, Beaumont R, Croteau-Chonka DC, Winkler TW, Consortium G, Hattersley AT, Loos RJF, Hirschhorn JN, … Lindgren CM (2019). Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Human Molecular Genetics, 28(1), 166–174. 10.1093/hmg/ddy327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quanjer PH (2013). Lung function, race and ethnicity: A conundrum. European Respiratory Journal, 41(6), 1249–1251. 10.1183/09031936.00053913 [DOI] [PubMed] [Google Scholar]
- Quanjer PH (2015). Lung function, genetics and socioeconomic conditions. The European Respiratory Journal, 45(6), 1529–1533. 10.1183/09031936.00053115 [DOI] [PubMed] [Google Scholar]
- Risch N, & Merikangas K (1996). The future of genetic studies of complex human diseases. Science, 273(5281), 1516–1517. 10.1126/science.273.5281.1516 [DOI] [PubMed] [Google Scholar]
- Ro, admap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, … Kellis M (2015). Integrative analysis of 111 reference human epigenomes. Nature, 518(7539), 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij FJA, Qayyum R, Smith AV, Zhou Y, Trompet S, Tanaka T, Keller MF, Chang L-C, Schmidt H, Yang M-L, Chen M-H, Hayes J, Johnson AD, Yanek LR, Mueller C, Lange L, Floyd JS, Ghanbari M, Zonderman AB, … Ganesh SK (2017). Genome-wide trans-ethnic meta-analysis identifies seven genetic loci influencing erythrocyte traits and a role for RBPMS in erythropoiesis. American Journal of Human Genetics, 100(1), 51–63. 10.1016/j.ajhg.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossiter CE, & Weill H (1974). Ethnic differences in lung function: Evidence for proportional differences. International Journal of Epidemiology, 3(1), 55–61. 10.1093/ije/3.1.55 [DOI] [PubMed] [Google Scholar]
- Rüeger S, McDaid A, & Kutalik Z (2018). Evaluation and application of summary statistic imputation to discover new height-associated loci. PLOS Genetics, 14(5), e1007371 10.1371/journal.pgen.1007371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J, Katz SA, Fegley RW, & Tockman MS (1988). Sex and race differences in the development of lung function. American Review of Respiratory Disease, 138(6), 1415–1421. 10.1164/ajrccm/138.6.1415 [DOI] [PubMed] [Google Scholar]
- Shahin MH, Conrado DJ, Gonzalez D, Gong Y, Lobmeyer MT, Beitelshees AL, Boerwinkle E, Gums JG, Chapman A, Turner ST, Cooper-DeHoff RM, & Johnson JA (2018). Genome-wide association approach identified novel genetic predictors of heart rate response to β-blockers. Journal of the American Heart Association, 7(5), 10.1161/JAHA.117.006463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D (2013). Overview of admixture mapping. Current Protocols in Human Genetics, 76(1), 1.23.1–1.23.8. 10.1002/0471142905.hg0123s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung H-K, Naranjo S, Acemel RD, … Nóbrega MA (2014). Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature, 507(7492), 371–375. 10.1038/nature13138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobota RS, Shriner D, Kodaman N, Goodloe R, Zheng W, Gao Y-T, Edwards TL, Amos CI, & Williams SM (2015). Addressing population-specific multiple testing burdens in genetic association studies, Annals of Human Genetics (79, pp. 136–147. 2 10.1111/ahg.12095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear ML, Hu D, Pino-Yanes M, Huntsman S, Eng C, Levin AM, Ortega VE, White MJ, McGarry ME, Thakur N, Galanter J, Mak ACY, Oh SS, Ampleford E, Peters SP, Davis A, Kumar R, Farber HJ, Meade K, … Burchard EG (2019). A genome-wide association and admixture mapping study of bronchodilator drug response in African Americans with asthma. The Pharmacogenomics Journal, 19(3), 249–259. 10.1038/s41397-018-0042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, DeLisle RK, Gold L, Pezer M, Lauc G, El-Din Selim MA, Mook-Kanamori DO, Al-Dous EK, Mohamoud YA, Malek J, Strauch K, … Graumann J (2017). Connecting genetic risk to disease end points through the human blood plasma proteome. Nature Communications, 8, 14357 10.1038/ncomms14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange O (2018). GNU Parallel 2018. Ole Tange. 10.5281/zenodo.1146014 [DOI] [Google Scholar]
- Thakur N, Oh SS, Nguyen EA, Martin M, Roth LA, Galanter J, Gignoux CR, Eng C, Davis A, Meade K, LeNoir MA, Avila PC, Farber HJ, Serebrisky D, Brigino-Buenaventura E, Rodriguez-Cintron W, Kumar R, Williams LK, Bibbins-Domingo K, … Burchard EG (2013). Socioeconomic status and childhood asthma in urban minority youths. The GALA II and SAGE II studies. American Journal of Respiratory and Critical Care Medicine, 188(10), 1202–1209. 10.1164/rccm.201306-1016OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. (2015). A global reference for human genetic variation. Nature, 526(7571), 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Global Asthma Network. (2018). The Global Asthma Report 2018 http://www.globalasthmareport.org/foreword/summaries.php [Google Scholar]
- The International HapMap 3 Consortium. (2010). Integrating common and rare genetic variation in diverse human populations. Nature, 467(7311), 52–58. 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Ellinghaus E, Stuart PE, Capon F, Knight J, Tejasvi T, Kang HM, Allen MH, Lambert S, Stoll SW, Weidinger S, Gudjonsson JE, Koks S, Kingo K, Esko T, Das S, Metspalu A, Weichenthal M, … Elder JT (2015). Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nature Communications, 6, 7001 10.1038/ncomms8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanger J, Clausen JL, Coates A, Pedersen OF, Brusasco V, Burgos F, Casaburi R, Crapo R, Enright P, van der Grinten CPM, Gustafsson P, Hankinson J, Jensen R, Johnson D, Macintyre N, McKay R, Miller MR, Navajas D, Pellegrino R, & Viegi G (2005). Standardisation of the measurement of lung volumes. The European Respiratory Journal, 26(3), 511–522. 10.1183/09031936.05.00035005 [DOI] [PubMed] [Google Scholar]
- Whelton P.aulK, Carey Robert M, Aronow Wilbert S, Casey Donald E, Collins Karen J, Dennison Himmelfarb Cheryl, DePalma Sondra M, Gidding S.amuel, Jamerson Kenneth A, Jones Daniel W, MacLaughlin E.ricJ., Paul M.untner, Bruce O.vbiagele, Smith Sidney C, Spencer Crystal C, Stafford Randall S, Taler Sandra J, Thomas Randal J, Williams Kim A, … Wright Jackson T (2018). 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: A report of the american college of cardiology/american heart association task force on clinical practice guidelines. Hypertension, 71(6), e13–e115. 10.1161/HYP.0000000000000065 [DOI] [PubMed] [Google Scholar]
- White MJ, Risse-Adams O, Goddard P, Contreras MG, Adams J, Hu D, Eng C, Oh SS, Davis A, Meade K, Brigino-Buenaventura E, LeNoir MA, Bibbins-Domingo K, Pino-Yanes M, & Burchard EG (2016). Novel genetic risk factors for asthma in African American children: Precision medicine and the SAGE II Study. Immunogenetics, 68(6–7), 391–400. 10.1007/s00251-016-0914-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock EP, Williams SB, Gold R, Smith PR, & Shipman SA (2005). Screening and interventions for childhood overweight: A summary of evidence for the US Preventive Services Task Force. Pediatrics, 116(1), e125–e144. 10.1542/peds.2005-0242 [DOI] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant graphics for data analysis. Springer-Verlag; http://ggplot2.org [Google Scholar]
- Wickham H, & Garrett G (2017). R for Data Science. O’Reilly Media, Inc. https://r4ds.had.co.nz/ [Google Scholar]
- Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, Highland HM, Patel YM, Sorokin EP, Avery CL, Belbin GM, Bien SA, Cheng I, Cullina S, Hodonsky CJ, Hu Y, Huckins LM, Jeff J, Justice AE, … Carlson CS (2019). Genetic analyses of diverse populations improves discovery for complex traits. Nature, 570(7762), 514–518. 10.1038/s41586-019-1310-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss AB, Sofer T, Lee MK, Terzikhan N, Nguyen JN, Lahousse L, Latourelle JC, Smith AV, Bartz TM, Feitosa MF, Gao W, Ahluwalia TS, Tang W, Oldmeadow C, Duan Q, de Jong K, Wojczynski MK, Wang X-Q, Noordam R, … London SJ (2018). Multiethnic meta-analysis identifies ancestry-specific and cross-ancestry loci for pulmonary function. Nature Communications, 9(1), 1–15. 10.1038/s41467-018-05369-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, & Visscher PM (2011). GCTA: A tool for genome-wide complex trait analysis. American Journal of Human Genetics, 88(1), 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zaitlen NA, Goddard ME, Visscher PM, & Price AL (2014). Advantages and pitfalls in the application of mixed-model association methods. Nature Genetics, 46(2), 100–106. 10.1038/ng.2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yao S, Iliopoulou BP, Han X, Augustine MM, Xu H, Phennicie RT, Flies SJ, Broadwater M, Ruff W, Taube JM, Zheng L, Luo L, Zhu G, Chen J, & Chen L (2013). B7–H5 costimulates human T cells via CD28H. Nature Communications, 4, 2043 10.1038/ncomms3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Lee PH, Chaffin MD, Chung W, Loh P-R, Lu Q, Christiani DC, & Liang L (2018). A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nature Genetics, 50(6), 857–864. 10.1038/s41588-018-0121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


