Abstract
Genotyping for CYP2C19 no function alleles to guide antiplatelet therapy after percutaneous coronary intervention (PCI) improves clinical outcomes. Although results for the increased function CYP2C19*17 allele are also reported, its clinical relevance in this setting remains unclear. A collaboration across nine sites examined antiplatelet therapy prescribing and clinical outcomes in 3,342 patients after implementation of CYP2C19-guided antiplatelet therapy. Risk of major atherothrombotic and bleeding events over 12 months after PCI were compared across CYP2C19 metabolizer phenotype and antiplatelet therapy groups by proportional hazards regression. Clopidogrel was prescribed to a similar proportion of CYP2C19 normal (84.5%), rapid (82.9%) and ultrarapid metabolizers (80.6%) (P=0.360). Clopidogrel-treated normal metabolizers (20.4 events/100 patient-years; adjusted hazard ratio [HR] 1.00, 95% confidence interval [CI] 0.75–1.33, P=0.993) and clopidogrel-treated rapid or ultrarapid metabolizers (19.1 events/100 patient-years; adjusted HR 0.95, 95% CI 0.69–1.30, P=0.734) exhibited no difference in major atherothrombotic events compared to patients treated with prasugrel or ticagrelor (17.6 events/100 patient-years). In contrast, clopidogrel-treated intermediate and poor metabolizers exhibited significantly higher atherothrombotic event risk compared to prasugrel/ticagrelor-treated patients (adjusted HR 1.56, 95% CI 1.12–2.16, P=0.008). When comparing clopidogrel-treated rapid or ultrarapid metabolizers to normal metabolizers, no difference in atherothrombotic (adjusted HR 0.97, 95% CI 0.73–1.29, P=0.808) or bleeding events (adjusted HR 1.34, 95% CI 0.83–2.17, P=0.224) were observed. In a real-world setting of genotype-guided antiplatelet therapy, the CYP2C19*17 allele did not significantly impact post-PCI prescribing decisions or clinical outcomes. These results suggest the CYP2C19 *1/*17 and *17/*17 genotypes have limited clinical utility to guide antiplatelet therapy after PCI.
Keywords: clopidogrel, CYP2C19, pharmacogenomics, ultrarapid metabolizer, percutaneous coronary intervention, cardiovascular events, antiplatelet therapy
INTRODUCTION
Approximately 480,000 percutaneous coronary interventions (PCIs) are performed annually in the United States (U.S.).1 Dual antiplatelet therapy with aspirin and a P2Y12 inhibitor (clopidogrel, prasugrel, or ticagrelor) is indicated following PCI to reduce the risk of major atherothrombotic events.2
Clopidogrel, a prodrug requiring bioactivation by the CYP2C19 enzyme, remains the most widely prescribed P2Y12 inhibitor.3 CYP2C19 no function alleles, most notably *2, are common and significantly impair clopidogrel active metabolite formation, platelet inhibition, and clinical effectiveness after PCI.4,5 In contrast, CYP2C19 genotype does not impact the clinical effects of prasugrel or ticagrelor,6,7 which exhibit more consistent antiplatelet effects, superior efficacy in acute coronary syndrome (ACS) patients in the absence of CYP2C19 genotyping, and higher bleeding risk compared to clopidogrel.8,9 Accumulating evidence has demonstrated that genotyping and prescribing prasugrel or ticagrelor in CYP2C19 no function allele carriers after PCI improves clinical outcomes.10–13
The increased function CYP2C19*17 allele (rs12248560) is carried by approximately 30% of the U.S. population, with individuals carrying one *17 allele and one normal function allele (*1/*17: rapid metabolizers, RM) or two *17 allele copies (*17/*17: ultrarapid metabolizers, UM) exhibiting increased CYP2C19 expression and metabolic function compared to normal metabolizers (NM).5,14 The CYP2C19*17 allele has been associated with increased active metabolite formation, enhanced inhibition of platelet aggregation, higher bleeding risk, and lower risk for ischemic events with clopidogrel.15–19 However, other studies have reported no significant association between the *17 allele and clopidogrel pharmacokinetics, pharmacodynamics, and ischemic and bleeding outcomes after accounting for the *2 allele.7,20–22 It has been postulated that the observed relationship between the *17 allele and clopidogrel’s antiplatelet effects may be due to absence of the no function *2 allele rather than presence of the increased function *17 allele.5,20 Thus, the impact of the CYP2C19*17 allele on clopidogrel clinical effectiveness and safety remains unclear.
Multiple institutions have successfully implemented genotype-guided antiplatelet therapy into clinical workflows.23,24 While *17 allele results are typically reported in addition to no function alleles on clinically implemented CYP2C19 tests,25 distinction of CYP2C19 UMs and RMs from NMs to guide antiplatelet therapy prescribing decisions is not currently recommended due to insufficient evidence.5 The primary objective of this study was to investigate the impact of the CYP2C19 UM or RM phenotypes on antiplatelet therapy selection and clinical outcomes in PCI patients who underwent CYP2C19 genotyping in real-world clinical settings.
METHODS
Study Design and Population.
This was an expansion of a previously described multi-center investigation of CYP2C19 genotype-guided antiplatelet therapy post-PCI.10 Seven institutions (University of Florida, Gainesville; University of North Carolina, Chapel Hill; University of Maryland, Baltimore; University of Alabama, Birmingham; University of Pittsburgh; University of Illinois, Chicago; and Indiana University) contributed data for 1,815 patients for the original analysis. The expanded dataset includes additional data for 1,002 patients from the original sites plus data for 525 patients from two additional sites (University of Pennsylvania and University of Florida, Jacksonville, as described26,27). All sites had implemented clinical CYP2C19 testing with genotype results returned via the electronic health record (EHR) for consideration during antiplatelet therapy prescribing, and participated in the NIH-funded Implementing Genomics in Practice (IGNITE) Network Pharmacogenetics Working Group.23,28 Data collection was approved by the Institutional Review Board at each site.
The study design was pragmatic, such that delivery of the genotype intervention was part of clinical care, the ultimate decision to order genetic testing and choice of drug therapy was left to the discretion of the prescriber, and clinically meaningful outcome data were collected from the EHR.29 All patients from each site ≥18 years of age who underwent PCI, CYP2C19 genotyping, and received a P2Y12 inhibitor (clopidogrel, prasugrel, or ticagrelor) after PCI were included, regardless of length of follow-up. A total of 3,342 patients across the nine institutions met these criteria and were included in the analysis.
CYP2C19 Genotyping and Phenotyping.
Genotyping was performed at each institution in a Clinical Laboratory Improvement Amendments (CLIA)-licensed laboratory. All sites genotyped for the CYP2C19*2 and *3 no function alleles and the *17 increased function allele, with additional rare alleles genotyped at five institutions (TABLE S1). As summarized in TABLE S2, CYP2C19 metabolizer phenotype was assigned according to standardized Clinical Pharmacogenetics Implementation Consortium (CPIC) recommendations: UM (two increased function alleles); RM (one increased and one normal function allele); NM (absence of any tested increased or no function allele), intermediate metabolizer (IM, one no function allele), or poor metabolizer (PM, two no function alleles).30 Notably, patients with one no function and one increased function allele (e.g., *2/*17) were classified as IMs. Alternative antiplatelet therapy, consisting of prasugrel or ticagrelor in the absence of contraindications, was recommended over clopidogrel for IMs and PMs in accordance with CPIC recommendations.5 In NMs, RMs and UMs, antiplatelet therapy selection was left to the discretion of the prescriber.
Data Abstraction and Endpoints.
Demographic, clinical, genotype, and medication data were manually abstracted from the EHR at each site through review of patient encounters, including the index PCI hospitalization and subsequent hospitalizations and outpatient visits, using a common data collection form, as described.10 The index PCI (baseline) was defined as the PCI performed in association with CYP2C19 genotyping.
The occurrence of cardiovascular and bleeding clinical events for up to 12 months following the index PCI (baseline) were determined. Events were identified based on physician-reported diagnoses abstracted from the cardiac catheterization laboratory report, hospital discharge summary notes, or outpatient clinical notes of the EHR. Antiplatelet therapy and dose at the time of each event, and at the last follow-up within 12 months, was documented. The primary outcome was major atherothrombotic events, defined as the composite of death, myocardial infarction (MI), stent thrombosis, ischemic stroke, or hospitalization for unstable angina, consistent with a recent clinical trial of genotype-guided antiplatelet therapy.31 The secondary outcome was clinically significant bleeding events, defined as Global Use of Strategies to Open Occluded Arteries (GUSTO) moderate (requiring blood transfusion but not resulting in hemodynamic compromise) or severe/life-threatening (intracerebral hemorrhage or bleeding resulting in hemodynamic compromise requiring treatment).32
Statistical Analysis.
Data were curated and aggregated at the University of Florida, Gainesville. Continuous data are presented as mean±standard deviation or median (interquartile range), and categorical data are presented as count (%), unless otherwise indicated. Antiplatelet therapy was dichotomized (clopidogrel or prasugrel/ticagrelor) for analysis. Due to the small number of UMs and PMs, CYP2C19 metabolizer phenotype was categorized into three groups for the primary analysis: UM or RM, NM, and IM or PM. Baseline demographic and clinical factors were compared across CYP2C19-antiplatelet therapy groups using Student’s t-test, chi-square, or Fisher’s exact test.
The time to occurrence of an atherothrombotic or bleeding event within 12 months after the index PCI (baseline) was calculated in each patient. Patients who did not experience an event were censored at the time of last encounter in which P2Y12 inhibitor treatment was documented. Event rates were reported as the number of events per 100 patient-years of follow-up. The relationship between P2Y12 inhibitor therapy, CYP2C19 metabolizer phenotype, and time to occurrence of the primary and secondary clinical outcomes was evaluated by Cox proportional hazards regression, as described.10,33 Kaplan-Meier curves were generated, and the log-rank test was used to compare the cumulative risk of an event across groups.
In order to examine the impact of CYP2C19 metabolizer status on clopidogrel clinical effectiveness relative to alternative antiplatelet therapy, the primary analysis compared outcomes across four CYP2C19-antiplatelet therapy groups: clopidogrel-UM or RM, clopidogrel-NM, clopidogrel-IM or PM, and prasugrel or ticagrelor (regardless of CYP2C19 phenotype). Because of the known absence of an association between CYP2C19 metabolizer status and either prasugrel or ticagrelor clinical effectiveness,6,7 and guideline recommendations for use of prasugrel or ticagrelor as first-line therapy in ACS patients undergoing PCI,2 all patients receiving prasugrel or ticagrelor were combined into a single alternative therapy reference group (alternative-combined). Secondary analyses evaluated outcomes across CYP2C19 metabolizer status within the stratum of patients treated with clopidogrel. Clopidogrel-treated NMs served as the reference group and were initially compared to clopidogrel-treated UM/RMs and clopidogrel-treated IM/PMs. An additional analysis compared outcomes across clopidogrel-treated UMs (*17/*17), RMs (e.g., *1/*17), NMs (*1/*1), IMs carrying the CYP2C19*17 allele (e.g., *2/*17), IMs not carrying the *17 allele (e.g., *1/*2), and PMs (e.g., *2/*2).
Because of the nonrandomized design, analyses were completed after adjusting for age, gender, race, and baseline covariates that differed across CYP2C19-antiplatelet groups. Covariate adjusted hazard ratio (HR) and 95% confidence intervals (CIs) for each between-group comparison were calculated. Multivariable models were created by selecting candidate covariates, described in TABLE 1, with the criterion of P<0.10 to enter the model. In the primary analysis, covariates included in the adjusted model were age (continuous), gender, race (white, black, or other), body mass index (continuous), ACS indication for PCI, drug-eluting stent at index PCI, diabetes, hypertension, chronic kidney disease (defined as an estimated glomerular filtration rate <60 ml/min), peripheral vascular disease, atrial fibrillation, heart failure, current cancer, prior stent, prior stroke or transient ischemic attack (TIA), prior gastrointestinal or intracerebral hemorrhage, and statin at discharge. In the secondary clopidogrel stratum analysis, covariates included in the adjusted model were age, gender, race, ACS indication for PCI, chronic kidney disease, peripheral vascular disease, prior MI, prior stent, prior stroke or TIA, prior gastrointestinal or intracerebral hemorrhage, and beta-blocker at discharge.
Table 1.
Study Population Characteristics at the Time of Index PCI
| All Patients | Alternative (combined)* | Clopidogrel UM or RM | Clopidogrel NM | Clopidogrel IM or PM | P-valu^ | P-value# | |
|---|---|---|---|---|---|---|---|
| N | 3342 | 933 | 810 | 1123 | 476 | ||
| Age (years) | 63 ± 12 | 60 ± 11 | 64 ± 12 | 64 ± 12 | 64 ± 12 | <0.001 | 0.636 |
| Age ≥ 70 | 957 (29) | 176 (19) | 252 (31) | 378 (34) | 151 (32) | <0.001 | 0.466 |
| Female | 1075 (32) | 250 (27) | 289 (36) | 368 (33) | 168 (35) | <0.001 | 0.357 |
| Race | |||||||
| White | 2448 (73) | 692 (74) | 603 (74) | 830 (74) | 323 (68) | 0.007 | 0.003 |
| Black | 659 (20) | 171 (18) | 168 (21) | 204 (18) | 116 (24) | ||
| Other | 235 (7) | 70 (7) | 39 (5) | 89 (8) | 37 (8) | ||
| BMI (kg/m2) | 30 ± 6 | 30 ± 6 | 30 ± 6 | 30 ± 6 | 30 ± 7 | 0.078 | 0.366 |
| Current Smoker | 969 (29) | 288 (31) | 231 (29) | 308 (27) | 142 (30) | 0.364 | 0.609 |
| PCI Indication | |||||||
| ACS | 2290 (69) | 689 (74) | 541 (67) | 742 (66) | 318 (67) | <0.001 | 0.932 |
| STEMI | 628 (19) | 247 (26) | 129 (16) | 192 (17) | 60 (13) | ||
| Non-STEMI | 939 (28) | 275 (29) | 230 (28) | 297 (26) | 137 (29) | ||
| Unstable Angina | 723 (22) | 167 (18) | 182 (23) | 253 (23) | 121 (26) | ||
| Non-ACS | 1052 (31) | 244 (26) | 269 (33) | 381 (34) | 158 (33) | ||
| Drug-eluting stent at index PCI | 2868 (86) | 819 (88) | 706 (87) | 942 (84) | 401 (84) | 0.035 | 0.117 |
| Diabetes | 1354 (41) | 317 (34) | 343 (42) | 486 (43) | 208 (44) | <0.001 | 0.874 |
| Hypertension | 2704 (81) | 700 (75) | 675 (83) | 927 (83) | 402 (84) | <0.001 | 0.642 |
| Dyslipidemia | 2317 (69) | 632 (68) | 579 (71) | 772 (69) | 334 (70) | 0.360 | 0.429 |
| CKD† | 963 (29) | 216 (23) | 231 (29) | 347 (31) | 169 (36) | <0.001 | 0.034 |
| PVD | 335 (10) | 67 (7) | 83 (10) | 118 (11) | 67 (14) | <0.001 | 0.072 |
| Heart Failure | 533 (16) | 115 (12) | 138 (17) | 191 (17) | 89 (19) | 0.004 | 0.687 |
| Atrial Fibrillation | 316 (9) | 52 (6) | 98 (12) | 108 (10) | 58 (12) | <0.001 | 0.143 |
| Cancer | 184 (6) | 39 (4) | 54 (7) | 68 (6) | 23 (5) | 0.097 | 0.409 |
| Prior MI | 866 (26) | 235 (25) | 191 (24) | 301 (27) | 139 (29) | 0.127 | 0.070 |
| Prior Stent | 770 (23) | 193 (21) | 172 (21) | 286 (25) | 119 (25) | 0.027 | 0.083 |
| Prior Stroke or TIA | 348 (10) | 53 (6) | 91 (11) | 131 (12) | 73 (15) | <0.001 | 0.069 |
| Prior GI Bleed or ICH | 111 (3) | 26 (3) | 18 (2) | 40 (4) | 27 (6) | 0.007 | 0.005 |
| Discharge Meds | |||||||
| Aspirin | 3269 (98) | 918 (98) | 790 (98) | 1097 (98) | 464 (97) | 0.552 | 0.961 |
| Statin | 3143 (94) | 897 (96) | 751 (93) | 1046 (93) | 449 (94) | 0.009 | 0.532 |
| ACEi or ARB | 2245 (67) | 639 (68) | 557 (69) | 732 (65) | 317 (67) | 0.289 | 0.257 |
| Beta-blocker | 2852 (85) | 794 (85) | 696 (86) | 971 (86) | 391 (82) | 0.152 | 0.072 |
| Anticoagulant | 321 (10) | 57 (6) | 94 (12) | 115 (10) | 55 (12) | <0.001 | 0.573 |
| Proton Pump Inhibitor | 1049 (31) | 277 (30) | 250 (31) | 377 (34) | 145 (30) | 0.259 | 0.322 |
Data are presented as n (%) or mean ± standard deviation
All patients receiving prasugrel or ticagrelor, regardless of CYP2C19 phenotype, were combined into a single alternative therapy reference group
P-value for comparison across four antiplatelet therapy-CYP2C19 phenotype groups: alternative (combined), clopidogrel-UM or RM, clopidogrel-NM, clopidogrel-IM or PM
P-value for comparison across three CYP2C19 phenotype groups in clopidogrel-treated patients: clopidogrel-UM or RM, clopidogrel-NM, clopidogrel-IM or PM
Chronic kidney disease was defined as estimated glomerular filtration rate <60 ml/min
ACEi = angiotensin-converting enzyme inhibitor; ACS = acute coronary syndrome; ARB = angiotensin receptor blocker; BMI = body mass index; CKD = chronic kidney disease; GI = gastrointestinal; ICH = intracranial hemorrhage; MI = myocardial infarction; PCI = percutaneous coronary intervention; PVD = peripheral vascular disease; STEMI = ST-segment elevation MI; TIA = transient ischemic attack
All analyses were performed using R statistical software (version 3.6; http://www.r-project.org/). P-values <0.05 were considered statistically significant.
RESULTS
Study Population.
Baseline characteristics of the 3,342 patients included in the study are summarized in TABLE 1. The mean age was 63±12 years, 32.2% were female, and 19.7% were African-American. Comorbidities such as diabetes (40.5%), chronic kidney disease (28.8%), and atrial fibrillation (9.5%) were common. Overall, 2,290 patients (68.5%) had an ACS indication for PCI. Most patients received a drug eluting stent (85.8%) and were prescribed aspirin at discharge (97.8%) in addition to a P2Y12 inhibitor.
CYP2C19 Metabolizer Status and Antiplatelet Therapy.
Among the study population, 144 (4.3%), 837 (25.0%), 1,329 (39.8%), 934 (27.9%), and 98 (2.9%) were classified as a CYP2C19 UM, RM, NM, IM, and PM, respectively, based on their CYP2C19 genotype. The frequency of each genotype is summarized in TABLE S2.
Clopidogrel (72.1%) was the most commonly prescribed P2Y12 inhibitor maintenance therapy, followed by prasugrel (17.0%) and ticagrelor (10.9%). Prasugrel or ticagrelor was prescribed to a significantly higher proportion of IM/PMs (53.9%) compared to NMs (15.5%) and UM/RMs (17.4%) (P<0.001, FIGURE 1), with 75 of 98 (76.5%) PMs and 481 of 934 (51.5%) IMs prescribed an alternative antiplatelet therapy. Among the IMs, 52.6% of *17 allele non-carriers (e.g., *1/*2) and 48.3% of *17 allele carriers (e.g., *2/*17) were prescribed prasugrel or ticagrelor (P=0.256). Among patients without a CYP2C19 no function allele, clopidogrel 75 mg/day was prescribed to a similar proportion of NMs, RMs and UMs (84.5% vs. 82.9% vs. 80.6%, P=0.360).
Figure 1. Study Population by CYP2C19 Metabolizer Phenotype and Antiplatelet Therapy.
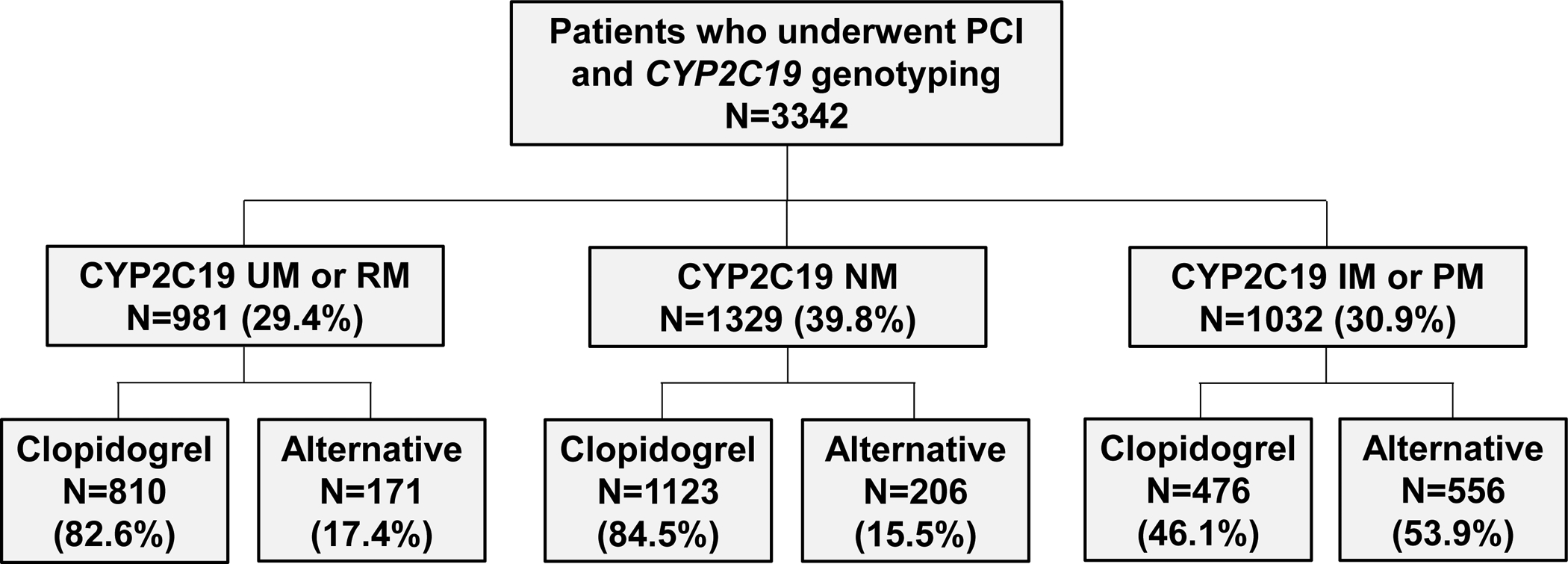
Flow diagram describing the number of study participants by CYP2C19 Metabolizer Phenotype (ultrarapid or rapid metabolizers [UM or RM], normal metabolizers [NM], intermediate or poor metabolizers [IM or PM] by the prescribed P2Y12 inhibitor therapy during follow-up (clopidogrel or alternative therapy).
There were baseline differences between antiplatelet therapy groups in age, race, indication for PCI, prevalence of multiple comorbidities including, diabetes, chronic kidney disease, peripheral vascular disease, atrial fibrillation, stroke, bleeding, and use of oral anticoagulants (TABLE 1). These differences were absent or diminished, and were only significant for race, chronic kidney disease, and prior bleeding, when comparing across CYP2C19 metabolizer status among patients prescribed clopidogrel. Aspirin use was not different across groups.
Clinical Outcomes.
During a median follow-up of 6.3 (1.0–11.0) months after PCI, 353 patients (10.6%) experienced a major atherothrombotic event. The risk of the primary outcome by CYP2C19 metabolizer status and antiplatelet therapy is shown in FIGURE 2. Compared to patients treated with prasugrel or ticagrelor, IMs or PMs treated with clopidogrel exhibited a significantly higher risk of major atherothrombotic events (17.6 vs. 33.7 events per 100 patient-years, respectively; adjusted HR 1.56, 95% CI 1.12–2.16, P=0.008). In contrast, clopidogrel-treated CYP2C19 NMs (20.4 events per 100 patient-years; adjusted HR 1.00, 95% CI 0.75–1.33, P=0.993) and clopidogrel-treated UMs or RMs (19.1 events per 100 patient-years; adjusted HR 0.95, 95% CI 0.69–1.30, P=0.734) exhibited no difference in risk for a major atherothrombotic event compared to patients treated with prasugrel or ticagrelor (TABLE 2).
Figure 2. Cardiovascular outcomes following PCI by CYP2C19 metabolizer status and antiplatelet therapy.
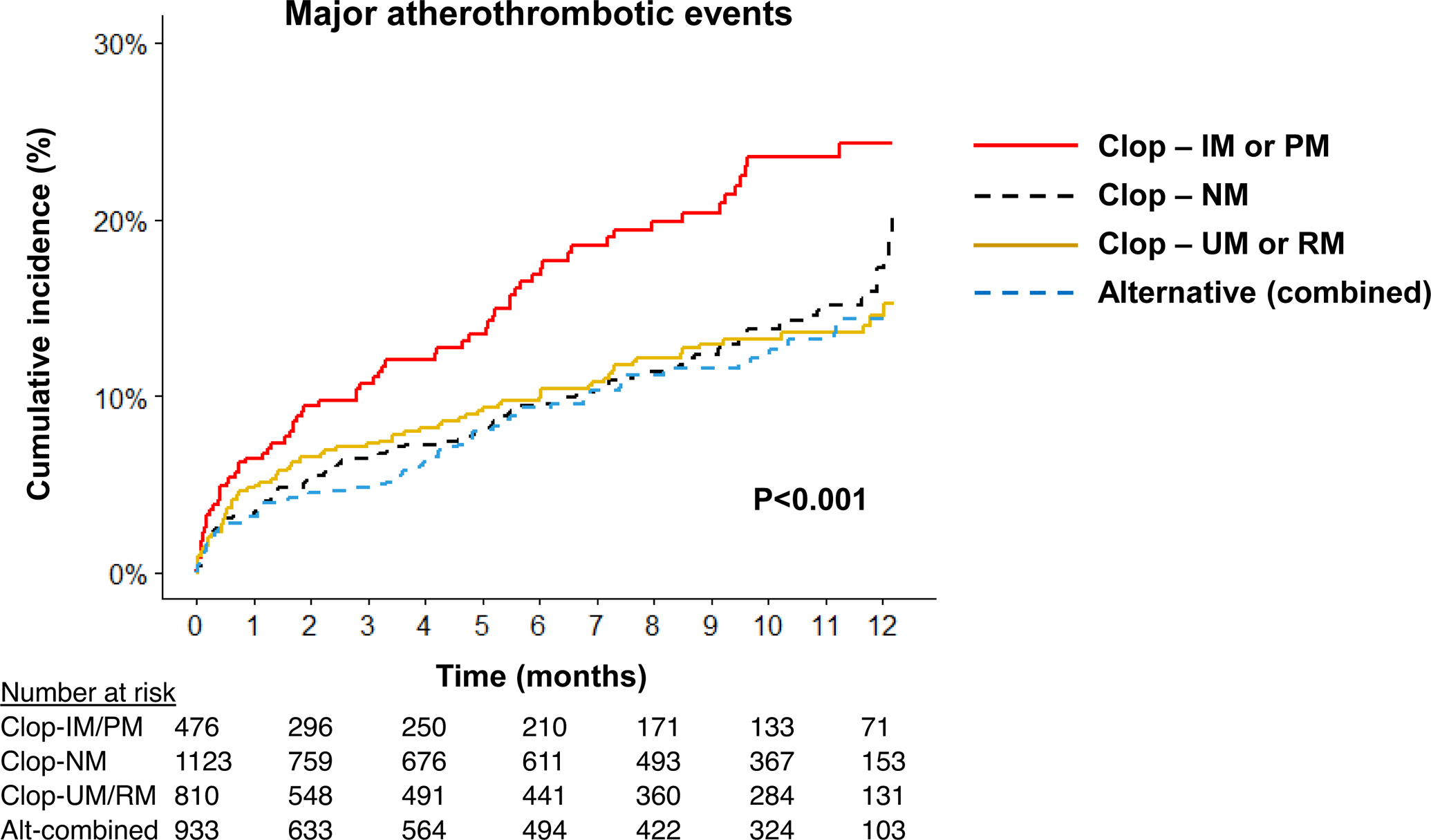
Kaplan-Meier curve for cumulative major atherothrombotic event incidence over 12 months after PCI. Data are shown across four groups: ultrarapid or rapid metabolizers prescribed clopidogrel (Clop-UM or RM); normal metabolizers prescribed clopidogrel (Clop-NM); intermediate or poor metabolizers prescribed clopidogrel (Clop-IM or PM); alternative antiplatelet therapy regardless of CYP2C19 metabolizer status (Alt-combined). The unadjusted log rank P-value for outcomes across the four groups is provided.
Table 2.
Cardiovascular and Bleeding Outcomes by CYP2C19 Metabolizer Status and Antiplatelet Therapy
| Clinical outcome by P2Y12 inhibitor - CYP2C19 phenotype | Event No. (%)* | Event rate (per 100pt-yrs)^ | Unadjusted HR (95% CI) | P-value | Adjusted HR (95% CI) | P-value |
|---|---|---|---|---|---|---|
| Major atherothrombotic events | ||||||
| Clopidogrel - UM or RM | 80 (9.9%) | 19.1 | 1.09 (0.80–1.48) | 0.576 | 0.95 (0.69–1.30) | 0.734 |
| Clopidogrel - NM | 117 (10.4%) | 20.4 | 1.16 (0.88–1.54) | 0.300 | 1.00 (0.75–1.33) | 0.993 |
| Clopidogrel - IM or PM | 72 (15.1%) | 33.7 | 1.86 (1.36–2.55) | <0.001 | 1.56 (1.12–2.16) | 0.008 |
| Alternative (combined)† | 84 (9.0%) | 17.6 | Reference | Reference | ||
| Clinically significant bleeding events | ||||||
| Clopidogrel - UM or RM | 34 (4.2%) | 7.9 | 1.04 (0.66–1.65) | 0.871 | 0.96 (0.60–1.54) | 0.865 |
| Clopidogrel - NM | 35 (3.1%) | 6.0 | 0.78 (0.49–1.23) | 0.287 | 0.68 (0.42–1.08) | 0.104 |
| Clopidogrel - IM or PM | 16 (3.5%) | 7.3 | 0.93 (0.52–1.67) | 0.812 | 0.77 (0.43–1.40) | 0.391 |
| Alternative (combined)† | 39 (4.1%) | 7.7 | Reference | Reference |
Data are presented as the number (%) of patients in each group who experienced the event over 12 months of follow-up after the index PCI.
The event rate was calculated as the number of events per 100 patient-years of follow-up.
All patients receiving prasugrel or ticagrelor, regardless of CYP2C19 phenotype, were combined into a single alternative therapy reference group.
In patients treated with clopidogrel, IM/PMs exhibited a significantly higher risk of major atherothrombotic events compared to NMs (adjusted HR 1.56, 95% CI 1.16–2.10, P=0.003). In contrast, no difference was observed between clopidogrel-treated UM/RMs versus NMs (adjusted HR 0.97, 95% CI 0.73–1.29, P=0.808). The impact of the CYP2C19*17 allele on these associations with major atherothrombotic events was evaluated (FIGURE 3). No difference in event rate was observed across clopidogrel-treated UMs (*17/*17), RMs (*1/*17), and NMs (18.5, 19.3, and 20.4 events per 100 patient-years, respectively; TABLE S3). The risk of an atherothrombotic event was higher in clopidogrel-treated PMs and IMs who did not carry the *17 allele (e.g., *1/*2) compared to NMs (adjusted HR 1.72, 95% CI 1.25–2.37, P<0.001). The subset of IMs who carried the *17 allele (e.g., *2/*17), however, did not exhibit a significantly higher risk of atherothrombotic events compared to NMs (adjusted HR 1.15, 95% CI 0.67–1.97, P=0.613). When comparing IMs who carried the *17 allele to IMs without the *17 allele, risk of an atherothrombotic event was not significantly different (25.7 vs. 37.9 events per 100 patient-years, respectively; unadjusted HR 0.69, 95% CI 0.39–1.23; P=0.211; adjusted HR 0.67, 95% CI 0.37–1.21, P=0.182).
Figure 3. Cardiovascular events following PCI by CYP2C19 metabolizer and CYP2C19*17 allele status in patients treated with clopidogrel.
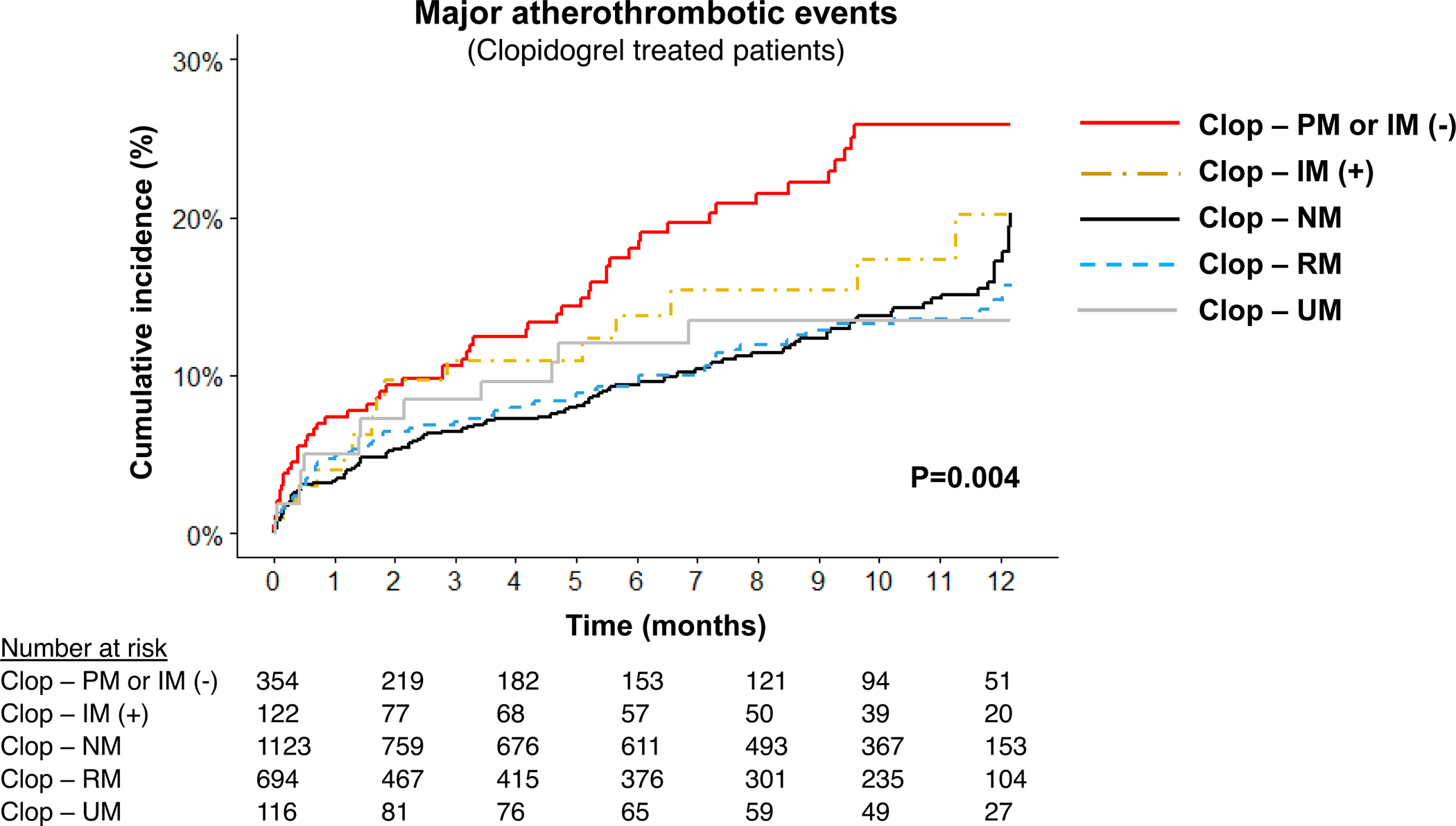
Kaplan-Meier curve for cumulative major atherothrombotic event incidence over 12 months after PCI in the stratum of patients treated with clopidogrel (n=2409). Data are shown across five groups: ultrarapid metabolizers (Clop-UM); rapid metabolizers (Clop-RM); normal metabolizers (Clop-NM); intermediate metabolizers carrying the CYP2C19*17 allele (e.g., *2/*17) (Clop-IM[+]); poor metabolizers or intermediate metabolizers not carrying the *17 allele (e.g., *1/*2) (Clop-PM or IM[−]). Due to the very low number of CYP2C19 PMs prescribed clopidogrel, IM(−) and PM were combined for analysis. The unadjusted log rank P-value for outcomes across the five groups is provided.
The secondary outcome of clinically significant bleeding was documented in 124 (3.7%) patients during the follow-up period. Overall, there was no difference in risk of developing a clinically significant bleeding event across groups (FIGURE 4, TABLE 2). In patients treated with clopidogrel, no difference in bleeding risk was observed between CYP2C19 UM/RMs and NMs (7.9 vs. 6.0 events per 100 patient-years, respectively; adjusted HR 1.34, 95% CI 0.83–2.17, P=0.224) or between clopidogrel-treated UMs and RMs (7.3 and 8.0 events per 100 patient-years, respectively; TABLE S3).
Figure 4. Bleeding outcomes following PCI by CYP2C19 metabolizer status and antiplatelet therapy.

Kaplan-Meier curves for cumulative clinically significant bleeding event incidence over 12 months after PCI. Data are shown across four groups, as described in Figure 2: Clop-UM or RM, Clop-NM, Clop-IM or PM, and Alt-combined. The number of patients in each group was slightly different than for the atherothrombotic endpoint as patients were stratified based on P2Y12 therapy prescribed at the time of the bleeding event or last follow-up. The unadjusted log rank P-value for outcomes across the four groups is provided.
DISCUSSION
The current study investigated the impact of the CYP2C19*17 allele on antiplatelet therapy selection and outcomes following clinical implementation of CYP2C19 genotype-guided antiplatelet therapy across multiple centers in a real-world setting. Results illustrated that clopidogrel was prescribed at a similar rate in CYP2C19 UMs, RMs, and NMs after PCI, and that presence of the *17 allele did not influence the frequency of alternative antiplatelet therapy selection in CYP2C19 IMs (e.g. those with the *2/*17 genotype). Clopidogrel-treated UMs or RMs exhibited no difference in risk for a major atherothrombotic or clinically significant bleeding event compared to NMs over 12 months after PCI. In addition, cardiovascular and bleeding outcomes in clopidogrel-treated UMs or RMs and clopidogrel-treated NMs were not significantly different when compared to patients prescribed prasugrel or ticagrelor. Collectively, our results demonstrate that the CYP2C19*17 allele does not significantly influence antiplatelet therapy selection or clinical outcomes in a real-world setting and suggest that use of the CYP2C19 UM and RM phenotypes to guide antiplatelet therapy prescribing after PCI is likely of limited clinical utility.
Accumulating data demonstrate that use of CYP2C19 genotyping to guide selection of prasugrel or ticagrelor in CYP2C19 IMs and PMs after PCI lowers the risk of major atherothrombotic events compared with conventional treatment strategies without increasing bleeding risk.10–13,34,35 As a consequence, multiple institutions have successfully implemented CYP2C19 genotype-guided antiplatelet therapy into clinical workflows.23,24,26,27,36–40 The increased function *17 allele, in addition to the *2 and *3 no function alleles, are recommended as tier 1 variants for inclusion on clinically implemented CYP2C19 genotype tests by the Association for Molecular Pathology.25 However, the impact of the *17 allele on clopidogrel clinical effectiveness and safety, and the optimal P2Y12 inhibitor to use in *17 allele carriers, has remained unclear.
Several studies have reported that CYP2C19*17 allele carriers have increased active metabolite formation, enhanced inhibition of platelet aggregation, higher bleeding risk, and lower risk for ischemic events with clopidogrel compared to non-carriers.15–19 In a study of 1,524 PCI patients, CYP2C19*17 allele carriers had lower on-treatment platelet reactivity and 85% greater bleeding risk at 30 days compared to non-carriers, indicating enhanced responsiveness to clopidogrel treatment.15 Lower on-treatment platelet reactivity values were similarly shown in a study of chronic clopidogrel treatment post-PCI.16 In a study of 928 patients with MI, *17 allele carriers exhibited a 22% lower risk of clinically-driven target vessel revascularization and major ischemic events at one year compared to non-carriers.17 Importantly, these studies did not account for the impact of linkage disequilibrium between *2 and *17 allele defining variants by adjusting for the presence of the *2 allele.
Because the increased function T allele of *17 occurs on the same haplotype as the normal function G allele of *2, it has been postulated that the observed *17 allele associations with clopidogrel response may be due to absence of the no function *2 allele rather than presence of the increased function *17 allele.5,20 Indeed, multiple studies have reported no significant association between the *17 allele and clopidogrel pharmacokinetics, pharmacodynamics, and outcomes after accounting for the *2 allele.7,20–22 Notably, in a pharmacokinetic-pharmacodynamic study performed in the Amish population, the *17 allele was marginally associated with higher clopidogrel active metabolite levels and lower post-treatment platelet aggregation, but the association was abrogated after adjustment for the *2 allele.20 A retrospective genetic analysis of 3,391 clopidogrel-treated patients in the International Clopidogrel Pharmacogenomics Consortium further demonstrated that the relationship between the *17 allele and enhanced inhibition of platelet aggregation was no longer significant after adjusting for the *2 allele.22 In our pragmatic investigation of patients that underwent clinical CYP2C19 genetic testing, while clopidogrel-treated IM/PMs (no function allele carriers) had a significantly higher risk of major atherothrombotic events compared to NMs (*1/*1) over 12 months after PCI, clopidogrel-treated UMs (*17/*17) or RMs (*1/*17) exhibited no difference in atherothrombotic or bleeding events compared to NMs. Our study did not investigate clopidogrel pharmacokinetics or pharmacodynamics or clopidogrel dose-responses, and thus is limited by a lack of direct mechanistic insight into the observed clinical outcomes. However, our outcome findings in a real-world setting accounted for all tested alleles through use of CYP2C19 metabolizer phenotypes,5 are consistent with these prior retrospective genotyping studies that adjusted for the *2 allele,20,22 and further demonstrate that the *17 allele does not significantly alter clopidogrel effectiveness post-PCI after accounting for CYP2C19 no function alleles.
In accordance with current CPIC recommendations,5,30 the combination of one no function and one increased function allele (e.g., *2/*17) was assigned the IM phenotype across sites. In a secondary analysis that distinguished IMs with and without the *17 allele, we observed that presence of the *17 allele in IMs did not influence antiplatelet therapy prescribing. As hypothesized, clopidogrel-treated PMs and IMs without the *17 allele (e.g., *1/*2) exhibited a significantly higher risk of atherothrombotic events compared to NMs. In contrast, the atherothrombotic event rate in the subset of IMs who carried the *17 allele (e.g., *2/*17) was not significantly higher compared to NMs. A study evaluating the joint impact of each allele on clopidogrel’s antiplatelet effects demonstrated that patients with a *2/*17 genotype had on-treatment platelet aggregation values in between those seen in *1/*2 and *1/*17 patients, indicating a potential gene-dose effect.16 Although presence of the increased function *17 allele could partially diminish the higher risk of atherothrombotic events imparted by the no function *2 allele in clopidogrel-treated patients, other studies have reported no difference in platelet reactivity between *1/*2 and *2/*17.41 Moreover, we observed no significant difference in atherothrombotic events between the *1/*2 and *2/*17 genotypes. Thus, our data support current CPIC guidelines that classify patients with the *2/*17 genotype as an IM and recommend prasugrel or ticagrelor after PCI unless contraindicated.5,30 Due to the relatively low number of IMs carrying a *17 allele (n=122) in our study, statistical power for this comparison was limited, confidence intervals were wide, and the results of this secondary analysis should be interpreted with caution. Further study in a larger population is needed to more confidently determine whether the *17 allele significantly influences clopidogrel effectiveness in IMs and whether phenotypic distinction of IMs should be considered in antiplatelet therapy prescribing decisions.
Our study also demonstrated that clopidogrel-treated UMs, RMs and NMs exhibited no difference in risk for a major atherothrombotic or clinically significant bleeding event when compared to patients prescribed prasugrel or ticagrelor. These results are consistent with outcomes from the recent POPular Genetics trial in patients with ST-elevation MI.11 The trial specifically demonstrated that a genotype-guided approach, with CYP2C19 IMs and PMs treated with ticagrelor or prasugrel and all others (NMs, RMs, and UMs) treated with clopidogrel 75 mg/day, was non-inferior to universal use of ticagrelor or prasugrel for risk of developing major cardiovascular or bleeding events. The genotype-guided arm also exhibited significantly lower risk for bleeding events, which was driven by a reduction in minor bleeding. These results, together with our data, demonstrate that clopidogrel use at standard doses in patients without a CYP2C19 no function allele is safe and effective after PCI. These data lend additional evidence in support of the current CPIC guideline recommendations,5 and strongly suggest that in the absence of contraindications any P2Y12 inhibitor can be used after PCI in CYP2C19 NMs, RMs and UMs.
Data from our study were generated from patients receiving genotype-guided antiplatelet therapy in a real-world clinical setting, and thus offer greater generalizability and compliment data generated from clinical trials with more rigid eligibility criteria. For example, our study population had greater racial diversity and higher prevalence of comorbidities such as diabetes, chronic kidney disease, and history of stroke compared to recent clinical trials of biomarker-guided antiplatelet therapy.11,31,42 Our study also included 31% non-ACS patients for whom the impact of CYP2C19 genotype on clopidogrel outcomes has been reported to be less than in ACS patients.43,44 Notably, in the setting of real-world implementation, genotype testing and dissemination of results must be integrated into the workflow without the extra resources afforded in the setting of a clinical trial. Clinical trials often allow for genetic samples to be collected and processed efficiently with changes to therapy dictated by protocols. In the real-world setting, clinicians may choose whether to act on genotype without a protocol directing their actions. In contrast to the recent POPular Genetics and TAILOR-PCI clinical trials of CYP2C19-guided antiplatelet therapy, neither of which considered the CYP2C19*17 allele in their genotype-guided treatment protocols,11,31 our study sites reported *17 allele results in the EHR. This enabled our study to evaluate impact of the *17 allele on treatment outcomes, and to investigate for the first time whether return of *17 allele results influenced the antiplatelet therapy prescribing decision in a real-world setting. These results, however, cannot be extrapolated to other CYP2C19 substrates.45,46
Several limitations to this analysis should be noted. First, because this was a pragmatic study, randomization was not performed, and antiplatelet selection was based on individual prescriber discretion. Outcomes could have therefore been impacted by the baseline differences between CYP2C19-antiplatelet therapy groups. Covariate-adjusted and stratified analyses were conducted to lessen these effects. Although associations of similar magnitude were observed following adjustment, residual confounding may remain. Second, the analysis is based on medications prescribed, but there are no available data on antiplatelet therapy adherence or pharmacodynamic effects. Moreover, the frequency CYP2C19 inhibitor use was not systematically evaluated. Thus, we cannot rule out whether differences in antiplatelet therapy adherence, aspirin responsiveness, or drug-drug interactions across groups influenced the observed outcomes. Third, clinical events were not independently adjudicated. While the manual collection of EHR data, use of common data forms, and experience of the study teams increased rigor and data quality, it is possible that some clinical events may have been mis-categorized. Events such as deaths or bleeding may have also been missed if they were not recorded in the EHR or occurred at a different healthcare system. Finally, our focus on clinically actionable bleeding (GUSTO moderate or severe/life-threatening bleeding) yielded a low number of bleeding events, and the analysis of bleeding risk in CYP2C19 UMs or RMs was likely underpowered to detect differences between drug-phenotype groups.
In summary, our data from real-world implementation of CYP2C19-guided antiplatelet therapy demonstrate that the increased function *17 allele is not associated with atherothrombotic or bleeding outcomes after PCI. These data add to the existing body of evidence showing no significant impact of the *17 allele on clopidogrel’s antiplatelet effects or clinical effectiveness after accounting for *2 allele, and support current CPIC guidelines that provide similar antiplatelet therapy prescribing recommendations for CYP2C19 NMs, RMs and UMs after PCI.5
Supplementary Material
Table S3. Cardiovascular and Bleeding Outcomes by CYP2C19 Metabolizer and CYP2C19*17 Allele Status in Patients Treated with Clopidogrel (n=2409)
Table S2. CYP2C19 Genotype Frequencies and Metabolizer Phenotype Assignment
Table S1. CYP2C19 Genotyping Platform and Alleles Detected at Each Institution
Study Highlights.
• What is the current knowledge on the topic?
The increased function CYP2C19*17 allele confers the rapid metabolizer (RM) and ultra-rapid metabolizer (UM) phenotypes and has inconsistently been associated with increased bioactivation, antiplatelet effects, and clinical effectiveness of clopidogrel. Thus, it remains unclear whether the *17 allele is clinically actionable to guide antiplatelet therapy prescribing.
• What question did this study address?
This multi-site pragmatic study examined the impact of the CYP2C19*17 allele on major atherothrombotic and bleeding events in 3,342 patients who underwent percutaneous coronary intervention (PCI) and clinical CYP2C19 genotyping.
• What does this study add to our knowledge?
In a real-world setting of CYP2C19 genotype-guided antiplatelet therapy, the CYP2C19*17 allele does not influence antiplatelet therapy prescribing decisions or clinical outcomes in PCI patients once the CYP2C19*2 and other no function alleles are accounted for.
• How might this change clinical pharmacology or translational science?
These findings support current pharmacogenetic guidelines that provide similar antiplatelet therapy prescribing recommendations after PCI for patients with the CYP2C19 UM, RM and normal metabolizer phenotypes.
Funding:
Work supported by the National Institutes of Health (NIH) grants U01 HG007269 (JAJ, LHC, JDD, YC, KJC, YG, CWM), U01 HG007775 (ALB), and U01 HG007762 (TCS, RPK) as part of the NIH IGNITE network. Additional support provided by: NIH T32 HG008958 (CDT), K01 HL141690 (CWM), NIH grants U01 GM074492 and U01 HL105198 (both part of the NIH Pharmacogenomics Research Network), and by substantial institutional support from the University of Florida and its Clinical Translational Science Institute (grants UL1 TR000064 and UL1 TR001427) (LHC, JAJ, JDD, YC, KJC, YG, CWM); NIH UL1 TR002489 (CRL, KEW, GAS); Penn Center for Precision Medicine at the Perelman School of Medicine at the University of Pennsylvania (ST, JG); NIH R01 HL092173, K24 HL133373, UL1 TR000165 and Hugh Kaul Precision Medicine Institute at the University of Alabama, Birmingham (NAL); NIH UL1 TR0000005, the University of Pittsburgh/UPMC Institute for Precision Medicine, American Society of Health System Pharmacists, American Heart Association grant 17MCPRP33400175, and by an Anonymous Donor (PEE, JCC, JMS); NIH U54 MD010723 (JCL).
Footnotes
Conflict of interest/disclosure: F.F. receives honoraria from AstraZeneca and Sanofi. R.P.K. receives research funding from Idorsia and consulting fees from Haemonetics. T.C.S. receives consulting fees from Indiana University Health. J.G. receives honoraria from Astra Zeneca and Inari Medical. K.E.W is a member of the Association for Molecular Pathology Pharmacogenomics working group. D.J.A. has received consulting fees or honoraria from Abbott, Amgen, Aralez, AstraZeneca, Bayer, Biosensors, Boehringer Ingelheim, Bristol-Myers Squibb, Chiesi, Daiichi-Sankyo, Eli Lilly, Haemonetics, Janssen, Merck, PhaseBio, PLx Pharma, Pfizer, Sanofi, and The Medicines Company and has received payments for participation in review activities from CeloNova and St. Jude Medical. D.J.A. also declares that his institution has received research grants from Amgen, AstraZeneca, Bayer, Biosensors, CeloNova, CSL Behring, Daiichi-Sankyo, Eisai, Eli Lilly, Gilead, Janssen, Matsutani Chemical Industry Co., Merck, Novartis, Osprey Medical, Renal Guard Solutions, and the Scott R. MacKenzie Foundation. JAJ is a consultant for United Health Group. All other authors declared no competing interests for this work.
References
- (1).Virani SS et al. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation 141, e139–e596 (2020). [DOI] [PubMed] [Google Scholar]
- (2).Capodanno D, Alfonso F, Levine GN, Valgimigli M & Angiolillo DJ ACC/AHA Versus ESC Guidelines on Dual Antiplatelet Therapy: JACC Guideline Comparison. J Am Coll Cardiol 72, 2915–31 (2018). [DOI] [PubMed] [Google Scholar]
- (3).Dayoub EJ et al. Trends in Platelet Adenosine Diphosphate P2Y12 Receptor Inhibitor Use and Adherence Among Antiplatelet-Naive Patients After Percutaneous Coronary Intervention, 2008–2016. JAMA Intern Med 178, 943–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mega JL et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 304, 1821–30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Scott SA et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94, 317–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mega JL et al. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation 119, 2553–60 (2009). [DOI] [PubMed] [Google Scholar]
- (7).Wallentin L et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet 376, 1320–8 (2010). [DOI] [PubMed] [Google Scholar]
- (8).Wallentin L et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med 361, 1045–57 (2009). [DOI] [PubMed] [Google Scholar]
- (9).Wiviott SD et al. TRITON-TIMI Investigators. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med 357, 2001–15 (2007). [DOI] [PubMed] [Google Scholar]
- (10).Cavallari LH et al. Multi-site Investigation of outcomes with implementation of CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. JACC Cardiovasc Interv 11, 181–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Claassens DMF et al. A Genotype-Guided Strategy for Oral P2Y12 Inhibitors in Primary PCI. N Engl J Med 381, 1621–31 (2019). [DOI] [PubMed] [Google Scholar]
- (12).Klein MD, Williams AK, Lee CR & Stouffer GA Clinical Utility of CYP2C19 Genotyping to Guide Antiplatelet Therapy in Patients With an Acute Coronary Syndrome or Undergoing Percutaneous Coronary Intervention. Arterioscler Thromb Vasc Biol 39, 647–52 (2019). [DOI] [PubMed] [Google Scholar]
- (13).Notarangelo FM et al. Pharmacogenomic Approach to Selecting Antiplatelet Therapy in Acute Coronary Syndromes: PHARMCLO trial. J Am Coll Cardiol 71, 1869–77 (2018). [DOI] [PubMed] [Google Scholar]
- (14).Sim SC et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 79, 103–13 (2006). [DOI] [PubMed] [Google Scholar]
- (15).Sibbing D et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation 121, 512–8 (2010). [DOI] [PubMed] [Google Scholar]
- (16).Sibbing D et al. Isolated and interactive impact of common CYP2C19 genetic variants on the antiplatelet effect of chronic clopidogrel therapy. J Thromb Haemost 8, 1685–93 (2010). [DOI] [PubMed] [Google Scholar]
- (17).Tiroch KA et al. Protective effect of the CYP2C19 *17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J 160, 506–12 (2010). [DOI] [PubMed] [Google Scholar]
- (18).Gross L et al. Genotype-Phenotype Association and Impact on Outcomes following Guided De-Escalation of Anti-Platelet Treatment in Acute Coronary Syndrome Patients: The TROPICAL-ACS Genotyping Substudy. Thromb Haemost 118, 1656–67 (2018). [DOI] [PubMed] [Google Scholar]
- (19).Saiz-Rodriguez M et al. Influence of CYP2C19 Phenotype on the Effect of Clopidogrel in Patients Undergoing a Percutaneous Neurointervention Procedure. Clin Pharmacol Ther 105, 661–71 (2019). [DOI] [PubMed] [Google Scholar]
- (20).Lewis JP et al. The CYP2C19*17 variant is not independently associated with clopidogrel response. J Thromb Haemost 11, 1640–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Danielak D et al. Influence of genetic co-factors on the population pharmacokinetic model for clopidogrel and its active thiol metabolite. Eur J Clin Pharmacol 73, 1623–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lewis JP et al. Pharmacogenomic polygenic response score predicts ischaemic events and cardiovascular mortality in clopidogrel-treated patients. Eur Heart J Cardiovasc Pharmacother 6, 203–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Empey PE et al. Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy. Clin Pharmacol Ther 104, 664–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Luzum JA et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and Metrics of Pharmacogenetic Implementations Across Diverse Healthcare Systems. Clin Pharmacol Ther 102, 502–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Pratt VM et al. Recommendations for Clinical CYP2C19 Genotyping Allele Selection: A Report of the Association for Molecular Pathology. J Mol Diagn 20, 269–76 (2018). [DOI] [PubMed] [Google Scholar]
- (26).Tuteja S et al. Prospective CYP2C19 Genotyping to Guide Antiplatelet Therapy Following Percutaneous Coronary Intervention: A Pragmatic Randomized Clinical Trial. Circ Genom Precis Med 13, e002640 (2020). [DOI] [PubMed] [Google Scholar]
- (27).Cavallari LH et al. Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention. J Transl Med 16, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cavallari LH et al. The IGNITE Pharmacogenetics Working Group: An Opportunity for Building Evidence with Pharmacogenetic Implementation in a Real-World Setting. Clin Transl Sci 10, 143–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sox HC & Lewis RJ Pragmatic Trials: Practical Answers to “Real World” Questions. JAMA 316, 1205–6 (2016). [DOI] [PubMed] [Google Scholar]
- (30).Caudle KE et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 19, 215–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Pereira NL et al. Clopidogrel Pharmacogenetics. Circ Cardiovasc Interv 12, e007811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Mehran R et al. Standardized bleeding definitions for cardiovascular clinical trials: a consensus report from the Bleeding Academic Research Consortium. Circulation 123, 2736–47 (2011). [DOI] [PubMed] [Google Scholar]
- (33).Martin J et al. Frequency and clinical outcomes of CYP2C19 genotype-guided escalation and de-escalation of antiplatelet therapy in a real-world clinical setting. Genet Med 22, 160–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Moon JY et al. Role of genetic testing in patients undergoing percutaneous coronary intervention. Expert Rev Clin Pharmacol, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sibbing D et al. Updated Expert Consensus Statement on Platelet Function and Genetic Testing for Guiding P2Y12 Receptor Inhibitor Treatment in Percutaneous Coronary Intervention. JACC Cardiovasc Interv 12, 1521–37 (2019). [DOI] [PubMed] [Google Scholar]
- (36).Weitzel KW et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet 166C, 56–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Shuldiner AR et al. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. Am J Med Genet C Semin Med Genet 166C, 76–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Peterson JF et al. Physician response to implementation of genotype-tailored antiplatelet therapy. Clin Pharmacol Ther 100, 67–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Harada S et al. Precision Medicine at the University of Alabama at Birmingham: Laying the Foundational Processes Through Implementation of Genotype-Guided Antiplatelet Therapy. Clin Pharmacol Ther 102, 493–501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lee CR et al. Clinical Outcomes and Sustainability of Using CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. Circ Genom Precis Med 11, e002069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Rossi JS et al. Clopidogrel dose adjustment after outpatient screening for CYP2C19 variant alleles: a pilot study. Pharmacogenomics 15, 915–23 (2014). [DOI] [PubMed] [Google Scholar]
- (42).Sibbing D et al. Guided de-escalation of antiplatelet treatment in patients with acute coronary syndrome undergoing percutaneous coronary intervention (TROPICAL-ACS): a randomised, open-label, multicentre trial. Lancet 390, 1747–57 (2017). [DOI] [PubMed] [Google Scholar]
- (43).Kim HS et al. CYP2C19 poor metabolizer is associated with clinical outcome of clopidogrel therapy in acute myocardial infarction but not stable angina. Circ Cardiovasc Genet 6, 514–21 (2013). [DOI] [PubMed] [Google Scholar]
- (44).Johnson JA, Roden DM, Lesko LJ, Ashley E, Klein TE & Shuldiner AR Clopidogrel: a case for indication-specific pharmacogenetics. Clin Pharmacol Ther 91, 774–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Moriyama B et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP2C19 and Voriconazole Therapy. Clin Pharmacol Ther 102, 45–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).El Rouby N, Lima JJ & Johnson JA Proton pump inhibitors: from CYP2C19 pharmacogenetics to precision medicine. Expert Opin Drug Metab Toxicol 14, 447–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Cardiovascular and Bleeding Outcomes by CYP2C19 Metabolizer and CYP2C19*17 Allele Status in Patients Treated with Clopidogrel (n=2409)
Table S2. CYP2C19 Genotype Frequencies and Metabolizer Phenotype Assignment
Table S1. CYP2C19 Genotyping Platform and Alleles Detected at Each Institution


