Abstract
The diagnosis of Mendelian disorders following uninformative exome and genome sequencing remains a challenging and often unmet need. Following uninformative exome and genome sequencing of a family quartet including two siblings with suspected mitochondrial disorder, RNA sequencing (RNAseq) was pursued in one sibling. Long-read amplicon sequencing was used to determine and quantify transcript structure. Immunoblotting studies and quantitative proteomics were performed to demonstrate functional impact. Differential expression analysis of RNAseq data identified significantly decreased expression of the mitochondrial OXPHOS complex I subunit NDUFB10 associated with a cryptic exon in intron 1 of NDUFB10, that included an in-frame stop codon. The cryptic exon contained a rare intronic variant that was homozygous in both affected siblings. Immunoblot and quantitative proteomic analysis of fibroblasts revealed decreased abundance of complex I subunits, providing evidence of isolated complex I deficiency. Through multi-omic analysis we present data implicating a deep intronic variant in NDUFB10 as the cause of mitochondrial disease in two individuals, providing further support of the gene-disease association. This study highlights the importance of transcriptomic and proteomic analyses as complementary diagnostic tools in patients undergoing genome-wide diagnostic evaluation.
Keywords: Aberrant splicing, Mitochondrial Disease, Genomics, Proteomics, RNA sequencing
Mitochondrial diseases are genetic disorders with a primary defect in oxidative phosphorylation (OXPHOS), an important cellular energy generating system (Frazier, Thorburn, & Compton, 2019; Gorman et al., 2016). The majority of mitochondrial disease-associated genes are related to OXPHOS biogenesis (185 of 289 genes) (Frazier et al., 2019). OXPHOS requires the assembly of five unique enzyme complexes, complex I – complex V, with complex I (NADH:ubiquinone reductase) being the largest of these with 44 unique subunits and more than 15 assembly factors (Stroud et al., 2016), 27 of which have been associated with mitochondrial disease (Frazier et al., 2019; Sazanov, 2015). Most subunits are nuclear-encoded, however seven subunits are encoded on the mitochondrial genome. While the rate of gene discovery has accelerated substantially in the genomic era, diagnosis can still remain challenging in some cases. For example, pathogenic DNA variants in non-coding regions can often be difficult to identify using standard genome analysis approaches.
In this study we describe the use of RNA sequencing (RNAseq) and quantitative proteomic analysis in a sibling pair with a clinical presentation suggestive of mitochondrial disease (hearing loss, neurological deterioration, and early death). Through transcriptome analysis we detected aberrant splicing in NDUFB10 (MIM# 603843), with markedly decreased expression of this gene relative to controls. We present transcriptomic and proteomic data implicating a biallelic deep intronic variant as the cause of complex I deficiency in these individuals. Variants in NDUFB10, a complex I subunit, have previously been reported in only one individual, an infant with fatal lactic acidosis and cardiomyopathy and decreased complex I activity in multiple tissues (Friederich et al., 2016).
Individuals 1 (I-1) and 2 (I-2) are siblings, born of fourth-degree consanguineous parents of Pakistani origin. There is one unaffected sibling and two paternal cousins with hearing impairment, but otherwise no family history of neurologic disease. A full clinical summary is provided in Supp. Table S1.
All research activities were performed under institutional ethics approval from The Royal Children’s Hospital Melbourne (HREC/16/RCHM/150) and written informed consent was provided by both parents. Clinical data and neuroimaging results were reviewed and abstracted from the medical record.
Both affected siblings presented in the first year of life and both had bilateral sensorineural hearing loss diagnosed at 7 weeks and 9 months, respectively. I-1 had a flexion deformity of the left hand and hypoplasia of the left thumb, suggestive of a radial ray anomaly (Supp. Figure S1:1C) while his younger sibling had a milder defect. By nine months I-1 had made developmental gains in gross and fine motor skills but was still below the 3rd centile in all growth parameters. He was admitted at 11 months due to increased episodes of apnea requiring intubation, and died from complications of his underlying disease. I–2 had a normal perinatal period and made developmental gains without regression and at seven months was able to sit independently, roll over, transfer objects, vocalize, and was attentive to objects and sounds. Independent sitting was lost on follow-up at nine months of age. At 14 months he was admitted to the intensive care unit due to episodes of bradycardia and persistent apnea and died.
Laboratory investigations performed for I-1 including urine organic acids and glycosaminoglycans, plasma very long chain fatty acids, amino acids and serum transferrin isoforms, were all normal. Plasma lactate levels were mildly elevated in both siblings at (3.5 mmol/L and 3.6 mmol/L, respectively [normal range 1.0–1.8 mmol/L]). Respiratory chain enzyme activities were normal in fibroblasts of I-1 in skeletal muscle, Complex I activity was normal relative to total protein but borderline low (~40% residual activity) relative to citrate synthase and Complex II. Mitochondrial complex I activity was also borderline low in liver relative to citrate synthase (~50% residual activity while Complex II, III, and IV activities were in the range 87 to 120%). A muscle biopsy showed no significant histological pathology.
Extensive genetic testing performed on both siblings was unable to identify a molecular diagnosis. Chromosomal microarray and breakage analysis performed in I-1 was negative. Exome sequencing of both affected siblings and biological parents did not identify any phenotype-relevant variants. Testing for common mitochondrial genome point mutations was also negative. Finally, genome sequencing was undertaken on the affected siblings and both parents by the Broad Center for Mendelian Genomics. Extensive analysis and multidisciplinary review of the data did not identify any plausible causative candidate variants.
Human whole transcriptome sequencing on RNA derived from fibroblasts of I-1 was performed by the Genomics Platform at the Broad Institute of MIT and Harvard. The transcriptome product combines poly(A)-selection of mRNA transcripts with a strand-specific cDNA library preparation. Libraries were sequenced on the HiSeq 2500 platform to generate 50 million 2 × 100 nt reads. Following alignment to the Human Reference Genome Build 38 using STAR (Version 2.7.3a)(Dobin et al., 2013), duplicate reads were masked with Picard MarkDuplicates and quantification was performed using FeatureCounts from the R Subread package (Version 1.34.7)(Liao, Smyth, & Shi, 2019). Gene level differential expression analysis was performed using the DESeq2 package (Version 1.25.9)(Love, Huber, & Anders, 2014).
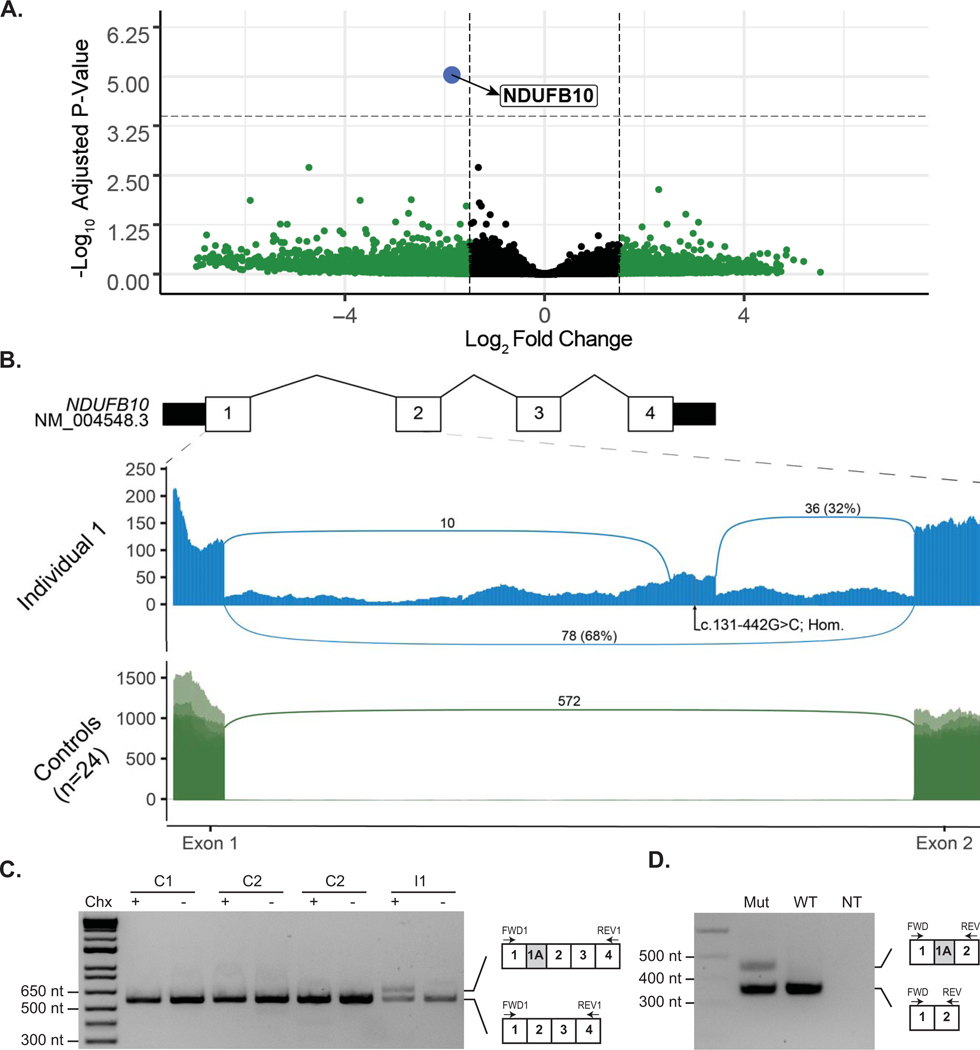
Differential expression analysis identified a greater than three-fold reduction in expression of the nuclear encoded mitochondrial gene NDUFB10 relative to 24 similarly sequenced controls (Log2 Fold Change −1.86; p-adj = 9.058787e-06 [Figure 1A; Supp. Table S6]). Inspection of the reads mapping to NDUFB10 identified two novel splicing events consistent with the inclusion of a 94-nt cryptic exon (referred to as exon 1A) within intron 1 of NDUFB10. Neither of the novel junctions were observed in any of the 24 control samples and no transcripts utilizing either junction are present in either the Gencode v32 or Refseq human transcript databases. Approximately 32% of junction-spanning reads using the exon 2 acceptor splice site support the inclusion of the cryptic exon in I-1. In addition to the cryptic exon, reads mapping along the length of intron 1 were enriched in I-1 relative to controls, suggesting that intron 1 may be retained in some transcripts. The inclusion of exon 1A would result in a frameshift p.(Arg43fs*32), while intron retention would be predicted to introduce a premature termination sequence p.(Arg43fs*135) to this transcript, with both events expected to be subjected to nonsense-mediated decay (NMD) leading to an overall reduced level of NDUFB10 in this individual.
Figure 1. Identification of reduced expression and aberrant splicing in Intron 1 of NDUFB10 in RNA sequencing data from Individual 1.
(A) Analysis of RNA sequencing data revealed significantly decreased expression of NDUFB10 in the affected individual compared to 24 similarly sequenced controls. Dashed lines represent a Log2 Fold Change (L2FC) cut-off of 1.5 or −1.5 on the x-axis and by the log transformed adjusted p-value on the y axis. Details of each gene meeting these thresholds are provided in Supp. Table S6. NDUFB10 is labeled in blue and can be seen as a clear outlier with a L2FC of −1.86 and p-adj = 9.06e-06. Genes colored in green were significant based on their L2FC values as described above while genes color in grey were not significant for their adjusted p-value or for the L2FC value. (B) Sashimi plot depicting junction spanning reads and read depth at each nucleotide from exon 1 to exon 2 of NDUFB10. RNA sequencing data from fibroblasts of Individual 1 (I-1) are shown in the top panel. Two novel splice junctions can be seen, suggesting the presence of a cryptic exon extending between chr16: 1,960,658 – 1,960,751, in intron 1. The position of the homozygous variant, NM_004548.3:c.131–442G>C (NC_000016.10:g.1960711G>C), is shown near the center of the putative cryptic exon. The lower panel displays an overlay of 24 similarly sequenced fibroblast control samples. No reads supporting either of the novel splice junctions were detected in any of the control samples. (C) RT-PCR of NDUFB10 from fibroblast RNA from Individual 1 (I-1) and three control lines (C1, C2 and C3) with (+) or without (−) treatment with cycloheximide (Chx). (D) A mini-gene splicing assay containing the c.131–442G>C was prepared and amplification of RNA from both the mutant and wild-type plasmids by RT-PCR using primers between exon 1 and exon 2 of NDUFB10 yielded an amplicon consistent with the size expected from reference NDUFB10 splicing. In the mutant plasmid, a second larger amplicon was present. This amplicon, approximately 94-nt in size, is absent in the wild-type plasmid and is consistent in size with aberrant splicing event observed on RNA sequencing data.
To confirm the presence of a cryptic exon, we used reverse transcription polymerase chain reaction (RT-PCR) to amplify transcripts from fibroblast RNA using primers situated in exon 1 and exon 4 of NDUFB10 (Supp. Table S2). An amplicon consistent with the expected size of 563-nt was generated from I-1 as well as fibroblasts from three healthy controls (Figure 1C). An additional larger amplicon consistent with the inclusion of the cryptic exon observed in the RNA sequencing data was amplified from I-1, but only when fibroblasts were treated with cycloheximide to inhibit NMD, consistent with the hypothesis that this transcript is subject to degradation. The larger amplicon was not detectable in any of the three control lines.
To confirm the sequence and genomic location of the cryptic exon observed by RNA sequencing we used an Oxford Nanopore Flongle to sequence full length reads of the RT-PCR amplicons from cycloheximide treated fibroblast RNA. Over 5,000 reads were generated from I-1 and three healthy controls. The reads were processed using the Full-Length Alternative Isoform analysis of RNA (FLAIR) (Tang et al., 2020) analysis tool to identify and quantitate the isoforms present in each RT-PCR. An isoform containing the cryptic exon identified by RNA sequencing was present in 24.5% of reads from I-1 (Supp. Figure S2). No nanopore reads were identified that supported the retention of intron 1 that was observed in the RNA sequencing data from I-1. However, amplification of transcripts retaining intron 1 may be disfavored by RT-PCR due to their increased length which could explain their absence from the sequencing library.
With these data in mind, we reviewed the genome sequencing data to identify variants consistent with recessive inheritance in NDUFB10. In intron 1 there were 17 variants that were homozygous in both affected individuals; of these 16 could be excluded on the basis of high population allele frequency (> 0.05) and previously observed homozygotes seen in a reference allele database (Supp. Table S7). The only remaining variant, NM_004548.3:c.131–442G>C, sits within the cryptic exon (Figure 1B) and was found to be homozygous in both affected siblings and heterozygous in the unaffected parents. This variant is absent in available population allele frequency databases.
To determine if the c.131–442G>C variant was responsible for the observed NDUFB10 splicing defect we generated a minigene splicing reporter by cloning 2.1 kb of genomic sequence, including exon 1 to exon 3 of NDUFB10, into the mammalian expression vector pEYFP-C1. Primer directed mutagenesis was used to generate a version of the plasmid containing the c.131–442G>C variant before transfection into human embryonic kidney (HEK293T) cells (Supp. Table S2 and S3). Amplification of RNA transcribed from both the mutant and wild-type plasmids by RT-PCR using primers between exon 1 and exon 2 of NDUFB10 yielded an amplicon consistent with the size expected from reference NDUFB10 splicing (Figure 1D [Supp. Table S4]). A second approximately 100-nt larger amplicon was generated only by the mutant plasmid, consistent with the aberrant splicing observed by RNA sequencing in I-1.
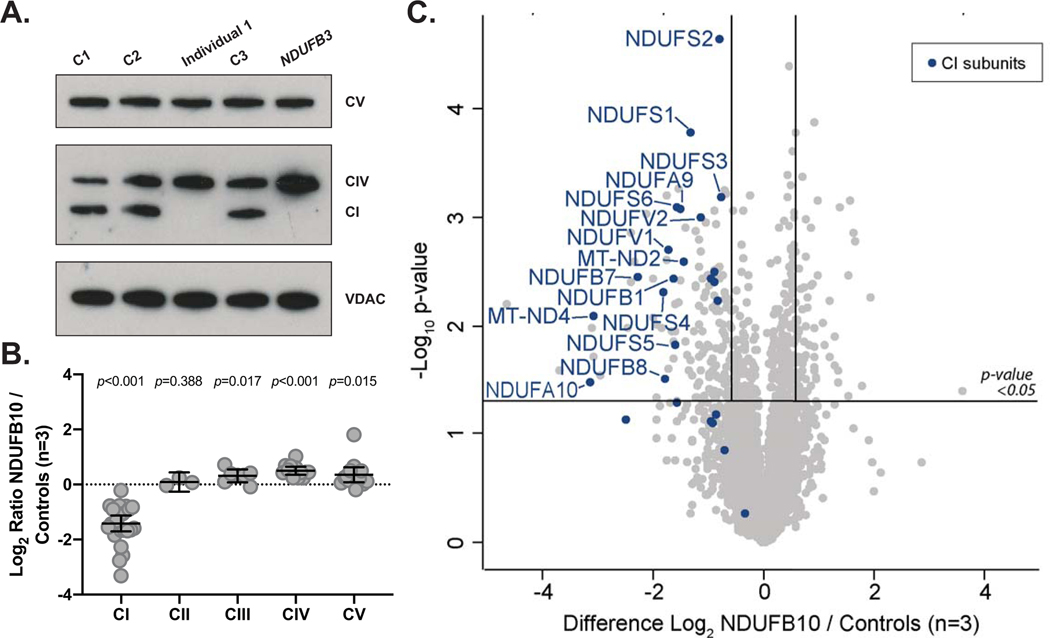
We sought to determine if the aberrant NDUFB10 splicing affects the overall stability of complex I or other mitochondrial respiratory chain complexes. Immunoblotting was performed using an antibody against one subunit of each of the five mitochondrial OXPHOS complexes on cell lysates isolated from a fibroblast sample from I-1, an affected patient control with known complex I deficiency due to a homozygous variant in NDUFB3 (MIM# 603839) (Calvo et al., 2012) and three control samples. This revealed an absence of the complex I subunit NDUFB8 and normal levels of the remaining mitochondrial OXPHOS complexes relative to controls (Figure 2A), consistent with an isolated Complex I defect.
Figure 2. Evidence of isolated complex I deficiency in Individual 1.
(A) SDS-PAGE and immunoblotting of protein isolated from fibroblasts and probed with antibodies specific for mitochondrial OXPHOS proteins reveals an absence of complex I subunit NDUFB8 in Individual 1 (I-1) from the family in this study (Lane 3) relative to controls (Lane 1, 2, 4) similar to a patient with known complex I defect due to variants in NDUFB3 previously reported in Calvo et al. (2012). Levels of COX2 (complex IV; CIV) and ATP5A (complex V; CV) and VDAC1 as the loading control are also shown. Separate panels represent individual images of the same gel with different exposure times set to 30 seconds for CV (Top Panel), 60 seconds for CI/CIV (Middle Panel), and 20 seconds for VDAC (Bottom Panel). (B) Analysis of OXPHOS protein levels detected using quantitative proteomics in Individual 1 (I-1), represented as log2 ratio of the mean of NDUFB10 and controls. The middle bar represents the mean value, while the upper and lower bars represent the 95% confidence interval of the mean value. Each dot represents a single protein. (C) Volcano plot depicting quantitative proteomics data revealing decreased levels of complex I subunits in Individual 1 (I-1) compared to controls. The horizontal line within the volcano plot represents a significance value of p=0.05 whereas the vertical lines represent fold-change of +/−1.5. Complex I subunits are depicted in blue.
Having identified an isolated complex I defect due to transcription of a cryptic exon in NDUFB10, quantitative proteomic analysis was then performed on fibroblasts from I-1 relative to three individual controls to determine the effects of the mutation on the cellular proteome. We detected over 5,000 unique proteins and quantified the changes in 4,200. While NDUFB10 was readily detected in all three of the controls, it was only detected in one of the three replicates from the patient and with a greater than 4 fold reduction in abundance (Supp. Table S8 and S9). Moreover, the abundance of the majority of complex I subunits were decreased in I-1 relative to controls while the levels of other mitochondrial respiratory chain complex subunits remained relatively unchanged (Figure 2B and C). These results are largely consistent with our previously reported study of a HEK293T cell line gene-edited to lack expression of NDUFB10 (Stroud et al., 2016), suggesting the effect of the variant in I-1 leads to similar defects in assembly of the complex (Supp. Figure S3).
In this study, we used RNA sequencing and quantitative proteomic analysis to identify the underlying etiology of disease in an extensively investigated family with previously inconclusive exome and genome sequencing. RNA sequencing identified aberrant splicing leading to significantly reduced expression, guiding re-analysis of genome sequencing to a deep intronic variant in NDUFB10. Through SDS-PAGE and immunoblotting and subsequently quantitative proteomics, we were able to identify that the molecular defect caused by this variant resulted in destabilization of complex I in a similar way to that observed in gene-edited HEK293T cells lacking NDUFB10. Importantly, HEK293T cells lacking NDUFB10 also exhibited no detectable complex I enzyme activity and they had severe defects in mitochondrial respiration (Stroud et al., 2016). The data provided here further support evidence that genetic variants in NDUFB10 result in defective assembly of complex I and clinically manifest as severe mitochondrial disease.
Following negative genome sequencing, RNA sequencing and quantitative proteomic analysis offer an intriguing opportunity for disease diagnosis. This family had been extensively investigated over a period of 10 years with biochemical screening, single-gene analysis, and exome sequencing followed by genome sequencing, without a molecular diagnosis being reached. In silico splicing prediction tools provided only moderate confidence predictions that the identified NM_004548.3:c.131–442G>C variant would alter splicing. SpliceAI (Jaganathan et al., 2019) gives a low confidence prediction (score=0.15) that the variant will favor the use of a donor splice site 40-nt 3’ of the variant, consistent with the position of the new donor splice site used by exon 1A. This highlights a specific challenge in variant interpretation, particularly in non-coding regions of the genome, where confidence is lacking in splicing predictions tools and functional annotation of the effect of genetic variants is unknown and suggests that RNA sequencing may be one approach to overcome these limitations. Recent studies have assessed the diagnostic yield of RNA sequencing in larger cohorts of affected individuals (Cummings et al., 2017; Frésard et al., 2019; Gonorazky et al., 2019; Kremer et al., 2017; van Eyk et al., 2018), suggesting that it can offer a complementary approach to existing DNA datasets and provide a further avenue for disease diagnosis.
Only a single case of mitochondrial disease associated with variants in NDUFB10 has previously been reported. Friederich et al. described an individual who presented at birth with a mitochondrial disorder characterized by severe lactic acidosis and cardiomyopathy and died in the first 28 hours of life. This study provides further evidence that loss of NDUFB10 function causes human disease, and variants in this gene should be considered as part of diagnostic practice.
Supplementary Material
Acknowledgements
The authors thank the affected individuals and their family for their participation in this research. We acknowledge the Bio21 Mass Spectrometry and Proteomics Facility (MMSPF) for the provision of instrumentation, training, and technical support.
Funding:
The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. This study was supported in part by the Leukodystrophy Flagship Massimo’s Mission, funded by the Medical Research Future Fund (ARG76368). We acknowledge funding from the National Health and Medical Research Council (NHMRC Project Grants 1164479 to AGC, DRT, JC, DAS, 1140906 to DAS; NHMRC Fellowships 1072662 to MBC, 1140851 to DAS and 1155244 to DRT). DHH is supported by a Melbourne International Research Scholarship and the Mito Foundation PhD Top-up Scholarship. Sequencing and analysis were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 and in part by National Human Genome Research Institute grant R01 HG009141.
Footnotes
Data availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Conflicts of Interest
MBC and RDP have received support from Oxford Nanopore Technolgies (ONT) to present their findings at scientific conferences. However, ONT played no role in study design, execution, analysis or publication. Otherwise, the authors report no conflicts of interest.
Web Resources:
gnomAD (Karczewski et al., 2020)
OMIM (“Online Mendelian Inheritance in Man, OMIM®,”)
SpliceAI (Jaganathan et al., 2019)
Accession Numbers
ClinVar - RCV001093633.1
Supplemental Data
A supplemental text file has been provided and includes a full description of methods used in this study.
References:
- Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, . . . Mootha VK (2012). Molecular Diagnosis of Infantile Mitochondrial Disease with Targeted Next-Generation Sequencing. Sci Transl Med, 4(118), 118ra110–118ra110. doi: 10.1126/scitranslmed.3003310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings BB, Marshall JL, Tukiainen T, Lek M, Donkervoort S, Foley AR, . . . MacArthur DG (2017). Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med, 9(386). doi: 10.1126/scitranslmed.aal5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, & Jha S. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Thorburn DR, & Compton AG (2019). Mitochondrial energy generation disorders: genes, mechanisms, and clues to pathology. Journal of Biological Chemistry, 294(14), 5386–5395. doi: 10.1074/jbc.R117.809194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frésard L, Smail C, Ferraro NM, Teran NA, Li X, Smith KS, . . . Care4Rare Canada C. (2019). Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nature Medicine. doi: 10.1038/s41591-019-0457-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederich MW, Erdogan AJ, Coughlin CR II, Elos MT, Jiang H, O’Rourke CP, . . . Riemer (2016). Mutations in the accessory subunit NDUFB10 result in isolated complex I deficiency and illustrate the critical role of intermembrane space import for complex I holoenzyme assembly. Human Molecular Genetics, 26(4), 702–716. doi: 10.1093/hmg/ddw431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonorazky HD, Naumenko S, Ramani AK, Nelakuditi V, Mashouri P, Wang P, . . . Dowling JJ (2019). Expanding the Boundaries of RNA Sequencing as a Diagnostic Tool for Rare Mendelian Disease. Am J Hum Genet, 104(3), 466–483. doi: 10.1016/j.ajhg.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R, . . . Turnbull DM (2016). Mitochondrial diseases. Nat Rev Dis Primers, 2, 16080. doi: 10.1038/nrdp.2016.80 [DOI] [PubMed] [Google Scholar]
- Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, . . . Farh KK-H (2019). Predicting Splicing from Primary Sequence with Deep Learning. Cell, 176(3), 535–548.e524. doi: 10.1016/j.cell.2018.12.015 [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, . . . MacArthur DG (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. bioRxiv, 531210. doi: 10.1101/531210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer LS, Bader DM, Mertes C, Kopajtich R, Pichler G, Iuso A, . . . Prokisch H. (2017). Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun, 8, 15824. doi: 10.1038/ncomms15824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, & Shi W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res, 47(8), e47. doi: 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Online Mendelian Inheritance in Man, OMIM®. Retrieved from https://omim.org/
- Sazanov LA (2015). A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol, 16(6), 375–388. doi: 10.1038/nrm3997 [DOI] [PubMed] [Google Scholar]
- Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, . . . Ryan MT (2016). Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature, 538(7623), 123–126. doi: 10.1038/nature19754 [DOI] [PubMed] [Google Scholar]
- Tang AD, Soulette CM, van Baren MJ, Hart K, Hrabeta-Robinson E, Wu CJ, & Brooks AN (2020). Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nature Communications, 11(1), 1438. doi: 10.1038/s41467-020-15171-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eyk CL, Corbett MA, Gardner A, van Bon BW, Broadbent JL, Harper K, . . . Gecz J. (2018). Analysis of 182 cerebral palsy transcriptomes points to dysregulation of trophic signalling pathways and overlap with autism. Translational Psychiatry, 8(1), 88. doi: 10.1038/s41398-018-0136-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.