Abstract
Acoustofluidics, the fusion of acoustics and microfluidic techniques, has recently seen increased research attention across multiple disciplines due in part to its capabilities in contactless, biocompatible, and precise manipulation of micro-/nano-objects. Herein, a bimodal signal amplification platform which relies on acoustofluidics-induced enrichment of nanoparticles is introduced. The dual-function biosensor can perform sensitive immunofluorescent or surface-enhanced Raman spectroscopy (SERS) detection. The platform functions by using surface acoustic waves to concentrate nanoparticles at either the center or perimeter of a glass capillary; the concentration location is adjusted simply by varying the input frequency. The immunofluorescence assay is achieved by concentrating fluorescent analytes and functionalized nanoparticles at the center of the microchannel, thereby improving the visibility of the fluorescent output. By modifying the inner wall of the glass capillary with plasmonic Ag nanoparticle-deposited ZnO nanorod arrays and focusing analytes toward the perimeter of the microchannel, SERS sensing using the same device setup is achieved. Nanosized exosomes are used as a proof-of-concept to validate the performance of the acoustofluidic bimodal biosensor. With its sample-enrichment functionality, bimodal sensing, short processing time, and minute sample consumption, the acoustofluidic chip holds great potential for the development of lab-on-a-chip based analysis systems in many real-world applications.
Keywords: acoustofluidics, biosensors, immunofluorescent detection, lab-on-a-chip, surface-enhanced Raman spectroscopy
1. Introduction
Miniaturized biosensors have been recognized as formidable analytical devices for efficient detection of biomarkers such as cells, microorganisms, and biomolecules. These sensors play a critical role in monitoring and maintaining the health of human, animal, and plant communities, as well as aiding in the analysis of environmental and food safety factors.[1-4] To date, numerous biosensor platforms that allow for the detection of analytes in different source matrices have been developed, such as enzyme-linked immunosorbent assays, optical absorbance, chemiluminescence, radioactivity, and electrochemical assays.[5-8] Most of these techniques are still hindered by long processing times, large sample consumption, and/or extensive labeling procedures. These limitations often render these techniques impractical for many applications in which the concentration of analytes is low, or a high detection speed is required.
As an advancement of these methods, microfluidic approaches have recently been developed to improve biosensor technology by virtue of their low sample consumption and fast response times.[9-14] One area of microfluidic technology which has shown potential for the development of precision biosensors is acoustofluidics, which integrates acoustics and microfluidics. Acoustic waves are well-known for their versatility, biocompatibility, ease of integration, and precision in manipulating micro-/nano-particles, such as cells, bacteria, and exosomes.[15-19] These advantages have been leveraged to significantly advance the field of analytical assays.[20] Specifically, acoustofluidic methods present unique features for analytical applications, such as simple operation with gentle forces to move suspended objects, minute sample consumption for serial long-term monitoring, adjustable throughputs for both fundamental research and practical applications, and low power/instrument requirements for device miniaturization and outstanding component integration.[21-24] However, current acoustofluidic biosensor devices are limited to single modality, and the technology has rarely been utilized in the sensing and detection of subcellular structures.[19,20,25,26] Single-modality biosensors only measure one unique sensing parameter, such as fluorescence. When used in real-world applications, single-function biosensors often have limited versatility and accuracy, preventing widespread adoption. Therefore, there is still an unmet need for developing a flexible, sensitive, and simple lab-on-a-chip acoustofluidic system toward the sensing of biological targets (especially nanoscale analytes) with two or more sensing modalities.
Herein, we present an acoustofluidics-assisted bimodal sensing platform that can perform immunofluorescent and surface-enhanced Raman spectroscopy (SERS) based detection. In this platform, we used acoustofluidics to concentrate silica nanoparticles that have been coated with the target of interest at different locations of square-shaped glass capillaries for signal amplification. Immunoassay enhancement was realized by concentrating fluorescent analytes and functionalized nanoparticles toward the center of the glass capillary, significantly enhancing the output fluorescent signal. By simply adjusting the input frequencies, analytes which are bound to silica nanoparticles can also be focused on the edge of the capillary microchannel to achieve interaction with plasmonic ZnO─Ag nanorod arrays coated onto the device's surface. This dual-functionality can be achieved with a variety of biological samples, with the mechanism remaining the same for either immunofluorescent or SERS based detection. We validated the performance of our acoustofluidic biosensor using exosomes (30–150 nm) as a proof-of-concept. We were able to successfully detect the presence of these biomarkers in real-time even at minute concentrations (as low as tens of exosomes μL−1), using both of our sensing modalities. With its advantages in simplicity, versatility, and flexibility, our acoustofluidic biosensing platform may shed light on the development of advanced on-chip methodologies for many practical applications.
2. Results and Discussion
2.1. Working Concept of Acoustofluidics-Assisted Biosensors
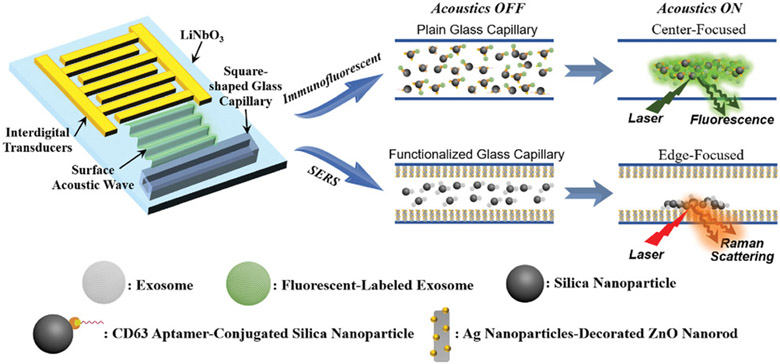
Figure 1 shows a schematic illustration of the acoustofluidics-assisted biosensing platform. The acoustofluidic device is composed of a transparent piezoelectric lithium niobate (LiNbO3) substrate with patterned interdigital transducers (IDTs) and a square-shaped glass capillary bonded to the substrate. When a radio frequency signal is applied to the IDTs, surface acoustic waves (SAWs) are generated and propagate toward the glass capillary. As SAWs encounter the sample liquid confined inside the capillary microchannel, they will generate longitudinal leakage waves and cause pressure fluctuations within the liquid. These pressure fluctuations result in lateral acoustic radiation forces, which drive suspended particles to either pressure nodes (minimum pressure regions) or antinodes (maximum pressure regions), depending on the particle and fluid properties.[22,27-30]
Figure 1.
Schematic diagram showing the working mechanism of the acoustofluidic biosensor (Objects are not drawn to scale). The bimodal platform functions by using surface acoustic waves to concentrate target of interest at either the center or perimeter of a glass capillary.
Here, we use acoustofluidic biosensors to analyze exosomes. Exosomes are membrane-bound phospholipid vesicles that have a typical size range of 30–150 nm and carry valuable molecular information, such as nucleic acids and proteins that could directly reflect the cells of origin for precise disease treatment.[31-34] They are a promising tool for health monitoring and the diagnosis of human diseases.[35] To enable exosome concentration and detection, we bind silica nanoparticles to exosomes for use as a sensing assistant due to their high surface-to-volume ratio and good biocompatibility.[36] As a result, acoustofluidics-induced aggregation and exosome enrichment by nanoparticles can enhance our biosensor's performance by driving exosomes which are bound to silica nanoparticles to desired locations for achieving different sensing modalities. Enriching biological particles at the center of the fluid chamber enables enhanced immunofluorescent identification by concentrating the signal from individual particles in one location, whereas, focusing biological particles at an edge near a plasmonic surface enables sensitive SERS recognition (Figure 1).
2.2. Acoustofluidic Device Fabrication and Enrichment Demonstration
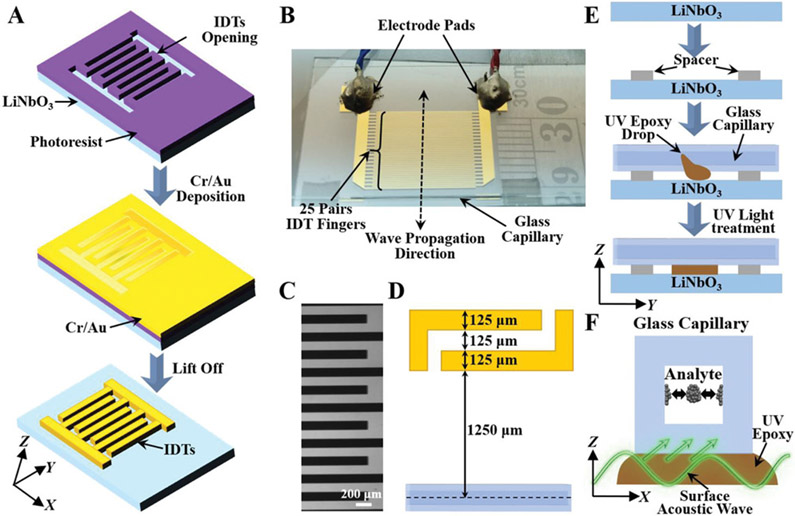
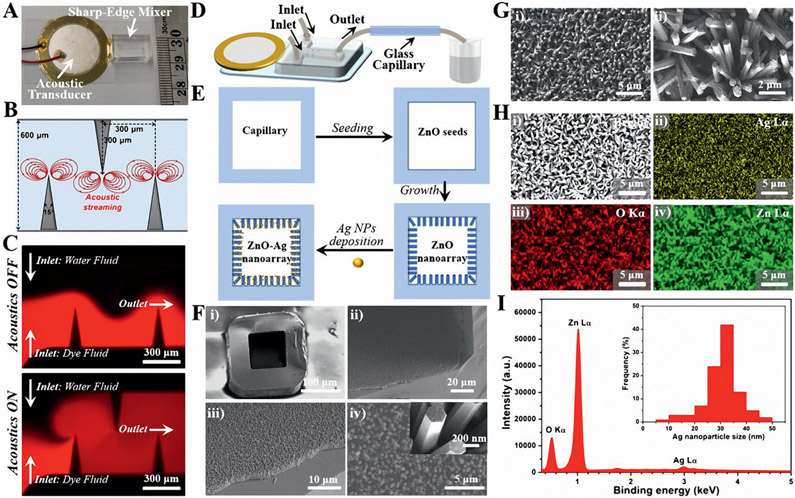
To generate SAWs for sample enrichment and biosensing, a LiNbO3 wafer was employed as the transparent substrate for IDT deposition by photolithography and e-beam evaporation techniques (Figure 2A,B). Two IDTs containing 25 electrodes each were patterned onto the LiNbO3 substrate. The width of each electrode and the gap distance between two adjacent electrodes are both 125 μm (Figure 2C-D), corresponding to a working wavelength of ≈200 μm (at a frequency of ≈7.5 MHz) inside the capillary microchannel. A square-shaped borosilicate hollow glass tube with an inner diameter and outer diameter of 100 and 200 μm, respectively, was bonded to the LiNbO3 substrate and aligned parallel to the electrodes (Figure 2E); this capillary was chosen due the fact that its inner diameter roughly matches the half wavelength of the acoustic wave, improving the concentration effect. When a radio frequency signal is applied, SAWs will be generated and travel along the X-direction. Then the acoustic wave can propagate into the glass capillary via the epoxy layer and generate an acoustic radiation force to concentrate biological particles suspended in the sample fluid (Figure 2F).[28,37,38]
Figure 2.
Development of the SAW-based acoustofluidic enrichment platform. A) Schematic of the fabrication process showing the formation of IDTs on a piezoelectric LiNbO3 substrate for surface acoustic wave generation. Objects are not drawn to scale. B) A photograph of the SAW-based acoustofluidic device used in the experiments with a ruler for scale. C) Structural pattern of IDTs under optical microscope. D) Design parameters of SAW-based acoustofluidic enrichment setup. E) Schematic diagram showing the integration process of the glass capillary onto the LiNbO3 substrate. F) Schematic of surface acoustic wave propagation for sample enrichment inside the glass capillary.
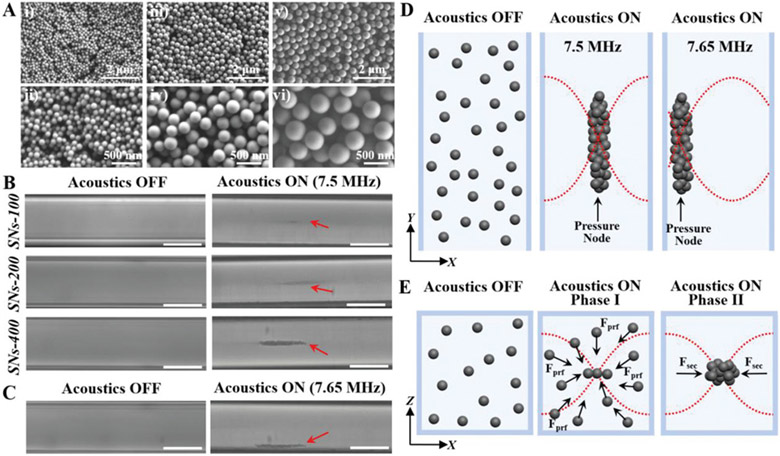
To demonstrate the sensing capabilities of nanoparticles and to examine the relationship between particle size and sensing performance, we first performed experiments to concentrate different-sized silica nanoparticles using our SAW-based acoustofluidic platform. Solid silica nanoparticles were synthesized by a modified Stöber method using tetraethyl orthosilicate (TEOS) as a silica precursor and NH3•H2O as a catalyst in a mixed solvent of water and ethanol.[39] The sizes of the nanoparticles are conveniently controlled by the molar ratios of the reaction mixture. As shown in Figure 3A, well-defined silica nanoparticles with relatively uniform particle sizes and smooth surfaces can be synthesized on a large scale and imaged under a scanning electron microscope (SEM). By increasing the molar ratios of TEOS and NH3•H2O, the particle sizes of the silica nanoparticles were gradually increased. Statistical size distributions of 200 silica nanoparticles under SEM were further analyzed (Figure S1, Supporting Information). The results showed that all three types of silica nanoparticles exhibit narrow size distributions with mean diameters and standard deviations of 94 ± 5, 206 ± 9, and 395 ± 12 nm, which are denoted as SNs-100, SNs-200, and SNs-400, respectively. The sensing performance of each size of silica nanoparticles was then analyzed using our SAW-based acoustofluidic device. As shown in Figure 3B, after injecting silica nanoparticle solutions (0.5 μL, 200 μg mL−1) into the glass capillary, no aggregation was observed in the absence of acoustics within 2 min, suggesting that the particles were well suspended in the solution. On the other hand, when the IDTs were driven with an applied voltage and frequency of 10 V and 7.5 MHz, respectively, silica nanoparticles were successfully focused into the center of the glass capillary and formed a straight-line parallel to the bonded glass capillary. The larger the particle size, the more obvious the concentration spot and the shorter time necessary for concentration. In addition, by changing the applied frequency from 7.5 to 7.65 MHz, it was found that the particles can be focused at the edge of the glass capillary (Figure 3C), indicating tunable settings for multimodal biosensing applications.
Figure 3.
Demonstration of the acoustofluidic enrichment of different-sized silica nanoparticles inside the glass capillary. A) SEM images of SNs-100, SNs-200, and SNs-400 with particle sizes around 100 nm (i, ii), 200 nm (iii, iv), and 400 nm (v, vi), respectively. B) Top view of particle solution inside the glass capillary when the acoustics is OFF and ON at 7.5 MHz. Scale bar = 100 μm. C) Top view of solution with SNs-400 when the acoustics is OFF (left) and ON by changing the applied frequency from 7.5 to 7.65 MHz (right). Scale bar = 100 μm. D) Schematic illustration showing the use of acoustics for directed transport of silica nanoparticles. E) Typical acoustic particle trapping configuration when interdigital transducers emit sound in a direction perpendicular to the flow. Fprf and Fsec indicate the primary radiation forces and the secondary forces, respectively.
To unveil the behavior of particles inside the microchannel, we analyzed the motion of nanoparticles by examining the acoustic radiation force and the viscous drag force (the gravity force and the buoyant force are not calculated since they are similar in magnitude but opposite in direction, and thus nearly balanced over a short period of analysis time in the fluid).[27,29,40,41] In the acoustic field, the viscous drag force (Fv) and the acoustic primary radiation force (Fr) on the particle can be expressed as[27,29,40-42]
| (1) |
| (2) |
| (3) |
Equation (1) expresses the viscous drag force on a particle related to its inherent properties, in which η, r, and υ correspond to the medium viscosity, particle radius, and the relative velocity, respectively. Equation (2) shows the primary acoustic force exerted on a particle, which depends on several parameters including acoustic pressure amplitude (P0), compressibility of medium (βm), particle volume (Vp), wavelength (λ), wave number (k), and distance from a pressure node (x). Importantly, the location at which the particles are focused is dictated by the value of the acoustic contrast factor (φ) defined in Equation (3), where the density and compressibility of the particles and the medium are denoted as (ρp, βp) and (ρm, βm), respectively. Particles with higher densities and lower compressibilities relative to the fluid tend to focus along the pressure nodes (φ>0), whereas particles with the opposite properties tend to focus along the pressure antinodes (φ<0). Given the relatively higher density and lower compressibility of silica particles than those of the aqueous solution in this study, the positive acoustic contrast property of silica nanoparticles renders them to focus along the pressure nodes (Figure 3D).[29,43] Specifically, when the transducer is active, a standing wave will be generated in the capillary microchannel and create pressure nodes, where the primary radiation force can drive particulate objects of the fluid to aggregate. Once a cluster is formed, the secondary radiation force and the lateral component of the primary radiation force will keep the objects together and counteract the drag force on the cluster while new objects can be transported to the pressure nodes (Figure 3E).[44,45] In addition, all silica particles in the experiments have the same density and compressibility, but are different in size. Since the viscous drag force and the acoustic radiation force are proportional to the particle radius (r) and the particle volume (r3), respectively, larger-sized particles should experience much stronger net forces and thus move faster toward the pressure nodes than smaller ones, which theoretically supports the observations from Figure 3B. Furthermore, the wavelength of the acoustic signal is given by λ = υ/f, where υ and f indicate the acoustic velocity and frequency, respectively. Since the acoustic velocity is fixed (≈1495 m s−1),[46] the wavelength of the acoustic waves can be altered by changing the applied frequency. Therefore, by changing the frequency from 7.5 to 7.65 MHz in the experiments, the focused region of silica particles can be relocated from the center to the edge of the glass capillary (Figure 3B-D).
2.3. Signal Amplification of Immunofluorescent Assay via the Acoustofluidics-Assisted Biosensor
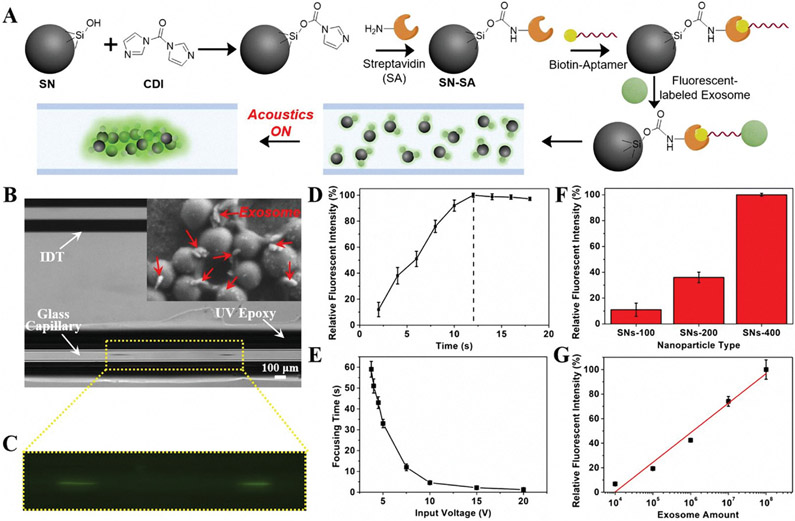
To realize an enhanced immunoassay of exosomes, we used our acoustofluidic device for signal enhancement. As demonstrated above, our device enables the concentration of different-sized silica nanoparticles with a simple setup in a contactless and rapid manner. To validate the feasibility of the acoustofluidics-based immunoassay, we functionalized silica nanoparticles with exosome-specific aptamers for recognition and concentration of exosomes. As shown in Figure 4A, streptavidin molecules were first immobilized on the silica surface directly by the aid of N,N’-carbonyldiimidazole (CDI). The CDI was used to activate the silanol groups on the silica particle surface, promoting the conjugation process to the primary amine groups of streptavidin molecules.[47,48] The resultant particles were then modified with biotinylated CD63 aptamer, containing a sequence of 32 bases (CACCCCACCTCGCTCCCGTGACACTAATGCTA),[49] for specific recognition of exosomes. As such, if fluorescent-labeled exosomes are present in a solution that is mixed with aptamer-conjugated silica particles, they will be captured by the silica particles. After applying acoustic waves, the exosome-bound particles can be concentrated at the center of a glass capillary for fluorescent analysis. The presence of silica particles significantly enhances the fluorescent signals as compared to randomly distributed signals (Figure 4A).
Figure 4.
Acoustofluidics-assisted immunofluorescent detection of exosomes. A) Schematic diagram showing the immunoassay process. B) Top view microscope image of solution with exosome-conjugated SNs-400 when the SAW was ON. The inset is a SEM image showing the interactions of exosomes with SNs-400. C) Enlarged image of solution when excited by a 365 nm laser. D) Relative fluorescent intensity changes of exosomes-conjugated SNs-400 as enrichment time increased when using an input voltage of 7.5 V. The dashed line indicates the focusing time when the fluorescent intensity reached the maximum level. E) Focusing time changes of exosomes-conjugated SNs-400 as the input voltage increased. F) Comparison of different-sized nanoparticles for immunofluorescent detection of exosomes. G) Relative fluorescent intensity of exosomes-conjugated SNs-400 at different concentrations of exosomes and the fitted linear relationship between exosome quantity and fluorescent intensity (γ = 24.12x – 96.2, R2 = 0.973). The concentrations of exosomes and silica particles in (B–F) were 106 exosomes μL−1 and 0.2 μg μL−1, respectively. The driving voltage and frequency are 10 V and 7.5 MHz, respectively, in these tests unless otherwise stated.
To reveal the immunofluorescent sensing performance of our acoustofluidic platform, we utilized a standard ExoStd human urine exosome sample that was labeled with green fluorescence from BioVision (Figures S2 and S3, Supporting Information). After mixing aptamer-conjugated silica nanoparticles (SNs-400) with fluorescent-labeled exosomes for 10 min, we injected the mixture into the glass capillary and applied a voltage and frequency of 10 V and 7.5 MHz, respectively. As shown in Figure 4B, particle aggregation was observed at the center of the glass capillary. When excited by the fluorescent light (≈488 nm), strong green fluorescent signals were detected (Figure 4C), suggesting the rapid and efficient capture of exosomes by CD63 aptamer-conjugated silica nanoparticles. Comparatively, when the acoustics was off, there was no obvious particle aggregation observed under the microscope (Figure S4, Supporting Information), and the fluorescent signal was uniformly distributed across the capillary microchannel (Figure S5, Supporting Information). In addition, when silica particles were mixed with only phosphate buffer solution (PBS) (i.e., absence of exosomes), no fluorescent signal was detected (Figure S6, Supporting Information). These results not only successfully demonstrated fluorescent signal enhancement by our acoustofluidic platform, but also showed a straightforward and simple method for detecting the presence of the target molecules without introducing any unnecessary manual steps such as centrifugation, washing, and pipetting.
To further validate the immunofluorescent assays, we examined a series of experimental parameters and quantified the relative fluorescent intensity under different conditions. The focusing time was first evaluated with CD63 aptamer-conjugated SNs-400 and fluorescent-labeled exosomes at an input voltage of 7.5 V (Figure 4D). Since the exosomes which are bound to silica nanoparticles were gradually concentrated until reaching a maximum level at the center of the glass capillary, the fluorescent intensity was therefore continuously increased to a near-steady value. This interval of increasing fluorescent intensity represents the focusing time. We also examined the effect of applied voltage on the focusing time. As shown in Figure 4E, the higher the input voltage, the shorter the focusing time. It is noted that our acoustofluidic device can achieve successful concentration with an input voltage as low as ≈3.75 V, though the focusing time was prolonged to roughly 1 min due to a reduction of acoustic radiation efficiency. In addition, the effect of particle size on the immunofluorescent sensing performance was investigated. As shown in Figure 4F, larger-sized particles generate a stronger fluorescent signal owing to the relatively higher acoustic radiation force, which is also in agreement with the results from Figure 3B. The fluorescent intensity of the SNs-400 exosome sensing substrate is nearly 10 times higher than that of the SNs-100 particles. Furthermore, we tested the immunofluorescent performance of the acoustofluidic setup with different quantities of exosomes from 104 to 108 (Figure 4G). The fluorescent signal was found to be linearly related to the quantities of exosomes (R2 = 0.973), and a limit of detection of 1.3 × 103 exosomes μL−1 was achieved, revealing the excellent performance of such acoustofluidics-assisted immunoassay (Table S1, Supporting Information). With a sample volume of only 0.5 μL required for analysis, such an acoustofluidic platform demonstrates promising potential in point-of-care applications.
2.4. Acoustofluidic Engineering of 3D Plasmonic ZnO─Ag Nanoarray
To realize the SERS sensing assay of exosomes with enhanced optical scattering, we generated Ag nanoparticles-coated ZnO nanorod array on the inner surface of the square-shaped glass capillary. To ensure relatively uniform patterning of the plasmonic nanoarray during fabrication, rapid and efficient mixing of reactants is required.[50-54] Here, we utilized an acoustofluidic sharp-edge mixer with two inlets and one outlet for mixing reactants (Figure 5A; and Figure S7, and see the Supporting Information for the optimization process of device parameters). Upon the actuation of the acoustic transducer with a frequency of 4.25 kHz, the sharp-edge structures oscillate. These oscillations can generate a pair of counter-rotating vortices in the fluid (acoustic streaming) around the tip of each sharp-edge (Figure 5B). COMSOL simulation results confirm that no acoustic streaming pattern is observed when the acoustic transducer is OFF, whereas, the acoustic streaming phenomenon arises around the tips of the sharp-edges when the transducer is activated (Figure S8, Supporting Information). To visualize the mixing performance of the acoustofluidic sharp-edge mixer, we injected Rhodamine B dye solution into one inlet and water into the other (see the Supporting Information for the details of operating conditions). As shown in Figure 5C, when the acoustic transducer is OFF, no obvious mixing was observed as revealed by the clear side-by-side laminar flow profile. In contrast, when the acoustic transducer is ON, rapid and homogeneous mixing of the dye and water fluids was achieved even only flowing through the first two sharp-edges (Figure S9, and see the Supporting Information for the mixing details).
Figure 5.
Acoustofluidic synthesis of functional plasmonic nanoarray inside square-shaped glass capillaries. A) A photograph of an acoustofluidic sharp-edge mixer device with a ruler for scale. B) The design parameters and the proposed acoustic streaming pattern of the sharp-edge micromixer. C) Fluorescent images for comparing the mixing performance of water fluid and dye fluid (Rhodamine B) when the acoustic transducer is OFF (top) and ON (bottom). D) Schematic diagram showing the operation of the acoustofluidic sharp-edge mixer for plasmonic nanoarray synthesis. E) Schematic workflow showing the synthesis process of the plasmonic nanoarray inside the glass capillary. F) SEM images showing the structures of the ZnO nanoarray. The inset in (F-iv) indicates the smooth surface of the ZnO nanorods. G) SEM images showing the structures of the ZnO─Ag nanoarray. H) SEM image (i) and the corresponding element mapping profiles of Ag ii), O iii), and Zn iv). I) EDS spectrum of ZnO─Ag nanoarray, and the inset is the size distributions of Ag from 200 random particles under TEM.
Since the acoustofluidic sharp-edge mixer drastically enhances the mass transport across the microchannel by perturbing the bulk fluids and breaking the interface of the laminar fluids, it provides great potential for engineering functional 3D plasmonic nanoarrays inside a square-shaped glass capillary. As shown in Figure 5D, the acoustofluidic sharp-edge mixer was employed to mix two reactant fluids and coat plasmonic nanoarrays on the inner wall of the glass capillary. A two-step seeding and growth approach was first adopted to pattern a ZnO nanorod array inside a capillary, and then Ag nanoparticles were deposited onto the nanorods (Figure 5E). To seed the ZnO layer, the two inlet fluids, one containing zinc acetate and the other sodium hydroxide, were pumped to fill the capillary. Meanwhile, the acoustic transducer was activated with an applied voltage and frequency of 10 V and 4.25 kHz, respectively. The growth step of the ZnO nanoarray was similar to the seeding step except that the reactants were replaced by zinc nitrate and hexamethylenetetramine. As shown in Figure 5F, a relatively dense and uniform ZnO nanorod array can be successfully patterned inside the square-shaped glass capillary due to the efficient mixing at both the seeding and the growth steps (Figure S10, Supporting Information). The length and diameter of the ZnO nanorods are around 3 μm and 200 nm, respectively, and the very smooth surface of the nanorod structures can be observed (Figure 5F-iv). However, it is noted that there was no regular ZnO array formed in the absence of acoustics (Figure S11, Supporting Information) and that prolonging the reaction time during the growth step can significantly change the structural morphology from nanorod to nanosheet (Figure S12, Supporting Information). To construct the plasmonic nanoarray, we deposited Ag nanoparticles on the ZnO nanorods by replacing the reactants with silver nitrate and sodium borohydride in the acoustofluidic sharp-edge mixer. As shown in Figure 5G, SEM images demonstrate that the nanorod structures were well maintained after Ag particle deposition, but the smooth surface of the ZnO nanorods became coarse. After scratching samples from the inner wall of the glass capillary, TEM characterization further confirmed that a large number of small-sized nanoparticles had adhered to the surface of the ZnO nanorods (Figure S13, Supporting Information). Element mapping profiles verified the existence of Ag, O, and Zn elements and also showed the intensive distribution of Ag nanoparticles on ZnO nanorods (Figure 5H). Energy dispersive X-ray spectroscopy (EDS) analysis further revealed that a 5.8 wt% Ag content was adhered to the ZnO nanoarray, and that the sizes of the Ag nanoparticles were mainly distributed in the size range of 20–40 nm in diameter (Figure 5I).
2.5. Signal Amplification of SERS Assay via the Acoustofluidics-Assisted Biosensor
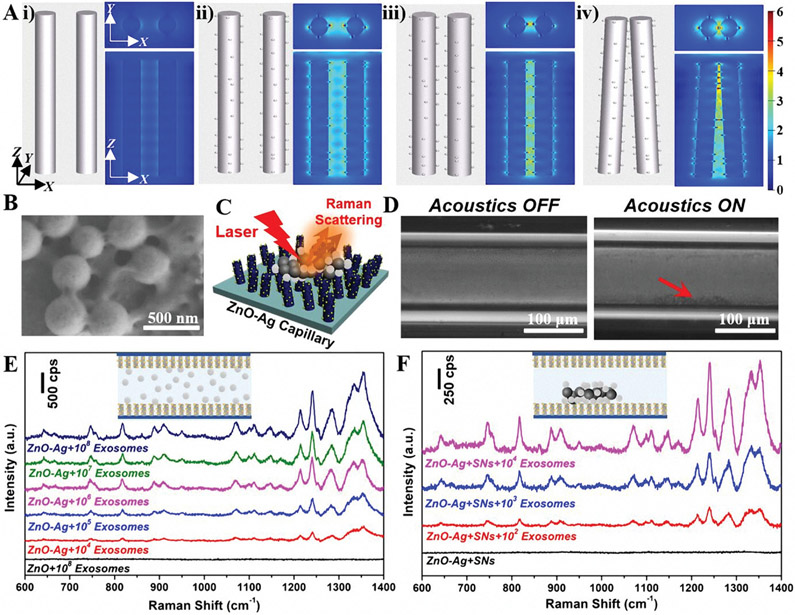
To validate the structural design of the 3D plasmonic nanoarray for SERS sensing, finite-difference time-domain (FDTD), a numerical tool for modeling computational electrodynamics, was utilized.[55] The electromagnetic field distribution was simulated over two identical ZnO nanorods and ZnO─Ag nanorods with different patterns. The electromagnetic field enhancement is minimal on parallel pristine ZnO nanorods without Ag nanoparticles (Figure 6A-i). Comparatively, electromagnetic field enhancement starts to appear when Ag nanoparticles are attached to the surface of ZnO nanorods due to the excitation of the localized surface plasmon resonance on the Ag metal surface (Figure 6A-ii). In addition, the electromagnetic field is further magnified if the ZnO─Ag nanorods approach each other (Figure 6A-iii). Moreover, since the ZnO─Ag nanorod array inside the capillary is not in a parallel-aligned pattern (as evident from Figure 5), we also simulated a model where the ZnO─Ag nanorods lean toward each other. The results reveal a dramatically enhanced electromagnetic field, especially from the neighboring regions (Figure 6A-iv), arising from the attractive “hot-spots” in the field intensity.[56, 57] As a result, the 3D ZnO─Ag inside the glass capillary induces numerous SERS “hot-spots” among neighboring Ag nanoparticles on both the same and adjacent ZnO nanorods (Figure S14, Supporting Information). This finding not only highlights the importance of structural design when applying plasmonic materials to biosensing applications, but also reveals the great potential of our 3D plasmonic nanoarray system for serving as an excellent SERS substrate.
Figure 6.
Acoustofluidics-assisted SERS detection of exosomes. A) FDTD simulations showing the electromagnetic field distributions of different nanoarray structures. i) pristine nanorods with a gap distance of 200 nm with limited electromagnetic field enhancement; ii) ZnO─Ag nanorods with a gap distance of 200 nm showing strong electromagnetic field enhancement; iii) ZnO─Ag nanorods with a gap distance of 100 nm showing further magnified electromagnetic field enhancement; iv) Tilted plasmonic nanorods with ends clustered together showing dramatically enhanced electromagnetic field. B) SEM images of SNs-400 after interaction with exosomes under mild sonication treatment. C) Schematic diagram showing the nanoparticle-assisted SERS sensing mechanism of the ZnO─Ag capillary toward exosomes. D) Top view microscope image of solution with exosomes which are bound to silica nanoparticles (SNs-400) when the SAW was OFF and ON (10 V, 7.65 MHz). E) SERS spectra of exosomes with concentrations ranging from ≈104 to 108 μL−1 using ZnO─Ag capillary without the assistance of silica particles (10 V, 7.65 MHz). F) SERS spectra of exosomes with concentrations ranging from ≈102 to 104 μL−1 using the nanoparticle-assisted acoustofluidic method (10 V, 7.65 MHz).
To develop sensitive acoustofluidics-assisted SERS biosensors, we adopted a nanoparticle-assisted strategy that makes exosomes bind to silica particles for manipulation via SAWs. Before testing exosomes, we found that nanoparticles can be concentrated at different locations of the decorated capillary by simply adjusting the applied frequency (Figure S15, Supporting Information), further confirming the robust capability of our acoustofluidic bimodal system. When mixing exosomes and nanoparticles under a mild sonication treatment, the viscosity of the exosome membrane significantly decreases and thus create cavities in the exosomal lipid bilayer, promoting attachment to nanoparticles.[58-60] As shown in the SEM images of Figure 6B, after treatment, the smooth particle surface of and clear boundaries among the silica nanoparticles disappeared, indicating the presence of an irregular layer of organic vesicles on the particle surface.[61] These results indicate the successful attachment of exosomes to silica particles, enabling enhanced acoustic focusing and SERS sensing of exosome analytes at the edge locations (Figure 6C). To demonstrate this, we injected the mixture of exosomes which are bound to silica nanoparticles (SNs-400) into a ZnO─Ag-coated square-shaped glass capillary. As shown in Figure 6D, there was a relatively uniform distribution of exosomes which are bound to silica nanoparticles in the absence of acoustics; whereas, an obvious agglomeration can be observed at the edge of the glass capillary when the acoustics are activated with an applied voltage and frequency of 10 V and 7.65 MHz, respectively.
To demonstrate the SERS sensing performance of our acoustofluidics-assisted plasmonic nanoarray system, we measured the SERS spectra of standard human urine exosomes with and without the assistance of nanoparticles using a Raman spectrophotometer. For the label-free SERS detection of exosomes (without the assistance of nanoparticles), a 0.5 μL sample with different concentrations (≈104–108 exosomes μL−1 that were determined by nanoparticle tracking analysis, Figure S16, Supporting Information) was directly injected into the ZnO─Ag capillary. As shown in Figure 6E, in the absence of Ag nanostructures (i.e., ZnO nanorod array only), there were no observable SERS peaks, which is also well in agreement with our simulation results (Figure 6A). However, many sharp peaks were observed using the plasmonic nanoarray-coated capillary at a concentration as low as 104 exosomes μL−1 (Figure 6E), which can be attributed to the significantly enhanced Raman scattering induced by the localized surface plasmon resonance effect in the proximity of Ag nanostructures.[62] The major peaks shown in the Raman spectra are located at 642 cm−1 (weak, protein), 667 cm−11 (weak, nucleic acid), 749 cm−1 (medium, nucleic acid), 819 cm−1 (medium, protein), 889 cm−1 (medium, tryptophan), 910 cm−1 (medium, protein), 950 cm−1 (weak, lipid, protein), 1070 cm−1 (medium, lipid), 1111 cm−1 (weak, phenylalanine), 1148 cm−1 (weak, fatty acids), 1167 cm−1 (weak, protein), 1214 cm−1 (strong, protein), 1241 cm−1 (strong, lipid), 1286 cm−1 (strong, lipid, protein), 1339 cm−1 (strong, phospholipid), and 1356 cm−1 (strong, lipid).[63-65] These typical peaks mainly come from proteins, nucleic acids, and lipids, which are the main components of exosomes. In addition, without the presence of silica nanoparticles, there was no obvious difference in the intensity of Raman peaks between the acoustic ON and OFF status, suggesting the relatively uniform distribution of exosomes (Figure 6E-inset). Comparatively, exosomes which are bound to silica nanoparticles, even at a very low concentration, can be easily focused on the inner wall of a glass capillary (Figure 6D), which may significantly improve the SERS sensing performance. As shown in Figure 6F, there were no apparent peaks when only silica particles were present.[66-68] However, after we injected a mixture of exosomes which are bound to silica nanoparticles (SNs-400) into the ZnO─Ag-coated capillary and applied the acoustic signals, the intensity of all SERS peaks was dramatically increased because of the effective nanoparticle-assisted enrichment of exosome samples closer to the “hot-spots” of the plasmonic nanoarray. In addition, the SERS peaks were observed even at concentrations as low as ≈102 exosomes μL−1. To further demonstrate the applicability of our acoustofluidic platform, we tested human plasma-derived exosomes that were isolated using our previously developed acoustofluidic exosome isolation technique.[69] A linear detection range of 102–108 exosomes μL−1 was achieved and the detection limit was shown to be nearly 20 exosomes μL−1 (Figure S17, Supporting Information). These results not only successfully demonstrate the efficient label-free SERS detection of exosomes using a plasmonic nanoarray capillary, but also validate the greatly enhanced performance of acoustofluidics-assisted nanoparticle-enabled SERS sensing system (Table S1, Supporting Information).
3. Conclusion
In summary, we developed an acoustofluidics-assisted nanoparticle-enabled signal amplification platform as immunofluorescent and SERS bimodal exosome biosensors. The SAW-based acoustofluidic device was used to concentrate different-sized silica nanoparticles (100–400 nm) at the center or perimeter locations of a glass capillary by simply adjusting the input frequency. The larger the particle size, the faster the focusing time. The center-focused immunofluorescent assay was achieved by concentrating CD63 aptamer-conjugated silica particles and fluorescently-labeled exosomes. The immunoassay was realized within just a few seconds by applying a voltage of 10 V and above, but it can also be operated with an input voltage as low as 3.75 V. Larger-sized silica particles provide a more readable fluorescent signal, and a linear detection range from 104 to 108 exosomes μL−1 was obtained. Edge-focused SERS sensing was demonstrated by driving exosomes which are bound to silica nanoparticles toward the edge of a plasmonic nanoarray-coated glass capillary. An acoustofluidic sharp-edge mixer device designed for ultrafast and active mixing was utilized to engineer the Ag nanoparticles-deposited ZnO nanoarray inside the square-shaped glass capillary. FDTD simulations theoretically demonstrated the greatly enhanced electromagnetic field distribution of 3D ZnO─Ag plasmonic structural patterns. Label-free SERS sensing can recognize 104 exosomes μL−1, whereas, our acoustofluidics-assisted nanoparticle-enabled strategy can significantly amplify the signal for sensitive detection of exosomes at as low as 102 exosomes μL−1. In addition, a detection limit of ≈20 exosomes μL−1 was achieved from human plasma-derived exosome samples. Given its unique features such as its high sensitivity, fast response time, simple configuration, minute sample consumption, and multiple modalities, our acoustofluidic device holds great promise for advancing the development of point-of-care analysis and diagnosis platforms across a variety of biomedical applications.
4. Experimental Section
Materials and Reagents:
128° Y-Cut X-propagating lithium niobate (LiNbO3) substrate was obtained from Red Optronics. Square borosilicate hollow glass tubes with an inner diameter and outer diameter of 100 and 200 μm, respectively, were obtained from VitroCom. TEOS, ammonium hydroxide (NH3•H2O, 25%), 200-proof ethanol, N,N’-carbonyldiimidazole (CDI), streptavidin, dimethyl sulfoxide (DMSO), Rhodamine B, potassium permanganate (KMnO4), 1-butanol, zinc nitrate (Zn(NO3)2) hexahydrate, zinc acetate (Zn(OAc)2) dihydrate, hexamethylenetetramine (HMTA), sodium hydroxide (NaOH), 1H,1H,2H,2H-Perfluorooctyl trichlorosilane (PFOCTS), silver nitrate (AgNO3), and sodium borohydride (NaBH4) were purchased from Sigma-Aldrich. Polydimethylsiloxane (PDMS, Sylgard 184 kit) was obtained from Dow Corning. Biotinylated CD63 aptamer with a sequence of 32 bases (CACCCCACCTCGCTCCCGTGACACTAATGCTA) was provided by Integrated DNA Technologies. Acoustic transducer was obtained from PUI Audio (AB2720B-LW100-R). Fluorescent polystyrene beads (1 μm) were provided from Bangs Laboratory. ExoStd human urine fluorescent exosome and ExoStd human urine exosome standard samples were purchased from BioVision. Human plasma from a healthy donor was bought from ZenBio.
IDT Device Fabrication:
The acoustofluidic enrichment device used in the experiments is composed of a LiNbO3 substrate with IDTs patterned on top surface and a square-shaped glass capillary that is bonded via UV epoxy. Two IDT structures that have 25 pairs of electrodes were fabricated on a LiNbO3 substrate with an identical 125 μm spacing distance between two adjacent electrodes. Briefly, the LiNbO3 wafer was first patterned with photoresist, two metal layers (Cr/Au, 50/500 Å) were then deposited on the wafer by a photolithography process and an e-beam evaporation process successively. A lift-off process was finally used to remove the photoresist and the attached metals to obtain the IDTs for acoustic wave generation. The glass capillary was aligned parallel to the IDTs above the LiNbO3 substrate and bonded by exposure of UV epoxy to UV light for 20 min. To keep the glass capillary level during the bonding process, adhesive tape with a thickness of ≈100 μm was first placed on the LiNbO3 substrate as a spacer. Then, a small volume of UV epoxy was dripped between the glass capillary and the LiNbO3 substrate. After filling the gap between the glass capillary and the substrate via the capillary effect, the UV epoxy was exposed to UV light to cause it to solidify.
Sharp-Edge Device Fabrication:
The acoustofluidic mixer is composed of a glass coverslip, an acoustic transducer, and a PDMS channel with four sharp-edge structures. In brief, after designing the sharp-edge pattern with AutoCAD software, a film mask was obtained to fabricate the master silicone mold using standard photolithography. After treatment with PFOCTS, the PDMS replica was obtained by pouring PDMS precursor onto the silicone mold. The inlet and outlet of PDMS channel were punched to bond with the cover glass through oxygen plasma treatment. Finally, an acoustic transducer was bonded near PDMS to make the acoustofluidic mixer device.
Devices Setup and Operation:
Both acoustofluidic devices were driven by sinewave signals from Tektronix AFG3011C function generator. IDTs-based acoustofluidic enrichment device was mounted on the stage of an inverted microscope (Eclipse Ti-U, Nikon, Japan), which is equipped with a CCD camera for recording the motion of the particles inside the glass capillary. For the acoustofluidic sharp-edge mixer device, the reagents were delivered to the microchannels by BD Bioscience syringes, which were operated by an external automated neMESYS syringe pump.
Synthesis of Silica Nanoparticles:
Different-sized silica nanoparticles were synthesized by a modified Stöber method using TEOS as a silica precursor and NH3•H2O as a catalyst.[39] Briefly, NH3•H2O was added into a mixture of water and ethanol with stirring for 10 min. TEOS was then added with vigorous stirring for 2 h at room temperature. The size of silica nanoparticles was controlled by the molar ratios of the reaction mixture. Here, the molar ratios of the reaction mixture (H2O:CH3CH2OH:NH3•H2O:TEOS) for the synthesis of SNs-100, SNs-200, and SNs-400 are 1400:850:27:7, 1400:850:40:14, and 1400:850:60:25, respectively. Different-sized silica nanoparticles were collected by centrifugation and then washed several times with ethanol and water. The solid products were dried at 65 °C for further use.
Immunofluorescent Assay:
Streptavidin molecules were first immobilized on the silica surface directly by the aid of CDI without any further intermediate chemical modifications.[47,48] Typically, silica nanoparticles (10 mg) and CDI (1 mg) were dispersed into anhydrous DMSO (20 mL). After stirring for 2 h at room temperature, the CDI-activated silica nanoparticles were collected by centrifugation and washed with several times with DMSO and PBS (pH 7.4). The particles were redispersed into PBS (10 mL) and then streptavidin (0.2 mg) was added. The mixture was vigorously stirred for 12 h at 4 °C. After centrifugation and thoroughly rinsing with PBS several times, the solid samples were mixed with biotinylated aptamer (10 × 10−9 m) and incubating at 4 °C for 30 min for preparing aptamer-conjugated silica nanoparticles. For the immunoassay, the quantification of exosomes was determined by using Nanosight LM10 nanoparticle tracking analysis (NTA, Malvern Panalytica). In general, fluorescent-labeled exosome suspension (10 μL) was mixed with aptamer-conjugated silica nanoparticles (10 μL). The mixture was placed at room temperature for 10 min with gentle shaking to finish the binding, and then an aliquot of the solution was injected into glass capillary for fluorescent analysis. The relative fluorescent intensity was analyzed by ImageJ software (NIH, Bethesda, MD).
Development of Plasmonic ZnO─Ag Nanoarray-Coated Glass Capillary:
The construction of plasmonic ZnO─Ag nanoarray-coated glass capillary is relied on the acoustofluidic sharp-edge mixer device. Specifically, ZnO nanoarray was first engineered inside glass capillary via a two-step seeding and growth approach.[70,71] The glass capillary was activated by freshly prepared KMnO4 (5 × 10−3 m, containing 50 μL 1-butanol per 20 mL solution) at 65 °C for 15 min. ZnO seeds were then patterned using one inlet containing Zn(OAc)2 (2.5 × 10−3 m) and the other NaOH (5 × 10−3 m). Both were operated at the same flow rate of 1 μL min−1. During the pumping process, the acoustofluidic sharp-edge mixer device was driven with an applied voltage and frequency of 10 V and 4.25 kHz, respectively. After placed at 75 °C for 1 h, the capillary was transferred to the growth stage using aqueous solutions of Zn(NO3)2 (2 × 10−3 m) and HMTA (2 × 10−3 m) and a reaction temperature of 90 °C. When the reaction was complete after 30 min, 3D ZnO-patterned capillary was cleaned with water and dried at 65 °C.
Plasmonic ZnO─Ag nanoarray was then realized using a similar acoustofluidic setting as mentioned above by a direct reduction of AgNO3 with NaBH4. In brief, two inlet flows, one containing AgNO3 (1 × 10−3 m) and the other NaBH4 (3 × 10−3 m), were delivered into the sharp-edge device with an flow rate, applied voltage, and frequency of 1 μL min−1, 10 V, and 4.25 kHz, respectively. After continuously reacted for 30 min, the capillary device was rinsed thoroughly, and the resulting black-colored capillary was dried for future use.
Materials Characterization:
The size and morphology of silica nanoparticles and nanoarray materials inside microchannel were investigated using a Thermo Scientific Apreo SEM. Square-shaped glass capillaries were broken into ≈0.5 cm pieces before characterization. EDS equipped on SEM was used to evaluate local composition distribution of the nanoarray samples. FEI Tecnai G2 Twin TEM was employed to characterize the samples that were scratched from the inner wall of glass capillary.
FDTD Simulation of Electromagnetic Field Enhancement:
The FDTD method was utilized to simulate the electromagnetic field distribution of nanoarray inside glass capillary. The plasmonic nanostructures were simplified as ZnO cylinders functionalized by spherical Ag nanoparticles. As revealed by SEM, ZnO cylinders with a height of 3000 nm and a radius of 100 nm were used to imitate the ZnO nanorods. Ag nanoparticles with a radius of 15 nm were randomly placed on the ZnO rods. For reflecting the practical sensing conditions, a plane wave was utilized to illuminate the whole structure immersed in a water solution.
SERS Sensing Assay:
In general, ExoStd human urine exosome standard samples (10 μL) with different concentrations that were determined by NTA system were first mixed with silica nanoparticles (10 μL). The quantity ratio of exosomes to nanoparticles was kept at around 1:1. The mixture was placed at room temperature for 30 min with mild sonication (240 W, 6 cycles of 30 s on/off for 3 min with a 2 min cooling period between each cycle[58,59]) for evaluating the SERS sensing performance of ZnO─Ag capillary. The analytes (0.5 μL) were injected into the capillary microchannel containing plasmonic ZnO─Ag nanoarrays. Human plasma-derived exosomes, which were isolated by the previously developed acoustic method,[69] were treated by a similar way with silica particles before SERS analysis. The concentrations of exosomes were determined by Nanosight NTA analysis. A Horiba Jobin Yvon LabRAM Aramis Raman spectrophotometer was used to collect Raman scattering spectra. A diode laser was used for excitation at 785 nm, and the accumulation time was 15 s.
Statistical Analysis:
NTA results were obtained as means and standard errors of means of five independent measurements. All immunoassay and SERS tests were evaluated at least three times, and the data were presented as means and standard deviations.
Supplementary Material
Acknowledgements
The authors acknowledge support from the National Institutes of Health (Grant Nos. R01GM135486, UG3TR002978, and R43AG063643) and National Science Foundation (Grant No. ECCS-1807601). Z.P., P.L., and L.S. acknowledge the financial support from the China Scholarship Council (CSC).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
T.J.H. has co-founded a start-up company, Ascent Bio-Nano Technologies Inc., to commercialize technologies involving acoustofluidics and acoustic tweezers.
References
- [1].Turner APF, Science 2000, 290, 1315. [DOI] [PubMed] [Google Scholar]
- [2].Baeumner AJ, Anal. Bioanal. Chem 2003, 377, 434. [DOI] [PubMed] [Google Scholar]
- [3].Khater M, de la Escosura-Muñiz A, Merkoçi A, Biosens. Bioelectron 2017, 93, 72. [DOI] [PubMed] [Google Scholar]
- [4].Neethirajan S, Tuteja SK, Huang S-T, Kelton D, Biosens. Bioelectron 2017, 98, 398. [DOI] [PubMed] [Google Scholar]
- [5].Quesada-González D, Merkoçi A, Biosens. Bioelectron 2015, 73, 47. [DOI] [PubMed] [Google Scholar]
- [6].Borisov SM, Wolfbeis OS, Chem. Rev 2008, 108, 423. [DOI] [PubMed] [Google Scholar]
- [7].Ronkainen NJ, Halsall HB, Heineman WR, Chem. Soc. Rev 2010, 39, 1747. [DOI] [PubMed] [Google Scholar]
- [8].Roda A, Mirasoli M, Michelini E, Fusco MD, Zangheri M, Cevenini L, Roda B, Simoni P, Biosens. Bioelectron 2016, 76, 164. [DOI] [PubMed] [Google Scholar]
- [9].Contreras-Naranjo JC, Wu H-J, Ugaz VM, Lab Chip 2017, 17, 3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hao N, Zhang JXJ, Sep. Purif. Rev 2018, 47, 19. [Google Scholar]
- [11].Hao N, Zhang JXJ, Biomicrofluidics 2019, 13, 051501. [Google Scholar]
- [12].Im H, Lee K, Weissleder R, Lee H, Castro CM, Lab Chip 2017, 17, 2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gholizadeh S, Shehata Draz M, Zarghooni M, Sanati-Nezhad A, Ghavami S, Shafiee H, Akbari M, Biosens. Bioelectron 2017, 91, 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hao N, Nie Y, Zhang JXJ, Int. Mater. Rev 2018, 63, 461. [Google Scholar]
- [15].Ozcelik A, Rufo J, Guo F, Gu Y, Li P, Lata J, Huang TJ, Nat. Methods 2018, 15, 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Melde K, Mark AG, Qiu T, Fischer P, Nature 2016, 537, 518. [DOI] [PubMed] [Google Scholar]
- [17].Tian Z, Yang S, Huang P-H, Wang Z, Zhang P, Gu Y, Bachman H, Chen C, Wu M, Xie Y, Huang TJ, Sci. Adv 2019, 5, eaau6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ahmed D, Ozcelik A, Bojanala N, Nama N, Upadhyay A, Chen Y, Hanna-Rose W, Huang TJ, Nat. Commun 2016, 7, 11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Barani A, Paktinat H, Janmaleki M, Mohammadi A, Mosaddegh P, Fadaei-Tehrani A, Sanati-Nezhad A, Biosens. Bioelectron 2016, 85, 714. [DOI] [PubMed] [Google Scholar]
- [20].Xie Y, Bachman H, Huang TJ, Trends Anal. Chem 2019, 117, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang SP, Lata J, Chen C, Mai J, Guo F, Tian Z, Ren L, Mao Z, Huang PH, Li P, Yang S, Huang TJ, Nat. Commun 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shi J, Ahmed D, Mao X, Lin S-CS, Lawit A, Huang TJ, Lab Chip 2009, 9, 2890. [DOI] [PubMed] [Google Scholar]
- [23].Ozcelik A, Nama N, Huang P-H, Kaynak M, McReynolds MR, Hanna-Rose W, Huang TJ, Small 2016, 12, 5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guo F, Mao Z, Chen Y, Xie Z, Lata JP, Li P, Ren L, Liu J, Yang J, Dao M, Suresh S, Huang TJ, Proc. Natl. Acad. Sci. USA 2016, 113, 1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xu T, Luo Y, Liu C, Zhang X, Wang S, Anal. Chem 2020, 92, 7816. [DOI] [PubMed] [Google Scholar]
- [26].Zhu Q, Xu T, Song Y, Luo Y, Xu L, Zhang X, Biosens. Bioelectron 2020, 158, 112185. [DOI] [PubMed] [Google Scholar]
- [27].Shi J, Huang H, Stratton Z, Huang Y, Huang TJ, Lab Chip 2009, 9, 3354. [DOI] [PubMed] [Google Scholar]
- [28].Mao Z, Li P, Wu M, Bachman H, Mesyngier N, Guo X, Liu S, Costanzo F, Huang TJ, ACS Nano 2017, 11, 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shields CW, Sun D, Johnson KA, Duval KA, Rodriguez AV, Gao L, Dayton PA, López GP, Angew. Chem., Int. Ed 2014, 53, 8070. [DOI] [PubMed] [Google Scholar]
- [30].Yeo LY, Friend JR, Annu. Rev. Fluid Mech 2014, 46, 379. [Google Scholar]
- [31].Théry C, Zitvogel L, Amigorena S, Nat. Rev. Immunol 2002, 2, 569. [DOI] [PubMed] [Google Scholar]
- [32].Kowal J, Tkach M, Théry C, Curr. Opin. Cell Biol 2014, 29, 116. [DOI] [PubMed] [Google Scholar]
- [33].Colombo M, Raposo G, Théry C, Annu. Rev. Cell Dev. Biol 2014, 30, 255. [DOI] [PubMed] [Google Scholar]
- [34].Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R, Nature 2015, 523, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pegtel DM, Gould SJ, Annu. Rev. Biochem 2019, 88, 487. [DOI] [PubMed] [Google Scholar]
- [36].Hao N, Li LF, Tang FQ, Int. Mater. Rev 2017, 62, 57. [Google Scholar]
- [37].Antfolk M, Muller PB, Augustsson P, Bruus H, Laurell T, Lab Chip 2014, 14, 2791. [DOI] [PubMed] [Google Scholar]
- [38].Wu Z, Jiang H, Zhang L, Yi K, Cui H, Wang F, Liu W, Zhao X, Zhou F, Guo S, Lab Chip 2019, 19, 3922. [DOI] [PubMed] [Google Scholar]
- [39].Stöber W, Fink A, Bohn E, J. Colloid Interface Sci 1968, 26, 62. [Google Scholar]
- [40].Ren L, Yang S, Zhang P, Qu Z, Mao Z, Huang P-H, Chen Y, Wu M, Wang L, Li P, Huang TJ, Small 2018, 14, 1801996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Johnson LM, Gao L, ShieldsIV C, Smith M, Efimenko K, Cushing K, Genzer J, López GP, J. Nanobiotechnol 2013, 11, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wu M, Mao Z, Chen K, Bachman H, Chen Y, Rufo J, Ren L, Li P, Wang L, Huang TJ, Adv. Funct. Mater 2017, 27, 1606039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vakarelski IU, Li EQ, Abdel-Fattah AI, Thoroddsen ST, Colloids Surf., A 2016, 506, 138. [Google Scholar]
- [44].Li P, Huang TJ, Anal. Chem 2019, 91, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Evander M, Nilsson J, Lab Chip 2012, 12, 4667. [DOI] [PubMed] [Google Scholar]
- [46].Yuan Z, Jiang H, Med. Phys 2012, 39, 6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen J, Duncan B, Wang Z, Wang L-S, Rotello VM, Nugen SR, Nanoscale 2015, 7, 16230. [DOI] [PubMed] [Google Scholar]
- [48].Gopinath SCB, Awazu K, Fujimaki M, Shimizu K, Mizutani W, Tsukagoshi K, Analyst 2012, 137, 3520. [DOI] [PubMed] [Google Scholar]
- [49].Yu X, He L, Pentok M, Yang H, Yang Y, Li Z, He N, Deng Y, Li S, Liu T, Chen X, Luo H, Nanoscale 2019, 11, 15589. [DOI] [PubMed] [Google Scholar]
- [50].Nama N, Huang P-H, Huang TJ, Costanzo F, Lab Chip 2014, 14, 2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nama N, Huang P-H, Huang TJ, Costanzo F, Biomicrofluidics 2016, 10, 024124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Huang P, Xie Y, Ahmed D, Rufo J, Nama N, Chen Y, Chan CY, Huang TJ, Lab Chip 2013, 13, 3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Doinikov AA, Gerlt MS, Dual J, Phys. Rev. Lett 2020, 124, 154501. [DOI] [PubMed] [Google Scholar]
- [54].Doinikov AA, Gerlt MS, Pavlic A, Dual J, Microfluid. Nanofluid 2020, 24, 32. [Google Scholar]
- [55].Zeng Z, Liu Y, Wei J, Trends Anal. Chem 2016, 75, 162. [Google Scholar]
- [56].Hao N, Liu P, Bachman H, Pei Z, Zhang P, Rufo J, Wang Z, Zhao S, Huang TJ, ACS Nano 2020, 14, 6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Xie Y, Yang S, Mao Z, Li P, Zhao C, Cohick Z, Huang P-H, Huang TJ, ACS Nano 2014, 8, 12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O, Hingtgen SD, Kabanov AV, Batrakova EV, Nanomedicine 2016, 12, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sancho-Albero M, Encabo-Berzosa MDM, Beltrán-Visiedo M, Fernández-Messina L, Sebastián V, Sánchez-Madrid F, Arruebo M, Santamaría J, Martín-Duque P, Nanoscale 2019, 11, 18825. [DOI] [PubMed] [Google Scholar]
- [60].Sancho-Albero M, Navascués N, Mendoza G, Sebastián V, Arruebo M, Martín-Duque P, Santamaría J, J. Nanobiotechnol 2019, 17, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zabeo D, Cvjetkovic A, Lässer C, Schorb M, Lötvall J, Höög JL, J. Extracell. Vesicles 2017, 6, 1329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mayer KM, Hafner JH, Chem. Rev 2011, 111, 3828. [DOI] [PubMed] [Google Scholar]
- [63].Yan Z, Dutta S, Liu Z, Yu X, Mesgarzadeh N, Ji F, Bitan G, Xie Y-H, ACS Sens. 2019, 4, 488. [DOI] [PubMed] [Google Scholar]
- [64].Lee C, Carney RP, Hazari S, Smith ZJ, Knudson A, Robertson CS, Lam KS, Wachsmann-Hogiu S, Nanoscale 2015, 7, 9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Otto C, van den Tweel TJJ, de Mul FFM, Greve J, J. Raman Spectrosc 1986, 17, 289. [Google Scholar]
- [66].Volovšek V, Furić K, Bistričić L, Leskovac M, Macromol. Symp 2008, 265, 178. [Google Scholar]
- [67].Alessi A, Agnello S, Buscarino G, Gelardi FM, J. Non-Cryst. Solids 2013, 362, 20. [Google Scholar]
- [68].Alessi A, Agnello S, Buscarino G, Gelardi FM, J. Raman Spectrosc 2013, 44, 810. [Google Scholar]
- [69].Wu M, Ouyang Y, Wang Z, Zhang R, Huang P-H, Chen C, Li H, Li P, Quinn D, Dao M, Suresh S, Sadovsky Y, Huang TJ, Proc. Natl. Acad. Sci. USA 2017, 114, 10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hao N, Xu Z, Nie Y, Jin C, Closson AB, Zhang M, Zhang JXJ, Chem. Eng. J 2019, 378, 122222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hao N, Zhang M, Zhang JXJ, Biomater. Sci 2020, 8, 1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.