Abstract
Dementia of Alzheimer’s Type (DAT) is associated with devastating and irreversible cognitive decline. Predicting which patients with mild cognitive impairment (MCI) will progress to DAT is an ongoing challenge in the field. We developed a deep learning model to predict conversion from MCI to DAT. Structural magnetic resonance imaging scans were used as input to a three-dimensional convolutional neural network (3D-CNN). The 3D-CNN was trained using transfer learning; in the source task, normal control and DAT scans were used to pre-train the model. This pre-trained model was then re-trained on the target task of classifying which MCI patients converted to DAT. Our model resulted in 82.4% classification accuracy at the target task, outperforming current models in the field. Next, we visualized brain regions that significantly contribute to the prediction of MCI conversion using an occlusion map approach. Contributory regions included the pons, amygdala, and hippocampus. Finally, we showed that the model’s prediction value is significantly correlated with rates of change in clinical assessment scores, indicating that the model is able to predict an individual patient’s future cognitive decline. This information, in conjunction with the identified anatomical features, will aid in building a personalized therapeutic strategy for individuals with MCI.
Keywords: Convolutional Neural Network, Dementia of Alzheimer’s Type, Magnetic resonance imaging, Mild Cognitive Impairment, Predictive Modeling
1. Introduction
Dementia of Alzheimer’s Type (DAT) is a common and severe neurodegenerative disorder (Alzheimer's Association, 2019; Heun et al., 1997). Mild Cognitive Impairment (MCI), which is characterized by noticeable cognitive decline, precedes DAT and 10%-12% of individuals with MCI convert to DAT every year (Petersen, 2000). Predicting patients who will progress from MCI to DAT is important for patient care as well as in patient selection for clinical trials aimed at treating and preventing Alzheimer’s disease (Roberson and Mucke, 2006). However, current diagnostic tools for predicting conversion to DAT rely heavily on clinical interviews and neuropsychological evaluations, and may not be sensitive to the earliest changes required to predict future disease development. Thus, new methodology is needed in order to better predict disease progression.
With the development of computational methods such as machine learning and deep learning, there is increased utility of biomarker-based diagnosis for disease prediction. Numerous computational methodologies have been proposed to tackle the problem of predicting which MCI patients will convert to DAT (MCI-Converters or MCI-C) vs. those who do not (MCI-Non-Converters or MCI-NC) (Basaia et al., 2019; Cheng et al., 2015; Li, H. et al., 2014; Suk et al., 2017). Of those published, reported accuracy of models is around 75% to 80%. There are, however, several limitations of existing studies. First, many failed to assess their model using a separate, independent test dataset. It is important to randomly reserve a portion of the whole data set to be included in the independent test set (Kuhn and Johnson, 2013). This is the gold standard practice in the field to evaluate a model’s effectiveness and generalizability (Russell and Norvig, 2016), particularly in the absence of feature visualization. In addition, previous research has relied on specific features (e.g., cortical thickness, hippocampal volume) extracted from raw data to train the model (Zheng and Casari, 2018). This approach assumes that the chosen feature is the most informative and may neglect important information inherent in the raw data.
CNN is a deep-learning approach that has evolved in recent years to produce better classification performance and feature visualization than conventional machine learning methods across several fields (Borji et al., 2019). A CNN trained on raw, whole brain data can automatically extract the important imaging features and can offer insights beyond current methods in predicting disease progression. An end-to-end system, which places the model’s input and output on each end of the model, requires minimal or no feature extraction, producing features that are not biased. This approach has not yet been actively implemented to predict conversion from MCI to DAT.
In the present study, we implemented an end-to-end 3D-CNN model with transfer learning (Torrey and Shavlik, 2010) to classify MCI-NC vs. MCI-C patients using structural magnetic resonance images (sMRI). Transfer learning improves the model’s performance by training the model through two classification tasks: the source task and the target task. At the source task, the model is pre-trained using visual information similar to that used in the target task. Through this task, the model learns generic knowledge that will be helpful in target classification. At the target task, the model is re-trained with the resource that is directly relevant to the classification objective using the previously established generic knowledge.
The present study aimed to predict which individuals with MCI converted (MCI-C) and did not covert (MCI-NC), using a CNN model that has been first trained on sMRI scans of healthy individuals (NC) and those with suspected Alzheimer’s disease (DAT). Using the terminology described above, we used NC and DAT scans in the source task. The model learns features that most strongly distinguish healthy from diseased brains. The generic knowledge obtained from the source task is transferred to the target task in which scans from patients with MCI are used. The model is then re-trained with MCI-NC and MCI-C patients scans, to extract features that can predict conversion to DAT. Previous research suggests that the classification task of NC vs. DAT is similar to the classification task of MCI-NC vs. MCI-C (Coupé et al., 2012; Da et al., 2014; Young et al., 2013) and has been used to pre-train machine learning models in previous studies (Basaia et al., 2019; Cheng et al., 2015). In this project, we utilized a classification task of NC vs. DAT as the source task for transfer learning to our target task model.
Furthermore, we utilized a novel occlusion map method (Zeiler and Fergus, 2014) to visualize the features significantly contributing to our model. Finally, we demonstrate the model’s clinical relevance through the association of the model’s prediction output to rate of cognitive decline.
2. Materials and Methods
2.1. Subjects
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD).
The source task utilized 1406 DAT and 2084 NC scans from 1080 subjects. At the source task only, scans from multiple timepoints are included, if available. In the target task, we examined MCI-C patients with a conversion time of up to 3 years (longer conversion times are examined later) to MCI-NC patients with a clinical diagnosis that remains MCI for a duration of at least 3 years. MCI subjects with a duration of MCI less than 3 years without conversion were excluded due to the potential possibility of near-future conversion. This resulted in 228 MCI-C patients and 222 MCI-NC patients.
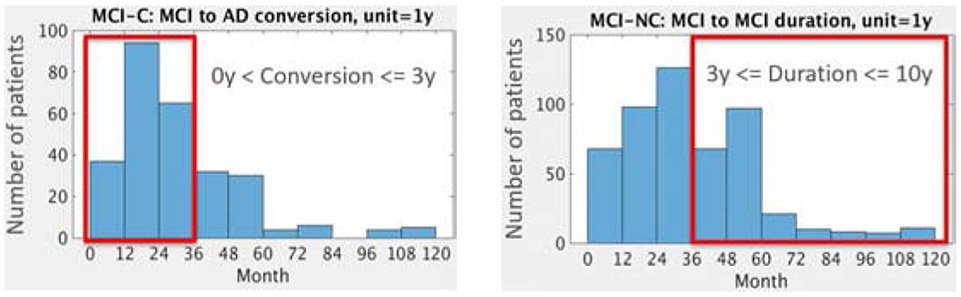
Included in the target task is the single timepoint sMRI scan at which an individual first received a diagnosis of MCI. Group differences in demographic and clinical history information were evaluated with one-way analyses of variance and chi-square tests. Demographic information and clinical scores for the sample are shown in Table 1. The distributions of conversion time of MCI-C patients and duration of diagnosis for MCI-NC patients are shown in Figure 1. Additional clinical information is provided in Supplemental Table 11.
Table 1.
Demographic and Clinical Information within Subjects for the Source and Target Tasks
| Source Task | Target Task | |||||
|---|---|---|---|---|---|---|
| NC | DAT | p value | MCI-NC | MCI-C | p value | |
| N total | 2084 | 1406 | -- | 222 | 228 | -- |
| Age | 76.49 (±5.92) | 76.18 (±7.22) | ns | 72.25 (±−7.32) | 74.18 (±6.96) | p < .05 |
| % Male | 49.80% | 60.10% | ns | 63.10% | 57.00% | ns |
| Education | 16.35 (±2.74) | 15.35 (±2.90) | p < .05 | 15.97 (±−2.85) | 15.87 (±−2.78) | ns |
| CDRSB | 0.09 (±0.30) | 5.22 (±2.41) | p < .05 | 1.18 (±−0.63) | 1.97 (±−0.98) | p < .05 |
| ADAS11 | 5.56 (±2.85) | 20.47 (±7.85) | p < .05 | 8.61 (±−3.41) | 13.17 (±−5) | p < .05 |
| ADAS13 | 8.7 (±1.32) | 31.03 (±9.43) | p < .05 | 13.77 (±−5.33) | 21.27 (±−6.04) | p < .05 |
| MMSE | 29.04 (±1.21) | 22.31 (±3.68) | p < .05 | 28 (±−1.69) | 26.77 (±−1.72) | p < .05 |
Results are reported as mean ± standard deviation. Age and education are reported in years. CDR=Clinical Dementia Rating Scale; ADAS11=Alzheimer’s Disease Assessment Scale 11; ADAS13=Alzheimer’s Disease Assessment Scale 13; MMSE=Mini Mental State Exam.
Figure 1.
Distribution of MCI-C (N=277) and MCI-NC (N=514) patients according to years until conversion and duration of diagnosis, respectively: Upper histogram red box: MCI-C patients who converted to DAT within 3 years were selected for the target task (N=228). As a comparison to this group, lower histogram red box: MCI-NC patients whose duration of MCI diagnosis is at least 3 years (N=222) are included in this study.
2.2. Structural MRI data setup for transfer learning
1.5T and 3T sMRI data were downloaded from ADNI. (Detailed MRI scanner protocols for T1-weighted sequences by vendor are available online: http://adni.loni.usc.edu/methods/documents/mri-protocols/). Preprocessing included skull-stripping (Wang et al., 2011), re-orientation, cropping and padding. This resulted in images with 158 x 196 x 170 voxels. The FMRIB Software Library (FSL; https://fsl.fmrib.ox.ac.uk) was then used to correct intensity inhomogeneity by using an N3 algorithm (Sled et al., 1998) and to co-register the scans to the Montreal Neurological Institute (MNI) 152 atlas by using affine linear alignment.
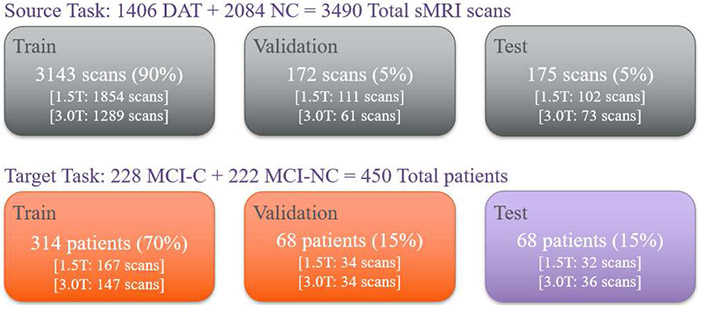
For the source task, DAT and NC scans were randomly selected and divided into train, validation, and test sets (Figure 2). To provide diverse generic knowledge, 90% of the data (3143 scans) were assigned to the train set while the validation and test set each contained 5% of the data (172 and 175 scans respectively). ANOVAs were calculated to confirm that groups within the train, validation, and test set did not differ significantly in demographic and clinical characteristics: sex, race, ethnicity, marital status, age, years of education, clinical scores, and genetic information1.
Figure 2.
Graphical layout of data division for the source task and target task. The number of scans and patients used for train, validation, and test set are shown. The percentage in parenthesis indicates the ratio of data size compared to the whole dataset, i.e., either source or target data set. The number of 1.5T and 3.0T scans are also shown in brackets.
For the target task, MCI-C and MCI-NC scans were randomly split into training, validation, and test sets by following the conventional ratio of 70% vs. 15% vs. 15% (314, 68, and 68 scans, respectively). ANOVAs were also calculated to confirm the no significant group difference between train, validation, and test set. To avoid data leakage (Wen et al., 2020), which exposes the information of the test set to the train and validation set, thereby falsely producing a higher test set classification accuracy, a single time point scan was used for each subject. The test portion of the target task was also ensured to be fully independent from the data used in both the source task and the training/validation portion of the target task. Therefore, no subjects in the target task test set overlapped with the rest of the samples. This step has been overlooked in previous research and is crucial for avoiding biased learning and increasing the generalizability of the model.
2.3. Architecture of Convolutional Neural Network
A base model for transfer learning was developed by benchmarking Residual Network 50 (ResNet50) (He et al., 2016). ResNet50 is composed of 5 residual blocks. The first block contains one convolutional and pooling layer, and the following blocks consist of 3, 4, 6, and 3 bottleneck layers, respectively. Each bottleneck layer has 3 convolutional layers interconnected through a skip connection that can smooth the loss landscape and is beneficial in achieving global optima (Li et al., 2018; Orhan and Pitkow, 2017). Each convolutional layer receives representations from the previous layers and transforms them to the deeper level of feature maps. These feature maps then contribute to the model’s classification decision.
ResNet50, however, has over 23 million trainable parameters, which is complex enough to cause high variance for the MCI-C vs. MCI-NC classification task. Therefore, we tailored ResNet50 to this task by reducing the number and width of convolutional layers. The resulting model had narrower and shorter network architecture than ResNet50 and was named ResNet29 (Figure 3). The number of filters of the convolutional layer in the first convolutional block was reduced from 64 to 32. The number of bottleneck layers in the following residual blocks were reduced from 3, 4, 6, and 3 to 2, 2, 2, and 2, respectively. Finally, one additional residual block which consists of one bottleneck layer was added at the end. The number of filters of each residual block was divided by 4. In the end, the model has about 4 million trainable parameters.
Figure 3.
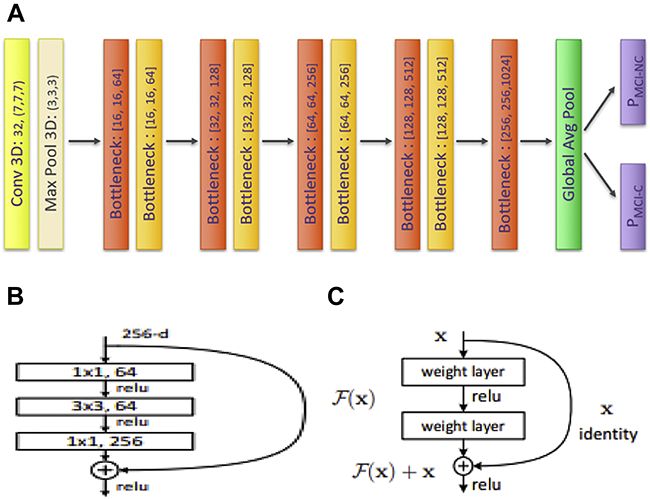
(a). Architecture of Convolutional Neural Network (CNN). The original ImageNet Model, i.e., ResNet50 was scaled down by narrowing and shortening the model. Figure 3. (b) Bottleneck layers were set to reduce the model’s complexity and thereby improve the classification performance (He et al., 2016). Figure 3. (c) Skip connection was used to enable the model to reach a global optima (He et al., 2016).
ResNet29 was developed as an end-to-end binary classification model. The model produces two prediction scores: the probability that the scan is classified as an MCI-C subject and the inverse probability that the scan is an MCI-NC subject. The sum of these two prediction scores is always one; If the prediction score for MCI-C is higher than the prediction score for MCI-NC, then the model decides the given brain scan is from an MCI-C patient. Similarly, if the prediction score for MCI-NC is higher than the prediction score for MCI-C, then the model predicts the given brain scan belongs to the MCI-NC patient group.
All codes were built in Python Keras as a TensorFlow backend. Experiments were conducted by using 4 NVIDIA P100 Pascal (12G HBM2 memory). The training time for the source and target task were 9 and 3 hours, respectively.
2.4. Hyperparameters
Hyperparameters are variables set before training which determine the network structure and how the network is trained. We evaluated multiple hyperparameters with the objective of improving classification accuracy. At the source task, the model was trained with a cyclically changing learning rate to avoid the model being stuck in local optima and to promote the model to reach to the global optima (Loshchilov and Hutter, 2016). The maximum learning rate and minimum learning rate were set as 1e-2 and 1e-4. The learning rate was cyclically changing through the entire epoch of 75 with a unit epoch of 25. To reduce overfitting, ridge regression and weight constraint with the value of 4e-4 and 2 were used throughout every convolutional layer, and a batch normalization layer was also used (Ioffe and Szegedy, 2015). To prevent gradient exploding, gradient clipping was set as 1 (Philipp et al., 2018).
The model and the weight matrix obtained from NC vs. DAT classification task were transferred to the target task of classifying MCI-NC vs. MCI-C. At the target task, the first 127 out of 155 layers were frozen, which resulted in 2,767,106 trainable parameters. The model was retrained with a cyclically changing learning rate from 1e-3 to 1e-5 with a unit epoch of 25 through the entire epoch of 125. Ridge regression, weight constraint, and gradient clipping were set as 7e-4, 2, and 1, and a batch normalization layer was also used.
All convolutional layers were initialized with ‘he_normal’ (He et al., 2015). Additionally, the ‘elu’ activation function, proposed by Clevert et al., (2015), was used with the intention of increasing training speed. Finally, the output layer used the ‘softmax’ activation function, which produces the output probabilities between 0 and 1, with the sum of the probabilities been equal to 1 (Nwankpa et al., 2018). Categorical cross entropy was used as a loss function and stochastic gradient descent was used as an optimizer.
2.5. Feature Visualization Method: Occlusion Map
Feature visualization was completed using an occlusion method implemented on all sMRI scans that had been included in the test set of the target task. Following model training, each sMRI scan was fed into the model with a 2x2x2 voxel patch (intensity 0) ‘occluded’. The patch position was iterated through each voxel with a stride of 2 of the whole 3D brain. Predictions scores were extracted from the model of each iterated occluded brain and the prediction score (for either class, MCI-C or MCI-NC) was recorded at the occluded brain region. This visualization creates a heatmap of brain regions that significantly alter the model prediction.
A degree of change in prediction score due to the occluded portion represents the importance of that region for the model’s classification decision. The brain regions where the prediction score decreased when occluded vs. un-occluded were colored as blue (Blue occlusion map). In contrast, the brain regions where the prediction score increased when occluded were colored as red (Red occlusion map). The blue regions contribute to a higher prediction score of the predicted class in the un-occluded image, and the red regions contribute to produce higher prediction score of the class not predicted in the un-occluded image. To the best of our knowledge, this method has yet to be implemented for classifying MCI-NC vs. MCI-C.
2.6. Relating Mean Intensity Values of Gray Matter beneath the Occlusion Maps to Neuropsychological and Cerebrospinal Fluid Measures
To validate the model, we calculated the Mean Intensity values of Gray Matter Beneath the Occlusion Map (MIGMBO). Atrophy in gray matter (both cortex and deep nuclei) is related to the accumulation of amyloid beta plaques and neurofilament tangles (Bejanin et al., 2017; Jack et al., 2019; Sepulcre et al., 2016). To identify the meaningful information that contributes to the conversion from MCI to DAT, MIGMBO for all patients in the test set of the target task were calculated and regressed with measures of clinical change, neuropsychological test performance, and cerebrospinal fluid (CSF) markers.
For clinical measures, we included the rate of change in the Clinical Dementia Rating-Sum of Boxes (CDRSB), Alzheimer’s Disease Assessment Scale – cognitive 11 item (ADAS 11) and cognitive 13 item (ADAS 13), Mini Mental State Exam (MMSE), Rey Auditory Verbal Learning Test (RAVLT) – RAVLT Immediate, RAVLT Learning, RAVLT Forgetting, RAVLT Percent Forgetting, and Functional Activities Questionnaire (FAQ) (Folstein et al., 1975; Mayo, 2012; Rosen et al., 1984; Samtani et al., 2014; Schmidt, 1996; Skinner et al., 2012). For CSF measures, we included Amyloid Beta (Aβ), Tau, Phosphorylated Tau (P-Tau), Aβ/Tau, and Aβ/P-Tau (Detailed description of csf acquisition protocols can be found on the ADNI website: http://adni.loni.usc.edu/data-samples/data-types/).
The output of our generated occlusion maps were divided into three bins based on strength of change in the model’s prediction, and each bin was used as a predictor for change in variables listed above in a multiple regression analysis. The ‘low’ occlusion bin are those in which there was only a small change in the prediction score, i.e., a change within one standard deviation. The ‘medium’ occlusion bin are those in which the prediction score changed between one and two standard deviations. The ‘high’ occlusion bin are those in which prediction score changed greater than two standard deviations.
To determine the gray matter regions that contribute to the DAT progression, the blue occlusion map for MCI-NC predicted patients and the red occlusion map for MCI-C predicted patients were used. The blue occlusion map of MCI-NC patients identifies the brain regions that make the MCI-NC brain scan look more similar to the MCI-C brain scan. The red occlusion map of MCI-C patients shows the brain regions that make the MCI-C brain scan look more dissimilar to the MCI-NC brain scan. Therefore, the brain regions covered by these occlusion maps provide information about DAT progression.
The other occlusion map, i.e., the red occlusion map for MCI-NC and the blue occlusion map for MCI-C, represent the brain regions that are indicative of MCI-NC that does not progress to DAT. Therefore, these regions were not used to examine brain regions that associate conversion to DAT to clinical and CSF measures.
2.7. Relating CNN’s prediction score to Neuropsychological Measures
To evaluate the clinical validity of the 3D-CNN model, we examined the prediction score from the earliest sMRI scan of an MCI patient to the rate of cognitive decline. Out of 514 MCI-NC and 277 MCI-C subjects throughout the whole conversion and duration years (Figure 1), patients whose brain scan were used in training/validating the model were excluded. This resulted in a sample of 323 MCI-NC and 86 MCI-C patients. The longitudinal clinical scores (i.e., CDRSB, ADAS11, ADAS13, MMSE, RAVLT – RAVLT Immediate, RAVLT Learning, RAVLT Forgetting, RAVLT Percent Forgetting, and FAQ) from the first MCI-diagnosed time point to the end of clinical history were used to obtain the month-wise rate of change in clinical assessments scores. Pearson’s correlation between the CNN prediction score from the baseline sMRI scan and the clinical scores’ month-wise rate of change obtained through the first to the last clinical history were also examined.
3. Results
3.1. 3D-CNN classification results
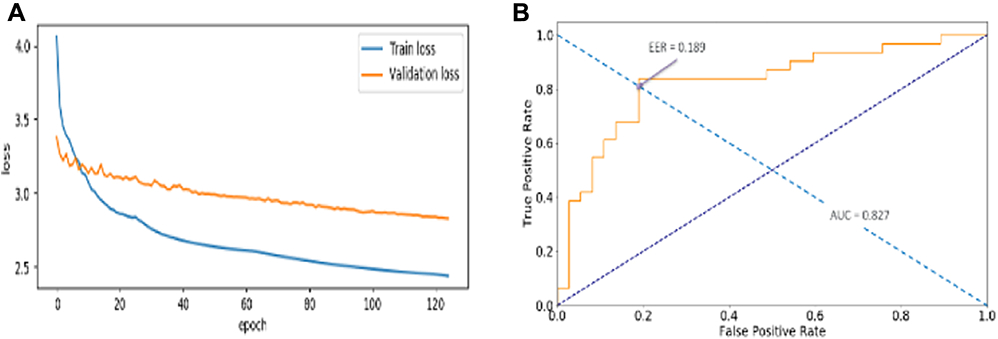
Classifying MCI-C vs. MCI-NC through transfer learning with a base model of ResNet29 was successful. The loss value of training and validation set decreased throughout the training epochs (Figure 4a). This indicates that the model was well optimized with a set of well-defined hyperparameters. It produced a test set classification accuracy of 82.4% and 0.827 Area Under the Curve (AUC) as well as 0.189 Equal Error Rate (EER) value (Figure 4b).
Figure 4.
Loss history of train and validation data (a) and classification performance (b), i.e., Area Under the Curve (AUC) and Equal Error Rate (EER) on test data. Train and validation loss continuously decreased along the epochs, indicating that the model was learning. The weight matrix that was restored and used to evaluate the test classification accuracy was where the validation loss showed the minimum. Test classification accuracy reported 82.4%. AUC and EER value are 0.827 and 0.189, respectively.
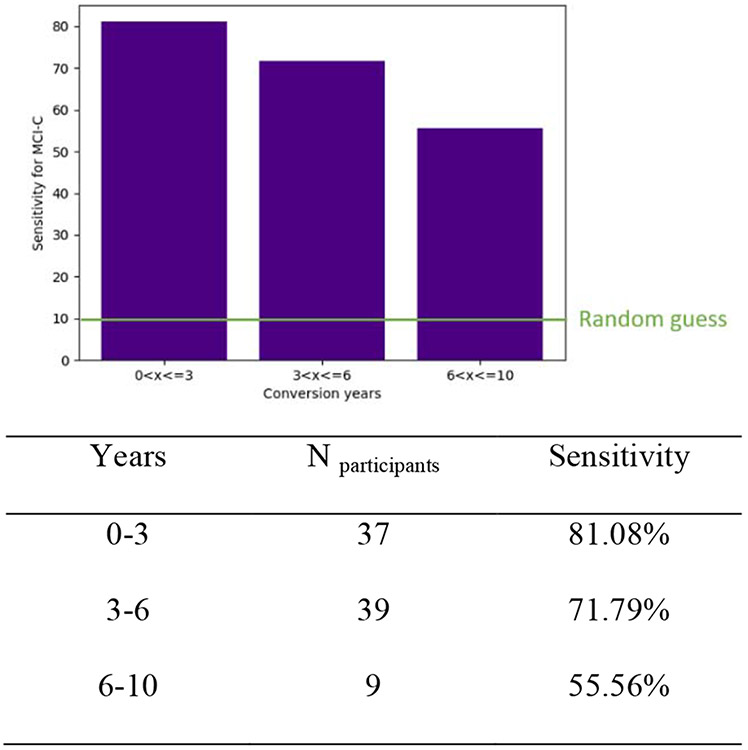
The test set was composed of MCI-C patients whose conversion time was between 0 and 3 years. To further look at the models’ prediction performance over a longer conversion time, a separate MCI-C dataset with a conversion time longer than 3 years was used. In conversion times from 0 to 3 years, 3 to 6 years, and 6 to 10 years, there were 37, 39, and 9 MCI-C subjects, and the same model and its weight matrix were implemented to predict these patients. The model’s sensitivity for these three groups were 81.08%, 71.79%, and 55.56%, respectively. The results showed that prediction score decreases with longer conversion times (Figure 5).
Figure 5.
The sensitivity to predict patients with conversion years from 0 to 10. The random guess is 10% as it is the chance of one out of 10 different conversion years.
Furthermore, we provide the model’s separate accuracy on 1.5T and 3.0T sMRI scanner (Table 2.). For 1.5T scanner, 25 out of 32 scans are correctly predicted and report 78.13% accuracy. For 3.0T scanner, 31 out of 36 scans are correctly predicted at 86.11% accuracy.
Table 2.
Prediction accuracy on 1.5T and 3.0T scans in the test set.
| Scanner | N total | N correctly predicted | Accuracy (%) |
|---|---|---|---|
| 1.5T | 32 | 25 | 78.13 |
| 3.0T | 36 | 31 | 86.11 |
3.2. Feature Visualization
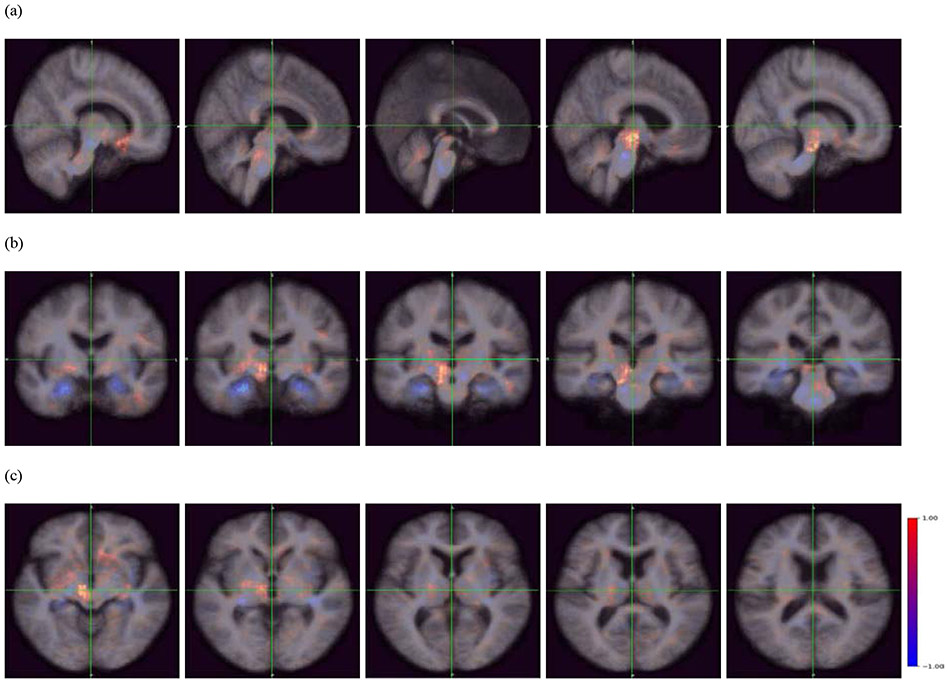
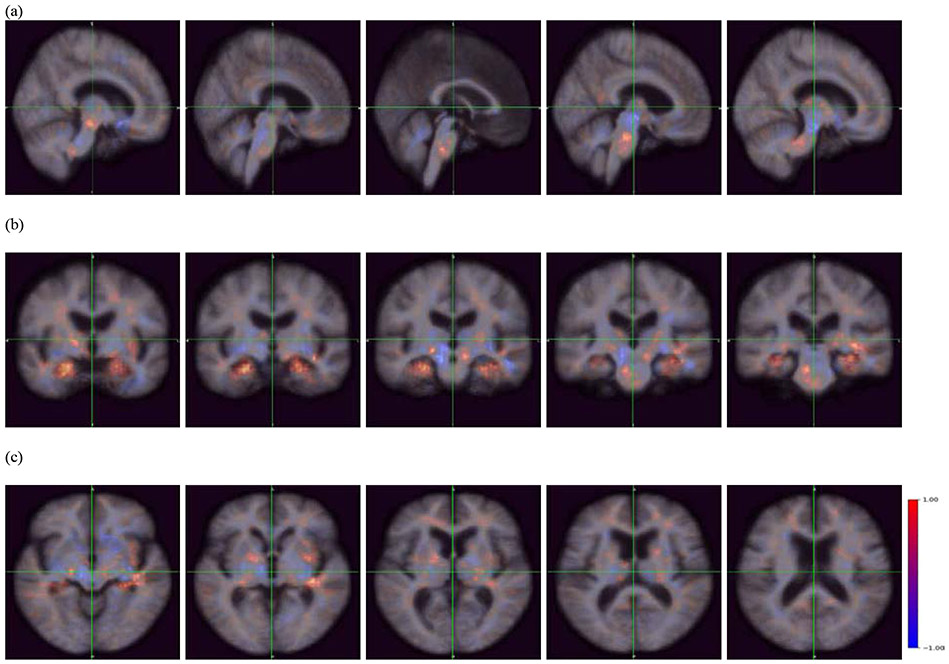
Using occlusion mapping, we identified structural features recognized by the model. As seen in Figures 5 and 6, the occlusion of the hippocampus, parahippocampal gyrus, amygdala, and pons increased the probability score for MCI-C; the hippocampus, parahippocampal gyrus, amygdala, and pons were covered by the blue occlusion map for MCI-NC and the red occlusion map for MCI-NC. On the other hand, the occlusion of the nucleus accumbens, caudate nucleus, globus pallidus, thalamus, cerebellum, and midbrain increased the probability score for MCI-NC; these regions were covered by the red occlusion map for MCI-NC and the blue occlusion map for MCI-C. We note that the occlusion maps for MCI-NC and MCI-C are complementary.
Figure 6.
Occlusion maps (a) sagittal plane, (b) coronal plane, and (c) transverse plane across all correctly predicted MCI-NC patients. The red color indicates a higher prediction score, whereas the blue color indicates a lower prediction score. The blue regions indicate importance in predicting MCI-NC, and include the pons, amygdala, hippocampus, and parahippocampal gyrus.
3.3. Relating MIGMBO to rate of change in Neuropsychological (Clinical) Measures
The MIGMBO score from the high occlusion bin predicted the rate of change in CDRSB, ADAS11, ADAS13, MMSE and FAQ score at a significance level of 0.05 (Table 3.). MIGMBO was negatively correlated with the rate of MMSE decline and positively correlated with the rate of increase in CDRSB, ADAS11, ADAS13, and FAQ score. In contrast, the MIGMBO score did not show a significant correlation with rate of change in RAVLT scores.
Table 3.
Correlation between MIGMBO and rate of cognitive decline: Red MCI-C and Blue MCI-NC
| Clinical Measures |
constant |
Low x1 |
Medium x2 |
High x3 |
N | R2 | F-statistic | p-value |
|---|---|---|---|---|---|---|---|---|
| CDRSB | 1.181 (0.771) |
0.046 (0.677) |
−0.073 (0.507) |
***0.007 (0.000) |
68 | 0.275 | 8.07 | 0.0001 |
| ADAS11 | −2.795 (−0.627) |
0.303 (0.943) |
−0.282 (−0.882) |
***0.014 (3.251) |
68 | 0.156 | 3.95 | 0.0120 |
| ADAS13 | −4.007 (−0.770) |
0.405 (1.080) |
−.372 (−0.994) |
***0.018 (3.529) |
68 | 0.182 | 4.74 | 0.0048 |
| MMSE | 3.735 (1.475) |
0.013 (0.073) |
−0.048 (−0.264) |
***−0.008 (−3.402) |
68 | 0.204 | 5.47 | 0.0021 |
| RAVLT Immediate | 4.529 (0.783) |
0.008 (0.016) |
−0.060 (−0.144) |
−0.008 (−1.476) |
68 | 0.053 | 1.19 | 0.3215 |
| RAVLT Learning | −3.392 (−1.938) |
0.137 (0.282) |
−0.088 (0.486) |
−0.002 (0.233) |
68 | 0.084 | 1.95 | 0.1306 |
| RAVLT Forgetting | −0.365 (−0.164) |
−0.176 (−1.092) |
0.183 (1.144) |
−0.003 (−1.398) |
68 | 0.039 | 0.86 | 0.4645 |
| RAVLT Percent Forgetting | 16.104 (0.585) |
**−4.596 (0.024) |
**4.382 (0.031) |
−0.019 (0.493) |
68 | 0.079 | 1.84 | 0.1493 |
| FAQ | 5.463 (1.333) |
−0.046 (−0.156) |
−0.057 (−0.194) |
***0.018 (4.551) |
68 | 0.260 | 7.51 | 0.0002 |
Notes: Figures in parentheses are t statistics.
p<.10
p<0.05
p<0.01. MIGMBO score could predict the rate of change in CDRSB, ADAS11, ADAS13, MMSE, and FAQ score. MIGMBO score from the most significant occlusion map are important in predicting rate of cognitive decline.
3.4. Relating MIGMBO to CSF Measures
MIGMBO scores showed strong correlation with Aβ, Aβ/Tau, and Aβ/P-Tau with a p-value below 0.05, indicating statistical significance (Table 4.). For relating Aβ/Tau and Aβ/P-Tau, all predictors showed a significant relationship. The low, medium, and high occlusion maps all played a crucial role in predicting the dependent variables. With Tau and P-Tau, MIGMBO showed p-values of 0.0683 and 0.0707, respectively, which approximate statistical significance.
Table 4.
Correlation between MIGMBO and CSF Measures: Red MCI-C and Blue MCI-NC
| CSF Measures |
constant |
Low x1 |
Medium x2 |
High x3 |
N | R2 | F-statistic | p-value |
|---|---|---|---|---|---|---|---|---|
| Aβ |
***4029.0 (3.0) |
***480.7 (3.4) |
***−507.3 (−3.5) |
**−2.3 (−2.2) |
26 | .494 | 7.15 | 0.00159 |
| Tau | −791.4 (−1.3) |
−76.5 (−1.2) |
87.3 (1.3) |
0.8 (1.6) |
29 | .244 | 2.69 | 0.0683 |
| P-Tau | −93.1 (−1.3) |
−10.1 (−1.3) |
11.3 (1.5) |
0.1 (1.6) |
29 | .241 | 2.65 | 0.0707 |
| Aβ/Tau |
***30.8 (4.1) |
***3.3 (4.2) |
***−3.6 (−4.4) |
***−0.0 (−2.9) |
26 | .634 | 12.7 | 4.98e-05 |
| Aβ/P-Tau |
***388.0 (4.4) |
***42.1 (4.5) |
***−45.3 (−4.7) |
***−0.2 (−3.0) |
26 | .665 | 14.6 | 1.92e-05 |
Notes: Figures in parentheses are t statistics.
p<.10
p<0.05
p<0.01. MIGMBO score could predict accumulation of Aβ, and ratio of Aβ/Tau and Aβ/P-Tau. All three predictors, i.e., MIGMBO score from the least, medium, and the most significant occlusion map, contribute to the prediction.
3.5. Relating CNN-based prediction score to rate of Cognitive Decline
The CNN-based prediction score at the first MCI-diagnosed timepoint showed significant correlation with the rate of change in CDRSB, FAQ, MMSE, and RAVLT forgetting (Table 5.). The CNN prediction score was positively correlated with the rate of change in the CDRSB and FAQ scores and was negatively correlated with the rate of change in the MMSE and RAVLT forgetting scores. On the other hand, RAVLT immediate, RAVLT learning, ADAS11, and ADAD13 did not show a significant correlation with the 3D-CNN-based prediction scores.
Table 5.
Correlation between CNN prediction score and clinical assessment scores’ rate of change
| CDRSB | ADAS11 | ADAS13 | MMSE | RAVLT Immediate |
RAVLT Learning |
RAVLT Forgetting |
RAVLT Percent Forgetting |
FAQ | |
|---|---|---|---|---|---|---|---|---|---|
| First MCI sMRI (N = 409) | ***0.264 | 0.062 | 0.039 | ***−0.146 | 0.000 | *0.089 | **−0.117 | −0.078 | ***0.243 |
| Baseline sMRI (N = 409) | ***0.242 | 0.008 | 0.005 | ***−0.177 | −0.011 | *0.094 | **−0.106 | ***−0.143 | ***0.144 |
Correlation between CNN prediction score from first MCI-diagnosed sMRI scan and clinical assessment scores' rate of change (the first row). CNN prediction score is positively correlated with the rate of change in CDRSB and FAQ score and negatively correlated with rate of change in MMSE and RAVLT Forgetting score. Correlation between CNN-based score from the baseline sMRI scan and clinical assessment scores' rate of change. CNN prediction score is positively correlated with the rate of change in CDRSB and FAQ score and negatively correlated with the rate of change in MMSE, RAVLT Forgetting, and RAVLT percent Forgetting (the second row).
p<.10
p<0.05
p<0.01.
The prediction score produced by the baseline sMRI scan also showed significant correlation with the rate of cognitive decline (Table 5). It was positively correlated with the rate of change in CDRSB and the FAQ score, and negatively correlated with MMSE, RAVLT Forgetting, and RAVLT Percent Forgetting scores.
4. Discussion
Leveraging ADNI data, we aimed to predict MCI conversion to DAT using a CNN model trained on sMRI scans of healthy individuals and those with DAT. In so doing, we developed ResNet29, an end-to-end 3D-CNN which trained, through transfer learning of these sMRI scans of healthy vs. DAT subjects, to predict MCI patients that either remained stable in their diagnosis or progressed to DAT. Our model achieved this with an 82.4% accuracy, and also showed the most significant prediction increase from random guess, 31.7%.
ResNet 29 trained through a novel transfer learning meets the level of complexity that is required to interpret the heterogeneous nature of DAT development. Most biomarkers of DAT, including atrophy on sMRI scans, are known to nonlinearly worsen with increased disease severity (Jack Jr et al., 2013). We recognize this is a limitation of machine learning models which are trained on the final stage of disease outcomes. However, we note that our base model, ResNet29, is constructed with a series of convolutional layers so that it may extract complex patterns through a series of non-linear transformations. The model learns generic knowledge through NC and DAT scans during the source task so that it is finely optimized to determine the degree (expressed as a probability of belonging to the MCI-C) to which a subject’s baseline scan resembles a DAT scan. We additionally note that our model is designed to individually classify ultimate disease outcome, rather than reflect the nuances of disease progression.
Compared to previous studies (Table 6), our model achieved the highest accuracy in classifying MCI-NC from MCI-C. Li, H. and colleagues (2014) used a Random Forest method with weak hierarchical lasso feature selection to achieve 74.8% classification accuracy using 161 MCI-NC and 132 MCI-C sMRI scans. Cheng et al. (2015) produced 79.4% classification accuracy by using Domain Transfer Feature Selection (DTFS) and Domain Transfer Sample Selection (DTSS) for extracting features and Support Vector Machine (SVM) model for classifying 43 MCI-NC and 56 MCI-C patients. Similarly, Suk et al. (2017) had 74.8% classification accuracy in classifying 226 MCI-NC and 167 MCI-C patients by using a 2D-CNN based on 93 regions of interest (ROIs) as features2. By using 3D-CNN, Basaia and colleagues (2019) showed 74.9% classification accuracy in classifying 533 MCI-NC and 280 MCI-C patients based on gray matter tissue probability maps, and Yee and colleagues (2020) recorded 74.7% accuracy in classifying 871 MCI-NC and 362 MCI-C scans.
Table 6.
Summary of MCI-C vs. MCI-NC classification research
| Biomarker | Conversion time (years) |
Random guess (%) |
Accuracy (%) | Increase (%) | |
|---|---|---|---|---|---|
| Proposed model | sMRI | 3 | 50.7 | 82.4 | 31.7 |
| Yee et al., 2020 | FDG | 3 | 70.6 | 74.7 | 4.1 |
| Basaia et al., 2019 | sMRI | 3 | 65.6 | 74.9 | 9.3 |
| Suk et al., 2017 | sMRI, Clinical Score | 1.5 | 57.5 | 74.8 | 17.3 |
| Cheng et al., 2015 | sMRI, PET, CSF | 2 | 56.6 | 79.4 | 22.8 |
| Li et al., 2014 | MRI, Meta features3 | 4 | 54.9 | 74.8 | 19.9 |
While most previous research did not use an independent test set, Basaia and colleagues (2019) assigned a relatively small portion (10%) of the whole dataset as a test set to verify the model’s generalized performance. The most effective splitting ratio of the training, validation, and test sets is still under discussion, although we set the ratio as 70:15:15 which is traditionally accepted and successfully demonstrated the generalizability of our model.
The ability to predict DAT conversion based on a single time point MRI is advantageous for the clinical field. While some previous studies include multimodal biomarkers in their prediction models, such as positron emission tomography (PET) and cerebrospinal fluid (CSF) biomarkers of disease (Cheng et al., 2015), our model outperformed these models with high accuracy by using a single time point sMRI scan. sMRI is often included in routine assessment of those at risk for AD. It is less expensive than other imaging scans and is minimally invasive, therefore reducing patient risk. Our model showed 8% higher accuracy in prediction with 3T than 1.5T sMRI scans. This finding is consistent with the known accuracy advantages of higher resolution images for CNN (Sabottke & Spieler, 2020). Nevertheless, the accuracy of 78% with 1.5T scanners, which are more common in hospital settings, is still high, indicating that our model is clinically implementable. In comparison to studies using combined modalities, our model produces more accurate predictions on DAT progression with less economic burden and infection risk to the patients.
Several factors contributed to the improvement of the classification performance by our model. It was largely empowered by the architecture of our deep learning model, specifically tailored for the MCI-C vs. MCI-NC classification task. These factors include our novel transfer learning pipeline using healthy vs. AD subjects that can produce diverse generic knowledge while avoiding data leakage, along with various engineering techniques such as a cyclically changing learning rate (Loshchilov and Hutter, 2016). Finally, we carefully tuned a set of hyperparameters including the type of activation layers, number of convolutional layers and their size and learning rates, through numerous experimental condition until the model achieved the highest accuracy reported here. Further, these identified sets of hyperparameters that produced the best prediction results were validated using the test set which also showed the highest accuracy from the numerous experiments: 82.4%.
In addition, our model was provided with a whole 3D brain scan without specification of any particular feature for training. Previous studies have limited the input resource through feature engineering. For example, studies that selected gray matter as the feature for model training (Basaia et al., 2019) did not consider CSF space or white matter changes known to also play an important role in DAT and the pathological process of Alzheimer’s disease (Jack Jr et al., 2010; Li, X. et al., 2014; Weiler et al., 2015). Additionally, Cheng et al. (2015) selected subjectively defined “useful” features using DTFS and DTSS. Machines trained with these samples could be biased and thus may not be generalizable to independent populations. Therefore, unlike feature engineering which limits the information able to be learned based on a researcher’s pre-assumption of what may be important in classification, the presented model learned from every possible feature available in the image.
It should be noted that the conversion times in previous studies range from 1.5 (Suk et al., 2017) to 4 years (Li, H. et al., 2014), while one of the latest experiments uses a 3-year conversion time window (Basaia et al., 2019). We chose this conversion time for the present study in order to directly compare performance to the most current research. Further, setting the conversion time at 3 years provided a well-balanced data set between MCI-NC (N=222) and MCI-C (N=228) (Buda et al., 2018). It allowed unbiased learning by the model on MCI-C and MCI-NC patients’ brains.
To the best of our knowledge, a deep learning model that can identify anatomical brain regions critical for predicting the conversion from MCI to DAT has not been demonstrated previously. Feature visualization methods are able to highlight regions in an input image with strong influence on the classification decision. It is important as it enables us to understand and validate the reasoning that has driven the model’s classification. Especially in the study of neurodegenerative disease, it is critical to explain the behavior of a machine/deep learning model to elucidate the neuroimaging biomarkers that contribute to conversion from MCI to DAT. State-of-the-art visualization techniques include Gradient Class Activation Map (Grad-CAM) and Guided Gradient CAM (Guided-Grad-CAM) (Selvaraju et al., 2017; Yee et al., 2020). However, in the medical field, these methods are unable visualize the features that contribute to disease-negative samples (Ardila et al., 2019). Further, feature maps that directly contribute to the classification decision often have too low resolution to show fine structural features within the brain.
An occlusion method to feature visualization avoids these problems and produces finer feature maps (Zeiler and Fergus, 2014). We implemented an occlusion method and identified key brain structures that contribute to DAT conversion. The occlusion method is critical in this research as the occlusion patch could represent structural alteration. A major strength of the presented model is that the input is naïve to specified brain regions. As the model uses whole 3D sMRI scan as an input without limiting itself to predefined ROIs or features that are obtained from feature engineering, the occlusion map, too, examined the level of contribution at the voxel level (2x2x2) in the progression of disease. The results that the occlusion map shows are completely driven by statistical calculations from the ResNet29.
The blue occlusion map presented brain regions that decreased the probability of being MCI-NC or MCI-C patients when such structural alteration occurs. For example, the hippocampus was covered by the blue occlusion map for MCI-NC-predicted patients. Thus, when information from the hippocampus was missing (occluded), the model recognized MCI-NC-predicted scans as more similar to an MCI-C scan; the probability for MCI-NC was decreased while the probability for MCI-C was increased. Therefore, morphological alteration in the hippocampus contributes to the DAT classification. This aligns with our understanding of the importance of the hippocampus during MCI stages and progression into DAT (Ferrarini et al., 2009; Gupta et al., 2019; Lee et al., 2020; Li et al., 2007). In contrast, the thalamus was covered by the blue occlusion map for MCI-C-predicted patients---meaning that when information from the thalamus was missing in the model, MCI-C-predicted patients looked more similar to MCI-NC patients. Therefore, as far as we can measure, structural change in the thalamus does not promote the DAT development within a 3-year window.
Many subcortical white matter and deep gray structures were detected as features. As both MCI-C and MCI-NC patients have MCI, they do not yet manifest significant cortical atrophy on sMRI. These patients experience cognitive decline related to atrophy in these regions. (For DAT patients, cortical regions are recognized as a feature (Supplementary Figure S14)).
For the quantitative voxel analysis, we segmented subcortical regions by using FMRIB’s Integrated Registration and Segmentation Tool and count the number of voxels of the blue occlusion map, the red occlusion map, and whole brain structure. The sizes of two groups’ subcortical structures presented similarly; they did not show a difference between their size at the significance level of 0.1. Surprisingly, however, the blue occlusion map was dominant in the hippocampus, amygdala, and pons for MCI-NC patients, while was dominant in the nucleus accumbens, caudate nucleus, globus pallidus, and thalamus for MCI-C patients (Table 7). Therefore, within a 3-year time window until DAT diagnosis, structural changes in the hippocampus, amygdala, and pons promote DAT development, rather than the nucleus accumbens, caudate nucleus, globus pallidus, putamen, and thalamus. We note that the occlusion patch color (black) used in the occlusion map did not alter the visualization results, as we found identical results using a white colored patch.
Table 7.
Mean number of voxels beneath the occlusion map
| N Blue occlusion map | N Red occlusion map | N Total | ||||||
|---|---|---|---|---|---|---|---|---|
| MCI-NC | MCI-C | MCI-NC | MCI-C | MCI-NC | MCI-C | |||
| Brain Stem | *** | |||||||
| 13927±4200 | 17025±2814 | 17051±3728 | 16218±2332 | 30979±7256 | 33247±3501 | |||
| Accumbens | *** | *** | ||||||
| 305±279 | 526±276 | 655±303 | 357±255 | 960±324 | 883±266 | |||
| Amygdala | *** | *** | ||||||
| 2608±1422 | 1303±1096 | 955±991 | 2261±1228 | 3562±1053 | 3564±830 | |||
| Caudate | *** | |||||||
| 4567±1157 | 5408±1330 | 5056±1632 | 4721±1047 | 9623±2221 | 10129±1637 | |||
| Hippocampus | *** | *** | ||||||
| 7218±3455 | 3505±2633 | 3122±2373 | 6128±2768 | 10340±2882 | 9632±1686 | |||
| Pallidus | *** | *** | ||||||
| 1655±749 | 2174±697 | 2363±695 | 1899±622 | 4017±1121 | 4073±606 | |||
| Putamen | 5293±1717 | 5680±1336 | 5164±1837 | 4918±1312 | 10457±3086 | 10598±1890 | ||
| Thalamus | *** | |||||||
| 6878±1901 | 8210±1836 | 9585±2203 | 8577±2066 | 16463±3361 | 16787±1884 | |||
Notes: The number shows mean ± standard deviation. The symbol * shows p-value from t-statistics, which indicate the difference between the number of voxels in the occlusion map for MCI-C and MCI-NC patients
p<.10
p<0.05
p<0.01. For MCI-NC patients, the blue occlusion map is dominant in the amygdala and hippocampus. For MCI-C patients, the blue occlusion map is dominant in the accumbens, caudate, pallidus, and thalamus. There is no significant difference in the whole volume of brain structure between MCI-NC and MCI-C patients.
Brain structures recognized by our deep learning model were consistent with previous research. Previous research has been published that structural alteration of subcortical brain structures reflects DAT progression. Many research studies indicate that morphological changes in the hippocampus and amygdala (Ball et al., 1985; Convit et al., 1993; Gupta et al., 2019; Lee et al., 2020; Lehericy et al., 1994; Li et al., 2007; Poulin et al., 2011; Zanchi et al., 2017) are significant. Also, the pons was recognized as a significant biomarker in predicting AD progression. Olivieri et al. (2019) suggested that structural alteration occurs in the pons before AD develops ieri et al., 2019).
To further provide content validity on regions identified by the occlusion map, we used the mean intensity value of gray matter beneath the three occlusion maps. Gray matter is known to be associated with the biomarkers of AD pathology (Bejanin et al., 2017; Jack et al., 2019; Sepulcre et al., 2016). Therefore, by showing the relationship between MIGMBOs and rate of cognitive decline, verified that our occlusion maps captured clinically meaningful brain regions. The MIGMBO score of the high occlusion map showed positive correlation with rate of change in CDRSB, ADAS11, ADAS13, and FAQ and negative correlation with rate of change in MMSE. Therefore, the higher the mean intensity of gray matter is, CDRSB, ADAS11, ADAS13, and FAQ scores increase and MMSE scores decrease more quickly.
Also, by showing a significant correlation with MIGMBO and accumulation of Aβ, Aβ/Tau and, Aβ/P- Tau, we confirmed that the model’s prediction aligns with neuropathologic markers of AD by solely utilizing information from a sMRI scan. All three occlusion maps, which were split based on their degree of prediction change, could be a useful resource in predicting Aβ, Aβ/Tau and, Aβ/P-Tau.
Finally, we showed that the prediction scores from our model were related to worsening of neuropsychological performance measures over time. Since all scans received a prediction score of being classified as MCI-C, we calculated the Pearson’s correlation between this score and the rate of cognitive decline for all the patients with MCI. The rate of change in CDRSB and FAQ were positively correlated with an MCI-C classification. This indicates that as the confidence in a scan being classified as MCI-C increases, the faster the increase in CDRSB and FAQ score. Additionally, rate of change in MMSE, RAVLT Forgetting, and RAVLT Percent Forgetting were negatively correlated with the predicted MCI-C score. This indicates that these scores decrease more quickly as the confidence that the scan should be classified as MCI-C increases.
Aside from the sMRI scan from the first MCI-diagnosed time point, we used the baseline sMRI scan of all MCI-C and MCI-NC patients in showing these correlations. Therefore, regardless of conversion time and duration time of MCI-C and MCI-NC patients, the model could predict the future cognitive decline of an individual patient by solely utilizing a sMRI scan from the first visit to the clinic. Considering that we do not know which patients will suffer from cognitive deficit in clinical practice, these results provide evidence that the model could be used to foretell future cognitive decline.
In future research, we plan to include all subcortical brain structures, including sub-structures of the brain stem and cerebellum, as well as parcellated cortical regions to predict the DAT progression throughout different conversion years of MCI-C patients and duration years of MCI-NC patients. This regression model could show the contribution of each brain region in promoting conversion to DAT and improve personalized preventive medicine.
In conclusion, the current clinical evaluation protocols cannot accurately predict which patients with MCI will progress to DAT (Ward et al., 2013). An automated classification system for MCI-NC vs. MCI-C, such as the method presented in this study, offers promise for informing the clinical prognosis of these patients. Furthermore, the methods presented here will be useful for identifying which patients would benefit most from participating in clinical trials by providing individualized information on the disease progression, i.e., brain regions that cause cognitive deficit and future cognitive decline. Our methods not only produced the highest performance in the field, but also avoided problems previously neglected such as data shortage, high variance, and data leakage. Our research showed high accuracy in predicting conversion as well as novel visualization features, both critical to advancing our understanding of conversion from MCI to DAT, and personalized preventive medicine.
Supplementary Material
Figure 7.
Occlusion maps (a) sagittal plane, (b) coronal plane, and (c) transverse plane across all correctly predicted MCI-C patients. The red color indicates a higher prediction score, whereas the blue color indicates a lower prediction score. The blue regions indicate importance in predicting MCI-C, and include the midbrain, nucleus accumbens, caudate nucleus, cerebellum, globus pallidus, and thalamus.
Highlights.
Deep learning shows the highest accuracy in predicting Dementia of Alzheimer’s Type
Novel transfer learning that improves performance, but avoids data leakage problem
Occlusion map identifies brain regions progressing to Dementia of Alzheimer’s Type
Deep learning’s prediction score can foretell an individual’s cognitive decline
Structural features of brain are associated with pathology of Alzheimer’s Disease
Acknowledgements
This research was funded by grant AG055121 and AG045333 from the National Institute on Aging, and by grants from Brain Canada, CIHR, NSERC and Compute Canada.
ADNI data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
This research was funded by the following grants from the National Institute on Aging: AG055121 and AG045333, and by grants from Brain Canada, CIHR, NSERC and Compute Canada.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
This manuscript has nothing to disclose for actual or potential conflict and interest.
Clinical scores and genetic information include CDR, ADAS11, ADAS13, MMSE, RAVLT immediate, RAVLT learning, RAVLT forgetting, RAVLT percent forgetting, FAQ, APGN1, APGN2, APOE2, APOE3, and APOE4.
93 ROI for each sMRI and PET, and 3 features from CSF are used.
MRI features indicates average cortical thickness, standard deviation in cortical thickness, volumes of cortical parcellations, volumes of specific white matter parcellations, and the total surface area of the cortex. And Meta features includes demographic, genetic information, baseline cognitive scores, and lab tests. 305 MRI features and 52 Meta features are used.
References
- Alzheimer's Association. 2019 Alzheimer's disease facts and figures. Alzheimers Dement 2019;15(3):321–87. [Google Scholar]
- Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019;25(6):954–61. [DOI] [PubMed] [Google Scholar]
- Ball M, Hachinski V, Fox A, Kirshen A, Fisman M, Blume W, Kral V, Fox H, Merskey H. A new definition of Alzheimer's disease: a hippocampal dementia. Lancet 1985;325(8419):14–6. [DOI] [PubMed] [Google Scholar]
- Basaia S, Agosta F, Wagner L, Canu E, Magnani G, Santangelo R, Filippi M, Initiative AsDN. Automated classification of Alzheimer's disease and mild cognitive impairment using a single MRI and deep neural networks. NeuroImage Clin 2019;21:101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejanin A, Schonhaut DR, La Joie R, Kramer JH, Baker SL, Sosa N, Ayakta N, Cantwell A, Janabi M, Lauriola M. Tau pathology and neurodegeneration contribute to cognitive impairment in Alzheimer’s disease. Brain 2017;140(12):3286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borji A, Cheng M-M, Hou Q, Jiang H, Li J. Salient object detection: A survey. Comput Vis Media 2019:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buda M, Maki A, Mazurowski MA. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw 2018;106:249–59. [DOI] [PubMed] [Google Scholar]
- Cheng B, Liu M, Zhang D, Munsell BC, Shen D. Domain transfer learning for MCI conversion prediction. IEEE Trans Biomed Eng 2015;62(7):1805–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevert DA, Unterthiner T, Hochreiter S. Fast and accurate deep network learning by exponential linear units (elus). arXiv preprint arXiv:151107289 2015. [Google Scholar]
- Convit A, De Leon M, Golomb J, George A, Tarshish C, Bobinski M, Tsui W, De Santi S, Wegiel J, Wisniewski H. Hippocampal atrophy in early Alzheimer's disease: anatomic specificity and validation. Psychiatr Q 1993;64(4):371–87. [DOI] [PubMed] [Google Scholar]
- Coupé P, Eskildsen SF, Manjón JV, Fonov VS, Pruessner JC, Allard M, Collins DL, Initiative AsDN. Scoring by nonlocal image patch estimator for early detection of Alzheimer's disease. NeuroImage Clin 2012;1(1):141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da X, Toledo JB, Zee J, Wolk DA, Xie SX, Ou Y, Shacklett A, Parmpi P, Shaw L, Trojanowski JQ. Integration and relative value of biomarkers for prediction of MCI to AD progression: spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. NeuroImage Clin 2014;4:164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarini L, Frisoni GB, Pievani M, Reiber JH, Ganzola R, Milles J. Morphological hippocampal markers for automated detection of Alzheimer's disease and mild cognitive impairment converters in magnetic resonance images. J Alzheimers Dis 2009;17(3):643–59. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975;12(3):189–98. [DOI] [PubMed] [Google Scholar]
- Gupta Y, Lee KH, Choi KY, Lee JJ, Kim BC, Kwon GR, Dementia NRCf, Initiative AsDN. Early diagnosis of Alzheimer’s disease using combined features from voxel-based morphometry and cortical, subcortical, and hippocampus regions of MRI T1 brain images. PLoS One 2019;14(10):e0222446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification, Proceedings of the IEEE international conference on computer vision 2015:1026–34. [Google Scholar]
- He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition, Proceedings of the IEEE conference on computer vision and pattern recognition 2016:770–8. [Google Scholar]
- Heun R, Mazanek M, Atzor K-R, Tintera J, Gawehn J, Burkart M, Gänsicke M, Falkaic P, Stoeter P. Amygdala-hippocampal atrophy and memory performance in dementia of Alzheimer type. Dement Geriatr Cogn Disord 1997;8(6):329–36. [DOI] [PubMed] [Google Scholar]
- Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:150203167 2015. [Google Scholar]
- Jack CR Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol 2010;9(1):119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Wiste HJ, Therneau TM, Weigand SD, Knopman DS, Mielke MM, Lowe VJ, Vemuri P, Machulda MM, Schwarz CG. Associations of amyloid, tau, and neurodegeneration biomarker profiles with rates of memory decline among individuals without dementia. JAMA 2019;321(23):2316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M, Johnson K, Applied predictive modeling. Springer, 2013. [Google Scholar]
- Lee S, Lee H, Kim KW. Magnetic resonance imaging texture predicts progression to dementia due to Alzheimer disease earlier than hippocampal volume. J Neurosci 2020;45(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehericy S, Baulac M, Chiras J, Pierot L, Martin N, Pillon B, Deweer B, Dubois B, Marsault C. Amygdalohippocampal MR volume measurements in the early stages of Alzheimer disease. Am J Neuroradiol 1994;15(5):929–37. [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu Y, Gong P, Zhang C, Ye J, Initiative ADN. Hierarchical interactions model for predicting Mild Cognitive Impairment (MCI) to Alzheimer's Disease (AD) conversion. PLoS One 2014;9(1):e82450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu Z, Taylor G, Studer C, Goldstein T. Visualizing the loss landscape of neural nets, Adv Neural Inf Process Syst. 2018:6389–99. [Google Scholar]
- Li S, Shi F, Pu F, Li X, Jiang T, Xie S, Wang Y. Hippocampal shape analysis of Alzheimer disease based on machine learning methods. Am J Neuroradiol 2007;28(7):1339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loshchilov I, Hutter F. Sgdr: Stochastic gradient descent with warm restarts. arXiv preprint arXiv:160803983 2016. [Google Scholar]
- Mayo AM. Use of the Functional Activities Questionnaire in older adults with dementia. Try This: Best Practices in Nursing Care to Older Adults with Dementia 2012;13. [Google Scholar]
- Nwankpa C, Ijomah W, Gachagan A, Marshall S. Activation functions: Comparison of trends in practice and research for deep learning. arXiv preprint arXiv:181103378 2018. [Google Scholar]
- Olivieri P, Lagarde J, Lehericy S, Valabrègue R, Michel A, Macé P, Caillé F, Gervais P, Bottlaender M, Sarazin M. Early alteration of the locus coeruleus in phenotypic variants of Alzheimer’s disease. Ann Clin Trans Neurol 2019;6(7):1345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhan AE, Pitkow X. Skip connections eliminate singularities. arXiv preprint arXiv:170109175 2017. [Google Scholar]
- Petersen R Mild cognitive impairment: transition between aging and Alzheimer's disease. Neurologia 2000;15(3):93–101. [PubMed] [Google Scholar]
- Philipp G, Song D, Carbonell JG. Gradients explode-deep networks are shallow-resnet explained. 2018. [Google Scholar]
- Poulin SP, Dautoff R, Morris JC, Barrett LF, Dickerson BC, Initiative AsDN. Amygdala atrophy is prominent in early Alzheimer's disease and relates to symptom severity. Psychiatry Res Neuroimaging 2011;194(1):7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Mucke L. 100 years and counting: prospects for defeating Alzheimer's disease. Science 2006;314(5800):781–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer's disease. Am J Psychiatry 1984. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Norvig P. Artificial intelligence: a modern approach. Malaysia: Pearson Education Limited, 2016, p.^pp. [Google Scholar]
- Sabottke CF, Spieler BM. The effect of image resolution on deep learning in radiography. Radiology: Artificial Intelligence 2020;2(1):e190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samtani MN, Raghavan N, Novak G, Nandy P, Narayan VA. Disease progression model for clinical dementia rating–sum of boxes in mild cognitive impairment and Alzheimer’s subjects from the Alzheimer’s disease Neuroimaging initiative. Neuropsychiatr Dis Treat 2014;10:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Rey auditory verbal learning test: A handbook Western Psychological Services Los Angeles, CA, 1996. [Google Scholar]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-cam: Visual explanations from deep networks via gradient-based localization, Proceedings of the IEEE international conference on computer vision 2017:618–26. [Google Scholar]
- Sepulcre J, Schultz AP, Sabuncu M, Gomez-Isla T, Chhatwal J, Becker A, Sperling R, Johnson KA. In vivo tau, amyloid, and gray matter profiles in the aging brain. J Neurosci 2016;36(28):7364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J, Carvalho JO, Potter GG, Thames A, Zelinski E, Crane PK, Gibbons LE, Initiative AsDN. The Alzheimer’s disease assessment scale-cognitive-plus (ADAS-Cog-Plus): an expansion of the ADAS-Cog to improve responsiveness in MCI. Brain Imaging Behav 2012;6(4):489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998;17(1):87–97. [DOI] [PubMed] [Google Scholar]
- Suk HI, Lee SW, Shen D, Initiative AsDN. Deep ensemble learning of sparse regression models for brain disease diagnosis. Med Image Anal 2017;37:101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey L, Shavlik J. Transfer learning, Handbook of research on machine learning applications and trends: algorithms, methods, and techniques. IGI global, 2010:242–64. [Google Scholar]
- Wang Y, Nie J, Yap P-T, Shi F, Guo L, Shen D. Robust deformable-surface-based skull-stripping for large-scale studies, International Conference on Medical Image Computing and Computer-Assisted Intervention Springer, 2011: 635–42. [DOI] [PubMed] [Google Scholar]
- Ward A, Tardiff S, Dye C, Arrighi HM. Rate of conversion from prodromal Alzheimer's disease to Alzheimer's dementia: a systematic review of the literature. Dement Geriatr Cogn Dis Extra 2013;3(1):320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler M, Agosta F, Canu E, Copetti M, Magnani G, Marcone A, Pagani E, Balthazar MLF, Comi G, Falini A. Following the spreading of brain structural changes in alzheimer’s disease: a longitudinal, multimodal MRI study. J Alzheimers Dis 2015;47(4):995–1007. [DOI] [PubMed] [Google Scholar]
- Wen J, Thibeau-Sutre E, Diaz-Melo M, Samper-González J, Routier A, Bottani S, Dormont D, Durrleman S, Burgos N, Colliot O. Convolutional Neural Networks for Classification of Alzheimer's Disease: Overview and Reproducible Evaluation. Med Image Anal 2020:101694. [DOI] [PubMed] [Google Scholar]
- Yee E, Popuri K, Beg MF, Initiative AsDN. Quantifying brain metabolism from FDG-PET images into a probability of Alzheimer's dementia score. Hum Brain Mapp 2020;41(1):5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S, Initiative AsDN. Accurate multimodal probabilistic prediction of conversion to Alzheimer's disease in patients with mild cognitive impairment. NeuroImage Clin 2013;2:735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchi D, Giannakopoulos P, Borgwardt S, Rodriguez C, Haller S. Hippocampal and amygdala gray matter loss in elderly controls with subtle cognitive decline. Front Aging Neurosci 2017;9:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiler MD, Fergus R. Visualizing and understanding convolutional networks, European conference on computer vision Springer, 2014, 818–3 [Google Scholar]
- Zheng A, Casari A, Feature engineering for machine learning: principles and techniques for data scientists. " O'Reilly Media, Inc." 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.